Abstract
Angiogenesis is a critical process in repair of tissue injury that is regulated by a delicate balance between pro- and antiangiogenic factors. In disease states associated with impaired angiogenesis, we identified that miR-135a-3p is rapidly induced and serves as an antiangiogenic microRNA (miRNA) by targeting endothelial cell (EC) p38 signaling in vitro and in vivo. MiR-135a-3p overexpression significantly inhibited EC proliferation, migration, and network tube formation in matrigel, whereas miR-135-3p neutralization had the opposite effects. Mechanistic studies using transcriptomic profiling, bioinformatics, 3′-UTR reporter and miRNA ribonucleoprotein complex -immunoprecipitation assays, and small interfering RNA dependency studies revealed that miR-135a-3p inhibits the p38 signaling pathway in ECs by targeting huntingtin-interacting protein 1 (HIP1). Local delivery of miR-135a-3p inhibitors to wounds of diabetic db/db mice markedly increased angiogenesis, granulation tissue thickness, and wound closure rates, whereas local delivery of miR-135a-3p mimics impaired these effects. Finally, through gain- and loss-of-function studies in human skin organoids as a model of tissue injury, we demonstrated that miR-135a-3p potently modulated p38 signaling and angiogenesis in response to VEGF stimulation by targeting HIP1. These findings establish miR-135a-3p as a pivotal regulator of pathophysiological angiogenesis and tissue repair by targeting a VEGF-HIP1-p38K signaling axis, providing new targets for angiogenic therapy to promote tissue repair.—Icli, B., Wu, W., Ozdemir, D., Li, H., Haemmig, S., Liu, X., Giatsidis, G., Cheng, H. S., Avci, S. N., Kurt, M., Lee, N., Guimaraes, R. B., Manica, A., Marchini, J. F., Rynning, S. E., Risnes, I., Hollan, I., Croce, K., Orgill, D. P., Feinberg, M. W. MicroRNA-135a-3p regulates angiogenesis and tissue repair by targeting p38 signaling in endothelial cells.
Keywords: VEGF, diabetic wounds, human organoid
Angiogenesis, or the formation of new blood vessels from preexisting ones, is a critical component of the dynamic process of tissue repair in ischemic cardiovascular events and in disease states associated with impaired microvascular disease such as diabetes mellitus (DM) (1–3). In response to tissue injury, proangiogenic growth factors stimulate the activation of endothelial cells (ECs) to proliferate, migrate, and generate new primary capillaries from existing ones. Impairment of such processes can result in maladaptive tissue repair in a wide array of clinically relevant disease states such as diabetic wound healing, myocardial infarction (MI), or peripheral artery disease (4). In this context, a better understanding of the angiogenic signaling pathways associated with impaired tissue repair may offer novel therapeutic approaches to improve clinical outcomes.
Growing insight into the role of angiogenic dysfunction associated with poor tissue repair has incited efforts to promote therapeutic angiogenesis to improve cardiovascular outcomes. Over the years, preclinical investigations have demonstrated the capacity of proangiogenic growth factors to generate potent angiogenic responses accompanied by functional recovery in various animal models, such as hindlimb and myocardial ischemia. Therefore, numerous phase I and phase II clinical trials have endeavored to administer proangiogenic growth factors including VEGF, fibroblast growth factor (FGF), hepatocyte growth factor (HGF), hypoxia-inducible factor-1a (HIF-1a), and developmental endothelial locus-1 (Del-1) to patients with peripheral arterial disease or refractory angina in efforts to augment angiogenesis in these subjects (5–8). However, these trials largely failed to demonstrate significant beneficial effects of these interventions (9), and this has been ascribed to a variety of factors including heterogeneity of patient selection, challenges with dosing and delivery, and more stringent diabetic environments. Thus, therapeutic angiogenesis has not been implemented in clinical practice to date (10).
Emerging studies suggest that failures in therapeutic angiogenesis may not be due to a deficiency of these angiogenic growth factors. Indeed, several investigations have demonstrated that levels of circulating growth factors such as VEGF, basic FGF (bFGF), Tie2, and angiopoietin-2 are elevated in patients and animal models of MI and peripheral arterial disease, underscoring that growth factor deficiency is likely not the primary cause of defective angiogenesis in states of ischemia (11–15). Rather, impairment of downstream signaling may be the predominant mechanism governing poor neovascularization in ischemic cardiovascular disease or tissue injury in DM. Canonically, growth factors such as VEGF activate a broad range of pathways including Akt (protein kinase B), endothelial nitric oxide synthase (eNOS), and p38 MAPK to promote EC functions that are necessary for successful angiogenesis. In particular, p38 phosphorylation and signaling have been shown to promote angiogenesis in response to myocardial damage, cardiac remodeling, and peripheral tissue ischemia (16, 17). Collectively, these data suggest that correction of pathways downstream of growth factor activation can rescue proangiogenic responses and improve therapeutic angiogenesis.
MicroRNAs (miRNAs) are evolutionarily conserved, small, single-stranded, noncoding RNAs that are capable of suppressing target gene expression and have emerged as important regulators of the angiogenic response in a variety of pathophysiological processes (18–20). For example, miRNAs such as miR-17-92, -217, -34, and -26a are able to confer antiangiogenic effects, whereas miRNAs such as miR-7-5p, -18a, -31, -155, -201, and -424 can generate proangiogenic responses (21–29). Thus, the identification of relevant miRNAs and their target genes may hold promise in rescuing the impaired angiogenic signaling that characterizes pathologic tissue repair and ischemic disease states.
In this report, we identify miR-135a-3p as a novel, antiangiogenic regulator of VEGF-p38 signaling in ECs by targeting huntingtin-interacting protein 1 (HIP1). Neutralization of miR-135a-3p rapidly induced angiogenesis and improved diabetic wound healing in mice and human skin organoids, whereas miR-135a overexpression impaired these effects. Together, these findings reveal a novel therapeutic strategy to overcome impaired angiogenic signaling and for rapidly stimulating angiogenesis that may be applicable for maladaptive tissue reparative states such as diabetes or ischemic cardiovascular disease.
MATERIALS AND METHODS
Cell culture and transfection
HUVECs were passaged <5 times and were used for all experiments. For transfection studies, HUVECs were cultured overnight before being transfected with Lipofectamine 2000 Transfection Reagent (Thermo Fisher Scientific, Waltham, MA, USA). For reporter studies, HUVECs were transfected with 400 ng of the indicated reporter constructs and 200 ng β-galactosidase expression plasmids. Each reading of luciferase activity was normalized to the β-galactosidase activity or total protein read for the same lysate.
Real-time quantitative PCR
HUVECs [stimulated in the presence or absence of VEGF (25 ng/ml), bFGF (25 ng/ml), thrombospondin (TSP)1 (0.1, 1, 10 μg/ml), TSP2 (0.1, 1, 10 μg/ml), or MC1568 (10 ng/ml)], mouse tissues, and human organoids were suspended in Trizol reagent (Thermo Fisher Scientific) and total RNA and miRNA were isolated per manufacturers’ instructions. Human skin and plasma were isolated using the Total RNA Purification Kit (Norgen Biotek, Thorold, ON, Canada) per manufacturers’ instructions. Reverse transcriptions were performed by using miScript Reverse Transcription Kit from Qiagen (Germantown, MD, USA) (218061). Either QuantiTect SYBR Green RT-PCR Kit (204243) or miScript SYBR Green PCR Kit (218073) from Qiagen was used for real-time quantitative PCR (qPCR) analysis with the AriaMx RT-PCR System (Agilent Technologies, Santa Clara, CA, USA) following the manufacturer’s instructions. Gene-specific primers were used to detect human and mouse HIP1. Samples were normalized to endogenous human and mouse glyceraldehyde 3-phosphate dehydrogenase (GAPDH). To amplify mature miRNA sequences, miScript primer assays for Hs_miR-135a-3p_1 (MS00042259) from Qiagen were utilized. The samples were normalized to endogenous 5S RNA. The fold changes were calculated by ΔΔCt method. Significance was determined by a 2-tailed Student’s t test, P < 0.05. The primer sequences for HIP1 were 5′-CGAGCAGTTCGACAAGACCC-3′ (forward) and 5′-GTGTGCCCAGAATGATGCG-3′ (reverse).
Western blot analysis
HUVECs transfected with miR-135a-3p mimic, miR-135a-3p inhibitor, nonspecific (NS) miRNA controls, HIP1 small interfering RNA (siRNA), or control siRNA were cultured for 72 h. Mouse tissue was harvested per experimental protocol and homogenized at 30/s for 5 min using Tissuelyser II (Qiagen). Human organoids transfected with miR-135a-3p mimic, miR-135a-3p inhibitor, or NS miRNA controls were cultured for 72 h, treated with VEGF (50 ng/ml) accordingly, and homogenized at 30/s for 5 min. Total cellular protein was isolated by RIPA buffer (50 mM Tris-HCL pH 7.4, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS) supplemented with protease inhibitors (Roche, Basel, Switzerland). Cell or tissue debris was removed by centrifugation at 12,500 rpm for 15 min. Protein quantification was performed using a BCA kit (Thermo Fisher Scientific). Lysates were separated using 5–14% Mini-Protean TGX Precast Gels (Bio-Rad, Hercules, CA, USA), transferred to PVDF membranes, and subjected to Western blotting using antibodies against HIP1 from Proteintech; and P-Akt, Pan Akt, P-p44/42 MAPK, P44/42 MAPK, P-eNOS, eNOS, P-p38 MAPK, p38 MAPK, GADPH, and β-actin from Cell Signaling Technology (Danvers, MA, USA). Horseradish peroxidase-conjugated goat anti-rabbit or mouse antibody (Santa Cruz Biotechnology, Dallas, TX, USA) was used at 1:2000 dilution. ECL assay was performed per manufacturers’ instructions (RPN2132; GE Healthcare, Waukesha, WI, USA). Three or more biologic replicates were performed for each experiment. Significance was determined by a 2-tailed Student’s t test, P < 0.05.
Tube-like network formation on matrigel (in vitro)
Matrigel (BD Biosciences, San Jose, CA, USA) basement membrane matrix was added to 96-well culture plates and incubated at 37°C until gelation occurred. Matrigel was not supplemented with additional growth factors. Network tube formation was assessed 6–10 h postplating and quantitated by counting the number of tubes formed per high power field as previously described (29–31).
Chemotaxis assays
Migration assay was performed using ChemTX Multiwell System (Neuro Probe, Gaithersburg, MD, USA). The number of cells migrating to the lower chamber (EC growth medium supplemented with 50 ng/ml VEGF or bFGF as indicated) was counted using a hemocytometer after 6–8 h (32).
Bromodeoxyuridine assay
HUVECs transfected with miR-135-3p mimic, miR-135-3p inhibitor, or NS negative controls were cultured for 5 d. Cell proliferation was measured using the bromodeoxyuridine (BrdU) ELISA assay (Roche) as described by the manufacturer.
MicroRNA ribonucleoprotein complex immunoprecipitation
MiRNA ribonucleoprotein complex (miRNP) -immunoprecipitation (MiRNP-IP) was performed as previously described (33). Briefly, Myc-tagged Ago-2 (a kind gift from G. Hannon, Cold Spring Harbor, NY, USA) was cotransfected with either miR-615-5p or miRNA negative control in HUVECs. Cells were washed in ice cold PBS, released by scraping, and lysed in buffer (10 mM Tris-HCl pH 7.5, 10 mM NaCl, 2 mM EDTA, 0.5% Triton X-100, 100 U/ml of RNasin Plus (Promega, Madison, WI, USA) supplemented with 1 time protease inhibitor (Roche). The lysed cell solution was adjusted to a final NaCl concentration of 150 mM prior to centrifugation. One-twentieth of the supernatant volume was collected in Trizol for use as an extract control. The remaining portion of the supernatant was precleared with Protein A/G UltraLink Resin (Pierce, Rockford, IL, USA), to which 2 μg anti-c-myc antibody was added and the mixture was allowed to incubate overnight at 4°C; the following day Protein A/G UltraLink Resin was added. After 4 h of mechanical rotation at 4°C, the agarose beads were pelleted and washed 4 times in wash buffer (50 mM Tris-HCl pH 7.5, 150 mM NaCl, 0.05% Triton X-100). Finally, 1 ml of Trizol was added into the beads and RNA was isolated. Total RNA was reverse transcribed into cDNA for real-time qPCR analysis.
Scratch assay for EC migration
Wound healing assays was performed using Culture-Insert 2-Well 35-mm µ-Dishes (Ibidi). HUVECs transfected with miR-135a-3p mimic, miR-135a-3p inhibitor, NS miRNA controls, HIP1 siRNA, or control siRNA were cultured for 60 h in 12-well plates and plated at 21 K cells per well into the µ-Dishes. Inserts were lifted at 72 h after transfection, and cells were imaged using an Eclipse TE2000-U inverted microscope (Nikon, Tokyo, Japan) at 2 and 4 times over time to assess for wound closure. Three technical replicates were performed per condition.
In vivo miR-135a-3p inhibition or overexpression and mouse experiments
Animal protocols were approved by the Laboratory Animal Care at Harvard Medical School and Brigham and Women’s Hospital (BWH). For mouse dermal wound studies, male, 8–10 wk old, db/db mice (The Jackson Laboratory, Bar Harbor, ME, USA) were used for local intradermal injections of either scrambled control LNA-anti-miR or LNA-anti-miR-135-3p (Exiqon, Seoul, South Korea) at 0.63 mg/kg 48 and 24 h prior to surgery. On d 0, dorsal full-thickness skin wounds (1 cm2) were generated and covered with semiocclusive dressing (Tegaderm). Images of the wounds were immediately acquired after surgery (d 0) and on d 9 following the removal of the Tegaderm dressing. Mice were euthanized 9 d after surgery and the 1 × 1-cm2 sections of skin surrounding the wound were excised down to fascia. Angiogenesis in wounds was analyzed by mouse CD31 staining (DIA-310; Dianova, Pine Bush, NY, USA) and DAPI (H21492; Thermo Fisher Scientific) of the paraffin-embedded wound sections. Goat Anti-Mouse IgG H&L (Alexa Fluor 488; 150113; Abcam, Cambridge, MA, USA) was used as secondary antibody. Relative CD31 expression was measured in the wound edge and quantified using ImageJ (Bethesda, MD, USA) (28). Granulation tissue thickness was measured on d 9 using hematoxylin and eosin–stained sections obtained from the center of the wound. Granulation tissue thickness is defined as the distance of intact tissue from the bottom of the epidermis to the top of the subcutaneous fat layer and will be quantified using ImageJ. Fluorescent images were acquired by the Olympus Fluoview FV1000 confocal microscope.
Human plasma and skin samples
BWH cohort
We examined EDTA plasma samples from prospectively enrolled patients that underwent cardiac catheterization at BWH. Plasma was isolated from whole blood. Control patients were defined as without clinically significant coronary atherosclerosis (<20% stenosis in any epicardial coronary artery determined by angiography) and had no elevation of cardiac biomarkers. Patients with acute coronary syndrome (ACS) were defined as acute atherothrombotic coronary artery occlusion resulting in either a non-ST-elevation MI (with >70% occlusion of an epicardial artery) or an ST-elevation MI (complete occlusion of an epicardial coronary artery determined by angiography) with elevation of cardiac biomarkers. The study was approved by the Institutional Review Board–approved protocol at BWH. Written informed consent was obtained from participants or their appropriate surrogates. Anonymized plasma samples were generated from blood collected in EDTA-containing tubes at the time of the procedure and stored at −80°C. Total RNA was isolated from plasma using the Total RNA Purification Kit from Norgen Biotek and reverse transcription and real-time qPCR was performed as previously described.
Fering Heart Biopsy Study 2
We examined EDTA plasma and skin specimens (taken from thoracic surgical incisions) from nondiabetic and diabetic patients included in the Fering Heart Biopsy Study 2 biobank. Fering Heart Biopsy Study 2 enrolled prospectively consecutive patients undergoing coronary artery bypass surgery in the Fering Heart Clinic in Norway between 2011 and 2017. Diabetes was defined as diabetes diagnosis registered in medical records. The study was approved by the Regional Ethics Committee in Norway. Written informed consent was obtained from participants or their appropriate surrogates. Skin samples were preserved in Allprotect tissue reagent (Qiagen) and stored at −80°C. Later, they were homogenized for real-time qPCR analyses. Plasma samples were generated from fasting blood collected in EDTA-containing containers before coronary artery bypass grafting, and stored at small aliquots at −80°C. No thawing of the specimens was allowed prior to the actual RNA analyses.
Generation of human skin organoids
Full-thickness circular (6-mm) human skin organoids were taken from surgical skin samples, and 3-mm full-thickness wounds were created as previously described (34). Briefly, human skin organoids were then embedded in collagen I matrix and maintained in DMEM (Thermo Fisher Scientific) supplemented with 10-mm 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid, 50 μg/ml ascorbic acid, 100 μm adenine, 0.5 μm hydrocortisone, 0.1 nm cholera toxin, 100 μU/ml penicillin, and 10 μg/ml streptomycin (MilliporeSigma, Burlington, MA, USA). The ex vivo organ cultures were cultured at the air-liquid interface and maintained in the cell culture incubator at 37°C with 5% CO2. The media were changed every other day. The viability of cultured explants was validated by histologic evaluation. Human skin organoids were transfected at the indicated time points using 30 nM miR-135a-3p mimics (miR-135-3pm) or NS controls or 100 nM miR-135a-3p inhibitors (miR-135-3pi) or NS controls. Neovascularization was measured at d 3 and 7 to determine miR-135a-3p effects on wound healing. Angiogenesis in human paraffin embedded sections was analyzed by CD31 staining (28364; Abcam) and DAPI (H21492; Thermo Fisher Scientific). Alexa 647 conjugated donkey anti-rabbit antibody was used for secondary antibody (711-605-152; Jackson ImmunoResearch Laboratories, West Grove, PA, USA). Relative CD31 expression was measured in the entire cross section of the human organoid and quantified using ImageJ (28).
Statistical analysis
Data are presented as means ± sem. All in vitro and in vivo experiments are representative of 3 independent experiments unless indicated otherwise. Sample sizes for mouse and human organoid studies were chosen based upon pilot studies or similar well-characterized studies in the literature. There were no inclusion or exclusion criteria used. Data were subjected to an unpaired, 2-sided Student’s t test or 1-way ANOVA with Bonferroni correction for multiple comparisons, and P < 0.05 was considered statistically significant.
RESULTS
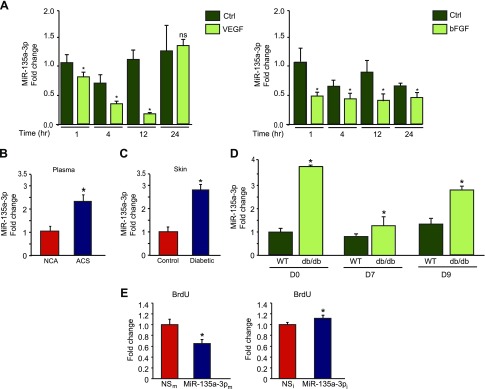
We undertook an miRNA microarray profiling study to identify miRNAs that regulate angiogenesis in response to tissue injury. Using plasma from human subjects with ACS with coronary angiograms bearing >70% stenotic lesions compared with non-ACS human subjects with coronary angiograms with lesions <20% stenosis, we identified an miRNA, miR-135a-3p, with significantly increased expression compared to controls (data not shown). Furthermore, miR-135-3p was highly expressed in ECs compared to several other miRNAs identified from this sample set (Supplemental Fig. S1A), prompting further investigation of this miRNA. In response to proangiogenic stimuli VEGF and bFGF, miR-135a-3p expression was significantly reduced by VEGF (25 ng/ml) and bFGF (25 ng/ml) over 24 h (Fig. 1A), whereas antiangiogenic stimuli such as TSP1 and TSP2 significantly increased miR-135-3p expression (Supplemental Fig. S1B). Costimulation of ECs with VEGF and TSP1 resulted in the loss of VEGF-mediated suppression of miR-135-3p expression (Supplemental Fig. S1C). VEGF is known to repress target genes via a histone deacetylase (HDAC) class IIa mechanism (35, 36). Pretreatment of ECs with the HDAC Class IIa inhibitor MC-1568 blocked the VEGF repression of miR-135-3p, suggesting that VEGF-mediated repression of miR-135-3p was mediated in part by HDAC Class IIa (Supplemental Fig. S1D). Using a larger cohort of human subjects with ACS and coronary angiograms bearing >70% stenotic lesions compared to non-ACS human subjects with coronary angiograms with lesions <20% stenosis, we verified that expression of miR-135a-3p was increased by 2.4 fold (Fig. 1B). In addition, circulating levels of miR-135a-3p were increased by 2.8-fold in the skin of patients with DM (Fig. 1C). We also examined miR-135a-3p expression in response to tissue injury from dermal wounds generated by punch biopsy of diabetic db/db mice. MiR-135a-3p expression increased up to 3.8-fold higher in db/db mice compared to wild-type mice over the course of 9 d postwounding (Fig. 1D). Together, these data suggest that miR-135a-3p expression is differentially regulated by pro- and antiangiogenic stimuli in ECs and maladaptive tissue reparative states such as diabetes or ischemic cardiovascular disease, thereby providing the rationale that targeting this miRNA may facilitate the induction of angiogenesis.
Figure 1.
MiR-135a-3p is regulated by proangiogenic stimuli and inhibits EC growth. A) Real-time qPCR analysis of miR-135-3p expression in response to VEGF (25 ng/ml) and bFGF (25 ng/ml) in HUVECs. *P < 0.01 compared to control (Ctrl). B) Circulating miR-135a-3p levels are increased in plasma from human subjects with ACS (n = 20) compared to subjects with normal coronary angiograms (NCA) (n = 40). *P < 0.05 compared to normal coronary angiogram. C) MiR-135a-3p expression is increased in skin of patients with diabetes (n = 13) compared to nondiabetic controls (n = 10). *P < 0.05 compared to controls. D) Wild-type (WT) and db/db mice underwent punch-biopsy wounding of the dorsal skin, and wounds were collected for qPCR analyses for miR-135a-3p on the indicated days after wounding. E) HUVECs transfected with NSm, miR-135a-3pm, miR inhibitor negative control (NSi), or miR-135a-3p inhibitor (miR-135a-3pi) were subjected to BrdU cell proliferation assay. Ns, nonsignificant. All data represent means ± sem. *P < 0.05 compared to controls.
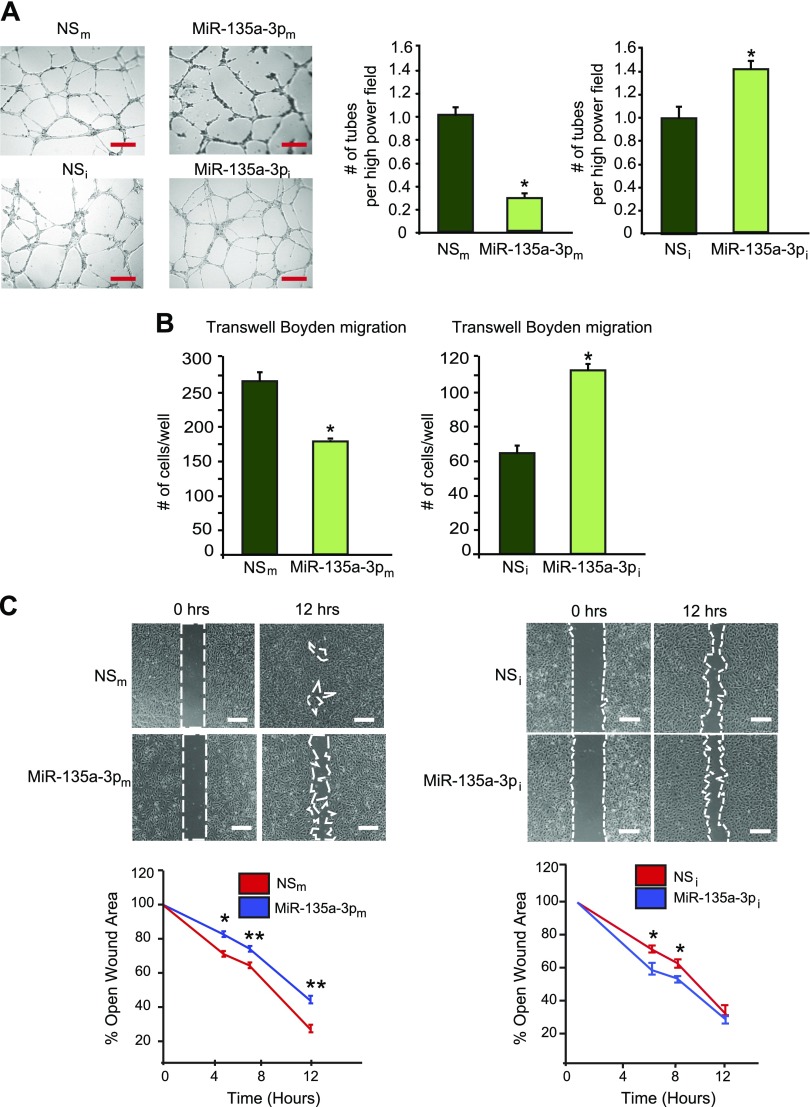
To gain insights into the significance of miR-135a-3p on endothelial angiogenic functions, we performed gain- and loss-of-function experiments. As shown in Fig. 1E, overexpression of miR-135-3pm in HUVECs inhibited cell growth by 54%, whereas miR-135-3pi (complementary antagonist) increased EC growth by 17%. Expression of miR-135-3p in ECs in response to miR-135-3p overexpression or inhibition was verified by RT-qPCR (Supplemental Fig. S1E, F). In addition, vascular network formation assays in matrigel showed that overexpression of miR-135a-3p inhibited network tube formation (Fig. 2A, top) by 68%, whereas miR-135a-3p inhibition significantly increased tube formation in matrigel in vitro by 40% (Fig. 2A, bottom). Similarly, overexpression of miR-135a-3p decreased EC migration in response to the proangiogenic stimulus VEGF (50 ng/ml) by 38%, compared to the NS control, whereas miR-135a-3p inhibition potently increased migration by 69% in response to VEGF compared to the NS control (Fig. 2B). Interestingly, whereas bFGF (50 ng/ml) only modestly increased EC migration by ∼10%, overexpression of miR-135-3p significantly inhibited the bFGF-induced EC migration (Supplemental Fig. S1G). Finally, similar to its regulation of EC growth and migration, overexpression of miR-135a-3p decreased wound closure in EC scratch assays, whereas inhibition of miR-135a-3p promoted wound closure compared to the NS control (Fig. 2D). Taken together, these data indicate that miR-135a-3p inhibited EC angiogenic functions in vitro.
Figure 2.
MiR-135a-3p inhibits proangiogenic functions in ECs in vitro. HUVECs transfected with NSm, miR-135a-3pm, miR inhibitor negative control (NSi), or miR-135a-3p inhibitor (miR-135a-3pi) were subjected to tube-like network formation in matrigel (A); EC migration in transwell Boyden chambers (B); scratch assay (C); n = 6/group. All data represent means ± sem. Scale bars: 150 µm (A); 100 µm (C). *P < 0.05 compared to NS, **P < 0.001 compared to NS.
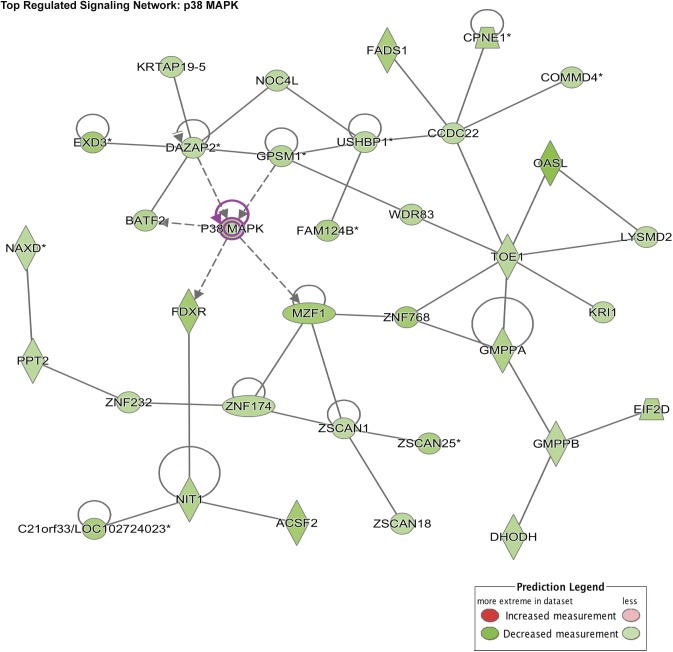
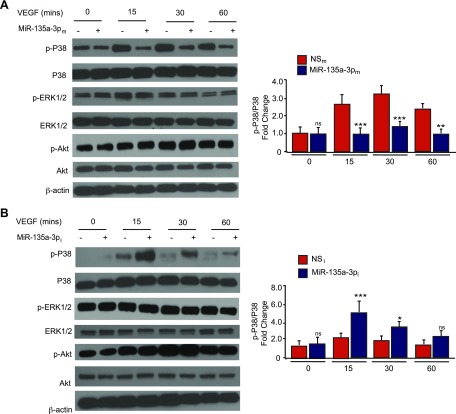
To identify potential target signaling pathways of miR-135a-3p, we performed a microarray gene chip profiling approach from HUVECs overexpressing miR-135a-3p followed by gene ontology (GO) analyses. Interestingly, cellular growth and proliferation was predicted to be within the top 10 biologic pathways to be regulated by miR-135a-3p (Fig. 3), supporting our findings in EC angiogenic functional assays (Figs. 1E and 2). In addition, p38 was identified as the top regulated signaling pathway by miR-135a-3p in ECs (Tables 1 and 2). Because of the relatively modest effects of bFGF compared to VEGF in EC functional assays (Supplemental Fig. S1G), we used VEGF as the prototypical treatment for subsequent EC signaling pathways. In alignment with the GO analyses, miR-135a-3p overexpression potently reduced p38 phosphorylation by 40 and 62% in response to 15 and 30 min, respectively, of VEGF (50 ng/ml) treatment in HUVECs (Fig. 4A), whereas miR-135a-3p inhibition increased p38 phosphorylation by 40% under basal conditions and up to 2.2 fold in response to 15, 30, and 60 min of VEGF (50 ng/ml) stimulation (Fig. 4B). Furthermore, this miR-135a-3p regulation was specific to p38 signaling and not other signaling pathways including Akt and ERK1/2 (Fig. 4; Supplemental Fig. S1).
Figure 3.
Bioinformatics and miR-135a-3p gene profiling predicts p38 as a targeted signaling pathway. Gene ontology and GSEA predicted p38 MAPK signaling pathway to be the top regulated signaling network regulated by miR-135a-3p.
TABLE 1.
Gene ontology analysis of 1687 genes repressed by miR-135a-3p overexpression in endothelial cells identified from transcriptomic profiling
| GO ID | Molecular and cellular function | P | Gene (n) |
|---|---|---|---|
| GO:000628194 | DNA replication and repair | 3.06E−05 | 94 |
| GO:0007049 | Cell cycle | 2.03E−04 | 186 |
| GO:0008219 | Cell death and survival | 3.13E−04 | 262 |
| GO:0008219 | Cellular development | 3.54E−04 | 21 |
| GO:0009986 | Cell surface | 6.33E−04 | 183 |
| GO:0008283 | Cellular growth and proliferation | 1.34E−03 | 16 |
| GO:0048869 | Molecular transport | 1.38E−03 | 74 |
| GO:0007267 | Cell-to-cell signaling interaction | 2.21E−03 | 42 |
| GO:0016043 | Cellular assembly and organization | 3.17E−03 | 76 |
| GO:0006928 | Cellular movement | 3.17E−03 | 24 |
TABLE 2.
The p38 MAPK signaling pathway is predicted to be the top regulated signaling network regulated by miR-135a-3p from gene set enrichment analyses
| Signaling network | Score | Gene (n) |
|---|---|---|
| p38 MAPK | 42 | 34 |
| FAK signaling | 42 | 34 |
| GRK2 | 42 | 34 |
| VEGF | 37 | 32 |
| Pdi | 37 | 32 |
| NF-кB | 35 | 31 |
| JNK | 35 | 31 |
| GRB2 | 35 | 31 |
| Smad1/5/8 | 35 | 31 |
| PDGF | 35 | 31 |
FAK, focal adhesion kinase; GRB2, growth factor receptor-bound protein 2; GRK2, G protein-coupled receptor kinase; Pdi, protein disulfide isomerase; PDGF, platelet-derived growth factor.
Figure 4.
MiR-135a-3p regulates the expression of downstream P38 signaling in ECs. HUVECs transfected with NSm or miR-135a-3pm (A) or miR inhibitor negative control (NSi) or miR-135a-3p inhibitor (miR-135a-3pi) (B), and stimulated with VEGF (50 ng/ml) at the given time points were subjected to Western analysis using antibodies to p-p38, p38, p-ERK1/2, ERK1/2, p-Akt, Akt, and β-actin (n = 3–5 experiments). Ns, nonsignificant. All data represent means ± sem. Differences among groups were analyzed by using 1-way ANOVA. *P < 0.05, **P < 0.005, ***P < 0.001 compared to controls.
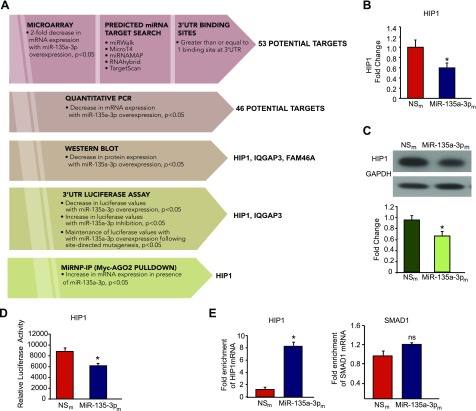
Using a step-wise approach involving a combination of bioinformatics and prediction algorithms (e.g., miRWalk, Micro T4, miRNAMAP, RNAhybrid, and TargetScan), transcriptomic profiling, 3′-UTR reporter assays, and miRNP-IP studies, we sought to identify and validate miR-135a-3p target genes (Fig. 5A). Of the 53 potential target genes that were reduced on microarray profiling by at least 2-fold and also contained at least one potential binding site in the 3′-UTR, 46 genes showed significantly decreased mRNA expression in HUVECs overexpressing miR-135a-3p (Fig. 5A). Seven genes could not be detected by RT-qPCR (data not shown). Of these, overexpression of miR-135a-3p in HUVECs subsequently decreased the mRNA and protein expression (Fig. 5B, C) and 3′-UTR activity (Fig. 5D) of 1 gene – HIP1. In addition, deletion of the miR-135a-3p binding site on the HIP1 3′UTR blocked the ability of miR-135-3p to repress the activity of this 3′UTR reporter (Supplemental Fig. S2). Next, we performed Argonaute2 (AGO2) miRNP-IP studies to assess whether HIP1 mRNA is enriched in the RNA-induced silencing complex following miR-135a-3p overexpression in HUVECs. An ∼8-fold enrichment of HIP1 mRNA was observed after AGO2 miRNP-IP in the presence of miR-135a-3p, as compared with the miRNA negative control. AGO2 miRNP-IP did not enrich the mRNA for SMAD1, a gene that was not predicted to be a miR-135a-3p target (Fig. 5E).
Figure 5.
HIP1 is a bona fide target of miR-135a-3p in ECs. A) Discovery and validation of miR-135a-3p target genes. HUVECs transfected with NSm and miR-135a-3pm were subjected to microarray gene profiling. Potential gene targets were further narrowed down by sequential use of bioinformatics and prediction algorithms, real-time qPCR, Western blot analyses, 3′-UTR reporter studies, and miRNP-IP analysis. B, C) HUVECs transfected with NSm or miR-135a-3pm were subjected to real-time qPCR for HIP1 expression (B) or Western blot analyses using antibodies to HIP1 and GAPDH (n = 3 experiments) (C). D) Luciferase activity of HIP1 3′-UTR normalized to total protein was quantified in HUVECs transfected with NSm or miR-135a-3pm (n = 3 experiments). E) MiRNP-IP analysis of enrichment of HIP1 mRNA in HUVECs transfected with NSm or miR-135a-3pm. RT-qPCR was performed to detect HIP1 or SMAD1. *P < 0.01. ns, nonsignificant. Results are representative of n = 3 replicates/group and 2 independent experiments. All data represent means ± sem.
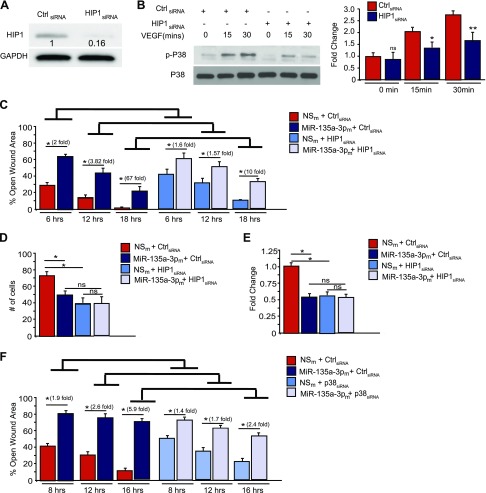
To further investigate the functionality of these 2 target genes in ECs, we undertook an siRNA silencing approach of HIP1 (Fig. 6A). HIP1 siRNA-mediated knockdown successfully “phenocopied” the effects of miR-135a-3p overexpression on p38 phosphorylation (Fig. 6B) and on EC migration using modified Boyden migration chambers (Supplemental Fig. S3A) and EC scratch assays (Supplemental Fig. S3B). To explore whether the miR-135a-3p-mediated inhibitory effects on EC proliferation were dependent on HIP1, we performed siRNA knockdown studies using EC scratch assays in the presence and absence of miR-135-3pm and quantified EC wound closure. In the absence of HIP1, miR-135a-3p overexpression had markedly impaired ability to inhibit EC wound closure [Fig. 6C: miR-135-3pm+ control siRNA group vs. miR-135-3pm + HIP1siRNA group at 6 h (2- vs. 1.6-fold, respectively); at 12 h (3.82- vs. 1.57-fold, respectively); and at 18 h (67- vs. 10-fold, respectively)]. Because the siRNA-mediated knockdown of the target gene HIP1 is significantly more efficient (Fig. 6A, ∼85% knockdown) compared to miRNA-135-3p mimic overexpression (Fig. 5C, ∼30%), we would anticipate that the miR negative control (NSm) + HIP1 siRNA group would have higher open wound areas compared to the NSm + control siRNA group—and indeed this is the case as quantified: [NSm + HIP1 siRNA group vs. NSm + control siRNA group at 6 h (41 vs. 28%, respectively), 12 h (32 vs. 13%, respectively), and 18 h (11 vs. 4%, respectively)]. Thus, these data highlight that the miR-135a-3p-mediated effects are dependent in part on HIP1 for EC wound closure in scratch assays. To investigate for miR-135-3p dependency upon HIP1 in other EC functional assays, we explored EC proliferation (BrdU) and migration studies. As demonstrated in Fig. 6D, the ability of miR-135-3pm to inhibit EC proliferation as quantified by BrdU was significantly impaired (NSm+control siRNA group vs. miR-135-3pm + HIP1 siRNA group: 34 vs. 1.1%, respectively). Similarly, the ability of miR-135-3pm to inhibit EC migration as quantified by transwell Boyden chamber assays was also significantly impaired (Fig. 6E: NSm+control siRNA group vs. miR-135-3pm + HIP1 siRNA group: 48 vs. 2%, respectively). These studies highlight, the functional importance of this target. Moreover, to assess for miR-135a-3p dependent effects on p38K signaling, we also performed siRNA knockdown studies for p38K in EC scratch assays. As shown in Fig. 6F, compared to siRNA control, the ability of miR-135a-3p to inhibit wound closure was significantly attenuated in the presence of p38K siRNA knockdown (miR-135-3pm+ control siRNA group vs. miR-135-3pm + p38 siRNA group at 6 h (1.9- vs. 1.4-fold, respectively); at 12 h (2.6- vs. 1.7-fold, respectively); and at 18 h (5.9- vs. 2.4-fold, respectively). Collectively, these data indicate that HIP1 is a direct target of miR-135-3p in ECs where increased levels of miR-135a-3p facilitates reduced HIP1 expression and p38K signaling, an effect restraining EC growth and angiogenesis.
Figure 6.
SiRNA-mediated knockdown of HIP1 recapitulates miR-135-3p functional effects in ECs. A, B) HUVECs were transfected with siRNA to HIP1 or scrambled control (Ctrl) siRNA. Protein expression was determined by Western analysis under baseline conditions (A) or in response to VEGF (50 ng/ml) treatment (B) using antibodies to HIP1, p-P38, P38, and GAPDH (n = 2 experiments). *P < 0.05, **P < 0.001. C–E) HUVECs were transfected with siRNA to HIP1 or scrambled Ctrl siRNA in the presence of NSm or miR-135a-3pm. EC functional angiogenic assays were performed for scratch assay (C), Boyden transwell migration (D), or proliferation by BrdU assay (E). *P < 0.01. F) HUVECs were transfected with siRNA to p38K or siRNA control in the presence of NSm or miR-135a-3pm and EC scratch assays were performed. Ns, nonsignificant. Results are representative of n = 3 replicates/group. All data represent means ± sem. Differences among groups were analyzed by using 1-way ANOVA. *P < 0.01.
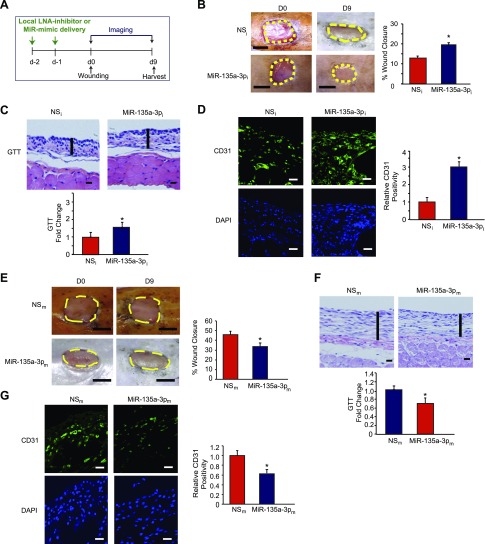
Diabetes is a complex disease and impairs multiple components of wound healing including inflammation, matrix deposition, and angiogenesis (28, 37). Impaired angiogenesis is a hallmark in the pathogenesis of such wounds associated with significant complications such as limb amputations (38–41). MiR-135a-3p inhibition promoted various aspects of EC angiogenesis in vitro (Figs. 1E and 2). Therefore, we explored the effect of inhibiting miR-135a-3p on angiogenesis in a diabetic db/db model of dermal wound healing generated by punch biopsy of the skin on the dorsal suface of the mice (Fig. 7A). Local delivery of LNA-anti-miR-135a-3p (MiR-135a-3pi) not only significantly improved wound closure (Fig. 7B) but also increased granulation tissue thickness (GTT) by 1.6-fold (Fig. 7C) and robustly induced angiogenesis as measured by CD31 by 3-fold (Fig. 7D) compared to scrambled NS control anti-miRs (NSi). In contrast, overexpression of miR-135a-3p significantly delayed wound closure by 61% (Fig. 7E), decreased GTT by 32% (Fig. 7F), and reduced angiogenesis in wounds by 37% compared to mice that received local intradermal delivery of control LNA-anti-miRs (Fig. 7G). Thus, targeting miR-135a-3p induced angiogenesis and wound healing under diabetic conditions.
Figure 7.
Local delivery of LNA-anti-miR-135a-3p promotes wound healing in db/db mice. A) After 2 local injections in mice of LNA-anti-miR-135a-3p (MiR-135a-3pi) or scrambled NS control LNA-anti-miRs (NSi) (n = 11–12/group), mice underwent dorsal skin wounding. B–D) Wound analyses included: wound closure areas (B), GTT (C), and confocal immunofluorescence staining for CD31 (D). E–G) After 2 local injections in mice of MiR-135a-3 PM or scrambled NS control miRs (NSm) (n = 10/group), mice underwent dorsal skin wounding. Wound analyses included: wound closure areas (E), GTT (F), and confocal immunofluorescence staining (G) for CD31. All data represent means ± sem. Scale bars: 5 mm (B, E); 500 μm (C, F); 50 μm (D, G). *P < 0.05.
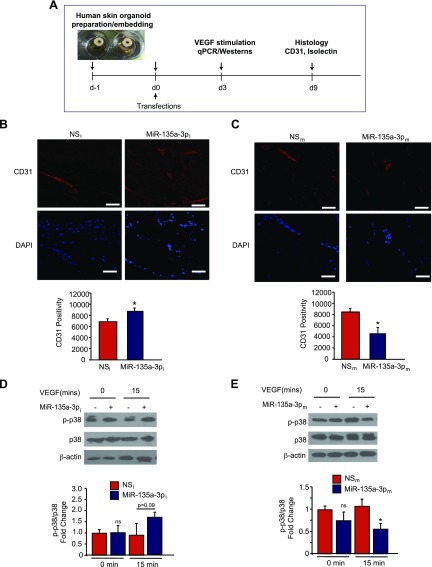
In order to evaluate whether neutralization of miR-135a-3p regulates angiogenesis in response to tissue injury in human tissues, we developed a modified human skin organoid assay. The model system consists of a 6-mm circular full-thickness punch biopsy where in the middle of each biopsy a 3-mm full-thickness wound is created using a 3-mm punch biopsy. The human skin organoid can be maintained at the air-liquid interface in culture for several days (Fig. 8A) (34). We used an NS-Cy3 miRNA control and verified transduction of human skin organoids (data not shown). To study the ability of miR-135a-3p to regulate angiogenesis in human skin organoids, we transduced human skin organoids with LNA-anti-miR-135a-3p (MiR-135a-3pi) or scrambled NS control anti-miRs (NSi) or MiR-135a-3pm or scrambled NS control anti-miRs (NSm) (Fig. 8A). In accordance with our in-vivo findings in mice, inhibition of miR-135a-3p induced angiogenesis as measured by CD31 by ∼33% (Fig. 8B) in human skin organoids, whereas overexpression of miR-135a-3p reduced angiogenesis by ∼57% (Fig. 8C). Additionally, inhibition of miR-135a-3p showed a nonsignificant increased trend (P = 0.09) in p38 phosphorylation in response to 15 mins of VEGF (50 ng/ml) stimulation by ∼1.7 fold (Fig. 8D). In contrast, overexpression of miR-135a-3p significantly decreased p38 phosphorylation in response to 15 mins of VEGF stimulation by 47% in human skin organoids (Fig. 8E).
Figure 8.
Inhibition of miR-135a-3p promotes angiogenesis in human skin organoids. A) Punch biopsies of human skin were embedded into a collagen matrix, transfected with miR inhibitor negative control (NSi), miR-135a-3p inhibitor (miR-135a-5pi), NSm, or miR-135a-3pm, and cultured for indicated number of days. B, C) Human skin organoids were transduced with the indicated miRNAs and cultured for 9 d followed by confocal immunofluorescence staining for CD31. D, E) Human skin organoids (n = 3–6) were transduced with the indicated miRNAs and cultured for 3 d. Human skin organoids were treated with VEGF (50 ng/ml) followed by Western blot analyses for p-p38, p38, and β-actin at the indicated times. All data represent means ± sem. Scale bars: 50 μm (B); 25 μm (C). *P < 0.05 (1-way ANOVA).
Finally, we investigated the effects of miR-135a-3p inhibition or overexpression on HIP1 in human skin organoids. Similar to our in-vitro findings in ECs, overexpression of miR-135a-3p in human skin organoids potently decreased HIP1 mRNA (Supplemental Fig. S4A, left) and protein expression (Supplemental Fig. S4B, left), whereas the miR-135a-3p inhibitor increased both HIP1 mRNA and protein expression (Supplemental Fig. S4A, B right). In summary, our in-vitro and in-vivo findings indicate that increased miR-135a-3p expression adversely affects angiogenesis in response to tissue injury, and its neutralization can potently promote angiogenesis in mice and human tissues.
DISCUSSION
Angiogenesis is a critical step after tissue injury. Indeed, impaired angiogenesis can lead to maladaptive tissue repair in a range of disease states such as diabetic wound healing and ischemic cardiovascular disease. Among EC functional properties contributing to ineffective angiogenesis, reduced cell proliferation and migration may significantly impact therapeutic angiogenesis (4, 42–46). Interestingly, initial treatment strategies that mostly focused on delivery of recombinant growth factors such as VEGF, bFGF, HGF, HIF-1a, Del-1, among others, have shown outright failure or at best limited efficacy (6, 7, 47–51). One possibility for the lack of robust angiogenic responsiveness to these growth factors therapeutically is the concept of “angiogenic resistance,” which is due to impaired growth factor signaling. Accumulating studies support this hypothesis, including the findings that these angiogenic growth factors are actually increased (not deficient) in subjects with diabetes or ischemic cardiovascular disease states (11, 14). For example, compared to human subjects with intermittent limb claudication, subjects with critical limb ischemia have higher levels of VEGF-A, angiopoietin-1, and Tie-2. Furthermore, intermittent claudicants had higher levels of these factors than controls, suggesting that higher ischemic burden is associated with higher circulating levels of proangiogenic growth factors (15). Moreover, experimental rodent models show that despite sufficiency of proangiogenic growth factors (e.g., VEGF) in diabetic limbs, there is markedly reduced activation of downstream signaling (52). Therefore, mechanisms that may underlie “angiogenic resistance” because of impaired angiogenic signaling may provide a more direct strategy to improve therapeutic angiogenesis.
MicroRNAs can regulate signaling pathways via multiple targets. Not only are they highly expressed in ECs but also their expression and targets may vary in different disease settings or in response to pathophysiological stimuli making them potentially attractive candidates with therapeutic interest (46, 53). MiRNAs can regulate key downstream regulators of angiogenic signaling pathways. For instance, we previously identified that the expression of miR-26a, an antiangiogenic miRNA, was increased under acute MI in mice and in human subjects with ACS (29) as well as in skin wounds of db/db mice (28). We also identified that its neutralization could effectively promote angiogenesis by targeting Smad1, an effect promoting downstream effects of the bone morphogenic protein signaling pathway (28).
In this study, we identified a new miRNA, miR-135a-3p, acting as a negative modulator of angiogenesis under pathologic conditions such as diabetes and wound healing by regulating the p38 MAPK signaling pathway (schema; Supplemental Fig. S5). In addition, we identified a previously unknown target of miR-135-3p—HIP1. Gain- and loss-of-function studies in ECs demonstrate that miR-135a-3p serves as an antiangiogenic miRNA. MiR-135a-3p overexpression significantly impaired EC angiogenic functions such as EC growth, migration, and matrigel network tube formation, whereas its inhibition had the opposite effects. Delivery of miR-135a-3p locally to skin wounds of db/db mice not only improved diabetic wound healing in mice but also significantly promoted angiogenesis. In human skin organoids, inhibition of miR-135a-3p significantly increased angiogenesis and p38 phosphorylation. Overexpression of miR-135a-3p had the opposite effects in both diabetic wounds in mice and in human skin organoids. Interestingly, using a combination of transcriptomic profiling and gene set enrichment analyses (GSEA), we found that the top pathway targeted by miR-135a-3p was p38 MAPK signaling. In support, miR-135a-3p selectively regulates p38 MAPK signaling pathway in response to VEGF stimulation and not the ERK1/2 or Akt pathways (Fig. 4 and Supplemental Fig. S1).
Activation of the EC p38 MAPK signaling pathway has been implicated in critical components of angiogenesis including EC migration and proliferation. For example, p38-regulated/activated kinase can induce p38 MAPK signaling to promote EC migration (54). Another study showed that p38 MAPK signaling pathway participates in EC migration by regulating urokinase plasminogen activator expression (55). Sheer stress-induced activation of p38 MAPK through the VEGF receptor 2 promoted angiogenesis in rat skeletal muscle (56). Interestingly, inhibition of bFGF-mediated activation of p38 MAPK signaling was shown to increase endothelial growth in chick chorioallantoic membrane assays, but the vessels displayed abnormal features of hyperplasia, suggesting a duality in the involvement of p38 in angiogenesis (57). We focused on the p38 MAPK signaling pathway as it emerged as a top pathway regulated by miR-135-3p from GSEA and the known contribution of this pathway in VEGF signaling and in wound healing (56, 58). Although the role of miR-135-3p on other top signaling pathways identified from GSEA, such as focal adhesion kinase signaling, has not been explored, our data strongly indicate that miR-135-3p regulates VEGF signaling in a p38K MAPK-dependent manner. Our findings that miR-135a-3p overexpression inhibits EC p38 MAPK phosphorylation, migration, and proliferation, whereas miR-135a-3p has the opposite effects, support the notion that p38 MAPK activation promotes proangiogenic functions in ECs (Figs. 2 and 4). Moreover, miR-135a-3p’s effects on EC wound closure were markedly impaired with siRNA-mediated knockdown of p38K. Furthermore, siRNA-mediated knockdown of HIP1 (Fig. 6B) phenocopied miR-135a-3p’s inhibitory effect on p38 MAPK signaling pathway in response to VEGF further supporting the notion that p38 MAPK can positively regulate the angiogenic functionality of ECs.
HIP1, a cofactor in clathrin-mediated vesicle trafficking, is a novel target of miR-135a-3p that we identified in this study. Interestingly, HIP1 is expressed in tumor cells of epithelial, colon, and prostate cancer and is associated with cell survival (59). Its expression is considered a marker of relapse in patients with prostate cancer (59). Overexpression of HIP1 promotes tumor formation and alters receptor trafficking of EGF and transferrin (60). Furthermore, HIP1 was shown to be overexpressed and physically associated with EGFR in glial brain tumors (61). Our study established a previously unknown role of HIP1 in ECs. MiR-135a-3p through binding to the 3′UTR of HIP1 can regulate HIP1 expression, EC proliferation, and migration (Figs. 5 and 6, and Supplemental Fig. S3). In addition, miR-135a-3p’s ability to modulate EC wound closure (Fig. 6C), EC migration (Fig. 6D), and proliferation (Fig. 6E) was significantly blocked in the absence of HIP1. The mechanism by which HIP1 regulates p38K phosphorylation is not completely understood. However, the target of rapamycin complex 1 signaling pathway is positively regulated by p38 signaling (62). Another study conducted in budding yeast showed target of rapamycin promoted the activity of HIP1 (63). HIP1 also regulates Rac1 signaling in rats in the setting of rheumatoid arthritis; HIP1 knockdown reduced Rac1 activation (64). Rac1 is known to mediate p38 MAPK signaling in NIH 3T3 cells whereby Ras- and Rac1-mediated p38 and JNK signals are required for Stat3 transcriptional activity induced by the Src oncoprotein (65). Collectively, these reports provide potential links for HIP1 regulation of p38 MAPK phosphorylation. Further studies will be required to understand this relationship.
Despite optimal medical therapy, management of diabetic wound healing remains relatively stagnant in human subjects. Impaired angiogenesis is a major contributor to the pathologic remodeling. Manipulation of miRNAs, for example using LNA-anti-miR-135-3p, to improve angiogenesis provides new opportunities for therapeutic intervention. Given the growing number of miRNAs implicated in either angiogenesis or diabetic wound healing, it will be of future interest to explore whether targeting a combination of miRNAs involved in divergent angiogenic signaling pathways using LNA-anti-miR-based therapeutics provides a more effective means to achieve prohealing properties. Because LNA-anti-miRs can be locally injected, this type of therapeutic intervention may facilitate wound healing in different phases of wound healing without systemic effects. Future investigation on the role of miRNAs using innovative gain- and loss-of-function studies by local intradermal delivery in both diabetic mouse models of wound healing and in human skin organoid preparations may help to close the preclinical gap in translating these findings closer to the clinic.
In summary, our study highlights miR-135a-3p as an antiangiogenic miRNA that is dynamically regulated in response to proangiogenic stimuli in vitro and in vivo. These findings establish miR-135-3p as a new link between VEGF-mediated activation of p38 MAPK signaling pathway by targeting HIP1. Inhibition of miR-135-3p increased angiogenesis under diabetic conditions in animal tissue injury models and in response to injury using human skin organoids. Because the expression of miR-135-5p is increased in disease states associated with impaired angiogenic responses, these findings provide the rationale for therapeutic neutralization of miR-135-3p to overcome “angiogenic resistance” and promote tissue repair in conditions such as diabetic wound healing or ischemic cardiovascular disease.
Supplementary Material
This article includes supplemental data. Please visit http://www.fasebj.org to obtain this information.
ACKNOWLEDGMENTS
The authors thank Lay-Hong Ang (Beth Israel Deaconess Medical Center, Boston, MA, USA) for confocal microscopy technical assistance, and the Harvard Digestive Disease Center and U.S. National Institutes of Health (NIH) National Institute of Diabetes and Digestive and Kidney Diseases Grant P30DK034854 This work was supported by the NIH National Heart, Lung, and Blood Institute Grants HL115141, HL117994, and HL134849, and NIH National Institute of General Medical Sciences Grant GM115605 (to M.W.F.); the Arthur K. Watson Charitable Trust (to M.W.F.); the Dr. Ralph and Marian Falk Medical Research Trust (Bank of America, N.A., Trustee; to M.W.F.); the American Heart Association Grant 18SFRN33900144 (to M.W.F.); the American Diabetes Association Grant 1-16-JDF-046 (to B.I.); the Watkins Discovery Award (to B.I.); the Lerner Young Investigator Award (to B.I.); a Tübitak Predoctoral Scholarship (to D.O.); and a grant from South-Eastern Regional Health Authorities and from Norwegian Women’s Public Health Association, Norway (to I.H.). The authors declare no conflicts of interest.
Glossary
- ACS
acute coronary syndrome
- Akt
protein kinase B
- BrdU
bromodeoxyuridine
- DM
diabetes mellitus
- EC
endothelial cell
- eNOS
endothelial nitric oxide synthase
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- HDAC
histone deacetylase
- HIP1
huntingtin-interacting protein 1
- MI
myocardial infarction
- miRNA
microRNA
- MiRNP
miRNA ribonucleoprotein
- NS
nonspecific
- qPCR
quantitative PCR
- siRNA
small interfering RNA
Footnotes
This article includes supplemental data. Please visit http://www.fasebj.org to obtain this information.
AUTHOR CONTRIBUTIONS
M. W. Feinberg conceived the hypothesis; B. Icli and M. W. Feinberg designed research; B. Icli, W. Wu, D. Ozdemir, H. Li, S. Haemmig, X. Liu, G. Giatsidis, S. N. Avci, M. Kurt, and N. Lee carried out the experiments; R. B. Guimaraes, A. Manica, J. F. Marchini, S. E. Rynning, I. Risnes, I. Hollan, K. Croce, and D. P. Orgill contributed critical reagents; B. Icli, W. Wu, D. Ozdemir, H. Li, X. Liu, I. Hollan, and M. W. Feinberg analyzed and interpreted the data; and B. Icli, W. Wu, and M. W. Feinberg wrote the manuscript.
REFERENCES
- 1.Krishna S. M., Moxon J. V., Golledge J. (2015) A review of the pathophysiology and potential biomarkers for peripheral artery disease. Int. J. Mol. Sci. 16, 11294–11322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Falanga V. (2005) Wound healing and its impairment in the diabetic foot. Lancet 366, 1736–1743 [DOI] [PubMed] [Google Scholar]
- 3.Prabhu S. D., Frangogiannis N. G. (2016) The biological basis for cardiac repair after myocardial infarction: from inflammation to fibrosis. Circ. Res. 119, 91–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Potente M., Gerhardt H., Carmeliet P. (2011) Basic and therapeutic aspects of angiogenesis. Cell 146, 873–887 [DOI] [PubMed] [Google Scholar]
- 5.Kusumanto Y. H., van Weel V., Mulder N. H., Smit A. J., van den Dungen J. J., Hooymans J. M., Sluiter W. J., Tio R. A., Quax P. H., Gans R. O., Dullaart R. P., Hospers G. A. (2006) Treatment with intramuscular vascular endothelial growth factor gene compared with placebo for patients with diabetes mellitus and critical limb ischemia: a double-blind randomized trial. Hum. Gene Ther. 17, 683–691 [DOI] [PubMed] [Google Scholar]
- 6.Grossman P. M., Mendelsohn F., Henry T. D., Hermiller J. B., Litt M., Saucedo J. F., Weiss R. J., Kandzari D. E., Kleiman N., Anderson R. D., Gottlieb D., Karlsberg R., Snell J., Rocha-Singh K. (2007) Results from a phase II multicenter, double-blind placebo-controlled study of Del-1 (VLTS-589) for intermittent claudication in subjects with peripheral arterial disease. Am. Heart J. 153, 874–880 [DOI] [PubMed] [Google Scholar]
- 7.Powell R. J., Simons M., Mendelsohn F. O., Daniel G., Henry T. D., Koga M., Morishita R., Annex B. H. (2008) Results of a double-blind, placebo-controlled study to assess the safety of intramuscular injection of hepatocyte growth factor plasmid to improve limb perfusion in patients with critical limb ischemia. Circulation 118, 58–65 [DOI] [PubMed] [Google Scholar]
- 8.Rajagopalan S., Mohler E. R., III, Lederman R. J., Mendelsohn F. O., Saucedo J. F., Goldman C. K., Blebea J., Macko J., Kessler P. D., Rasmussen H. S., Annex B. H. (2003) Regional angiogenesis with vascular endothelial growth factor in peripheral arterial disease: a phase II randomized, double-blind, controlled study of adenoviral delivery of vascular endothelial growth factor 121 in patients with disabling intermittent claudication. Circulation 108, 1933–1938 [DOI] [PubMed] [Google Scholar]
- 9.Jones W. S., Annex B. H. (2007) Growth factors for therapeutic angiogenesis in peripheral arterial disease. Curr. Opin. Cardiol. 22, 458–463 [DOI] [PubMed] [Google Scholar]
- 10.Iyer S. R., Annex B. H. (2017) Therapeutic angiogenesis for peripheral artery disease: lessons learned in translational science. JACC Basic Transl. Sci. 2, 503–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hojo Y., Ikeda U., Zhu Y., Okada M., Ueno S., Arakawa H., Fujikawa H., Katsuki T., Shimada K. (2000) Expression of vascular endothelial growth factor in patients with acute myocardial infarction. J. Am. Coll. Cardiol. 35, 968–973 [DOI] [PubMed] [Google Scholar]
- 12.Kawamoto A., Kawata H., Akai Y., Katsuyama Y., Takase E., Sasaki Y., Tsujimura S., Sakaguchi Y., Iwano M., Fujimoto S., Hashimoto T., Dohi K. (1998) Serum levels of VEGF and basic FGF in the subacute phase of myocardial infarction. Int. J. Cardiol. 67, 47–54 [DOI] [PubMed] [Google Scholar]
- 13.Li J., Brown L. F., Hibberd M. G., Grossman J. D., Morgan J. P., Simons M. (1996) VEGF, flk-1, and flt-1 expression in a rat myocardial infarction model of angiogenesis. Am. J. Physiol. 270, H1803–H1811 [DOI] [PubMed] [Google Scholar]
- 14.Nakajima K., Tabata S., Yamashita T., Kusuhara M., Arakawa K., Ohmori R., Yonemura A., Higashi K., Ayaori M., Nakamura H., Ohsuzu F. (2004) Plasma vascular endothelial growth factor level is elevated in patients with multivessel coronary artery disease. Clin. Cardiol. 27, 281–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Findley C. M., Mitchell R. G., Duscha B. D., Annex B. H., Kontos C. D. (2008) Plasma levels of soluble Tie2 and vascular endothelial growth factor distinguish critical limb ischemia from intermittent claudication in patients with peripheral arterial disease. J. Am. Coll. Cardiol. 52, 387–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cannizzo C. M., Adonopulos A. A., Solly E. L., Ridiandries A., Vanags L. Z., Mulangala J., Yuen S. C. G., Tsatralis T., Henriquez R., Robertson S., Nicholls S. J., Di Bartolo B. A., Ng M. K. C., Lam Y. T., Bursill C. A., Tan J. T. M. (2018) VEGFR2 is activated by high-density lipoproteins and plays a key role in the proangiogenic action of HDL in ischemia. FASEB J. 32, 2911–2922 [DOI] [PubMed] [Google Scholar]
- 17.Rose B. A., Yokota T., Chintalgattu V., Ren S., Iruela-Arispe L., Khakoo A. Y., Minamisawa S., Wang Y. (2017) Cardiac myocyte p38α kinase regulates angiogenesis via myocyte-endothelial cell cross-talk during stress-induced remodeling in the heart. J. Biol. Chem. 292, 12787–12800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Caputo M., Saif J., Rajakaruna C., Brooks M., Angelini G. D., Emanueli C. (2015) MicroRNAs in vascular tissue engineering and post-ischemic neovascularization. Adv. Drug Deliv. Rev. 88, 78–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kir D., Schnettler E., Modi S., Ramakrishnan S. (2018) Regulation of angiogenesis by microRNAs in cardiovascular diseases. Angiogenesis 21, 699–710 [DOI] [PubMed] [Google Scholar]
- 20.Bonauer A., Boon R. A., Dimmeler S. (2010) Vascular microRNAs. Curr. Drug Targets 11, 943–949 [DOI] [PubMed] [Google Scholar]
- 21.Suárez Y., Fernández-Hernando C., Yu J., Gerber S. A., Harrison K. D., Pober J. S., Iruela-Arispe M. L., Merkenschlager M., Sessa W. C. (2008) Dicer-dependent endothelial microRNAs are necessary for postnatal angiogenesis. Proc. Natl. Acad. Sci. USA 105, 14082–14087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ghosh G., Subramanian I. V., Adhikari N., Zhang X., Joshi H. P., Basi D., Chandrashekhar Y. S., Hall J. L., Roy S., Zeng Y., Ramakrishnan S. (2010) Hypoxia-induced microRNA-424 expression in human endothelial cells regulates HIF-α isoforms and promotes angiogenesis. J. Clin. Invest. 120, 4141–4154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bernardo B. C., Gao X. M., Winbanks C. E., Boey E. J., Tham Y. K., Kiriazis H., Gregorevic P., Obad S., Kauppinen S., Du X. J., Lin R. C., McMullen J. R. (2012) Therapeutic inhibition of the miR-34 family attenuates pathological cardiac remodeling and improves heart function. Proc. Natl. Acad. Sci. USA 109, 17615–17620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fasanaro P., D’Alessandra Y., Di Stefano V., Melchionna R., Romani S., Pompilio G., Capogrossi M. C., Martelli F. (2008) MicroRNA-210 modulates endothelial cell response to hypoxia and inhibits the receptor tyrosine kinase ligand Ephrin-A3. J. Biol. Chem. 283, 15878–15883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Menghini R., Casagrande V., Cardellini M., Martelli E., Terrinoni A., Amati F., Vasa-Nicotera M., Ippoliti A., Novelli G., Melino G., Lauro R., Federici M. (2009) MicroRNA 217 modulates endothelial cell senescence via silent information regulator 1. Circulation 120, 1524–1532 [DOI] [PubMed] [Google Scholar]
- 26.Doebele C., Bonauer A., Fischer A., Scholz A., Reiss Y., Urbich C., Hofmann W. K., Zeiher A. M., Dimmeler S. (2010) Members of the microRNA-17-92 cluster exhibit a cell-intrinsic antiangiogenic function in endothelial cells. Blood 115, 4944–4950 [DOI] [PubMed] [Google Scholar]
- 27.Nicoli S., Standley C., Walker P., Hurlstone A., Fogarty K. E., Lawson N. D. (2010) MicroRNA-mediated integration of haemodynamics and Vegf signalling during angiogenesis. Nature 464, 1196–1200; erratum: 467, 356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Icli B., Nabzdyk C. S., Lujan-Hernandez J., Cahill M., Auster M. E., Wara A. K., Sun X., Ozdemir D., Giatsidis G., Orgill D. P., Feinberg M. W. (2016) Regulation of impaired angiogenesis in diabetic dermal wound healing by microRNA-26a. J. Mol. Cell. Cardiol. 91, 151–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Icli B., Wara A. K., Moslehi J., Sun X., Plovie E., Cahill M., Marchini J. F., Schissler A., Padera R. F., Shi J., Cheng H. W., Raghuram S., Arany Z., Liao R., Croce K., MacRae C., Feinberg M. W. (2013) MicroRNA-26a regulates pathological and physiological angiogenesis by targeting BMP/SMAD1 signaling. Circ. Res. 113, 1231–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Melero-Martin J. M., Bischoff J. (2008) Chapter 13. An in vivo experimental model for postnatal vasculogenesis. Methods Enzymol. 445, 303–329 [DOI] [PubMed] [Google Scholar]
- 31.Wara A. K., Croce K., Foo S., Sun X., Icli B., Tesmenitsky Y., Esen F., Rosenzweig A., Feinberg M. W. (2011) Bone marrow-derived CMPs and GMPs represent highly functional proangiogenic cells: implications for ischemic cardiovascular disease. Blood 118, 6461–6464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wara A. K., Foo S., Croce K., Sun X., Icli B., Tesmenitsky Y., Esen F., Lee J. S., Subramaniam M., Spelsberg T. C., Lev E. I., Leshem-Lev D., Pande R. L., Creager M. A., Rosenzweig A., Feinberg M. W. (2011) TGF-β1 signaling and Krüppel-like factor 10 regulate bone marrow-derived proangiogenic cell differentiation, function, and neovascularization. Blood 118, 6450–6460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sun X., Icli B., Wara A. K., Belkin N., He S., Kobzik L., Hunninghake G. M., Vera M. P., Blackwell T. S., Baron R. M., Feinberg M. W.; MICU Registry (2012) MicroRNA-181b regulates NF-κB-mediated vascular inflammation. J. Clin. Invest. 122, 1973–1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Balaji S., Moles C. M., Bhattacharya S. S., LeSaint M., Dhamija Y., Le L. D., King A., Kidd M., Bouso M. F., Shaaban A., Crombleholme T. M., Bollyky P., Keswani S. G. (2014) Comparison of interleukin 10 homologs on dermal wound healing using a novel human skin ex vivo organ culture model. J. Surg. Res. 190, 358–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sacilotto N., Chouliaras K. M., Nikitenko L. L., Lu Y. W., Fritzsche M., Wallace M. D., Nornes S., García-Moreno F., Payne S., Bridges E., Liu K., Biggs D., Ratnayaka I., Herbert S. P., Molnár Z., Harris A. L., Davies B., Bond G. L., Bou-Gharios G., Schwarz J. J., De Val S. (2016) MEF2 transcription factors are key regulators of sprouting angiogenesis. Genes Dev. 30, 2297–2309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nebbioso A., Dell’Aversana C., Bugge A., Sarno R., Valente S., Rotili D., Manzo F., Teti D., Mandrup S., Ciana P., Maggi A., Mai A., Gronemeyer H., Altucci L. (2010) HDACs class II-selective inhibition alters nuclear receptor-dependent differentiation. J. Mol. Endocrinol. 45, 219–228 [DOI] [PubMed] [Google Scholar]
- 37.Ozdemir D., Feinberg M. W. (2018) MicroRNAs in diabetic wound healing: pathophysiology and therapeutic opportunities. [E-pub ahead of print] Trends Cardiovasc. Med. doi: 10.1016/j.tcm.2018.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Erba P., Ogawa R., Ackermann M., Adini A., Miele L. F., Dastouri P., Helm D., Mentzer S. J., D’Amato R. J., Murphy G. F., Konerding M. A., Orgill D. P. (2011) Angiogenesis in wounds treated by microdeformational wound therapy. Ann. Surg. 253, 402–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Greene A. K., Puder M., Roy R., Arsenault D., Kwei S., Moses M. A., Orgill D. P. (2006) Microdeformational wound therapy: effects on angiogenesis and matrix metalloproteinases in chronic wounds of 3 debilitated patients. Ann. Plast. Surg. 56, 418–422 [DOI] [PubMed] [Google Scholar]
- 40.Peng C., Chen B., Kao H. K., Murphy G., Orgill D. P., Guo L. (2011) Lack of FGF-7 further delays cutaneous wound healing in diabetic mice. Plast. Reconstr. Surg. 128, 673e–684e [DOI] [PubMed] [Google Scholar]
- 41.Song G., Nguyen D. T., Pietramaggiori G., Scherer S., Chen B., Zhan Q., Ogawa R., Yannas I. V., Wagers A. J., Orgill D. P., Murphy G. F. (2010) Use of the parabiotic model in studies of cutaneous wound healing to define the participation of circulating cells. Wound Repair Regen. 18, 426–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sun L., Bai Y., Du G. (2009) Endothelial dysfunction--an obstacle of therapeutic angiogenesis. Ageing Res. Rev. 8, 306–313 [DOI] [PubMed] [Google Scholar]
- 43.Goligorsky M. S. (2005) Endothelial cell dysfunction: can’t live with it, how to live without it. Am. J. Physiol. Renal Physiol. 288, F871–F880 [DOI] [PubMed] [Google Scholar]
- 44.Avogaro A., Albiero M., Menegazzo L., de Kreutzenberg S., Fadini G. P. (2011) Endothelial dysfunction in diabetes: the role of reparatory mechanisms. Diabetes Care 34 (Suppl 2), S285–S290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Goveia J., Stapor P., Carmeliet P. (2014) Principles of targeting endothelial cell metabolism to treat angiogenesis and endothelial cell dysfunction in disease. EMBO Mol. Med. 6, 1105–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chamorro-Jorganes A., Araldi E., Suárez Y. (2013) MicroRNAs as pharmacological targets in endothelial cell function and dysfunction. Pharmacol. Res. 75, 15–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Belch J., Hiatt W. R., Baumgartner I., Driver I. V., Nikol S., Norgren L., Van Belle E.; TAMARIS Committees and Investigators (2011) Effect of fibroblast growth factor NV1FGF on amputation and death: a randomised placebo-controlled trial of gene therapy in critical limb ischaemia. Lancet 377, 1929–1937 [DOI] [PubMed] [Google Scholar]
- 48.Creager M. A., Olin J. W., Belch J. J., Moneta G. L., Henry T. D., Rajagopalan S., Annex B. H., Hiatt W. R. (2011) Effect of hypoxia-inducible factor-1alpha gene therapy on walking performance in patients with intermittent claudication. Circulation 124, 1765–1773 [DOI] [PubMed] [Google Scholar]
- 49.Bergers G., Hanahan D. (2008) Modes of resistance to anti-angiogenic therapy. Nat. Rev. Cancer 8, 592–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Welti J., Loges S., Dimmeler S., Carmeliet P. (2013) Recent molecular discoveries in angiogenesis and antiangiogenic therapies in cancer. J. Clin. Invest. 123, 3190–3200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee R., Channon K. M., Antoniades C. (2012) Therapeutic strategies targeting endothelial function in humans: clinical implications. Curr. Vasc. Pharmacol. 10, 77–93 [DOI] [PubMed] [Google Scholar]
- 52.Hazarika S., Dokun A. O., Li Y., Popel A. S., Kontos C. D., Annex B. H. (2007) Impaired angiogenesis after hindlimb ischemia in type 2 diabetes mellitus: differential regulation of vascular endothelial growth factor receptor 1 and soluble vascular endothelial growth factor receptor 1. Circ. Res. 101, 948–956 [DOI] [PubMed] [Google Scholar]
- 53.Landskroner-Eiger S., Moneke I., Sessa W. C. (2013) miRNAs as modulators of angiogenesis. Cold Spring Harb. Perspect. Med. 3, a006643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yoshizuka N., Chen R. M., Xu Z., Liao R., Hong L., Hu W. Y., Yu G., Han J., Chen L., Sun P. (2012) A novel function of p38-regulated/activated kinase in endothelial cell migration and tumor angiogenesis. Mol. Cell. Biol. 32, 606–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yu J., Bian D., Mahanivong C., Cheng R. K., Zhou W., Huang S. (2004) p38 Mitogen-activated protein kinase regulation of endothelial cell migration depends on urokinase plasminogen activator expression. J. Biol. Chem. 279, 50446–50454 [DOI] [PubMed] [Google Scholar]
- 56.Gee E., Milkiewicz M., Haas T. L. (2010) p38 MAPK activity is stimulated by vascular endothelial growth factor receptor 2 activation and is essential for shear stress-induced angiogenesis. J. Cell. Physiol. 222, 120–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Matsumoto T., Turesson I., Book M., Gerwins P., Claesson-Welsh L. (2002) p38 MAP kinase negatively regulates endothelial cell survival, proliferation, and differentiation in FGF-2-stimulated angiogenesis. J. Cell Biol. 156, 149–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Medicherla S., Wadsworth S., Cullen B., Silcock D., Ma J. Y., Mangadu R., Kerr I., Chakravarty S., Luedtke G. L., Dugar S., Protter A. A., Higgins L. S. (2009) p38 MAPK inhibition reduces diabetes-induced impairment of wound healing. Diabetes Metab. Syndr. Obes. 2, 91–100 [PMC free article] [PubMed] [Google Scholar]
- 59.Rao D. S., Hyun T. S., Kumar P. D., Mizukami I. F., Rubin M. A., Lucas P. C., Sanda M. G., Ross T. S. (2002) Huntingtin-interacting protein 1 is overexpressed in prostate and colon cancer and is critical for cellular survival. J. Clin. Invest. 110, 351–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rao D. S., Bradley S. V., Kumar P. D., Hyun T. S., Saint-Dic D., Oravecz-Wilson K., Kleer C. G., Ross T. S. (2003) Altered receptor trafficking in huntingtin interacting protein 1-transformed cells. Cancer Cell 3, 471–482 [DOI] [PubMed] [Google Scholar]
- 61.Bradley S. V., Holland E. C., Liu G. Y., Thomas D., Hyun T. S., Ross T. S. (2007) Huntingtin interacting protein 1 is a novel brain tumor marker that associates with epidermal growth factor receptor. Cancer Res. 67, 3609–3615 [DOI] [PubMed] [Google Scholar]
- 62.Cully M., Genevet A., Warne P., Treins C., Liu T., Bastien J., Baum B., Tapon N., Leevers S. J., Downward J. (2010) A role for p38 stress-activated protein kinase in regulation of cell growth via TORC1. Mol. Cell. Biol. 30, 481–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ma N., Liu Q., Zhang L., Henske E. P., Ma Y. (2013) TORC1 signaling is governed by two negative regulators in fission yeast. Genetics 195, 457–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Laragione T., Brenner M., Lahiri A., Gao E., Harris C., Gulko P. S. (2018) Huntingtin-interacting protein 1 (HIP1) regulates arthritis severity and synovial fibroblast invasiveness by altering PDGFR and Rac1 signalling. Ann. Rheum. Dis. 77, 1627–1635 [DOI] [PubMed] [Google Scholar]
- 65.Turkson J., Bowman T., Adnane J., Zhang Y., Djeu J. Y., Sekharam M., Frank D. A., Holzman L. B., Wu J., Sebti S., Jove R. (1999) Requirement for Ras/Rac1-mediated p38 and c-Jun N-terminal kinase signaling in Stat3 transcriptional activity induced by the Src oncoprotein. Mol. Cell. Biol. 19, 7519–7528 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.