Abstract
Despite recent advances in structural definition of GPCR–G protein complexes, the basis of receptor selectivity between G proteins remains unclear. The Gα12 and Gα13 subtypes together form the least studied group of heterotrimeric G proteins. G protein–coupled receptor 35 (GPR35) has been suggested to couple efficiently to Gα13 but weakly to Gα12. Using combinations of cells genome-edited to not express G proteins and bioluminescence resonance energy transfer–based sensors, we confirmed marked selectivity of GPR35 for Gα13. Incorporating Gα12/Gα13 chimeras and individual residue swap mutations into these sensors defined that selectivity between Gα13 and Gα12 was imbued largely by a single leucine-to-isoleucine variation at position G.H5.23. Indeed, leucine could not be substituted by other amino acids in Gα13 without almost complete loss of GPR35 coupling. The critical importance of leucine at G.H5.23 for GPR35–G protein interaction was further demonstrated by introduction of this leucine into Gαq, resulting in the gain of coupling to GPR35. These studies demonstrate that Gα13 is markedly the most effective G protein for interaction with GPR35 and that selection between Gα13 and Gα12 is dictated largely by a single conservative amino acid variation.—Mackenzie, A. E., Quon, T., Lin, L.-C., Hauser, A. S., Jenkins, L., Inoue, A., Tobin, A. B., Gloriam, D. E., Hudson, B. D., Milligan, G. Receptor selectivity between the G proteins Gα12 and Gα13 is defined by a single leucine-to-isoleucine variation.
Keywords: GPCR, GPR35, G protein barcode, genome editing
Recent years have seen enormous advances in knowledge of the structural organization of members of the GPCR superfamily (1–3), with many examples now available of inactive state structures of rhodopsin-like, class A GPCRs. Moreover, although less abundant, a significant number of active state or active state–like structures have also been described (4). A common distinction between inactive and active states is the reorientation of the intracellular facing elements of the transmembrane helices, particularly of transmembrane domain VI (5). It has long been appreciated that the C-terminal α5 helix of G protein α subunits engages with the active state receptor and that this is a crucial step toward triggering guanine nucleotide exchange on the G protein α subunit to initiate steps that result in regulation of signal transduction cascades (6, 7). Although still limited in number (8–11), structural complexes of an active state GPCR and a G protein have provided validation of such models that were developed over many years. Such a role of the extreme C-terminal region of the G protein α subunit in engaging the agonist-occupied receptor had been predicted. For example, the site of pertussis toxin–catalyzed ADP-ribosylation, which prevents effective interactions between GPCRs and Gαi-family G proteins, is the Cys residue located 4 aa from the C-terminal tail in each of these Gα subunits (12), whereas the molecular basis of the uncoupled mutation of Gαs that also prevents interaction with appropriate GPCRs is a single amino acid alteration 6 residues from the C-terminal tail (12). Moreover, the capacity to produce chimeric G proteins that alter GPCR interaction selectivity by providing as few as the C-terminal 5–10 aa of a G protein α subunit have been integral to such understanding (12).
Partially for convenience of classification, GPCRs are often designated as being coupled primarily to members of one of the 4 families, Gαs, Gαi, Gαq/Gα11, and Gα12/Gα13, of heterotrimeric G proteins. In many cases, this is well justified because the receptor in question clearly signals predominantly in a manner consistent with engagement with members of only one of the 4 groups. However, in many other cases studies using in vitro expression systems indicate a broader G protein–coupling profile with, in certain examples, a degree of interaction with virtually all G proteins tested (13). More importantly, a capacity to activate G proteins from more than 1 G protein family is also true for many receptors expressed natively, and this may dictate distinct physiologic outcomes. For example, although free fatty acid receptor 4 promotes secretion of the incretin glucagon-like peptide 1 by activation of pertussis toxin–insensitive G proteins of the Gαq/11 family (14), regulation of release of the satiety hormone ghrelin requires activation of one or more pertussis toxin–sensitive Gαi-family G protein (15). Equally, although free fatty acid receptor 2 acts to counter lipolysis in mouse white adipose tissue via a pertussis toxin–sensitive mechanism (16), effects of this receptor on secretion of glucagon-like peptide 1 are instead mediated by Gαq/11-family G proteins (16). Although such examples are clearly defined, there is often less understanding of the importance of selective interactions of a receptor with different members from within one of the G protein families and little insight into the molecular basis of such selectivity, and there are currently no comparative structures of a single GPCR in complex with 2 different G proteins.
Although clearly involved in GPCR-mediated cytoskeletal organization or reorganization and the consequences thereof, the least studied of the Gα protein family is the 2-member Gα12/Gα13 subgroup (17, 18). Although it is activated by many GPCRs, relatively little has been published on these because of a lack of selective inhibitors, and assays to measure their activation are challenging (19). However, although Gα12 and Gα13 are generally coexpressed, mouse knock-out studies demonstrate that they are not interchangeable (17), and certain GPCRs appear to couple selectively to Gα12, Gα13, or both. An interesting case in point is G protein–coupled receptor 35 (GPR35) (20). Although officially an orphan receptor, in that suggestions of its endogenous activator or activators remain controversial (20–24), a wide range of surrogate agonists are available (20, 25). Although coupling to Gα12/Gα13 is well established, the basis of potential selectivity in so doing is not. Herein we address 2 key questions: Does GPR35 selectively activate Gα13 over Gα12, and, if so, what is the molecular basis of this difference?
MATERIALS AND METHODS
Materials for cell culture were from MilliporeSigma (Burlington, MA, USA) or Thermo Fisher Scientific (Waltham, MA, USA). Polyethylenimine linear MW-25000 was from Polysciences (Warrington, PA, USA). Zaprinast, lodoxamide, pamoic acid, and a specific GPR35 antagonist (CID2745687) were purchased from commercial sources. Compound 1 {4-[(Z)-[(2Z)-2-(2-fluorobenzylidene)-4-oxo-1,3-thiazolidin-5-ylidene]methyl]benzoic acid} is described in Neetoo-Isseljee et al. (25). Bufrolin [5,6-dimethyl-2-nitro-1H-indene-1,3(2H)-dione] is described in Mackenzie et al. (26). 6-bromo-8-(4-methoxybenzamido)-4-oxo-4H-chromene-2-carboxylic acid (PSB-13253) (27) was a gift from Christa Muller and Dominik Thimm (University of Bonn, Bonn, Germany). In all cases the GPR35a splice variant of human GPR35 (hGPR35) was used.
Generation of bioluminescence resonance energy transfer systematic protein affinity strength modulation sensors
Systematic protein affinity strength modulation (SPASM) sensors consisting of the receptor of interest [hGPR35a or mouse GPR35 (mGPR35)] were generated based on previously reported Förster resonance energy transfer SPASM sensors (28). These sensors consist of a single construct of receptor, fused at its C terminus to mCitrine, followed by an ER/K α helical linker (29), the bioluminescent protein Nanoluc (Promega, Madison, WI, USA), and to a peptide corresponding to the final 27 aa of Gα12, Gα13, or Gαq. To ensure flexibility within the sensor unit, (GlySerGly)4 linkers were included to separate each element—mCitrine, ER/K linker, Nanoluc, G peptide—of the sensor. Sensors were cloned using PCR and a seamless-end homology cloning approach (Thermo Fisher Scientific), generating constructs that did not contain restriction enzyme sites separating the various sensor elements. All constructs were fully sequenced prior to use.
Cell culture
Clones of cells genome-edited to lack expression of Gαq/Gα11, Gα12/Gα13 or each of Gαq/Gα11/Gα12/Gα13 were derived from parental human embryonic kidney 293 (HEK293) cells as previously described (30). Along with HEK293-T (T antigen) cells and Flp-In T-REx 293 cells harboring various sensor constructs these were grown in DMEM (MilliporeSigma) supplemented with 10% fetal bovine serum, 2 mM l-glutamine, and 1% penicillin/streptomycin. Cells were incubated in a humidified CO2 incubator at 37°C.
Bioluminescence resonance energy transfer studies using SPASM sensors
Flp-In T-REx 293 cell lines stably harboring the SPASM sensor of interest were seeded into poly-d-lysine coated Greiner white 96-well plates. Doxycycline (100 ng/ml) was added 3–4 h after seeding to induce expression of the sensor, and cells were incubated overnight at 37°C in 5% CO2. For transiently expressed constructs, HEK293-T cells were seeded to obtain 60% confluence the following day. The cells were then transfected with 30 ng DNA and 180 ng polyethylenimine per well 2 d prior to the experiment. Thirty minutes before the assay, cells were washed with HBSS buffer containing 10 mM HEPES and incubated in the same buffer at 37°C. Because bioluminescence resonance energy transfer (BRET) provides a ratiometric signal and herein the constructs are single polypeptides, outcomes are anticipated to be independent of expression levels.
Kinetic studies
Coelenterazine-h was added to each well to give a final concentration of 5 μM 15 min prior to the read. Each well was read for 30 s before addition of a ligand and read for a further 90 s before any second addition. Wells were read throughout a 6-min window using a PheraStar plate reader (BMG Labtech, Cary, NC, USA) for 0.5-s simulations with filters for wavelengths of 475 nm for Nanoluc luminescence and 535 nm for mCitrine emission. The raw value for 535 nm was divided by the 450-nm value to obtain a BRET ratio. This value was then divided by the baseline value of the first 30 s of read before the addition of any ligand to obtain a fold change over baseline. This value for the vehicle-only additions for each cell type was subsequently subtracted from the corresponding treatment to give the final value used for analysis.
Endpoint assay
Coelenterazine-h was added to each well to give a final concentration of 2.5 μM 5 min prior to the addition of ligands. Wells were read 5 min after addition of ligands and hence 10 min after addition of coelenterazine-h. Each well was read on a ClarioStar plate reader for 0.5 s consecutively at wavelengths of 450 ± 45 nm for the Nanoluc luminescence and 550 ± 45 nm for mCitrine emission. The raw value for 550 nm was divided by the 450 nm value to obtain a BRET ratio. This value was subsequently divided by the value of the vehicle-only addition to obtain a final fold change over baseline value.
BRET studies using full-length G protein α subunits
The Nanoluc luciferase coding sequence was inserted in the human Gα13 sequence immediately following residue Arg128, with (GlySerGly)4 linkers on either side. This position has previously been verified as maintaining Gα13 function in Förster resonance energy transfer studies that introduced a fluorophore at this location (31). HEK293 cells genome-edited to lack expression of both Gα12 and Gα13 were transiently transfected 2 d prior to the experiment to coexpress hGPR35 tagged at the C terminus with enhanced yellow fluorescent protein (eYFP) and the indicated Nanoluc-containing Gα13 protein. BRET assays were conducted as described above. Subsequent studies employed this base Nanoluc-containing Gα13 construct in which residues G.H5.22 (Gln) and G.H5.23 (Leu) were altered to the corresponding residues of Gα12 (G.H5.22, Asp) or (G.H5.23, Ile).
β-Arrestin-2 interaction studies
These employed a BRET-based assay. HEK293-T cells were transfected transiently to coexpress hGPR35-eYFP and β-arrestin-2 tagged with Renilla luciferase (32). Agonist-induced proximity between these 2 proteins generated enhanced BRET signal upon addition of the luciferase substrate coelenterazine-h.
High content imaging/internalization studies
These were performed essentially as described by Mackenzie et al. (26). Briefly, Flp-In T-REx 293 cells harboring the hGPR35-SPASM sensor of interest were seeded into poly(d-lysine)–coated black clear-bottom 96-well plates at a density of 80,000 cells per well. Receptor expression was induced via the addition of doxycycline (100 ng/ml) 6 h after seeding. Twenty-four hours later, cells were washed twice with serum-free medium and incubated with ligand for 45 min at 37°C, before being fixed with paraformaldehyde (4% v/v). Cells were washed with PBS and incubated with 10 μg/ml Hoechst nuclear stain at 37°C for 30 min to allow determination of cell number. Receptor internalization was quantified using an ArrayScan II high content imager (Thermo Fisher Scientific), which detected the location of the mCitrine fluorescent protein component of the sensor as it was trafficked to endocytic recycling compartments.
TGF-α shedding assay
TGF-α shedding in response to activation of hGPR35 was assessed as previously described (30, 33). Briefly, parental HEK293 cells or cells of clones genome-edited to lack expression of various G protein α subunit combinations (30, 34, 35) were seeded onto 6-well plates (5 × 105 cells per well) 24 h prior to transfection. Cells were transfected with 0.5 μg of a plasmid containing an alkaline phosphatase (AP) fusion protein of TGF-α, AP-TGF-α, and 0.1 μg hGPR35. The next day, transfected cells were trypsinized, washed with HBSS, and reseeded on to 96-well plates 30 min prior to treatment with ligands. Cells were then treated with agonists and incubated at 37°C for 1 h. After such treatment, conditioned medium was transferred into other 96-well plates. A solution of 10 mM paranitrophenylphosphate, 40 mM Tris-HCl (pH 9.5), 40 mM NaCl, and 10 mM MgCl2 was added to both cell and conditioned medium plates, and the absorbance was measured at 405 nm before and after a 60-min incubation at 25°C. TGF-α shedding was calculated as the percentage increase of optical density at 405 nm in conditioned medium plate/overall total increase of optical density at 405 nm (from both conditioned medium and cell plates).
Modeling studies
We first investigated sequence-based selectivity determinants in Gα13 by evolutionary conservation for G.H5.23 with its orthologs and paralogs as previously described in Flock et al. (36). Structure-based sequence alignments were performed using the GPCR Database numbering for GPCRs and the Common Gα Numbering scheme for G proteins (36, 37). In this scheme, G is the G protein, H5 is the α5 helix, and the numerical value is the position from the start of that helix (e.g., residue 23), hence G.H5.23. Structural inspections were performed on the receptor-G protein complexes of β2-adrenoceptor-Gαs (Protein Data Bank: 3SN6) and µ-opioid receptor-Gαi1 structure (Protein Data Bank: 6DDE). Structures were prepared using the protein preparation wizard in Maestro (Schrödinger, New York, NY, USA) using default settings and including addition of missing residues followed by H-bond assignment. Interface contacts between G.H5.23 and the receptor (including backbone interactions) were calculated using Arpeggio using default settings (maximum range of interaction set to 5.0 Å) (38). Superpositioning, rotation calculations, and visualizations were performed using PyMol (PyMol Molecular Graphics System, v.2.0; Schrödinger).
RESULTS
hGPR35 can interact with G12/G13 but not with Gq/G11
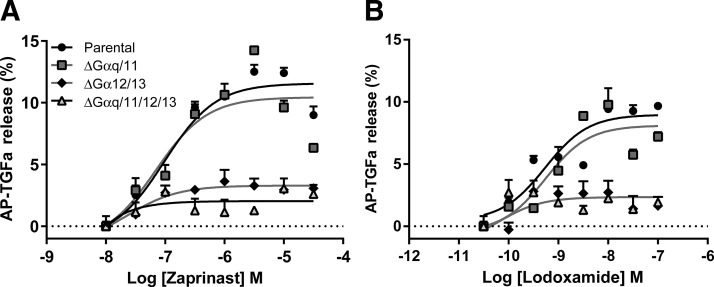
To assess the ability of hGPR35 to interact productively with G proteins of the Gα12/Gα13 and Gαq/Gα11 families, we employed HEK293 cells, because they express all 4 of these G proteins (30), and a TGF-α shedding assay (30, 33) because it has previously been established that the TGF-α shedding endpoint is promoted by receptors that stimulate activation of any combination of Gq, G11, G12, and G13 (30). Following transient cointroduction of hGPR35 and an AP-tagged form of TGF-α (AP-TGF-α) into parental HEK293 cells, the receptor was stimulated with either the most widely used GPR35 agonist zaprinast (39) (Fig. 1A) or the recently described high-potency agonist lodoxamide (26) (Fig. 1B). These both promoted, in a concentration-dependent manner, cleavage of AP-TGF-α and its release from the surface of the cells. In HEK293 cells that had been genome-edited using clustered regularly interspaced short palindromic repeat (CRISPR)–Cas9 (CRISPR-associated protein-9) (30, 34, 35) to eliminate expression of all 4 of Gαq, Gα11, Gα12, and Gα13, shedding of AP-TGF-α was no longer evident upon stimulation of hGPR35 (Fig. 1). Elimination of only a combination of Gα12 and Gα13 from HEK293 cells also prevented either of these hGPR35 active agonists from promoting AP-TGF-α cleavage (Fig. 1). By contrast, in a clone of HEK293 cells that had been genome-edited to eliminate expression of only Gαq and Gα11 (34), the 2 phosphoinositidase C–linked G proteins that are expressed by these cells, and which, therefore, still expressed Gα12 and Gα13, both zaprinast and lodoxamide promoted shedding of AP-TGF-α as effectively, and with equal potency, as in the parental HEK293 cells (Fig. 1). In combination with the lack of effect of activation of hGPR35 in the Gα12/Gα13-null cells, this indicates that hGPR35 does not interact productively with Gαq/Gα11-family G proteins.
Figure 1.
hGPR35 promotes shedding of an AP-tagged form of TGF-α via activation of G12/13 but not via Gq/G11. The ability of varying concentrations of either zaprinast (A) or lodoxamide (B) to promote shedding of an AP-tagged form of TGF-α was assessed following cotransfection of hGPR35 and AP-TGF-α into each of parental HEK293 cells (circles) or clones of HEK293 cells that had been genome-edited to lack expression of Gα12 + Gα13 (diamonds), Gαq + Gα11 (squares), or a combination of Gαq, Gα11, Gα12, and Gα13 (triangles). Basal levels of AP-TGF-α release were subtracted. Data represent means ± sd in triplicates from each group of a single experiment representative of 3 performed.
Development and characterization of SPASM sensors
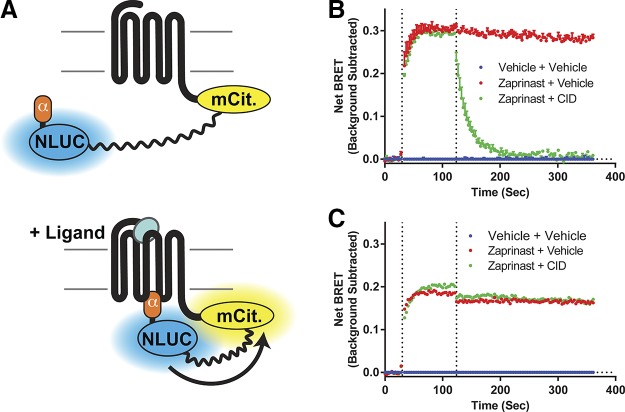
To provide improved quantification and to assess the relative degree of interaction of hGPR35 with Gα12 and Gα13 we generated a pair of GPCR– G protein SPASM sensors. In these the C-terminal tail of hGPR35 was linked in-frame to the C-terminal 27 aa of either G12 or G13 α subunits via a sequence that incorporated a BRET sensor that separates the luciferase Nanoluc from the fluorescent protein mCitrine with a 10 nm flexible linker (Fig. 2A). Based on similar Fluorescence Resonance Energy Transfer–based SPASM sensors for other GPCRs (28, 40) and structural insights into the mechanisms of interactions between GPCRs and G proteins α subunits (7–12) we anticipated that following construct expression agonist-induced engagement of the G protein segment with the receptor would result in enhanced BRET signal. We cloned these sensors into Flp-In T-REx 293 cells and, following doxycycline-induced expression, studied initially the kinetics of response of the Gα13-containing sensor upon addition of zaprinast. Zaprinast produced a rapid increase in BRET that was maintained over a period of at least 6 min (Fig. 2B). CID2745687 has been described as a human ortholog specific antagonist of GPR35 (41, 42). Addition of CID2745687 after the zaprinast-induced BRET signal had reached a steady, maximal level rapidly reversed the effect of zaprinast at hGPR35-Gα13 (Fig. 2B). It is well appreciated that rodent orthologs of GPR35 display distinctly different ligand pharmacology than hGPR35 (20), but whether rodent forms of the receptor couple to the same G proteins as hGPR35 is uncertain. To assess this question, we generated equivalent mGPR35-Gα12 and -Gα13 sensors. Following similar stable expression in Flp-In T-REx 293 cells, zaprinast, which is also an effective agonist at mGPR35 (20), once again produced a large BRET signal at the Gα13-sensor that was maintained over time (Fig. 2C). Addition of CID2745687, however, failed to reverse the effect of zaprinast at mGPR35-Gα13 (Fig. 2C). This is consistent with previous in vitro studies showing that CID2745687 is a high-affinity antagonist at hGPR35 but has little affinity for mGPR35 (42). This suggests that reported effects of CID2745687 in cells and tissues from rodents (43, 44) probably reflect off-target effects of the compound rather than being mediated by GPR35.
Figure 2.
Kinetics of activation and deactivation of GPR35-Gα sensors. A) A diagram illustrates the nature of the GPR35–G protein sensors used and why G protein engagement induced by binding of an agonist ligand to the receptor is anticipated to result in enhanced BRET signal as the Nanoluc (NLUC) and mCitrine (mCit) components move closer together. B, C) Flp-In T-REx 293 cells stably harboring hGPR35-Gα13 (B) or mGPR35-Gα13 (C) were treated with doxycycline to induce construct expression. Kinetic studies were then performed to assess changes in BRET signal over time. Zaprinast (red) (EC80 16 μM for human and 1 μM for mouse) or vehicle (blue) (HBSS with DMSO) were added at time 30 s (first vertical line) and maintained for a further 330 s. In certain studies, the human-specific GPR35 antagonist CID2745687 (CID) (green) (10 μM) was added at 120 s (second vertical line).
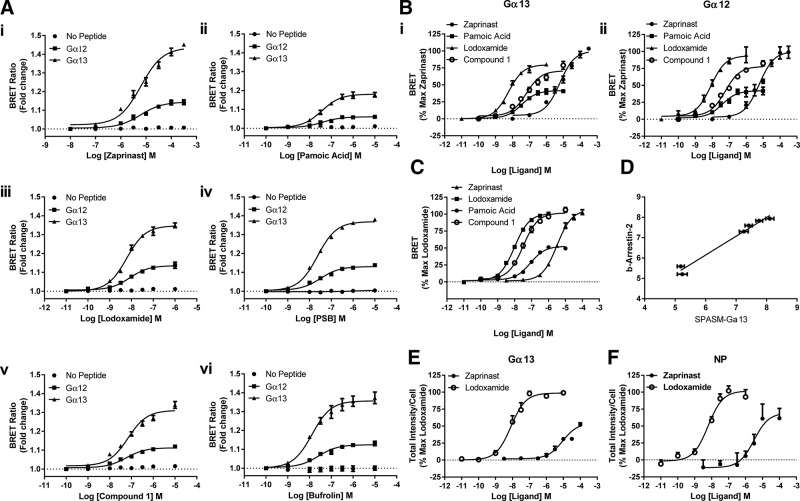
A wide range of previously characterized ligands with agonism at hGPR35, including zaprinast (39) (Fig. 3Ai), pamoic acid (41) (Fig. 3Aii), lodoxamide (26) (Fig. 3Aiii), PSB-13253 (27) (Fig. 3Aiv), compound 1 (25) (Fig. 3Av), and bufrolin (26) (Fig. 3Avi), added to cells expressing hGPR35-Gα13 all produced robust and concentration-dependent increases in BRET signal. This was also the case for hGPR35-Gα12 (Fig. 3A). Although the measured EC50 of each of the agonists tested was very similar for activation of Gα12 and Gα13 (Table 1), in every case the maximal response of the Gα13-based sensor was markedly higher (Fig. 3A). To demonstrate the critical role of the G protein C-terminal sequence in promoting the observed increase in BRET signal, we also generated a control no-peptide (NP) hGPR35-SPASM construct that simply lacked such a C-terminal sequence. Following its stable expression and induction in Flp-In T-REx 293 cells, none of the agonists was able to enhance the basal BRET signal of the NP sensor (Fig. 3A). In further support of the ability of these hGPR35-SPASM sensors to report the pharmacological characteristics of GPR35 agonists appropriately, as illustrated in Fig. 3Bi, ii, pamoic acid, although potent, clearly acted as a partial agonist compared with the other ligands at both the Gα13- and Gα12-containing sensors. This is fully consistent with findings using other assay endpoints, including binding of [35S]GTPγS and receptor internalization (32). Of equal importance in further validating use of the SPASM sensors, the potency measures for activation of the Gα13-containing sensor by different agonists were highly correlated (r2 = 0.98) with values obtained in BRET-based hGPR35–β-arrestin-2 interaction assays (Fig. 3C, D) that have provided the most widely used approach to identify and characterize ligands at GPR35 (25, 26, 41, 42). Interaction with a β-arrestin is an important step in agonist-induced internalization of many GPCRs (34) including GPR35. The G protein–containing sensor constructs retained the capacity to be internalized from the surface of their host Flp-In T-REx 293 cells upon exposure to either the high-potency agonist lodoxamide or the lower-potency agonist zaprinast, as assessed using a high-content imaging platform that detects the intracellular location of the mCitrine fluorescent protein component of the sensors (Fig. 3E), and this was also the case for the equivalent hGPR35-NP sensor (Fig. 3F).
Figure 3.
Validation and demonstration of effectiveness of hGPR35-Gα12 and hGPR35-Gα13 SPASM sensors. A) Flp-In T-REx 293 cells stably harboring hGPR35-Gα13 (squares), hGPR35-Gα12 (triangles), or hGPR35-NP (circles) were treated with doxycycline to induce construct expression; such cells were then treated with varying concentrations of zaprinast (i), pamoic acid (ii), lodoxamide (iii), PSB-13253 (PSB) (iv), compound 1 (v), or bufrolin (vi) for 5 min, and alteration in BRET signal was compared with basal. B) Data from A for the 4 denoted agonists are replotted, with maximal response to zaprinast defined as 100%. For both hGPR35-Gα13 (i) and hGPR35-Gα12 (ii), pamoic acid acted as a partial agonist compared with the other ligands. C) The same ligands as shown in B were used to assess interactions between hGPR35 and β-arrestin-2 in a BRET-based assay following transient cotransfection of hGPR35-eYFP and β-arrestin-2–Renilla luciferase into HEK293T cells. D) Correlation of potency of all 6 ligands shown in A in assays using the hGPR35-Gα13 sensor and the hGPR35a–β-arrestin-2 interaction assay is shown. E, F) Although the construction of the SPASM sensors adds a substantial molecular construct to the C-terminal tail of hGPR35, it does not prevent effective agonist-induced internalization of the constructs from the surface of Flp-In T-REx 293 cells expressing such constructs: hGPR35-Gα13 (E), hGPR35-NP (F).
TABLE 1.
Potency of ligands at hGPR35-Gα12 and hGPR35-Gα13 sensors
| Ligand | hGPR35-Gα12 pEC50 ± sem | hGPR35-Gα13 pEC50 ± sem |
|---|---|---|
| Zaprinast | 5.22 ± 0.10 | 5.20 ± 0.07 |
| Pamoic acid | 7.40 ± 0.15 | 7.44 ± 0.09 |
| Lodoxamide | 8.08 ± 0.09 | 8.22 ± 0.06 |
| PSB-13253 | 7.49 ± 0.10 | 7.63 ± 0.04 |
| Compound 1 | 7.33 ± 0.09 | 7.16 ± 0.08 |
| Bufrolin | 7.59 ± 0.11 | 7.89 ± 0.10 |
Data represent the mean ± sem from at least 3 independent experiments; p, negative logarithm.
The molecular basis for selective coupling to Gα13
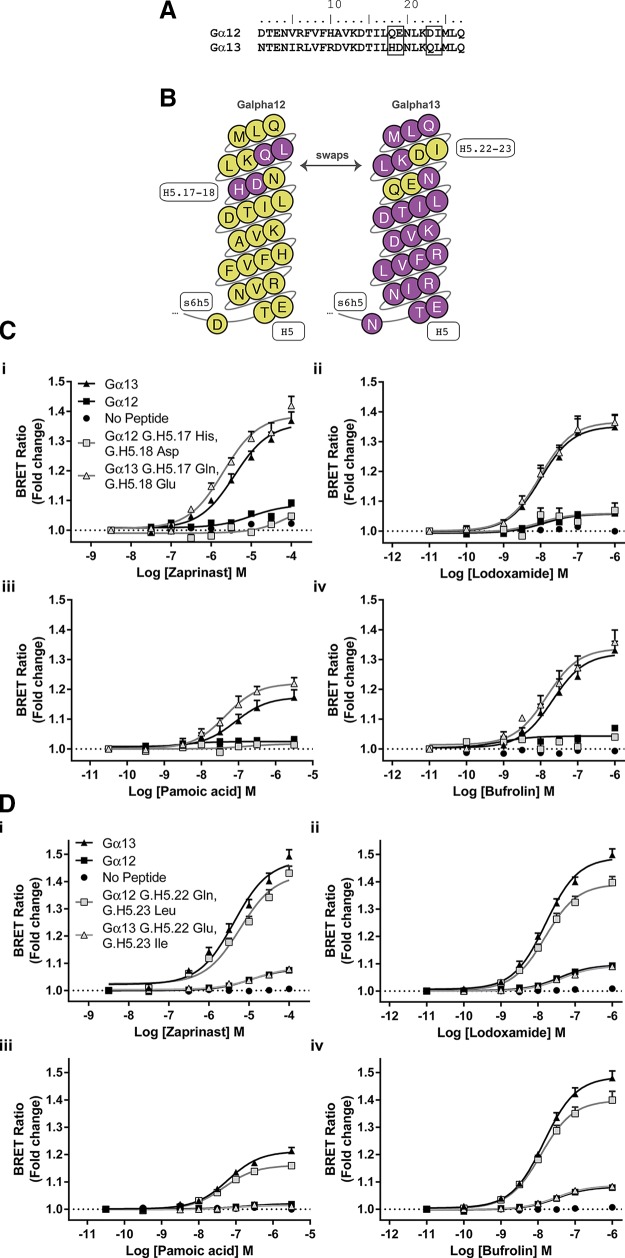
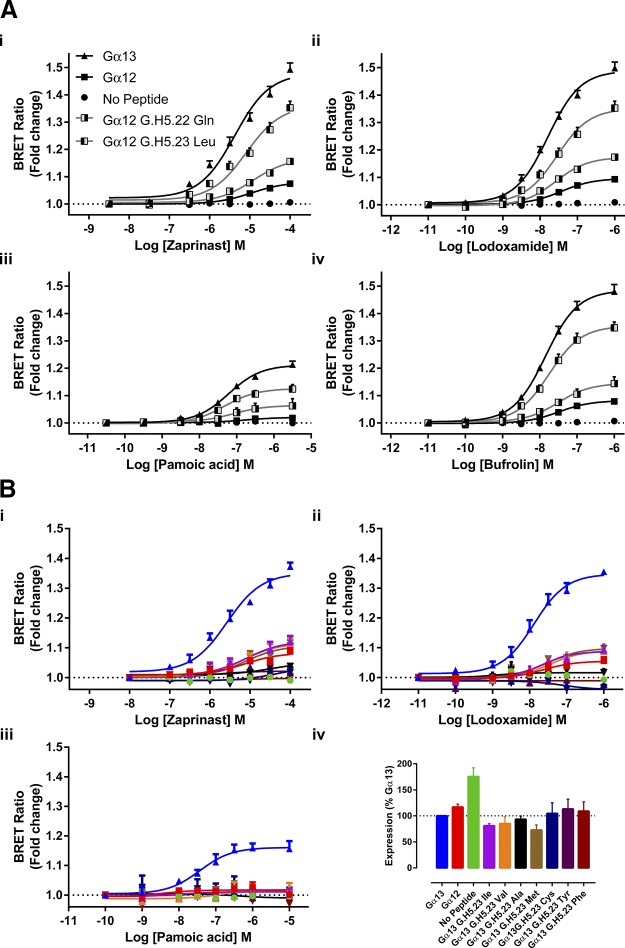
The marked difference in effectiveness of agonists at the hGPR35-Gα13 sensor and the hGPR35-Gα12 sensor, coupled with earlier evidence that hGPR35 selectively interacts with Gα13 over Gα12 (32), led us to assess the molecular basis for this difference. Studies with chimeric G protein α subunits have shown that substitution of between 5 and 10 aa at the extreme C-terminal end of a G protein, which is within the α5 helix, is sufficient to switch GPCR selectivity (12). Within this region there are only 4 aa differences between Gα13 and Gα12 (Fig. 4A, B). These differences are a pair of residues located at positions −10 [Common Gα Numbering system (35, 36) G.H5.17] and −9 (G.H5.18) and a pair of amino acids at positions −5 (G.H5.22) and −4 (G.H5.23) (Fig. 4B). Initially we swapped His and Asp at positions G.H5.17 (His) and G.H5.18 (Asp) of Gα13 for the equivalent amino acids, Gln and Glu, in Gα12 and vice versa in the context of the hGPR35 sensors. This had little effect on function compared with the wild type sequences (Fig. 4C), and this was the case for all hGPR35 agonists that we assessed (Fig. 4C). However, when we exchanged the residues Asp and Ile at positions G.H5.22 (Asp) and G.H5.23 (Ile) of Gα12 into Gα13 to replace Gln and Leu, activation by agonists resulted in as limited BRET signal as with full-length Gα12 (Fig. 4D). The opposite was true when Gln and Leu from Gα13 were used to replace Asp and Ile in the Gα12 sequence. This generated a sensor with substantial gain of function that was now almost equivalent to the full sequence from Gα13 (Fig. 4D). Once again, this was the case no matter whether zaprinast, lodoxamide, pamoic acid, or bufrolin was employed as the agonist (Fig. 4D).
Figure 4.
G12/G13 selectivity for hGPR35 resides within the C-terminal 10 aa. A) The C-terminal 27 aa of Gα12 and Gα13 are shown with amino acids that differ within the last 10 aa boxed. B) The Common Gα Numbering system (36, 37) is used to highlight these differences and their positions within the C-terminal G protein α5 helix. C, D) hGPR35-SPASM sensors were constructed in which residues at positions G.H5.17 and G.H5.18 (C) or G.H5.22 and G.H5.23 (D) were swapped between Gα12 and Gα13. Following stable expression and induction in Flp-In T-REx 293 cells, the ability of each of zaprinast (i), lodoxamide (ii), pamoic acid (iii), and bufrolin (iv) to enhance BRET signals was compared with the effect of these ligands at the hGPR35-Gα13, hGPR35-Gα12, and hGPR35-NP sensors.
We extended these studies by replacing only a single residue at a time. Remarkably, given the physiochemical similarities of Leu and Ile, substitution of Leu (G.H5.23) for Ile in Gα12 generated an hGPR35-SPASM sensor that responded almost as well to GPR35 agonists as the full sequence from Gα13 (Fig. 5A). By contrast, substitution of Asp (G.H5.22) by Gln in Gα12, although it produced an enhanced response to each of zaprinast, lodoxamide, pamoic acid, and bufrolin compared with the wild type Gα12 sequence (Fig. 5A), resulted in a much more limited effect compared with the introduction of Leu (G.H5.23) in place of Ile. Finally, in this context we assessed whether Leu at position G.H5.23 was the only amino acid that could provide effective coupling between hGPR35 and a Gα13-based G protein. This was the case; alteration of this residue to any of Val, Ala, Met, Cys, Tyr, or Phe generated a sensor that was not activated more effectively than either G.H5.23 Ile Gα13 or full-length Gα12 (Fig. 5B), and once more this was true when assessing a range of chemically distinct GPR35 agonist ligands (Fig. 5B). These amino acids were selected because they cover the identity of residue G.H5.23 across mammalian G protein α subunits. Although BRET provides a ratiometric signal that is anticipated to be independent of construct expression level, we directly examined the relative expression levels of each of the G.H5.23 point mutant forms of the Gα13- and Gα12-containing GPR35 sensors used in these transient expression experiments by directly measuring luciferase activity (Fig. 5B). This examination showed that the Gα12 sensor construct was expressed at a very similar level as the Gα13 sensor construct, although it generated a very limited signal compared with the Gα13 sensor, and that alteration of residue G.H5.23 in the Gα13 sensor across the range of amino acids introduced had no substantial effect on construct expression levels. The GPR35-NP sensor was expressed at rather higher levels than the others, and this may indicate that the G protein segment of the other constructs reduces expression.
Figure 5.
G12/G13 selectivity for hGPR35 is defined predominantly by a single leucine/isoleucine variation. A) In experiments akin to Fig. 4, hGPR35-G protein sensors were constructed and expressed in which single amino acids at positions G.H5.22 and G.H5.23 were swapped between Gα12 and Gα13, and the effect of the denoted ligands [zaprinast (i), lodoxamide (ii), pamoic acid (iii), and bufrolin (iv)] were compared with results generated using the hGPR35-Gα13, hGPR35-Gα12, and hGPR35-NP sensors. B) Position G.H5.23 (Leu) in Gα13 was altered to a number of other amino acids, and functionality was assessed in response to the noted GPR35 agonists [zaprinast (i), lodoxamide (ii), and pamoic acid (iii)], with parallel responses of hGPR35-Gα13, hGPR35-Gα12, and hGPR35-NP sensors recorded as controls. Direct measures of luciferase activity in cells transiently expressing these constructs defined their relative expression levels (iv).
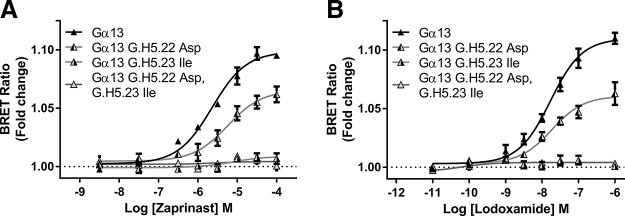
This selectivity is maintained in full-length Gα13
To ensure that outputs from the SPASM sensor studies would correlate with effects observed with the corresponding full-length G proteins, Gα13 was next modified by insertion of Nanoluc at residue position 128. This construct was cotransfected with hGPR35-eYFP into HEK293 cells lacking both Gα12 and Gα13, and BRET was measured as a surrogate of receptor-G protein interaction following addition of varying concentrations of zaprinast (Fig. 6A) or lodoxamide (Fig. 6B). Subsequent studies employed this base G protein construct with residues G.H5.22 (Gln), G.H5.23 (Leu), or both altered to the corresponding residues of Gα12 (G.H5.22, Asp) or (G.H5.23, Ile) (Fig. 6). The results using these variants were very similar to those obtained using the equivalent SPASM sensors. The largest signal was achieved using wild type full-length Gα13 (Fig. 6). Replacement of G.H5.22 (Gln) by Asp reduced interactions between receptor and G protein to a small extent, whereas replacement of G.H5.23 (Leu) by Ile or a combination of replacement of G.H5.22 (Gln) and G.H5.23 (Leu) by Asp and Ile resulted in forms of full-length Gα13 that were unable to interact effectively with GPR35 in an agonist-dependent manner (Fig. 6). This was the case when either zaprinast (Fig. 6A) or lodoxamide (Fig. 6B) was used as agonist.
Figure 6.
The importance of residue G.H5.23 in Gα13 sensors is maintained in the full-length G protein sequence. hGPR35-eYFP was coexpressed in HEK293 cells with forms of full-length Gα13 that either were wild type, had residue G.H5.22 converted to Asp, had residue G.H5.23 converted to Ile, or contained both of these alterations. Nanoluc had been introduced into all of the forms of Gα13 to provide a potential BRET pairing with the eYFP-tagged receptor. Cells were then stimulated with varying concentrations of either zaprinast (A) or lodoxamide (B). Data are shown as means ± sd (n = 3).
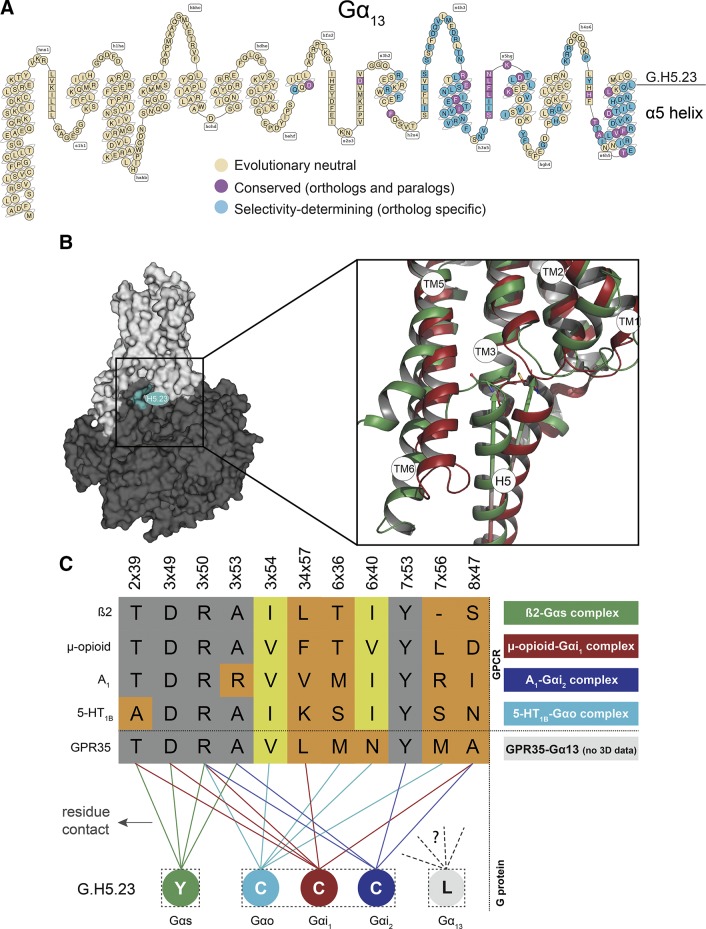
Computational analysis
The G.H5.23 position is not conserved among the human Gα subtype paralogs (7 Gα subtypes with Cys, 2 with Ile, 4 with Tyr, 1 with Phe, and 1 with Leu) but is highly conserved among the Gα12 and Gα13 orthologs. This renders it part of the selectivity-determining G protein barcode (36) (Fig. 7A). Residue positions in this barcode represent the determinants of GPCR–G protein selectivity, as deduced from evolutionary conservation (conserved in orthologs but not in paralogs) (36). Given the limited structural coverage of receptors and complexes to model a GPR35-Gα13 complex, we looked at the currently available receptor–G protein complexes of µ-opioid receptor–Gαi1 (45), β2-adrenoceptor–Gαs (8), adenosine A1 receptor–Gαi2 (46), and 5-HT1A–Gαo1 (47) to attempt to rationalize the importance of G.H5.23 for G13 function. A comparison between these structures reveals differences in the position of transmembrane receptor helix VI and in the orientation between the G protein’s α5 helix domain (∼20° rotation) (Fig. 7B). For the β2-adrenoceptor–Gαs complex, G.H5.23 (Tyr) has hydrophobic contacts with positions 2×39, 3×49, 3×50, and 3×53 of the receptor, whereas G.H5.23 (Cys) contacts 2×39, 3×49, 3×50, 34×57 (intracellular loop 2), and 8×47 in the µ-opioid–Gαi1; 3×50, 3×54, 6×36, 6×40, and 7×53 in 5-HT1B–Gαo; and 3×50, 3×53, 7×53, and 8×47 in adenosine A1–Gαi2 (Fig. 7C). The distinct binding patterns of the available structures suggest there must be a further distinct binding mode for GPR35-Gα13. Thus, it is plausible that this specific binding pose is highly constrained in space and hence only Leu has the correct balance of hydrophobicity, flexibility, isotropic surface area, and electrostatic properties.
Figure 7.
Modeling studies. A) Snake-like diagram of Gα13 with the G protein barcode highlighting evolutionary neutral, conserved, and selectivity-determining positions (barcode cutoff 0.96). B) Location of G.H5.23 (cyan) in the receptor-G protein complex (left) and comparison of the α5 helix domain C termini of Gαs (green) and Gαi1 (red) (right). Arrows indicate the rotation differences between the α5 helix domains. Comparison of interface contacts and contacting residues between recently published GPCR–G protein structures are shown, as well as for the GPR35 G12/G13 selectivity-determining position G.H5.23. C) Alignment of G.H5.23 contacting receptor residue positions (gray: conserved; yellow: differing). This suggests a structurally yet-to-be-defined, alternative binding mode and contact profile for the G12/G13 family subtypes and their receptor coupling partners.
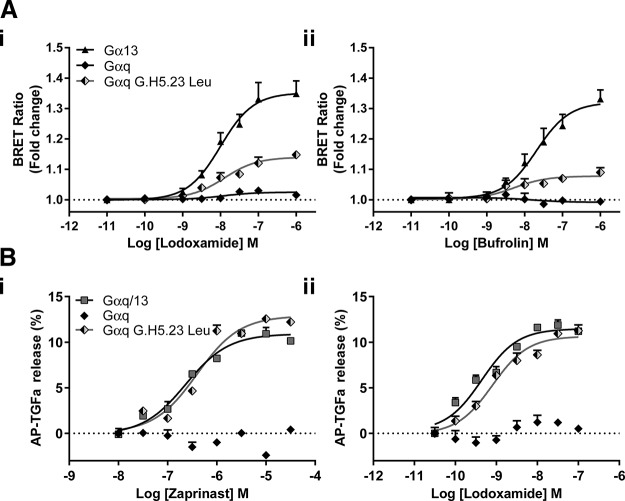
Introduction of G.H5.23 Leu into Gαq allows coupling to GPR35
It is intriguing, therefore, that although a GPR35-Gαq SPASM sensor was not activated by GPR35 agonists (Fig. 8A), replacement of G.H5.23 (Tyr in Gαq) by Leu also generated a sensor for GPR35 that provided a substantial level of functionality (Fig. 8A). To also extend this to the context of full-length G protein α subunits, we generated a Tyr-Leu mutation at G.H5.23 in Gαq and introduced this variant, along with hGPR35, into HEK293 cells genome-edited to lack each of Gαq, Gα11, Gα12, and Gα13. Now, in TGF-α shedding studies, both zaprinast and lodoxamide were able to promote release of AP-TGF-α in a concentration-dependent manner (Fig. 8B) and with potency for the 2 agonists that was not significantly different from that observed in either parental HEK293 or Gαq plus Gα11 knock-out HEK293 cells (Fig. 1). Moreover, the effectiveness of TGF-α shedding induced by either of these GPR35 agonists when activating G.H5.23 Leu Gαq was equivalent to that observed when we instead introduced into the genome-edited cells lacking each of Gαq, Gα11, Gα12, and Gα13 a chimeric form of Gαq/Gα13 in which all of the C-terminal 6 aa were derived from Gα13 (Fig. 8B).
Figure 8.
Replacement of Gαq G.H5.23 Tyr by Leu allows engagement of Gαq by agonist-occupied hGPR35. A) The ability of varying concentrations of either lodoxamide (i) or bufrolin (ii) to enhance BRET at hGPR35-SPASM sensors containing the C-terminal 27 aa of Gαq (filled diamonds), G.H5.23Leu Gαq (shaded diamonds), or Gα13 (triangles) was assessed. B) The ability of varying concentrations of either zaprinast (i) or lodoxamide (ii) to promote shedding of an AP-tagged form of TGF-α was assessed following cotransfection of hGPR35 and AP-TGF-α with either full-length Gαq (solid diamonds), G.H5.23 Leu Gαq (shaded diamonds), or chimeric Gαq/Gα13 containing the C-terminal hexapeptide of Gα13 (squares) into HEK293 cells that had been genome-edited to lack expression of a combination of Gαq, Gα11, Gα12, and Gα13. Basal levels of AP-TGF-α release were subtracted. Data represent means ± sd in triplicates from each group of a single experiment that is representative of 3 performed.
DISCUSSION
Although many GPCRs display a level of promiscuity in coupling to several families of heterotrimeric G proteins, there is little known about potential receptor selectivity between more closely related members of each of the 4 broad, generic families. Detailed studies on the Gs, Gi, and Gq families reflect their direct roles in the regulation of levels of secondary messengers and that toxins and chemical tools that disrupt signaling via members of these families are available and are used widely (12, 48). By contrast, the G12/G13 group is much less studied, although these play central roles in cytoskeletal organization and signaling via Rho-family small monomeric G proteins. It is also clear that many GPCRs can and do cause activation of these G proteins, but direct, easy-to-measure assays of their stimulation are not broadly available. Certain GPCRs do, however, appear to display considerable selectivity for G12/G13 over other G proteins, and in the case of the nominally orphan receptor GPR35 it has been suggested that there is even marked selectivity for G13 over G12 (49). To assess this in a controlled and potentially quantitative manner, we established a range of SPASM sensors (28, 40) in which hGPR35a was linked to the C-terminal 27 aa of various G protein α subunits via a flexible linker in which receptor interaction with the G protein segment results in enhanced BRET signal. A variant NP form, lacking the G protein segment, did not result in altered BRET with addition of various agonists at hGPR35, demonstrating that the G protein segment was indeed required for signal alteration. Moreover, in agreement with studies conducted using a TGF-α shedding assay performed in cells from which various G protein α subunits had been removed via genome editing, an hGPR35-Gαq SPASM sensor did not respond to GPR35 agonist ligands.
A number of key outcomes were produced. First, using a sensor containing hGPR35 and the C-terminal 27 aa of Gα13, a large increase in BRET signal was generated following the addition of a range of chemically distinct agonists. This sensor provided an appropriate measure of ligand potency because the potency profile in this assay was highly correlated with outcomes from an hGPR35–β-arrestin-2 interaction assay that has been widely used to identify novel agonists at this receptor (26, 27, 41, 42). Moreover, it also provided a suitable estimate of agonist efficacy because pamoic acid clearly functioned as a partial agonist, as previously defined in a range of other assays. A further key observation that validated the use of the sensors was that although a sensor containing mGPR35 was also activated effectively by zaprinast, which has similar potency at mGPR35 and hGPR35 (20), CID2745687, which is a human-specific antagonist of GPR35 (20, 42), was unable to reverse the effect of zaprinast at the mGPR35-containing sensor. It is important to highlight that although hGPR35 and mGPR35 are poorly conserved overall in their sequences, they do engage effectively with the same G protein in Gα13. Secondly, although the agonists tested are able to also activate Gα12 in such a sensor construct, the maximal signal of all GPR35 agonists tested was substantially lower than for Gα13. Because this assay generates a ratiometric signal, variations in response would not be anticipated to be related to expression level. Moreover, direct measures of the fluorescence of the mCitrine component of the BRET sensor indicated very similar expression levels of hGPR35-Gα13 and hGPR35-Gα12. As such, these outcomes indicated either that hGPR35 interacts substantially less effectively with Gα12 than with Gα13 or that marked variations in the orientation of these interactions alter the proximity of the Nanoluc-mCitrine BRET pair within the sensor. Although it is impossible to separate these possibilities conclusively, the fact that alteration of a single amino acid (G.H5.23) between Leu and Ile produced signal-level switches close to those observed for the full sequences of Gα13 and Gα12 strongly favors the effect as being an intrinsic difference in interaction of the receptor between the 2 proteins. Moreover, replacement of Leu at position G.H5.23 in the sequence of Gα13 with any amino acid we tried resulted in a loss of agonist-induced BRET to a level at or below those obtained with the Gα12 sequence. A sensor containing the C-terminal 27 aa of Gαq/Gα11 (these 2 G proteins are identical over this region) showed no agonist-induced signal, but, remarkably, simple replacement of Tyr G.H5.23 by Leu generated a substantial level of interaction, and this was also observed in the TGF-α shedding assay when we introduced Leu in place of Tyr in full-length Gαq.
Although GPCR and cognate G protein structure determination is advancing rapidly (8, 45–47), there are little direct data on how much difference might be expected for a receptor in complex with 2 different G proteins. Van Eps et al. (50) have suggested differences in the ways in which 2 distinct GPCRs interact with their cognate G proteins, but the outcomes are predictions rather than direct comparisons. Kleinau et al. (51) have taken a mutagenic approach to address selective interactions of the thyrotropin receptor with Gs and with Gq, but these studies focused on the receptor rather than the G protein. A study with a larger direct relevance used the SPASM approach also employed herein to assess differences in interactions between receptors with Gαs and Gαq, 2 still markedly different G proteins from different families (40). Furthermore, an extensive computational analysis on the potential basis for GPCR–G protein selectivity by Flock et al. (36) explored a potential barcode for receptor selectivity within the amino acid sequences at the C-terminal region of different G proteins. However, this remains challenging to interpret fully in the absence of a broader range of atomic-level structures and provided no insights into potential selectivity between Gα13 and Gα12, perhaps in part because these are the G proteins for which the most limited GPCR interaction profiles have been reported.
We have also taken advantage of the most recent data on natural genetic variations in the human population from the Genome Aggregation Database, consisting of 123,136 exome sequences (52). Strikingly, no natural mutations have been observed in any individual at G.H5.23 in Gα12 or Gα13, which suggests that G.H5.23 in G12/G13 is under strong selection in the human population. Interestingly, a Gα13 G.H5.23 Leu374-Ile mutation and a Gα12 G.H5.23 Ile378-Ser mutation have been reported in the Catalogue of Somatic Mutations in Cancer (53). In the case of the Gα13 G.H5.23 Leu374-Ile mutation, our study suggests this is linked to reduced function in response to GPR35 activation, and it will be interesting to explore if this is also the case for other Gα13-interacting receptors. GPR35 has been reported to be up-regulated in breast cancer tissue compared with normal adjacent tissue (54), but the overall significance of this remains uncertain.
Taken together, Gα13 Leu374 (G.H5.23) seems to be the single most relevant residue for G protein selectivity, at least for GPR35. However, the molecular details of this phenomenon are yet to be elucidated in structural and mutational studies (on the receptor side) to pinpoint the G12/G13 specific receptor interface partners of G.H5.23.
ACKNOWLEDGMENTS
D.E.G. thanks the Novo Nordisk Foundation for Supporting Grant NNF17OC0031226 and the European Research Council for Starting Grant 639125. A.E.M. thanks the U.K. Medical Research Council for an Industrial Collaborative Awards in Science and Engineering Studentship, and B.D.H. was supported by a Glasgow University Leadership Fellowship. These studies were supported by U.K. Biotechnology and Biosciences Research Council Grants BB/P000649/1 (to G.M.) and BB/P00069X/1 (to A.B.T.), Japanese Society for the Promotion of Science Grant-in-Aid for Scientific Research (KAKANHI) 17K08264 (to A.I.), and Japan Agency for Medical Research and Development Grant PRIME JP17gm5910013 (to A.I.). The authors declare no conflicts of interest.
Glossary
- AP
alkaline phosphatase
- BRET
bioluminescence resonance energy transfer
- CID2745687
GPR35 antagonist
- eYFP
enhanced yellow fluorescent protein
- GPR35
G protein–coupled receptor 35
- HEK293
human embryonic kidney 293
- hGPR35
human GPR35
- mGPR35
mouse GPR35
- NP
no peptide
- PSB-13253
6-bromo-8-(4-methoxybenzamido)-4-oxo-4H-chromene-2-carboxylic acid
- SPASM
systematic protein affinity strength modulation
AUTHOR CONTRIBUTIONS
A. B. Tobin, B. D. Hudson, and G. Milligan designed the research; A. E. Mackenzie, T. Quon, L.-C. Lin, and L. Jenkins performed the research; A. S. Hauser, A. Inoue, and D. E. Gloriam contributed new reagents or analytic tools; A. E. Mackenzie, T. Quon, L.-C. Lin, A. S. Hauser, and L. Jenkins analyzed the data; B. D. Hudson and G. Milligan wrote the manuscript; and all authors approved the final manuscript.
REFERENCES
- 1.Thal D. M., Vuckovic Z., Draper-Joyce C. J., Liang Y. L., Glukhova A., Christopoulos A., Sexton P. M. (2018) Recent advances in the determination of G protein-coupled receptor structures. Curr. Opin. Struct. Biol. 51, 28–34 [DOI] [PubMed] [Google Scholar]
- 2.Cooke R. M., Brown A. J., Marshall F. H., Mason J. S. (2015) Structures of G protein-coupled receptors reveal new opportunities for drug discovery. Drug Discov. Today 20, 1355–1364 [DOI] [PubMed] [Google Scholar]
- 3.Grisshammer R. (2017) New approaches towards the understanding of integral membrane proteins: a structural perspective on G protein-coupled receptors. Protein Sci. 26, 1493–1504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carpenter B., Tate C. G. (2017) Active state structures of G protein-coupled receptors highlight the similarities and differences in the G protein and arrestin coupling interfaces. Curr. Opin. Struct. Biol. 45, 124–132 [DOI] [PubMed] [Google Scholar]
- 5.Manglik A., Kruse A. C. (2017) Structural basis for G protein-coupled receptor activation. Biochemistry 56, 5628–5634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Preininger A. M., Meiler J., Hamm H. E. (2013) Conformational flexibility and structural dynamics in GPCR-mediated G protein activation: a perspective. J. Mol. Biol. 425, 2288–2298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moreira I. S. (2014) Structural features of the G-protein/GPCR interactions. Biochim. Biophys. Acta 1840, 16–33 [DOI] [PubMed] [Google Scholar]
- 8.Rasmussen S. G., DeVree B. T., Zou Y., Kruse A. C., Chung K. Y., Kobilka T. S., Thian F. S., Chae P. S., Pardon E., Calinski D., Mathiesen J. M., Shah S. T., Lyons J. A., Caffrey M., Gellman S. H., Steyaert J., Skiniotis G., Weis W. I., Sunahara R. K., Kobilka B. K. (2011) Crystal structure of the β2 adrenergic receptor-Gs protein complex. Nature 477, 549–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liang Y. L., Khoshouei M., Glukhova A., Furness S. G. B., Zhao P., Clydesdale L., Koole C., Truong T. T., Thal D. M., Lei S., Radjainia M., Danev R., Baumeister W., Wang M. W., Miller L. J., Christopoulos A., Sexton P. M., Wootten D. (2018) Phase-plate cryo-EM structure of a biased agonist-bound human GLP-1 receptor-Gs complex. Nature 555, 121–125 [DOI] [PubMed] [Google Scholar]
- 10.Liang Y. L., Khoshouei M., Radjainia M., Zhang Y., Glukhova A., Tarrasch J., Thal D. M., Furness S. G. B., Christopoulos G., Coudrat T., Danev R., Baumeister W., Miller L. J., Christopoulos A., Kobilka B. K., Wootten D., Skiniotis G., Sexton P. M. (2017) Phase-plate cryo-EM structure of a class B GPCR-G-protein complex. Nature 546, 118–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carpenter B., Nehmé R., Warne T., Leslie A. G., Tate C. G. (2016) Structure of the adenosine A(2A) receptor bound to an engineered G protein. Nature 536, 104–107; erratum: 538, 542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Milligan G., Kostenis E. (2006) Heterotrimeric G-proteins: a short history. Br. J. Pharmacol. 147 (Suppl 1), S46–S55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brown A. J., Goldsworthy S. M., Barnes A. A., Eilert M. M., Tcheang L., Daniels D., Muir A. I., Wigglesworth M. J., Kinghorn I., Fraser N. J., Pike N. B., Strum J. C., Steplewski K. M., Murdock P. R., Holder J. C., Marshall F. H., Szekeres P. G., Wilson S., Ignar D. M., Foord S. M., Wise A., Dowell S. J. (2003) The Orphan G protein-coupled receptors GPR41 and GPR43 are activated by propionate and other short chain carboxylic acids. J. Biol. Chem. 278, 11312–11319 [DOI] [PubMed] [Google Scholar]
- 14.Hirasawa A., Tsumaya K., Awaji T., Katsuma S., Adachi T., Yamada M., Sugimoto Y., Miyazaki S., Tsujimoto G. (2005) Free fatty acids regulate gut incretin glucagon-like peptide-1 secretion through GPR120. Nat. Med. 11, 90–94 [DOI] [PubMed] [Google Scholar]
- 15.Engelstoft M. S., Park W. M., Sakata I., Kristensen L. V., Husted A. S., Osborne-Lawrence S., Piper P. K., Walker A. K., Pedersen M. H., Nøhr M. K., Pan J., Sinz C. J., Carrington P. E., Akiyama T. E., Jones R. M., Tang C., Ahmed K., Offermanns S., Egerod K. L., Zigman J. M., Schwartz T. W. (2013) Seven transmembrane G protein-coupled receptor repertoire of gastric ghrelin cells. Mol. Metab. 2, 376–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bolognini D., Moss C. E., Nilsson K., Petersson A. U., Donnelly I., Sergeev E., König G. M., Kostenis E., Kurowska-Stolarska M., Miller A., Dekker N., Tobin A. B., Milligan G. (2016) A novel allosteric activator of free fatty acid 2 receptor displays unique Gi-functional bias. J. Biol. Chem. 291, 18915–18931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Worzfeld T., Wettschureck N., Offermanns S. (2008) G(12)/G(13)-mediated signalling in mammalian physiology and disease. Trends Pharmacol. Sci. 29, 582–589 [DOI] [PubMed] [Google Scholar]
- 18.Arthofer E., Hot B., Petersen J., Strakova K., Jäger S., Grundmann M., Kostenis E., Gutkind J. S., Schulte G. (2016) WNT stimulation dissociates a frizzled 4 inactive-state complex with Gα12/13. Mol. Pharmacol. 90, 447–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Siehler S. (2009) Regulation of RhoGEF proteins by G12/13-coupled receptors. Br. J. Pharmacol. 158, 41–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Milligan G. (2011) Orthologue selectivity and ligand bias: translating the pharmacology of GPR35. Trends Pharmacol. Sci. 32, 317–325 [DOI] [PubMed] [Google Scholar]
- 21.Maravillas-Montero J. L., Burkhardt A. M., Hevezi P. A., Carnevale C. D., Smit M. J., Zlotnik A. (2015) Cutting edge: GPR35/CXCR8 is the receptor of the mucosal chemokine CXCL17. J. Immunol. 194, 29–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Park S. J., Lee S. J., Nam S. Y., Im D. S. (2018) GPR35 mediates lodoxamide-induced migration inhibitory response but not CXCL17-induced migration stimulatory response in THP-1 cells; is GPR35 a receptor for CXCL17? Br. J. Pharmacol. 175, 154–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Binti Mohd Amir N. A. S., Mackenzie A. E., Jenkins L., Boustani K., Hillier M. C., Tsuchiya T., Milligan G., Pease J. E. (2018) Evidence for the existence of a CXCL17 receptor distinct from GPR35. J. Immunol. 201, 714–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shore D. M., Reggio P. H. (2015) The therapeutic potential of orphan GPCRs, GPR35 and GPR55. Front. Pharmacol. 6, 69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Neetoo-Isseljee Z., MacKenzie A. E., Southern C., Jerman J., McIver E. G., Harries N., Taylor D. L., Milligan G. (2013) High-throughput identification and characterization of novel, species-selective GPR35 agonists. J. Pharmacol. Exp. Ther. 344, 568–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.MacKenzie A. E., Caltabiano G., Kent T. C., Jenkins L., McCallum J. E., Hudson B. D., Nicklin S. A., Fawcett L., Markwick R., Charlton S. J., Milligan G. (2014) The antiallergic mast cell stabilizers lodoxamide and bufrolin as the first high and equipotent agonists of human and rat GPR35. Mol. Pharmacol. 85, 91–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Funke M., Thimm D., Schiedel A. C., Müller C. E. (2013) 8-Benzamidochromen-4-one-2-carboxylic acids: potent and selective agonists for the orphan G protein-coupled receptor GPR35. J. Med. Chem. 56, 5182–5197 [DOI] [PubMed] [Google Scholar]
- 28.Malik R. U., Dysthe M., Ritt M., Sunahara R. K., Sivaramakrishnan S. (2017) ER/K linked GPCR-G protein fusions systematically modulate second messenger response in cells. Sci. Rep. 7, 7749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sivaramakrishnan S., Spudich J. A. (2011) Systematic control of protein interaction using a modular ER/K α-helix linker. Proc. Natl. Acad. Sci. USA 108, 20467–20472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sergeev E., Hansen A. H., Bolognini D., Kawakami K., Kishi T., Aoki J., Ulven T., Inoue A., Hudson B. D., Milligan G. (2017) A single extracellular amino acid in Free Fatty Acid Receptor 2 defines antagonist species selectivity and G protein selection bias. Sci. Rep. 7, 13741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mastop M., Reinhard N. R., Zuconelli C. R., Terwey F., Gadella T. W. J., Jr., van Unen J., Adjobo-Hermans M. J. W., Goedhart J. (2018) A FRET-based biosensor for measuring Gα13 activation in single cells. PLoS One 13, e0193705; erratum: 13, e0195649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jenkins L., Alvarez-Curto E., Campbell K., de Munnik S., Canals M., Schlyer S., Milligan G. (2011) Agonist activation of the G protein-coupled receptor GPR35 involves transmembrane domain III and is transduced via Gα13 and β-arrestin-2. Br. J. Pharmacol. 162, 733–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Inoue A., Ishiguro J., Kitamura H., Arima N., Okutani M., Shuto A., Higashiyama S., Ohwada T., Arai H., Makide K., Aoki J. (2012) TGFα shedding assay: an accurate and versatile method for detecting GPCR activation. Nat. Methods 9, 1021–1029 [DOI] [PubMed] [Google Scholar]
- 34.Alvarez-Curto E., Inoue A., Jenkins L., Raihan S. Z., Prihandoko R., Tobin A. B., Milligan G. (2016) Targeted elimination of G proteins and Arrestins defines their specific contributions to both intensity and duration of G protein-coupled receptor signaling. J. Biol. Chem. 291, 27147–27159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Milligan G., Inoue A. (2018) Genome editing provides new insights into receptor-controlled signalling pathways. Trends Pharmacol. Sci. 39, 481–493 [DOI] [PubMed] [Google Scholar]
- 36.Flock T., Hauser A. S., Lund N., Gloriam D. E., Balaji S., Babu M. M. (2017) Selectivity determinants of GPCR-G-protein binding. Nature 545, 317–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pándy-Szekeres G., Munk C., Tsonkov T. M., Mordalski S., Harpsøe K., Hauser A. S., Bojarski A. J., Gloriam D. E. (2018) GPCRdb in 2018: adding GPCR structure models and ligands. Nucleic Acids Res. 46, D440–D446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jubb H. C., Higueruelo A. P., Ochoa-Montaño B., Pitt W. R., Ascher D. B., Blundell T. L. (2017) Arpeggio: a web server for calculating and visualising interatomic interactions in protein structures. J. Mol. Biol. 429, 365–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Taniguchi Y., Tonai-Kachi H., Shinjo K. (2006) Zaprinast, a well-known cyclic guanosine monophosphate-specific phosphodiesterase inhibitor, is an agonist for GPR35. FEBS Lett. 580, 5003–5008 [DOI] [PubMed] [Google Scholar]
- 40.Semack A., Sandhu M., Malik R. U., Vaidehi N., Sivaramakrishnan S. (2016) Structural elements in the Gαs and Gαq C termini that mediate selective G protein-coupled receptor (GPCR) signaling. J. Biol. Chem. 291, 17929–17940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhao P., Sharir H., Kapur A., Cowan A., Geller E. B., Adler M. W., Seltzman H. H., Reggio P. H., Heynen-Genel S., Sauer M., Chung T. D., Bai Y., Chen W., Caron M. G., Barak L. S., Abood M. E. (2010) Targeting of the orphan receptor GPR35 by pamoic acid: a potent activator of extracellular signal-regulated kinase and β-arrestin2 with antinociceptive activity. Mol. Pharmacol. 78, 560–568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jenkins L., Harries N., Lappin J. E., MacKenzie A. E., Neetoo-Isseljee Z., Southern C., McIver E. G., Nicklin S. A., Taylor D. L., Milligan G. (2012) Antagonists of GPR35 display high species ortholog selectivity and varying modes of action. J. Pharmacol. Exp. Ther. 343, 683–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Berlinguer-Palmini R., Masi A., Narducci R., Cavone L., Maratea D., Cozzi A., Sili M., Moroni F., Mannaioni G. (2013) GPR35 activation reduces Ca2+ transients and contributes to the kynurenic acid-dependent reduction of synaptic activity at CA3-CA1 synapses. PLoS One 8, e82180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tsukahara T., Hamouda N., Utsumi D., Matsumoto K., Amagase K., Kato S. (2017) G protein-coupled receptor 35 contributes to mucosal repair in mice via migration of colonic epithelial cells. Pharmacol. Res. 123, 27–39 [DOI] [PubMed] [Google Scholar]
- 45.Koehl A., Hu H., Maeda S., Zhang Y., Qu Q., Paggi J. M., Latorraca N. R., Hilger D., Dawson R., Matile H., Schertler G. F. X., Granier S., Weis W. I., Dror R. O., Manglik A., Skiniotis G., Kobilka B. K. (2018) Structure of the µ-opioid receptor-Gi protein complex. Nature 558, 547–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Draper-Joyce C. J., Khoshouei M., Thal D. M., Liang Y. L., Nguyen A. T. N., Furness S. G. B., Venugopal H., Baltos J. A., Plitzko J. M., Danev R., Baumeister W., May L. T., Wootten D., Sexton P. M., Glukhova A., Christopoulos A. (2018) Structure of the adenosine-bound human adenosine A1 receptor-Gi complex. Nature 558, 559–563 [DOI] [PubMed] [Google Scholar]
- 47.García-Nafría J., Nehmé R., Edwards P. C., Tate C. G. (2018) Cryo-EM structure of the serotonin 5-HT1B receptor coupled to heterotrimeric Go. Nature 558, 620–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kamato D., Mitra P., Davis F., Osman N., Chaplin R., Cabot P. J., Afroz R., Thomas W., Zheng W., Kaur H., Brimble M., Little P. J. (2017) Gaq proteins: molecular pharmacology and therapeutic potential. Cell. Mol. Life Sci. 74, 1379–1390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jenkins L., Brea J., Smith N. J., Hudson B. D., Reilly G., Bryant N. J., Castro M., Loza M. I., Milligan G. (2010) Identification of novel species-selective agonists of the G-protein-coupled receptor GPR35 that promote recruitment of β-arrestin-2 and activate Gα13. Biochem. J. 432, 451–459 [DOI] [PubMed] [Google Scholar]
- 50.Van Eps N., Altenbach C., Caro L. N., Latorraca N. R., Hollingsworth S. A., Dror R. O., Ernst O. P., Hubbell W. L. (2018) Gi- and Gs-coupled GPCRs show different modes of G-protein binding. Proc. Natl. Acad. Sci. USA 115, 2383–2388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kleinau G., Jaeschke H., Worth C. L., Mueller S., Gonzalez J., Paschke R., Krause G. (2010) Principles and determinants of G-protein coupling by the rhodopsin-like thyrotropin receptor. PLoS One 5, e9745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hauser A. S., Chavali S., Masuho I., Jahn L. J., Martemyanov K. A., Gloriam D. E., Babu M. M. (2018) Pharmacogenomics of GPCR drug targets. Cell 172, 41–54.e19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Forbes S. A., Beare D., Boutselakis H., Bamford S., Bindal N., Tate J., Cole C. G., Ward S., Dawson E., Ponting L., Stefancsik R., Harsha B., Kok C. Y., Jia M., Jubb H., Sondka Z., Thompson S., De T., Campbell P. J. (2017) COSMIC: somatic cancer genetics at high-resolution. Nucleic Acids Res. 45, D777–D783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Guo Y. J., Zhou Y. J., Yang X. L., Shao Z. M., Ou Z. L. (2017) The role and clinical significance of the CXCL17-CXCR8 (GPR35) axis in breast cancer. Biochem. Biophys. Res. Commun. 493, 1159–1167 [DOI] [PubMed] [Google Scholar]