Summary
Corpus callosum malformations are associated with a broad range of neurodevelopmental diseases. We report that de novo mutations in MAST1 cause mega-corpus-callosum syndrome with cerebellar hypoplasia and cortical malformations (MCC-CH-CM) in the absence of megalencephaly. We show that MAST1 is a microtubule associated protein, that is predominantly expressed in post-mitotic neurons, and is present in both dendritic and axonal compartments. We further show that Mast1 null animals are phenotypically normal, whereas the deletion of a single amino acid (L278del) recapitulates the distinct neurological phenotype observed in patients. In animals harboring Mast1 microdeletions we find that the PI3K/AKT3/mTOR pathway is unperturbed, whereas Mast2 and Mast3 levels are diminished, indicative of a dominant negative mode of action. Finally, we report that de novo MAST1 substitutions are present in patients with autism and microcephaly, raising the prospect that mutations in this gene give rise to a spectrum of neurodevelopmental diseases.
Graphical Abstract

eTOC:
Tripathy et al. show that mutations in MAST1, a microtubule associated protein, cause a syndrome characterised by an enlarged corpus callosum. They attribute this thickening to an increase in the number of contralateral projections despite extensive cortical apoptosis.
Introduction
The bilateral integration of sensory, motor and cognitive inputs is mediated by the corpus callosum, the largest white matter tract of the brain (Paul et al., 2007). A broad array of neurodevelopmental disorders are known to be associated with malformations of this structure. For instance, corpus callosum agenesis has been reported in patients with microcephaly, lissencephaly, polymicrogyria, and has been described in patients with autism (Parrini et al., 2016). Mouse and human genetic studies have provided insight into the molecular machinery that is required for the development of this important anatomical feature (Edwards et al., 2014). It has been shown that the midline crossing of post-mitotic neurons requires transcription factors such as Satb2 (Britanova et al., 2008), cell adhesion molecules such as L1-CAM (Demyanenko et al., 1999), guidance molecules such as Netrin1 and the semaphorins (Niquille et al., 2009; Serafini et al., 1996), as well as cytoskeletal proteins including Map1b and the β-tubulin Tubb3 (Meixner et al., 2000; Tischfield et al., 2010). Collectively these molecules specify the fate of neurons destined to traverse the cerebral hemispheres, guiding their leading process to the correct destination.
While thinning of the corpus callosum is relatively common, in rare instances patients present with a thickening of this myelinated structure (Edwards et al., 2014; Marsh et al., 2017). To date, this phenotype has been reported in patients with neurofibromatosis and megalencephaly-polymicrogyria-mega-corpus-callosum syndrome (DiMario et al., 1999; Gohlich-Ratmann et al., 1998). These diseases are associated with a generalised enlargement in brain size, driven by activation of the PI3K/AKT3/mTOR pathway attributable to mutations in PIK3R2 and NF1 (Johannessen et al., 2005; Nguyen et al., 2016; Terrone et al., 2016). As mutations in these genes do not account for all cases, it is apparent that our understanding of the molecular pathology that underlies mega corpus callosum syndromes is incomplete (Hengst et al., 2010). Here, we present a cohort of patients with an enlarged corpus callosum in the absence of megalencephaly, harboring mutations in the uncharacterized microtubule associated protein MAST1.
Results
Identification of MAST1 Mutations
As part of an ongoing endeavor to identify genetic variants associated with structural brain malformations we undertook whole exome sequencing on 7 patient-parent trios, where the affected individual presented with a striking enlargement of the corpus callosum. This led to the identification of de novo mutations in the gene MAST1 (microtubule associated serine threonine kinase 1) in 6 of the 7 affected individuals (Figure 1A–T). Each of these patients presented with a hyperplastic corpus callosum (particularly over the genu and mid-body), cerebellar hypoplasia, ventricular dilation, and impaired motor and verbal performance (Table 1). Four patients presented with gyral simplification (P2 (Pierson et al., 2008), P3, P5, P6) three with dysplastic longitudinal gyri (P1, P5, P6) and one (P4) with periventricular nodules of white matter. We refer to this syndrome as mega-corpus-callosum with cerebellar-hypoplasia and cortical malformations (MCC-CH-CM).
Figure 1. Patients with MAST1 mutations.
Selected magnetic resonance images from patients P1 (A-D), P2 (E-H), P3 (I,J), P4 (K,L), P5 (M-P) and P6 (Q-T) in the midline sagittal plane (A, E, I, K, M), parasagittal plane (Q), and axial planes through the brainstem (F, N), lateral ventricles (B, C, G, J, L, O, R, S, T), and high convexities (D, H, P). All patients have a cortical malformation or dysgyria characterized by diffuse undersulcation, shallow sulci (arrowheads in B, C, G, J, L, O, R, S, T point to selected more obvious areas), and in the more severely affected mildly thick cortex (G, J; thus consistent with mild lissencephaly). While diffused, the cortical malformation appears most severe in the posterior frontal and perisylvian regions. The lateral ventricles are mildly to moderately enlarged, and the corpus callosum is abnormally thick (arrows in A, E, I, K, M, Q), accompanied by mildly thick white matter. The brainstem especially the pons is mildly (E) or moderately (A, I, K, M, Q) small, and in at least one child a prominent ventral midline cleft of the pons is seen (arrow in N). Available axial images through the high convexity of the cerebral hemispheres showed very dysplastic, longitudinally oriented gyral pattern (long arrows in D, H, P). (U) Schematic representation of the MAST1 genomic locus shows the position of the mutations identified in patients P1–P6. (V) The MAST1 protein consists of a domain of unknown function (DUF1908, shown in red), a kinase domain (shown in yellow) and a PDZ domain (shown in blue). The amino acid boundaries of each of the domains are shown. (W-Z) Autoradiograph showing the results of the microtubule binding assay with Mast1. Murine Mast1 was radiolabelled (35S) by in vitro transcription and translation in rabbit reticulocyte lysate, before incubation with a porcine microtubule extract in the presence or absence of microtubule associated proteins (MAPs) (W,X). Following microtubule polymerization in the presence of Taxol, pelleted microtubules were analyzed by polyacrylamide gel electrophoresis (PAGE) and the ratio of pelleted radiolabeled-mMast1 to TnT input determined. This experiment revealed a decrease of binding of Mast1 to microtubules in the absence of MAPs (X; n=3 technical replicates; two-tailed unpaired t-test; t4=3.265, P<0.05). Patient mutations were introduced into mMast1, radiolabelled by in vitro transcription and translation (TnT), and microtubule binding assessed (Y,Z). As each mutant peptide was radiolabeled, the ratio of microtubule bound Mast1 (in the pelleted fraction), to total radiolabelled Mast1 was determined. Comparison of pelleted wild-type Mast1 to the K276del mutation shows a significant alteration in microtubule binding, and a similar trend for the L278del. (Z; n=6–9 repeated experiments; one-way ANOVA with Dunnett’s multiple comparison; WT vs K276del P<0.05). * shows P<0.05; ** shows P<0.01; *** shows P<0.001; **** shows P<0.0001. Error bars show mean +/− the standard error of the mean.
Table 1.
Clinical summary of patients with MCC-CH-CM.
| Patient | P1 | P2 | P3 | P4 | P5 | P6 |
|---|---|---|---|---|---|---|
| Mutation (chr19) | 19:1295867 7delGAG | chr19:129627 98delGAA | chr19:12962804 delGTT | 19:12975903G >A | 19:12975903G>A | 19:12975903 G>A |
| Mutation (NM_014975.2) | Glu194del c.580_582del | Lys276del c.825_827del | Leu278del c.831_833del | Gly517Ser c.1549G>A | Gly517Ser, c.1549G>A | Gly517Ser, c.1549G>A |
| CADD v1.3 | 16,71 | 17,35 | 21,4 | 27,9 | 27,9 | 27,9 |
| Inheritance | de novo | de novo | de novo | de novo | de novo | de novo |
| Ethnicity | Caucasian | Caucasian | Caucasian | Moroccan | Caucasian | Caucasian |
| Geographic Ancestry | Italian | Hungarian | French | Moroccan | Unknown | French |
| Sex | Male | Male | Female | Female | Female | Female |
| Age at last evaluation | 9 years | 11.5 years | 6 years | 6 years | 10 years | 1.5 year |
| OFC* at birth | Unknown | 36.5 cm (50–75th %) | Unknown | Unknown | Unknown | 35cm (50th) |
| OFC* at last evaluation | 52 cm | 53 cm (25–50th %) | 51 cm | 52 cm (+0.6 SD) | 49.2 cm at 2y9m (50%) | 46.5 cm at 18 m |
| Cortical dysgenesis | Dysplastic longitudinal gyri; subependym al heterotopias. | Extensive undersulcated gyral pattern over frontal, temporal and perisylvian regions dysplastic longitudinal gyri | Tubulinopathy-like dysgyria with mildly thick cortex and diffuse very shallow sulci | Periventricular lesions | Diffuse subtle dysgyria, radial/shallow sulci with some deep infolds; dysplastic longitudinal gyri | Subtle frontal dysgyria, dysplastic longitudinal gyri |
| Basal ganglia | Normal | Poorly developed | Normal | Normal | Normal | normal |
| Cerebellum | Vermis hypoplasia (++); counterclock wise rotation of cerebellar vermis | Mild diffuse cerebellar hypoplasia (+) with prominent sulci | Hypoplasia (++) | Hypoplasia (++) | Vermis hypoplasia (++) and mild hemispheric hypoplasia | Vermis hypoplasia (++), and mild hemispheric hypoplasia |
| Brainstem | Hypoplasia (++) | Small pons (+) | Hypoplasia (++) | Pontine hypoplasia (++) | Severe brainstem hypoplasia with a very small pons (++), thin midline cleft, long and mildly enlarged medulla | Severe brainstem hypoplasia (++), small pons with relative sparing of its buldging |
| Corpus callosum | Hyperplasia, mostly over genu and anterior body; (++) | Very thick and dysplastic corpus callosum (mega corpus callosum); (++) | Hyperplasic; (++) | Hyperplasia, mostly over genu and anterior body; (+) | Thick corpus callosum; (+) | Thick corpus callosum; (+) |
| Ventricular Dilation | Enlarged 4th ventricle (+) | Enlarged 3rd ventricles (++) | Enlarged 3rd ventricles (++) | Enlarged 4th ventricle (++) | Enlarged 3rd and 4th ventricles (++) | (+) |
| Cognitive abilities | Moderate cognitive impairment | Intellectual Disability | Severe encephalopathy | Severe intellectual disability | Global developmental delay | Global developmental delay |
| Verbal abilities | Poor | Non-verbal | Non verbal | Non verbal | Non-verbal | Only some sounds (vowels) |
| Motor abilities | Clumsy stumbling gate; generalized hypotonia | Generalized hypotonia, mild truncal ataxia | Severe Hypotonia | Able to stand with support, hyperreflexia | She can sit up by herself with assistance from her arms; able to roll. | Hypotonic, sitting: with cyphosis, and only with support on arms; able to roll. |
| Other | Short stature, paroxysmal EEG abnormalities (spike-wave complexes) with no clinical seizures | Infantile onset epilepsy, oculomotor apraxia | None | Short stature, strabismus | Seizures (onset 6y), history of aspiration | Head thrusts suggestive of oculomotor anomality. |
| Other de novo or novel variants | None | Inherited a novel silent mutation in TUBGCP5: chr15 (22851109) C>T (T457T) | None | None | None | None |
Through the Genematcher platform, four additional patients with de novo mutations in MAST1 were identified, who presented with either microcephaly accompanied by motor deficits (P7, P8) or autism spectrum disorder (P9, P10) (Figure S1A–C,D,E, Table S1) (Gilissen et al., 2014; Sobreira et al., 2015). In each case the mutation was verified by Sanger sequencing, was unreported in publicly available genome databases (e.g. dbSNP, 1000 genome, ExAC, see Table S2), and was in a highly conserved residue (Figure S1F–L) that was predicted to be deleterious when mutated (Table 1, Table S1, CADD score) (Kircher et al., 2014; Lek et al., 2016). We did not identify any unreported variants in our patient cohort with the exception of a silent mutation in TUBGCP5 (T457T) in P2 and a de novo mutation in NIMA related kinase 1 (NEK1) in P8 (Table 1 and Table S1). It should be noted that mutations in NEK1 have been previously associated with amyotrophic lateral sclerosis, cilial dysfunction and skeletal diseases, however, patient P8 did not present with symptoms consistent with these disorders (Kenna et al., 2016; Thiel et al., 2011). Three of the MCC-CH-CM associated variants in MAST1 were single amino acid deletions positioned in the hydrophobic core of a four-helix bundle in the domain of unknown function DUF1908 (P1: p.Glu194del; P2: p.Lys276del; P3: p.Leu278del) (Figure S1M) while the remainder (P4–P6: p.Gly517Ser) harboured a recurrent missense mutation in the kinase domain of the protein (Figure 1U–V). These microdeletions were not found in control individuals in the ExAC genome browser, however, a single microdeletion between the kinase and PDZ domains (Glu697del) has been reported (See Table S2).
MAST1 is a microtubule associated protein
Little is known of the function of the MAST family of proteins (MAST1–4). MAST2 was initially cloned from testes, and was shown to function as a kinase and interact with microtubules via other microtubule associated proteins (MAPs) (Walden and Cowan, 1993). To ascertain whether MAST1 associates with the microtubule cytoskeleton we performed an in vitro transcription and translation (TnT) reaction employing rabbit reticulocyte lysate with radiolabeled murine (m)Mast1 (which shares 94% sequence identity with human MAST1). The reaction products were then assayed for their ability to bind to microtubules in the absence or presence of MAPs. We found that mMast1 associates with Taxol-stabilized microtubules in a MAP-dependent manner (Figure 1W,X; n=3; P<0.05). Next, we asked whether the MAST1 mutations found in our patient cohort influence microtubule binding. We introduced each mutation into our pcDNA 3.1 vector, labelled Mast1 with 35S-methionine, and repeated the aforementioned microtubule binding assay. While translation efficiency was similar for all variants, we found that the K276 deletion significantly enhanced Mast1 binding to microtubules (Figure 1Y,Z; n=9; WT vs K276del P<0.05). Taken together these data show that Mast1 binds to microtubules in a MAP-dependent manner, and that mutations can perturb this interaction.
MAST1 is Expressed in Post-Mitotic Neurons
To gain insight into the role of MAST1 in neurodevelopment we analyzed its expression in the human and mouse brain during early developmental stages (Figure 2A and 2B). To this end we extracted mRNA from the developing murine brain (E10.5, E12.5, E14.5, E16.5, E18.5, P0 and P6), generated cDNA, and performed qPCR analysis. We observed that Mast1 expression begins at E12.5, peaks at E16.5, and decreases postnatally (Figure 2B). Consistent with this result qPCR analysis of human fetal brain cDNA showed moderate expression of MAST1 at gestation week 13 and 22 (Figure 2A). To determine which cells express Mast1 we performed immunohistochemistry on the murine cortex at E12.5, E14.5, E16.5 and P0 (n=3). We used a commercially available Mast1 antibody, having validated its specificity employing a Mast1 knockout animal (Figure S2G–L, Figure S3B–D). We observed staining in the developing cortical plate and intermediate zone at these time points (Figure 2C–F). Further analysis at E14.5 and E16.5 showed that Mast1 is located in the cytoplasm in Tuj positive post-mitotic neurons, but is largely absent from Sox2 positive progenitors and Tbr2 positive intermediate progenitors (Figure 2S–AP) (n=3). This expression pattern is mirrored in human cerebral organoids that were cultured for 64 days, where we again observed co-localisation with post mitotic markers (Figure S2A–F) (n=10 organoids) (Lancaster et al., 2013). Immunostaining on P0 mouse brain sections showed that Mast1 is present in Tuj positive corpus callosal fibers that cross the midline (n=3 animals) (Figure S2G–L). To gain further insight into the subcellular localisation of Mast1 we cultured primary P0 cortical neurons and P7 cerebellar granule neurons and performed immunostaining. We again validated the specificity of our Mast1 antibody for cell culture experiments employing a Mast1 KO animal (Figure S2S). We performed double staining with antisera specific for the axonal marker Tau or the dendritic marker Map2 (Kosik and Finch, 1987). We observed Mast1 staining throughout the soma, as well as in dendritic and axonal compartments in cortical neurons (Figure 2G–R, n=5) and in cerebellar granule neurons (Figure S2M–R, n=3). This staining had a punctate appearance, suggesting that Mast1 may associate with vesicular structures that are trafficked along the microtubule cytoskeleton. qPCR analysis of Mast1 expression in the adult mouse showed that it’s expression persists in all brain regions (albeit at much lower levels), and that it is present in the testes, liver and spleen at very low levels (n=3) (Figure S2T). These results are consistent with expression data that is available through online databases (e.g. Allen brain atlas, GTEx) (Consortium, 2013; Morris et al., 2010). Taken together these data show that Mast1 is predominantly expressed in post-mitotic neurons in the developing nervous system.
Figure 2. Mast1 expression in human and mouse embryonic brain.
(A) qPCR analysis reveals that MAST1 mRNA is expressed in the human developing fetal brain at gestational week (GW) 13 and the fetal frontal lobe at GW22. (B) qPCR analysis performed on mouse brain cDNA libraries from E10.5 to P6, show that Mast1 expression peaks at E16.5 in mice (n=3 animals per timepoint). (C-F) Immunohistochemistry employing a validated Mast1 antibody (see also Figure S2B–D) indicates staining in the post mitotic cortical plate and intermediate zone from E12.5 to P0 in mice. (G-R) Maximum projection images of cultured P0 cortical neurons at 5-DIV staining with the axonal marker Tau (G-L) and the dendritic marker Map2 (M-R), shows that Mast1 (I,L,O,R) is present in both axonal and dendritic compartments. Dashed boxes in G-I and M-O and expanded in J-L and P-R, respectively. (S-AP) Immunohistochemistry employing the progenitor marker Pax6 (S-U, AE-AG), intermediate progenitor marker Tbr2 (W-Y, AI-AK), and post mitotic marker Tuj (AA-AC, AM-AO) on E14.5 (S-AD) and E16.5 (AE-AP) murine sections reveal that Mast1 expression is restricted to post-mitotic neurons at these timepoints (PP: preplate; CP: cortical plate; MZ: marginal zone; IZ: intermediate zone; VZ: ventricular zone).
An Enlarged Corpus Callosum in a L278Del Mast1 Mouse Model
To investigate the effect of Mast1 mutations in vivo we exploited the power of the CRISPR-Cas9 genome editing system to generate a Mast1 knockout (KO) mouse and a Mast1 L278del (L278del) mouse employing guide RNAs targeted to exon 8 (Figure S3A) (Wang et al., 2013). We decided to recapitulate the mutation identified in patient P3 because of the striking callosal, cortical and cerebellar phenotypes. As homozygous L278del/del animals die shortly after birth we focused our initial analysis on homozygous Mast1 KO and heterozygous L278del/+ mice which are viable. We analyzed the thickness of the genu of the corpus callosum on rostral Nissl stained coronal sections (Bregma 0.86mm). While Mast1 KO animals were indistinguishable from wild type littermate controls (Figure S3E–G; n=5; +/+ v KO/KO; P>0.5), L278del/+ animals presented with a significantly thicker corpus callosum in comparison to wild-type littermate controls (Figure 3A–E; n=5; +/+ v L278del/+; P<0.001). High resolution volumetric magnetic resonance imaging (MRI) analysis confirmed an increased corpus callosum volume in L278del/+ animals (Figure 3H; n=3; +/+ v L278del/+; P<0.05), and revealed that the thickening was most prominent in the rostral regions encompassing the genu and the mid-body (Figure 3F–G). The enlarged corpus callosum was also present in Leu278/del mice aged 10 days (Figure S3K–M, n=5; +/+ v L278del/+; P<0.05). While off target mutations in Cas9-modified mice are rare (Iyer et al., 2015), we confirmed that this phenotype was not due to a linked mutation by undertaking whole exome sequencing for our L278del/+ mouse line. We identified no other non-synonymous mutations on chromosome 8 in this line (Table S3). Moreover, we generated a second independent L278del mouse line, and once again observed an enlarged corpus callosum (Figure S3H–J; +/+ v L278del/+_Line#7; n=3; P<0.01).
Figure 3. L278del mice have an enlarged corpus callosum associated with an increase in the number of callosal axons.
(A-D) Nissl-stained sections of 8-week old adult brains highlighting the thicker corpus callosum in L278del/+ animals compared to wild type littermates (black boxes in A,B expanded in C,D). Quantification of the thickness at the septum (E) reveals a significantly thicker corpus callosum in L278del/+ animals (n=5 animals per genotype; unpaired t-test; +/+ v L278del/+; t8=6.215, P<0.001). (F-G) MRI reconstructions of the corpus callosum in wild-type controls (F) and L278del/+ mice (G). The heat map reflects the thickness of the corpus callosum (blue: thinner region, red: thicker region). Note that in L278del/+ animals the genu and mid body region are most affected. (H) MRI volumetric quantification of the corpus callosum shows that this structure is significantly larger in L278del/+ animals compared to wild type littermates (n=3 animals per genotype; two-tailed unpaired t-test; +/+ v L278del/+; t4=4.233, P<0.05). (I-J) Electron microscopy images showing cross-sections of the corpus callosum at the midbody (region depicted with boxes in F and G). Myelinated axons can be clearly visualised. (K) Quantification of myelin thickness reveals no significant different between L278del/+ animals and wild type littermates (n=5 animals per genotype, >500 myelinated axons per animal; two-tailed unpaired t-test; t8=1.001, P>0.1). (L) Quantification of axonal caliber reveals no significant different between L278del/+ animals and wild type littermates (n=5 animals per genotype, >1500 myelinated axons per animal; +/+ v L278del/+; two-tailed unpaired t-test; t8=1.786, P>0.1, see also Figure S3N). (M-N) Assessment of the total number of myelinated axons within a 30μm wide box extending along the ventro-dorsal axis of the corpus callosum. (O) Quantification reveals a significant increase in axonal count in the L278del/+ animals in comparison to wild-type littermates (n=5 animals per genotype, 3 images analyzed per animal; two-tailed unpaired t-test; t8=4.095, P<0.01). * shows P<0.05; ** shows P<0.01; *** shows P<0.001; **** shows P<0.0001. Error bars show mean +/− the standard error of the mean.
We then asked whether the increase in the thickness of the corpus callosum of the L278del/+ mice was due to an increase in the thickness of myelinated axons. In light of our MRI studies, we focused our analysis on the mid body of the corpus callosum where we observe the most pronounced phenotype in L278del/+ mutants (Bregma −0.10). The corpus callosum of adult animals was microdissected, samples stained to enable clear visualisation of myelinated neurons crossing the midline, and images captured at high resolution (8900×) with transmission electron microscopy (Figure 3I–J) (Sturrock, 1980; West et al., 2015). We observed no significant difference in either myelin thickness (Figure 3K; n=5; n>500 axons per animal; +/+ v L278del/+; P>0.1) or axonal caliber (Figure 3L; n=5; n>1500 axons per animal; +/+ v L278del/+; P>0.1) when comparing littermate controls with L278del/+ animals. An analysis of the distribution of axon calibers in L278del/+ animals again revealed no significant difference in comparison to littermate controls (n= 5; +/+ v L278del/+; P>0.1 for all bins) (Figure S3U). Reflecting this an assessment of the G-ratio (axonal diameter/total fiber diameter), revealed no significant difference between genotypes (n=5, n>300 fibers animal, +/+ vs L278del/+; P>0.1) (Guy et al., 1989).
Given these results we asked whether more myelinated axons might cross the midline in L278del/+ animals. To assess this TEM images were acquired at a magnification of 710× along the dorso-ventral axis of the corpus callosum. These images (10–12 per animal) were tiled resulting in a single image that encompassed the entire cross section of the corpus callosum. A box 30μm wide was overlaid onto the structure, and the total number of myelinated axons counted manually (Figure 3M–N). Statistical analysis revealed a significant increase in the number of myelinated axons within that box in L278del/+ animals in comparison to littermate controls (Figure 3O, n=5 animals per genotype, n=3 images per animal, +/+ v L278del/+; P<0.005). Finally, we counted the number of Olig2 positive oligodendrocyctes and GFAP positive astrocyctes, in a box of fixed width that was overlaid onto coronal sections of the corpus callosum (Figure S3O–R). This revealed that in L278del/+ animals there are more oligodendrocytes and astrocyctes in comparison to wild-type controls (n=5; +/+ vs L278del/+; Olig2: P<0.05; GFAP: P<0.05) (Figure S3S,V). There was, however, no significant difference in the density of these cell types when comparing genotypes (n=5;+/+ vs L278del/+; Olig2: P>0.1; GFAP: P>0.05) (Figure S3T,W). Taken together these data show that our L278del/+ mouse model recapitulates the enlarged corpus callosum observed in affected patients, which we attribute to an increase in the number of myelinated axons crossing the midline.
Cortical and cerebellar hypoplasia in the L278del Mast1 Mouse Model
We extended our analysis of the L278del and KO mouse lines to the cortex and cerebellum, which are consistently affected in our MAST1 patient cohort. MRI volumetric analysis of adult animals revealed an overall reduction in cortical volume in L278del heterozygotes (Figure 4A–C; n=3; +/+ v L278del/+; P<0.05). We confirmed this finding by undertaking Nissl and NeuN staining, where we observed a significant reduction in cortical thickness in L278del animals that was most pronounced in caudal regions (Bregma −1.82mm; Figure 4D–F; Nissl: n=5; +/+ v L278del/+; P<0.001; NeuN: Figure S4A–D; n=5; +/+ v L278del/+; P<0.0001). We did observe a reduction in cortical thickness in the rostral motor cortex in L278del heterozygotes (Bregma 0.86 mm), however, this was not statistically significant (Figure S4E–G, n=5; +/+ v L278del/+; P>0.1). We assessed the thickness of the cortical layers in the affected caudal regions in L278del heterozygotes and littermate controls. We observed a significant reduction in the thickness of layer 5, marked by the transcription factor Er81 (Figure 4J–L; n=5; +/+ v L278del/+; P<0.005). We did not observe a significant difference in the thickness of Foxp2 positive layer VI neurons (Figure S4H–K; n=5; +/+ v L278del/+; P>0.1) (Ferland et al., 2003) or Cux1 positive granule cells (Figure 4G–I; n=5; +/+ v L278del/+; P>0.5) (Cubelos et al., 2015). Analysis of the caudal cortex at P10 by Nissl staining again revealed that it was thinner in L278del/+ mutants (Figure S4U, n=5; +/+ v L278del/+; P<0.01). Consistent with our previous findings adult Mast1 KO animals appeared to be phenotypically normal, without morphological or layering defects in the adult KO cortex (Figure S4A–D, H–T).
Figure 4. L278del mice have a reduction in cortical volume associated with an increase in neuronal apoptosis.
(A,B) MRI reconstructions of the cortex in adult animals reveals a reduction in cortical volume in L278del/+ mice in comparison to littermate controls. (C) Quantification of the MRI cortical volume (n=3 animals per genotype; two-tailed unpaired t-test; +/+ vs L278del/+; t4=2.902, P<0.05). (D-E) Nissl stained sections of the adult somatosensory cortex reveals a reduction in cortical thickness in L278del/+ adult mice in comparison to littermate controls. (F) Quantification of caudal cortical thickness (n=5 animals per genotype; two-tailed unpaired t-test; +/+ vs L278del/+; t8=5.417, P<0.001). (G-H) Labeling of Cux1-positive layer II-III and (J-K) Er81-positive layer 5 neurons in wild type and L278del/+ adult mice. (I) Quantification reveals no significant difference in Cux1 layer thickness when comparing genotypes but a significant reduction in the size of the Er81-positive layer (L) (n=5 animals per genotype; 2-way repeated measures ANOVA with a Bonferroni test for multiple comparisons; +/+ vs L278del/+; Cux1 layer P>0.1; Er81 layer P<0.01). (M-X) Representative Nissl (M-O) activated caspase-3 (P-U), and activated caspase 9 (V-X) stained P0 cortical sections of littermate controls (M,P,S,V), L278del/+ (N,Q,T,W), and L278del/del (O,R,U,X) animals. The white boxes in P,Q,R are expanded in S,T,U. The number of caspase-3 and 9 positive cells in the cortex of P0 animals was counted and averaged (3 sections per animal). There is a dose dependent increase in apoptotic cells in L278del/+ and L278del/del animals in comparison to littermate controls (see also Figure S4V). (Y) Quantitation of capase-3 staining and (Z) caspase-9 staining (n=5 animals per genotype; n=3 sections per animal; 2-way repeated measures ANOVA with a Bonferroni test for multiple comparison; Caspase-3: +/+ vs. L278del/+ P<0.01;’+/+ vs. L278del/del P<0.0001; L278del/+ vs. L278del/del P<0.001; Caspase-9: +/+ vs.L278del/+ P<0.05; ‘+/+ vs. L278del/del P<0.0001; L278del/+ vs. L278del/del P<0.01). * shows P<0.05; ** shows P<0.01; *** shows P<0.001; **** shows P<0.0001. Error bars show mean +/−the standard error of the mean.
MRI volumetric analysis of the cerebellum revealed a severe reduction in L278del heterozygotes (Figure 5A–C; n=3; +/+ v L278del/+; P=0.0005). Nissl staining as well as NeuN and calbindin immunohistochemistry, showed that the foliation and lamination of the cerebellum was unaffected in L278del heterozygotes, however, the granule and the molecular cell layer appeared thinner in mutants (Figure 5D–I). We calculated the density of granule cells as well as the linear density of Purkinje cells, allowing us to estimate the total number of these cell types in mid sagittal sections. While the density of both cell types was similar in L278del mutants in comparison to littermate controls (Figure S5A–B), we observed a significant reduction in the total number of granule cells per section (Figure 5J; n=5; +/+ v L278del/+; P<0.05). We also observed a reduction in the total number of Purkinje cells per section, but this reduction was not statistically significant (Figure 5K; n=5; +/+ v L278del/+; P>0.5). Volumetric analysis of the colliculi, putamen, thalamus and olfactory bulbs revealed no significant difference between wild-type littermates and L278del heterozygotes (Figure S5C; n=3; +/+ v L278del/+; P>0.5 for all regions). There was a reduction in the volume of the hippocampus in L278del heterozygotes, but this difference was not statistically significant (Figure S5C; n=3; +/+ v L278del/+; P=0.34).
Figure 5. L278del mice have a hypoplastic cerebellum.
(A-B) MRI reconstructions of the cerebellum from wild type littermates (A) and L278del/+ (B) adult animals. (C) Quantification reveals a significant reduction in cerebellar volume (mm3) in L278del/+ animals (n=3 animals per genotype; two-tailed unpaired t-test; +/+ vs. L278del/+; t4=10.16, P<0.001). (D-G) Nissl stained sagittal sections of 8-week old cerebellum confirms the reduction in cerebellar size and indicates that lamination within the cerebellum is normal in L278del/+ animals. (H-I) Immunostaining employing the neuronal marker NeuN (shown in red) and the Purkinje cell marker Calbindin (shown in green) revealed a reduction in the thickness of the granule cell layer and molecular layer in L278del/+ animals. (J-K) Estimates of the total number of granule cells (J) and Purkinje cells (K) per a midsaggital section in L278del/+ animals and littermate controls (n=5 animals per genotype; two-way ANOVA with a Bonferroni correction; +/+ vs. L278del/+; granule cell counts: P<0.05; Purkinje cell counts: P>0.5, see also Figure S5A,B). (L-Q) Immunohistochemistry employing sera for activated Caspase3 reveals an increase in apoptosis in the L278del/+ and L278del/del animals in the developing cerebellum at P0. The white boxes in L,N,P are expanded in M,O,Q, respectively. (R) Quantification of capase3 staining (n=3 animals per genotype; one-way ANOVA with Tukey multiple comparison; +/+ vs. L278del/+ P<0.01; +/+ vs. L278del/del P<0.001; L278del/+ vs. L278del/del P<0.01). * shows P<0.05; ** shows P<0.01; *** shows P<0.001; **** shows P<0.0001. Error bars show mean +/− the standard error of the mean.
L278Del Mast1
To determine if the smaller cortex and cerebellum in our L278del mutant mice is a result of neuronal cell death during development, we performed an activated caspase 3 staining. We detected a dose-dependent increase in the number of apoptotic cells in the P0 cortex of wild type versus heterozygous and homozygous L278del mice (Figure 4P-U,Y, AB; n=5; +/+ vs L287del/+ P<0.005; +/+ vs L278del/del P<0.0001; L278del/+ vs L278del/del P=0.0001). Consistent with our anatomical analysis of adult animals we found that apoptosis was more severe in the caudal regions of the P0 cortex in the L278del/+ and L278del/del animals (Figure S4V, n=5; rostral vs caudal: L278del/+ P<0.001; L278del/L278del P<0.0001). At P10 and in 8 week old adult mice apoptosis in L278del/+ mice was comparable to wildtype controls (Figure S4W–Z, n=5; +/+ vs. 278del/+; P10, P>0.1; Adult: P>0.1). An analysis of activated caspase 3 staining in the P0 cerebellum revealed a large increase in L278del/+ and L278del/del animals in comparison to wildtypes (Figure 5L–R) (n=3; +/+ vs L287del/+ P<0.01; +/+ vs L278del/del P=0.0001; L278del/+ vs L278del/del P<0.005). To ascertain whether this cell death is associated with intrinsic suicide pathways, we stained P0 cortical sections with sera that targets activated Caspase 9, an upstream regulator of caspase 3 that is associated with mitochondrial mediated apoptosis (Hyman and Yuan, 2012). We observed a dose-dependent increase in the number of activated caspase 9 positive cells in the cortex of heterozygous and homozygous L278del mice (Figure 4V-X,Z; n=5; +/+ vs. 278del/+ P<0.05; +/+ vs. L278del/del P<0.0001; L278del/+ vs. L278del/del P<0.01). Intriguingly, this was not associated with the upregulation of the tumour suppressor p53 (Figure S4AA-AF), which has been associated with a range of stressful cellular events including DNA damage, defects in cell cycle progression, as well as oxidative and nutritional stress (Szybinska and Lesniak, 2017). Taken together these data show that the L278del mutation results in a hypoplastic cortex and cerebellum, a phenotype that is associated with neuronal apoptosis mediated by activated caspases 3 and 9, but not p53 upregulation.
The L278
Pathway
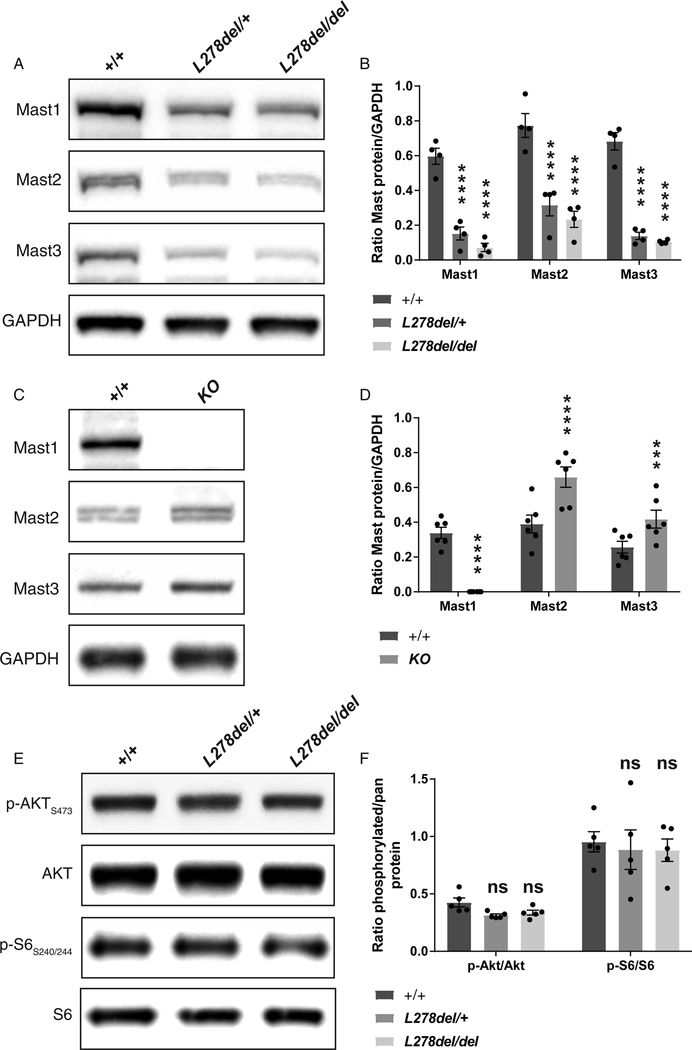
Next, we exploited these mouse models to investigate the underlying molecular impairment associated with MAST1 mutations. Western blot analysis of P0 brain lysates, showed a complete absence of Mast1 protein in KO animals (n=6) with an unexpected dose dependent reduction in Mast1 levels in the L278del line (Figure 6 A, n=4, Mast1: +/+ vs L278del/+ P<0.0001; +/+ vs L278del/Ldel P<0.0001). This absence of protein in the Mast1 KO animals correlates with a dramatic reduction in mRNA levels, but this is not the case in L278del mice where Mast1 transcript levels are statistically similar in +/+, L278del/+, and L278del/del animals (Figure S6 A, D). We assessed whether the reduction in Mast1 protein levels would influence the levels of other MAST family members. Relying on western blot analysis, we observed a striking reduction in the levels of Mast2 and Mast3 in our L278del animals (Figure 6 A–B; n=4; Mast2: +/+ vs L278del/+ P<0.0001; +/+ vs L278del/del P<0.0001; Mast3: +/+ vs L278del/+ P<0.0001; +/+ vs L278del/del P<0.0001). This was not attributable to changes in the mRNA levels of Mast2 and Mast3 which were similar in +/+, L278del/+, and L278del/del animals (Figure S6 B–C). In contrast in our Mast1 KO animals we observed a signficant increase in the levels of Mast2 and Mast3 (Mast2: +/+ vs KO P<0.0001; Mast3: +/+ vs KO P<0.001), again with no change in the mRNA levels (Figure S6E–F). These data imply that the L278del mutation in Mast1 acts by a dominant negative mode of action whereas a form of post-transcriptional compensation occurs in the Mast 1 KO line.
Figure 6. The L278del mutation influences Mast1/2/3 protein levels, but does not activate the PI3K/AKT3/mTOR pathway.
(A-B) Western blot analysis of Mast1, Mast2 and Mast3 on P0 cortical lysates from L278del animals. Quantification reveals a dramatic reduction of Mast1, Mast2 and Mast3 protein levels in L278del heterozygotes and homozygotes in comparison to littermate controls (n=4 animals per genotype; 2-way repeated measures ANOVA with a Bonferroni correction; Mast1: +/+ vs L278del/+ P<0.0001; +/+ vs L278del/del P<0.0001; Mast2: +/+ vs L278del/+ P<0.0001; +/+ vs L278del/del P<0.0001; Mast3: +/+ vs L278del/+ P<0.0001; +/+ vs L278del/del P<0.0001, see also Figure S6–C). (C-D) Western blot analysis of Mast1, Mast2 and Mast3 on brain lysates from Mast1 KO animals. While Mast1 is absent, there is a significant increase in the levels of Mast2 and Mast3 when compared to littermate controls (n=6 animals per genotype; 2-way repeated measures ANOVA with a Bonferroni test for multiple comparison; Mast1: +/+ vs KO P<0.0001; Mast2: +/+ vs KO P<0.0001; Mast3: +/+ vs KO P<0.001, see also Figure S6D–F). (E-F) Levels of phosphorylated AKT and ribosomal S6 proteins, indicators of activation of PI3K/AKT/m-TOR pathway, are not significantly different in wild type and L278del P0 cortex (n=5 animals per genotype; 2-way repeated measures ANOVA with a Bonferroni test for multiple comparison; +/+ vs L278del/+ AKT P>0.5; +/+ vs L278del/del AKT P>0.5; L278del/+ vs L278del/del AKT P>0.5; +/+ vs L278del/+ rpS6 P>0.5; +/+ vs L278del/L278del rpS6 P>0.5; L278del/+ vs L278del/Ldel rpS6 P>0.5). * shows P<0.05; ** shows P<0.01; *** shows P<0.001; **** shows P<0.0001. Error bars show mean +/− the standard error of the mean.
Finally, as previous studies have implicated activation of the PI3K/AKT3/mTOR pathway in corpus callosum hyperplasia associated with megalencephaly, we assessed the phosphorylation state of AKT and rpS6 in our L278del mice (Figure 6 E) (Broix et al., 2016; Poduri et al., 2012; Riviere et al., 2012; Terrone et al., 2016). We observed no difference in the levels of p-AKTS473 and p-rpS6 S240/244 when comparing heterozygous and homozygous L278del brain lysates with wild type controls (Figure 6 F; n=5 animals; +/+ vs L278del/+ AKT P>0.5; +/+ vs L278del/del AKT P>0.5; L278del/+ vs L278del/del AKT P>0.5; +/+ vs L278del/+ rpS6 P>0.5; +/+ vs L278del/del rpS6 P>0.5; L278del/+ vs L278del/del rpS6 P>0.5).
Discussion
Here, we report that de novo mutations in MAST1 cause MCC-CH-CM, a disease characterised by a striking enlargement of the corpus callosum, cerebellar hypoplasia and cortical malformations. For patients with this constellation of phenotypes mutations in MAST1 appear to be the primary genetic cause, as we observed MAST1 variants in 6 of the 7 patients that we studied. We have shown, that MAST1 is expressed predominantly in post-mitotic neurons in the developing nervous system, and that it is present in the soma as well as dendritic and axonal compartments. We demonstrate that a L278del mouse line recapitulates the enlarged corpus callosum and cerebellar hypoplasia observed in patients, while the Mast1 KO line lacks any morphological defects. We report extensive cortical apoptosis in our L278del mice that is mediated by activated caspases 3 and 9, but not p53 upregulation. We report that this apoptosis varies along the rostro-caudal axis of the cortex, being most severe in caudal regions where the enlarged corpus callosum is least pronounced. Our results show that unlike other syndromes associated with an enlarged corpus callosum activation of the PI3K/AKT3/mTOR pathway does not appear to be the underlying pathogenic driver (Terrone et al., 2016).
What molecular mechanism underlies MCC-CH-CM? We have shown that MAST1 binds to microtubules in a MAP-dependent manner, but this binding is only altered in the case of the K276del mutation. It therefore seems unlikely that alterations in microtubule affinity cause the spectrum of phenotypes we observe in patients with MMC-CH-CM. Analysis of our L278del mouse model has shown a dramatic reduction in the levels of Mast 2 and 3 protein. This result suggests that pathogenic microdeletions in Mast1 act by a dominant negative mechanism, and that the Mast proteins (like other AGC kinases) may physically interact (Leroux et al., 2017). Notably, three of the deletions detected in patients were located in the domain of unknown function (DUF1608), which adopts a 4-helix bundle structure in solution (PDB 2M9x). Since the helices pack together to form a hydrophobic core, it is expected that the Glu194del, Lys276del, and Leu278del microdeletions would disrupt this structure by changing the register of the alpha helices. It is therefore conceivable that these MAST1 microdeletions exert a dominant negative effect by titrating out functional endogenous MAST proteins and/or targeting them for degradation. The consequence of the reduction in Mast1/2/3 levels is likely to be complex, influencing the phosphorylation of multiple targets. Little is known of the targets of the MAST family, however, Andrade and colleagues have recently shown that in mature dopaminergic neurons in the striatum, MAST3 phosphorylates ARPP-16 at Ser46, which in turn inhibits the serine-threonine phosphatase PP2A (Andrade et al., 2017). Mast2 has been reported to interact with β2-syntrophin, but its targets are otherwise undefined (Lumeng et al., 1999). Future experiments that define and validate the targets and binding partners of the MAST family in the developing brain will provide valuable insight on this front.
Why do our L278del mice have a thicker corpus callosum? We report that our L278del mice present with an enlarged corpus callosum that is most severe in the midbody and genu, and manifests despite an increase in cortical apoptosis. Electron microscopic analysis has revealed that this phenotype is not attributable to either an increase in the caliber of myelinated neurons, or the thickness of the myelin sheath itself, but is associated with an increase in the number of axons that project to the contralateral hemisphere. Such a phenotype may result from: (1) an increase in the branching of callosal fibers; (2) the formation of multiple axons originating from the soma of projection neurons; or (3) a change in the fate of differentiating neurons during embryogenesis resulting in an increase in the number of callosal projection neurons at the expense of sub-cortical projection neurons (Baranek et al., 2012; Britanova et al., 2008).
Finally, we report the presence of de novo MAST1 substitutions in patients with severe autism and microcephaly. Notably, a de novo P500L mutation in MAST1 has also been reported in a patient presenting with cerebral palsy (McMichael et al., 2015). This suggests that mutations in MAST1 give rise to a spectrum of neurodevelopmental diseases. Unlike, those mutations that we describe in patients with MCC-CH-CM these variants are all substitutions, which might account for the different clinical phenotypes. At this stage we are not able to comment on the functional effect of these substitutions, or on the impact of the ~750 coding variants reported in the ExAC control genome database. In light of our work it is conceivable that mutations in MAST2 and MAST3, both of which are expressed in the developing and adult brain, might also result in neurological disease (Garland et al., 2008). In conclusion, we have defined the genetic lesion that causes MCC-CH-CM, and revealed the importance of the MAST family in global brain development.
STAR Methods
Contact for Reagent and Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Dr. David Keays (david.keays@imp.ac.at).
Experimental Model and Subject Details
Animals
Mice were maintained on a 12:12 light dark cycle and food was available ad libitum. Mice of ages E10.5, E12.5, E14.5, E16.5, P0, P6, 8–10 weeks were used in this study. Animals were not subject to randomization and allocated to experimental groups according to their genotypes as littermates regardless of the sex. For experiments with mice of ages E10.5, E12.5, E14.5, E16.5, P0, P6 the sex was not determined. For generating CRISPR mouse lines, female BL6/CBA F1 mice were used, whereas all backcrosses were performed using C57/BL6 mice to the 7th generation. All procedures were carried out according to legal requirements and covered by an approved license (M58/006093/2011/14).
Human studies
Informed consent was obtained from all patients included in this study. Details of each patient and the condition is provided in Table 1 and Supplementary Table 1. This study was conducted within approved ethical frameworks University of Oxford (08/MRE09/55); Stanford University Institutional Review Board (28362); University of Alabama at Birmingham (X130201001); and at the Seattle Children’s Research Institute (IRB protocol #13291); and by the Commissie Mensgebonden Onderzoek Regio Arnhem-Nijmegen (NL36191.091.11).
Cell Lines, primary cultures, microbe strains
For generating cerebral organoids, feeder free H9 human ES cells were obtained from Wicell (WA09), having a normal karyotype, female sex, and no contaminants. These cells were maintained using the commercially available mTESR media (Stem cell technologies #05850) at 37°C with 5%CO2 levels. These cells were not authenticated. Primary cell cultures were generated from P0 and P7 cortex or cerebellum. The sex of these pups was not determined. The primary neurons were cultured in Neurobasal media with B27, Penstrep and L-glutamine supplements at 37°C with 5%CO2 levels. The Escherichia coli DH5alpha strain was used for cloning purposes.
Method Details
Exome Sequencing
Patients with structural brain phenotypes and their parents were recruited in accordance with existing ethical frameworks and internal review boards. DNA from blood was extracted using standard methods and subject to exome sequencing. We exploited capture arrays (such as the Agilent Human All exon 50Mb array) to capture exonic genomic DNA, which was then subject to next generation paired end sequencing (Illumina). Bioinformatic filtering employed platforms such as the Genome Analysis Toolkit (GATK), and excluded variants with a population frequency greater than 1%. For this purpose, we relied on available genomic databases including dbSNP and the 1000 Genome server. Sanger sequencing was employed to confirm any putative de novo variants. This study was conducted within an approved ethical framework (08/MRE09/55). For mouse exome sequencing genomic DNA was extracted from the tails of BL6J, CBA, and L278del mice and then subjected to Illumina library preparation and captured on the Agilent SureSelect mouse (51Mb) array. The samples were sequenced on an Illumina HiSeq2500 with 100–125 nucleotides paired-end reads followed by data quality analysis using FASTQC. Reads were mapped to the mouse genome (GRCm38, mm10) using BWA (Li and Durbin, 2010). The BAM files were used as an input to UnifiedGenotyper module from the GATK-lite toolkit (v2.3). (McKenna et al., 2010) Variants were called and dbSNP variants were used as known sites, according to GATK Best Practices recommendations (DePristo et al., 2011). Finally, Variants were annotated by SnpEff v4.1 (Cingolani et al., 2012). Sanger sequencing was employed to confirm putative de novo variants on chromosome 8, potentially linked to the L278del mutation (Table S2).
qPCR
The following tissues were dissected from C57/BL6 mice: embryonic brains at stages E10.5, E12.5, E14.5, E16.5, E18.5 (undetermined sex); brains from early postnatal mice at P0, P6 (undetermined sex); the brain regions (cortex, cerebellum, hippocampus, striatum, midbrain, colliculi, hypothalmus, brain stem, spinal cord, olfactory bulbs) and organs (liver, spleen, heart, muscle, testis, kidney, lung) from male adult littermates (n=3). To ascertain Mast1, Mast2 and Mast3, mRNA levels in L278del and KO animals, cortices from littermate homozygote, heterozygote and wild type P0s were obtained (undetermined sex) (n=4). Tissue samples were snap frozen before total RNA extraction and cDNA synthesis (SuperScript III First-Strand Synthesis System, Invitrogen, 18080–051). We used SYBR green on a Bio-Rad Cycler together with intro spanning primers to amplify murine Mast1 (mMAST1_q PCR_F/G CAAG GTGTACAG CAG TATGG, mMAST1_qPCR _R/TGGGTCCCGCTTGCTG). For qPCR on human fetal tissue (hMAST1_qPCR _F/GGTGCATCTGGAGGAACAG, hMAST1_qPCR_R/GATGGTATCGAAGTCATTCTCCC) we performed technical triplicates on two different cDNA libraries (Biochain; C1244051, C1244035) again using intro spanning primers. In addition, we amplified three control genes (Pgk1, Tfrc, and Hprt). For each control gene technical triplicates were performed, and the geometric mean of the Ct values for the three control genes calculated. This geometric mean was then used to obtain the delta Ct value by subtracting it from the average (of triplicate runs) Ct value for Mast1. The relative mRNA levels were then obtained by taking into account the efficiency of the qPCR primers (See (Braun et al., 2010)).
Generation of CRISPR Mice
Female BL6/CBA F1 mice (3–4 weeks old) were superovulated according to standard protocols. Zygotes were isolated from donor females at the day of the coagulation plug (=E0.5). Surrounding cumulus cells were removed by incubation of the cumulus-zygote-complexes in hyaluronidase solution (at ~0.3mg/ml). The injection mix (50ng/μl Cas9 mRNA + 50ng/μl guide RNA + 200ng/μl L278del-ssODN repair template) was injected into the cytoplasm of zygotes. Injected zygotes were incubated for at least 15 minutes in a CO2 incubator at 37°C. Zygotes that survived microinjection were transferred the same day into the oviducts of pseudopregnant recipient females (=0.5 days post coitum). Resultant pups were genotyped by PCR amplification of chromosome 8 followed by sequencing using primers GGAGAGGGTCTTACTTGCTT and TCTTCTGGGTTGAATTCCTA, and backcrossed to C57/BL6.
MRI Analysis
MRI analysis was performed as described previously (Breuss et al., 2016). Briefly, eight-week old littermates were perfused with 0.9% NaCl and 4% PFA supplemented with 10% ProHance Solution (Bracco Imaging Group, 4002750). Images were then acquired with a 15.2 T Biospec horizontal bore scanner (Bruker BioSpin, Ettlingen, Germany). Brain regions of interest were manually segmented, blinded to the genotype, using Amira 5.6 (Visualization Science Group), relying on a mouse brain atlas, and the volumes obtained.
Electron Microscopy Studies
Mice aged 8–9 weeks (n=5) were sacrificed and brain tissue prepared for electron microscopy (Korogod et al., 2015). Briefly, animals were perfused employing a constant flow of 7 mL/min with2.5% glutaraldehyde and 2% paraformaldehyde in phosphate buffer (0.1 M, pH 7.4). Following dissection of the brain, 80-μm thick coronal slices were prepared on a vibratome (Leica VT 1000S). Matching slices were selected from the medial region of each brain (Bregma −0.10), the corpus callosum microdissected, and then post fixed for 40 min in 1% osmium tetroxide in cacodylate buffer (0.1 M, pH 7.4). Samples were then washed twice for 5 min in ddH20 and stained with 1% uranyl acetate for 40 min before being dehydrated in a graded acetone series and embedded in Epon resin. Ultrathin sections were prepared (70nm), mounted on 50 mesh grids and post-stained with uranyl acetate (10mins, 2% in water) and with Reynolds’ lead citrate (5 min). To assess differences in myelin thickness and axon, internal diameter images were captured at 8900× using a FEI Morgagni 268D (FEI Company) 100 kV transmission electron microscope operating at 80 kV. A customized script written in Definiens Architect XD (Version:2.7.0; Build 60765×64) was employed to ascertain axonal surface area and myelin thickness determined using ImageJ. To ascertain the number of axons crossing the midline, images were acquired at 710× along the dorsa-ventral axis. These images (approx 10–12) per animal were then tiled employing iTEM, resulting in a single image. A box measuring 30μm along the x axis was then overlaid onto the structure, and the total number of myelinated axons counted manually. Three stitched images were prepared per animal and all analysis was performed blind to genotype. To determine the G-ratio of axons crossing the midline we measured the total diameter of fibers (>300 per animal) and the axonal diameter (without myelin) on high resolution EM images. The G-ratio was then calculated (axon diameter/fiber diameter) (Guy et al., 1989).
Generation of organoids
Human cerebral organoids were generated from H9 human ES cells (Wicell, WA09) as previously described (Lancaster and Knoblich, 2014)_ENREF_14. 64 days-old organoids were then embedded in Neg-50 Medium (Richard-Allan Scientific) and sliced into 20 μm sections in a cryostat. The slides were dried overnight before storing at −20°C and stained as described below.
Nissl Staining and Immunohistochemistry
Brains were removed at E12.5, E14.5, E16.5, E18.5, P0 and drop-fixed in 4% PFA overnight followed by dehydration in 30% sucrose. Brains were embedded in Neg-50 Medium (Richard-Allan Scientific) and sliced into 12 μm sections in a cryostat. Adults brains were recovered from the animals after perfusion with 0.9% NaCl and 4% PFA. These brains were post-fixed, dehydrated and sliced using a sledge microtome into 40 μm sections. For Nissl staining, sections were washed in PBS, and bathed in Cresyl Violet for 3 min [0.25% Cresyl Violet acetate (Sigma, C5042) dissolved in distilled water with ten drops of glacial acetic acid per 100 ml of solution]. After briefly washing in PBS, slides were dehydrated in an alcohol series (30%, 70%, 96% and 100% ethanol, 2 min each), xylol (twice, 2 min each), mounted with DPX mountant (Fluka, 44581) and left overnight at room temperature. Images were acquired using the transmitted Mirax slide scanner (Zeiss).
Immunofluorescence experiments were performed as previously described (Breuss et al., 2016; Breuss et al., 2015). Where necessary antigen retrieval was performed on sections by slow heating of the slides in antigen retrieval buffer (Vector, H-3301) up to 90°C, followed by gradual cooling at room temperature for 20 minutes. Primary antibody in 0.1%−0.3% Triton-X100/PBS with 4% donkey serum as the blocking agent, was incubated overnight at 4°C. The following concentrations of primary antibodies were used: 1:300 Mast1 (Santa Cruz, sc-55851), 1:100 Mast1 (Santa Cruz, sc-373845) 1:300 cleaved-Caspase3 (Cell Signaling, #9661), 1:300 cleaved-Caspase9 (Cell Signaling #9509), 1:300 Pax6 (Covance, PRB-278P), 1:300 Tbr2 (Abcam, ab23345), 1:1000 Tuj (Covance, MMS-435P), 1:300 NeuN (Millipore, MAB377), 1:100 Cux1 (Santa Cruz, sc-6327), 1:1000 Er81 (Abcam, ab36788), 1:300 FoxP2 (Abcam, ab16046), 1:250 Calbindin (Millipore, AB1778), 1:1000 Olig2 (Millipore, AB9610), GFAP 1:500 (Dako, Z0334), 1:300 Map2 (Abcam, ab24640), 1:300 Tau (Abcam, ab61493), 1:1000 p53 (Leica, P53-CM5P). The next day, sections were washed in PBS and a species-specific secondary antibody (Molecular Probes, A-10037, A-11057, A-10042, A-21206; 1:500) was applied for 1 h in blocking solution at 4°C, followed by Hoechst 33342 staining (1:2000 in PBS) for 5 min. Subsequently the slides were mounted with Fluorescent Mounting Medium (Dako, S302380) and dried overnight at 4°C before imaging. Images were taken using a laser scanning confocal microscope (LSM 780 Zeiss).
Quantification of Histological Data
To determine the thickness of the corpus callosum in Nissl-stained sections the septum in the genu region was measured three times and an average determined (Bregma co-ordinates −0.86mm). To determine the cortical thickness six measurements were obtained for rostral (Bregma co-ordinates 0.86 mm) and caudal regions (Bregma co-ordinates −1.82mm) and averaged for each animal. To assess the thickness of various cortical layers, matched caudal immunostained (Foxp2, Cux1, Er81) sections were measured four times in the region of the somatosensory cortex and averaged for each animal. To assess the levels of apoptosis in the cortex of P0 animals Caspase-3 and Caspase-9 positive puncta were counted in 3 matched sections (spanning anatomical co-ordinates 3.75mm to 4.0mm (Paxinos G, 2007)), and the number averaged per animal. To compare rostral and caudal cortical apoptosis at P0 caspase-3 positive punctae were counted in a box measuring 1000 × 300 μm2, in rostral sections (n=2 sections per animal, co-ordinates 2.3–2.5mm (Paxinos G, 2007)) and caudal sections (n=2 sections per animal, co-ordinates: 3.75–4.0mm (Paxinos G, 2007). To assess levels of apoptosis in the P0 cerebellum, caspase-3 positive positive puncta were counted in 2 matched sections per animal (spanning anatomical co-ordinates 7mm to 7.25mm (Paxinos G, 2007)) and the number averaged per section. To quantify the number and density of oligodendrocyctes and astrocyctes within the corpus callosum coronal sections from 8 week old animals were stained with Olig2 and GFAP. A box measuring 770μm wide was placed over the septum of the corpus callosum extending from the ventral to dorsal edges, and positively stained cells were manually counted. To quantitate the number of Purkinje and granule cells in the cerebellum, free-floating, 40 μm, sagittal midline cerebellar sections were stained with anti-sera for Calbindin and NeuN, followed by counterstaining with DAPI (n=2 sections per animal). Folium III was imaged and all Purkinje cells in this folium were counted allowing determination of the Purkinje cell linear density (cells/mm of Purkinje cell layer). The total number of Purkinje cell cells per section was then estimated by measuring the entire length of the Purkinje cell layer. To estimate the number of granule cells per section 4 images per animal were captured within the granule cell layer of folium III, and the density of granule cells/mm2 of granule layer (GL) was determined. The total number of cells per section was then estimated by measuring the entire surface area of the GL and mutilpying it by the average density for that animal. All quantifications were done using ImageJ software and performed blinded to the genotype.
Immunoblotting
Brains were snap frozen in liquid nitrogen and stored at −80°C. Protein lysates were prepared from frozen tissue samples, homogenized in chilled lysis buffer (20 mM TrisHCl pH 7.5, 100 mM NaCl, 10% glycerol, 1% Triton X-100) supplemented with protease and phosphatase inhibitors (Pierce #88668). Using a Tungsten Carbide Bead the tissue was lysed in a Qiagen Tissue Lyser (2 × 1min, 20Hz), incubated at 4°C for 1h, and then centrifuged twice for 20 min. Protein concentration of the collected supernatant was measured using the Pierce BCA Protein Assay Kit (#23225). Protein lysates were run on NuPAGE protein gels, blotted onto nitrocellulose membranes and blocked in 5% skimmed milk/TBST. Primary antibodies [1:2000 GAPDH (Millipore, MAB374); 1:1000 Mast1 (Proteintech, 13305–1-AP); 1:300 Mast2 (Santa Cruz, sc-377198); 1:500 Mast3 (Novus; NBP1–82993); 1:1000 pan-Akt (Cell Signaling Technology #9272); 1:1000 Phospho-Akt (Ser473) (Cell Signaling Technology #4060); 1:500 S6 Ribosomal Protein (Cell Signaling Technology #5548); 1:1000 Phospho-S6 Ribosomal Protein (Ser240/244) (Cell Signaling Technology #2215)] were incubated overnight in blocking solution at 4°C. The following day, the blot was incubated with the corresponding HRP conjugated secondary antibody (abcam) for 1 hour at room temperature. The signal was detected on Amersham Hyperfilm™ ECL film using the ECL™ Western Blotting Detection Reagents kit (GE healthcare, RPN2106). Western blots were quantified using ImageJ.
Primary neuron culture
Cortical neurons were prepared from brains of C57/BL6 or Mast1 KO P0 mice while P7 mice were used for cerebellar neuronal cultures. In short, the brain region of interest was isolated, and the meninges carefully removed. The tissue was treated with 0.25% trypsin solution in a 37°C water bath for 15 mins, followed by three washes in HBSS (20mM HEPES, 2mM CaCl2, 5.4mM KCl, 1mM MgCl2, 1mM NaPi, 5.6mM glucose, pH7.3). The trypsinised tissue was titurated in plating media (DMEM, 10%Horse serum, 200mM L-glutamin, 100mM sodium pyruvate), first using a P1000 pipette and then a P200 pipette. The cells were passed through a 70μm strainer and the resulting cell suspension counted. Neurons plated at ~1 × 105 cells per well in a 24-well plate containing poly-L-lysine coated coverslips. After 3–4 hours the medium was changed to Neurobasal containing B27, L-glutamine and PenStrep supplements. After 4–5 days in culture the coverslips were fixed in 4% PFA and stained with anti-Map2, anti-Tau and anti-Mast1 antibodies.
In vitro transcription and translation of Mast1 and microtubule-binding assay
These experiments were performed as previously described (Keays et al., 2007; Walden and Cowan, 1993). Briefly, the full-length murine Mast cDNA was cloned into the pcDNA 3.1+ vector (Invitrogen) driven by the T7 promoter. The different patient mutations screened were introduced by site-directed mutagenesis using the QuikChange Lightning Multi Site-Directed Mutagenesis Kit (Agilent #210518) kit. Plasmids (20 ng/μL final concentration) were used to drive expression in rabbit reticulocyte lysate (TnTl T7 Coupled Reticulocyte Lysate System, Promega) containing 35S-methionine (specific activity: 1000 Ci/mMol; Perkin Elmer). Reactions were incubated at 30°C for 90 min. Prior to co-polymerization reactions, the TnT reaction product was cleared by centrifugation at 60,000g for 10 min at 4°C. MAP rich porcine brain tubulin (Cytoskeleton Inc. #ML116) was depolymerized in tubulin buffer (0.1 M PIPES buffer pH 6.9, 1 mM MgCl2 and 0.5 mM EGTA) by incubation on ice. Co-polymerization reactions were set up by adding the 35S-labeled Mast1 to MAP rich tubulin (1mg/ml), in tubulin buffer containing 1 mM GTP, 2 mM AMP-PNP and 20 μM taxol. To investigate Mast1 binding to microtubules in either the presence or absence of MAPs, the labeled Mast1 reaction was distributed equally between MAP rich and 99% pure porcine brain tubulin (Cytoskeleton Inc. #T240). All co-polymerization reactions were incubated at 30 °C for 30 min to promote microtubul e polymerization and binding of associated proteins. Reaction products were then loaded onto cushions (0.20 mL) containing 1 M sucrose in tubulin buffer, 0.5 mM GTP, 1 mM AMP-PNP and 10 μM taxol and centrifuged at 60,000g for 15 min at 30°C. All centrifugations were done in a Beckman TL-100.3 rotor. Proteins in the pellet were solubilized in SDS loading dye and separated in a 7% SDS-PAGE. Radioactivity content of the microtubule pellets obtained was determined following an overnight exposure to a high-sensitive film (KODAK BioMax MR film).
Quantification and Statistical Analyses
All statistical analysis was executed using GraphPad Prism software package (v7.0c). Numbers (n) of animals or replica for experiments are indicated in the text and the statistical test used indicated in individual figure legends. Statistical tests used in this study include the Student’s T-test, one-way ANOVA with Dunnett’s or Tukey’s multiple comparisons test, and two-way repeated measures ANOVA with Bonferroni’s multiple comparisons test. Samples and animals were not subject to randomization, but were assigned to experimental groups based on their genotype. Shapiro-Wilk or Kolmogorov-Smirnov tests were applied to assess whether assumptions of normality were met, and corrected for multiple comparisons were necessary. A summary of the statistical tests applied for each experiment, the n numbers, and the results obtained are shown in Table S4.
Data and Software Availablitity
In accordance with ethical frameworks and our commitment to patient confidentiality we have not deposited the primary exome sequencing data generated in this study in a community repository.
Supplementary Material
Table S2 - Coding variants reported in MAST1 in the ExAC browser, related to Table1. The effect of the variant and allele frequency is shown.
Table S4: Summary of statistical analysis, related to STAR Methods.
Highlights:
Mast1 associates with the microtubule cytoskeleton in a MAP-dependent manner.
Mast1 is expressed in postmitotic neurons but not neuronal progenitors.
Mast1 Leu278del mice have an enlarged corpus callosum and smaller cerebellum.
More axons cross the midline in Mast1 Leu278del mice despite cortical apoptosis.
Acknowledgements
D.A.K. wishes to thank the Austrian Science Foundation (FWF, P21092 and I2681-B27). We wish to acknowledge the support of Boehringer Ingelheim who support basic science at the IMP. We wish to thank the CAUSES study, specifically Shelin Adam, Christele Du Souich, Jane Gillis, Alison Elliott, Anna Lehman, Jill Mwenifumbo, Tanya Nelson, Clara Van Karnebeek, and Jan Friedman. We would also thank Prof. Dr. Bwee Tien Poll and Dr. Saskia Maas, for including the patient P4 in the study and providing clinical information. T.M.P. is supported by the Diana and Steve Marienhoff Fashion Industries Guild Endowed Fellowship in Pediatric Neuromuscular Diseases. G.M. is generously supported by the National Institute of Neurological Disorders and Stroke (NINDS) (K08NS092898) and Jordan’s Guardian Angels. We are indebted to Dr. Vissers for her help with patient P8. J.C.T. and A.P. are funded by Oxford NIHR Biomedical Research Centre. N.J.C acknowledges the support of the N.I.H (R01GM097376).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests
The authors declare no competing interests.
References
- Andrade EC, Musante V, Horiuchi A, Matsuzaki H, Brody AH, Wu T, Greengard P, Taylor JR, and Nairn AC (2017). ARPP-16 Is a Striatal-Enriched Inhibitor of Protein Phosphatase 2A Regulated by Microtubule-Associated Serine/Threonine Kinase 3 (Mast 3 Kinase). The Journal of neuroscience : the official journal of the Society for Neuroscience 37, 2709–2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baranek C, D ittrich M, Parthasarathy S, Bonnon CG, Britanova O, Lanshakov D, Boukhtouche F, Sommer JE, Colmenares C, Tarabykin V, and Atanasoski S (2012). Protooncogene Ski cooperates with the chromatin-remodeling factor Satb2 in specifying callosal neurons. Proceedings of the National Academy of Sciences of the United States of America 109, 3546–3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britanova O, de Juan Romero C, Cheung A, Kwan KY, Schwark M, Gyorgy A, Vogel T, Akopov S, M itkovski M, Agoston D, et al. (2008). Satb2 is a postmitotic determinant for upper-layer neuron specification in the neocortex. Neuron 57, 378–392. [DOI] [PubMed] [Google Scholar]
- Broix L, Jagline H, E LI, Schmucker S, Drouot N, Clayton-Smith J, Pagnamenta AT, Metcalfe KA, Isidor B, Louvier UW, et al. (2016). Mutations in the HECT domain of NEDD4L lead to AKT-mTOR pathway deregulation and cause periventricular nodular heterotopia. Nature genetics 48, 1349–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium GT (2013). The Genotype-Tissue Expression (GTEx) project. Nature genetics 45, 580–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubelos B, Briz CG, Esteban-Ortega GM, and Nieto M (2015). Cux1 and Cux2 selectively target basal and apical dendritic compartments of layer II-III cortical neurons. Developmental neurobiology 75, 163–172. [DOI] [PubMed] [Google Scholar]
- Demyanenko GP, Tsai AY, and Maness PF (1999). Abnormalities in neuronal process extension, hippocampal development, and the ventricular system of L1 knockout mice. The Journal of neuroscience : the official journal of the Society for Neuroscience 19, 4907–4920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiMario FJ Jr., Ramsby GR, and Burleson JA (1999). Brain morphometric analysis in neurofibromatosis 1. Archives of neurology 56, 1343–1346. [DOI] [PubMed] [Google Scholar]
- Edwards TJ, Sherr EH, Barkovich AJ, and Richards LJ (2014). Clinical, genetic and imaging findings identify new causes for corpus callosum development syndromes. Brain : a journal of neurology 137, 1579–1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferland RJ, Cherry TJ, Preware PO, Morrisey EE, and Walsh CA (2003). Characterization of Foxp2 and Foxp1 mRNA and protein in the developing and mature brain. The Journal of comparative neurology 460, 266–279. [DOI] [PubMed] [Google Scholar]
- Garland P, Quraishe S, French P, and O’Connor V (2008). Expression of the MAST family of serine/threonine kinases. Brain research 1195, 12–19. [DOI] [PubMed] [Google Scholar]
- Gilissen C, Hehir-Kwa JY, Thung DT, van de Vorst M, van Bon BW, Willemsen MH, Kwint M, Janssen IM, Hoischen A, Schenck A, et al. (2014). Genome sequencing identifies major causes of severe intellectual disability. Nature 511, 344–347. [DOI] [PubMed] [Google Scholar]
- Gohlich-Ratmann G, Baethmann M, Lorenz P, Gartner J, Goebel HH, Engelbrecht V, Christen HJ, Lenard HG, and Voit T (1998). Megalencephaly, mega corpus callosum, and complete lack of motor development: a previously undescribed syndrome. American journal of medical genetics 79, 161–167. [DOI] [PubMed] [Google Scholar]
- Guy J, Ellis EA, Kelley K, and Hope GM (1989). Spectra of G ratio, myelin sheath thickness, and axon and fiber diameter in the guinea pig optic nerve. The Journal of comparative neurology 287, 446–454. [DOI] [PubMed] [Google Scholar]
- Hengst M, Tucke J, Zerres K, Blaum M, and Hausler M (2010). Megalencephaly, mega corpus callosum, and complete lack of motor development: delineation of a rare syndrome. American journal of medical genetics Part A 152A, 2360–2364. [DOI] [PubMed] [Google Scholar]
- Hyman BT, and Yuan J (2012). Apoptotic and non-apoptotic roles of caspases in neuronal physiology and pathophysiology. Nature reviews Neuroscience 13, 395–406. [DOI] [PubMed] [Google Scholar]
- Iyer V, Shen B, Zhang W, Hodgkins A, Keane T, Huang X, and Skarnes WC (2015). Off-target mutations are rare in Cas9-modified mice. Nature methods 12, 479. [DOI] [PubMed] [Google Scholar]
- Johannessen CM, Reczek EE, James MF, Brems H, Legius E, and Cichowski K (2005). The NF1 tum or suppressor critically regulates TSC2 and mTOR. Proceedings of the National Academy of Sciences of the United States of America 102, 8573–8578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenna KP, van Doormaal PT, Dekker AM, Ticozzi N, Kenna BJ, Diekstra FP, van Rheenen W, van Eijk KR, Jones AR, Keagle P, et al. (2016). NEK1 variants confer susceptibility to amyotrophic lateral sclerosis. Nature genetics 48, 1037–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher M, Witten DM, Jain P, O’Roak BJ, Cooper GM, and Shendure J (2014). A general framework for estimating the relative pathogenicity of human genetic variants. Nature genetics 46, 310–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosik KS, and Finch EA (1987). MAP2 and tau segregate into dendritic and axonal domains after the elaboration of morphologically distinct neurites: an immunocytochemical study of cultured rat cerebrum. The Journal of neuroscience : the official journal of the Society for Neuroscience 7, 3142–3153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster MA, Renner M, M artin CA, Wenzel D, Bicknell LS, Hurles ME, Homfray T, Penninger JM, Jackson AP, and Knoblich JA (2013). Cerebral organoids model human brain development and microcephaly. Nature 501, 373–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lek M, Karczewski KJ, M inikel EV, Samocha KE, Banks E, Fennell T, O’Donnell-Luria AH, Ware JS, Hill AJ, Cummings BB, et al. (2016). Analysis of protein-coding genetic variation in 60,706 humans. Nature 536, 285–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leroux AE, Schulze JO, and Biondi RM (2017). AGC kinases, mechanisms of regulation and innovative drug development. Seminars in cancer biology. [DOI] [PubMed] [Google Scholar]
- Lumeng C, Phelps S, Crawford GE, Walden PD, Barald K, and Chamberlain JS (1999). Interactions between beta 2-syntrophin and a family of microtubule-associated serine/threonine kinases. Nature neuroscience 2, 611–617. [DOI] [PubMed] [Google Scholar]
- Marsh AP, Heron D, Edwards TJ, Quartier A, Galea C, Nava C, Rastetter A, Moutard ML, Anderson V, Bitoun P, et al. (2017). Mutations in DCC cause isolated agenesis of the corpus callosum with incomplete penetrance. Nature genetics 49, 511–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMichael G, Bainbridge MN, Haan E, Corbett M, Gardner A, Thompson S, van Bon BW, van Eyk CL, Broadbent J, Reynolds Cv et al. (2015). Whole-exome sequencing points to considerable genetic heterogeneity of cerebral palsy. Molecular psychiatry 20, 176–182. [DOI] [PubMed] [Google Scholar]
- Meixner A, Haverkamp S, Wassle H, Fuhrer S, Thalhammer J, Kropf N, Bittner RE, Lassmann H, Wiche G, and Propst F (2000). MAP1B is required for axon guidance and Is involved in the development of the central and peripheral nervous system. The Journal of cell biology 151, 1169–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris JA, Royall JJ, Bertagnolli D, Boe AF, Burnell JJ, Byrnes EJ, Copeland C, Desta T,Fischer SR, Goldy J, et al. (2010). Divergent and nonuniform gene expression patterns in mouse brain. Proceedings of the National Academy of Sciences of the United States of America 107, 19049–19054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen LS, Schneider T, Rio M, Moutton S, Siquier-Pernet K, Verny F, Boddaert N, Desguerre I, Munich A, Rosa JL, et al. (2016). A nonsense variant in HERC1 is associated with intellectual disability, megalencephaly, thick corpus callosum and cerebellar atrophy. European journal of human genetics : EJHG 24, 455–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niquille M, Garel S, Mann F, Hornung JP, Otsmane B, Chevalley S, Parras C, Guillemot F, Gaspar P, Yanagawa Y, and Lebrand C (2009). Transient neuronal populations are required to guide callosal axons: a role for semaphorin 3C. PLoS biology 7, e1000230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrini E, Conti V, Dobyns WB, and Guerrini R (2016). Genetic Basis of Brain Malformations. Molecular syndromology 7, 220–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul LK, Brown WS, Adolphs R, Tyszka JM, Richards LJ, Mukherjee P, and Sherr EH (2007). Agenesis of the corpus callosum: genetic, developmental and functional aspects of connectivity. Nature reviews Neuroscience 8, 287–299. [DOI] [PubMed] [Google Scholar]
- Paxinos GHG, Watson C, Koutcherov Y, Wang H (2007). Atlas of the Developing Mouse Brain (London: Academic Press; ). [Google Scholar]
- Pierson TM, Zimmerman RA, Tennekoon GI, and Bonnemann CG (2008). Mega-corpus callosum, polymicrogyria, and psychomotor retardation: confirmation of a syndromic entity. Neuropediatrics 39, 123–127. [DOI] [PubMed] [Google Scholar]
- Poduri A, Evrony GD, Cai X, Elhosary PC, Beroukhim R, Lehtinen MK, Hills LB, Heinzen EL, Hill A, Hill RS, et al. (2012). Somatic activation of AKT3 causes hemispheric developmental brain malformations. Neuron 74, 41–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riviere JB, Mirzaa GM, O’Roak BJ, Beddaoui M, Alcantara D, Conway RL, St-Onge J, Schwartzentruber JA, Gripp KW, Nikkel SM, et al. (2012). De novo germline and postzygotic mutations in AKT3, PIK3R2 and PIK3CA cause a spectrum of related megalencephaly syndromes. Nature genetics 44, 934–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serafini T, Colamarino SA, Leonardo ED, Wang H, Beddington R, Skarnes WC, and Tessier-Lavigne M (1996). Netrin-1 is required for commissural axon guidance in the developing vertebrate nervous system. Cell 87, 1001–1014. [DOI] [PubMed] [Google Scholar]
- Sobreira N, Schiettecatte F, Valle D, and Hamosh A (2015). GeneMatcher: A Matching Tool for Connecting Investigators with an Interest in the Same Gene. Hum Mutat 36, 928–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturrock RR (1980). Myelination of the mouse corpus callosum. Neuropathology and applied neurobiology 6, 415–420. [DOI] [PubMed] [Google Scholar]
- Szybinska A, and Lesniak W (2017). P53 Dysfunction in Neurodegenerative Diseases - The Cause or Effect of Pathological Changes? Aging and disease 8, 506–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terrone G, Voisin N, Abdullah Alfaiz A, Cappuccio G, Vitiello G, Guex N, D’Amico A, James Barkovich A, Brunetti-Pierri N, Del Giudice E, and Reymond A (2016). De novo PIK3R2 variant causes polymicrogyria, corpus callosum hyperplasia and focal cortical dysplasia. European journal of human genetics : EJHG 24, 1359–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel C, Kessler K, Giessl A, Dimmler A, Shalev SA, von der Haar S, Zenker M, Zahnleiter D, Stoss H, Beinder Ev et al. (2011). NEK1 mutations cause short-rib polydactyly syndrome type majewski. American journal of human genetics 88, 106–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tischfield MA, Baris HN, Wu C, Rudolph G, Van Maldergem L, He W, Chan WM, Andrews C, Demer JL, Robertson RL, et al. (2010). Human TUBB3 mutations perturb microtubule dynamics, kinesin interactions, and axon guidance. Cell 140, 74–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walden PD, and Cowan NJ (1993). A novel 205-kilodalton testis-specific serine/threonine protein kinase associated with microtubules of the spermatid manchette. Molecular and cellular biology 13, 7625–7635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, and Jaenisch R (2013). One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 153, 910–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West KL, Kelm ND, Carson RP, and Does MD (2015). Quantitative analysis of mouse corpus callosum from electron microscopy images. Data in brief 5, 124–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S2 - Coding variants reported in MAST1 in the ExAC browser, related to Table1. The effect of the variant and allele frequency is shown.
Table S4: Summary of statistical analysis, related to STAR Methods.