Abstract
Three previously unidentified Gram‐positive anaerobic coccoid bacteria, strains KhD‐2T, KHD4T, and Kh‐D5T, isolated from a vaginal swab, were characterized using the taxonogenomics concept. The phylogenic analysis, phenotypic characteristics, and genotypic data presented in this report attest that these three bacteria are distinct from previously known bacterial species with standing in nomenclature and represent three new Peptoniphilus species. Strain KhD‐2T is most closely related to Peptoniphilus sp. DNF00840 and Peptoniphilus harei (99.7% and 98.2% identity, respectively); strain KHD4T to Peptoniphilus lacrimalis (96%) and strain Kh‐D5T to Peptoniphilus coxii (97.2%). Strains KhD‐2T, KHD4T, and Kh‐D5T DNA G+C contents are, respectively, 34.23%, 31.87%, and 49.38%; their major fatty acid was C16:0 (41.6%, 32.0%, and 36.4%, respectively). We propose that strains KhD‐2T (=CSUR P0125 = DSM 101742), KHD4T (=CSUR P0110 = CECT 9308), and Kh‐D5T (=CSUR P2271 = DSM 101839) be the type strains of the new species for which the names Peptoniphilus vaginalis sp. nov., Peptoniphilus raoultii sp. nov., and Peptoniphilu pacaensis sp. nov., are proposed, respectively.
Keywords: bacterial vaginosis, culturomics, human microbiota, Peptoniphilus pacaensis, Peptoniphilus raoultii, Peptoniphilus vaginalis, taxogenomics
1. INTRODUCTION
Since the 1800s, physicians and researchers investigate the vaginal bacterial community using both cultivation and culture‐independent methods (Pandya et al., 2017; Srinivasan et al., 2016). To date, many species from the vaginal microbiota have been identified. The healthy vaginal flora is associated to a biotope rich in Lactobacilli species (Li, McCormick, Bocking, & Reid, 2012). The vaginal microbiota has a beneficial relationship with its host and can also impact women's health, that of their partners as well as their neonates (Lepargneur & Rousseau, 2002; Srinivasan & Fredricks, 2008). A depletion of vaginal Lactobacilli can lead to bacterial vaginosis (BV). This disease is a dysbiosis that may be associated to sexually transmitted infections as well as miscarriage and preterm birth in pregnant women (Afolabi, Moses, & Oduyebo, 2016; Martin & Marrazzo, 2016).
A microbial culturomics study exploring the bacterial community of the vaginal econiche flora in healthy women and patients suffering from bacterial vaginosis enabled the isolation of three Gram‐positive‐staining, anaerobic, and coccoid bacteria in the vaginal discharge of a woman with bacterial vaginosis (Lagier et al., 2015, 2016). These bacteria exhibited phylogenetic and phenotypic proximity to species of the Peptoniphilus genus. Created after the division of Peptostreptococcus genus into five genera (Ezaki et al., 2001), the Peptoniphilus genus belonging to the Peptoniphilaceae family that regroup members of the genera Peptoniphilus, Parvimonas, Murdochiella, Helcococcus, Gallicola, Finegoldia, Ezakiella, Anaerosphaera, and Anaerococcus (Johnson, Whitehead, Cotta, Rhoades, & Lawson, 2014; Patel et al., 2015). The Peptoniphilus genus is currently made of 16 valid published species (http://www.bacterio.net/peptoniphilus.html ). These bacteria employ amino acids and peptone as a major energy sources (Ezaki et al., 2001). They are mainly cultivated from diverse human samples such as sacral ulcer, vaginal discharge, as well as ovarian, peritoneal, and lacrymal gland abscesses (Ezaki et al., 2001; Li et al., 1992; Ulger‐Toprak, Lawson, Summanen, O'Neal, & Finegold, 2012).
Herein, we describe the isolation and taxonogenomic characterization (Fournier, Lagier, Dubourg, & Raoult, 2015) of strains KhD‐2T, KHD4T, and Kh‐D5T as type strains of three new Peptoniphilus species for which the names Peptoniphilus vaginalis sp. nov. (=CSUR P0125, =DSM 101742), Peptoniphilus raoultii sp. nov. (=CSUR P0110, =CECT 9308), and Peptoniphilus pacaensis sp. nov. (=CSUR P2271, =DSM 101839), are proposed, respectively. All the three strains were cultivated from the vaginal swab of the same patient.
2. MATERIALS AND METHODS
2.1. Samples and ethics
The vaginal specimen from a French 33‐year‐old woman with bacterial vaginosis was sampled at Hospital Nord in Marseille (France) in October 2015 using a Sigma Transwab (Medical Wire, Corsham, United Kingdom). Bacterial vaginosis was diagnosed as previously described (Menard, Fenollar, Henry, Bretelle, & Raoult, 2008). The patient had not received any antibiotic for several months. The local IFR48 ethics committee in Marseille (France) authorized the study (agreement number: 09‐022). In addition, the patient gave her signed informed consent.
2.2. Bacterial strain isolation and identification
After sampling, the specimen was preincubated in a blood culture bottle (Becton‐Dickinson Diagnostics, Le Pont‐de‐Claix, France). The blood culture bottle was enriched with 3 ml of sheep blood (bioMérieux, Marcy l'Etoile, France) and 4 ml of rumen fluid, filter‐sterilized through a 0.2 μm pore filter (Thermo Fisher Scientific, Villebon‐sur‐Yvette, France). Various preincubation periods (1, 3, 7, 10, 15, 20, and 30 days) were tested. Then, 50 μl of the supernatant were inoculated on both Colistin‐nalidixic acid (CNA) used for selective enrichment of Gram‐positive bacteria and trypticase soy agar plates used for cultivation of nonfastidious and fastidious microorganisms (both BD Diagnostics), and then incubated for 4 days under anaerobic conditions at 37°C. Isolated colonies were purified and subsequently identified by matrix‐assisted laser‐desorption/ionization time‐of‐flight (MALDI‐TOF) mass spectrometry with a Microflex spectrometer (Bruker, Leipzig, Germany) that compared the new spectra with those present in the library (Bruker database and URMITE database, constantly updated), as previously reported (Seng et al., 2009). If the score was >1.99, the bacterium was considered as identified at the genus level (score between 2.0 and 2.299) or species level (score from 2.3 to 3.0). When the score was <1.7, no identification was considered reliable. The 16S rRNA sequence of unidentified isolates was obtained using an ABI Prism 3130xl Genetic Analyzer capillary sequencer (Applied Biosystems, Bedford, MA, USA), as previously described (Morel et al., 2015; Seng et al., 2009). Finally, the sequences were compared to the NCBI nr database using the BLAST algorithm (https://blast.ncbi.nlm.nih.gov/Blast.cgi). If the 16S rRNA sequence similarity value was lower than 98.7%, the isolate was considered as a putative new species (Kim, Oh, Park, & Chun, 2014; Stackebrandt & Ebers, 2006; Yarza et al., 2014).
2.3. Phylogenetic analysis
The 16S rRNA sequences of isolates not identified using mass spectrometry and those of members of the family Peptoniphilaceae with standing in nomenclature (downloaded from the nr database) were aligned using CLUSTALW (Thompson, Higgins, & Gibson, 1994) with default setting. The phylogenetic inferences were performed using both the neighbor‐joining and maximum‐likelihood methods with the software MEGA version 6 (Tamura, Stecher, Peterson, Filipski, & Kumar, 2013).
2.4. Phenotypic characteristics
For each new isolate, cell morphology was visualized using optical and electron microscopy. Oxidase, catalase, motility, sporulation tests, as well as Gram stain were performed as already reported (Murray, Baron, Jorgensen, Landry, & Pfaller, 2007). Cells were fixed for electron microscopy for at least 1 hour at 4°C with 2.5% glutaraldehyde in a 0.1 mol L−1 cacodylate buffer. One drop of cell suspension was deposited for about 5 min on a glow‐discharged formvar carbon film on 400‐mesh nickel grids (FCF400‐Ni, EMS). The grids were dried on a blotting paper. Then, the cells were negatively stained at room temperature for 10 s with a 1% ammonium molybdate solution in filtered water. Micrographs were obtained using a Tecnai G20 Cryo (FEI) transmission electron microscope operated at 200 keV.
In order to characterize the best growth conditions of each isolate, bacteria were inoculated on 5% sheep blood‐enriched Columbia agar (bioMérieux) incubated at various atmospheres (aerobic, anaerobic, and microaerophilic) and temperatures (56, 42, 37, 28, and 25°C) (Mishra, Lagier, Nguyen, Raoult, & Fournier, 2013). Several salinity (NaCl concentrations of 0%, 5%, 15%, and 45%) and pH (5, 6, 6.5, 7, and 8.5) conditions were also tested.
Biochemical analyses were realized using various strips (API ZYM, API 20A, and API 50CH) according to the manufacturer's instructions (bioMérieux) (Avguštin, Wallace, & Flint, 1997; Durand et al., 2017). The tests were performed in anaerobic chamber. The strips were incubated there for 4, 24, and 48 hr, respectively.
For the analysis of cellular fatty acid methyl ester (FAME), gas chromatography/mass spectrometry (GC/MS) was achieved. All three isolates were grown anaerobically at 37°C on 5% sheep blood‐enriched Columbia agar (bioMérieux). For each isolate, after 2 days of incubation, two aliquots with roughly 25–70 mg of bacterial biomass per tube were prepared. FAME preparation and GC/MS analyses were performed as already reported (Dione et al., 2016; Sasser, 2006). FAMEs were separated with an Elite 5‐MS column and monitored by MS (Clarus 500‐SQ 8 S, Perkin Elmer, Courtaboeuf, France). A spectral database search was done with MS Search 2.0 operated using the standard reference database 1A (NIST, Gaithersburg, USA) as well as the FAMEs mass spectral database (Wiley, Chichester, UK).
The susceptibility of all three isolates was tested for 11 antibiotics: amoxicillin (0.16–256 μg/ml), benzylpenicillin (0.002–32 μg/ml), ceftriaxone (0.002–32 μg/ml), ertapenem (0.002–32 μg/ml), imipenem (0.002–32 μg/ml), amikacin (0.16–256 μg/ml), erythromycin (0.16–256 μg/ml), metronidazole (0.16–256 μg/ml), ofloxacin (0.002–32 μg/ml), rifampicin (0.002–32 μg/ml), and vancomycin (0.16–256 μg/ml). Minimal inhibitory concentrations (MICs) were estimated using E‐test strips (bioMérieux) and according to EUCAST recommendations (Citron, Ostovari, Karlsson, & Goldstein, 1991; Matuschek, Brown, & Kahlmeter, 2014).
2.5. Genome sequencing and analyses
After a pretreatment of 2 hr at 37°C using lysozyme, the genomic DNAs (gDNAs) of strains KhD‐2T, KHD4T, and Kh‐D5T were extracted using the EZ1 biorobot and EZ1 DNA Tissue kit (Qiagen). An elution volume of 50 μl was obtained for each sample. The gDNAs were quantified by a Qubit assay (Life technologies, Carlsbad, CA, USA) at 74.2, 22.4, and 16.4 ng/μl, respectively. Genomic sequencing of each strain was performed with a MiSeq sequencer (Illumina Inc, San Diego, CA, USA) and the Mate Pair strategy.
The Mate Pair library was prepared with the Nextera Mate Pair guide (Illumina) using 1.5 μg of gDNA. The gDNA samples were fragmented and tagged using a Mate Pair junction adapter (Illumina). Then, the fragmentation pattern was validated using a DNA 7500 labchip on an Agilent 2100 BioAnalyzer (Agilent Technologies Inc, Santa Clara, CA, USA). No size selection was done. Thus, 537, 600, and 480.7 ng of tagmented fragments were, respectively, circularized. Circularized DNAs were mechanically cut to smaller fragments using Optima on a bimodal curve at 507 and 1,244 bp for KhD‐2T, 975 and 1,514 bp for KHD4T, and 609 and 999 bp for Kh‐D5T on the Covaris device S2 in T6 tubes (Covaris, Woburn, MA, USA). The libraries profiles were visualized on a High Sensitivity Bioanalyzer LabChip (Agilent Technologies Inc, Santa Clara, CA, USA) and the final concentrations libraries were determined. Then, the libraries were normalized at 2 nmol L−1, pooled, denatured, diluted at 15 pmol L−1, loaded onto the reagent cartridge, and onto the instrument. Sequencing was performed in a single 39‐hr run in a 2 × 250‐bp.
The genome assembly was performed with a pipeline that enabled to create an assembly with various software such as Velvet (Zerbino & Birney, 2008), Spades (Bankevich et al., 2012), and Soap Denovo (Luo et al., 2012), on trimmed data with MiSeq and Trimmomatic (Bolger, Lohse, & Usadel, 2014) software or untrimmed data with only MiSeq software. In order to reduce gaps, GapCloser was used (Luo et al., 2012). Phage contamination was searched (blastn against Phage Phix174 DNA sequence) and eliminated. Finally, scaffolds with sizes under 800 bp and scaffolds with a depth value lower than 25% of the mean depth were identified as possible contaminants and removed. The best assembly was considered by using several criteria including number of scaffolds, N50, and number of N. Spades gave the best assembly for the three studied strains with depth coverage of 518x.
Prodigal was used to predict open reading frames (ORFs) (Hyatt et al., 2010) using default parameters. However, the predicted ORFs were excluded if they spanned a sequencing gap region (containing Ns). The predicted bacterial protein sequences were analyzed as previously reported (Alou et al., 2017). tRNA genes were found using the tRNAScan‐SE tool (Lowe & Eddy, 1997), while RNAmmer was used to find ribosomal RNAs (Lagesen et al., 2007). Phobius was used to predict lipoprotein signal peptides and the number of transmembrane helices (Käll, Krogh, & Sonnhammer, 2004). ORFans were identified when the BLASTP search failed to provide positive results (E‐value smaller than 1e−03 for ORFs with a sequence size larger than 80 aa or an E‐value smaller than 1e−05 for ORFs with a sequence length smaller than 80 aa), as previously reported (Alou et al., 2017). For genomic comparison, the closest species with validly published names in the 16S RNA phylogenetic tree were identified with the Phylopattern software (Gouret, Thompson, & Pontarotti, 2009). The complete genome, proteome, and ORFeome sequences were retrieved for each selected species in NCBI. An annotation of the entire proteome in order to define the distribution of functional classes of predicted genes according to the COG classification of their predicted protein products was performed as already reported (Alou et al., 2017). Annotation and comparison processes were done using the DAGOBAH software as previously described (Alou et al., 2017; Gouret et al., 2005, 2011). Finally, in order to evaluate the genomic similarity between the genomes, we determined two previously described parameters: average amino acid identity (AAI) based on the overall similarity between two genomic datasets of proteins available at (http://enve-omics.ce.gatech.edu/aai/index) and digital DNA–DNA hybridization (dDDH) (Auch, von Jan, Klenk, & Göker, 2010; Meier‐Kolthoff, Auch, Klenk, & Göker, 2013; Alou et al., 2017; Rodriguez & Konstantinidis, 2014; Chun et al., 2018).
3. RESULTS
3.1. Strain identification and phylogenetic analysis
The MS identification of the three bacteria, secluded, respectively, after 24 hr (strains KhD‐2T and KHD4T) and 15 days (Kh‐D5T) of preincubation, failed. This suggested that these isolates were not in the database and may be unknown species. Pairwise analysis of 16S rRNA sequences attested that strain KhD‐2T exhibited 92.8% and 87.4% sequence similarities with strains KHD4T and Kh‐D5T, respectively, and strains KHD4T and Kh‐D5T had an 88.7% identity. BLASTN sequence searches demonstrated that the three strains were related to the genus Peptoniphilus, suggesting that each strain represented a new species within this genus. Strain KhD‐2T exhibited a 16S rRNA similarity of 99.7% with Peptoniphilus sp. strain DNF00840 (GenBank KQ960236) over 1,842 bp and 98.2% with Peptoniphilus harei (GenBank NR_026358.1) over 1,488 bp. Strain KHD4T exhibited a 16S rRNA similarity of 96% with Peptoniphilus lacrimalis (GenBank NR_041938.1) over 1,489 bp. Finally, strain Kh‐D5T exhibited a 16S rRNA similarity of 97.2% with Peptoniphilus coxii (GenBank NR_117556.1) over 1,491 bp (Figure 1). As these percentage similarities were under the threshold of 98.7% established to delineate new species (Kim et al., 2014; Stackebrandt & Ebers, 2006; Yarza et al., 2014), strains KhD‐2T, KHD4T, and Kh‐D5T were considered as representative strains of putative new Peptoniphilus species. The names P. vaginalis sp. nov., P. raoultii sp. nov., and P. pacaensis sp. nov. are, respectively, proposed.
Figure 1.
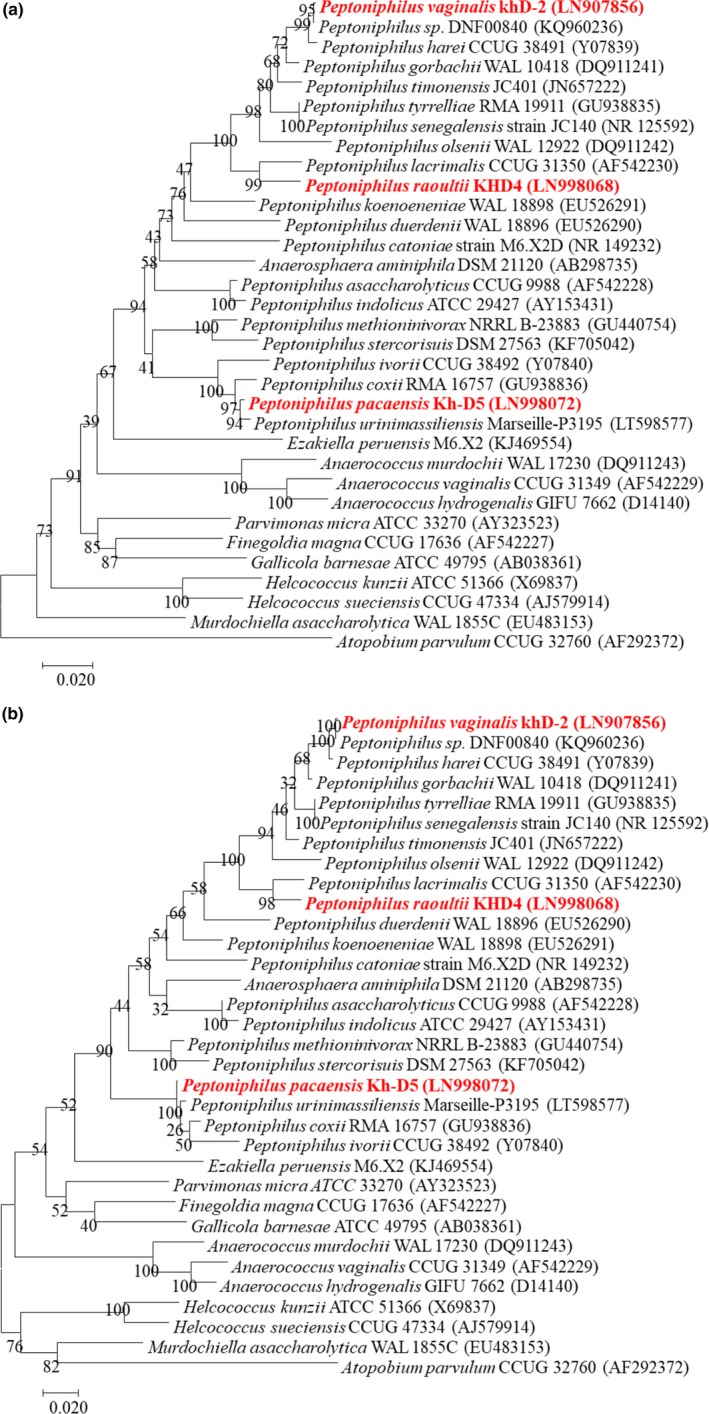
Phylogenetic analysis based on the 16S RNA gene sequence highlighting the position of Peptoniphilus vaginalis strain KhD‐2T, Peptoniphilus raoultii strain KHD4T, and Peptoniphilus pacaensis strain Kh‐D5T relative to other closely related strains. GenBank accession numbers are indicated in parentheses. Sequences were aligned using Muscle v3.8.31 with default parameters and, phylogenetic inferences were performed using the neighbor‐joining (a) and maximum‐likelihood (b) methods with the software MEGA version 6. The scale bar represents a 2% nucleotide sequence divergence
The reference MALDI‐TOF MS spectra of our isolates were added in our database (http://www.mediterranee-infection.com/article.php?laref=256&titre=urms-database) and then compared to those of other Peptoniphilus spp. (Figure 2).
Figure 2.
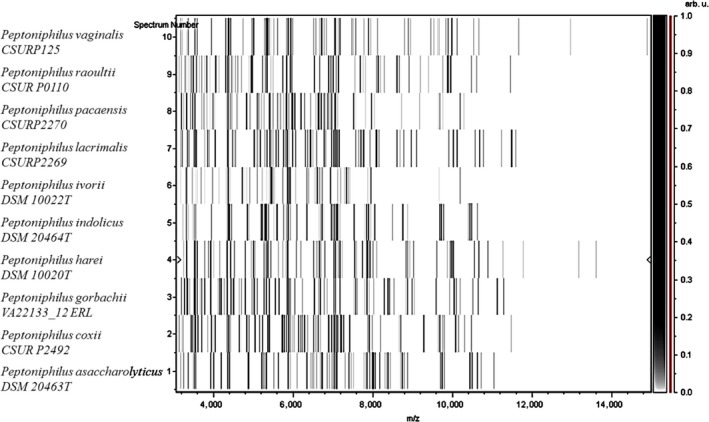
Gel view comparing strains KhD‐2T, KHD4T, and Kh‐D5T to other species within the genus Peptoniphilus. The gel view displays the raw spectra of loaded spectrum files arranged in a pseudo‐gel‐like look. The x‐axis records the m/z value. The left y‐axis displays the running spectrum number originating from subsequent spectra loading. The peak intensity is expressed by a gray scale scheme code. The right y‐axis indicates the relation between the color of a peak and its intensity, in arbitrary units. Displayed species are indicated on the left
3.2. Phenotypic features
Cells from all three novel strains (KhD‐2T, KHD4T, and Kh‐D5T) were Gram‐ ‐positive cocci (mean diameter of 0.6–0.7 μm for each). After 4 days of incubation, colonies on blood agar were grey and circular, and all had a diameter ranging from 1 to 2 mm. For all the three strains, growth occurred only in anaerobic atmosphere. Besides, optimal growth occurred at 37°C, with a pH between 6.5 and 8.5, and a NaCl concentration lower than 5%. They exhibited no catalase, oxidase, and urease activities. Using API 20A strips, all tests including aesculin, arabinose, cellobiose, gelatin, glucose, glycerol, indole, lactose, maltose, mannitol, mannose, raffinose, rhamnose, saccharose, sorbitol, trehalose, urease, and xylose were negative for strains KHD4T and Kh‐D5T, whereas for strain KhD‐2T, indole formation was positive, and gelatin was hydrolyzed. API ZYM strips showed that the three isolates exhibited positive reactions for acid phosphatase, esterase, and Naphthol‐AS‐BI‐phosphohydrolase. In addition, strains KhD‐2T and KHD4T had N‐acetyl‐β‐glucosaminidase and leucine arylamidase activities. In contrast, an alkaline phosphatase activity was observed for strains KhD‐2T and Kh‐D5T. All other remaining tests including valine arylamidase, lipase, cystine arylamidase, trypsin, galactosidase, glucosidase, β‐glucuronidase, α‐mannosidase, and α‐fucosidase were negative. Using API 50CH strips, all three isolates fermented ribose, tagatose, and potassium‐5‐ketogluconate. However, they did not ferment adonitol, aesculin, arabinose, arabitol, cellobiose, dulcitol, erythritol, fructose, fucose, galactose, glucose, glycerol, glycogen, inulin, lyxose, inositol, mannose, mannitol, maltose, melibiose, potassium gluconate, potassium‐2‐ketogluconate, salicine, saccharose, sorbitol, sorbose, trehalose, melezitose, raffinose, rhamnose, starch, turanose, xylitol, and xylose. Table 1 displayed the phenotypic differences between these bacteria and other Peptoniphilus spp.
Table 1.
Compared phenotypic characteristics of Peptoniphilus vaginalis strain KhD‐2T, Peptoniphilus raoultii strain KHD4T, Peptoniphilus pacaensis strain Kh‐D5T, and other closely related Peptoniphilus species. Data were obtained from the original descriptions of species
| Properties | P. vaginalis | P. raoultii | P. pacaensis | P. harei | P. lacrimalis | P. coxii | P. duerdenii | P. indolicus | P. asaccharolyticus |
|---|---|---|---|---|---|---|---|---|---|
| Cell diameter (μm) | 0.66 | 0.7 | 0.7 | 0.5–1.5 | 0.5–0.7 | <0.7 | ≥0.7 | 0.7–1.6 | 0.5–1.6 |
| % G+C | 34.23 | 31.87 | 49.38 | 34.44 | 30.22 | 44.62 | 34.24 | 31.69 | 32.30 |
| Major fatty acid (%) | C16:00 (41.6) | C16:00 (32) | C16:00 (36.4) | C16:00 (31.2) | C16:00 (27.7) | C16:00 (49.9) | C16:00 (33) | C16:00 (19.4) | C18:2ω6 (22.0) |
| Production of | |||||||||
| Alkaline phosphatase | + | − | + | − | − | − | − | + | + |
| Indole | + | − | − | + | − | − | + | + | − |
| Catalase | − | − | − | + | na | − | − | − | − |
| Urease | − | − | − | − | − | − | − | − | − |
| β‐galactosidase | − | − | − | − | − | − | − | − | − |
| N‐Acetyl‐β‐glucosaminidase | + | + | − | na | na | − | − | na | na |
| Acid from | |||||||||
| Ribose | + | + | + | − | − | − | − | − | − |
| d‐fructose | + | − | − | − | − | − | − | − | − |
| Habitat | Human vagina | Human vagina | Human vagina | Human sacral ulcer | Human eyes | Human specimens | Human vagina | Summer mastitis of cattle | Human vagina |
+, positive; −, negative; v, variable and na (not available) data.
The fatty acid composition of the three strains was as following: strain KhD‐2T contained saturated acid C16:0 (41.6%) and C14:0 (14.7%); unsaturated acids were also detected (Table 2); strains KHD4T and Kh‐D5T contained C16:0 (32% and 36%, respectively), C18:2ω6 (26% and 24%, respectively), and C18:1ω9 (26% and 21%, respectively) (Table 2). These fatty acid results were likened to those of related species in Table 2 (Johnson et al., 2014; Rooney, Swezey, Pukall, Schumann, & Spring, 2011). Strain KhD‐2T can be distinguished from its nearest neighbor P. harei by the production of C14:0 (14.7% vs. 4.4%). Strain KHD4T can be distinguished from its closest related species P. lacrimalis by the presence of fatty acids: C14:0, C17:0 iso 3‐OH, and anteiso‐C17:0. Finally, strain Kh‐D5T showed a fairly similar profile with its neighbors P. coxii and Peptoniphilus ivorii with some differences such as the presence of antesio‐C5:0, only in strain Kh‐D5T (4.5%), of iso‐C5:0 in P. coxii (5.5%), and C17:0 iso 3‐OH and antesio‐C17:0, solely in P. ivorii (7.7% and 3.8%, respectively). Besides, the three strains were sensitive to amoxicillin, benzylpenicillin, ceftriaxone, ertapenem, imipenem, metronidazole, rifampicin, and vancomycin, but resistant to amikacin, erythromycin, and ofloxacin (Table 3).
Table 2.
Cellular fatty acid profiles (%) of strains KhD‐2T, KHD4T, and Kh‐D5T compared with other Peptoniphilus species
| Fatty acids | Name | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| C4:00 | Butanoic acid | TR | − | − | − | − | − | − | − | − | − |
| iso‐C5:0 | 3‐Methyl‐butanoic acid | − | − | − | − | − | 5.5 | − | − | − | − |
| anteiso‐C5:0 | 2‐Methyl‐butanoic acid | TR | − | 4.5 | − | − | − | − | − | − | − |
| C10:0 | Decanoic acid | − | − | TR | TR | − | − | 2.8 | TR | − | − |
| C12:0 | Dodecanoic acid | TR | − | TR | − | TR | TR | − | 1.2 | TR | 2.3 |
| C13:0 | Tridecanoic acid | TR | − | − | − | − | − | − | − | − | − |
| C14:0 | Tetradecanoic acid | 14.7 | TR | 4.9 | 4.4 | 2.9 | 8.6 | 4.4 | 12.6 | 4.4 | 5.4 |
| C14:1ω5 | 9‐Tetradecenoic acid | TR | − | − | − | − | − | − | − | − | − |
| C15:0 | Pentadecanoic acid | 1.1 | TR | TR | − | − | 1.4 | − | − | − | − |
| C16:0 | Hexadecanoic acid | 41.6 | 32.0 | 36.4 | 32.1 | 27.7 | 49.9 | 33.0 | 19.4 | 29.5 | 14.4 |
| C16:0 9,10‐methylene | 2‐Hexyl‐cyclopropaneoctanoic acid | − | TR | − | − | − | − | − | − | − | − |
| C16:1ω5 | 11‐Hexadecenoic acid | TR | − | − | − | − | − | − | − | − | − |
| C16:1ω7 | 9‐Hexadecenoic acid | 6.2 | 1.0 | TR | 1.0 | 3.2 | − | − | − | 1.0 | 3.9 |
| C16:1ω9 | 7‐Hexadecenoic acid | TR | − | − | − | − | − | − | 3.6 | − | − |
| C17:0 | Heptadecanoic acid | TR | TR | TR | − | − | − | − | − | − | − |
| C17:0 iso 3‐OH | 3‐Hydroxy‐heptadecanoic acid | − | − | − | 6.0 | 3.0 | − | − | − | 7.7 | ‐ |
| anteiso‐C17:0 | 14‐Methyl‐hexadecanoic acid | TR | − | − | 4.2 | 1.8 | − | − | 2.6 | 3.8 | 1.6 |
| C17:1ω7 | 10‐Heptadecenoic acid | TR | − | − | − | − | − | − | − | − | − |
| C18:0 | Octadecanoic acid | 3.9 | 8.8 | 3.6 | 7.2 | 11.2 | 13.1 | 16.2 | 2.5 | 4.8 | 9.4 |
| C18:1ω7 | 11‐Octadecenoic acid | 4.8 | 3.7 | 2.0 | 1.9 | 3.5 | − | − | 3.5 | 2.6 | − |
| C18:1ω9 | 9‐Octadecenoic acid | 12.1 | 25.8 | 21.2 | 17.0 | 25.7 | 17.3 | 22.6 | 6.2 | 11.4 | 20.2 |
| C18:2ω6 | 9,12‐Octadecadienoic acid | 12.0 | 26.4 | 24.4 | 17.0 | 13.6 | 3.2 | 21.1 | 13.0 | 24.0 | 22.0 |
Strains: 1, P. vaginalis strain KhD‐2T; 2, P. raoultii strain KHD4T; 3, P. pacaensis strain Kh‐D5T; 4, Peptoniphilus harei DSM 10020T; 5, P. lacrimalis DSM 7455T; 6, P. coxii CSUR 2492T; 7, P. uerdenii WAL 18896T; 8, P. indolicus DSM 20464T, 9, P. ivorii CCUG 38492T and 10, P. asaccharolyticus CCUG 9988T. Strains 1, 2, 3, and 6 data are from this study and strains 4, 5, 7 to 9, data come from Rooney et al., 2011 and Johnson et al., 2014. Predominant products are shown in bold; TR, trace amounts < 1%; −, not detected.
Table 3.
Minimal inhibitory concentrations (MIC μg/μl) of antibiotics for P. vaginalis strain KhD‐2T, P. raoultii strain KHD4T, and P. pacaensis strain Kh‐D5T
| Antibiotics | Concentration (μg/ml) | P. vaginalis strain KhD‐2T | P. raoultii strain KHD4T | P. pacaensis strain Kh‐D5T |
|---|---|---|---|---|
| Amoxicillin | 0.016–256 | 0.032 | 0.016 | 0.016 |
| Benzylpenicillin | 0.002–32 | 0.094 | 0.002 | 0.002 |
| Ceftriaxone | 0.002–32 | 0.064 | 0.064 | 0.064 |
| Ertapenem | 0.002–32 | 0.002 | 0.003 | 0.002 |
| Imipenem | 0.002–32 | 0.004 | 0.002 | 0.002 |
| Metronidazole | 0.016–256 | 0.125 | 0.032 | 0.032 |
| Rifampicin | 0.002–32 | 0.002 | 0.002 | 0.002 |
| Vancomycin | 0.016–256 | 0.094 | 0.094 | 0.094 |
| Amikacin | 0.016–256 | >256 | >256 | >256 |
| Erythromycin | 0.016–256 | 1 | 2 | 2 |
| Ofloxacin | 0.002–32 | >256 | >256 | 2 |
3.3. Genome characteristics
Strains KhD‐2T, KHD4T, and Kh‐D5T exhibited genomes sizes of 1,877,211, 1,623,601, and 1,851,572 bp long, respectively (Figure 3). The genome characteristics were detailed in Table 4. The repartition of genes into the 25 general COG categories was represented in Table 5 and Figure 4. When compared to other Peptoniphilus species, the three strains had genome sizes, G+C contents and total gene counts in the same range (Table 6, Figure 5). Although, base composition varies widely among bacterial species, the genes within a given genome are relatively similar in G+C content with the exception of recently acquired genes. As a matter of fact, DNA sequences acquired by horizontal transfer often bear unusual sequence characteristics and can be distinguished from ancestral DNA notably by a distinct G+C content (Lawrence & Ochman, 1997). The region between 100,000 and 600,000 bp of the chromosome from strain KhD‐5T showed a high variation in G+C content (Figure 3). Thus, 43 genes putatively acquired by horizontal gene transfer were identified in this region, including 25 genes specific for strain KhD‐5T and 18 genes shared with strain Peptoniphilus urinimassiliensis. Consequently, the presence of these genes may play a role in the significant difference in genomic G+C content observed between strain KhD‐5T and other compared Peptoniphilus species as well as the similar genomic G+C content observed between strain KhD‐5T and P. urinimassiliensis.
Figure 3.
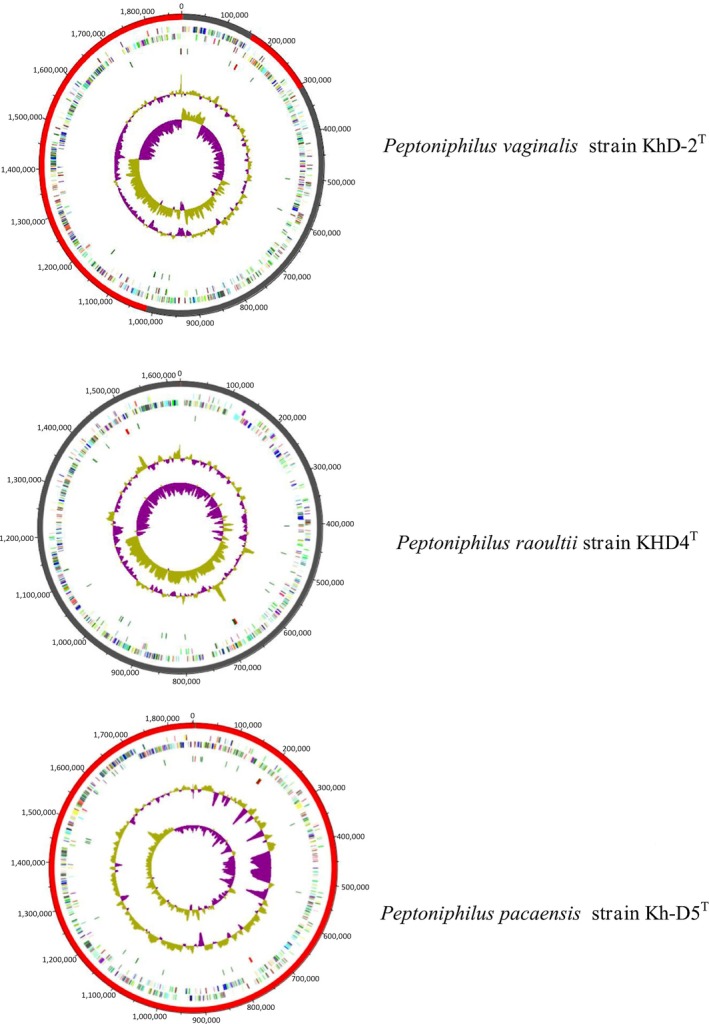
Graphical circular map of the three genomes. From outside to the center: Contigs (red/gray), COG category of genes on the forward strand (three circles), genes on forward strand (blue circle), genes on the reverse strand (red circle), COG category on the reverse strand (three circles), G+C content
Table 4.
Nucleotide and gene count levels of the genomes
| P. raoultii | P. vaginalis | P. vaginalis | ||||
|---|---|---|---|---|---|---|
| Attribute | Value | % of totala | Value | % of totala | Value | % of totala |
| Size (bp) | 1,623,601 | 100% | 1,877,211 | 100% | 1,851,572 | 100% |
| G+C content (bp) | 517,506 | 31.87% | 642,534 | 34.22% | 914,357 | 49.38% |
| Coding region (bp) | 1,467,557 | 90.39% | 1,692,527 | 90.16 | 3,579,496 | 85.07% |
| Total genes | 1,624 | 100% | 1,780 | 100% | 1,801 | 100% |
| RNA genes | 42 | 2.59% | 40 | 2.35% | 54 | 3.00% |
| Protein‐coding genes | 1,520 | 93.60% | 1,698 | 95.39% | 1,699 | 94.34% |
| Genes with function prediction | 1,222 | 75.25% | 1,375 | 77.24% | 1,323 | 73.45% |
| Genes assigned to COGs | 1,048 | 65.53% | 1,204 | 67.64% | 1,175 | 65.24% |
| Genes with peptide signals | 162 | 9.97% | 169 | 9.49% | 231 | 12.83% |
| Genes with transmembrane helices | 349 | 21.49% | 403 | 22.64% | 414 | 22.98% |
The total is based on either the size of the genome in base pairs or the total number of protein coding genes in the annotated genome.
Table 5.
Number of genes associated with the 25 general COG functional categories
| P. vaginalis | P. raoultii | P. pacaensis | |||||
|---|---|---|---|---|---|---|---|
| Code | Value | % value | Value | % value | Value | % value | Description |
| J | 170 | 9.70 | 170 | 10.69 | 171 | 9.78 | Translation |
| A | 0 | 0 | 0 | 0 | 0 | 0 | RNA processing and modification |
| K | 75 | 4.28 | 63 | 3.96 | 78 | 4.46 | Transcription |
| L | 64 | 3.65 | 65 | 4.09 | 63 | 3.60 | Replication, recombination, and repair |
| B | 0 | 0 | 0 | 0 | 0 | 0 | Chromatin structure and dynamics |
| D | 20 | 1.14 | 18 | 1.13 | 23 | 1.31 | Cell cycle control, mitosis, and meiosis |
| Y | 0 | 0 | 0 | 0 | 0 | 0 | Nuclear structure |
| V | 61 | 3.48 | 40 | 2.51 | 60 | 2.97 | Defense mechanisms |
| T | 44 | 2.51 | 43 | 2.70 | 52 | 3.64 | Signal transduction mechanisms |
| M | 50 | 2.85 | 50 | 3.14 | 55 | 3.14 | Cell wall/membrane biogenesis |
| N | 7 | 0.39 | 7 | 0.44 | 8 | 0.45 | Cell motility |
| Z | 0 | 0 | 0 | 0 | 0 | 0 | Cytoskeleton |
| W | 3 | 0.17 | 3 | 0.18 | 2 | 0.11 | Extracellular structures |
| U | 15 | 0.85 | 16 | 1.00 | 15 | 0.85 | Intracellular trafficking and secretion |
| O | 58 | 3.31 | 51 | 3.20 | 54 | 3.08 | Posttranslational modification, protein turnover, chaperones |
| X | 68 | 3.88 | 22 | 1.38 | 44 | 2.51 | Mobilome: prophages, transposons |
| C | 83 | 4.74 | 66 | 4.15 | 75 | 4.29 | Energy production and conversion |
| G | 40 | 2.28 | 47 | 2.95 | 48 | 2.74 | Carbohydrate transport and metabolism |
| E | 115 | 6.56 | 105 | 6.60 | 112 | 6.40 | Amino acid transport and metabolism |
| F | 57 | 3.25 | 52 | 3.27 | 58 | 3.31 | Nucleotide transport and metabolism |
| H | 71 | 4.05 | 52 | 3.27 | 84 | 4.80 | Coenzyme transport and metabolism |
| I | 56 | 3.19 | 53 | 3.33 | 45 | 2.57 | Lipid transport and metabolism |
| P | 68 | 3.88 | 48 | 3.02 | 69 | 3.94 | Inorganic ion transport and metabolism |
| Q | 19 | 1.08 | 18 | 1.13 | 11 | 0.62 | Secondary metabolites biosynthesis, transport, and catabolism |
| R | 111 | 6.33 | 107 | 6.73 | 98 | 5.60 | General function prediction only |
| S | 62 | 3.54 | 51 | 3.20 | 71 | 4.06 | Function unknown |
| ‐ | 547 | 31.23 | 541 | 34.04 | 573 | 32.78 | Not in COGs |
Figure 4.
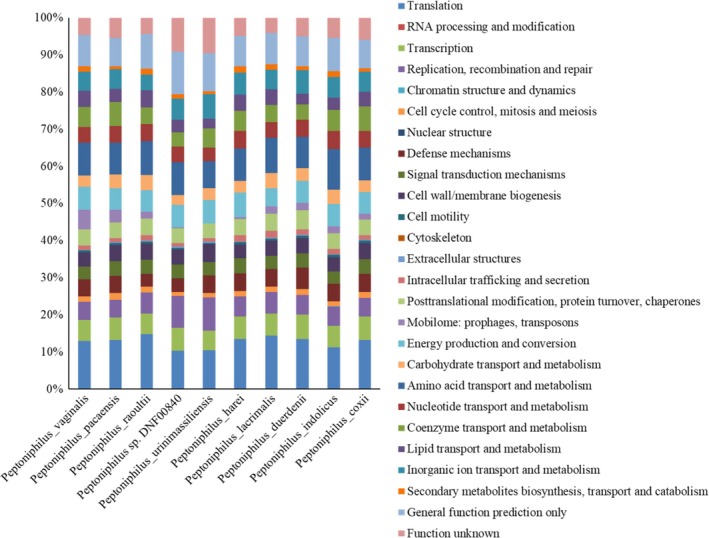
Distribution of functional classes of predicted genes according to the clusters of orthologous groups of proteins of P. vaginalis strain KhD‐2T, P. raoultii strain KHD4T, and P. pacaensis strain Kh‐D5T among other species
Table 6.
Genome comparison of closely related species to P. vaginalis strain KhD‐2T, P. raoultii strain KHD4T, and P. pacaensis strain Kh‐D5T
| Species | INSDC identifiera | Size (Mbp) | G+C Percent | Gene Content | Number of contigs | N50 Value |
|---|---|---|---|---|---|---|
| P. vaginalis KhD‐2 T | FXLP00000000 | 1.88 | 34.2 | 1,791 | 5 | 707,77 |
| P. raoultii KHD4 T | FMWM00000000 | 1.62 | 31.9 | 1,631 | 2 | 1,62 |
| P. pacaensis Kh‐D5 T | FLQT00000000 | 1.85 | 49.4 | 1,802 | 3 | 1,84 |
| Peptoniphilus sp. DNF00840 | LSDH00000000 | 1.88 | 34.3 | 1,671 | 91 | 50,04 |
| Peptoniphilus urinimassiliensis Marseille‐P3195 | FTPC00000000 | 1.82 | 49.7 | 1,770 | 5 | 563,37 |
| Peptoniphilus harei ACS‐146‐V‐Sch2b | AENP00000000 | 1.84 | 34.4 | 1,749 | 32 | 111,2 |
| Peptoniphilus lacrimalis CCUG 31350 | ARKX00000000 | 1.85 | 30.2 | 1,785 | 22 | 190,04 |
| Peptoniphilus duerdenii WAL 18896 | AEEH00000000 | 2.12 | 34.2 | 1,963 | 61 | 96,77 |
| Peptoniphilus indolicus ATCC 29427 | AGBB00000000 | 2.24 | 31.7 | 2,145 | 302 | 11,79 |
| Peptoniphilus coxii RMA 16757 | LSDG00000000 | 1.84 | 44.6 | 1,783 | 48 | 103,89 |
| Peptoniphilus asaccharolyticus DSM 20463 | FWWR00000000 | 2.23 | 32.3 | 2,054 | 17 | 1,358,172 |
INSDC: International Nucleotide Sequence Database Collaboration. Text and values in bold have been used to highlight new species.
Figure 5.
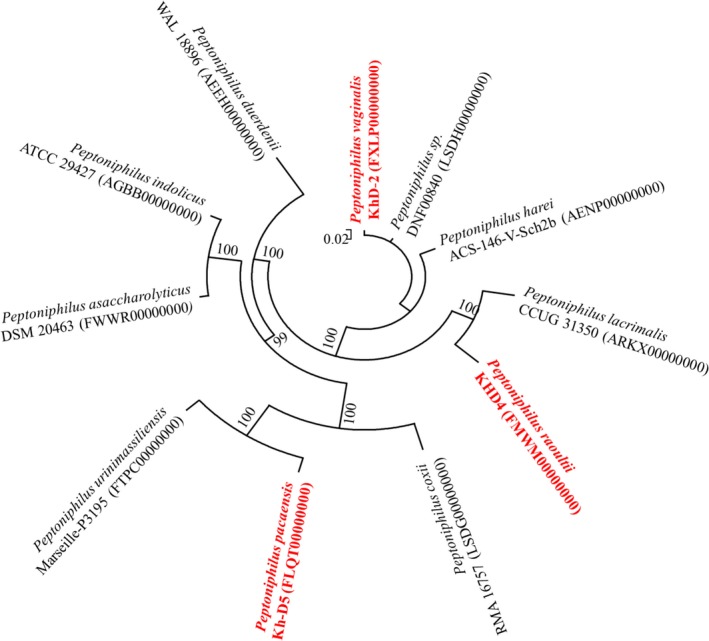
Phylogenetic tree based on whole genome sequence showing the position of P. vaginalis strain KhD‐2T, P. raoultii strain KHD4T, and P. pacaensis strain Kh‐D5T relative to their nearest neighbors. GenBank accession numbers are indicated in parentheses. Sequences were aligned using Mugsy software, and phylogenetic inferences were performed using the maximum likelihood method with the software FastTree. The scale bar represents a 2% nucleotide sequence divergence
The dDDH values ranked from 20.1% ± 2.3% between P. harei and P. duerdenii to 56.4% ± 2.75% between P. lacrimalis and P. urinimassiliensis (Table 7). When comparing the three new strains to other Peptoniphilus species, strain KhD‐2T exhibited dDDH values ranging from 22.7% ± 2.4% with Peptoniphilus indolicus to 47.3% ± 2.55% with P. coxii; dDDH values from strain KHD4T ranged from 19.0% ± 2.25% with P. harei to 44.3% ± 2.55% with P. coxii; and strain Kh‐D5T exhibited dDDH values ranging from 20.7% ± 2.35% with P. coxii to 45.0% ± 2.60% with P. urinimassiliensis (Table 7). Furthermore, the AAI values ranged from 51.3% between P. coxii and P. indolicus to 84.0% between P. indolicus and Peptoniphilus asaccharolyticus (Table 8). Comparing the three new isolates to their neighbors, strain KhD‐2T shared AAI values ranging from 51.5% with P. urinimassiliensis to 92.9% with P. harei, AAI values of strain KHD4T ranging from 50.9% with P. urinimassiliensis to 70.6% with P. lacrimalis, and strain Kh‐D5T exhibited AAI values ranging from 50.2% with P. asaccharolyticus to 92.9% with P. urinimassiliensis (Table 8). According to the fact that the threshold of dDDH and AAI values for distinguishing different species are 70% and 95%–96%, respectively (Chun et al., 2018; Klappenbach et al., 2007; Meier‐Kolthoff et al., 2013; Richter & Rosselló‐Móra, 2009; Rodriguez‐R & Konstantinidis, 2014), these data confirm the classification of strains KhD‐2T, KHD4T, and Kh‐D5T in distinct species.
Table 7.
dDDH values obtained by comparison of all studied genomes using GGDC, Formula 2 (DDH Estimates Based on Identities/HSP length)a
| P. vaginalis strain KhD‐2T | P. raoultii strain KHD4T | P. pacaensis strain Kh‐D5T | P. urini‐massiliensis | P. harei | P. lacrimalis | P. duerdenii | P. indolicus | P. coxii | P. asaccharolyticus | |
|---|---|---|---|---|---|---|---|---|---|---|
| P. vaginalis | 100 ± 00 | 22.9 ± 2.35 | 40.0 ± 2.50 | 35.3 ± 2.50 | 45.8 ± 2.60 | 25.6 ± 2.40 | 32.0 ± 2.45 | 22.7 ± 2.40 | 47.3 ± 2.55 | 33.20 ± 2.45 |
| P. raoultii | 100 ± 00 | 29.8 ± 2.45 | 40.5 ± 2.50 | 19.0 ± 2.25 | 20.4 ± 2.30 | 36.4 ± 2.55 | 22.2 ± 2.35 | 44.3 ± 2.55 | 28.40 ± 2.45 | |
| P. pacaensis | 100 ± 00 | 45.0 ± 2.60 | 42.0 ± 2.55 | 41.9 ± 2.55 | 38.7 ± 2.50 | 27.3 ± 2.45 | 20.7 ± 2.35 | 29.30 ± 2.45 | ||
| P. urinimassiliensis | 100 ± 00 | 32.9 ± 2.50 | 56.4 ± 2.75 | 42.9 ± 2.50 | 33.0 ± 2.45 | 20.1 ± 2.30 | 32.30 ± 2.45 | |||
| P. harei | 100 ± 00 | 34.3 ± 2.50 | 39.2 ± 2.50 | 20.1 ± 2.30 | 36.2 ± 2.45 | 33.30 ± 2.45 | ||||
| P. lacrimalis | 100 ± 00 | 39.3 ± 2.50 | 25.1 ± 2.40 | 40.6 ± 2.50 | 31.90 ± 2.45 | |||||
| P. duerdenii | 100 ± 00 | 24.3 ± 2.35 | 38.2 ± 2.50 | 32.80 ± 2.50 | ||||||
| P. indolicus | 100 ± 00 | 44.0 ± 2.55 | 26.70 ± 2.45 | |||||||
| P. coxii | 100 ± 00 | 35.40 ± 2.45 | ||||||||
| P. asaccharolyticus | 100 ± 00 |
The confidence intervals indicate the inherent uncertainty in estimating DDH values from intergenomic distances based on models derived from empirical test data sets (which are always limited in size).
Table 8.
AAI values obtained by comparison of all studied genomes
| P. raoultii strain KHD4T | P. pacaensis strain Kh‐D5T | P. urini‐massiliensis | P. harei | P. lacrimalis | P. duerdenii | P. indolicus | P. coxii | P. asaccharolyticus | |
|---|---|---|---|---|---|---|---|---|---|
| P. vaginalis | 62.7 | 51.2 | 51.5 | 92.9 | 61.5 | 57.0 | 55.9 | 53.2 | 57.9 |
| P. raoultii | 50.0 | 50.9 | 61.6 | 70.6 | 56.2 | 55.4 | 52.5 | 56.8 | |
| P. pacaensis | 92.9 | 51.8 | 51.2 | 51.8 | 50.4 | 74.1 | 50.2 | ||
| P. urinimassiliensis | 52.0 | 52.7 | 52.2 | 51.4 | 73.4 | 51.3 | |||
| P. harei | 64.2 | 58.5 | 56.4 | 51.7 | 58.5 | ||||
| P. lacrimalis | 58.0 | 55.9 | 51.8 | 57.1 | |||||
| P. duerdenii | 54.7 | 53.1 | 57.0 | ||||||
| P. indolicus | 51.3 | 84.0 | |||||||
| P. coxii | 51.2 |
4. DISCUSSION
The aim of this study was to investigate, using culturomics, the vaginal flora of a woman with bacterial vaginosis. Indeed, bacterial vaginosis is a gynecologic disorder marked by a perturbation of the vaginal microbiota equilibrium with a loss of commensal Lactobacillus spp. and their replacement with anaerobic bacteria including Atopobium vaginae, Bacteroides spp., Mobiluncus spp., Prevotella spp., and numerous Gram‐positive anaerobic cocci (Bradshaw et al., 2006; Onderdonk, Delaney, & Fichorova, 2016; Shipitsyna et al., 2013). Gram‐positive anaerobic cocci were associated to various infections (Murdoch, 1998). They represent about 24%–31% of anaerobic bacteria cultivated in clinical specimens (Murdoch, Mitchelmore, & Tabaqchali, 1994). In this present study, three novel Gram‐positive‐staining, anaerobic cocci (KhD‐2T, KHD4T, and Kh‐D5T) were cultured in the vaginal discharge of a patient suffering from bacterial vaginosis. These bacteria exhibited sufficient MALDI‐TOF MS profiles, 16S rRNA sequence, phenotypic, and genomic differences with Peptoniphilus species to be regarded as representative strains of three new species within this genus. Currently, this genus contains 16 species with validly published names. Most of them have been observed in human clinical specimens (Ezaki et al., 2001).
Data from phylogenetic analysis and genomic comparison exhibited the heterogeneity of this genus and revealed that strain KhD‐2T and Peptoniphilus sp. DNF00840T share 99.79% 16S rRNA gene sequence similarity, an ANI value of 96.83% and 75.0% of dDDH. In fact, to differentiate bacterial species, thresholds lower than 98.7%, 94%, and 70% were delimited for 16S rRNA sequence identity, ANI, and dDDH values, respectively. Therefore, the obtained values suggest that the two strains (KhD‐2T and Peptoniphilus sp. DNF00840T) belong to the same species. Unlike other Peptoniphilus spp., strains KhD‐2T, KHD4T, and Kh‐D5T ferment ribose and tagatose. The study of their genomes revealed that strain Kh‐D2T had 75 genes associated to carbohydrate metabolism, including 4 genes (1 rbsA gene, 2 rbsR genes, and 1 rpiB gene) encoding proteins involved in fermentation of ribose; the genome from strain KHD4T contained 61 genes associated to carbohydrate metabolism of which one rpiB gene is involved in fermentation of ribose; and strain KhD‐5T had 58 genes associated to carbohydrate metabolism with 3 genes implicated in ribose fermentation (2 rpiB genes and 1 rbsK) and 1 gene encoding a tagatose biphosphate aldolase enzyme involved in tagatose fermentation. In addition, the genomes of strains Kh‐D2T, KHD4T, and KhD‐5T also had 25 genes (5 genes encoding proteins responsible for the degradation of histidine, 1 of lysine, 2 of threonine, 12 of methionine, and 5 of arginine), 20 genes (5 of histidine, 1 of lysine, 1 of threonine, 7 of methionine, and 6 of arginine), and 21 genes (14 which degraded methionine, 6 for arginine and 1 for lysine), associated to amino acid degradation, respectively.
Finally, we propose that strains KhD‐2T, KHD4T, and Kh‐D5T are type strains of P. vaginalis sp. nov., P. raoultii sp. nov., and P. pacaensis sp. nov., respectively.
4.1. Description of P. vaginalis sp. nov
Peptoniphilus vaginalis (va.gi.na'lis. L. n. fem. gen. vaginalis from the feminine organ vagina; vaginalis pertaining to the vagina).
Gram‐stain—positive. Coccus‐shaped bacterium with a mean diameter of 0.66 μm. Peptoniphilus vaginalis sp. nov. is a mesophilic bacterium; its optimal growth occurs at temperature 37°C, a pH ranking from 6.5 to 8.5, and a NaCl concentration lower than 5%. Colonies are circular, translucent, gray, and have a diameter of 1–1.5 mm on Columbia agar. Cells are strictly anaerobic, not motile, and non‐spore‐forming. Catalase, oxidase, and urease activities are negative. Nitrate reduction is also negative nevertheless indole production is positive. P. vaginalis shows positive enzymatic activities for acid phosphatase, alkaline phosphatase, esterase, esterase lipase, leucine arylamidase, Naphthol‐AS‐BI‐phosphohydrolase, and N‐acetyl‐β‐glucosaminidase. P. vaginalis ferments fructose, potassium 5‐ketogluconate, ribose, and tagatose. C16:0, C14:0, C18:1ω9, and C18:2ω6 are its main fatty acids. Strain KhD‐2T is sensitive to amoxicillin, benzylpenicillin, ceftriaxone, imipenem, ertapenem, metronidazole, rifampicin, and vancomycin but resistant to amikacin, erythromycin, and ofloxacin. Its 1,623,601‐bp genome contains 34.23% G+C. In EMBL‐EBI, the 16S rRNA gene sequence is deposited under accession number LN907856 and the draft genome sequence under accession number FXLP00000000. The type strain of Peptoniphilus vaginalis sp. nov. is strain KhD‐2T (=CSUR P0125 = DSM 101742), which was cultured from the vaginal discharge of a woman suffering from bacterial vaginosis.
4.2. Description of P. raoultii sp. nov
Peptoniphilus raoultii (ra.oul'ti.i. N. L. masc. gen. n. raoultii of Raoult, to honor French scientist Professor Didier Raoult for his outstanding contribution to medical microbiology).
Gram‐stain—positive. Coccus‐shaped bacterium with a mean diameter of 0.7 μm. Peptoniphilus raoultii sp. nov. is a mesophilic bacterium; its optimal growth occurs at temperature 37°C, a pH ranking from 6.5 to 8.5, and a NaCl concentration lower than 5%. Colonies are circular, translucent, gray, and have a diameter of 1–1.5 mm on Columbia agar. Cells are strictly anaerobic, not motile, and non‐spore‐forming. Catalase, oxidase, urease, indole, and nitrate activities are negative. P. raoultii exhibits positive enzymatic activities for acid phosphatase, esterase, esterase lipase, leucine arylamidase, Naphthol‐AS‐BI‐phosphohydrolase, and N‐acetyl‐β‐glucosaminidase. P. raoultii ferments potassium 5‐ketogluconate, ribose, and tagatose. C16:0, C18:2ω6, and C18:1ω9 are its main fatty acids. Strain KHD4T is sensitive to amoxicillin, benzylpenicillin, ceftriaxone, imipenem, ertapenem, metronidazole, rifampicin, and vancomycin but resistant to amikacin, erythromycin, and ofloxacin. The genome is 1,877,211 bp long and contains 31.87% G+C. In EMBL‐EBI, the 16S rRNA gene sequence is deposited under accession number LN998068 and the draft genome sequence under accession number FMWM00000000. Strain KHD4T (=CSUR P0110 = CECT 9308) is the type strain of P. raoultii sp. nov., which was cultured from the vaginal discharge of a woman suffering from bacterial vaginosis.
4.3. Description of P. pacaensis sp. nov
Peptoniphilus pacaensis (pa.ca.en'sis N. L. gen. masc. n. pacaensis, from the acronym PACA, of Provence‐Alpes‐Côte d'Azur, the region where the type strain was first cultured and characterized).
Gram‐stain—positive. Coccus‐shaped bacterium with a mean diameter of 0.7 μm. Peptoniphilus pacaensis sp. nov. is a mesophilic bacterium; its optimal growth occurs at temperature 37°C, a pH ranking from 6.5 to 8.5, and a NaCl concentration lower than 5%. Colonies are circular, translucent, gray, and have a diameter of 1–1.5 mm on Columbia agar. Cells are strictly anaerobic, not motile, and non‐spore‐forming. Catalase, oxidase, urease, indole, and nitrate activities are negative. P. pacaensis shows positive enzymatic activities for alkaline phosphatase, acid phosphatase, esterase, esterase lipase, and Naphthol‐AS‐BI‐phosphohydrolase. P. pacaensis ferments potassium 5‐ketogluconate, ribose, and tagatose. C16:0, C18:2ω6, and C18:1ω9 are its main fatty acids. Strain Kh‐D5T is sensitive to amoxicillin, benzylpenicillin, ceftriaxone, imipenem, ertapenem, metronidazole, rifampicin, and vancomycin but resistant to amikacin, erythromycin, and ofloxacin. Its genome is 1,851,572 bp long with a 49.38% G+C content. In EMBL‐EBI, the 16S rRNA gene sequence is deposited under accession number LN998072 and the draft genome sequence under accession number FLQT00000000. The type strain of P. pacaensis sp. nov. is strain Kh‐D5T (=CSUR P2270 = DSM 101839), which was cultured from the vaginal discharge of a woman suffering from bacterial vaginosis.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
ACKNOWLEDGEMENTS
The authors thank Frederic Cadoret for administrative assistance and the Xegen Company (www.xegen.fr) for automating the genomic annotation process.
Diop K, Diop A, Michelle C, et al. Description of three new Peptoniphilus species cultured in the vaginal fluid of a woman diagnosed with bacterial vaginosis: Peptoniphilus pacaensis sp. nov., Peptoniphilus raoultii sp. nov., and Peptoniphilus vaginalis sp. nov. MicrobiologyOpen. 2019;8:e661 10.1002/mbo3.661
Méditerranée Infection and the National Research Agency under the program “Investissements d'avenir”, reference ANR‐10‐IAHU‐03, supported this study.
REFERENCES
- Afolabi, B. B. , Moses, O. E. , & Oduyebo, O. O. (2016). Bacterial vaginosis and pregnancy outcome in Lagos, Nigeria. Open Forum Infectious Diseases, 3, ofw030 10.1093/ofid/ofw030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alou, M. T. , Rathored, J. , Michelle, C. , Dubourg, G. , Andrieu, C. , Armstrong, N. , … Fournier, P. E. (2017). Inediibacterium massiliense gen. nov., sp. nov., a new bacterial species isolated from the gut microbiota of a severely malnourished infant. Antonie van Leeuwenhoek, 110, 737–750. 10.1007/s10482-017-0843-5 [DOI] [PubMed] [Google Scholar]
- Auch, A. F. , von Jan, M. , Klenk, H.‐P. , & Göker, M. (2010). Digital DNA‐DNA hybridization for microbial species delineation by means of genome‐to‐genome sequence comparison. Standards in Genomic Sciences, 2, 117–134. 10.4056/sigs.531120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avguštin, G. , Wallace, R. J. , & Flint, H. J. (1997). Phenotypic diversity among ruminal isolates of Prevotella ruminicola: Proposal of Prevotella brevis sp. nov., Prevotella bryantii sp. nov., and Prevotella albensis sp. nov. and redefinition of Prevotella ruminicola . International Journal of Systematic and Evolutionary Microbiology, 47, 284–288. [DOI] [PubMed] [Google Scholar]
- Bankevich, A. , Nurk, S. , Antipov, D. , Gurevich, A. A. , Dvorkin, M. , Kulikov, A. S. , … Pyshkin, A. V. (2012). SPAdes: A new genome assembly algorithm and its applications to single‐cell sequencing. Journal of Computational Biology, 19, 455–477. 10.1089/cmb.2012.0021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger, A. M. , Lohse, M. , & Usadel, B. (2014). Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics, 30, 2114–2120. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradshaw, C. S. , Tabrizi, S. N. , Fairley, C. K. , Morton, A. N. , Rudland, E. , & Garland, S. M. (2006). The association of Atopobium vaginae and Gardnerella vaginalis with bacterial vaginosis and recurrence after oral metronidazole therapy. Journal of Infectious Diseases, 194, 828–836. 10.1086/506621 [DOI] [PubMed] [Google Scholar]
- Chun, J. , Oren, A. , Ventosa, A. , Christensen, H. , Arahal, D. R. , da Costa, M. S. , … Trujillo, M. E. (2018). Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. International Journal of Systematic and Evolutionary Microbiology, 68, 461–466. 10.1099/ijsem.0.002516 [DOI] [PubMed] [Google Scholar]
- Citron, D. M. , Ostovari, M. I. , Karlsson, A. , & Goldstein, E. J. (1991). Evaluation of the E test for susceptibility testing of anaerobic bacteria. Journal of Clinical Microbiology, 29, 2197–2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dione, N. , Sankar, S. A. , Lagier, J. C. , Khelaifia, S. , Michele, C. , Armstrong, N. , … Fournier, P. E. (2016). Genome sequence and description of Anaerosalibacter massiliensis sp. nov. New Microbes and New Infections, 10, 66–76. 10.1016/j.nmni.2016.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand, G. A. , Pham, T. , Ndongo, S. , Traore, S. I. , Dubourg, G. , Lagier, J. C. , … Million, M. (2017). Blautia massiliensis sp. nov., isolated from a fresh human fecal sample and emended description of the genus Blautia. Anaerobe, 43, 47–55. 10.1016/j.anaerobe.2016.12.001 [DOI] [PubMed] [Google Scholar]
- Ezaki, T. , Kawamura, Y. , Li, N. , Li, Z.‐Y. , Zhao, L. , & Shu, S. (2001). Proposal of the genera Anaerococcus gen. nov., Peptoniphilus gen. nov. and Gallicola gen. nov. for members of the genus Peptostreptococcus . International Journal of Systematic and Evolutionary Microbiology, 51, 1521–1528. 10.1099/00207713-51-4-1521 [DOI] [PubMed] [Google Scholar]
- Fournier, P. E. , Lagier, J. C. , Dubourg, G. , & Raoult, D. (2015). From culturomics to taxonomogenomics: A need to change the taxonomy of prokaryotes in clinical microbiology. Anaerobe, 36, 73–78. 10.1016/j.anaerobe.2015.10.011 [DOI] [PubMed] [Google Scholar]
- Gouret, P. , Paganini, J. , Dainat, J. , Louati, D. , Darbo, E. , Pontarotti, P. , & Levasseur, A. (2011). Integration of evolutionary biology concepts for functional annotation and automation of complex research in evolution: The multi‐agent software system DAGOBAH In Pontarotti P. (Ed.), Evolutionary biology – concepts, biodiversity, macroevolution and genome evolution (pp. 71–87). Berlin Heidelberg: Springer; 10.1007/978-3-642-20763-1 [DOI] [Google Scholar]
- Gouret, P. , Thompson, J. D. , & Pontarotti, P. (2009). PhyloPattern: Regular expressions to identify complex patterns in phylogenetic trees. BMC Bioinformatics, 10, 298 10.1186/1471-2105-10-298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouret, P. , Vitiello, V. , Balandraud, N. , Gilles, A. , Pontarotti, P. , & Danchin, E. G. (2005). FIGENIX: Intelligent automation of genomic annotation: Expertise integration in a new software platform. BMC Bioinformatics, 6, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyatt, D. , Chen, G. L. , LoCascio, P. F. , Land, M. L. , Larimer, F. W. , & Hauser, L. J. (2010). Prodigal: Prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics, 11, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, C. N. , Whitehead, T. R. , Cotta, M. A. , Rhoades, R. E. , & Lawson, P. A. (2014). Peptoniphilus stercorisuis sp. nov., isolated from a swine manure storage tank and description of Peptoniphilaceae fam. nov. International Journal of Systematic and Evolutionary Microbiology, 64, 3538–3545. 10.1099/ijs.0.058941-0 [DOI] [PubMed] [Google Scholar]
- Käll, L. , Krogh, A. , & Sonnhammer, E. L. (2004). A combined transmembrane topology and signal peptide prediction method. Journal of Molecular Biology, 338, 1027–1036. 10.1016/j.jmb.2004.03.016 [DOI] [PubMed] [Google Scholar]
- Kim, M. , Oh, H.‐S. , Park, S.‐C. , & Chun, J. (2014). Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. International Journal of Systematic and Evolutionary Microbiology, 64, 346–351. 10.1099/ijs.0.059774-0 [DOI] [PubMed] [Google Scholar]
- Klappenbach, J. A. , Goris, J. , Vandamme, P. , Coenye, T. , Konstantinidis, K. T. , & Tiedje, J. M. (2007). DNA–DNA hybridization values and their relationship to whole‐genome sequence similarities. International Journal of Systematic and Evolutionary Microbiology, 57, 81–91. [DOI] [PubMed] [Google Scholar]
- Lagesen, K. , Hallin, P. , Rodland, E. A. , Staerfeldt, H.‐H. , Rognes, T. , & Ussery, D. W. (2007). RNAmmer: Consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Research, 35, 3100–3108. 10.1093/nar/gkm160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagier, J. C. , Hugon, P. , Khelaifia, S. , Fournier, P. E. , La Scola, B. , & Raoult, D. (2015). The rebirth of culture in microbiology through the example of culturomics to study human gut microbiota. Clinical Microbiology Reviews, 28, 237–264. 10.1128/CMR.00014-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagier, J. C. , Khelaifia, S. , Alou, M. T. , Ndongo, S. , Dione, N. , Hugon, P. , … Durand, G. (2016). Culture of previously uncultured members of the human gut microbiota by culturomics. Nature Microbiology, 12, 16203 10.1038/nmicrobiol.2016.203 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Lawrence, J. G. , & Ochman, H. (1997). Amelioration of bacterial genomes: Rates of change and exchange. Journal of Molecular Evolution, 44, 383–397. 10.1007/PL00006158 [DOI] [PubMed] [Google Scholar]
- Lepargneur, J. P. , & Rousseau, V. (2002). Protective role of the Doderleïn flora. Journal de Gynecologie, Obstetrique et Biologie de la Reproduction, 31, 485–494. [PubMed] [Google Scholar]
- Li, N. , Hashimoto, Y. , Adnan, S. , Miura, H. , Yamamoto, H. , & Ezaki, T. (1992). Three new species of the genus Peptostreptococcus isolated from humans: Peptostreptococcus vaginalis sp. nov., Peptostreptococcus lacrimalis sp. nov., and Peptostreptococcus lactolyticus sp. nov. International Journal of Systematic and Evolutionary Microbiology, 42, 602–605. [DOI] [PubMed] [Google Scholar]
- Li, J. , McCormick, J. , Bocking, A. , & Reid, G. (2012). Importance of vaginal microbes in reproductive health. Reproductive Sciences, 19, 235–242. 10.1177/1933719111418379 [DOI] [PubMed] [Google Scholar]
- Lowe, T. M. , & Eddy, S. R. (1997). tRNAscan‐SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Research, 25, 955–964. 10.1093/nar/25.5.0955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, R. , Liu, B. , Xie, Y. , Li, Z. , Huang, W. , Yuan, J. , … Tang, J. (2012). SOAPdenovo2: An empirically improved memory‐efficient short‐read de novo assembler. Gigascience, 1, 18 10.1186/2047-217X-1-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, D. H. , & Marrazzo, J. M. (2016). The vaginal microbiome: Current understanding and future directions. Journal of Infectious Diseases, 214, S36–S41. 10.1093/infdis/jiw184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matuschek, E. , Brown, D. F. , & Kahlmeter, G. (2014). Development of the EUCAST disk diffusion antimicrobial susceptibility testing method and its implementation in routine microbiology laboratories. Clinical Microbiology & Infection, 20, O255–O266. 10.1111/1469-0691.12373 [DOI] [PubMed] [Google Scholar]
- Meier‐Kolthoff, J. P. , Auch, A. F. , Klenk, H. P. , & Göker, M. (2013). Genome sequence‐based species delimitation with confidence intervals and improved distance functions. BMC Bioinformatics, 14, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menard, J. P. , Fenollar, F. , Henry, M. , Bretelle, F. , & Raoult, D. (2008). Molecular quantification of Gardnerella vaginalis and Atopobium vaginae loads to predict bacterial vaginosis. Clinical Infectious Diseases, 47, 33–43. 10.1086/588661 [DOI] [PubMed] [Google Scholar]
- Mishra, A. K. , Lagier, J. C. , Nguyen, T. T. , Raoult, D. , & Fournier, P.‐E. (2013). Non contiguous‐finished genome sequence and description of Peptoniphilus senegalensis sp. nov. Standards in Genomic Sciences, 7, 370–381. 10.4056/sigs.3366764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morel, A. S. , Dubourg, G. , Prudent, E. , Edouard, S. , Gouriet, F. , Casalta, J. P. , … Raoult, D. (2015). Complementarity between targeted real‐time specific PCR and conventional broad‐range 16S rDNA PCR in the syndrome‐driven diagnosis of infectious diseases. European Journal of Clinical Microbiology and Infectious Diseases, 34, 561–570. 10.1007/s10096-014-2263-z [DOI] [PubMed] [Google Scholar]
- Murdoch, D. A. (1998). Gram‐positive anaerobic cocci. Clinical Microbiology Reviews, 11, 81–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murdoch, D. A. , Mitchelmore, I. J. , & Tabaqchali, S. (1994). The clinical importance of gram‐positive anaerobic cocci isolated at St Bartholomew's Hospital, London, in 1987. Journal of Medical Microbiology, 41, 36–44. 10.1099/00222615-41-1-36 [DOI] [PubMed] [Google Scholar]
- Murray, P. R. , Baron, E. J. , Jorgensen, J. H. , Landry, M. L. , & Pfaller, M. A. (2007). Manual of clinical microbiology, 9th ed. Washington, D.C: ASM Press. [Google Scholar]
- Onderdonk, A. B. , Delaney, M. L. , & Fichorova, R. N. (2016). The human microbiome during bacterial vaginosis. Clinical Microbiology Reviews, 29, 223–238. 10.1128/CMR.00075-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandya, S. , Ravi, K. , Srinivas, V. , Jadhav, S. , Khan, A. , Arun, A. , … Madhivanan, P. (2017). Comparison of culture‐dependent and culture‐independent molecular methods for characterization of vaginal microflora. Journal of Medical Microbiology, 66, 149–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel, N. B. , Tito, R. Y. , Obregón‐Tito, A. J. , O'Neal, L. , Trujillo‐Villaroel, O. , Marin‐Reyes, L. , … Lewis, C. M. Jr (2015). Ezakiella peruensis gen. nov., sp. nov. isolated from human fecal sample from a coastal traditional community in Peru. Anaerobe, 32, 43–48. 10.1016/j.anaerobe.2014.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter, M. , & Rosselló‐Móra, R. (2009). Shifting the genomic gold standard for the prokaryotic species definition. Proceedings of the National Academy of Sciences, 106, 19126–19131. 10.1073/pnas.0906412106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez‐R, L. M. , & Konstantinidis, K. T. (2014). Bypassing cultivation to identify bacterial species. Microbe, 9, 111–118. [Google Scholar]
- Rooney, A. P. , Swezey, J. L. , Pukall, R. , Schumann, P. , & Spring, S. (2011). Peptoniphilus methioninivorax sp. nov., a Gram‐positive anaerobic coccus isolated from retail ground beef. International Journal of Systematic and Evolutionary Microbiology, 61, 1962–1967. 10.1099/ijs.0.024232-0 [DOI] [PubMed] [Google Scholar]
- Sasser, M. (2006). Bacterial identification by gas chromatographic analysis of fatty acids methyl esters (GC‐FAME). New york, NY: MIDI, Technical Note. [Google Scholar]
- Seng, P. , Drancourt, M. , Gouriet, F. , La Scola, B. , Fournier, P. E. , Rolain, J. M. , & Raoult, D. (2009). Ongoing revolution in bacteriology: Routine identification of bacteria by matrix‐assisted laser desorption ionization time of flight mass spectrometry. Clinical Infectious Diseases, 49, 543–551. 10.1086/600885 [DOI] [PubMed] [Google Scholar]
- Shipitsyna, E. , Roos, A. , Datcu, R. , Hallén, A. , Fredlund, H. , Jensen, J. S. , … Unemo, M. (2013). Composition of the vaginal microbiota in women of reproductive age–sensitive and specific molecular diagnosis of bacterial vaginosis is possible? PLoS ONE, 8(4), e60670 10.1371/journal.pone.0060670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan, S. , & Fredricks, D. N. (2008). The human vaginal bacterial biota and bacterial vaginosis. Interdisciplinary Perspectives on Infectious Diseases, 2008, 1–22. 10.1155/2008/750479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan, S. , Munch, M. M. , Sizova, M. V. , Fiedler, T. L. , Kohler, C. M. , Hoffman, N. G. , … Fredricks, D. N. (2016). More easily cultivated than identified: Classical isolation with molecular identification of vaginal bacteria. Journal of Infectious Diseases, 214(Suppl 1), S21–S28. 10.1093/infdis/jiw192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stackebrandt, E. , & Ebers, J. (2006). Taxonomic parameters revisited: Tarnished gold standards. Microbiology Today, 33, 152. [Google Scholar]
- Tamura, K. , Stecher, G. , Peterson, D. , Filipski, A. , & Kumar, S. (2013). MEGA6: Molecular evolutionary genetics analysis version 6.0. Molecular Biology and Evolution, 30, 2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J. D. , Higgins, D. G. , & Gibson, T. J. (1994). CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position‐specific gap penalties and weight matrix choice. Nucleic Acids Research, 22, 4673–4680. 10.1093/nar/22.22.4673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulger‐Toprak, N. , Lawson, P. A. , Summanen, P. , O'Neal, L. , & Finegold, S. M. (2012). Peptoniphilus duerdenii sp. nov. and Peptoniphilus koenoeneniae sp. nov., isolated from human clinical specimens. International Journal of Systematic and Evolutionary Microbiology, 62, 2336–2341. 10.1099/ijs.0.031997-0 [DOI] [PubMed] [Google Scholar]
- Yarza, P. , Yilmaz, P. , Pruesse, E. , Glöckner, F. O. , Ludwig, W. , Schleifer, K. H. , … Rosselló‐Móra, R. (2014). Uniting the classification of cultured and uncultured bacteria and archaea using 16S rRNA gene sequences. Nature Reviews Microbiology, 12, 635–645. 10.1038/nrmicro3330 [DOI] [PubMed] [Google Scholar]
- Zerbino, D. R. , & Birney, E. (2008). Velvet: Algorithms for de novo short read assembly using de Bruijn graphs. Genome Research, 18, 821–829. 10.1101/gr.074492.107 [DOI] [PMC free article] [PubMed] [Google Scholar]


