Abstract
Inappropriate activation of sterol regulatory element-binding proteins (SREBPs) can lead to non-alcoholic fatty liver disease (NAFLD). To link chemical exposure to SREBP activity, a previously characterized gene expression biomarker (Rooney et al., 2019) was used to identify microarray comparisons from rat liver that exhibited significant positive or negative correlation to the biomarker. The effects of 620 chemicals on SREBP activity were examined across 9305 chemical-dose-time microarray comparisons. SREBP was found to be frequently modulated by chemical exposure with 54% of the chemicals affecting SREBP activity. Activators included inhibitors of cholesterogenesis that act to inhibit HMG-CoA reductase (statins) or inhibit Cyp51 (conazoles). Most chemical effects were transient, lasting usually no more than 2-4 days. Modulation of SREBP in most cases led to coordinated increases or decreases in lipogenic and cholesterogenic genes. However, 570 chemical exposure conditions were identified in which regulation was uncoupled. Most of these conditions affected cholesterogenic genes in the absence of parallel effects on lipogenic genes. Together, these findings show that SREBP is a frequent target of chemical exposure and expression of lipogenic and cholesterogenic genes can be uncoupled.
Keywords: sterol regulatory element binding protein, transcript profiling, steatosis, cholesterol, fatty acids, triglycerides, statins, TG-GATES, microarray compendium
INTRODUCTION
The synthesis of triglycerides and cholesterol is controlled by master regulators called sterol regulatory element-binding proteins (SREBPs). This family of three transcription factors regulates the expression of genes important for the uptake and synthesis of cholesterol, fatty acids, and phospholipids [1–3]. There are two mammalian SREBP genes, SREBF1 encoding SREBP1a and SREBP1c proteins that mainly regulate the expression of genes involved in lipogenesis, and SREBF2 encoding the SREBP2 protein that regulates the genes responsible for cholesterol synthesis [1, 4, 5]. The SREBP proteins are synthesized as inactive precursors bound to the membrane of the endoplasmic reticulum (ER) through two transmembrane domains in their C-termini [1]. Activation of SREBP requires proteolytic release of an N-terminal fragment encoding the basic helix-loop-helix leucine zipper transcription factor [1, 6]. In the presence of high cholesterol levels, proteolytic cleavage of SREBPs is blocked through a mechanism involving SREBP-cleavage activating protein (SCAP) and insulin-induced gene protein (INSIG). Cholesterol binds to SCAP, inducing a conformational change that promotes binding to ER-anchored INSIG, preventing Golgi transport and activation of SREBPs [7–10]. When the cholesterol concentration decreases, the SCAP/SREBP complex dissociates from INSIG, allowing vesicular transport to the Golgi where two proteases release the transcriptionally active N-terminal fragment of the SREBPs [1, 3, 9]. Once released, the mature SREBP enters the nucleus and binds as a dimer to SREBP sites (sterol regulatory elements; SREs) within promoters of target genes.
Activity of SREBPs and expression of their target genes is mechanistically linked to non-alcoholic fatty liver disease (NAFLD), the most common form of chronic liver disease. NAFLD encompasses a wide spectrum of phenotypes from simple triglyceride (TG) accumulation in hepatocytes (hepatic steatosis) to steatohepatitis (hepatic steatosis associated with inflammation) which can progress to fibrosis, cirrhosis, and in severe cases, hepatocarcinoma [11]. Steatosis occurs when there is an imbalance between lipid availability and lipid disposal. There are a number of potential sources of fatty acids that contribute to fatty liver development and can include de novo lipogenesis which in cases of alcohol consumption and infections can account for up to 25–30% of the liver TG [12, 13]. Although NAFLD has classically been associated with increased levels of hepatic triglycerides [14], the severity of NAFLD was associated with an accumulation of free cholesterol (FC) without a similar increment in cholesterol esters (CE) in NAFLD [15, 16]. In human livers, increased severity of NAFLD was associated with increased SREBP2 protein maturation and expression of SREBP2 target genes [15]. A role for the SREBP family members in NAFLD has been demonstrated in three mouse models which express individual constitutively-active SREBP family members. These models exhibit increased expression of lipogenic and cholesterogenic genes and increases in liver triglycerides and cholesterol [17–19]. Abolishing or suppressing the expression of SCAP led to decreases in steatosis due to high fat or high fructose diets as well in a genetic model of obesity (ob/ob mice) [20].
A growing number of chemicals cause steatosis in sub-acute and chronic studies that are carried out as part of safety evaluations of potential industrial chemicals and pharmaceutical drugs [21]. The importance of steatosis as an endpoint of regulatory significance is highlighted by the fact that the Environmental Protection Agency (EPA) Integrated Risk Information System (IRIS) has often used steatosis as the critical effect to determine chemical exposure limits in risk assessments [22]. There is little information that links chemical exposure, SREBP modulation, and steatosis. A small number of chemicals activate liver X receptor (LXR) subtypes and are thought to increase steatosis through increased expression of SREBP1c and regulated genes [23]. There are chemicals that activate SREBP indirectly by reducing cholesterol levels, and these include statin drugs that inhibit HMG-CoA reductase [24] and inhibitors of Cyp51 [25]. The statin drugs at least are generally not known to cause steatosis. In a screen of 210 chemicals in the mouse liver, a relatively high percentage (87 chemicals; 41%) were found to modulate SREBP [26]. A comprehensive assessment of the effects of a large number of chemicals on SREBP in the rat liver has not been carried out but would be useful to help link SREBP modulation to steatosis.
In our previous study, computational methods were developed to evaluate chemical effects on SREBP in the livers of mice, as assessed using microarray-derived gene lists [26]. The method hinged on a gene expression biomarker that was constructed using a genetical and genomic approach and required biomarker genes to be dependent on SREBP family members and SCAP for expression. Using a fold-change rank-based statistical test, the SREBP gene biomarker was found to be highly accurate in predicting activation or suppression of SREBP function in mouse tissues. In the present study, this computational approach was used to predict chemical modulation of SREBP in rat liver. The analysis provides novel insights into the interactions of drugs and environmentally-relevant chemicals with SREBP.
Methods
Strategy for identification of chemicals that alter SREBP function in rat tissues.
The methods used in this study have been thoroughly described in our companion paper [26]. In this study, a gene expression biomarker was built using gene expression profiles from the tissues of mice expressing transgenic constitutively-active forms of individual SREBP family members or from SCAP-null mice. Genes in the biomarker were consistently expressed in two or more of the transgenic SREBP (tSREBP) comparisons and required SCAP modulation for expression. The SREBP gene biomarker consisted of 99 genes and associated fold-change values, which corresponded to average differences across the tSREBP studies.
Attempts were originally made to create biomarkers specific for each SREBP subtype or isoform [26]. Biomarkers consisting of 47, 79 and 31 genes were built for SREBP1a, SREBP1c, and SREBP2, respectively using the procedures described in [26]. Very few genes related to lipid synthesis were included in the biomarkers. Out of the ~2600 biosets examined for SREBP activity in the mouse liver, the biomarkers for SREBP1a, SREBP1c, and SREBP2 only identified 27, 30, or 23 biosets, respectively, far less than the number identified using the SREBP biomarker (592 biosets). Thus, we concluded that there was little value in using these biomarkers for screening the compendium. These attempts highlight the fact that the SREBP family members regulate an overlapping set of genes.
A commercially available gene expression database was utilized in the studies. As of July 23, 2017, Illumina’s BaseSpace Correlation Engine (BSCE), previously known as NextBio, contained 21,190 studies and 132,200 biosets across 14 molecular data types carried out in 16 species. In the database each list (called a bioset) has been compared to all other biosets in the database using a fold-change rank-based statistical algorithm called the Running Fisher test, which allows an assessment of the overlap in regulated genes and whether those overlapping genes are correlated in a positive or negative manner [27]. In the present study, biosets from the livers and primary hepatocytes derived from rats were evaluated. Available information about each bioset was extracted from the BSCE database and used to populate a compendium of information about each of the experiments. Biosets were further annotated using information derived from the original Gene Expression Omnibus (GEO) submission and/or the original publication. To assess activation or suppression of SREBP function, the SREBP gene biomarker was uploaded to BSCE and compared to all biosets in the database using the Running Fisher algorithm. Results of the comparisons were exported and used to populate the annotated compendium with p-values of each comparison and direction of the correlation. We have previously used this analysis strategy to accurately identify factors that activate or suppress transcription factors in the mouse liver including AhR, CAR, Nrf2, PPARα and STAT5b [28–34], androgen receptor in prostate cancer cells [35], and ERα in MCF-7 cells [36].
Identification of differentially expressed genes in BSCE microarray datasets.
The methods used to identify differentially expressed genes have been summarized in our previous publication [29] and in detail in Kuperschmidt et al. [27].
Annotation of a rat liver gene expression compendium.
All of the biosets examining gene expression in rat liver, rat primary hepatocytes, or rat hepatocyte-derived cell lines were annotated for study characteristics allowing a systematic assessment of the effect of different factors on function. The list of descriptors provided for each of the biosets included study ID (i.e., source), name, classification of factor (e.g., chemical, diet, etc.), name of chemical or treatment, CAS# (where appropriate), sex, source of material (e.g., liver or hepatocyte), and microarray platform. Further details of each experiment not in our annotated compendium are available at GEO (http://www.ncbi.nlm.nih.gov/geo/) or ArrayExpress (http://www.ebi.ac.uk/arrayexpress/).
Identification of chemicals that induce fatty liver.
Pathological findings were taken from the Open TG-GATES [37] website of Pathological Items (http://dbarchive.biosciencedbc.jp/en/open-tggates/data-12.html). A number of histopathological findings were used that are linked to steatosis [38]. These included “Accumulation..foam.cell”, “Degeneration..fatty”, “Degeneration..vacuolar”, “Deposit..lipid”, and “Vacuolization..cytoplasmic”. Findings with minimal, slight, moderate, and severe grades in the liver were included in the analysis. Any findings that were flagged as occurring spontaneously were removed from the final analysis. Chemicals that altered one or more of the findings at one or more of the time-dose pairs were flagged as potentially causing steatosis.
Comparison of the SREBP biomarker to a microarray database.
Methods for comparisons between the biomarker and biosets in the compendium have been described in our previous paper [26]. Out of the 99 genes in the mouse biomarker, there were 7 that were not found in the rat genome (H2-D1, H2-K1, H2-K2, H2-Q10, H2-Q2, H2-Q4, H2-Q8) and thus were not used in the analysis.
Classification prediction of SREBP function in rat tissues.
Biosets from microarray experiments in which the SREBP activation state was known were manually curated from a total of 97 studies which were examined as they represented a mixture of potential actives and inactives for SREBP modulation. Studies were first examined in Gene Expression Omnibus (GEO) or ArrayExpress for linkage to the primary paper describing the study. A total of 24 submissions examined were not linked to a manuscript and no further attempts were made to identify the manuscript linked to the submission. A total of 57 submissions representing 847 biosets were linked to a publication but these studies did not have any linkage to SREBP based on searches within the manuscript for “SREBP”, “SREBF”, “fatty acid”, or “sterol”. Within these, a total of 189 biosets exhibited predicted biomarker activity and thus represented novel associations between the chemicals being measured and SREBP. The final 46 biosets from 16 studies could be linked directly or indirectly to SREBP activity. Only a small subset of these studies measured SREBP maturation by western blot. Most studies measured the expression of multiple lipogenic and/or cholesterogenic genes and most of these mentioned SREBP family members as transcription factors which control the expression of these genes. The tissues examined included liver, white fat, brown fat, adrenal gland, prostate, and oocytes. The number of biosets used to test for an increase in SREBP activity (i.e., SREBP activation) was 15 positives and 31 negatives. The number of biosets used to test for a decrease in SREBP activity (i.e., SREBP suppression) was 23 positives and 23 negatives. Unlike more traditional machine learning classification methods, optimal conditions for classification were not derived from gene behavior as the biomarker genes were fixed. In this and in our previous studies [28–32], the biomarkers were compared to known positives and negatives using the Running Fisher algorithm. Studies with the biomarkers for AhR, CAR, PPARα and STAT5b showed that a cutoff of Running Fishers algorithm p-value ≤ 10−4 resulted in a balanced accuracy for activation of 95%, 97%, 98%, and 99% for AhR, CAR, PPARα, and STAT5b, respectively [28–32]. This same cutoff resulted in a balanced accuracy of SREBP activation or SREBP suppression of 94% or 93%, respectively for mice [26], and a balanced accuracy of SREBP activation or SREBP suppression of 93% and 98%, respectively for rats (see Results).
Lipogenic and cholesterogenic gene expression.
The expression of lipogenic and cholesterogenic genes were compared to the SREBP biomarker predictions as described before [26]. Briefly, all genes used in the analysis were identified and confirmed for function based on pathway annotations in mouse genome informatics (http://www.informatics.jax.org/genes.shtml), PubGene (http://www.pubgene.org/), and KEGG (http://www.genome.jp/kegg/) as well as those characterized in Horton et al. [5]. The final list of 24 genes involved in lipogenesis included Acaa1a, Acaa1b, Acaca, Acacb, Acsf2, Acsf3, Acsl1, Acsl3, Acsl4, Acsl5, Acss2, Cyb5b, Elovl1, Elovl2, Elovl3, Elovl5, Elovl6, Elovl7, Fads1, Fads2, Fasn, Me1, Scd1, and Scd2. The final list of 21 genes involved in cholesterogenesis included Aacs, Acly, Cyp51, Dhcr24, Dhcr7, Fdft1, Fdps, Hmgcr, Hmgcs1, Hsd17b7, Idi1, Lss, Msmo1, Mvd, Mvk, Nsdhl, Pmvk, Sc4mol, Sc5d, Sqle, and Tm7sf2. Expression data for specific rat genes was based on the processed microarray experiments from BSCE. All expression values (fold-changes) for each gene were used to populate the compendium, allowing a direct comparison between changes in specific genes and SREBP predictions.
Biosets in which uncoupling was hypothesized to occur were identified by examination of the discordance in the regulation of the lipogenic and cholesterogenic genes which was defined in the following manner for four classes of conditions:
Class I: ≥ 40% of the cholesterogenic genes were up-regulated, ≤ 20% of the cholesterogenic genes were down-regulated, and ≤ 20% of the lipogenic genes were up-regulated;
Class II: ≥ 40% of the cholesterogenic genes were down-regulated, ≤ 20% of the cholesterogenic genes were up-regulated, and ≤ 20% of the lipogenic genes were down-regulated;
Class III: ≥ 40% of the lipogenic genes were up-regulated, ≤ 20% of the lipogenic genes were down-regulated, and ≤ 20% of the cholesterogenic genes were up-regulated;
Class IV: ≥ 40% lipogenic genes were down-regulated, ≤ 20% of the lipogenic genes were up-regulated, and ≤ 20% of the cholesterogenic genes were down-regulated.
These conditions provided reasonable thresholds for identification of conditions in which the expression of the genes were coupled or uncoupled. Although the thresholds used to find the biosets with uncoupling were minimally stringent (40% and 20% minimum), most of the biosets exhibited striking differences in the level of activation or suppression of the lipogenic and cholesterogenic genes. The average difference in % of up-regulated cholesterogenic genes minus % up-regulated lipogenic genes in Class I was 44%. The average difference in % of down-regulated cholesterogenic genes minus % down-regulated lipogenic genes in Class II was 43%. The average difference in % of up-regulated lipogenic genes minus % up-regulated cholesterogenic genes in Class III was 40%. The average difference in % down-regulated lipogenic genes minus % down-regulated cholesterogenic genes in Class IV was 32%.
Additional computational analyses.
Heat maps were generated using Eisen Lab Cluster and Treeview software (http://rana.lbl.gov/EisenSoftware.htm).
Results
In our previous study, we built and characterized a biomarker for detecting SREBP-dependent modulation in a mouse microarray compendium [26]. Microarray comparisons (called biosets) in which SREBP was modulated were identified using the fold-change rank-based Running Fisher algorithm which identifies biosets with significant positive or negative correlation to the biomarker. These methods reliably predicted activation or suppression of SREBP function in mouse tissues, resulting in balanced accuracies of 94% and 93%, respectively. In the present study, this screening strategy was applied to identify and characterize chemicals that alter SREBP activity in rat liver.
Given the conserved nature of SREBP functions and gene targets across species [39], the accuracy of the mouse biomarker to predict SREBP activity in rat tissues was determined. Classification was performed on a set of 46 biosets with known effects on SREBP derived from 6 rat tissues (liver, white fat, brown fat, adrenal gland, prostate, oocytes). For prediction of activation, the SREBP biomarker had an 87% sensitivity and a 100% specificity, with a balanced accuracy of 93% (Figure 1). For prediction of suppression, the SREBP biomarker had a 96% sensitivity and a 100% specificity, with a balanced accuracy of 98%. Overall, the procedures using the biomarker resulted in excellent balanced accuracies to detect SREBP activity in rat tissues.
Figure 1. The biomarker accurately predicts SREBP modulation in rat tissues.
The biomarker was compared to 46 biosets derived from 5 tissues with known SREBP behavior or behavior of SREBP-regulated genes. Red, true positives for activation; green, true positives for suppression; blue, false negatives for activation; yellow, false negative for suppression.
Chemical modulation of SREBP in the livers of rats
The effects of chemical exposure on SREBP activity was evaluated in a rat liver database consisting of 9305 microarray comparisons encompassing 617 chemicals. Most of the microarray comparisons came from the TG-GATES and Drug Matrix studies (GSE8858, GSE8251). To visualize the relationships between the expression of genes in the biomarker as a function of the Running Fisher algorithm p-value, each bioset comprised of statistically-filtered genes, was compared to the SREBP biomarker and sorted by −Log(p-value). Figure 2A (left side) shows that for biosets with a positive correlation to the biomarker, the smaller the p-value (larger the −Log(p-value)), the greater the visual similarity to the biomarker in both the direction and relative magnitude of the gene changes. There were 213 chemicals in 589 biosets that caused SREBP activation in rat liver or rat primary hepatocytes, and they included the HMG-CoA reductase inhibitors (statins) discussed below. In these biosets, there was consistent activation of the lipogenic and cholesterogenic genes in the biomarker. Figure 2A (right side) includes biosets that exhibited negative correlation to the biomarker, with the biosets on the farthest right exhibiting the smallest p-values for negative correlation. In general, these biosets exhibited a pattern of gene expression that was opposite to that of the SREBP biomarker. There were 200 chemicals in 595 biosets that caused SREBP suppression, and these biosets exhibited consistent decreases in the expression of lipogenic and cholesterogenic genes in the biomarker. The total number of chemicals that altered SREBP was 333 (54% of total), less than the total of those that activated or suppressed, because some chemicals both activated and suppressed SREBP under different exposure conditions.
Figure 2. SREBP modulation in the rat liver gene expression compendium.
A. (Top) Biosets derived from microarray comparisons of rat liver or rat hepatocyte comparisons were ordered based on their correlation to the SREBP biomarker using the −Log(p-value) of the Running Fisher test. Biosets with positive correlation to the biomarker are on the left and biosets with negative correlation to the biomarker are on the right. The dashed lines denote the cutoff p-value = 10-4. (Bottom) The heat map shows the expression of genes in the SREBP biomarker across the biosets. B, biomarker genes.
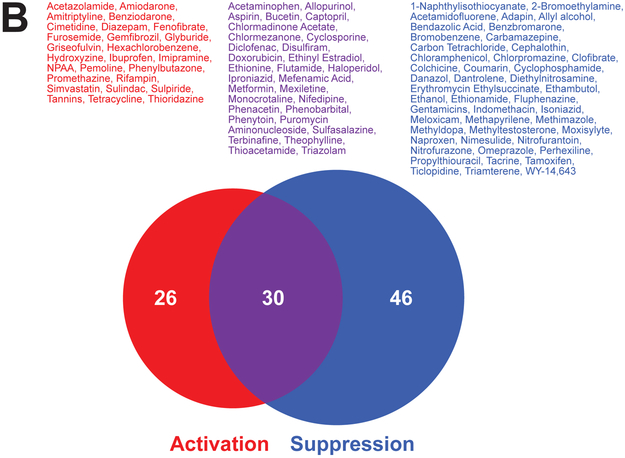
B. Distribution of chemicals that activated, suppressed, or both activated and suppressed SREBP in the TG-GATES study. Chemicals that fell into each of the groups are listed.
The changes in SREBP activity were examined in the TG-GATES study in which 134 chemicals were evaluated in the livers of rats at three doses and 8 time points [37]. The chemicals were divided into those that activate only, suppress only, and both activate and suppress at least at one of the 8 time points (3hr to 29d). There were more chemicals that suppressed SREBP than activated (Figure 2B). There were 30 chemicals that exhibited both activation and suppression at least at one dose level and time point with a total of 102 chemicals which modulated SREBP in any direction. The lists of chemicals and the exposure conditions which led to SREBP modulation are found in Supplemental File 1. It can be concluded that SREBP modulation by chemicals occurs often in the rat liver.
Temporal changes in SREBP activity in the TG-GATES study
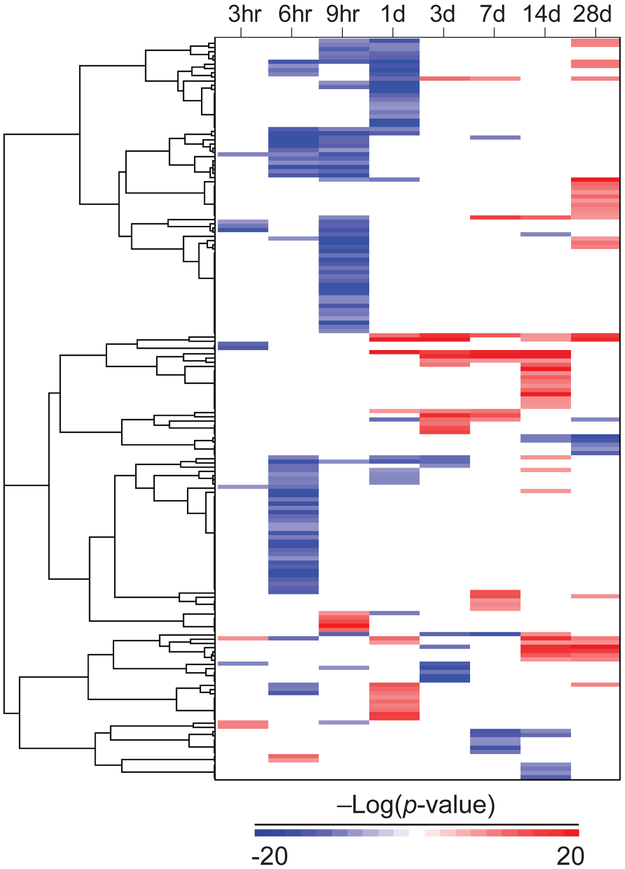
The temporal changes in SREBP activity were examined in the TG-GATES study [37]. Exposure to 99 chemicals in a total of 176 chemical-dose pairs was examined by one-dimensional clustering (Figure 3). The most striking feature was the transient nature of the changes in SREBP. There were only 26 chemical-dose pairs representing 23 chemicals that exhibited changes in more than two out of the 8 time points. The majority of exposures (149 out of the 176 chemical-dose pairs (85%)) led to SREBP modulation at only one or two of the time points. The largest cluster in the dendrogram were those in which there was suppression of SREBP between 6 and 24 hrs with no consistent changes at later time points. Another group of chemicals led to activation of SREBP at at least one time point between 1 and 28d. A smaller subset of chemicals caused suppression of SREBP between 8 and 28d.
Figure 3. Chemical modulation of SREBP in the livers of rats.
Modulation of SREBP across time in the TG-GATES study. Chemical-doses in which SREBP was modulated at at least one time point were clustered by one-dimensional hierarchical clustering (complete linkage, centered correlation). Only −Log(p-value)s that were significant are shown. Red, positive correlation. Blue, negative correlation.
The modulation of SREBP was compared to histopathology data from the same study. There were 13 chemical-dose pairs from 10 chemicals which led to increases in markers of steatosis at one or more time points. Out of these there were 5 chemical-dose pairs for 4 chemicals (amiodarone, hexachlorobenzene, hydroxyzine, tetracycline) which exhibited SREBP activation at 14 and 28d. The other chemical-dose pairs exhibited no obvious consistent pattern of SREBP modulation. Thus, based on this data, there did not appear to be a clear link between SREBP activation and steatosis.
SREBP modulation by inhibitors of HMG-CoA reductase
Statin drugs that inhibit the enzyme HMG-CoA reductase (encoded by Hmgcr, one of the biomarker genes) are well known to decrease cholesterol levels leading to SREBP2 activation [24]. The effects of seven statin drugs in 160 comparisons were examined to determine their effects on SREBP. Close to half of the exposures (82 biosets) led to significant positive correlations (Figure 4A, top). Surprisingly, there were 8 exposures (7 to atorvastatin and one to simvastatin) which led to significant SREBP suppression. Statin exposures in the rat liver led to consistent activation or suppression of cholesterogenic genes that paralleled the activation status of SREBP (Figure 4A, bottom). There were also parallel effects on lipogenic genes.
Figure 4. Activation of SREBP by chemical inhibition of cholesterol biosynthesis.
A. Activation of SREBP by statins. Seven statin compounds were examined in the livers of rats in 161 comparisons. (Top) The −log(p-value)s of the Running Fisher test were rank ordered. (Bottom) Heat map of the altered expression of genes in the SREBP biomarker, lipogenic genes and cholesterogenic genes.
B. Effect of atorvastatin on SREBP. Rats were treated with atorvastatin at 2.5 or 300 mg/kg each day and sacrificed at 6h, 1d, 3d, or 5d (derived from GSE8858).
C. Effect of simvastatin on SREBP. Rats were treated with 40, 120 or 400 mg/kg/day and sacrificed at the indicated times (derived from the TG-GATES study). Each chemical-time-dose was evaluated for effects on 24 lipogenic genes and 21 cholesterogenic genes which were expressed as percent changes of the total number of genes in each class. The asterisks show the conditions in which lipogenic and cholesterogenic gene expression was uncoupled.
SREBP modulation by individual statins were examined. Atorvastatin at the highest dose examined (300 mg/kg) caused significant activation of SREBP starting at day 1 (Figure 4B). At the lower dose tested (2.5 mg/kg), SREBP was significantly suppressed at 1 and 3d. The basis for the suppression is not known.
The effects of simvastatin were examined over 8 time points and three doses in the TG-GATES study (Figure 4C). The expression of 24 lipogenic genes and 21 cholesterogenic genes was examined. To simplify the analysis and make the results comparable between the two sets of genes, the results were presented as four values for each chemical-dose-time: 1) percent of lipogenic genes increased, 2) percent of lipogenic genes decreased, 3) percent of cholesterogenic genes increased, and 4) percent of cholesterogenic genes decreased. There was an initial activation at the lowest dose level and shortest time point; however, consistent activation did not occur at the highest dose until day 1. By 15 and 29 days, activation was no longer significant at any dose. Simvastatin exposure led to transient increases in cholesterogenic genes across all doses. At the highest dose there were also increases in lipogenic genes. There were few if any genes that decreased in expression throughout the treatment conditions. SREBP biomarker −Log(p-value)s increased in parallel with the increases in the cholesterogenic and lipogenic genes. Additionally, there were conditions in which cholesterogenic genes were increased in the absence of notable increases in lipogenic genes (asterisks). This uncoupling of the expression of these two classes of genes is described in detail below. The ability of HMG-CoA reductase and CYP51 inhibitors to activate SREBP in rat primary hepatocytes is discussed in Supplemental File 2. These studies highlight the previously unappreciated dose- and time-dependent effects of statins on SREBP.
Chemical modulation of cholesterogenic and lipogenic genes across time
The expression of SREBP target genes involved in lipogenesis and cholesterogenesis were examined across time and dose to determine relationships with the SREBP biomarker predictions. Chemicals that caused early suppression of SREBP (2-bromoethylamine, acetaminophen, 2-acetamidofluorene, bendazolic acid) were examined (Figure 5A). In each case, there were early and transient decreases in the expression of the cholesterogenic genes which exhibited a reproducible pattern across the three doses. In the case of 2-bromoethylamine, the decreases became more severe with increasing dose. The biomarker −Log(p-value)s decreased in parallel with the decreases in the genes. Thus, the transient suppression was observed across doses for most of these chemicals.
Figure 5. Chemical modulation of cholesterogenic and lipogenic genes across time.
The effects of chemical exposure on cholesterogenic and lipogenic genes were examined in the TG-GATES study. (Top panels) The −Log(p-value)s for the correlations between the SREBP biomarker and the indicated treatment. (Bottom panels) Effects of treatment on lipogenic and cholesterogenic genes is shown (see legend to Figure 4).
A. Chemicals that exhibited transient and early suppression of SREBP.
B. Activators of PPARα. The activation of PPARα is also shown as assessed using a gene expression biomarker that was characterized earlier (Rooney et al., 2018b).
C. Chemicals that activated SREBP and caused steatosis.
Four activators of the nuclear receptor peroxisome proliferator-activated receptor α (PPARα) were examined. Activation of PPARα was assessed using a rat PPARα gene expression biomarker characterized in an earlier study [33]. PPARα activation by clofibrate, fenofibrate, and WY occurred by 6 hrs irrespective of dose while the activation of PPARα by gemfibrozil did not occur until 9 hrs (Figure 5B). The activation remained sustained throughout the time of exposure. There were sustained increases in lipogenic genes that paralleled the activation of PPARα for all of the chemicals. All of the chemicals except gemfibrozil also exhibited decreases in cholesterogenic genes with subtle differences in the timing of the decreases. Gemfibrozil was the only PPARα agonist which caused increases in cholesterogenic genes which generally paralleled the increases in the lipogenic genes.
Lastly, four chemicals were examined which caused increases in markers of steatosis and increased SREBP activity (Figure 5C). Hexachlorobenzene, hydroxyzine, and tetracycline exposure led to increases in cholesterogenic genes to greater extents than lipogenic genes. Amiodarone caused about equal percentages of increases in lipogenic and cholesterogenic genes at the highest dose. However, given that SREBP was activated at only two time points, further work is needed to link SREBP activation with steatosis for these chemicals. In summary, these studies allow an assessment of the underlying molecular basis of SREBP biomarker predictions and highlight dose- and time-dependent changes in lipogenic and cholesterogenic genes, many of which are transient in nature.
Primary hepatocytes are a poor model for identification of chemical-induced modulation of SREBP.
High throughput screening often utilizes in vitro cell models to identify molecular targets of chemicals. Rat primary hepatocytes were evaluated to determine if they could be used as a surrogate to identify chemicals that cause modulation of SREBP in vivo. The analysis discussed in detail in Supplemental File 2, led to the conclusion that rat primary hepatocytes cannot accurately identify chemicals that modulate SREBP in vivo and are especially deficient at identifying chemicals that suppress SREBP.
Coordinated regulation of lipogenic and cholesterogenic genes after chemical exposure
In our previous study [26], we showed that the majority of treatments in the mouse liver resulted in coordinated expression of the lipogenic and cholesterogenic genes. We hypothesized that there would also be coordinated expression of these genes in rat liver. Each of the biosets was evaluated for percentage of genes that exhibited increased or decreased expression of the two classes of genes and then plotted relative to the rank-ordered –log(p-value) of the SREBP biomarker (Figure 6). Similar to the mouse liver, there was a striking concordance between increased expression of both lipogenic and cholesterogenic genes and activation of SREBP. Likewise, there was a striking concordance between decreased expression of both sets of genes and suppression of SREBP. Most of those biosets with no significant SREBP modulation assessed using the biomarker exhibited ≤ 20% of the genes altered in either direction. However, there were 261 biosets (~3% of 8121 biosets) which lacked significant SREBP modulation but exhibited modulation of either lipogenic and/or cholesterogenic genes ≥ 50%. Overall, these results indicate that there were only a relatively small number of biosets in which there were coordinated increases or decreases in these genes that were not predicted by the biomarker (discussed in greater detail below).
Figure 6. Concordant regulation of lipogenic and cholesterogenic genes in the liver.
A total of 9305 biosets derived from liver or hepatocyte comparisons were rank-ordered based on the –Log(p-value) of the correlation to the SREBP biomarker. The percentage of either lipogenic or cholesterogenic genes up-regulated (blue) or down-regulated (orange) are shown.
To further examine the relationships between the expression of the lipogenic and cholesterogenic genes, the expression of each group of genes was summarized as % genes with increased expression minus % genes with decreased expression and then plotted as lipogenic genes vs. cholesterogenic genes (Supplemental Figure 3). The plot shows that the vast majority of biosets exhibited coordinated increases or decreases in the expression of the two classes of genes. The correlation coefficient was 0.53. Overall, these results indicate that the two classes of genes are coordinately regulated under diverse chemical exposure conditions in the rat liver.
The number of biosets that exhibited coordinated changes in the lipogenic or cholesterogenic genes in the absence of significant changes in the biomarker were examined further. There were 8121 biosets that did not have any activity based on the biomarker (−log(p-value)s between 4 and −4), and of these, 6.8%, 3.2%, 1.3%, 0.46% or 0.07% exhibited 40%, 50%, 60%, 70%, or 80% of lipogenic and/or cholesterogenic genes increased or decreased. As there is no information about the biological impact of a partial response of these gene classes, it is difficult to determine what cutoff should be used. Regardless, the biomarker missed few biosets which exhibited coordinated changes in these genes.
The question was asked whether there were any biosets which exhibited significant modulation of SREBP but did not also have coordinated up or down expression of either the lipogenic or cholesterogenic genes. Only 29 out of the 1191 biosets (2.4%) which passed the threshold were identified in which there was SREBP biomarker activity but little or no coordinated expression of the lipogenic and cholesterogenic genes in the same direction (≤ 20% of the total number of lipogenic or cholesterogenic genes). There were no biosets in which there was SREBP biomarker activity and ≤ 10% of the total of either lipogenic genes or cholesterogenic genes altered in expression. Thus, the biosets in which SREBP is regulated are intimately linked to the coordinated expression of lipogenic and/or cholesterogenic genes.
Identification of chemicals that uncouple lipogenic and cholesterogenic gene regulation.
Similar to our previous study in the mouse liver, biosets were identified in which regulation of the two classes of genes were uncoupled. Uncoupling of gene expression (discussed in detail in the Methods) was defined as coordinated increases or decreases in one of the classes (≥ 40% of the genes) but, not the other (≤ 20% changes in the same direction in the second class of genes). Using these criteria, 570 biosets were identified that exhibited uncoupled regulation of the genes and included 277 and 293 biosets either predicted or not by the biomarker, respectively (Figure 7A). Most of the biosets which exhibited uncoupling (477; 83%) were those in which cholesterogenic genes were up- (Class I) or down-regulated (Class II) in the absence of parallel effects on lipogenic genes. In a minor number of these incidences, there was also opposite regulation of the lipogenic genes, especially for Class I biosets. Out of the biosets which exhibited changes in the lipogenic genes in the absence of parallel changes in the cholesterogenic genes (Class III and IV), most exhibited up-regulation of the lipogenic genes. A number of these Class III biosets also exhibited decreases in cholesterogenic genes. Given that predictions are dependent on the genes in the biomarker exhibiting coordinated parallel changes, it was not surprising that many of the biosets which exhibited opposite regulation of lipogenic and cholesterogenic genes did not result in significant biomarker activity (Figure 7A, bottom panel).
Figure 7. Identification of conditions in which lipogenic and cholesterogenic gene expression is uncoupled.
A. Biosets with uncoupled regulation of lipogenic and cholesterogenic genes were identified as described in the Methods. The biosets were grouped based on their type of regulation (Classes I-IV). Each group was first rank ordered based on the % of genes that define that class (I: increase in cholesterogenic genes; II: decrease in cholesterogenic genes; III: increase in lipogenic genes; IV: decrease in lipogenic genes). Each group was then rank ordered based on opposite expression of the other group of genes (e.g., Class I: decrease in lipogenic genes). (Top panel) Percent changes in the expression of the cholesterogenic and lipogenic genes. (Middle panel) Heat map showing the expression of the genes. (Bottom panel) Predictions of SREBP modulation based on the expression of the genes in the biomarker for each bioset.
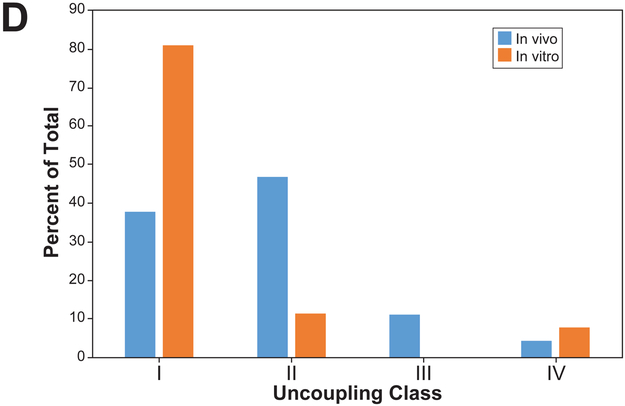
B. The distribution of the number of chemicals and number of biosets that fell into the four classes.
C. Number of biosets in each class across time of exposure.
D. Percentage of each of the classes derived from chemical treatment of rats or rat primary hepatocytes from the TG-GATES study.
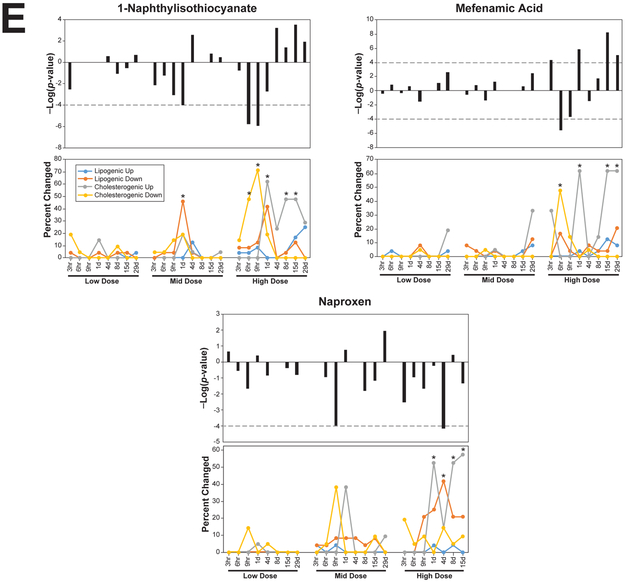
E. Examples of chemicals that exhibit uncoupling at four or five time points.
The distribution of the biosets and chemicals across the different classes is shown in Figure 7B. There were 302 biosets representing exposures to 152 chemicals in Class I biosets, 99 chemicals in 175 biosets in Class II, 13 chemicals in 55 biosets in Class III, and 31 chemicals in 38 biosets in Class IV. When the distribution of the uncoupled biosets was examined across time in the TG-GATES study, only Class II biosets were found at 3h and 6h (Figure 7C). Class II biosets were the most frequent class at 12h. At one day or later, the Class I biosets were found most often. These results indicate that for a number of chemicals, cholesterogenic genes and presumably SREBP2 were suppressed shortly after exposure in the absence of parallel effects on lipogenic genes.
The percentage of each of the classes found in vivo or in vitro were also compared using the biosets from the TG-GATES study. There was a greater percentage of uncoupled biosets in Class I in vitro compared to in vivo, while in vivo there were higher percentages of Classes II and III compared to in vitro (Figure 7D). The in vivo vs. in vitro distributions derived from DrugMatrix comparisons were similar to those examining other in vivo vs. in vitro studies in the compendium (data not shown).
Like modulation of SREBP, uncoupling appeared to be for the most part transient. There were 13 chemicals that uncoupled expression at two time points (benzbromarone, bucetin, clofibrate, danazol, dantrolene, ethionamide, indomethacin, nifedipine, phenacetin, rifampin, simvastatin, sulindac, theophylline), six chemicals that uncoupled at three time points (ethionine, hydroxyzine, phenobarbital, tetracycline, thioacetamide, ticlopidine), four chemicals that uncoupled at four time points (fenofibrate, mefenamic acid, naproxen, WY-14,643), and three chemicals that uncoupled at five time points (1-naphthylisothiocyanate, fenofibrate, WY-14,643). Examples of chemicals with extended uncoupling at four or five time points are shown in Figure 7E. Other examples are shown in Figure 4C, Figure 5. The extended uncoupling occurred most often with chemicals administered at the two highest doses (26 out of 28 chemical-dose pairs).
The biosets within each of the Classes were examined to determine if there were any similarities in the molecular targets of the chemicals that could be linked to regulation of the genes. The biosets in Class I included those from treatments by statins and CYP51 inhibitors. There were 16 biosets from treatments with statins (out of the 18 total that were uncoupled in any group). Most of these were exposures of 1d or earlier. Examples of the uncoupling are shown in which rats were treated with simvastatin for 4d (Figure 4C, asterisks). Examples of Class II uncoupling are shown in Figure 5A (asterisks) in which the expression of lipogenic genes was sometimes delayed by ~3–15 hrs after exposure to acetamidofluorene and bendazolic acid, indicating that effects on SREBP2 preceded those on SREBP1c. Ten out of 12 biosets representing exposures to CYP51 inhibitors were also in Class I. There were a number of chemicals known to induce steatosis in Class I and Class II (amitriptyline, carbon tetrachloride, coumarin, cyclosporine, diltiazem, disulfiram, ethanol, ethinyl estradiol, ethionamide, hydroxyzine, imipramine, lomustine, puromycin aminonucleoside, tetracycline) (chemicals from [41]). Almost all of the Class III biosets exhibited activation of PPARα (bezafibrate, clofibrate, diethylhexyl phthalate, fenofibrate, gemfibrozil, methyl salicylate, nafenopin, WY-14,643). Very few biosets from the other classes exhibited activation of PPARα. However, there were a number of PPARα activators in Class II which exhibited suppression of cholesterogenic and activation of lipogenic genes (bezafibrate, clofibrate, fenofibrate, nafenopin, WY-14,643). Identification of dose-times in which uncoupling occurred after exposure to the PPARα activators is shown in Figure 5B. The complete list of biosets which exhibited uncoupling is found in Supplemental File 1.
Discussion
Constitutive activation of SREBP family members is associated with fatty liver disease [4]. In the present study, a screen for chemicals that regulate SREBP in the rat liver was carried out to link chemical exposure, SREBP activation, and steatosis. The screen of ~9300 microarray comparisons of 620 chemicals identified 333 chemicals that modulated SREBP. This relatively high percentage of chemicals (54%) included more that suppressed than activated. Chemicals that activated included statin drugs and conazole fungicides that inhibit different steps in cholesterogenesis. The analysis revealed features of SREBP regulation not previously observed, at least not on this scale. 1) Most of the chemicals that affect SREBP do so in relatively short time windows of less than one day, especially those chemicals that suppress SREBP. 2) Sustained activation of SREBP more than 14 days was very rare. In fact only 2 chemicals caused activation for longer than 14 days. And 3) a chemical can both activate and suppress SREBP depending on the conditions of exposure. Most of the chemicals that alter the activity of SREBP also exhibited coordinated expression of lipogenic and cholesterogenic genes known to be principally regulated by SREBP1c or SREBP2, respectively. However, the analysis of the genes led to the identification of conditions that activate or suppress one set of genes in the absence of parallel effects on the other, implying that regulation of the SREBP isoforms/subtypes was uncoupled. Chemicals with different molecular targets fell into different classes of uncoupling; statins often regulated cholesterol but not lipogenic genes, while PPARα activators often regulated lipogenic but not cholesterogenic genes. Finally, a comparison of SREBP activity in vivo and in vitro found that rat primary hepatocytes are poor in vitro models to predict in vivo responses. In summary, the analysis not only identified a large number of chemicals that modulate SREBP in unique temporal patterns but highlighted a spectrum of coordinated or uncoupled expression responses in the lipogenic and cholesterogenic genes.
Chemicals that alter the activity of SREBP in the rat liver were identified using computational procedures described in our previous study [26]. The methods relied on a gene expression biomarker of SREBP- and SCAP-dependent genes and the Running Fisher test [27] to determine the significance in the expression in any overlapping genes between the biomarker and the microarray comparison being evaluated. Similar to the accuracy of the biomarker to detect modulation of mouse SREBP (94% or 93% for activation or suppression, respectively; [26]), the methods reliably predicted SREBP activation or suppression in rat tissues, with accuracies of 93% and 98%, respectively (Figure 1). Given the species conservation of the SREBP and regulated genes [39] including the 31 genes in the biomarker involved in lipogenesis and cholesterogenesis, it was not surprising that the mouse biomarker could be used to accurately predict modulation in rats. The high degree of accuracy is consistent with other studies from our lab supporting the contention that the Running Fisher test coupled with a set of genes identified through genomic and genetical approaches allows accurate identification of transcription factor modulation [28–32, 36].
The screen revealed that SREBP modulation by chemicals is a relatively common occurrence in rat liver (Figure 2). Approximately 13% of all biosets in the compendium exhibited SREBP modulation, with a greater number exhibiting SREBP suppression compared to activation and included a total of 331 chemicals (53% of all chemicals). This frequency of modulation was similar to that found in the mouse liver (41%; [26]) and included pharmaceutical and environmentally-relevant chemicals that induce therapeutic and toxic effects through a diverse set of molecular targets. When looking at the TG-GATES study of 134 chemicals evaluated at three doses and 8 time points, the percentage was even higher (76% of all chemicals). A novel finding was that many chemicals (30 chemicals) in the TG-GATES study both activated or suppressed SREBP at different times and doses of exposure. The frequent modulation of SREBP may reflect the ability of chemicals to alter the levels of regulatory molecules (e.g., fatty acids, triglycerides, glucose, cholesterol, insulin) [42] as well as the importance of SREBP and target genes in maintaining cellular and tissue homeostasis in the face of chemical stress.
Unique temporal patterns of SREBP modulation were observed in the TG-GATES study (Figure 3). Most chemicals that modulated SREBP did so within short windows of time despite daily exposure. The transient regulation of SREBP contrasts with the activation of PPARα (Figure 5B) as well as the aryl hydrocarbon receptor and the constitutive active receptor [43] which is sustained from ~6 hrs through the 29d time point. One of the more prominent patterns observed was the transient suppression of SREBP within the first 24 hrs of exposure. This pattern included a number of chemicals that are known to be cytotoxic including acetaminophen and thioacetamide (Figure 5A). Supporting the suppression of SREBP as assessed using the biomarker, there was consistent suppression of cholesterogenic and sometimes lipogenic genes across the three doses for each chemical. It is likely that many of these exposure conditions cause systemic toxicity due to their relatively high dose levels (hundreds of mg/kg per dose). The toxic response would be predicted to decrease appetite and stimulate a fasting response which includes mobilization of triglycerides in peripheral fat and their transport to the liver. Fasting is known to be a potent suppressor of SREBP. The timing of the suppression of cholesterol and lipogenic genes in the TG-GATES study overlaps with the disappearance of SREBP1 in nuclear extracts that occurs between 1 and 12 hrs after the start of fasting [44].
A smaller group of chemicals caused transient activation of SREBP. The activation usually lasted no more than 1–2 weeks. In contrast to the chemicals which caused suppression, most of the activation occurred after 24 hrs. Surprisingly, there were only 8 chemicals that stimulated activation of SREBP for more than two time points. They included ethionine, gemfibrozil, hydroxyzine, mefenamic acid, promethazine, simvastatin, sulfasalazine, and tetracycline. A number of chemicals were identified that increased histopathological indicators of steatosis (amiodarone, hexachlorobenzene, hydroxyzine, tetracycline) but their dependence on SREBP activation is tenuous as SREBP was only activated at 15d and 29d. While amiodorone and tetracycline have been shown to cause steatosis in other studies (e.g., [45]), further work is required to link steatosis with SREBP activation including studies that have exposure times longer than 29d.
The effects of different statins on SREBP modulation and target gene expression in vivo and in vitro were examined. Statin drugs inhibit HMG-CoA reductase, an early step in cholesterogenesis resulting in decreases in cellular cholesterol and subsequent activation of SREBP2. A comprehensive examination of multiple statins on activation of SREBPs and target genes has not been carried out. Most of the statin exposures in vivo led to activation of SREBP with somewhat less than half being statistically significant (Figure 4A). Surprisingly, like other chemicals in the TG-GATES study, atorvastatin caused both activation and suppression of SREBP depending on the dose. Treatments in which there was suppression in vivo came from rats treated with the two lower doses (2.5 and 5 mg/kg/day) while almost all of the highest dose exposures (300 mg/kg/day) led to activation (Figure 4B). The reasons for this divergence in regulation requires further study; it may be informative to determine relationships to changes in serum and liver lipid levels. Despite daily treatments of simvastatin, significant activation of SREBP and parallel increases in cholesterogenic genes were no longer observed 15d or later (Figure 4C). These results indicate that negative feedback mechanisms are in place to dampen the initial changes in activity of the SREBPs.
In our study of the mouse liver microarray compendium, we found that cholesterogenic and lipogenic gene expression is often coupled [26] despite the fact that individual SREBP family members are regulated by different nutritional cues [2, 42]. The extent of the coupling was also examined in the rat liver. In the majority of chemical exposure conditions, lipogenic and cholesterogenic genes were regulated in parallel. Out of the 1184 biosets which exhibited alteration in SREBP, most (907; 77%) exhibited coordinated increases or decreases in the expression of these genes (Figure 6). The extent of the coupling is identical to that found in the mouse liver (77% of the biosets exhibited coupling). These results imply that coordinated activation or suppression of SREBP family members is a common feature of regulation while conditions which lead to uncoupling are uncommon.
Clues to the molecular basis of this coupling come from a number of studies showing linkages between SREBP2 activation, generation of an oxysterol ligand of LXR, and LXR-mediated activation of the Srebf1 gene. Wong et al. [46] found that SREBP2 positively regulates LXR by generating oxysterol ligands presumably through increased cholesterogenesis. Expression of Srebf1c mRNA in rat hepatoma cells was found to require endogenous LXR ligands [47]. Rong et al. [48] found that SREBP2-dependent activation of cholesterogenic genes leads to increased levels of a proposed cholesterol intermediate or biosynthesis product that serves as a ligand for LXR, required for increased activation of the Srebf1 gene, SREBP1c expression, and activation of lipogenic genes. Indications that SREBP family members are regulated in parallel came from earlier studies in which overexpression of a constitutively active form of human SREBP2 in mice led to increases in expression of endogenous SREBP1c and regulated genes [17], while overexpression of constitutively active form of SREBP1a led to increased expression of endogenous SREBP2 and cholesterogenic genes [18]. Coupling cholesterol and lipogenesis is likely the cellular default important for efficient esterification of cholesterol, carried out by the enzyme ACAT which prefers oleic acid as a substrate [49]. Additionally, because cholesterol is a component of the lipid core of the VLDL particle, linking cholesterol and lipogenesis may be necessary for efficient VLDL production by the liver.
Chemicals were identified that uncoupled lipogenic and cholesterogenic gene expression (Figure 7 and 8), presumably through differences in activation or suppression of the SREBP subtypes/isoforms. Like in the mouse liver [26], most of the uncoupling led to effects on cholesterogenic genes in the absence of parallel effects on lipogenic genes. We had previously shown that many of these biosets were from microarray studies that examined factors known to modulate cholesterol levels through cholesterol availability, synthesis, or transport. For example, high cholesterol diets led to suppression of cholesterogenic genes in the absence of suppression of lipogenic genes while overexpression of human CYP7A1 which catalyzes the conversion of cholesterol to bile acids led to activation of cholesterogenic genes but not lipogenic genes [26]. While the underlying basis for most of uncoupling remains to be determined, some chemicals with well-defined molecular targets tended to fall into one of the four classes of uncouplers. The biosets in Class I included many from the livers of rats treated with statins and CYP51 inhibitors usually after short-term exposures of 1d or earlier. There were 16 biosets from treatments with statins (out of 18 total) in Class I. The example of simvastatin shows that the uncoupling is transient and depending on the dose, found at only one or two time points (Figure 4C). The chemicals which activated PPARα exhibited a unique pattern of early suppression of cholesterogenic genes followed by later activation of lipogenic genes (Class II or Class III uncouplers) (Figure 5B). The increases in the lipogenic genes may be due to the prior depletion of fatty acids through increases in β- and ω-fatty acid oxidation that occurs upon activation of PPARα [50]. The PPARα agonist fenofibrate regulates an overlapping set of genes with SREBP, dependent on SREBP1c [51]. SREBPs are suppressed indirectly by unsaturated fatty acids through suppression of LXR-mediated increases in SREBP1c expression or directly through interaction with INSIG protein [42]. The experimental conditions that led to uncoupling are a starting point for further experiments to determine their underlying molecular basis of regulation including effects on post-translational processing of the SREBP family members and nuclear localization.
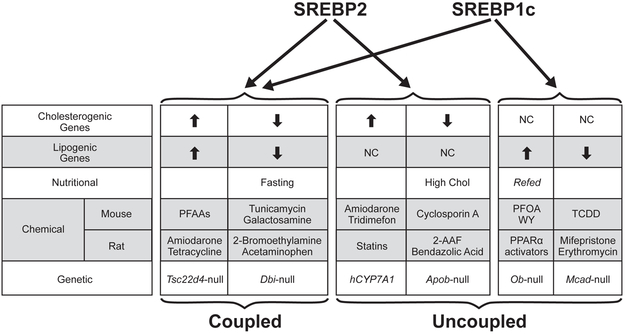
Figure 8. Summary of factors that modulate SREBP highlighting differences in targeted regulation of cholesterogenic and lipogenic genes.
Table provides examples of nutritional, chemical or genetic factors that impact the coupled (left) or uncoupled (right) expression of the cholesterogenic and lipogenic genes through modulation of SREBP family members. Chemicals and genes that modulate SREBP in mouse liver are from (26). The example of “Refed” is described in (40). Abbreviations: PFAAs, perfluoroalkyl acids; PFOA, perfluorooctanoic acid; WY, WY-14,643; TCDD, 2,3,7,8-tetrachlorodibenzodioxin; 2-AAF, 2-acetylaminofluorene.
In summary, a gene expression biomarker of SREBP- and SCAP-dependent genes was used to predict modulation of SREBP family members in the rat liver. In the screen of 9305 microarray comparisons, SREBP was often modulated by chemical exposure. In the majority of comparisons regardless of the chemical exposure conditions, lipogenic and cholesterogenic genes were coordinately up- or down-regulated. However, chemical exposure conditions were identified that led to uncoupling of the expression of these genes and thus presumably the regulation of the SREBP family members that control them.
Supplementary Material
Supplemental File 1. Excel document. Contains chemical exposure conditions in the livers of rats and rat hepatocytes 1) that modulate SREBP and 2) in which lipogenic and cholesterogenic gene expression is uncoupled.
Supplemental File 2. Word document. Contains the sections 1) “Activation of SREBP by cholesterogenesis inhibitors in rat primary hepatocytes” including Supplemental Figures 1A-D, 2) “Primary hepatocytes are a poor model for identification of chemical-induced modulation of SREBP” including Supplemental Figure 2A,B, and 3) Supplemental Figure 3 “The majority of biosets exhibit concordant regulation of lipogenic and cholesterogenic genes.”
Research Highlights.
Overactivity of sterol regulatory element-binding proteins (SREBPs) causes increases in lipid accumulation in the liver.
To identify chemicals that might cause steatosis in rats, a previously characterized SREBP biomarker was used.
The biomarker was very accurate at identifying conditions in rats that led to SREBP activation or suppression.
SREBP was found to be frequently modulated by diets and chemicals including statins in a microarray compendium.
Chemical exposure conditions were identified that led to uncoupling of lipogenic and cholesterogenic gene expression.
Acknowledgements
This study was carried out as part of the EPA Chemicals for Safety and Sustainability (CSS) steatosis AOP project. The information in this document has been funded by the U.S. Environmental Protection Agency. This study has been subjected to review by the National Health and Environmental Effects Research Laboratory and approved for publication. Approval does not signify that the contents reflect the views of the Agency, nor does mention of trade names or commercial products constitute endorsement or recommendation for use. I thank Drs. Guosheng Liang, Wolfgang Patsch and Selma Soyal for review of the manuscript, and Mr. Chuck Gaul for assistance in creating the figures.
References
- 1.Goldstein JL, DeBose-Boyd RA, and Brown MS, Protein sensors for membrane sterols. Cell, 2006. 124(1): p. 35–46. [DOI] [PubMed] [Google Scholar]
- 2.Jeon TI and Osborne TF, SREBPs: metabolic integrators in physiology and metabolism. Trends Endocrinol Metab, 2012. 23(2): p. 65–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nohturfft A and Zhang SC, Coordination of lipid metabolism in membrane biogenesis. Annu Rev Cell Dev Biol, 2009. 25: p. 539–66. [DOI] [PubMed] [Google Scholar]
- 4.Horton JD, Goldstein JL, and Brown MS, SREBPs: transcriptional mediators of lipid homeostasis. Cold Spring Harb Symp Quant Biol, 2002. 67: p. 491–8. [DOI] [PubMed] [Google Scholar]
- 5.Horton JD, et al. , Combined analysis of oligonucleotide microarray data from transgenic and knockout mice identifies direct SREBP target genes. Proc Natl Acad Sci U S A, 2003. 100(21): p. 12027–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang X, et al. , SREBP-1, a membrane-bound transcription factor released by sterol-regulated proteolysis. Cell, 1994. 77(1): p. 53–62. [DOI] [PubMed] [Google Scholar]
- 7.Adams CM, et al. , Cholesterol and 25-hydroxycholesterol inhibit activation of SREBPs by different mechanisms, both involving SCAP and Insigs. J Biol Chem, 2004. 279(50): p. 52772–80. [DOI] [PubMed] [Google Scholar]
- 8.Radhakrishnan A, et al. , Switch-like control of SREBP-2 transport triggered by small changes in ER cholesterol: a delicate balance. Cell Metab, 2008. 8(6): p. 512–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sun LP, et al. , Sterol-regulated transport of SREBPs from endoplasmic reticulum to Golgi: Insig renders sorting signal in Scap inaccessible to COPII proteins. Proc Natl Acad Sci U S A, 2007. 104(16): p. 6519–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang T, et al. , Crucial step in cholesterol homeostasis: sterols promote binding of SCAP to INSIG-1, a membrane protein that facilitates retention of SREBPs in ER. Cell, 2002. 110(4): p. 489–500. [DOI] [PubMed] [Google Scholar]
- 11.Neuschwander-Tetri BA and Caldwell SH, Nonalcoholic steatohepatitis: summary of an AASLD Single Topic Conference. Hepatology, 2003. 37(5): p. 1202–19. [DOI] [PubMed] [Google Scholar]
- 12.Diraison F, Moulin P, and Beylot M, Contribution of hepatic de novo lipogenesis and reesterification of plasma non esterified fatty acids to plasma triglyceride synthesis during non-alcoholic fatty liver disease. Diabetes Metab, 2003. 29(5): p. 478–85. [DOI] [PubMed] [Google Scholar]
- 13.Siler SQ, Neese RA, and Hellerstein MK, De novo lipogenesis, lipid kinetics, and whole-body lipid balances in humans after acute alcohol consumption. Am J Clin Nutr, 1999. 70(5): p. 928–36. [DOI] [PubMed] [Google Scholar]
- 14.Browning JD and Horton JD, Molecular mediators of hepatic steatosis and liver injury. J Clin Invest, 2004. 114(2): p. 147–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Min HK, et al. , Increased hepatic synthesis and dysregulation of cholesterol metabolism is associated with the severity of nonalcoholic fatty liver disease. Cell Metab, 2012. 15(5): p. 665–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Puri P, et al. , A lipidomic analysis of nonalcoholic fatty liver disease. Hepatology, 2007. 46(4): p. 1081–90. [DOI] [PubMed] [Google Scholar]
- 17.Horton JD, et al. , Activation of cholesterol synthesis in preference to fatty acid synthesis in liver and adipose tissue of transgenic mice overproducing sterol regulatory element-binding protein-2. J Clin Invest, 1998. 101(11): p. 2331–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shimano H, et al. , Overproduction of cholesterol and fatty acids causes massive liver enlargement in transgenic mice expressing truncated SREBP-1a. J Clin Invest, 1996. 98(7): p. 1575–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shimomura I, et al. , Insulin resistance and diabetes mellitus in transgenic mice expressing nuclear SREBP-1c in adipose tissue: model for congenital generalized lipodystrophy. Genes Dev, 1998. 12(20): p. 3182–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moon YA, et al. , The Scap/SREBP pathway is essential for developing diabetic fatty liver and carbohydrate-induced hypertriglyceridemia in animals. Cell Metab, 2012. 15(2): p. 240–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wahlang B, et al. , Toxicant-associated steatohepatitis. Toxicol Pathol, 2013. 41(2): p. 343–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaiser JP, Lipscomb JC, and Wesselkamper SC, Putative mechanisms of environmental chemical-induced steatosis. Int J Toxicol, 2012. 31(6): p. 551–63. [DOI] [PubMed] [Google Scholar]
- 23.Vinken M, The adverse outcome pathway concept: a pragmatic tool in toxicology. Toxicology, 2013. 312: p. 158–65. [DOI] [PubMed] [Google Scholar]
- 24.Konrad RJ, Troutt JS, and Cao G, Effects of currently prescribed LDL-C-lowering drugs on PCSK9 and implications for the next generation of LDL-C-lowering agents. Lipids Health Dis, 2011. 10: p. 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harding HP, et al. , Bioactive small molecules reveal antagonism between the integrated stress response and sterol-regulated gene expression. Cell Metab, 2005. 2(6): p. 361–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rooney J, Chorley B, Corton JC, A Gene Expression Biomarker Identifies Chemicals and Other Factors in the Mouse Liver That Modulate Sterol Regulatory Element Binding Protein (SREBP) Highlighting Differences in Targeted Regulation of Cholesterogenic and Lipogenic Genes. Submitted, Submitted. [Google Scholar]
- 27.Kupershmidt I, et al. , Ontology-based meta-analysis of global collections of high-throughput public data. PLoS One, 2010. 5(9). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oshida K, et al. , Identification of chemical modulators of the constitutive activated receptor (CAR) in a gene expression compendium. Nucl Recept Signal, 2015. 13: p. e002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oshida K, et al. , Screening a mouse liver gene expression compendium identifies modulators of the aryl hydrocarbon receptor (AhR). Toxicology, 2015. 336: p. 99–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oshida K, et al. , Identification of modulators of the nuclear receptor peroxisome proliferator-activated receptor alpha (PPARalpha) in a mouse liver gene expression compendium. PLoS One, 2015. 10(2): p. e0112655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oshida K, et al. , Disruption of STAT5b-Regulated Sexual Dimorphism of the Liver Transcriptome by Diverse Factors Is a Common Event. PLoS One, 2016. 11(3): p. e0148308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oshida K, Waxman DJ, and Corton JC, Chemical and Hormonal Effects on STAT5b-Dependent Sexual Dimorphism of the Liver Transcriptome. PLoS One, 2016. 11(3): p. e0150284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rooney J, et al. , Adverse outcome pathway-driven identification of rat liver tumorigens in short-term assays. Toxicol Appl Pharmacol, 2018. 356: p. 99–113. [DOI] [PubMed] [Google Scholar]
- 34.Rooney J, et al. , Activation of Nrf2 in the liver is associated with stress resistance mediated by suppression of the growth hormone-regulated STAT5b transcription factor. PLoS One, 2018. 13(8): p. e0200004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rooney JP, et al. , Identification of Androgen Receptor Modulators in a Prostate Cancer Cell Line Microarray Compendium. Toxicol Sci, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ryan N, et al. , Moving Toward Integrating Gene Expression Profiling Into High-Throughput Testing: A Gene Expression Biomarker Accurately Predicts Estrogen Receptor alpha Modulation in a Microarray Compendium. Toxicol Sci, 2016. 151(1): p. 88–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Igarashi Y, et al. , Open TG-GATEs: a large-scale toxicogenomics database. Nucleic Acids Res, 2015. 43(Database issue): p. D921–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thoolen B, et al. , Proliferative and nonproliferative lesions of the rat and mouse hepatobiliary system. Toxicol Pathol, 2010. 38(7 Suppl): p. 5s–81s. [DOI] [PubMed] [Google Scholar]
- 39.Osborne TF and Espenshade PJ, Evolutionary conservation and adaptation in the mechanism that regulates SREBP action: what a long, strange tRIP it’s been. Genes Dev, 2009. 23(22): p. 2578–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rooney J, Hill T, Qin C, Sistare FD, Corton JC, Adverse Outcome Pathway-Driven Identification of Rat Hepatocarcinogens in Short-Term Assays. Submitted. [DOI] [PubMed] [Google Scholar]
- 41.Sahini N, Selvaraj S, and Borlak J, Whole genome transcript profiling of drug induced steatosis in rats reveals a gene signature predictive of outcome. PLoS One, 2014. 9(12): p. e114085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shao W and Espenshade PJ, Expanding roles for SREBP in metabolism. Cell Metab, 2012. 16(4): p. 414–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rooney J, et al. , Adverse outcome pathway-driven identification of rat liver tumorigens in short-term assays. Toxicol Appl Pharmacol, 2018. 356: p. 99–113. [DOI] [PubMed] [Google Scholar]
- 44.Horton JD, et al. , Regulation of sterol regulatory element binding proteins in livers of fasted and refed mice. Proc Natl Acad Sci U S A, 1998. 95(11): p. 5987–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Antherieu S, et al. , Induction of vesicular steatosis by amiodarone and tetracycline is associated with up-regulation of lipogenic genes in HepaRG cells. Hepatology, 2011. 53(6): p. 1895–905. [DOI] [PubMed] [Google Scholar]
- 46.Wong J, Quinn CM, and Brown AJ, SREBP-2 positively regulates transcription of the cholesterol efflux gene, ABCA1, by generating oxysterol ligands for LXR. Biochem J, 2006. 400(3): p. 485–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.DeBose-Boyd RA, et al. , Expression of sterol regulatory element-binding protein 1c (SREBP-1c) mRNA in rat hepatoma cells requires endogenous LXR ligands. Proc Natl Acad Sci U S A, 2001. 98(4): p. 1477–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rong S, et al. , Expression of SREBP-1c Requires SREBP-2-mediated Generation of a Sterol Ligand for LXR in Livers of Mice. Elife, 2017. 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang H, et al. , Functional expression of a cDNA to human acyl-coenzyme A:cholesterol acyltransferase in yeast. Species-dependent substrate specificity and inhibitor sensitivity. J Biol Chem, 1997. 272(7): p. 3980–5. [DOI] [PubMed] [Google Scholar]
- 50.Kersten S, Integrated physiology and systems biology of PPARalpha. Mol Metab, 2014. 3(4): p. 354–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Oosterveer MH, et al. , Fenofibrate simultaneously induces hepatic fatty acid oxidation, synthesis, and elongation in mice. J Biol Chem, 2009. 284(49): p. 34036–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental File 1. Excel document. Contains chemical exposure conditions in the livers of rats and rat hepatocytes 1) that modulate SREBP and 2) in which lipogenic and cholesterogenic gene expression is uncoupled.
Supplemental File 2. Word document. Contains the sections 1) “Activation of SREBP by cholesterogenesis inhibitors in rat primary hepatocytes” including Supplemental Figures 1A-D, 2) “Primary hepatocytes are a poor model for identification of chemical-induced modulation of SREBP” including Supplemental Figure 2A,B, and 3) Supplemental Figure 3 “The majority of biosets exhibit concordant regulation of lipogenic and cholesterogenic genes.”