Abstract
Neural organoids have the potential to improve our understanding of human brain development and neurological disorders. However, it remains to be seen whether these tissues can model circuit formation with functional neuronal output. Here, we have adapted air-liquid interface culture to cerebral organoids leading to improved neuronal survival and axon outgrowth. The resulting thick axon tracts display various morphologies including long-range projection within and away from the organoid, growth cone turning, and decussation. Single-cell RNA-sequencing reveals various cortical neuronal identities, and retrograde tracing demonstrates tract morphologies that match proper molecular identities. These cultures exhibit active neuronal networks, and extracortical projecting tracts can innervate mouse spinal cord and evoke contractions of adjacent muscle in a manner dependent on intact organoid-derived innervating tracts. Overall, these results reveal a remarkable self-organization of corticofugal and callosal tracts with a functional output, providing new opportunities to examine relevant aspects of human CNS development and disease.
Keywords: Axon guidance, nerve tracts, in vitro, cerebral organoids, corticospinal tract, corpus callosum, brain and spinal cord injury, organoid
Introduction
Cerebral organoids1 have been shown to model early brain development with remarkable fidelity. Recent advances in this field have demonstrated their capacity to model neurogenesis1,2, neuronal migration and positioning3,4, and even response to sensory input5. However, later neuronal maturation is limited by insufficient oxygen and nutrient availability due to the lack of a blood supply. Recent advances have been made to improve survival either by providing growth factors such as BDNF5 or by transplanting neural organoids into a rodent host to allow for vascularization6. Not only was transplantation shown to result in unprecedented survival of cells due to effective blood perfusion, but cells within the organoid were demonstrated to exhibit neural connectivity with the host brain.
Such in vivo transplantations point to the possibility that cerebral organoids have the intrinsic potential to form functional connections, particularly if survival can be improved. However, this approach is tedious and requires specialist skills to perform and analyse transplants, making it difficult to perform more extensive characterization on multiple samples. Thus, a fully in vitro system that would improve nutrient supply and could be easily scaled up would be ideal for more extensive studies. While in vitro vascularisation of tissue engineered constructs is a high priority in the bioengineering field, as of yet this has not been successfully accomplished in a manner that maintains self-organization and tissue architecture.
In an effort to improve oxygen supply but retain the accessibility and scalability afforded in vitro, we tested an entirely different approach: application and adaptation of a classic method of organotypic slice culture7 at the air-liquid interface. We show that air-liquid interface cerebral organoids (hereafter referred to as ALI-COs) exhibit greatly improved survival and morphology with extensive axon outgrowths reminiscent of nerve tracts. Live imaging reveals a variety of axon guidance behaviors mimicking the dynamics seen in vivo. Furthermore, single-cell RNA-seq reveals separate clusters of intracortical and subcortical projection identities and we show that these correspond to the tract morphologies observed. Finally, through electrophysiological and co-culture studies, we demonstrate functionality of these tracts, which are even capable of eliciting coordinated muscle contractions in co-cultured mouse spinal cord-muscle explants. This approach is likely to be a useful new tool not only because of its ease, but also due to its utility in studying axon guidance, tract formation, and connectivity in a human system.
Results
Air-liquid interface culture of cerebral organoids leads to improved survival and maturation
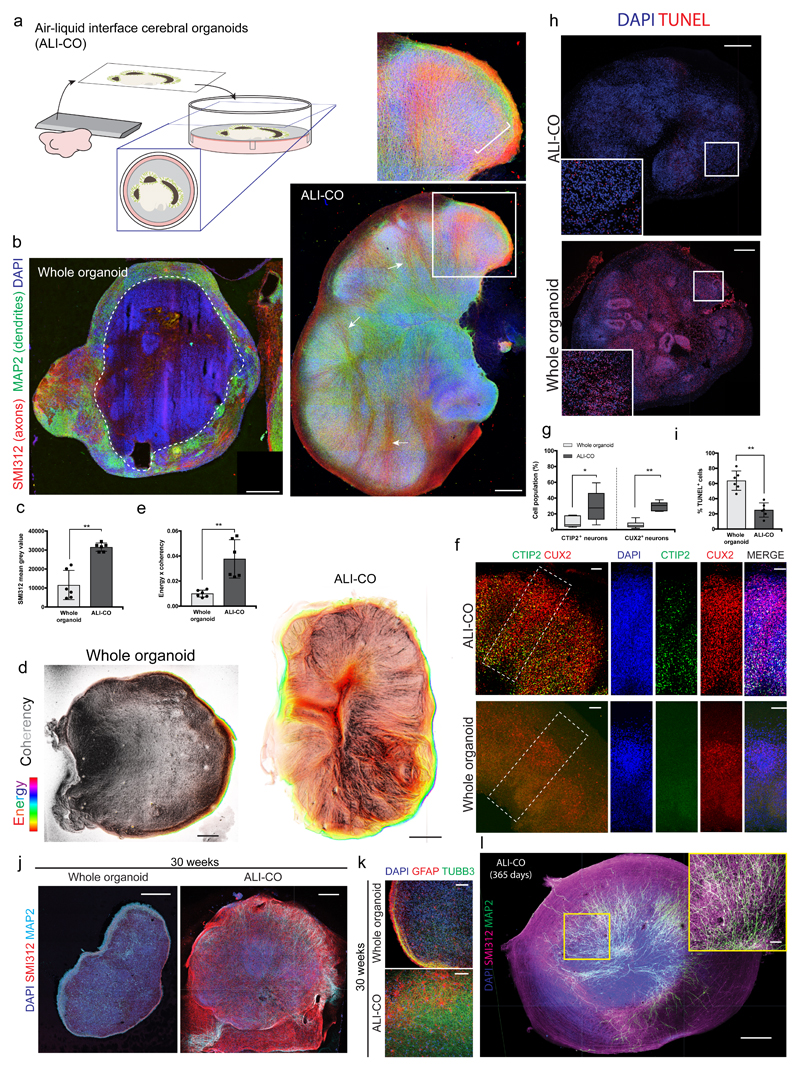
After optimizing sectioning and culture conditions (see Methods, Figure 1a), we found that ALI-COs displayed overall improved morphology compared with whole organoids (Figure 1b-e, Supplementary Fig. 1a) as well as increased numbers of cortical neuron populations, suggesting improved survival (Figure 1f, g). TUNEL assay further confirmed a decrease in cell death in ALI-COs (Figure 1h, i).
Figure 1. Culture at the air-liquid interface leads to improved neuronal survival and morphology.
a. Schematic of the culture paradigm as detailed in methods. b. Immunohistochemistry for the marker SMI312 (red) to stain axons, and MAP2 (green) for dendrites, on representative sections of whole versus ALI cerebral organoid cultures. ALI-CO age is 82 days total (sliced at 61 days plus 21 days at ALI). Whole organoid age is 90 days. Dashed line indicates the border of healthy neurons along the organoid surface. Inset shows a higher magnification of a lobule containing radially aligned neurons of the cortical plate (bracket) and arrows indicate SMI312 positive inward projecting tracts. c. Quantification of overall SMI312 levels reveals increased axon staining in ALI-COs **P=0.0022 Two-tailed Mann-Whitney test, n = 6 whole organoids from 2 independent batches (ages: 90-105d) and 6 ALI-COs from 2 independent batches (ages: 85-92d), error bars are S.D. d. Whole organoids exhibit less aligned axon staining as demonstrated by OrientationJ analysis (detailed in methods) of SMI312 staining (Supplementary Fig. 1a), while axons in ALI-COs are more aligned. Pixel brightness corresponds to coherency of aligned structures while hue corresponds to energy, where pixels with higher energy report less isotropic and more oriented structures23. Shown are representative images out of six samples from two independent batches each. e. The product of OrientationJ coherency and energy output levels was quantified in whole organoids compared with ALI-COs, demonstrating more aligned structures. **P<0.0022 Two-tailed Mann-Whitney test, n = 6 for each (same samples quantified as in c.), error bars are S.D. f. Staining for deep layer (CTIP2+) and upper layer (CUX2+) neurons reveals increased numbers of both populations in ALI-COs with a particularly strong effect on deep layer neurons where most of the staining for CTIP2 is unspecific in whole organoids at this stage (whole organoid age: 116 days, ALI-CO age: 120 days, of which 36 days ALI). g. ALI-COs display significantly higher numbers of CTIP2+ and CUX2+ neurons than whole organoids. Statistical analysis was carried out on a total of 6 whole organoids from three independent batches and 6 ALI-COs from two independent batches each with ages ranging between 98-105 days total in culture. *P=0.0411 and **P=0.0022 Two-tailed Mann-Whitney test, whiskers are min and max values, center line is median and limits are upper and lower quartiles. h. Representative TUNEL staining (red) in cryosections of whole organoids compared with ALI-COs used for quantifications in i. reveals increased cell death in whole organoids. Inset is higher magnification of outlined region. i. Quantification TUNEL+ cells in 6 whole organoids from three independent batches and 6 ALI-COs from two independent batches with ages ranging between 98-105 days total in culture show that whole organoids display significantly higher levels of cell death compared to ALI-COs. **P=0.0022 Two-tailed Mann-Whitney test, error bars are S.D. j. 30-week whole organoid compared with age-matched ALI-CO (142 days at the ALI, 210 days total) stained for axons (SMI312) and dendrites (MAP2) demonstrates continued survival and improved morphology in organoids kept at the ALI. Representative image of one of three ALI-COs stained compared with one whole organoid. k. Higher magnification of 30-week age matched whole and ALI-CO stained for glia (GFAP) and neurons (TUBB3) showing increased neurons and healthier astrocyte morphology in the ALI-CO. Representative image of one of three ALI-COs stained compared with one whole organoid. l. 1-year ALI-CO (275 days ALI) stained for axons (SMI312) and dendrites (MAP2) continues to display abundant healthy neurons with evident axon tracts. Three ALI-COs from one organoid were stained with similar morphologies. Scale bars: 500 μm in b, d, h, j, l; 100 μm in f, k, and l inset.
ALI-CO cultures could be maintained for many months and continued to display improved morphology and survival compared with whole organoids (Figure 1j, Supplementary Fig. 1b). Whole organoids in contrast exhibited a loss of neurons and an accumulation of reactive astrocytes, particularly along the surface of the organoid (Figure 1k, Supplementary Fig. 1c), similar to previous observations8. Astrocytes in ALI-COs exhibited a healthier morphology with numerous fine processes. Even at one year, ALI-COs still exhibited abundant healthy neurons with numerous axons and dendritic processes (Figure 1l).
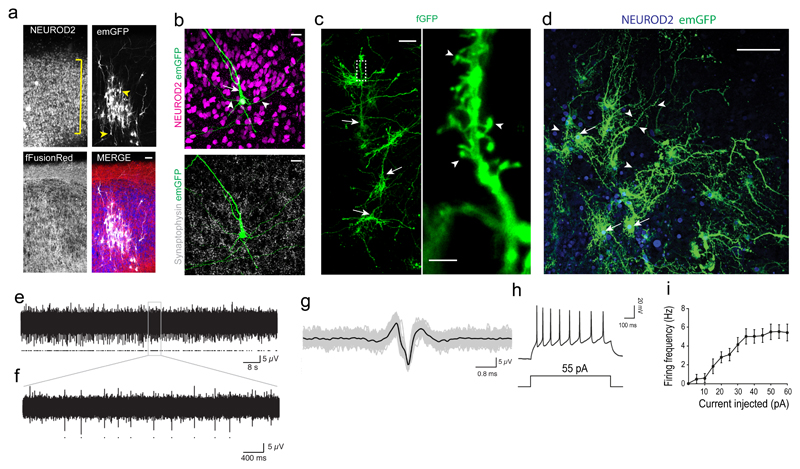
In order to examine neuronal morphology in more detail, we performed sparse labelling with GFP, which revealed aligned neurons of the cortical plate (Figure 2a) that displayed mature morphologies (Figure 2b) with complex dendrites and dendritic spines (Figure 2c). Neuronal morphology continued to mature with 1-year ALI-COs displaying highly complex dendritic architectures (Figure 2d, Supplementary Fig. 2a). Staining for pre- and post-synaptic markers demonstrated synapses decorating the dendrites (Supplementary Fig. 2b). We also observed various interneuron types and the presence of GABAergic synapses (Supplementary Fig. 2c-e). This suggests the various cortical neuron and synapse types are present to allow for functional circuit formation. Indeed, ALI-COs stained positive for the marker of neuronal activity c-Fos (Supplementary Fig. 2f). Multi-electrode array recordings (Supplementary Fig. 2g) revealed the presence of spontaneous neural activity (Figure 2e-g, Supplementary Fig. 2h), which was blocked by tetrodotoxin (TTX, Supplementary Fig. 2i-l). We also performed whole-cell patch clamp recordings of individual neurons demonstrating the ability to fire trains of action potentials upon positive current injection (Figure 2h, i). These findings suggest the acquisition of mature neuronal morphology and establishment of activity in ALI-COs.
Figure 2. ALI-CO cultures exhibit mature neuronal morphology and function.
a. Sparse labeling in an ALI-CO at 37 days at the ALI (80 days total) by Sendai-virus encoding emGFP (white) reveals radially aligned neurons (NEUROD2, blue) with complex dendritic architectures and pyramidal morphologies (arrowheads) within the aligned cortical plate (bracket). All cells contain fFusionRed (red) to visualize the overall tissue morphology. b. Higher magnification of a maximum intensity projection of a single emGFP (green) labeled neuron (NEUROD2+, magenta) displaying typical pyramidal morphology with primary dendrite (arrow) and basal dendrites (arrowheads). Synaptophysin (white) reveals extensive synaptic staining throughout. c. Electroporation of membrane targeted farnesylated GFP (fGFP, green) reveals complex dendritic architecture of radially aligned pyramidal neurons (arrows) in a maximum intensity projection with evident dendritic spines (arrowheads, inset) in an ALI-CO at 51 days at the ALI (143 days total) d. Sparse labeling of a 1-year ALI-CO (90 + 275 days ALI) with Sendai-emGFP labels several individual neurons (arrows) with highly complex dendritic architectures and abundant dendritic spines (arrowheads). Sparse labeling with emGFP and fGFP in a-d were performed on three ALI-COs from three organoids with similar results. e. Two minutes of spontaneous activity recorded from a single electrode of a multi-electrode array (MEA) in an ALI-CO at 54 days at the ALI (117 days total); detected action potentials marked with dots. f. Five-seconds from the same recording (expanded from grey box). g. Overlay of all detected spikes (grey, marked by dots in f) with mean waveform in black. h. Whole-cell patch clamp recordings of action potentials evoked by a 55pA current injection. i. Frequency-current (F-I) curve showing average action potential firing rate with increasing amplitude of current injection (error bars are S.E.M., n=13 cells from 7 ALI-COs of 3 independent organoid batches). Scale bars: 50 μm in a, c; 20 μm in b, 5 μm in c inset, 100 μm in d.
Long-term live imaging reveals axon outgrowth dynamics
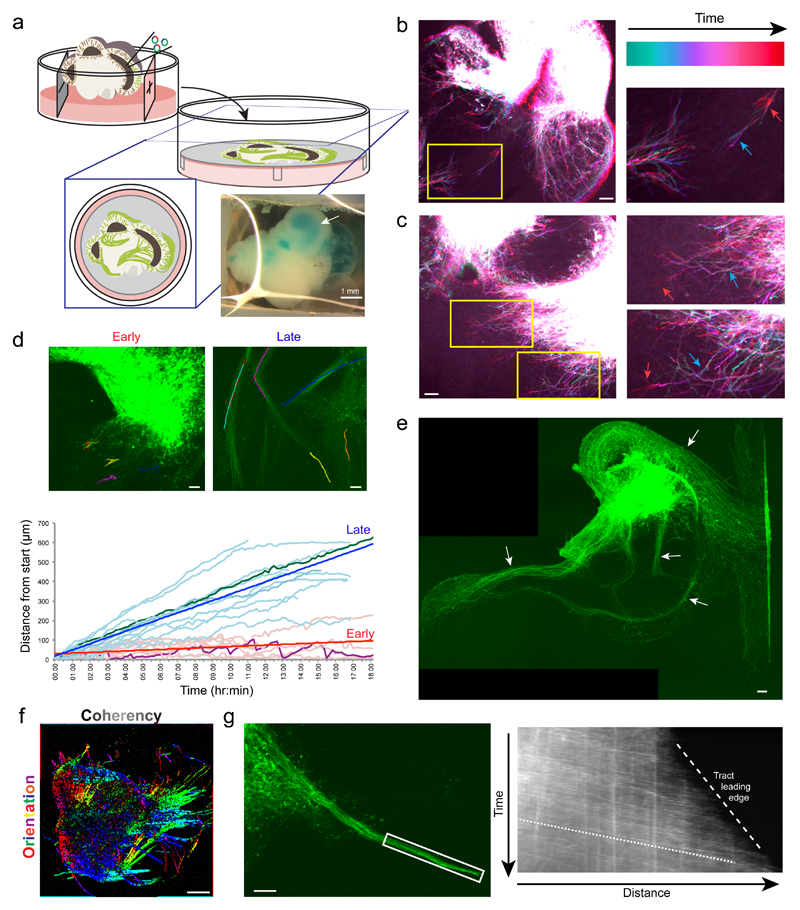
Because the slice culture is easily tractable for live imaging, we examined axon guidance in GFP-labelled neurons over extended time periods (Figure 3a, Supplementary Fig. 3a). Initial axon outgrowth was highly dynamic (Figure 3b, Supplementary Fig. 3b, Supplementary Videos 1, 2) while later axons projected in a directional fashion (Figure 3c, Supplementary Video 3) and with greater velocity within bundles of axons (Figure 3d, Supplementary Fig. 3c, Supplementary Videos 4, 5). These behaviours are reminiscent of the axon outgrowth dynamics of pioneer axons and follower axons of established tracts9. Indeed, ALI-COs displayed robust bundles of axons (Supplementary Fig. 3d) that became reinforced over time (Figure 3e) rather than randomly filling with axons in all directions as is more typically seen in vitro10. Overall, the resultant axon bundles showed specific orientations and a high degree of coherency that is typical of tract morphology (Figure 3f, Supplementary Fig. 3e).
Figure 3. Neurons of ALI-COs exhibit dynamic axon guidance behaviors.
a. Schematic of electroporation and preparation of ALI-COs for live imaging. Inset image shows an organoid after plasmid injection along with a blue dye (FastGreen) to visualize the injected ventricle (arrow). b. Temporal projection image pseudocolored by time (Supplementary Video 2) of an electroporated ALI-CO 5 days after placement at the ALI (69 days total age) showing dynamic axon outgrowth with growth cone retraction (blue arrow to red arrow in the higher magnification inset). Representative image shown out of 5 similarly staged and live imaged ALI-COs. c. Temporal projection image (Supplementary Video 3) after 9 days at the ALI (73 days total age) showing more directed axon outgrowth with progressive extension (blue arrow to red arrow in higher magnification insets). Representative image shown out of 3 similarly staged and live imaged ALI-COs. d. Tracing of individual growth cones over time from ALI-COs at the ALI for 2-5 days (early) and 14-24 days (late) reveals disparate behaviors and velocities (see Supplementary Videos 1, 4). Growth cones in later, established tracts exhibit a higher velocity (bold blue and red lines are average linear regression for each set of data), while earlier growth cones exhibit dynamic retractions (visible in a highlighted trace in purple compared with later highlighted in green). An example image of each is shown above with superimposed traces. Tracing was done on 9 growth cones (early) and 12 growth cones (late) from four organoids generated from two independent batches. e. Axon tracts after 18 days ALI culture (88 days total) demonstrating numerous dense bundles (arrows) with nonrandom projection pattern. Note the same overall pattern but with reinforced bundles compared with Supplementary Fig. 3d, which was taken 4 days earlier. Shown is a representative ALI-CO out of 7 similarly staged and imaged ALI-COs. f. Directional image analysis for the orientation (hue) and coherency (brightness) of aligned axon tracts (original image Supplementary Fig. 3e). Representative image shown out of 5 such directional analyses performed with similar results. g. Still image (left) and kymograph (right) of an extending tract (boxed region) (Supplementary Video 8). Note the higher velocity (shallow slope, dotted line) of incoming growth cones while the leading edge of the tract as a whole progresses more slowly (steep slope, dashed line). Representative results out of three experiments performed. Scale bars: 1 mm in a, 100 μm in b, c, d, e, g; 500 μm in f
Interestingly, these axon tracts could be observed to project within local regions, across the organoid over long distances, and even away from the organoid altogether (Supplementary Fig. 3f, g). Some tracts crossed in a manner similar to decussation (Supplementary Fig. 3h, Supplementary Video 6), with incoming growth cones retaining their directionality upon arrival at the intersection, while other growth cones and tracts could be seen exhibiting turning behaviors (Supplementary Fig. 3c, i, Supplementary Video 7). Finally, axon bundles could be seen extending as a whole, with incoming growth cones projecting at a fast pace, while the tract leading edge progressed much more slowly (Figure 3g, Supplementary Video 8).
ALI-COs exhibit axon tracts with diverse morphologies and the presence of endogenous guidance molecules
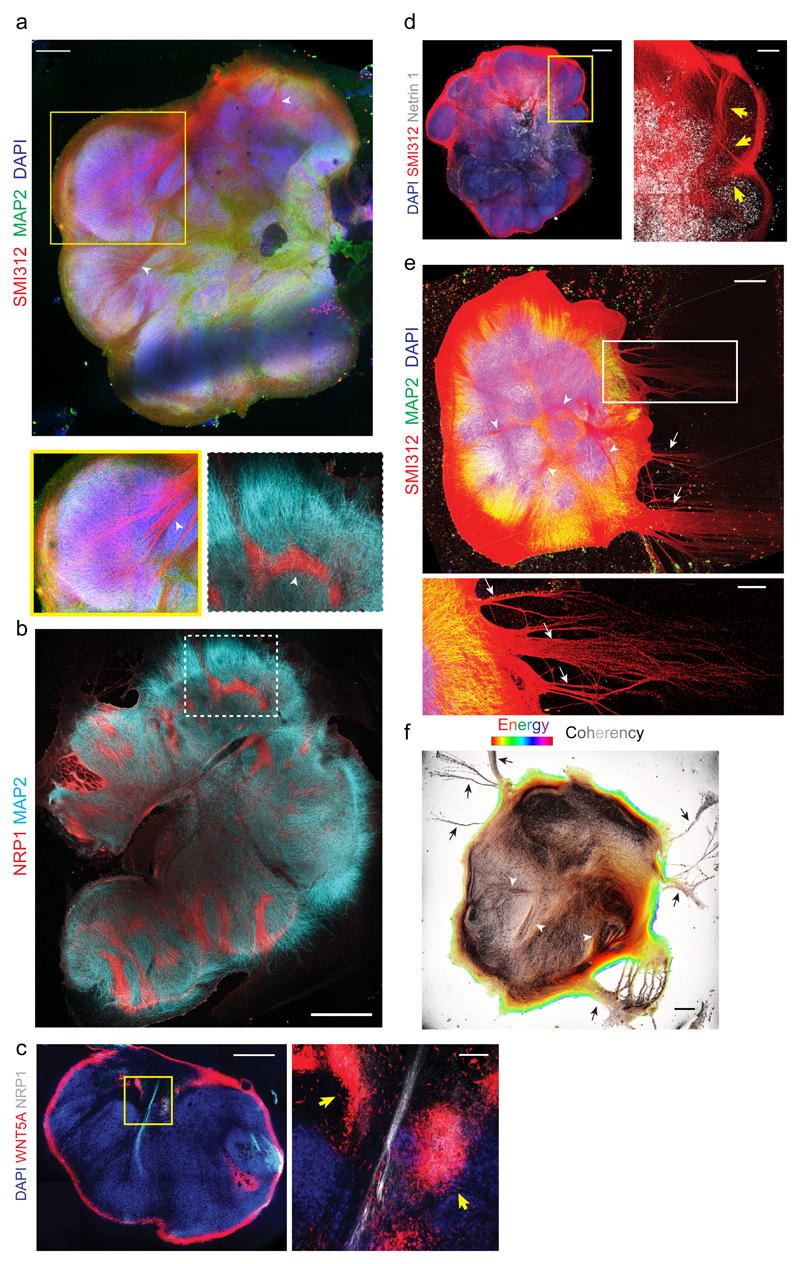
Live imaging results point to the establishment of diverse axon tract morphologies; however, these visible tracts were only a subset of axons labelled by electroporation. In order to visualize the full diversity of axon tracts, we stained mature ALI-COs with SMI312, a broad marker of axons, which could be seen originating within discrete lobules and merging to form dense axon bundles (Figure 4a) reminiscent of intracortical tracts of the CNS. To further investigate these internal projecting tracts, we performed staining for the specific marker of the developing corpus callosum, NRP111, which revealed a subset of ALI-CO tracts (Figure 4b) that often appeared thicker and more fibrous than NRP1-negative tracts (Supplementary Fig. 4a). These tracts, like those observed by live imaging, could often be observed turning, suggesting the presence of endogenous attractants or repulsants that may guide axon outgrowth in the establishment of ALI-CO tracts. To test this hypothesis, we performed staining for known guidance cues including WNT5A, Netrin, and EphrinB112. We could observe striking foci of WNT5A near and surrounding NRP1-positive tracts (Figure 4c) similar to the pattern described at the glial wedge in vivo that helps guide callosal tracts13. Furthermore, we could also observe a subset of NRP1 positive tracts that were also positive for the WNT receptor RYK (Supplementary Fig. 4b)14, as well as internally projecting tracts positive for EphrinB1 (Supplementary Fig. 4c). Finally, staining for the attractant Netrin-1 revealed large foci with numerous nearby inward projecting tracts oriented toward the Netrin-1 signal (Figure 4d).
Figure 4. ALI-COs exhibit diverse axon tract morphologies.
a. Staining for all axons (SMI312) and dendrites (MAP2) on an ALI-CO after 36 days at the ALI (85 days total) reveals thick bundles (arrowheads) that can be seen projecting within the organoid and merging to form large tracts (inset below, arrowhead). Representative image of seven ALI-COs stained with similar results. b. Staining for the marker of corpus callosum, NRP1, reveals several internal tracts that are positive and even appear to turn. MAP2 stains dendrites. Age: 49 days at the ALI, 120 days total. Representative image of four ALI-COs stained with similar results. c. Costaining for NRP1 and the callosal guidance factor WNT5A reveals discrete foci (yellow arrows) surrounding an NRP1+ internally projecting tract in an ALI-CO at 54 days at the ALI, 117 days total. Image at right is magnification of boxed region. Representative image of two ALI-COs stained. d. Staining for the axon attractant Netrin 1 reveals large areas of positivity with evident tracts (SMI312+, arrows) projecting inward and toward the Netrin 1 signal. Age: 32 days at the ALI, 81 days total. Representative image of two ALI-COs stained. e. Axon (SMI312) and dendrite (MAP2) staining of an ALI-CO with tract “escaping” from the main mass (arrows) after 34 days at the ALI (89 days total), in addition to internal projections (arrowheads). Representative image of seven ALI-COs stained with similar results. f. Analysis of axon alignment and coherency by OrientationJ analysis (detailed in methods) of SMI312 staining in a whole ALI-CO at 41 days at the ALI (89 days total). Pixel brightness corresponds to coherency while hue corresponds to energy, where pixels with higher energy report less isotropic and more oriented structures23. Representative analysis shown out of six ALI-COs analyzed with similar results. Scale bars: 500 μm in a, c, d, e, f, 1 mm in b, 100 μm in c inset, 200 μm in d inset and e inset.
In addition to internally projecting tracts, we observed outgrowth of axon tracts projecting away from the organoid altogether (Figure 4e, Supplementary Fig. 4d). Retrograde tracing with the lipophilic dye DiI (Supplementary Fig. 4b), as well as tract coherency/alignment analysis (Figure 4f), better revealed the morphology of these escaping tracts as well as the location of the responsible projection neurons (Supplementary Fig. 4e). These “escaping” tracts appeared most similar to subcortical projections.
scRNA-seq reveals the full repertoire of cortical neuron types
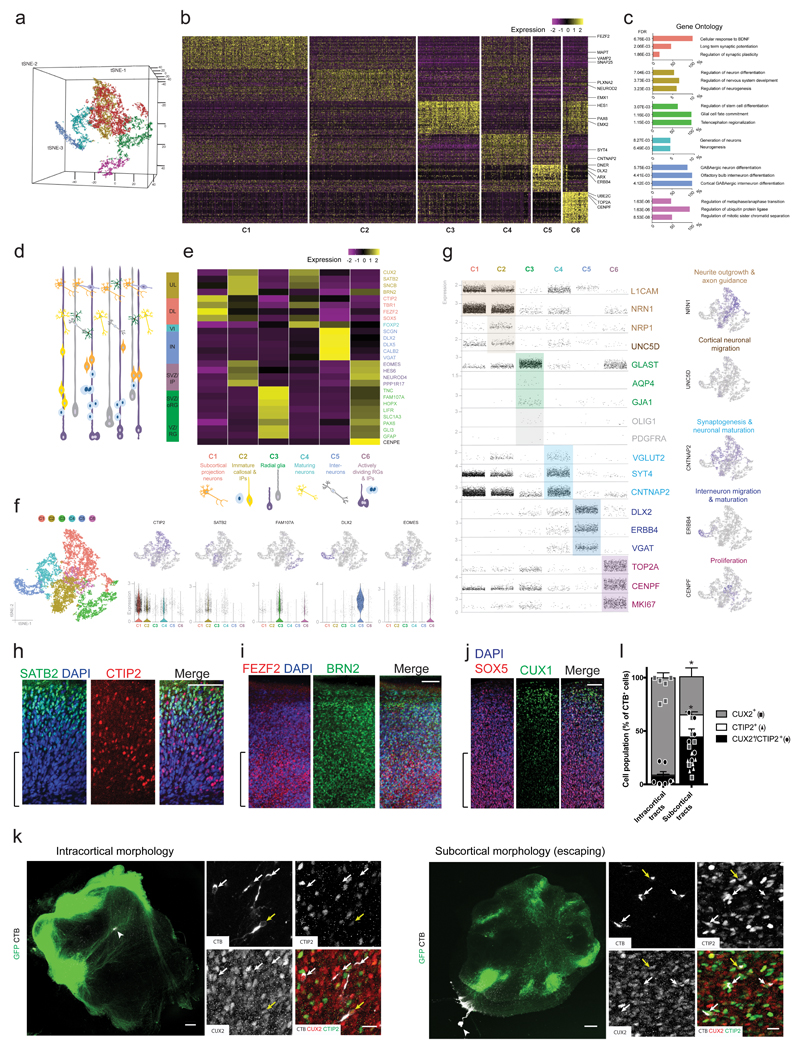
The presence of a variety of axon tract morphologies suggests that ALI-COs might display various neuron identities that could exhibit different projection behaviors. In order to test this possibility, we performed single-cell RNA-seq on ALI-COs to examine the full repertoire of cell types. We analysed six slices from three ALI-CO preparations with an average of 4,427 cells per sample, which were processed through the 10X single-cell genomics platform (see Methods). Unbiased clustering of cell populations was achieved by principle component analysis (PCA) using highly variable genes as input and was visualized following dimensionality reduction using tSNE embedding (Figure 5a). This resulted in the separation of six well-defined major clusters (C1-C6, Figure 5b).
Figure 5. ALI-COs contain diverse neuron identities that exhibit specific projection patterns.
a. Unbiased tSNE separation of clusters based on single-cell RNA-sequencing data of 13,280 cells from 6 ALI-COs of 3 organoids (sample and data acquisition detailed in methods) visualized on a 3D space, showing six main populations. b. Heatmap demonstrating scaled levels of the top 50 differentially expressed genes in the main clusters (C1-6) with example genes (shown at right). c. Histograms of the three top gene ontology (GO) biological process annotations that were defined on the basis of the highest fold-enrichment amongst the most significant terms (Fisher’s exact test with FDR multiple test correction above 0.1% by http://geneontology.org, p < 0.001, n = 50 top differentially expressed genes per cluster).). Colour coding of bars represent cluster identities. d. Schematic representation of cells residing in the different layers of the fetal human cortex: ventricular zone (VZ) containing radial glia (RG), subventricular zone (SVZ) containing outer/basal radial glia (oRG) and intermediate progenitors (IP), deep (DL) and upper cortical plate layers (UL) as well as interneurons (IN) and layer 6 neurons (VI). e. Heatmap showing the scaled mean expression levels of layer- and cell-type specific genes within the six unbiased clusters, identifying the major cell populations. The colour coding of layers, marker genes (right side) and cell types (bottom) corresponds with cluster identities (C1-C6, bottom). f. 2D tSNE plots of clusters (left side) and the distribution of cells expressing layer or cell-type marker genes across the main populations (upper row). Scatter plots below show the distribution of corresponding normalized gene expression values per cell within each cluster, and the violin plot is displayed where the proportion of cells expressing a given gene is the highest. Tails of the plot have been trimmed to represent the maxima and minima values of expression levels. The central hinge represents the median value for each cluster. The distribution in each cluster is based on the filtered and merged datasets deriving from the six organoid slice samples (n = 4191 cells for C1, 3565 cells for C2, 2068 cells for C3, 1658 cells for C4, 952 cells for C5, 846 cells for C6). g. Scatter plots demonstrating the normalized gene expression levels of genes per cell across the six main clusters with a focus on relevant neuronal, progenitor and glial functions. Feature plots (right hand column) demonstrate example genes expressed by populations confined to a particular cluster or by multiple cell populations. Colour coding represents functional gene associations. h. Staining for SATB2 reveals the presence of this upper layer marker and its distribution in a more superficial region, compared with CTIP2. Age: 20 days at the ALI, 90 days total. i. FEZF2+ deep-layer cells and BRN2+ superficial-layer cells are also present and exhibit appropriate distributions in an ALI-CO generated from another organoid. Age: 20 days at the ALI, 90 days total. j. Superficial CUX1+ cells are present in more superficial regions with SOX5+ cells in deeper locations. Age: 20 days at the ALI, 84 days total. Brackets in h-j denote the deeper regions of the ALI-CO cortical plate. h-j are representative images of a region of a single ALI-CO stained for each. k. Representative images of samples used for retrograde labeling by CTB microinjection (arrowheads indicate injection site) and quantification in l. reveals disparate identities contributing to distinct tract morphologies. Inset images are maximum intensity projections of Z-stacks used for quantifications. Individual channels and merge are shown. Yellow arrows indicate CTIP2+/CUX2+ cells and white arrows indicate CUX2 single positive (intracortical morphology, left) or CTIP2 single positive (subcortical escaping morphology, right) cells. l. Quantification of CTB+ cells indicate that tracts with internal projection morphology (left) traced back primarily to CUX2+ callosal projection identity cells while escaping subcortical morphology tracts traced back primarily to CTIP2+ identity cells, as well as CTIP2/CUX2 double positive cells. Six ALI-COs for each condition (intracortical and escaping) from four different organoids were labeled and all CTB-labeled cells across the entire depth of antibody penetration in whole ALI-CO slices were counted for CUX2 or CTIP2 staining. Statistics were performed only on comparisons of single positive cells because double-positive cells are of indeterminate identity. **P=0.0022 Two-tailed Mann-Whitney test, n = 6, error bars are S.E.M. Age: 33 days at the ALI, 97 days total. Scale bars: 100 μm in h, I, j; 300 μm in k, 20 μm in k inset.
GO term analysis (FDR > 0.1% with highest fold-enrichment) of the top 50 differentially expressed genes (Figure 5c) raised a possibility that developmental cell states define the main cell populations, reflecting stages of in vivo cortical development (Figure 5d). In support of this, pseudotime analyses (Supplementary Fig. 5a, b) and gene expression correlation (Supplementary Fig. 5c-e) revealed a similar co-expression pattern of progenitor zone and neuronal layer marker genes in the organoids to that shown for the age-matched fetal brain. Thus, we next compared the average expression of developing cortical cell-type-, cell state-, and region-specific marker genes (see Methods). Cluster identities better corresponded with cell-types and maturity (Figure 5e) than brain region specific markers (Supplementary Fig. 5f, g). This also supports previous findings that the enCOR method used as the starting material for ALI-COs preferentially generates a forebrain identity4.
Cell-type and maturity markers revealed a distinct population of deep layer subcortical projection neurons (C1: e.g. CTIP2, FEZF2), upper layer intracortical (callosal) projection neurons and intermediate progenitors (C2: e.g. SATB2, EOMES), ventricular and subventricular zone radial glial cells (C3: GFAP, FAM107A), more mature upper and deep layer neurons (C4: e.g. FOXP2, CUX2), interneurons (C5: e.g. DLX2) and actively dividing cells with intermediate progenitor and radial glia markers (C6: e.g. CENPE, EOMES, GLAST) (Figure 5d-f). Staining for upper layer and deep layer identities further supported the presence of a variety of projection types (Figure 5h-j). Interestingly, the overlapping cluster of upper layer neurons and intermediate progenitors suggests many of the upper layer neurons may still be quite immature, which would match previous findings that upper layer neurons are actively being generated at this time point (69-75 days) in organoid development8.
We then examined if the cluster forming cell types were associated with a gene expression profile that appropriately corresponds with their expected functional characteristics, such as axon-projection and neuronal circuit formation. Many of these were also present within the differentially expressed genes (Figure 5g). Genes associated with axon outgrowth/tract formation (L1CAM, NRN1) were expressed more abundantly and at increased levels in the C1-2 clusters formed by extra- and intracerebral projection neurons and to a lesser extent in the C4 population. The C4 cluster showed higher levels of more mature markers of excitatory and inhibitory synapse formation (VGLUT2, CNTNAP2, SYT4). The high relative expression of DLX2, DLX5, ERBB4 and VGAT in C5 further indicated a very distinct interneuron population and, together with histological data (Supplementary Fig. 2c-e), raised a possibility for the presence of early cortical circuits. Some cells with functionally relevant markers of astroglial maturity (AQP4, GJA1) and the oligodendrocyte-lineage (OLIG1, PDGFRA) were present in C3, but to a very low degree at this relatively early stage. The C6 progenitor population displayed high levels of transcripts indicative of cell proliferation (CENPF, TOP2A), corresponding with the amplifying cortex-forming populations. Overall there was a well-defined association between cell-types and their expected function suggested by the molecular profiles in each cluster (Figure 5f, g).
To test whether, as seen in vivo, the upper and deep layer neuron populations in ALI-COs project as tracts with distinct morphology, retrograde labelling with cholera toxin subunit B (CTB) was performed on escaping and internal axonal bundles with immunohistochemistry for both CTIP2, marking subcortical projecting identities, and CUX2, marking callosal projecting identities (Figure 5k, l). Whilst 91.1 ± 11.0% (S.D.) of neurons projecting into internal tracts stained specifically for the callosal marker CUX2, only 35.7 ± 19.9% of escaping tracts were positive for only this marker. Instead, 65.4 ± 24.3% of escaping tract projection neurons were positive for CTIP2 with 20.6 ± 6.4% being specifically stained for only this marker, compared with 1.1 ± 1.5% of internal projecting neurons. These data suggest the morphology of the tracts matches the correct molecular identity.
ALI-COs exhibit neural networks with functional output
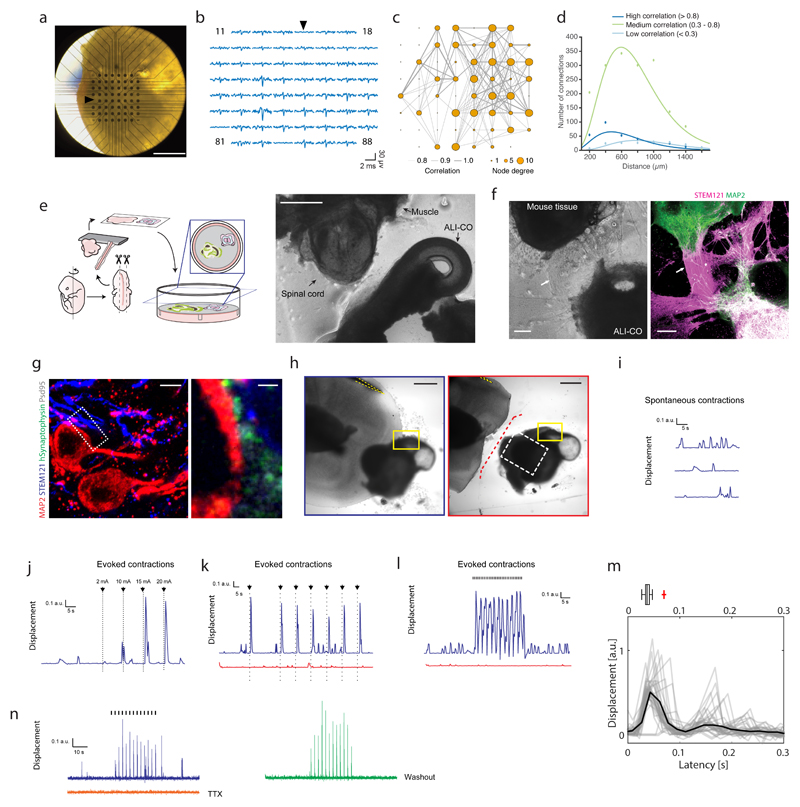
We sought to test the functionality of these tracts, focusing first on the internal projecting tracts. We employed 3D multi-electrode arrays (MEAs) to perform extracellular recordings (Figure 6a) and infer the functional connectivity within ALI-COs from analysis of correlated spontaneous activity15. ALI-COs showed network bursts (Figure 6b, Supplementary Fig. 6a, b) in which neurons near multiple electrodes across the MEA showed simultaneous bursts of action potentials, as seen in mature neuronal networks16. Comparison of the correlated activity revealed densely connected local networks (Figure 6c, Supplementary Fig. 6c) in which the highly correlated neuronal activity could be found within both short- and long-range connections between nodes (Figure 6d). Interestingly, while the connections appeared stronger over shorter distances, the highest correlated activity occurred at a greater distance than the distance between two electrodes (which are 200 μm apart) suggesting these are not simply nearest neighbour connections but that there is a degree of spatial specificity in the connections made. Overall, these findings point to specific spatial patterns of connectivity within ALI-CO cultures.
Figure 6. Functional connectivity in intracortical and extracortical projecting axon tracts.
a. Image of a 130 day ALI-CO (39 days ALI) upon transfer to the 3D microelectrode array. b. 3.4-millisecond-long traces of spontaneous activity in the 59 recording electrodes at the time of a network burst. Reference electrode location and trace are indicated by arrows in a. and b. c. Network plot showing functional connectivity between specific sites within the ALI-CO shown in a. Line thickness represents strength of correlation in the spontaneous activity recorded between connected nodes (59 recording electrodes of the MEA) in the network, while size of the node indicates the number of connections. Correlated activity was determined using the spike time tiling coefficient as detailed in methods. d. Distribution of distances between functionally connected nodes in c. shows short-, medium- and long-range connections within the ALI-CO. Peak of highest correlated connection distances occurs at approx. 400 μm, medium correlated at approx. 600 μm and lowly correlated at approx. 800 μm. e. Schematic diagram of the ALI-CO-mouse spinal cord co-culture with a representative image (right panel) taken just after sectioning (at 62 days of organoid culture). f. Example of innervation of mouse spinal cord by human ALI-CO derived tracts at 68 days after initiation of co-culture (total organoid age = 123 days). Brightfield image (left panel) shows overall positioning of the tissues, with evident axon tracts (arrows) projecting from the ALI-CO (lower right) to the mouse spinal cord (upper left). Staining (right panel) with the human specific marker STEM121 reveals these are ALI-CO derived, and MAP2 staining reveals mouse neurons within the spinal cord and also stains neurons of the human tissue, which is double positive. Images in e and f are representative of six such co-culture experiments performed. g. Immunofluorescence using a human specific antibody against the presynaptic marker Synaptophysin (SYP), STEM121, MAP2 and the post-synaptic marker Psd95 and high magnification imaging in the mouse spinal cord region of a mouse spinal cord-ALI-CO co-culture after 32 days (total organoid age = 84 days) reveals mature human-mouse synapses with human SYP juxtaposed to Psd95 along the surface of mouse Map2+ and STEM121+ neurons. Lower magnification image is a maximum intensity projection. Staining was performed on one sample out of six co-culture experiments. h. Brightfield images of 32-day (84 day total organoid age) human-mouse co-culture used for panels i-l. Images show pre- (outlined in blue) and post- (outlined in red) axotomy with a stimulation electrode (yellow dashed outline) placed on axon tracts leading from the organoid. Yellow box: region imaged for muscle contraction traces in i-l, white dashed box: region shown in Supplementary Fig. 6g, red dashed line: site of axotomy. Shown is a representative image of six co-culture experiments used for stimulation paradigms as shown in i-l, two of which included axotomy as shown, with similar results in all. i. Spontaneous muscle contractions recorded as displacement (see Methods) of the co-culture shown in h. Recording was made before stimulation of the tissue. j. Muscle displacement (contraction) amplitude in response to increasing amplitude of single current pulses (black arrows, 2 mA to 20 mA, 120 μs-long) at 30, 40, 50 and 60 s of the recording (Supplementary Video 11). k. Evoked muscle contractions (Supplementary Video 12) pre- (blue) and post- (red) axotomy in response to single current pulses (black arrows, 15 mA, 120 μs-long) applied at 30, 60, 75, 90, 105, 120, 135s during the recording (150 s total duration). l. Evoked muscle contractions (Supplementary Video 13) pre- (blue) and post-axotomy (red) in response to 30 s of 1 Hz TTL stimulation with 15 mA current pulses (black hash marks, 120 μs-long). Both spontaneous and evoked contractions were abolished by axotomy (Supplementary Video 14) in k and l, while only a few very low amplitude contractions are seen. m. Overlay of evoked muscle contraction waveforms elicited by repeated current pulses (15 mA, 160 μs-long, 0.6 Hz), and waveform average (black), showing the first peak at ~ 37 ms after stimulation. Reported above is a box-and-whiskers plot capturing the spread of the individual contraction events. n= 35 contraction events, center line is median, limits are quartiles, whiskers are min and max. Red cross is an outlier with another outlier recorded at 1 sec (not shown). Organoid age: 42 days at the ALI, 104 days total. n. Evoked muscle contractions to 30 s of repeated current pulses (blue trace, 0.4 Hz, 25 mA, 180 μs-long) in a 21-day human-mouse co-culture (89 days total organoid age) (Supplementary Fig. 6j) were abolished by application of 2 μM TTX (orange trace) and restored (green trace) following wash-out with warm media (8x) and 30-minute incubation at 37 °C. Scale bars: 1 mm in a; 500 μm in e, h; 200 μm in f; 5 μm in g; 1 μm in g inset.
We next examined the functionality of the escaping, extracortical projecting tracts. ALI-COs were co-cultured with sections of the spinal column dissected from embryonic mice in which the associated peripheral nerves and paraspinal muscles were still intact17 (Figure 6e). After 2-3 weeks in co-culture, dense axon tracts from the ALI-COs could be seen innervating the mouse spinal cord (Figure 6f) and synapses were visible between human projecting axons and neurons of the mouse spinal cord (Figure 6g). Live imaging of the mouse muscle tissue revealed sporadic concerted muscle contractions with an irregular periodicity (Figure 6h, i, Supplementary Fig. 6d, Supplementary Video 9). Importantly, these muscle contractions were coordinated and had high amplitude compared with the spontaneous uncoordinated twitches of individual muscle units that could be observed in the absence of innervation by human tracts (Supplementary Fig. 6e). In addition, the coordinated, high amplitude contractions of ALI-CO-innervated mouse tissue could be abolished by lesion of the ALI-CO axonal tracts, being replaced with low amplitude single muscle unit fibrillations (Supplementary Fig. 6d, Supplementary Video 10), pointing to specificity and dependence on ALI-CO tract innervation.
To test whether ALI-CO extracortical axon tracts could control the mouse paraspinal muscle contractions, we performed extracellular stimulation of the ALI-CO axon tracts and observed muscle response. A single brief stimulation of sufficient intensity of the axon tract could elicit a robust muscle contraction (Figure 6j, k, Supplementary Fig. 6f). Evoked muscle contractions were intensity-dependent such that larger stimulation currents increased the amplitude of the muscle contraction (Figure 6j, Supplementary Fig. 6f, Supplementary Video 11), and we could reliably drive the muscle contractions with repeated stimulation (Figure 6k, Supplementary Video 12), and at frequencies up to 1 Hz (Figure 6l, Supplementary Fig. 6f, Supplementary Video 13). The median latency of response from stimulation to beginning of muscle contraction was approximately 37 ms (with a measurement uncertainty of 17 ms, the time interval between consecutive imaging frames) (Figure 6m), which was remarkably similar to recorded latencies of the developing human descending motor pathway18, and is also consistent with a polysynaptic circuit. The latency also suggests electrical stimulation was not activating the muscle directly but relied on the organoid neural tract. Indeed, staining of the responding tissues revealed bundles projecting into the mouse spinal cord (Supplementary Fig. 6g, h), and lesion of this tract eliminated the ability to evoke muscle contractions (Figure 6h, k,l, Supplementary Fig. 6i, Supplementary Video 14). Finally, application of tetrodotoxin (TTX) to an intact ALI-CO-mouse spinal co-culture prevented evoked muscle contractions, which were restored after TTX washout (Figure 6n, Supplementary Fig. 6j). These findings provide evidence that ALI-CO tracts can produce functional output to an external target.
Discussion
We find that by culturing cerebral organoids at the ALI, the tissue remains healthy over a very long period of time (we have tested up to one year). Importantly, we performed sectioning of organoids after the establishment of the cortical plate as described previously4, thus first allowing proper tissue morphogenesis before ALI culture. In this way, the method is highly similar to slice culture typically performed on mid-neurogenesis stage fetal cortex19. The axon dynamics observed in ALI-COs is quite striking, and patterns that ensue suggest a high degree of intrinsic organization. The bundling behaviour further suggests proper tract formation, but importantly, it is distinct from the fasciculation sometimes seen in vitro where bundles form indiscriminately between all neighbouring clusters of neurons20. Furthermore, the presence of endogenous axon guidance cues along with turning behaviors points to similarities to in vivo development and supports the utility of ALI-COs for future studies of axon guidance through application of exogenous signals. Finally, studies with MEAs and explant co-culture point to functional connectivity within and between ALI-COs and external targets. We further show that tractotomy demonstrates specificity and can be used to study the effects of lesion.
These experiments are the first to our knowledge to show a functional output from a neural organoid. This opens up the possibility to perform studies of neural connectivity on a complete circuit, with both input and output, making it possible to examine various aspects of how neurons connect, circuit rewiring, and even more complex aspects of neuronal processing. Furthermore, the ALI-CO approach could be used to model human conditions affecting connectivity. For instance, it opens the door to the study of neurodevelopmental conditions of the corpus callosum, neuronal circuit imbalances seen in epilepsy, and other defects where connectivity is thought to play a role, such as in autism and schizophrenia. Finally, brain organoids are increasingly being used to study degenerative conditions21 and effects of environmental injury22, and ALI-COs could also be used to examine injury or degeneration specifically of axon tracts, such as spinal cord injury, white matter stroke, or amyotrophic lateral sclerosis. Our hope is that by modelling ever more closely specific features of the developing human brain, one day we will be closer to understanding these debilitating diseases.
Methods
Plasmid constructs
The integrating farnesylated GFP construct (pT2-CAG-fGFP, Addgene #108714) and the sleeping beauty transposase plasmid pCAGEN-SB100X as modified from pCMV-SB10025 (Addgene #34879) were used as previously described4. The integrating farnesylated FusionRed construct was obtained by replacing EGFP in pT2-CAG-fGFP with FusionRed. pT2-CAG-fGFP was linearized using MluI and EcoRI restriction enzymes. The FusionRed cassette was amplified by PCR from pCi-C-FusionRed-DEST (a gift from Harvey McMahon’s laboratory) using primers:
FW 5’-GTGCTGTCTCATCATTTTGGCAAAGAATTCATGGTGAGCGAGCTGATTAAGGAG-3’ and RV 5’- GAGGGTTCAGCTTACTCACGCGTGATTTACCTCCATCACCAGCGC-3’
The cassette was inserted into the linearized pT2-CAG-farnesyl backbone by Gibson assembly.
Human pluripotent stem cell culture and genome editing
H9 (female) and H1 (male) human embryonic stem cells (hESCs) (obtained from WiCell, and approved for use in this project by the U.K. Stem Cell Bank Steering Committee) were maintained in StemFlex (ThermoFisher, cat. #A3349401) on Matrigel (Corning, cat. #354230) coated plates and passaged twice a week using EDTA. Unless otherwise specified, organoids and ALI-COs shown were generated from H9 cells. For generation of the fFusionRed line the pCAGEN-SB100X (0.125 μg/ml) plasmid and the transposon donor plasmid pT2-CAG-fFusionRed (0.375 μg/ml) were transfected into H9 hESCs with Lipofectamine Stem (ThermoFisher, cat. #STEM00001). After ~10 days from transfection positive cells were harvested as a pool by fluorescence activated cell sorting on a MoFlo XDP cell sorter (Beckman Coulter).
Generation of cerebral organoids
Cerebral organoids were generated as previously described according to the enCOR method in order to reliably generate forebrain tissue and a proper cortical plate4. Briefly, 18,000 cells were plated with PLGA microfilaments prepared from Vicryl sutures. The original set of media described previously4 or alternatively the STEMdiff Cerebral Organoid Kit (Stem Cell Technologies) were used for organoid culture. From day 35 onward the medium was supplemented with 2% dissolved Matrigel basement membrane (Corning, cat. #354234) to achieve establishment of the cortical plate. Between day 40-60 of the protocol, plasmids were delivered to the organoids by injection and electroporation in the ventricles. Approximately 1-2 weeks after electroporation the organoids were processed for ALI-CO culture.
Electroporation of cerebral organoids
Glass microcapillaries (Drummond Scientific, 1-000-0500) were pulled using a P2000 micropipette puller (Suttern Instrument) with the following settings: heat – 550, filament – 5, velocity – 25, delay – 150, pull – 150. The microcapillaries were opened using dissecting scissors to obtain a tip taper of ~8-9 mm. A total of 5 μl of a 320 ng/μl plasmid solution (80 ng/μl pT2-CAG-fGFP and 240 ng/μl pCAGEN-SB100) was used for injection and electroporation. Electroporation settings were as previously described1.
Air-Liquid Interface Cerebral Organoid (ALI-CO) culture
ALI-CO cultures were prepared using a modified slice culture protocol26. Mature organoids (approx. 55-60 days old, in some cases up to 90 days) were harvested using a cut plastic P1000 pipette tip, washed in HBSS without Ca2+ and Mg2+ (ThermoFisher, cat. #14175095) and embedded in 3% low gelling temperature (LGT) agarose (Sigma, cat. #A9414) at ~40 °C in peel-a-way embedding molds (Sigma, cat. #E6032), typically 1-3 organoids per mold. The agarose blocks were incubated on ice for 10-15 min and processed on a Leica VT1000S vibrating microtome in cold HBSS. 300 μm-thick sections were collected onto Millicell-CM cell culture inserts (Millipore, cat. #PICM0RG50) in 6-well plates and left to equilibrate for one-hour at 37 °C in serum-supplemented slice culture medium (SSSCM): DMEM, 10% FBS, 0.5% (w/v) glucose, supplemented with penicillin-streptomycin and Fungizone. SSSCM was then replaced with serum-free slice culture medium (SFSCM): Neurobasal (Invitrogen, cat. #21103049), 1:50 (v/v) B27+A (Invitrogen, cat. #17504044), 0.5% (w/v) glucose, 1X (v/v) Glutamax supplemented with Antibiotic-Antimycotic (ThermoFisher, cat. #15240062). ALI-CO cultures were maintained in SFSCM at 37 °C and 5% CO2 with daily media changes. Media was provided only below the filter insert so that sections stayed at the ALI and were not submerged.
Live imaging and image analysis
For live imaging, ALI-COs were left to flatten for at least one day before extended imaging on a Zeiss LSM 780 or 710 confocal microscope with incubation chamber set at 7% CO2 and 37 °C. Time-lapse movies were taken at 10 minute intervals over several hours or days. Image analysis was performed in ImageJ. Temporal projection images were generated from time-lapse stacks using the Temporal-Color Code tool in FIJI. Axon growth cone tracing was done using MTrackJ27 with manual tracking of growth cones. Data was then exported and plotted as the distance from the start of the track over time. Linear regression was performed on all tracks and the average best fit line calculated for each group (late and early). Orientation analysis of GFP labeled tracts was done using the OrientationJ analysis plugin23 using Gaussian Gradient setting, with hue determined by the orientation of tracts, brightness determined by coherency, and saturation set at constant. For quantification of axons in ALI-COs compared with whole organoids, mean grey levels of SMI312 staining were calculated on the entire organoid section of normalized images. For axon tract morphology measurements of SMI312 staining, OrientationJ was used on Riesz Filter setting. Mean grey value was calculated from each of the coherency and energy image outputs, on a region of interest, which was the entire outlined organoid section. These grey values were multiplied to give energy x coherency values. Kymographs were generated using the MultiKymograph tool. For quantification of relative abundance of CTIP2+ and CUX2+ neurons in whole organoids compared to ALI-COs a total of 6 whole organoids and 6 ALI-COs were cryosectioned, stained and imaged. To control for bias and differences in the distribution of neurons in sections of whole organoids or ALI-COs, for each sample two images were acquired, one internal and one near the edge of the tissue. The same approach was taken for the acquisition of the TUNEL assay data. Cell counts were performed in NIS Elements Advanced Research using a quantification macro optimized for each type of quantification separately. Parameter optimization was done on a representative image randomly selected and all images were processed using a batch processing function. TUNEL+, CTIP2+ and CUX2+ cells were defined by the co-expression of DAPI and their respective marker, and their relative abundance was expressed as a ratio to the total number of DAPI+ nuclei.
Histological and immunohistochemical analysis
Organoids and ALI-COs were fixed in 4% PFA for 20 min at room temperature or overnight at 4 °C and washed 3 times in PBS. For cryostat processing, samples were incubated overnight in 30% sucrose. Embedding, cryosectioning and staining were performed as previously described28 on whole organoids or ALI-COs after removal from the cell culture insert by cutting the filter around the ALI-CO. For higher magnification staining cryosectioning of ALI-COs was necessary due to their thickness (>300 μm). Whole ALI-COs were stained using a modified protocol - all staining steps were done in permeabilisation buffer (0.25% Triton-X, 4% normal donkey serum in PBS) at 4 °C and their duration was extended as follows: permeabilisation – overnight, primary and secondary antibody incubation – 2 days, wash steps (3X) – 8 hours each. Primary antibodies used with corresponding dilutions were: chicken anti-MAP2 (Abcam, ab5392, 1:500), mouse anti-MAP2 (Chemicon, MAB3418, 1:300), rat anti-CTIP2 (Abcam, ab18465, 1:300), mouse anti-SATB2 (Abcam, ab51502, 1:200), rabbit anti-CUX2 (Abcam ab130395, 1:200), mouse anti-SMI312 (BioLegend, 837904, 1:500), mouse anti-c-Fos (EnCor, MCA-2H2, 1:100), mouse anti-Piccolo (PCLO) (Origene, TA326497, 1:100), mouse anti-STEM121 (Takara, Y40410, 1:500), sheep anti-human-Neuropilin-1 (NRP1) (R&D Systems, AF3870, 1:200), rabbit anti-Homer 1 (Synapti Systems, 160003, 1:100), rabbit anti-Psd95 (Abcam, ab18258, 1:500), mouse anti-human-Synaptophysin (EP10) (ThermoFisher, 14-6525-80, 1:200), goat anti-human-Synaptophysin (R&D Systems, AF5555, 1:100), chicken anti-GFP (ThermoFisher, A10262, 1:500), mouse anti-Bassoon (Enzo, SAP7F407, 1:200), rabbit anti-VGAT (Synaptic Systems, 131013, 1:1000), mouse anti-Calretinin (Swant, 63B, 1:500), rat anti-Somatostatin (Millipore, MAB354, 1:100), mouse anti-GAD67 (Millipore, MAB5406, 1:100), mouse anti-Ephrin-B1 (C-6) (Santa Cruz, sc-515264), mouse anti-WNT5A (A-5) (Santa Cruz, sc-365370), goat anti-BRN2 (C-20) (Santa Cruz, sc-6029), mouse anti-TUBB3 (BioLegend, #801202), rabbit anti-RYK (Abcam, ab5518), rabbit anti-Netrin 1 [EPR5428] (Abcam, ab126729), mouse anti-CUX1 [2A10] (Abcam, ab54583), rabbit anti-GFAP (Abcam, ab7260), rabbit anti-SOX5 (Abcam, ab94396). All primary antibodies are well-described and validated for immunohistochemistry in the literature and were checked for compatibility for human tissue based upon similarity to the human sequence. No novel antibodies were generated in this study. TdT-mediated dUTP-X nick end labelling (TUNEL) was performed using the In Situ Cell Death Detection Kit TMR red (Sigma, cat. #12156792910) as described in the supplier’s technical bulletin.
Cholera toxin subunit B (CTB), DiI and emGFP labelling
ALI-COs were visualised on an EVOS FL inverted microscope (ThermoFisher) and using a microinjection capillary <0.2 μl of 1 mg/ml AlexaFluor 647-conjugate CTB (ThermoFisher, C34778) or 1-4 DiI crystals (ThermoFisher, D3911) were applied to the target region. DiI tracing was performed multiple times but due to difficulty with dye uptake and low signal-to-noise ratio only in one experiment was tracing of sufficient quality for analyis. Similarly, to achieve sparse neuronal labeling, ALI-COs were injected with <0.2 μl of CytoTune emGFP Sendai fluorescence reporter (ThermoFisher, A16519). Four days after CTB and DiI labeling and 5 days after viral emGFP labelling the samples were fixed for histological analyses.
Whole-cell patch-clamp recordings
Organoid slices were placed in a submerged chamber and continuously superfused at room temperature with artificial cerebrospinal fluid containing (in mM): 119 NaCl, 2.5 KCl, 11 glucose, 26 NaHCO3, 1.25 NaH2PO4, 2.5 CaCl2 and 1.3 MgCl2 (pH 7.4), saturated with 5% CO2/95% O2. Whole cell patch clamp recordings were performed in current-clamp configuration using an Axon Multiclamp 700B amplifier (Molecular Devices, San Jose, CA) under a Slicescope (Scientifica, Uckfield, UK) equipped with a 40x objective lens (Olympus, Tokyo, Japan) and a WAT-902H analogue camera (Watec, Newburgh, NY). Signals were filtered at 1 kHz and sampled at 10 kHz with a Digidata 1550A (Molecular Devices, San Jose, CA) connected to a computer running pClamp10 (Molecular Devices, San Jose, CA). Pipettes were pulled from borosilicate glass capillaries (1.5 mm OD x 0.86 mm ID; Harvard Apparatus, Holliston, MA) and typical pipette resistance was 12-15 MΩ. Pipettes were filled with artificial intracellular solution containing (in mM): 145 K-gluconate, 5 MgCl2, 0.5 EGTA, 2 Na2ATP, 0.2 Na2GTP, 10 HEPES, adjusted to pH 7.2 with KOH (osmolarity: 280-290 mOsm). Resting membrane potential (RMP) was estimated in current clamp mode immediately after establishing whole cell configuration. Cells with RMP ≤ -50mV were selected for analysis (Supplementary Table). A series of current steps (800 msec) of increasing amplitude (5pA increments) were applied in current clamp mode to determine the frequency-current relationship.
Organoid cell dissociation for scRNAseq
Organoid tissue slices (two slices from an H9 day 53 organoid then cultured at the ALI for 22 days; and two slices from each of two H1 day 53 organoids then cultured at the ALI for 16 days) were transferred from the ALI into a conical tube (the two slices from the same organoid were pooled) containing Hibernate Medium (Thermo Fisher Scientific, A1247601) plus 1X B-27 Supplement (Thermo Fisher Scientific, 17504044). Gentle dipping helped remove any remaining agarose. The slices were then placed into a 10cm2 tissue culture dish and after washing twice with 1X dPBS (Sigma, D8537), transferred into a gentleMACS C Tube (Miltenyi, 130-093-237) containing 2 ml of Accumax (Sigma, A7089) solution. The C Tube was subsequently attached to the gentleMACS Octo Dissociator (Miltenyi) and ran with the recommended cell dissociation settings. The cell suspension was run through a 70μm strainer to remove any residual cell and debris clumps. A small volume of the strained cell suspension was removed for cell counts with the remaining being diluted 4-fold in 1X dPBS and then centrifuged at 200g for 5 minutes. The cell pellet was then resuspended in dPBS containing 0.04% BSA (Sigma, A9418) to give a final concentration of 206 cells/μl. This suspension was kept on ice for 30 minutes until being processed.
Single-cell RNA-seq library generation and sequencing
Single cell RNA-seq libraries were prepared as instructed by the manufacturer using the 10X Genomics Chromium Single Cell 3’ Library & Gel Bead Kit (10X Genomics, 120237) workflow. The cell suspension (34μl with a total of 7000 cells) was loaded onto a 10X Genomics Chromium Single Cell 3’ Chip with the appropriate amount of Mastermix. The cell capture rate for barcoding varied between 50-75%, resulting in 3500-4400 barcoded cells. The Chromium Controller was run according to the protocol producing the single cell gel beads in emulsion mixture. The reverse transcriptase reaction and subsequent amplification was carried out in a C1000 Touch Thermal Cycler (Biorad), with the cDNA being amplified for 12 cycles. Before sequencing, libraries were quality tested using the 2100 Bioanalyzer Instrument (Agilent) and their concentration was measured by qPCR. Samples were pooled together and sequenced on an Illumina Hiseq 4000 platform.
Single-cell data analysis
The single-cell RNA sequencing data analysis pipeline was constructed using the CellRanger, Seurat and Monocle software packages. The reads were aligned to the GRCh38 reference genome using STAR in CellRanger 2.1.1. This provided a gene expression matrix of 13,333 cells with a median of 2,320 genes and 29,086 mean reads per cell post-normalization. Quality of reads (exonic and intronic) were analysed by FastQC, showing 87.2% fraction reads in cells. Low quality ends were disregarded during the mapping process. Only reads that were uniquely mapped to the transcriptome were used for UMI counting in CellRanger. The read depth was normalized in the ‘Aggr’ function between the libraries of the samples. UMI (transcript) counts were normalized for each cell to the total counts. The values were multiplied by 10,000 and transformed into log-space. Genes expressed in a minimum of three cells and cells expressing between 200 and 5,000 genes with a maximum of 15% mitochondrial genes were opted in during the filtering process. Further data processing was performed using the Seurat 2.3.0 R package with recommended settings. This yielded a final object of 13,280 cells for the merged libraries, which was then scaled and normalized. Unbiased clustering was achieved by principle component analysis (PCA) using the highly variable genes that were defined by selecting standard deviation as the dispersion function in the ‘FindVariableGenes’ option (bin = 20 for the scaled analysis). PCElbowPLot analysis has guided selecting the maximum number of dimensions assisting the cluster separations process. This was followed by the application of the ‘FindClusters’ function. Clustering was driven by the recommended resolution of 0.16. Clusters were then visualised in 2D and 3D views based on tSNE separation in R. Robustness of cluster identity was determined comparing top differential genes for each cluster with a cut-off at 25% expression frequency within a population, which identified six well-defined clusters. Cell population identities were determined by gene enrichment analysis using cell type, layer, region, dorsoventral position and lobe specific gene sets obtained from databases (Allen Brain Atlas at http://human.brain-map.org) and published work1,5,8,29–33. The final cluster identities were then assigned based on the relative proportion of cells expressing the particular reference genes. GO term analysis was performed using the Gene Ontology Consortium online software (http://www.geneontology.org). Gene enrichment was defined by Fisher's Exact with FDR multiple test correction, and the top 3 biological process annotations for the enriched genes were presented on the basis of the highest fold-enrichment amongst the most significant terms (p < 0.001). To compare the developmental profile of the organoids and the fetal brain, we used the Monocle R-package to derive a pseudotime trajectory of gene expressions from the scRNA-seq data. The raw gene expression matrix of the 12 and 13-week old fetal brain24 were processed through the same quality control and filtering process in Seurat as our organoid derived dataset, which served as input to processing in Monocle. Expression levels across the established pseudotime trajectory were observed for the layer markers and visualised in heatmaps comparing data deriving from our organoids and the fetal brain (Supplementary Fig. 5a,b). To compare the representation of distinct developmental cell states in the 69-75-day old ALI-COs to that seen for three month old and 58-65-days old whole organoids in other studies5,31, Pearson correlation plots were generated for layer-specific gene expression profiles obtained from our dataset and published databases (GEO accession codes: GSE86153, GSE75140). For direct comparison the downloaded raw count matrices were uploaded onto Seurat, and filtering was standardized across the datasets. Cells with under 400 genes expressed were discarded, and an upper cap for number of genes analyzed was determined as performed for the analysis of our 69-75-day old ALI-COs. The analysis was carried out in Python Jupyter and visualized through Python Seaborn (Supplementary Fig. 5c-e).
Microelectrode array (MEA) recordings
ALI-COs used for electrophysiological recordings were kept in BrainPhys (STEMCELL Technologies, cat. # 05790) supplemented with Neurocult SM1 neuronal supplement (STEMCELL Technologies, cat. # #05793) for a minimum of 12 hours prior to recordings. Extracellular recordings of spontaneous activity in the organoid slices (n=10) were made using a MEA system (MEA1600, Multi Channel Systems). Organoid slices were transferred immediately prior to recording to a 3D grid MEA (60-3DMEA200/12iR-Ti-gr, 60 electrodes, 12 μm diameter, 200 μm spacing, with an internal reference electrode). Media was removed until a thin layer remained allowing the slice to settle on the 3D MEA grid. The temperature was maintained at 37 °C (TC01 controller and TCX-Control software, Multi Channel Systems). During some recordings, tetrodotoxin (1-2 μM in warm media) was applied to the slice using a Pasteur pipette, which was sufficient to block activity (n=3). The signal was sampled at 25 kHz and stored using the 64-channel data acquisition board (MC Card) and MC Rack software (both Multi Channel Systems). Any electrodes with noise fluctuations greater than 50 μV were grounded prior to recording. Data was exported as a binary file to Matlab (MathWorks) for analysis. The raw signal was bandpass filtered (third-order Butterworth, 600-8000 Hz) and a threshold 6 times the standard deviation above the background noise was used to detect extracellular spike waveforms in each channel with a 2 ms refractory period imposed after each detected spike. Correlated spontaneous activity was compared between electrodes using the spike time tiling coefficient34 with a synchronicity window (Δt) of 40 ms by translating the publicly available code in C (https://github.com/CCutts/Detecting_pairwise_correlations_in_spike_trains) to Matlab.
Mouse spinal cord-ALI-CO co-culture
For mouse embryo spinal cord dissection, C57BL/6 pregnant female mice (12.5 days-post-conception) were euthanized and uterine horns harvested. Mouse tissue was collected according to Schedule 1– Humane Methods of Euthanasia of Animals for tissue removal and as controlled under the ‘Animal (Scientific Procedures) Act 1986’ by a trained technician of the MRC-LMB animal facilities within the MRC Centre covered by the ‘Certificate of Designation’. All steps described from this point on were carried out in ice-cold PBS without Ca2+ and Mg2+. The uterine horns were cut between implantation sites to separate the embryos. Using precision tweezers (IDEAL-TEK, 5SA) the muscle layer, the decidua and remaining extra-embryonic tissue were removed to isolate individual embryos (E12.5). For isolation of spinal cords together with dorsal root ganglia and paraspinal muscles, 0.15 mm ∅ dissecting pins (Fine Science Tools, 2600215) were inserted in the head and pelvic-regions to stabilise the embryo. First, the embryo was placed face-down with straddled limbs, the skin overlying the spinal cord was peeled off and the limbs were removed by cutting the embryo along its length in a posterior-to-anterior fasion, at a distance of approximately 1 mm from the midline, on either side of the spinal cord, using 3 mm cutting edge spring scissors (CohanVannas, 15000-01). The embryo was then repositioned on its side and the internal organs were removed. Last the embryo was placed ventral side up, any remaining undesired tissue was removed including the head and tail.
The dissected mouse spinal cords with surrounding muscle tissue were incubated in ice-cold PBS until embedding. Typically, one organoid and two mouse tissues were first washed in ice-cold HBSS without Ca2+ and Mg2+, then washed and embedded in 3% low-gelling-temperature agarose at ~40°C. Prior to sectioning and before agarose polymerization, the organoid was positioned centrally at the bottom of the mold on top of a layer of solidified agarose. The mouse tissues were placed flat on either side with their roof plates facing inwards towards the organoid (~1-3 mm away). The organoid was oriented such that its longest axis would be parallel to the spinal cords, thus ensuring the maximum number of slices cut that included both organoid and mouse tissue. The block was oriented on the stage for sectioning along the axial plane of the spinal cords. Sectioning was performed as outlined above. Mouse-organoid co-culture slices were maintained on SFSC medium with daily media changes. After two-three weeks human tracts could be seen innervating mouse spinal cords.
ALI-CO stimulation and axotomy
Spontaneous contractions of mouse spinal muscles in the organoid-mouse co-cultures were typically observed after 20-30 days in co-culture using light microscopy and were recorded using a Nikon TE2000 equipped with Andor Neo sCMOS camera using the NIS Elements software for image acquisition. Images were acquired at ~2.3 Hz or ~60 Hz for estimation of latency. Data was acquired as either a .avi or .nd2 file and analyzed using a custom macro in ImageJ (NIH) to calculate the displacement of the tissue upon muscle contraction as a function of time. Muscle contractions evoked by extracellular stimulation of the organoid axonal tracts were elicited using a stainless steel electrode (57100, A-M Systems) connected to a constant current isolated stimulator (0.2-30 mA, 120-180 μs manually- or TTL-triggered pulses, model DS3, Digitimer). When operating the stimulator in TTL-triggered mode, TTL pulses of a given frequency (5 ms duration) were generated using an Arduino running on its internal clock.
For analyses of the latency, we simultaneously recorded the TTL triggers (which trigger the stimulator) and the camera “expose-out” TTL (which indicates when each frame is exposed) using an oscilloscope with a sampling rate of 200kHz (Picoscope 2406B). This allowed us to precisely (±5μs) measure the delay between the first frame of the movie and the previous TTL pulse, from which we could compute the time delay between each frame and the previous stimulation based on the hardware timestamps of each frame and the frequency of the TTL pulses.
Application of tetrodotoxin (TTX, 2 μM) abolished axon tract stimulation evoked muscle contractions after 15 minutes (n = 2). Washing the co-culture with warm media to remove the TTX restored the ability to evoke muscle contractions after 30 minutes in the incubator at 37 °C in one of the experiments. For axotomy, the filters were retrieved from the imaging vessels, placed on a plastic support, visualized on an inverted cell culture microscope at 10X magnification and the incision was performed using a microknife (FST, cat. #10316-14). In order to detect the muscle activity from acquired image sequences, the average of the difference between two consecutive frames for the region of interest (ROI) was computed. The resulting temporal signal was then decomposed into the sum of a baseline, stimulation and residual motion. The baseline was estimated as a 1D rolling ball and the stimulation peaks were detected as outliers (12 x SD) from the mean. The overall processing was implemented as a macro for ImageJ that was applied to a selected ROI. The fire TTL signal was used to synchronize the acquired images and the electrical stimulation signal, the latencies were computed using a Matlab script as the delay between each recorded stimulation TTL pulse and the time when the nearest residual motion went above 2 X S.D.
Code availability
All custom code for the MEA analysis in this paper is publicly available at https://github.com/Timothysit/organoids. The code used for latency analysis can be accessed at the following link: https://github.com/jboulanger/stimulation-motion.
Statistics
Statistical comparisons performed as stated in figure legends included Mann-Whitney unpaired two-tailed test, Fisher's Exact with FDR multiple test correction (FDR above 0.1%), Pearson correlation of layer-specific gene expression, and correlation of neural activity determined using the spike time tiling coefficient as detailed above. No statistical methods were used to pre-determine sample sizes due to the exploratory nature of the experiments, but our sample sizes are similar to those reported in previous publications4,8. In cases of different treatments (ALI-CO versus whole) organoid samples were randomly assigned to the two treatments by taking organoids from within the same culture vessels and randomly separating into whole organoid or ALI culture conditions. Because of the nature of the treatments being obviously different in terms of culture paradigm, data collection and analysis were not performed blind to the conditions of the experiments.
For further information regarding statistics, data and code availability, and reagents see Life Sciences Reporting Summary.
Supplementary Material
Acknowledgements
The authors would like to thank members of the Lancaster lab for helpful discussions, as well as members of the LMB mouse facility for help with timed matings and tissue collections. We also thank members of the Harvey McMahon laboratory for plasmids and the LMB light microscopy facility, in particular Ben Sutcliffe, for assistance with imaging. We are grateful to Paul Coupland and Stephane Ballereau (CRUK) for technical assistance and to Marco Galardini and Pedro Beltrao (EBI) for helping with computing resources. This work was supported by the Medical Research Council MC_UP_1201/9 (M.A.L.), European Research Council ERC STG 757710 (M.A.L.), Medical Research Council MR/P008658/1 (A.L.), Wellcome Trust ISSF_RRZC/115_RG89529 (A.L.), Newton Trust RRZC/115_RG89305(A.L.), MRC Clinician Scientist Fellowship (A.L.), grants from the Biotechnology and Biological Sciences Research Council (BBSRC) (O.P.), Medical Research Council MC_UP_1201/2 (M.T.), European Research Council ERC Starting Grant, STG 677029 (M.T.), Medical Research Council MC_UP_1201/13 (E.D.), and HFSP CDA (E.D.).
Footnotes
Data Availability Statement
The data that support the findings of this study are included in the supporting material with the paper. All scRNA-seq data has been deposited on GEO, accession number GSE124174. Raw data (e.g. raw images and electrophysiological recordings) is available upon request from the corresponding author.
Accession codes
scRNA-seq data available on GEO, accession number GSE124174.
Author contributions
S.L.G. planned and performed experiments, analyzed data, and wrote the manuscript. S.B.M. planned and performed MEA and stimulation experiments, analyzed data and wrote the manuscript. G.M.G. performed the organoid cell dissociation and provided quality control for the single-cell preparation, and L.M.D.W. analyzed the RNAseq data. L.M. performed whole-cell electrophysiology experiments and analyzed data under the supervision of M.T. T.S. analyzed MEA electrophysiology data. M.S. generated and maintained organoids and ALI-CO cultures, and analyzed data. J.B. analyzed time-lapse image data. E.D. performed and analyzed time-lapse imaging. O.P. supervised electrophysiology experiments and interpreted data. A.L. planned experiments, analyzed and interpreted data, and supervised G.M.G and L.M.D.W. M.A.L. conceived the project, planned and performed experiments, supervised the project, and wrote the manuscript.
Declaration of competing financial interests
The authors declare no competing financial interests.
References
- 1.Lancaster MA, et al. Cerebral organoids model human brain development and microcephaly. Nature. 2013;501:373–379. doi: 10.1038/nature12517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kadoshima T, et al. Self-organization of axial polarity, inside-out layer pattern, and species-specific progenitor dynamics in human ES cell-derived neocortex. Proc Natl Acad Sci U S A. 2013;110:20284–20289. doi: 10.1073/pnas.1315710110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Birey F, et al. Assembly of functionally integrated human forebrain spheroids. Nature. 2017;545:54–59. doi: 10.1038/nature22330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lancaster MA, et al. Guided self-organization and cortical plate formation in human brain organoids. Nat Biotechnol. 2017;35:659–666. doi: 10.1038/nbt.3906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Quadrato G, et al. Cell diversity and network dynamics in photosensitive human brain organoids. Nature. 2017;545:48–53. doi: 10.1038/nature22047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mansour AA, et al. An in vivo model of functional and vascularized human brain organoids. Nat Biotechnol. 2018;36:432–441. doi: 10.1038/nbt.4127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gähwiler BH, Capogna M, Debanne D, McKinney RA, Thompson SM. Organotypic slice cultures: a technique has come of age. Trends in Neurosciences. 1997;20:471–477. doi: 10.1016/s0166-2236(97)01122-3. [DOI] [PubMed] [Google Scholar]
- 8.Renner M, et al. Self-organized developmental patterning and differentiation in cerebral organoids. EMBO J. 2017 doi: 10.15252/embj.201694700. e201694700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bak M. Axon fasciculation and differences in midline kinetics between pioneer and follower axons within commissural fascicles. Development. 2003;130:4999–5008. doi: 10.1242/dev.00713. [DOI] [PubMed] [Google Scholar]
- 10.Polleux F, Snider W. Initiating and Growing an Axon. Cold Spring Harb Perspect Biol. 2010;2:a001925–a001925. doi: 10.1101/cshperspect.a001925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Piper M, et al. Neuropilin 1-Sema Signaling Regulates Crossing of Cingulate Pioneering Axons during Development of the Corpus Callosum. Cereb Cortex. 2009;19:i11–i21. doi: 10.1093/cercor/bhp027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chédotal A, Richards LJ. Wiring the brain: the biology of neuronal guidance. Cold Spring Harb Perspect Biol. 2010;2:a001917. doi: 10.1101/cshperspect.a001917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shu T, Richards LJ. Cortical Axon Guidance by the Glial Wedge during the Development of the Corpus Callosum. Journal of Neuroscience. 2001;21:2749–2758. doi: 10.1523/JNEUROSCI.21-08-02749.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keeble TR, et al. The Wnt Receptor Ryk Is Required for Wnt5a-Mediated Axon Guidance on the Contralateral Side of the Corpus Callosum. Journal of Neuroscience. 2006;26:5840–5848. doi: 10.1523/JNEUROSCI.1175-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schroeter MS, Charlesworth P, Kitzbichler MG, Paulsen O, Bullmore ET. Emergence of rich-club topology and coordinated dynamics in development of hippocampal functional networks in vitro. Journal of Neuroscience. 2015;35:5459–5470. doi: 10.1523/JNEUROSCI.4259-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cotterill E, Charlesworth P, Thomas CW, Paulsen O, Eglen SJ. A comparison of computational methods for detecting bursts in neuronal spike trains and their application to human stem cell-derived neuronal networks. Journal of Neurophysiology. 2016;116:306–321. doi: 10.1152/jn.00093.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Streit J, Spenger C, Lüscher HR. An Organotypic Spinal Cord-Dorsal Root Ganglion-Skeletal Muscle Coculture of Embryonic Rat. II. Functional Evidence for the Formation of Spinal Reflex Arcs In Vitro. European Journal of Neuroscience. 1991;3:1054–1068. doi: 10.1111/j.1460-9568.1991.tb00042.x. [DOI] [PubMed] [Google Scholar]
- 18.Koh TH, Eyre JA. Maturation of corticospinal tracts assessed by electromagnetic stimulation of the motor cortex. Arch Dis Child. 1988;63:1347–1352. doi: 10.1136/adc.63.11.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Daza RAM, Englund C, Hevner RF. Organotypic slice culture of embryonic brain tissue. CSH Protoc. 2007;2007 doi: 10.1101/pdb.prot4914. pdb.prot4914. [DOI] [PubMed] [Google Scholar]
- 20.Sorkin R, et al. Compact self-wiring in cultured neural networks. J Neural Eng. 2006;3:95–101. doi: 10.1088/1741-2560/3/2/003. [DOI] [PubMed] [Google Scholar]
- 21.Gonzalez C, et al. Modeling amyloid beta and tau pathology in human cerebral organoids. Mol Psychiatry. 2018;362:329. doi: 10.1038/s41380-018-0229-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qian X, et al. Brain-Region-Specific Organoids Using Mini-bioreactors for Modeling ZIKV Exposure. Cell. 2016;165 doi: 10.1016/j.cell.2016.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rezakhaniha R, et al. Experimental investigation of collagen waviness and orientation in the arterial adventitia using confocal laser scanning microscopy. Biomech Model Mechanobiol. 2012;11:461–473. doi: 10.1007/s10237-011-0325-z. [DOI] [PubMed] [Google Scholar]
- 24.Camp JG, et al. Human cerebral organoids recapitulate gene expression programs of fetal neocortex development. Proc Natl Acad Sci U S A. 2015;112:15672–15677. doi: 10.1073/pnas.1520760112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mátés L, et al. Molecular evolution of a novel hyperactive Sleeping Beauty transposase enables robust stable gene transfer in vertebrates. Nat Genet. 2009;41:753–761. doi: 10.1038/ng.343. [DOI] [PubMed] [Google Scholar]
- 26.Daza RAM, Englund C, Hevner RF. Organotypic slice culture of embryonic brain tissue. CSH Protoc. 2007;2007 doi: 10.1101/pdb.prot4914. pdb.prot4914. [DOI] [PubMed] [Google Scholar]
- 27.Meijering E, Dzyubachyk O, Smal I. Methods for cell and particle tracking. Imaging and Spectroscopic Analysis of Living Cells - Optical and Spectroscopic Techniques. 2012;504:183–200. doi: 10.1016/B978-0-12-391857-4.00009-4. [DOI] [PubMed] [Google Scholar]
- 28.Lancaster MA, Knoblich JA. Generation of cerebral organoids from human pluripotent stem cells. Nature Protocols. 2014;9:2329–2340. doi: 10.1038/nprot.2014.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhong S, et al. A single-cell RNA-seq survey of the developmental landscape of the human prefrontal cortex. Nature. 2018;555:524–528. doi: 10.1038/nature25980. [DOI] [PubMed] [Google Scholar]
- 30.Watanabe M, et al. Self-Organized Cerebral Organoids with Human-Specific Features Predict Effective Drugs to Combat Zika Virus Infection. Cell Rep. 2017;21:517–532. doi: 10.1016/j.celrep.2017.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pollen AA, et al. Molecular identity of human outer radial glia during cortical development. Cell. 2015;163:55–67. doi: 10.1016/j.cell.2015.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Preissl S, et al. Single nucleus analysis of the chromatin landscape in mouse forebrain development. bioRxiv. 2017:159137. doi: 10.1101/159137. [DOI] [Google Scholar]
- 33.Lake BB, et al. Neuronal subtypes and diversity revealed by single-nucleus RNA sequencing of the human brain. Science. 2016;352:1586–1590. doi: 10.1126/science.aaf1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cutts CS, Eglen SJ. Detecting pairwise correlations in spike trains: an objective comparison of methods and application to the study of retinal waves. J Neurosci. 2014;34:14288–14303. doi: 10.1523/JNEUROSCI.2767-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.