Abstract
The Wzx flippase is a critical component of the O‐antigen biosynthesis pathway, being responsible for the translocation of oligosaccharide O units across the inner membrane in Gram‐negative bacteria. Recent studies have shown that Wzx has a strong preference for its cognate O unit, but the types of O‐unit structural variance that a given Wzx can accommodate are poorly understood. In this study, we identified two Yersinia pseudotuberculosis Wzx that can distinguish between different terminal dideoxyhexose sugars on a common O‐unit main‐chain, despite both being able to translocate several other structurally‐divergent O units. We also identified other Y. pseudotuberculosis Wzx that can translocate a structurally divergent foreign O unit with high efficiency, and thus exhibit an apparently relaxed substrate preference. It now appears that Wzx substrate preference is more complex than previously suggested, and that not all O‐unit residues are equally important determinants of translocation efficiency. We propose a new “Structure‐Specific Triggering” model in which Wzx translocation proceeds at a low level for a wide variety of substrates, with high‐frequency translocation only being triggered by Wzx interacting with one or more preferred O‐unit structural elements found on its cognate O unit(s).
Keywords: flippase, lipopolysaccharide, O antigen, specificity, Wzx
1. INTRODUCTION
The O‐antigen component of lipopolysaccharide (LPS) is the outermost cell component in many Gram‐negative bacteria, and plays an important protective role that is best understood for pathogens (Joiner, 1988; Matamouros & Miller, 2015; Murray, Attridge, & Morona, 2006). O antigens consist of short repeating oligosaccharide units (O units), each with typically 3–8 sugars, which can vary enormously in composition, even within a species. For instance, there are at least 185 currently identified O antigens in Escherichia coli (DebRoy et al., 2016), 46 in Salmonella enterica (Liu et al., 2014) and 34 in Shigella (Liu et al., 2008). This diversity is likely driven by selective pressures arising from the highly antigenic nature of O antigen as a component of LPS.
The vast majority of heteropolymeric O antigens are synthesized via the Wzx/Wzy pathway (Kalynych, Morona, & Cygler, 2014; Reeves & Cunneen, 2009), with most of the responsible genes grouped together in a gene cluster at a conserved chromosomal location. O‐unit synthesis is initiated at the cytoplasmic face of the inner membrane (IM), where a phosphoglycosyl transferase attaches the first sugar phosphate (typically N‐acetylglucosamine phosphate (GlcNAc‐P) in the Enterobacteriaceae) onto the polyisoprenoid lipid carrier molecule undecaprenyl phosphate (UndP), generating a pyrophosphate (PP) linkage (Price & Momany, 2005). A series of glycosyltransferases (GTs) then sequentially add the remaining sugars onto the growing chain to form a complete UndPP‐linked O unit. The Wzx flippase then translocates these UndPP‐linked O units across the IM to the periplasmic face, where the Wzy polymerase assembles polymeric UndPP‐linked O antigen under the control of the chain‐length regulator Wzz. The WaaL ligase then removes the O antigen from UndPP and attaches it to the lipid A‐core component of LPS, which is synthesized separately (Raetz & Whitfield, 2002), and the complete LPS molecule is exported to the outer leaflet of the outer membrane via the Lpt pathway (Whitfield & Trent, 2014).
Wzx was first proposed to be the O‐unit flippase over 20 years ago (Liu, Cole, & Reeves, 1996), and this is now generally accepted given the absence of a credible alternative flippase candidate (Hong, Liu, & Reeves, 2018; Islam & Lam, 2013). While there is data supporting the role of Wzx in O‐unit translocation (Rick et al., 2003), definitive biochemical proof remains elusive. Wzx has long been overlooked as a key factor in the evolution of O‐antigen diversity, largely due to early studies concluding that despite the enormous amount of sequence diversity among different Wzx, they were specific only for the first O‐unit residue (Feldman et al., 1999; Marolda, Vicarioli, & Valvano, 2004). However it has since been demonstrated on several occasions that Wzx can exhibit a strong preference for residues beyond the first sugar (Hong et al., 2018). Furthermore, inefficient Wzx translocation of nonpreferred O‐unit substrates results in cytoplasmic O‐unit accumulation and sequestration of UndP, which in turn interferes with peptidoglycan synthesis, causing a loss of membrane integrity and ultimately cell lysis (Jorgenson & Young, 2016; Liu, Stent, Hong, & Reeves, 2015). It is now becoming clear that Wzx substrate specificity represents a key checkpoint in the O‐antigen biosynthesis pathway. In spite of this, our knowledge of the types of O‐unit structural features that can influence Wzx translocation efficiency are currently limited to the identity of the first sugar (Marolda et al., 2004) and the presence or absence of terminal side‐branch residues (Hong & Reeves, 2014; Liu et al., 2015).
A group of 14 closely‐related Yersinia pseudotuberculosis O‐antigen gene clusters with four unique wzx sequence types, which produce O units with one of six alternative main‐chains and one of seven alternative side‐branch residues (Kenyon, Cunneen, & Reeves, 2017), represent a rare opportunity to expand our understanding of Wzx substrate preference. Within this set, the O:1a, O:2a, and O:4b O units (Figure 1a) consist of three shared residues (GlcNAc, galactose (Gal) and 6‐deoxymannoheptose (6dManHep)) with identical linkages, and one of three different terminal dideoxyhexose (DDH) residues (paratofuranose (Parf), O:1a; abequose (Abep), O:2a; tyvelose (Tyvp), O:4b) attached via an α(1,3) (O:2a, O:4b) or β(1,3) (O:1a) linkage by different GTs (WbyA, O:2a/O:4b; WbyM, O:1a). All three O antigens share a common Wzy and polymerization linkage, where the 6dManHep and DDH residues become two‐sugar side‐branches. The corresponding O‐antigen gene clusters (Figure 1a) differ only in the genes responsible for DDH synthesis and attachment, and interestingly, contain one of two unrelated wzx genes (wzx 1, O:1a; wzx 2, O:2a/O:4b). These wzx are notably found in 12 of the 14 related gene clusters, and are exclusively associated with specific terminal DDH or DDH‐like residues (altrofuranose (Altf), Parf or ascarylose (Ascp) with wzx 1; Abep or Tyvp with wzx 2) on different O‐unit main‐chains (two main‐chains associated with both wzx, three main‐chains associated with only one wzx) (Kenyon et al., 2017) (Figure 1b). In this study we describe the cloning and manipulation of the Y. pseudotuberculosis O:1a, O:2a and O:4b O‐antigen gene clusters for expression in E. coli, and the use of these clones to conclusively demonstrate that both Wzx1 and Wzx2 show a strong preference for specific terminal DDH residues. Since both Wzx have several cognate O‐unit main‐chain structures, and several other Y. pseudotuberculosis Wzx are also shown to efficiently flip a foreign O unit in this study, we propose a new model for Wzx substrate preference by “Structure‐Specific Triggering” involving direct recognition of a small subset of O‐unit structural elements.
Figure 1.
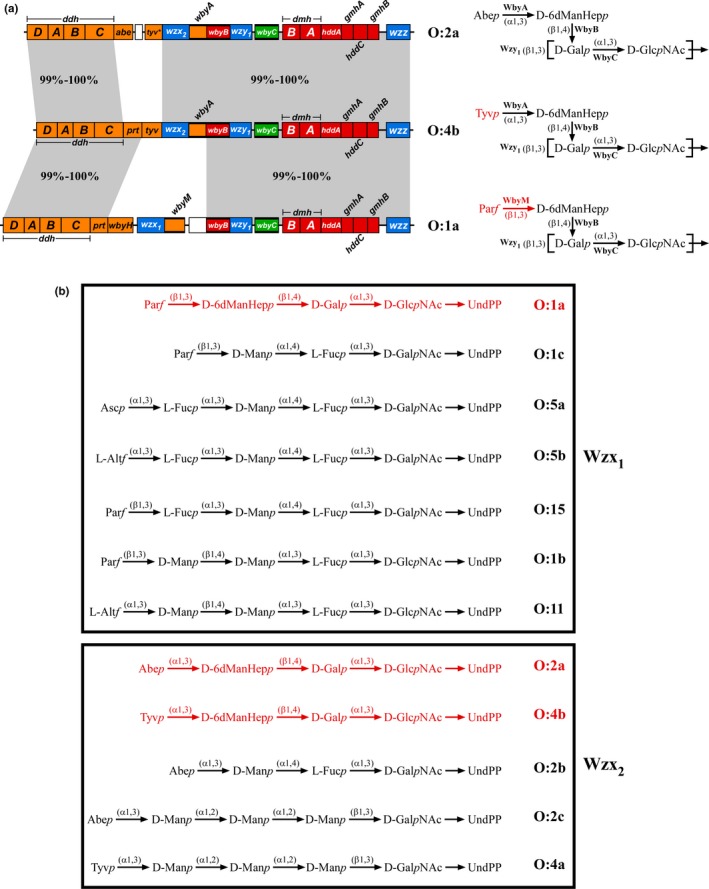
(a) The Y. pseudotuberculosis O:1a, O:2a and O:4b O‐antigen gene clusters (Pacinelli, Wang, & Reeves, 2002) and O‐unit structures (Kondakova et al., 2008, 2009, 2012). Gene clusters are drawn to scale on the left using the sequences from GenBank entries AF461768 to AF461770. Precursor sugar synthesis and glycosyltransferase (GT) genes are color‐coded based on their respective products: orange for the CDP‐dideoxyhexoses, red for CDP‐6‐deoxymannoheptose and green for UDP‐galactose, with the GT genes also indicated by bolded lines. The O‐antigen processing genes (wzx, wzy and wzz) are shown in blue, while remnant insertion sequence elements are indicated by the white boxes. Regions with high‐level nucleotide sequence identity (99–100%) between the gene clusters are shown within the gray shaded areas. The corresponding O‐unit structures are shown to the right of each gene cluster, along with the GTs responsible for each linkage. The square brackets denote the sites of the Wzy polymerization linkage. Residues, linkages and GTs on the O:1a and O:4b O units that differ from the O:2a O unit are shown in red. This figure is adapted from Kenyon et al. (2017). (b) Full range of Y. pseudotuberculosis O units translocated by the Wzx1 and Wzx2 flippases. O‐unit structures are shown without their polymerization linkages, as presented to Wzx prior to translocation. O units translocated by the same Wzx are grouped together into boxes, with the relevant Wzx indicated next to each box. The O units used in this study are shown in red
2. EXPERIMENTAL PROCEDURES
2.1. Strains and plasmids
The strains and plasmids used in this study are described in Table 1 and Table 2, respectively. E. coli and Y. pseudotuberculosis strains were grown in nutrient broth (10 g/L peptone, 5 g/L yeast extract, 5 g/L NaCl) at 37°C or 25°C, respectively. Gal‐free defined M9 media (6 g/L Na2HPO4, 3 g/L KH2PO4, 0.5 g/L NaCl, 1 g/L NH4Cl, 0.1 mM CaCl2, 1.6 mM MgSO4, 0.4% (v/v) glycerol, 20 mM sodium pyruvate, 3 g/L casamino acids, 8 μg/ml tryptophan, 1 μg/ml thiamine) was used when exogenous Gal was required to activate O‐antigen production. Agar was added (15 g/L) for solid growth media where required. Ampicillin (100 μg/ml), chloramphenicol (3 or 12.5 μg/ml), kanamycin (25 μg/ml) and/or streptomycin (1.6 mg/ml) were added as required for selection. S. cerevisiae strains were grown at 30°C in YPD broth (20 g/L peptone, 10 g/L yeast extract, 20 g/L glucose) or SD Ura˗ broth (6.7 g/L yeast nitrogen base (Sigma‐Aldrich), 1.92 g/L dropout supplement without uracil (Sigma‐Aldrich), 20 g/L glucose), with agar added (20 g/L) for solid growth media. Cycloheximide (2.5 μg/ml) was added to SD Ura˗ media for recombinant clone selection.
Table 1.
Strains used in this study
| Strain | Description | Source/Reference |
|---|---|---|
| CRY1‐2 | S. cerevisiae recombinational cloning strain; MATα, ura3Δ, cyh2 R | Raymond, Sims, and Olson (2002) |
| M85 | Y. pseudotuberculosis serotype O:2a | Kessler et al. (1991) |
| M444 | Y. pseudotuberculosis strain H892/87; serotype O:1a | Kessler, Haase, and Reeves (1993) |
| M451 | Y. pseudotuberculosis strain H713/86; serotype O:3 | Kessler et al. (1993) |
| M454 | Y. pseudotuberculosis strain H715/56; serotype O:4b | Kessler et al. (1993) |
| M460 | Y. pseudotuberculosis strain H720/86; serotype O:6 | Kessler et al. (1993) |
| M462 | Y. pseudotuberculosis strain H455/86; serotype O:7 | Kessler et al. (1993) |
| M2575 | Y. pseudotuberculosis strain 6088; serotype O:10 | Kenyon et al. (2011) |
| M2577 | Y. pseudotuberculosis strain MW864‐2; serotype O:12 | De Castro et al. (2013) |
| P5993 | E. coli DH10B ΔwecA::FRT | Liu et al. (2017) |
| P6124 | E. coli SΦ874 ΔgalE::FRT | This study |
| SΦ874 | E. coli lacZ4503 trp‐355 upp‐12 relA rpsL150 Δ(sbc‐rfb)86 | Neuhard and Thomassen (1976) |
Table 2.
Plasmids used in this study
| Plasmid | Descriptiona | Source/Reference |
|---|---|---|
| pPR981 | Low‐copy cosmid containing the entire M85 O:2a O‐antigen gene cluster. KanR, StrepR, SpecR | Kessler et al. (1991) |
| pPR1213 | SalI fragment of pPR981 containing the 3΄ half of the O:2a O‐antigen gene cluster cloned into pUC18. AmpR | Kessler et al. (1991) |
| pPR1214 | SalI fragment of pPR981 containing the 5΄ half of the O:2a O‐antigen gene cluster cloned into pUC18. AmpR | Kessler et al. (1991) |
| pPR2182 | The M85 wbyA gene cloned into pTrc99A. AmpR | This study |
| pPR2216 | The S. enterica LT2 abe gene cloned into pTrc99A. AmpR | Hong and Reeves (2015) |
| pPR2252 | Yeast‐E. coli shuttle vector with hooks from E. coli K‐12 galF and gnd and Y. pseudotuberculosis O:1b ddhD and wzz for recombinational cloning of O‐antigen gene clusters. AmpR, CmlR (E. coli); CyhS (yeast) | Liu et al. (2017) |
| pPR2272 | The M85 O:2a O‐antigen gene cluster cloned into pPR2252 (O:2a WT). CmlR | This study |
| pPR2283 | The M451 wzx 3 gene cloned into pWQ552. AmpR | This study |
| pPR2284 | The M2575 wzx 7 gene cloned into pWQ552. AmpR | This study |
| pPR2285 | The M2577 wzx 8 gene cloned into pWQ552. AmpR | This study |
| pPR2288 | The M85 wzx 2 gene cloned into pWQ552. AmpR | This study |
| pPR2289 | The M460 wzx 4 gene cloned into pWQ552. AmpR | This study |
| pPR2290 | The M462 wzx 5 gene cloned into pWQ552. AmpR | This study |
| pPR2293 | The M444 wzx 1 gene cloned into pWQ552. AmpR | This study |
| pPR2296 | pPR2272 with the wzx 2 gene replaced by an rpsL‐kan cassette (O:2a Δwzx). CmlR, KanR | This study |
| pPR2297 | pPR2272 with the abe‐tyv* region replaced by an rpsL‐kan cassette (O:2a Δabe). CmlR, KanR | This study |
| pPR2299 | pPR2272 with the wzx 2 gene replaced by the M444 wzx 1 gene (O:2a‐wzx 1). CmlR | This study |
| pPR2300 | pPR2272 with the abe‐tyv* region replaced by the M454 prt‐tyv region (O:4b WT). CmlR | This study |
| pPR2303 | pPR2272 with the wzx 2 gene replaced by the M451 wzx 3 gene (O:2a‐wzx 3). CmlR | This study |
| pPR2304 | pPR2272 with the wzx 2 gene replaced by the M460 wzx 4 gene (O:2a‐wzx 4). CmlR | This study |
| pPR2305 | pPR2272 with the wzx 2 gene replaced by the M462 wzx 5 gene (O:2a‐wzx 5). CmlR | This study |
| pPR2306 | pPR2272 with the wzx 2 gene replaced by the M2575 wzx 7 gene (O:2a‐wzx 7). CmlR | This study |
| pPR2307 | pPR2272 with the wzx 2 gene replaced by the M2577 wzx 8 gene (O:2a‐wzx 8). CmlR | This study |
| pPR2310 | pPR2300 with the wzx 2 gene replaced by an rpsL‐kan cassette (O:4b Δwzx). CmlR, KanR | This study |
| pPR2313 | pPR2300 with the wzx 2 gene replaced by the M444 wzx 1 gene (O:4b‐wzx 1). CmlR | This study |
| pPR2314 | pPR2272 with the abe‐tyv* region and wbyA gene replaced by the M444 prt‐wbyH region and wbyM gene, respectively (O:1a‐wzx 2). CmlR | This study |
| pPR2315 | pPR2272 with the abe‐tyv* and wzx 2 ‐wbyA regions replaced by the M444 prt‐wbyH and wzx 1 ‐wbyM regions, respectively (O:1a WT). CmlR | This study |
| pPR2320 | The M444 prt‐wbyH region cloned into pTrc99A. AmpR | This study |
| pPR2321 | The M454 prt‐tyv region cloned into pTrc99A. AmpR | This study |
| pPR2322 | pPR2300 with the tyv gene replaced by an rpsL‐kan cassette (O:4b Δtyv). CmlR, KanR | This study |
| pPR2323 | pPR2313 with the tyv gene replaced by an rpsL‐kan cassette (O:4b‐wzx 1 Δtyv). CmlR, KanR | This study |
| pPR2326 | pPR2315 with the prt‐wbyH region replaced by an rpsL‐kan cassette (O:1a Δprt‐wbyH). CmlR, KanR | This study |
| pPR2327 | pPR2315 with the wzx 1 gene replaced by an rpsL‐kan cassette (O:1a Δwzx). CmlR, KanR | This study |
| pTrc99A | Expression vector with pBR322 ori, IPTG‐inducible promoter (Ptrc), AmpR | Amann, Ochs, and Abel (1988) |
| pWQ552 | Expression vector with p15A ori, tetracycline‐inducible promoter (Ptet), AmpR | Willis and Whitfield (2013) |
Underlined descriptors indicate the plasmid names as shown in the main text. Abbreviations: ampicillin (AmpR), chloramphenicol (CmlR), kanamycin (KanR), spectinomycin (SpecR), streptomycin (StrepR) resistance; cycloheximide sensitivity (CyhS).
2.2. PCR and oligonucleotides
All PCR products were amplified with Q5 HF DNA Polymerase (New England Biolabs; NEB) and purified with the Isolate II PCR and Gel Kit (Bioline) as per the manufacturers’ instructions. All oligonucleotides used in this study are described in Supporting Information Table S1.
2.3. Strain manipulation
The SΦ874 ΔgalE strain (P6124) was generated, using the lambda‐RED recombinase system (Datsenko & Wanner, 2000), with the galE gene replaced by a kanamycin‐resistance cassette, which was subsequently excised by the FLP recombinase. Sanger sequencing (Australian Genome Research Facility; AGRF) was performed across the insertion junctions to confirm the gene knockout.
2.4. Single‐gene cloning and bacterial transformation
PCR products and vectors were double‐digested with the appropriate restriction enzymes (NEB high‐fidelity range), and digested vectors were dephosphorylated with recombinant shrimp alkaline phosphatase (NEB) under the standard conditions described by the manufacturer. Digested DNA was gel‐purified with the Wizard SV Gel and PCR Clean‐up Kit (Promega), and ligations were performed overnight at 16°C, using T4 DNA ligase (NEB) as per the manufacturer's instructions. Ligated DNA was transformed into CaCl2‐treated E. coli DH5α (Lederberg & Cohen, 1974), with putative clones isolated with the Purelink Quick Plasmid Miniprep Kit (Invitrogen). Sanger sequencing (AGRF) was used for confirmation of clones. Plasmids were transformed into the relevant E. coli strains by electroporation as described previously by Hong and Reeves (2014).
2.5. Cloning and manipulation of O‐antigen gene clusters
The “Operon Assembly Protocol”, described previously by Liu, Kenyon, Lee, and Reeves (2017), was used for assembly of the O:2a O‐antigen gene cluster onto the yeast‐E. coli Operon Assembly Vector (OAV; pPR2252) as a single‐copy clone. The OAV contains the Y. pseudotuberculosis O:2a ddhD and wzz “hook” sequences flanked by the E. coli K‐12 galF, JUMPStart and gnd sequences (Supporting Information Figure S3), meaning that expression of the cloned O‐antigen gene clusters are under the control of the K‐12 JUMPStart region (Bailey, Hughes, & Koronakis, 1997). Three DNA fragments were used to assemble the O:2a WT clone (Supporting Information Figure S3): the SalI fragments from the O:2a subclones pPR1213 and pPR1214, which contain the 3΄ and 5΄ halves of the O:2a gene cluster, respectively, and an overlapping ~4.4 kb HindIII fragment from pPR981, a multicopy cosmid clone of the O:2a O‐antigen gene cluster (Kessler, Brown, Romana, & Reeves, 1991). These fragments and the SmaI‐digested OAV were transformed into S. cerevisiae CRY1‐2, and recombinant clones were subsequently isolated and recovered in E. coli strain P5993 as per the standard OAP methodology. Functional testing of the O:2a WT clone (pPR2272) showed that it was producing a similar LPS profile to pPR981 when expressed in SΦ874 (Supporting Information Figure S4).
All subsequent O‐antigen gene cluster clones were generated by targeted gene knockouts and/or replacements using the modified version of the lambda‐RED recombinase system described by Hong, Cunneen, and Reeves (2012), where genes were replaced by an rpsL‐kan cassette under kanamycin selection, which was subsequently replaced by the gene of interest under high‐level streptomycin selection. All gene replacements were performed in P5993, which has an rpsL mutation conferring high‐level streptomycin resistance. Sanger sequencing (AGRF) was performed across the entire length of replacement genes or cassettes.
2.6. LPS extraction and visualization
Bacterial strains were subcultured 1:100 from overnight cultures and incubated, using the appropriate growth medium, selection and temperature, with shaking at 200 rpm. Overexpression of single‐gene clones was induced at OD600 ~ 0.3 with tetracycline (50 ng/ml) for pWQ552‐based clones or IPTG (1 mM) for pTrc99A‐based clones (Table 2). D‐Gal (0.2% (w/v)) was added at OD600 ~ 0.5 to activate O‐antigen production in ΔgalE strains, and cells were harvested at OD600 ~ 0.7.
LPS extractions were performed as described by McGrath and Osborn (1991), but with the modifications described by Liu et al. (2017). Samples were separated by tricine‐SDS‐PAGE on 13% (v/v) polyacrylamide gels, with loadings standardized relative to the densities of the lipid A‐core bands of each sample. LPS was visualized by silver staining (Brown, Romana, & Reeves, 1991) or transferred to a Hybond‐C Extra nitrocellulose membrane (Amersham Biosciences) for immunoblot analysis (Howland, 1996). Membranes were incubated with polyclonal anti‐Salmonella O2, O4 or O9 antisera (Statens Serum Institut), followed by goat‐anti‐rabbit‐IgG‐HRP‐conjugated antibody (Pierce). Chromogenic detection was performed with 0.3% (w/v) 4‐chloro‐1‐napthol, 0.075% (v/v) H2O2 and 10% (v/v) methanol in TBS‐Tween solution. Silver‐stained gels and Western blots were imaged in a ChemiDoc MP Imaging System (Bio‐Rad).
3. RESULTS
3.1. Cloning and expression of wild‐type Y. pseudotuberculosis O‐antigen gene clusters
Expression of a given O‐antigen gene cluster can often vary dramatically across different strains, due to the actions of unidentified or poorly understood regulatory factors (Lerouge & Vanderleyden, 2002). To facilitate meaningful comparisons between the Y. pseudotuberculosis O:1a, O:2a and O:4b O‐antigens and their Wzx flippases, we cloned the respective O‐antigen gene clusters and expressed them in a common genetic background. The E. coli strain SΦ874, which has a deletion of its entire O‐antigen gene cluster, was selected as the tester strain. An SΦ874 ΔgalE strain (P6124) was constructed to allow control of O‐antigen production by the presence or absence of exogenous Gal and avoid potential lethality, as the shared O‐unit main‐chain has Gal as the second sugar, and attachment of the second sugar is the first committed step in O‐antigen biosynthesis.
An O:2a WT clone (pPR2272) was constructed, using the Operon Assembly Protocol (Liu et al., 2017), resulting in the gene cluster being located on a plasmid with a mini‐F replicon, for which the copy number approximates the number of chromosomes per cell (Kline, 1985). The gene cluster is positioned downstream of the K‐12 O‐antigen regulatory system, including the JUMPStart sequence (Bailey et al., 1997), so expression is expected to be similar to normal chromosomal levels. An artificial O:4b WT clone (pPR2300) was then constructed by replacing the O:2a WT abe and degraded tyv genes (abe‐tyv*) with the O:4b prt‐tyv region (Figure 1a), while an artificial O:1a WT clone (pPR2315) was similarly constructed by replacing the O:2a WT abe‐tyv* and wzx 2 ‐wbyA regions with the O:1a prt‐wbyH and wzx 1 ‐wbyM regions (Figure 1a), respectively. As a result, the shared regions are identical across the three gene cluster clones. All three constructs were expressed in P6124, where their LPS profiles were found to be indistinguishable (Figure 2a, lanes 2, 4, 6), in stark contrast to the varied LPS profiles of the parental Y. pseudotuberculosis strains (Figure 2a, lanes 3, 5, 7), reinforcing the benefits of reconstructing the gene clusters and comparing them in the same strain. The identical O:2a and O:4b LPS profiles (Figure 2a, lanes 2, 4) also confirms that the O:2a Wzx2 (99% similar to the O:4b Wzx2) is equally efficient at flipping O:2a and O:4b O units. It is important to note that LPS from P6124 and the Y. pseudotuberculosis strains migrate at different rates due to differences in the composition of the LPS core oligosaccharides (Amor et al., 2000; Knirel & Anisimov, 2012).
Figure 2.
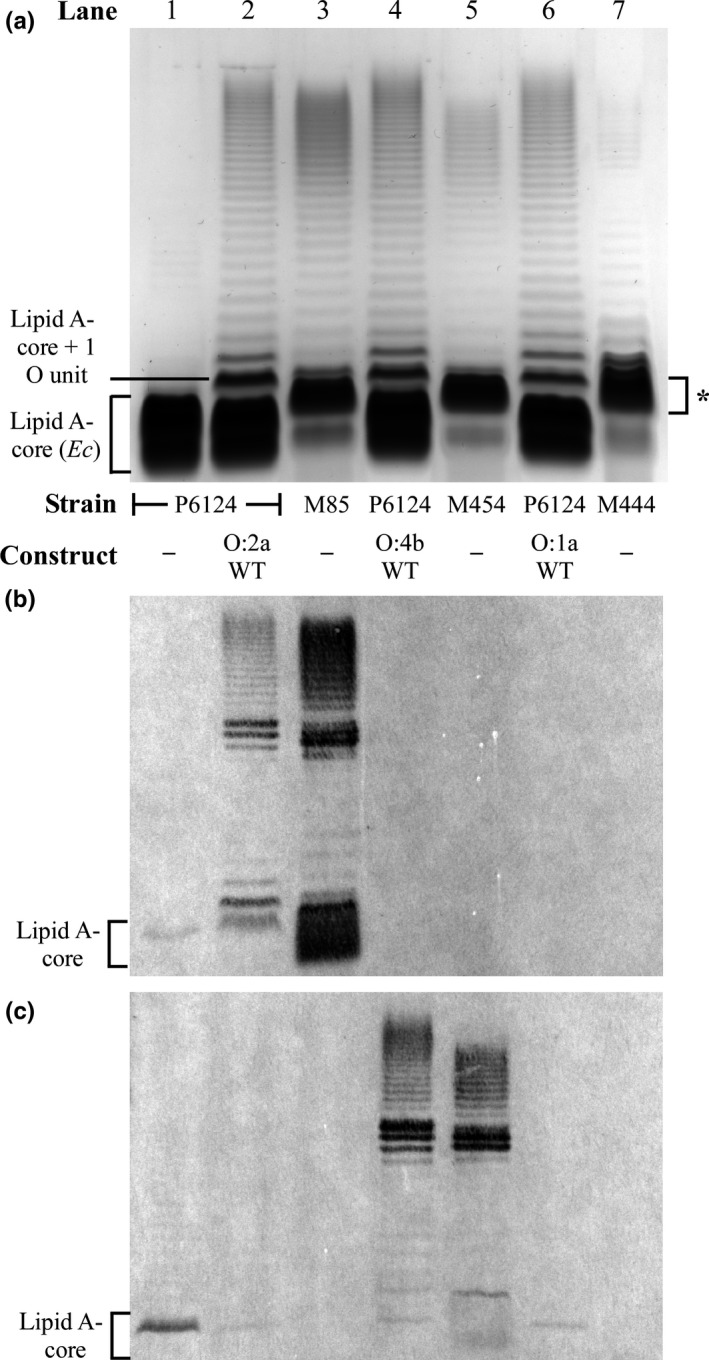
LPS profiles of the WT O:1a, O:2a and O:4b O‐antigen gene cluster clones. LPS samples were extracted from the indicated Y. pseudotuberculosis strains or P6124 harboring the indicated O‐antigen gene cluster constructs. The LPS was separated on a 13% (v/v) polyacrylamide gel by tricine‐SDS‐PAGE, followed by detection via silver staining (a) or transfer to a nitrocellulose membrane for immunoblotting with the S. enterica O4 (b) or O9 (c) antisera. The band corresponding to E. coli LPS without O antigen (lipid A‐core) is indicated to the left of the gel, while the equivalent band from Y. pseudotuberculosis is indicated by the asterisk on the right. The apparent location of these lipid A‐core only bands, which reacted nonspecifically with the antisera, are shown to the left of each membrane. The band corresponding to E. coli LPS with a single O unit (lipid A‐core + 1 O unit) is also shown on the gel
Immunoblotting was performed to confirm that the O:1a, O:2a and O:4b WT constructs were producing O antigen with the expected DDH structures. As expected, the Abep‐specific S. enterica O4 antiserum reacted with the P6124 + O:2a WT and M85 O:2a LPS (Figure 2b, lanes 2–3), while the Tyvp‐specific S. enterica O9 antiserum reacted with the P6124 + O:4b WT and M454 O:4b LPS (Figure 2c, lanes 4–5), confirming that both constructs are producing the expected O‐antigen structures. A weak nonspecific reaction was also visible with both antisera against the lipid A‐core LPS of P6124 (Figure 2b–c, lane 1). While there is no commercially available antiserum for detection of Parf residues, several indirect lines of evidence indicate that the O:1a WT construct is producing O:1a O antigen. The migration rate for P6124 + O:1a WT LPS is identical to its O:2a and O:4b counterparts (Figure 2a, lanes 2, 4, 6), suggesting the presence of a DDH residue. This residue is clearly not Abep or Tyvp, as neither P6124 + O:1a WT nor M444 O:1a LPS reacted with the O4 or O9 antisera (Figure 2b–c, lanes 6–7). Finally, these LPS samples also failed to react specifically with a paratose (Parp)‐specific S. enterica O2 antiserum (data not shown), leaving Parf as by far the most likely residue.
It is noteworthy that the O4 and O9 antisera both exhibited a stronger reaction with the parental Y. pseudotuberculosis O:2a and O:4b LPS than with the equivalent E. coli LPS (Figure 2b, lanes 2–3; Figure 2c, lanes 4–5), which did not reflect the different levels of O antigen as observed by silver staining (Figure 2a, lanes 2–5). A possible explanation for this difference may be that the bacteriophage‐encoded O‐antigen glucosylation system in E. coli K‐12 (Mann, Ovchinnikova, King, & Whitfield, 2015) is almost certainly adding a glucose (Glc) residue onto the GlcNAc of the O antigens. The presence of these Glc residues may be interfering with the interaction of the antisera with their target epitopes, resulting in a weakened immunoblot signal. However, this was not problematic, as the immunoblots were only intended to qualitatively identify the presence or absence of Abep and Tyvp residues, which was successfully achieved. Importantly, glucosylation of O antigens by this system occurs in the periplasm after Wzx translocation (Allison & Verma, 2000), and thus has no effect on our ability to interpret Wzx translocation efficiency.
3.2. Wzx1 and Wzx2 are both inefficient at flipping O units without a DDH residue
Removal of the terminal DDH residue from several S. enterica Gal‐initiated O antigens results in poor Wzx translocation (Hong et al., 2012; Liu et al., 2015, 2017). To assess this situation for the Y. pseudotuberculosis Wzx1 and Wzx2, we made O:2a Δabe (pPR2297) and O:1a Δprt‐wbyH (pPR2326) constructs that both produce the same O unit main‐chain without a DDH residue, but express wzx 2 or wzx 1, respectively.
These two constructs were transformed into P6124 and their LPS profiles analyzed by SDS‐PAGE. O‐antigen production from the O:2a Δabe construct (Figure 3, lane 3) was severely reduced relative to the O:2a WT construct (Figure 3, lane 2), and the LPS also migrated faster than WT, as expected for truncated O units. The O:1a Δprt‐wbyH construct similarly produced faster‐migrating LPS (Figure 3, lane 7), but with even lower O‐antigen levels than the O:2a Δabe construct (Figure 3, lane 3). Complementation of these two constructs with abe (pPR2216) or prt‐wbyH clones (pPR2320) fully restored WT O‐antigen levels and LPS migration rates (Figure 3, lanes 4, 8), confirming that the observed effects were not due to mutational polarity effects.
Figure 3.
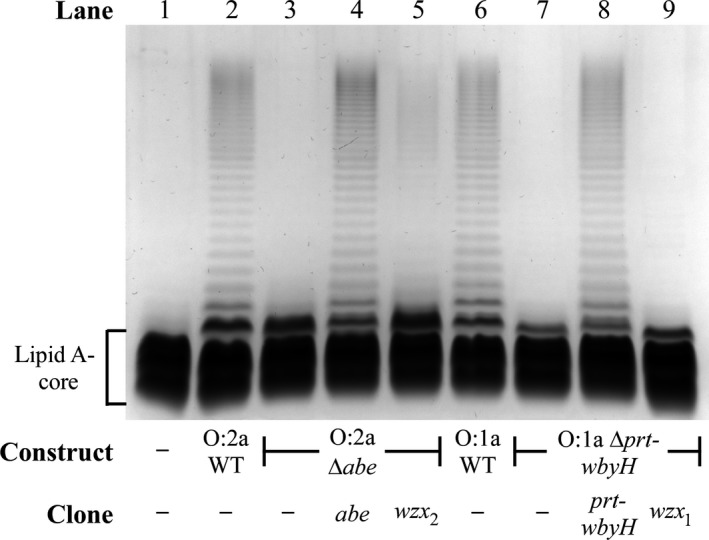
LPS profiles of the Y. pseudotuberculosis O‐antigen gene cluster clones expressing O antigen without a DDH residue. LPS samples were extracted from P6124 harboring the indicated O‐antigen gene cluster constructs and single‐gene clones, and separated on a 13% (v/v) polyacrylamide gel by tricine‐SDS‐PAGE, followed by detection via silver staining. The location of LPS without O antigen (lipid A‐core) is indicated
Wzx overexpression has previously been shown to improve the otherwise inefficient translocation of truncated O units (Hong & Reeves, 2014; Liu et al., 2015), so we overexpressed tetracycline‐inducible wzx 2 (pPR2288) and wzx 1 (pPR2293) clones alongside the O:2a Δabe and O:1a Δprt‐wbyH constructs, respectively. In both instances, O‐antigen levels increased (Figure 3, lanes 5, 9), with more detectable single‐O‐unit LPS for both constructs, and a small amount of long‐chain O antigen for the O:2a Δabe construct, but both remained well below WT levels (Figure 3, lanes 2, 6). Although this did confirm that both Wzx are inefficient at translocating the truncated O unit and that Wzx2 is more efficient than Wzx1, we cannot exclude the possibility that Wzy is also inefficient at polymerizing these truncated O units, making it impossible to determine the extent of Wzx inefficiency.
3.3. Wzx1 and Wzx2 show varied efficiency for flipping O‐units with alternative DDH residues
We next constructed O‐antigen gene cluster clones containing the alternative wzx (O:2a‐wzx 1 (pPR2299), O:4b‐wzx 1 (pPR2313) and O:1a‐wzx 2 (pPR2314)) to compare the translocation efficiencies of Wzx1 and Wzx2 with the alternative DDH structure(s) on a common O‐unit main‐chain. Equivalent wzx knockout clones (O:2a Δwzx (pPR2296), O:4b Δwzx (pPR2310) and O:1a Δwzx (pPR2327)) were also constructed to act as comparison controls.
These constructs were transformed into P6124 and their LPS profiles analyzed by SDS‐PAGE. Comparison of the O:1a, O:2a and O:4b Δwzx constructs with their WT counterparts revealed near‐complete loss of O‐antigen production (Figure 4, lanes 2–3, 5–6, 8–9), with the exception of a small amount of single‐O‐unit LPS that likely reflects background translocation by the E. coli colanic acid or enterobacterial common antigen flippases. The O:2a‐wzx 1 and O:4b‐wzx 1 constructs produced O‐antigen levels that were indistinguishable from their Δwzx counterparts (Figure 4, lanes 3–4, 6–7), demonstrating that Wzx1 is effectively unable to flip the O:2a and O:4b O units. The O:1a‐wzx 2 construct also had notably reduced O‐antigen levels relative to its WT counterpart (Figure 4, lanes 8, 10), demonstrating that while Wzx2 is clearly inefficient at flipping O:1a O units, this inefficiency is far less dramatic than observed for Wzx1 with the O:2a and O:4b O units (Figure 4, lanes 4, 7). When complemented with overexpressed cognate wzx clones, all wzx deletion and replacement constructs produced WT O‐antigen levels (Supporting Information Figure S1), confirming that the observed reductions in O antigen were exclusively due to the change in wzx. Overall, these results clearly demonstrate that both Wzx can distinguish between different terminal DDH residues with only minor differences in structure and/or linkage.
Figure 4.
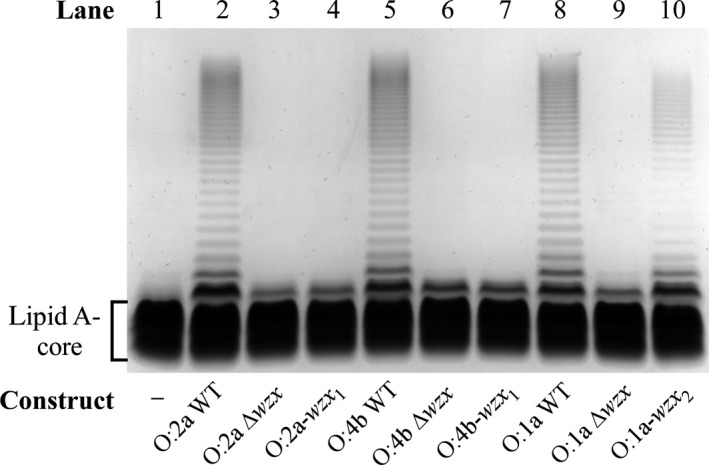
LPS profiles of O:1a, O:2a and O:4b O‐antigen gene cluster clones with the alternative wzx genes. LPS samples were extracted from P6124 harboring the indicated O‐antigen gene cluster constructs and separated on a 13% (v/v) polyacrylamide gel by tricine‐SDS‐PAGE, followed by detection via silver staining. The location of LPS without O antigen (lipid A‐core) is indicated
Our analysis was further expanded to explore whether Wzx1 or Wzx2 could efficiently translocate the equivalent O unit with an α(1,3)‐linked terminal Parp residue. This involved synthesis of a structure not found in nature, so the possibility of the terminal sugar GT or Wzy being inefficient at processing the O unit also had to be considered. Parp is the precursor sugar for Tyvp, so deletion of the tyv gene results in the synthesis of Parp exclusively, hence O:4b Δtyv (pPR2322) and O:4b‐wzx 1 Δtyv (pPR2323) clones were constructed. These two constructs were expressed in P6124 and their LPS profiles analyzed by SDS‐PAGE. The O‐antigen levels from the wzx 2‐expressing O:4b Δtyv construct (Figure 5a, lane 3) were significantly reduced relative to the O:4b WT construct (Figure 5a, lane 2), although substantial short‐chain and some long‐chain O antigen was still detectable. The low‐level O‐antigen produced by the O:4b‐wzx 1 Δtyv construct (Figure 5a, lane 7) was similar to the O:4b‐wzx 1 construct (Figure 5a, lane 6). Introduction of a prt‐tyv clone (pPR2321) reverted O‐antigen levels from both Δtyv constructs to that of their parental constructs (Figure 5a, lanes 4, 8).
Figure 5.
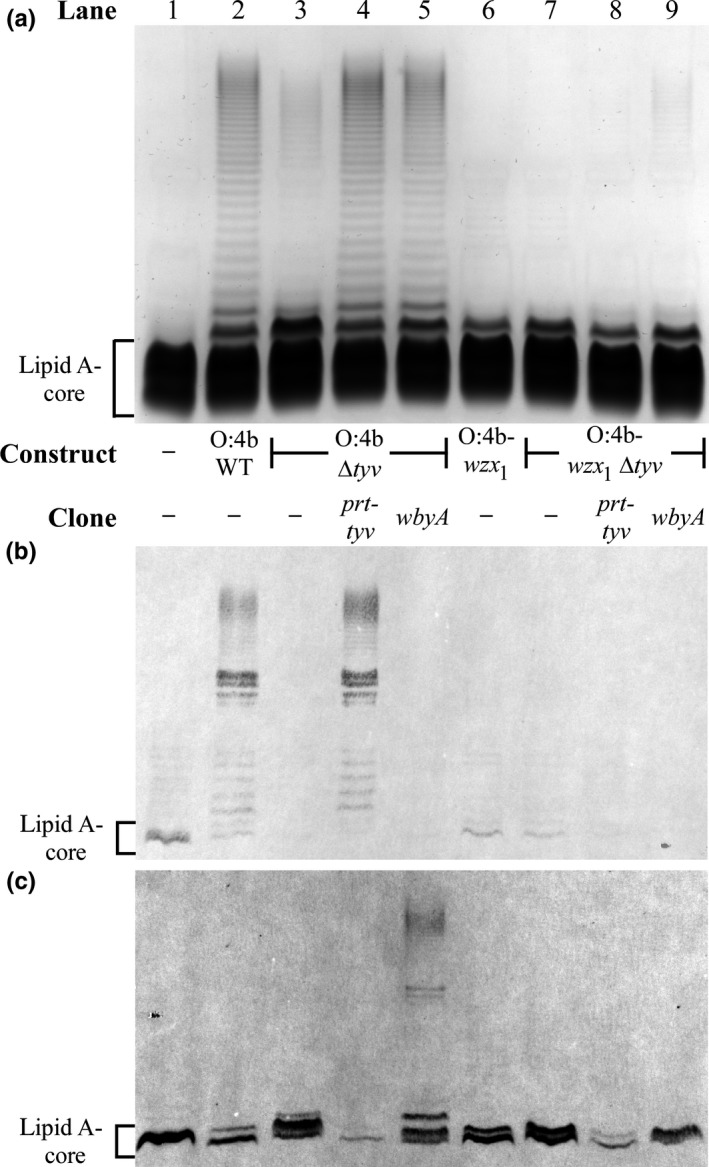
LPS profiles of Y. pseudotuberculosis O‐antigen gene cluster clones expressing O units with Parp. LPS samples were extracted from P6124 harboring the indicated O‐antigen gene cluster constructs and single‐gene clones, and separated on a 13% (v/v) polyacrylamide gel by tricine‐SDS‐PAGE, followed by detection via silver staining (a) or transfer to a nitrocellulose membrane for immunoblotting with the S. enterica O9 (b) or O2 (c) antisera. The location of LPS without O antigen (lipid A‐core), which reacted nonspecifically with the antisera, is indicated to the left of the gel and membranes
To determine if inefficient attachment of Parp by WbyA was a factor affecting O‐antigen production for the Δtyv constructs, an IPTG‐inducible wbyA clone (pPR2182) was introduced and overexpressed. For the wzx 2‐expressing O:4b Δtyv construct, this dramatically increased O‐antigen production (Figure 5a, lane 5) to levels that were indistinguishable from its WT counterpart (Figure 5a, lane 2). This indicates that WbyA is inefficient at Parp attachment, but once this inefficiency is overcome, Wzx2 can flip the available Parp‐containing O units with similar efficiency to its cognate O:2a and O:4b O units. In contrast, for the O:4b‐wzx 1 Δtyv construct, wbyA overexpression only minimally increased O‐antigen levels, including a very small amount of long‐chain O antigen (Figure 5a, lane 9). Hence it appears that Wzx1 is much less efficient than Wzx2 at translocating the Parp‐containing O units, but is more efficient than with the equivalent Abep‐ and Tyvp‐containing O units (Figure 4, lanes 4, 7).
Immunoblotting was used to confirm the predicted DDH residues on the O antigen from these constructs. O:4b WT and O:4b Δtyv + prt‐tyv LPS were both shown to contain Tyvp residues based on their strong reaction with the S. enterica O9 antiserum (Figure 5b, lanes 2, 4). Low‐level O9 background reactivity was also observed with the lipid A‐core band of several LPS samples (Figure 5b, lanes 1, 6, 7). The only specific (albeit weak) reaction detected with the Parp‐specific S. enterica O2 antiserum was with long‐chain LPS from O:4b Δtyv + wbyA (Figure 5c, lane 5), which confirmed that the O antigen produced by the O:4b Δtyv construct does contain Parp, and that Wzx2 can efficiently flip these O units following WbyA overproduction. The remaining LPS samples exhibited varying degrees of nonspecific lipid A‐core cross‐reactivity with the O2 antiserum, which in combination with the very low levels of O antigen produced by the O:4b‐wzx 1 Δtyv construct, prevented the positive identification of Parp‐containing O units from this construct (Figure 5c, lanes 7, 9). However, given that the same deletion in the O:4b Δtyv construct clearly resulted in the production of Parp‐containing O units (Figure 5c, lane 5), it is highly likely that the same is occurring for the O:4b‐wzx 1 Δtyv construct.
3.4. Several unrelated Wzx can efficiently translocate the O:2a O unit
We next attempted to compare the abilities of other Y. pseudotuberculosis Wzx flippases with more structurally divergent cognate O units to flip the O:2a O unit. The flippases selected were the O:3 Wzx3, O:6 Wzx4, O:7 Wzx5, O:10 Wzx7 and O:12 Wzx8, whose cognate O units all contain a DDH residue (Fig. S2). The wzx 3 and wzx 8 genes represent the other two wzx types found within the 14‐related Y. pseudotuberculosis O‐antigen gene clusters, whereas wzx 4 and wzx 5 are from unrelated gene clusters (Kenyon et al., 2017).
O:2a clones with the wzx 2 gene replaced by the alternative wzx (O:2a‐wzx 3 (pPR2303), O:2a‐wzx 4 (pPR2304), O:2a‐wzx 5 (pPR2305), O:2a‐wzx 7 (pPR2306) and O:2a‐wzx 8 (pPR2307)) were constructed and expressed in P6124, and their LPS profiles analyzed by SDS‐PAGE. In contrast to the O:2a‐wzx 1 construct (Figure 6a, lane 4), which produced O‐antigen levels indistinguishable from the O:2a Δwzx construct (Figure 6a, lane 3), all other wzx replacement constructs produced at least some detectable long‐chain O antigen (Figure 6a, lanes 5–9). The O‐antigen levels from the O:2a‐wzx 8 construct (Figure 6a, lane 9) were indistinguishable from the O:2a WT construct (Figure 6a, lane 2), while the O:2a‐wzx 4 construct (Figure 6a, lane 6) produced O‐antigen levels only slightly below WT. The O:2a‐wzx 3, O:2a‐wzx 5 and O:2a‐wzx 7 constructs (Figure 6a, lanes 5, 7, 8) had reduced O‐antigen levels not dissimilar from those observed previously for the O:1a‐wzx 2 construct (Figure 4, lane 10).
Figure 6.
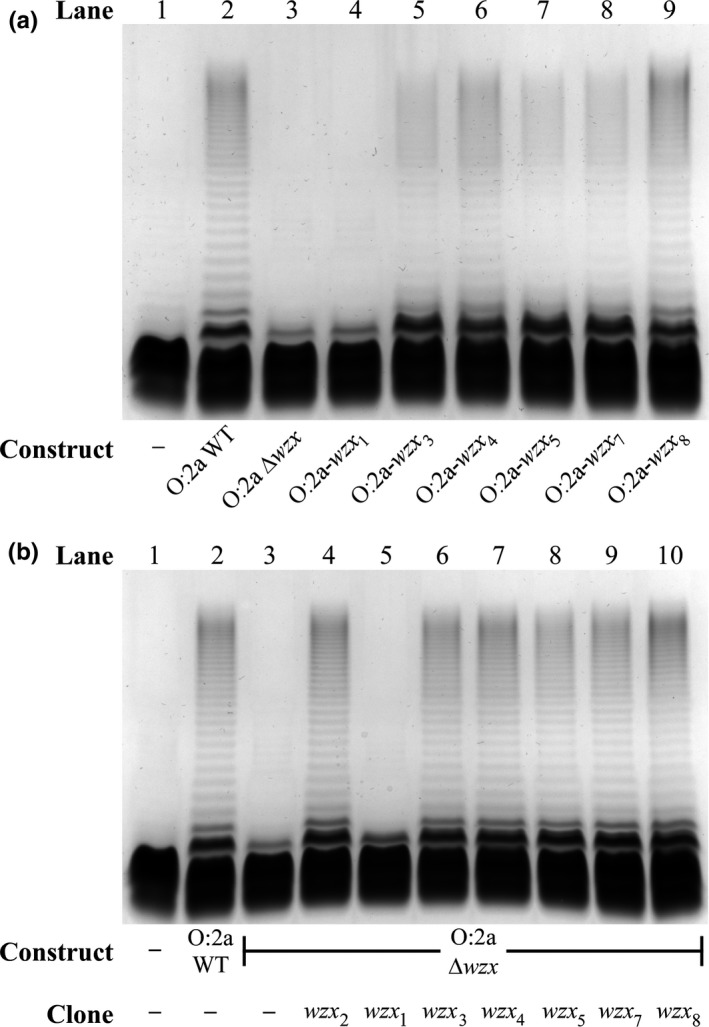
LPS profiles of the O:2a O‐antigen gene cluster clone containing alternative wzx genes. LPS samples were extracted from P6124 harboring the O:2a construct with wzx replaced (a) or the O:2a Δwzx construct alongside the indicated overexpressed wzx clones (b). Samples were separated on a 13% (v/v) polyacrylamide gel by tricine‐SDS‐PAGE, followed by detection via silver staining.
Overexpression of tetracycline‐inducible clones of each alternative wzx (wzx 1, pPR2293; wzx 3, pPR2283; wzx 4, pPR2289; wzx 5, pPR2290; wzx 7, pPR2284; wzx 8, pPR2285) restored O‐antigen production from the O:2a Δwzx construct (Figure 6b, lanes 6–10) to O:2a WT levels (Figure 6b, lane 2), except for wzx 1 (Figure 6b, lane 5), which was effectively indistinguishable from the O:2a Δwzx construct alone (Figure 6b, lane 3). This wzx 1 clone is known to be functional, as it can fully restore O‐antigen production from the O:1a Δwzx construct (Supporting Information Figure S1). It is apparent that Wzx1 is extremely inefficient at translocating the O:2a O unit, being less efficient than other Wzx flippases with cognate O units that are more structurally‐divergent from the O:2a O unit.
4. DISCUSSION
4.1. Wzx flippases can exhibit both strict and relaxed substrate preferences
Our understanding of the nature of Wzx substrate specificity has been continuously evolving over time, ranging from Wzx sequence diversity being seen as related to O‐unit diversity, through relaxed Wzx substrate specificity being driven only by the identity of the first O‐unit residue (Feldman et al., 1999; Marolda et al., 2004), to Wzx specificity extending along the length of its cognate O‐unit structure (Hong & Reeves, 2014; Hong et al., 2012). By undertaking a detailed analysis of the translocation efficiencies of Y. pseudotuberculosis Wzx with a series of closely related O‐unit structures (data summarized in Table 3), we have discovered that Wzx substrate preference can be more complex than either previously‐suggested model.
Table 3.
Comparison of Wzx repeat‐unit translocation efficiencies
| Repeat unit namea | Repeat unit structureb | Wzx (efficiency)c,d | Reference |
|---|---|---|---|
| Yp O:2a | Abep‐(α1→3)‐D‐6dManHepp‐(β1→4)‐D‐Galp‐(α1→3)‐D‐GlcpNAc→ |
Yp Wzx1 (˗) Yp Wzx 2 (+++) Yp Wzx3 (+) Yp Wzx4 (++) Yp Wzx5 (+) Yp Wzx7 (+) Yp Wzx8 (+++) |
This study |
| Yp O:4b | Tyvp‐(α1→3)‐D‐6dManHepp‐(β1→4)‐D‐Galp‐(α1→3)‐D‐GlcpNAc→ |
Yp Wzx1 (˗) Yp Wzx 2 (+++) |
|
| Yp O:1a | Parf‐(β1→3)‐D‐6dManHepp‐(β1→4)‐D‐Galp‐(α1→3)‐D‐GlcpNAc→ |
Yp
Wzx
1 (+++) Yp Wzx2 (++) |
|
| Yp O:4b‐Parp | Parp‐(α1→3)‐D‐6dManHepp‐(β1→4)‐D‐Galp‐(α1→3)‐D‐GlcpNAc→ |
Yp Wzx1 (+) Yp Wzx2 (+++) |
|
| Yp O:2a ΔAbep | D‐6dManHepp‐(β1→4)‐D‐Galp‐(α1→3)‐D‐GlcpNAc→ |
Yp Wzx1 (˗) Yp Wzx2 (+) |
|
| Se gp B | Abep‐(α1→3)‐D‐Manp‐(α1→4)‐L‐Rhap‐(α1→3)‐D‐Galp→ |
Se
Wzx
B (+++) Se WzxD (+++) |
Hong et al. (2012) |
| Se gp D | Tyvp‐(α1→3)‐D‐Manp‐(α1→4)‐L‐Rhap‐(α1→3)‐D‐Galp→ |
Se WzxB (+++) Se Wzx D (+++) Se WzxE (+) |
|
| Se gp E | D‐Manp‐(α1→4)‐L‐Rhap‐(α1→3)‐D‐Galp→ | Se WzxB (+) | |
| Se WzxD (+) | |||
| Se Wzx E (+++) | |||
| Se gp C2 | Abep‐(α1→3)‐L‐Rhap‐(α1→2)‐D‐Manp‐(α1→2)‐D‐Manp‐(α1→3)‐D‐Galp→ | Se Wzx C (+++) | Liu et al. (2015) |
| Se gp C2 ΔAbep | L‐Rhap‐(α1→2)‐D‐Manp‐(α1→2)‐D‐Manp‐(α1→3)‐D‐Galp→ | Se WzxC (+) | |
| Ec O1A | D‐ManpNAc‐(β1→2)‐L‐Rhap‐(α1→3)‐L‐Rhap‐(α1→3)‐L‐Rhap‐(β1→4)‐D‐GlcpNAc→ |
Ec
Wzx
O1A (+++) Ec WzxO2 (‐) Se WzxO42 (+++) |
Liu et al. (2017) |
| Ec O1A ΔManpNAc | L‐Rhap‐(α1→3)‐L‐Rhap‐(α1→3)‐L‐Rhap‐(β1→4)‐D‐GlcpNAc→ | Ec Wzx O1A (‐) | |
| Ec O16 |
OAc↓(2) D‐Galf‐(β1→6)‐D‐Glcp‐(α1→3)‐L‐Rhap‐(α1→3)‐D‐GlcpNAc→ |
Ec Wzx O16 (+++) | Hong and Reeves (2014) |
| Ec WzxO7 (++) | |||
| Ec WzxO111 (+) | |||
| Ec WzxO157 (‐) | |||
| Sf Wzx2a (‐) | |||
| Se WzxB (‐) | |||
| Ec O111 |
D‐Colp
(α1↓3) D‐Glcp‐(α1→4)‐D‐Galp‐(α1→3)‐D‐GlcpNAc→ (α1↑6) D‐Colp |
Ec
Wzx
O111 (+++) Ec WzxO16 (++) Ec WzxO7 (+) Sf Wzx2a (‐) |
|
| Ea amylovoran | Pyr‐(4,6)‐D‐Galp‐(α1→4)‐D‐GlcpA‐(β1→4)‐D‐Galp‐(α1→6)‐D‐Galp‐(β1→3)‐D‐Galp→ |
Ea
AmsL1 (+++) Ea AmsL2 (˗) |
Wang et al. (2012) |
| Ps stewartan | D‐Glcp‐(β1→6)‐D‐Galp‐(α1→4)‐D‐GlcpA‐(β1→4)‐D‐Galp‐(α1→6)‐D‐Glcp‐(β1→3)‐D‐Galp→ |
Ps Wzx1 (˗) Ps Wzx2 (+++) |
The prefixes indicate the source species for each structure: Erwinia amylovora (Ea), Escherichia coli (Ec), Pantoea stewartii (Ps), Salmonella enterica (Se), Shigella flexneri (Sf) or Yersinia pseudotuberculosis (Yp).
Repeat units are shown without their respective polymerization linkages, with residues displayed from last‐ to first‐sugar reading left‐to‐right, and the rightmost arrow denoting the undecaprenyl pyrophosphate linkage. All residue abbreviations are defined in‐text, except for D‐colitose (D‐Colp), D‐galactofuranose (D‐Galf), D‐glucopyranuronic acid (D‐GlcpA), D‐mannose (D‐Manp), D‐N‐acetylmannosamine (D‐ManpNAc), O‐acetyl (OAc) and L‐rhamnose (L‐Rhap).
The cognate Wzx for each repeat unit is bolded. Wzx translocation efficiencies for O units are ranked based on the amount of O antigen produced when Wzx is expressed at WT levels as follows: (˗) little to no detectable O antigen; (+) low‐level O‐antigen production with minimal long‐chain LPS; (++) moderate O‐antigen production with considerable long‐chain LPS that is below WT levels; (+++) O‐antigen production that is indistinguishable from WT levels.
The Wzx translocation efficiencies for the exopolysaccharides amylovoran and stewartan are only ranked as (˗) or (+++), as their production levels were only described as being “detectable” or “undetectable”.
Elimination of the terminal DDH residue dramatically affected the O‐unit translocation efficiency of Wzx1 and Wzx2 in this study (Figure 3), which was similarly observed for the Wzx of the S. enterica Gal‐initiated O‐antigen set (Hong et al., 2012; Liu et al., 2015). More importantly, both these Wzx could also distinguish, to varying degrees, changes to the structure and/or linkage of the terminal DDH residue on a common O‐unit main‐chain. Wzx1 was severely deficient at flipping O units with α(1,3)‐linked Abep, Tyvp or Parp residues, while Wzx2 was moderately deficient when translocating O units with a β(1,3)‐linked Parf residue (Table 3). Wang, Yang, and von Bodman (2012) previously proposed that two distinct Wzx pairs in Pantoea stewartii and Erwinia amylovora were required to translocate a pair of exopolysaccharides differing only by a Glc or pyruvate terminal side‐branch moiety, but did not directly confirm the role of each Wzx nor test the degree of specificity for each structure (Table 3). Hence, this study is the first to unambiguously demonstrate that Wzx substrate preference can be sensitive enough to be affected by minor alterations to the structure and/or linkage type of a single O‐unit residue.
In light of this high degree of specificity, it is surprising that Wzx1 and Wzx2 are both used for the translocation of several different O‐unit structures (Figure 1b). Wzx1 (94–95% similarity) translocates seven different O units with Parf, Altf or Ascp terminal sugars, while Wzx2 (99% similarity) translocates five different O units with Abep or Tyvp terminal sugars (Figure 7). Hence, both of these Wzx appear to be adapted to a defined set of terminal residues, rather than to entire O‐unit structures. However, other Wzx with more divergent cognate O‐unit structures were found to translocate the O:2a O unit with efficiencies ranging from moderate to indistinguishable from WT (Figure 6), indicating that the ability to translocate different O‐unit structures is common among the Y. pseudotuberculosis Wzx. Hong and Reeves (2014) also showed that several foreign Wzx were able to translocate the E. coli O16 and O111 O units with low‐to‐moderate efficiency (Table 3), indicating that partially‐relaxed Wzx substrate preference may be a widespread property.
Figure 7.
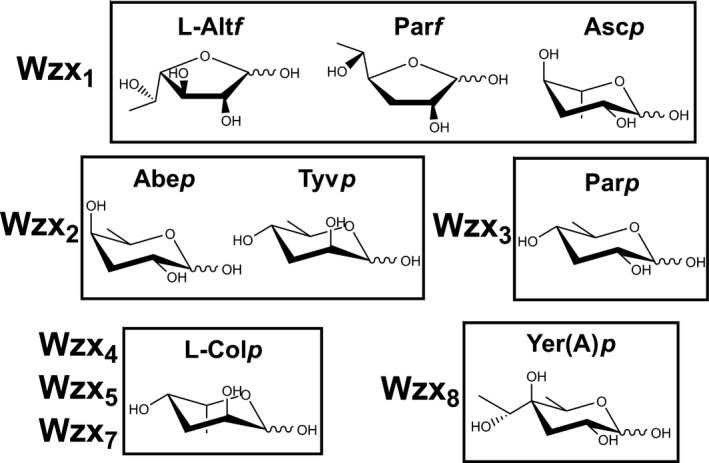
Structures of the DDH or DDH‐like terminal residues from Y. pseudotuberculosis O antigens. Residues grouped together into boxes are from O units flipped by the same Wzx, with the relevant Wzx indicated to the left of each box. Abbreviations: Abep, abequose; L‐Altf, L‐altrofuranose; Ascp, ascarylose; L‐Colp, L‐colitose; Parf, paratofuranose; Parp, paratose; Tyvp, tyvelose; Yer(A)p, yersiniose
4.2. A new model for Wzx substrate recognition
Wzx proteins belong to the polysaccharide transport (PST) family, one of 12 in the MOP (multidrug/oligosaccharide lipid/polysaccharide exporter) superfamily, and typically have 12 transmembrane segments (TMS). While there are currently no crystal structures available for Wzx or any other PST family member, the recently published structures of the related peptidoglycan flippase MurJ (Kuk, Mashalidis, & Lee, 2016) and Campylobacter oligosaccharide flippase PglK (Perez et al., 2015) have provided interesting insights. Both contain a central cavity that can be presented on either side of the IM, with transport proposed to occur by entry of the substrate into the cytoplasmic‐facing cavity conformation and exit via the periplasmic‐facing form, and it is likely that Wzx O‐unit translocation proceeds by a similar mechanism.
The combination of highly sensitive and relaxed substrate preference exhibited by Wzx1 and Wzx2 seems too complex to be explained by a simple lock‐and‐key model in which Wzx recognises the entire O‐unit structure. Rather, we propose “Structure‐Specific Triggering” as the mechanism, in which flipping is triggered by Wzx interacting with one or more specific structural elements on the complete O unit. We envisage specific sites within each Wzx that interact directly with the structural element(s) and trigger the translocation event. The full O‐unit structure would need to fit within the Wzx central cavity, and there may well be other critical requirements, such as a specific first sugar, but some changes to the composition and length of the O‐unit main‐chain have little effect on translocation efficiency for Wzx1 and Wzx2. Rapid translocation triggered by complete O units would mean that intermediates or unrelated foreign O units lacking the key structural element(s) are rarely flipped under normal circumstances. However, in cells unable to synthesize the complete O unit, other structural elements on the incomplete or foreign O units may occasionally interact with Wzx and trigger flipping at a reduced frequency. Overexpression of wzx can at least partially overcome this inefficiency by increasing the number of available Wzx proteins present within the cell to perform low‐frequency translocation; however, the capacity for wzx overexpression in nature is poorly understood. In the absence of the triggering residues, the increase in number of Wzx can compensate for the reduced translocation efficiency (Hong & Reeves, 2014; Liu et al., 2015). The fact that Wzx overexpression alone can dramatically improve O‐antigen production implies that Wzx does not need to be part of a multi‐protein complex to function as a flippase. However, this does not necessarily infer that Wzx cannot behave differently if it is in a complex, as has been observed for the E. coli K‐12 ECA Wzx (Marolda, Tatar, Alaimo, Aebi, & Valvano, 2006).
The structural elements responsible for triggering high‐frequency translocation in Wzx1 and Wzx2 appear to be the terminal DDH or DDH‐like residues. The ability of both Wzx to flip several different DDH residues suggests that specific structural elements are directly recognized by Wzx. For instance, the different preferences of the two Wzx might be explained by Abep, Tyvp and Parp all having a carbon‐6 positioned above the ring plane that is recognized by Wzx2, while the two‐carbon structure protruding from the furanose rings of Altf and Parf may be recognized by Wzx1 (Figure 7). A possible trigger for Wzx1 translocation by Ascp is harder to identify, however, the different orientation of its carbon‐6 relative to Abep, Tyvp and Parp, which cannot be flipped by Wzx1 (Table 3), may be important. Direct recognition of specific DDH structural elements may also explain the ability of Wzx4 and Wzx8 to flip the O:2a O unit with high efficiency. The cognate O:12 O unit of Wzx8 contains a terminal yersiniose (Yer(A)p), which is structurally identical to Abep outside of a two‐carbon branched‐chain on carbon‐4 (Figure 7), so both residues may interact equally well with the Wzx8 recognition site. The terminal residues of the Wzx4, Wzx5 and Wzx7 cognate O units are all colitose (Fig. 7), yet Wzx4 was much more efficient at flipping the O:2a O unit. This may be due to its cognate O:6 O unit also having a Yer(A)p residue (Fig. S2) that may be recognized by Wzx4, thus facilitating recognition of the O:2a Abep residue.
Despite the scarcity of closely related O‐antigen gene clusters and structures in most species, there is some supporting evidence for Structure‐specific Triggering being a broadly applicable model. The S. enterica O42 Wzx is known to be highly efficient in the translocation of the E. coli O1A O unit (Table 3), possibly due to its cognate O unit having the same terminal residue as O1A (Liu et al., 2017). The trisaccharide O units of Pseudomonas aeruginosa serotypes O2, O5, O16, O18, and O20 all share a common Wzx, but the only conserved sugar among the O units is the initial UndPP‐linked N‐acetylfucosamine (Islam & Lam, 2014). The remaining two residues on each O unit are both uronic acid sugars with only minor linkage variations, which may be the critical element required to trigger high‐frequency Wzx translocation. This example illustrates the importance of experimentally varying the O unit for studying Wzx specificity, which is necessary to prove a requirement for a specific trigger. Although additional functional and structural studies are still necessary, these known instances of diverse Wzx with both single‐residue specificity and a capacity to translocate a broader range of substrates suggest that our Structure‐Specific Triggering model does provide a foundation for resolving these seemingly disparate properties and developing a deeper understanding of the complex nature of Wzx‐substrate interactions.
CONFLICT OF INTEREST
MAL and PRR contributed to the conception and design of the study. MAL and PM contributed to the acquisition, analysis and interpretation of the data. MAL and PRR contributed to the writing of the manuscript. The authors declare that they have no conflicts of interest.
Supporting information
ACKNOWLEDGMENTS
Some of the work reported here was supported by Australian Research Centre Grant DP140104068. The wbyA clone was constructed by Dr Johanna Kenyon (Queensland University of Technology) while at the University of Sydney.
Liu MA, Morris P, Reeves PR. Wzx flippases exhibiting complex O‐unit preferences require a new model for Wzx–substrate interactions. MicrobiologyOpen. 2019;8:e655 10.1002/mbo3.655
REFERENCES
- Allison, G. E. , & Verma, N. K. (2000). Serotype‐converting bacteriophages and O‐antigen modification in Shigella flexneri . Trends in Microbiology, 8, 17–23. 10.1016/S0966-842X(99)01646-7 [DOI] [PubMed] [Google Scholar]
- Amann, E. , Ochs, B. , & Abel, K. J. (1988). Tightly regulated tac promoter vectors useful for the expression of unfused and fused proteins in Escherichia coli . Gene, 69, 301–315. 10.1016/0378-1119(88)90440-4 [DOI] [PubMed] [Google Scholar]
- Amor, K. , Heinrichs, D. E. , Frirdich, E. , Ziebell, K. , Johnson, R. P. , & Whitfield, C. (2000). Distribution of core oligosaccharide types in lipopolysaccharides from Escherichia coli . Infection and Immunity, 68, 1116–1124. 10.1128/IAI.68.3.1116-1124.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey, M. J. A. , Hughes, C. , & Koronakis, V. (1997). RfaH and the ops element, components of a novel system controlling bacterial transcription elongation. Molecular Microbiology, 26, 845–851. 10.1046/j.1365-2958.1997.6432014.x [DOI] [PubMed] [Google Scholar]
- Brown, P. K. , Romana, L. K. , & Reeves, P. R. (1991). Cloning of the rfb gene cluster of a group C2 Salmonella strain: Comparison with the rfb regions of groups B and D. Molecular Microbiology, 5, 1873–1881. 10.1111/j.1365-2958.1991.tb00811.x [DOI] [PubMed] [Google Scholar]
- Datsenko, K. A. , & Wanner, B. L. (2000). One‐step inactivation of chromosomal genes in Escherichia coli K‐12 using PCR products. Proceedings of the National Academy of Sciences, 97, 6640–6645. 10.1073/pnas.120163297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Castro, C. , Kenyon, J. J. , Cunneen, M. M. , Molinaro, A. , Holst, O. , Skurnik, M. , & Reeves, P. R. (2013). The O‐specific polysaccharide structure and gene cluster of serotype O:12 of the Yersinia pseudotuberculosis complex, and the identification of a novel l‐quinovose biosynthesis gene. Glycobiology, 23, 346–353. 10.1093/glycob/cws145 [DOI] [PubMed] [Google Scholar]
- DebRoy, C. , Fratamico, P. M. , Yan, X. , Baranzoni, G. , Liu, Y. , Needleman, D. S. , … Mwangi, M. (2016). Comparison of O‐antigen gene clusters of all O‐serogroups of Escherichia coli and proposal for adopting a new nomenclature for O‐typing. PLoS ONE, 11, e0147434. 10.1371/journal.pone.0147434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman, M. F. , Marolda, C. L. , Monteiro, M. A. , Perry, M. B. , Parodi, A. J. , & Valvano, M. A. (1999). The activity of a putative polyisoprenol‐linked sugar translocase (Wzx) involved in Escherichia coli O antigen assembly is independent of the chemical structure of the O repeat. Journal of Biological Chemistry, 274, 35129–35138. 10.1074/jbc.274.49.35129 [DOI] [PubMed] [Google Scholar]
- Hong, Y. , Cunneen, M. M. , & Reeves, P. R. (2012). The Wzx translocases for Salmonella enterica O‐antigen processing have unexpected serotype specificity. Molecular Microbiology, 84, 620–630. 10.1111/j.1365-2958.2012.08048.x [DOI] [PubMed] [Google Scholar]
- Hong, Y. , Liu, M. A. , & Reeves, P. R. (2018). Progress in our understanding of Wzx flippase for translocation of bacterial membrane lipid‐linked oligosaccharide. Journal of Bacteriology, 200, e00154–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong, Y. , & Reeves, P. R. (2014). Diversity of O‐antigen repeat unit structures can account for the substantial sequence variation of Wzx translocases. Journal of Bacteriology, 196, 1713–1722. 10.1128/JB.01323-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong, Y. , & Reeves, P. R . (2015). Model for the controlled synthesis of O‐antigen repeat units involving the WaaL ligase. MSphere, 1, e00074–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howland, J. L . (1996). Short protocols in molecular biology, third edition. p. 836. New York: Headington Hill Hall, John Wiley & Sons. [Google Scholar]
- Islam, S. T. , & Lam, J. S. (2013). Wzx flippase‐mediated membrane translocation of sugar polymer precursors in bacteria. Environmental Microbiology, 15, 1001–1015. 10.1111/j.1462-2920.2012.02890.x [DOI] [PubMed] [Google Scholar]
- Islam, S. T. , & Lam, J. S. (2014). Synthesis of bacterial polysaccharides via the Wzx/Wzy‐dependent pathway. Canadian Journal of Microbiology, 60, 697–716. 10.1139/cjm-2014-0595 [DOI] [PubMed] [Google Scholar]
- Joiner, K. A. (1988). Complement evasion by bacteria and parasites. Annual Review of Microbiology, 42, 201–230. 10.1146/annurev.mi.42.100188.001221 [DOI] [PubMed] [Google Scholar]
- Jorgenson, M. A. , & Young, K. D. (2016). Interrupting biosynthesis of O antigen or the lipopolysaccharide core produces morphological defects in Escherichia coli by sequestering undecaprenyl phosphate. Journal of Bacteriology, 198, 3070–3079. 10.1128/JB.00550-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalynych, S. , Morona, R. , & Cygler, M. (2014). Progress in understanding the assembly process of bacterial O‐antigen. FEMS Microbiology Reviews, 38, 1048–1065. 10.1111/1574-6976.12070 [DOI] [PubMed] [Google Scholar]
- Kenyon, J. J. , Cunneen, M. M. , & Reeves, P. R. (2017). Genetics and evolution of Yersinia pseudotuberculosis O‐specific polysaccharides: A novel pattern of O‐antigen diversity. FEMS Microbiology Reviews, 41, 200–217. 10.1093/femsre/fux002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenyon, J. J. , De Castro, C. , Cunneen, M. M. , Reeves, P. R. , Molinaro, A. , Holst, O. , & Skurnik, M. (2011). The genetics and structure of the O‐specific polysaccharide of Yersinia pseudotuberculosis serotype O:10 and its relationship with Escherichia coli O111 and Salmonella enterica O35. Glycobiology, 21, 1131–1139. 10.1093/glycob/cwr006 [DOI] [PubMed] [Google Scholar]
- Kessler, A. C. , Brown, P. K. , Romana, L. K. , & Reeves, P. R. (1991). Molecular cloning and genetic characterization of the rfb region from Yersinia pseudotuberculosis serogroup IIA, which determines the formation of the 3,6‐dideoxyhexose abequose. Journal of General Microbiology, 137, 2689–2695. 10.1099/00221287-137-12-2689 [DOI] [PubMed] [Google Scholar]
- Kessler, A. C. , Haase, A. , & Reeves, P. R. (1993). Molecular analysis of the 3,6‐dideoxyhexose pathway genes of Yersinia pseudotuberculosis serogroup IIA. Journal of Bacteriology, 175, 1412–1422. 10.1128/jb.175.5.1412-1422.1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kline, B. C. (1985). A review of mini‐F plasmid maintenance. Plasmid, 14, 1–16. 10.1016/0147-619X(85)90027-7 [DOI] [PubMed] [Google Scholar]
- Knirel, Y. A. , & Anisimov, A. P. (2012). Lipopolysaccharide of Yersinia pestis, the cause of plague: Structure, genetics, biological properties. Acta Naturae, 4, 46–58. [PMC free article] [PubMed] [Google Scholar]
- Kondakova, A. N. , Bystrova, O. V. , Shaikhutdinova, R. Z. , Ivanov, S. A. , Dentovskaya, S. V. , Shashkov, A. S. , …. Anisimov, A. P. (2009). Structure of the O‐antigen of Yersinia pseudotuberculosis O:4b. Carbohydrate Research, 344, 152–154. 10.1016/j.carres.2008.10.002 [DOI] [PubMed] [Google Scholar]
- Kondakova, A. N. , Bystrova, O. V. , Shashkov, A. S. , Lindner, B. , Creuzenet, C. , & Knirel, Y. A. (2008). Structural studies of the O‐antigens of Yersinia pseudotuberculosis O:2a and mutants thereof with impaired 6‐deoxy‐d‐manno‐heptose biosynthesis pathway. Carbohydrate Research, 343, 1383–1389. 10.1016/j.carres.2008.04.003 [DOI] [PubMed] [Google Scholar]
- Kondakova, A. N. , Sevillano, A. M. , Shaikhutdinova, R. Z. , Lindner, B. , Komandrova, N. A. , Dentovskaya, S. V. , … Knirel, Y. A. (2012). Revision of the O‐polysaccharide structure of Yersinia pseudotuberculosis O:1a; confirmation of the function of WbyM as paratosyltransferase. Carbohydrate Research, 350, 98–102. 10.1016/j.carres.2011.12.014 [DOI] [PubMed] [Google Scholar]
- Kuk, A. C. Y. , Mashalidis, E. H. , & Lee, S.‐Y. (2016). Crystal structure of the MOP flippase MurJ in an inward‐facing conformation. Nature Structural & Molecular Biology, 24, 171–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lederberg, E. M. , & Cohen, S. N. (1974). Transformation of Salmonella typhimurium by plasmid deoxyribonucleic acid. Journal of Bacteriology, 119, 1072–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerouge, I. , & Vanderleyden, J. (2002). O‐antigen structural variation: Mechanisms and possible roles in animal/plant‐microbe interactions. FEMS Microbiology Reviews, 26, 17–47. 10.1111/j.1574-6976.2002.tb00597.x [DOI] [PubMed] [Google Scholar]
- Liu, D. , Cole, R. A. , & Reeves, P. R. (1996). An O‐antigen processing function for Wzx (RfbX): A promising candidate for O‐unit flippase. Journal of Bacteriology, 178, 2102–2107. 10.1128/jb.178.7.2102-2107.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, M. A. , Kenyon, J. J. , Lee, J. , & Reeves, P. R. (2017). Rapid customised operon assembly by yeast recombinational cloning. Applied Microbiology and Biotechnology, 101, 4569–4580. 10.1007/s00253-017-8213-9 [DOI] [PubMed] [Google Scholar]
- Liu, B. , Knirel, Y. A. , Feng, L. , Perepelov, A. V. , Senchenkova, S. N. , Reeves, P. R. , & Wang, L. (2014). Structural diversity in Salmonella O antigens and its genetic basis. FEMS Microbiology Reviews, 38, 56–89. 10.1111/1574-6976.12034 [DOI] [PubMed] [Google Scholar]
- Liu, B. , Knirel, Y. A. , Feng, L. , Perepelov, A. V. , Senchenkova, S. N. , Wang, Q. , … Wang, L. (2008). Structure and genetics of Shigella O antigens. FEMS Microbiology Reviews, 32, 627–653. 10.1111/j.1574-6976.2008.00114.x [DOI] [PubMed] [Google Scholar]
- Liu, M. A. , Stent, T. L. , Hong, Y. , & Reeves, P. R . (2015). Inefficient translocation of a truncated O unit by a Salmonella Wzx affects both O‐antigen production and cell growth. FEMS Microbiology Letters, 362, fnv053. [DOI] [PubMed] [Google Scholar]
- Mann, E. , Ovchinnikova, O. G. , King, J. D. , & Whitfield, C. (2015). Bacteriophage‐mediated glucosylation can modify lipopolysaccharide O‐antigens synthesized by an ATP‐Binding Cassette (ABC) transporter‐dependent assembly mechanism. Journal of Biological Chemistry, 290, 25561–25570. 10.1074/jbc.M115.660803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marolda, C. L. , Tatar, L. D. , Alaimo, C. , Aebi, M. , & Valvano, M. A. (2006). Interplay of the Wzx translocase and the corresponding polymerase and chain length regulator proteins in the translocation and periplasmic assembly of lipopolysaccharide O antigen. Journal of Bacteriology, 188, 5124–5135. 10.1128/JB.00461-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marolda, C. L. , Vicarioli, J. , & Valvano, M. A. (2004). Wzx proteins involved in biosynthesis of O antigen function in association with the first sugar of the O‐specific lipopolysaccharide subunit. Microbiology, 150, 4095–4105. 10.1099/mic.0.27456-0 [DOI] [PubMed] [Google Scholar]
- Matamouros, S. , & Miller, S. I. (2015). S. Typhimurium strategies to resist killing by cationic antimicrobial peptides. Biochimica et Biophysica Acta, 1848, 3021–3025. 10.1016/j.bbamem.2015.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath, B. C. , & Osborn, M. J. (1991). Localisation of the terminal steps of O‐antigen synthesis in Salmonella typhimurium . Journal of Bacteriology, 173, 649–654. 10.1128/jb.173.2.649-654.1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray, G. L. , Attridge, S. R. , & Morona, R. (2006). Altering the length of the lipopolysaccharide O antigen has an impact on the interaction of Salmonella enterica serovar Typhimurium with macrophages and complement. Journal of Bacteriology, 188, 2735–2739. 10.1128/JB.188.7.2735-2739.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuhard, J. , & Thomassen, E. (1976). Altered deoxyribonucleotide pools in P2 eductants of Escherichia coli K‐12 due to deletion of the dcd gene. Journal of Bacteriology, 126, 999–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacinelli, E. , Wang, L. , & Reeves, P. R. (2002). Relationship of Yersinia pseudotuberculosis O antigens IA, IIA, and IVB: The IIA gene cluster was derived from that of IVB. Infection and Immunity, 70, 3271–3276. 10.1128/IAI.70.6.3271-3276.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez, C. , Gerber, S. , Boilevin, J. , Bucher, M. , Darbre, T. , Aebi, M. , … Locher, K. P. (2015). Structure and mechanism of an active lipid‐linked oligosaccharide flippase. Nature, 524, 433–438. 10.1038/nature14953 [DOI] [PubMed] [Google Scholar]
- Price, N. P. , & Momany, F. A. (2005). Modeling bacterial UDP‐HexNAc: Polyprenol‐P HexNAc‐1‐P transferases. Glycobiology, 15, 29R–42R. 10.1093/glycob/cwi065 [DOI] [PubMed] [Google Scholar]
- Raetz, C. R. H. , & Whitfield, C. (2002). Lipopolysaccharide endotoxins. Annual Review of Biochemistry, 71, 635–700. 10.1146/annurev.biochem.71.110601.135414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond, C. K. , Sims, E. H. , & Olson, M. V. (2002). Linker‐mediated recombinational subcloning of large DNA fragments using yeast. Genome Research, 12, 190–197. 10.1101/gr.205201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves, P. R. , & Cunneen, M. M. (2009). Biosynthesis of O‐antigen chains and assembly In Moran A. P., Holst O., Brennan P. J., & von Itzstein M. (Eds.), Microbial glycobiology: Structures, relevance and applications (pp. 319–335). Amsterdam: Elsevier. [Google Scholar]
- Rick, P. D. , Barr, K. , Sankaran, K. , Kajimura, J. , Rush, J. S. , & Waechter, C. J. (2003). Evidence that the wzxE gene of Escherichia coli K‐12 encodes a protein involved in the transbilayer movement of a trisaccharide‐lipid intermediate in the assembly of enterobacterial common antigen. Journal of Biological Chemistry, 278, 16534 10.1074/jbc.M301750200 [DOI] [PubMed] [Google Scholar]
- Wang, X. , Yang, F. , & von Bodman, S. B. (2012). The genetic and structural basis of two distinct terminal side branch residues in stewartan and amylovoran exopolysaccharides and their potential role in host adaptation. Molecular Microbiology, 83, 195–207. 10.1111/j.1365-2958.2011.07926.x [DOI] [PubMed] [Google Scholar]
- Whitfield, C. , & Trent, M. S. (2014). Biosynthesis and export of bacterial lipopolysaccharides. Annual Review of Biochemistry, 83, 99–128. 10.1146/annurev-biochem-060713-035600 [DOI] [PubMed] [Google Scholar]
- Willis, L. M. , & Whitfield, C. (2013). KpsC and KpsS are retaining 3‐deoxy‐d‐manno‐oct‐2‐ulosonic acid (Kdo) transferases involved in synthesis of bacterial capsules. Proceedings of the National Academy of Sciences, 110, 20753–20758. 10.1073/pnas.1312637110 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


