Abstract
Objective pain assessment is required for appropriate pain management in the clinical setting. However, clinical gold standard pain assessment is based on subjective methods. Automated pain detection from physiological data may provide important objective information to better standardize pain assessment. Specifically, electrodermal activity (EDA) can identify features of stress and anxiety induced by varying pain levels. However, notable variability in EDA measurement exists and research to date has demonstrated sensitivity but lack of specificity in pain assessment. In this paper, we use timescale decomposition (TSD) to extract salient features from EDA signals to identify an accurate and automated EDA pain detection algorithm to sensitively and specifically distinguish pain from no-pain conditions.
I. Introduction
Definitions of pain have attempted to specify essential features that characterize all forms of pain and distinguish them from other experiences [1]. The widely used and endorsed definition of pain promulgated by the International Association for the Study of Pain (1979) focuses on multidimensional distress, viz., “an unpleasant sensory and emotional experience associated with actual or potential tissue damage or described in terms of such damage” [2]. While there is disagreement on the necessary and sufficient features that should be included when defining pain (Williams and Craig, 2016 [1], argue for explicit recognition of cognitive and social features), there is common agreement that the sensory experience is dominated by emotional qualities.
A number of acute pain-related studies suggest EDA can provide an objective means of accessing emotional distress associated with pain [3–7] (Table 1). Schestatsky et al. [6] studied the correlation between heat pain perception and skin conductance (SC), exposing 22 healthy adult volunteers to three heat stimuli: light warmth, high warmth, and maximum warmth. A positive correlation between changes in sudomotor activity and temperature perception was found, wherein mean EDA was drastically higher during the pain phase when comparing pre-perception warmth to post-perception warmth. Loggia et al. [3] assessed heart rate (HR), SC, and verbal ratings in 39 healthy adult males and demonstrated that both HR and SC increased with more intense pain stimuli. Bradley et al. [7] used HR, blink reflexes, and EDA signals to identify pain in response to electrical shock in 72 healthy participants. In children, Erikson et al [4] reported that SC variables (i.e., basal level, number of waves, and mean wave amplitude) differentiated painful stimulation from no-pain tactile stimulation in newborn infants. Choo et al. [5] found that mean number of fluctuations in SC per second (NFSC) predicted severe pain (reported pain score greater than 7 on a 0 to 10 numerical rating scale) with 56.3% sensitivity and 78.4% specificity in school-aged children after surgery. Gruss et al. [15] used a fusion of EDA, electromyography, and electroencephalography through radial basis function SVM and forward selection algorithm. In that study, 85 participants were subjected to painful heat stimuli (baseline, pain threshold, two intermediate thresholds, and pain tolerance threshold). A significant accuracy rate of 90.94% was obtained using the fusion of those signals when pain tolerance threshold was classified compared to baseline.
TABLE 1.
Studies evaluating EDA responses to acute pain
| Author | Sample | Physiological Signals | ML* applied (Yes/No) |
|---|---|---|---|
| Loggia et al. [3] | Healthy males (19 to 34yrs) (N=39) | - EDA - HR |
No |
| Erikson et al. [4] | Healthy Infants (N=22) | - EDA | No |
| Choo et al. [5] | Children undergoing surgery (N=90) | - EDA - HR - Systolic blood pressure -Respiratory rate |
No |
| Schestatsky et al. [6] | Healthy adults (N=22) | -EDA | No |
| Bradley et al. [7] | Healthy adults (N=72) | - EDA - HR - Blink reflex |
No |
| Gruss et al. [15] | Healthy Adults (N=85) | - EDA - Electromyography - Electroencephalography |
Yes, Forward selection extraction followed by RBF SVM ACC: 90.94 % |
| Susam (this work) | Children following laparoscopic appendecto my (N=21) | - EDA | Yes, TSD Feature extraction followed by Linear SVM Average ACC: 77.66% |
Machine learning
While the aforementioned studies provide evidence for EDA as a sensitive measure of pain, there is notable variability in specificity estimates among them. One of the reasons for this inconsistency may relate to the fact that there are several signal characteristics to consider in EDA data. Tonic refers to slower moving, background characteristics of the signal. Phasic refers to faster fluctuating elements of the signal. Notable variation in tonic signals can be found even within a given individual, making it difficult to accurately detect and interpret [8]. Additionally, phasic EDA in response to a particular stimulus often has a latency period of 1–3 seconds [8]; and in the case of pain stimuli, the latency period may be confounded by anxiety and stress experienced in anticipation of the upcoming stimulus.
As a pioneering attempt to automatically detect pain using only EDA signals, and taking into account the aforementioned issues with tonic fluctuations and phasic response delays, we developed a framework using timescale decomposition (TSD) for statistical feature extraction from EDA signals and a linear support vector machine (SVM) based classifier to distinguish pain from no-pain conditions. This approach decomposes EDA signals into shifting windows of time to enable identification of short (phasic) and long-term (tonic) changes over the signal, with subsequent feature discovery leading to the creation of a pain detection algorithm. We also compare our pain detection results with the only existing EDA study that developed machine learning based classifiers for acute pain detection.
II. METHOD
A. Participants and Experimental Setup
Twenty-one neurotypical youth (16 males; 5 females) primarily Hispanic (71%) and with a median age of 11 years and a interquartile range of 10–15.5 years who had undergone laparoscopic appendectomy participated in a study examining automated assessment of children’s post-operative pain using video and body sensors [9]. Children and their parents were approached for participation after undergoing surgery and provided assent and parental consent prior to study evaluations.
Study participants underwent three study visits during which they were assessed for pain across the recovery period following laparoscopic appendectomy. Visit 1 occurred within 24 hours following surgery (inpatient arena), Visit 2 occurred one day later (inpatient arena), and Visit 3 occurred up to 42 days later (outpatient arena). Only data from Visits 1 and 3 were utilized in the analyses reported herein. At each study visit, EDA and behavioral reactions were recorded during manual abdominal pressure applied adjacent to the surgical incision site for a 10-second interval (hereby referred to as the pressure stimulus and equivalent to a clinical exam). EDA was collected with the Affectiva Q sensor, worn on the wrist of the arm without intravenous catheter placement, modified to collect fingertip EDA, and used gelled adhesive electrodes for signal collection. The Affectiva Q Sensor captures EDA, skin surface temperature, and 3D motion at 16Hz. The sensor was designed to work in real-world environments in an untethered, unobtrusive way and is housed in a durable plastic case. Skin surface temperature and 3D motion permit detection of confounding influences on electrodermal readings associated with changes in environmental temperature and increased physical activity. The Q sensor has been validated against FDA-approved commercial laboratory sensors and is highly correlated with gold-standard EDA measurement devices during cognitive, emotional, and physical demand tasks [10]. Youths scored their experienced pain during the pressure stimulus from 0 (no pain) to 10 (worst pain ever).
Prior to our data analyses, skin surface temperature and motion data were assessed, and EDA data quality was visually inspected by experts blind to experimental condition (pain vs. no pain). Data were removed if the signal did not match standard expectations for EDA based on recommendations from Dawson [11]. Selected high quality data were then smoothed using a 0.35 Hz FIR low pass filter. This filter setting was designed to remove high frequency components associated with artifacts such as movement, while maintaining all other signal components up to the fastest components of an electrodermal response [11]. After filtering, data were down sampled to 1 Hz to reduce analysis time and simplify interpretation of results. Finally, to account for baseline differences in EDA levels, data were normalized to have a mean of 0 using z-score normalization. Before feature extraction, EDA signals were trimmed to a fixed length of 30 seconds surrounding the time of noxious stimuli (comprising 10 seconds before, 10 seconds during, and 10 seconds after the pressure stimulus).
B. Feature Extraction
We utilized TSD to extract statistical features from EDA signals during periods of reported pain and no pain. TSD is a method designed to simultaneously measure short and long-term changes in time series data. Previously, Sips et al. [12] proposed a visual analytic approach that addresses the detection of interesting patterns in environmental time series data. Specifically, their work focused on detecting embedded patterns using comprehensive visual inspection. Compared to this existing study, the novelty of our technique is that we utilize TSD as a new feature extraction method for use with EDA signals. The procedure is a simple extension of the sliding window method, wherein time series data are segmented into consecutive or overlapping fixed length windows and a given metric is calculated (e.g., mean, standard deviation) on each segment. TSD calculates a given metric at all possible window lengths at all possible starting points, and systematically organizes results into a single matrix. The resulting triangular matrix is organized such that consecutive rows differ by a window length of 1, with progressively increasing window lengths.
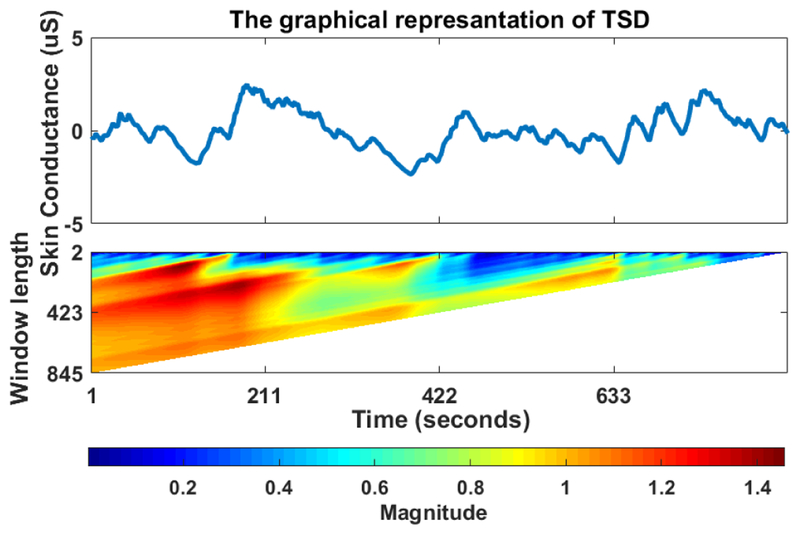
To capture temporal characteristics of our EDA data, we selected standard deviation (SD) as the TSD metric. To illustrate the TSD output, we took a normalized EDA signal (Figure 1 top) and computed a TSD of its SD consecutively without any overlap, then used a heat map to plot the resulting matrix (Figure 1 bottom). Heat maps are graphical techniques that hierarchically represent values in a matrix with colors, allowing simple visual inspection of large amounts of data [13]. Note that the TSD plot shows how SD changes depending on the timescale (window length) and starting point. Thus, graphical representations of TSD enables discovery of a wide range of potentially embedded patterns that are difficult to detect non-visually.
FIGURE 1.
The heat-map of timescale decomposition
To our knowledge, we are the first to apply TSD to EDA data in a new feature extraction overview. We completed TSD using SD on each high quality, normalized, filtered, down-sampled EDA signal. We then extracted features from each TSD matrix for use in machine learning classification. To generate these features, we computed the mean, SD, and entropy of each row of each TSD, and entered them into a single feature matrix. We utilized TSD on SD because in different time scales SD captures fluctuations in tonic response and delays in phasic responses. Specifically, TSD extracts statistical information about changes in SD and accordingly about the dynamic characteristics of tonic and phasic responses over different time scales.
C. Classification Scheme
The features obtained through TSD were normalized using z-score normalization. Then, leave one participant out cross validation (LOPO) was used to divide data into training and testing sets. After that, principal component analysis (PCA) was implemented. In this step, PCA was set to capture between 90% and 99% of the variance in the data, removing all other data. To avoid over-fitting, PCA was applied to a training set and then the subsequent testing feature vector was projected on principal components to generate reduced feature vectors. These reduced features were then used as input to linear support vector machines (Linear SVMs), with two discrete outputs: pain or no pain. That is, a binary classification scheme was used to distinguish EDA responses recorded under pain and no pain conditions. Finally, performance measurements over all PCA thresholds were averaged.
We first performed classification between pain and no-pain conditions based on visit number, where Visit 1 and Visit 3 EDA data were pain and no pain conditions, respectively. Visit 1 and Visit 3 data from the same participants (N=21) were used for this differentiation between pain and no-pain conditions. Of note, Visit 1 pain scores were typically greater than 1 (mean (SD) pain score = 4.9(2.5)) while pain scores at Visit 3 were 0–1 given complete clinical resolution after surgery (mean (SD): 0 (0.4)).
Next, using the accompanying pain scores obtained from participants as ground-truth for the pain condition, we then classified pain vs. no pain based on assigned pain score thresholds. We considered different pain score thresholds to determine pain and no pain conditions (e.g., if the pain score threshold was 4, EDA data from participants whose pain score >=4 in Visit 1 were considered as pain and EDA data from participants with pain scores <4 in Visit 3 were considered as no pain). Based on classification accuracies obtained for the different thresholds, we identified the best pain score threshold to categorize data as pain vs. no pain conditions.
III. RESULTS AND DISCUSSION
We performed two different analyses. In the first analysis, pain data and no pain data were classified solely based on visit number, where Visit 1 data were assigned pain data and Visit 3 data were assigned as no-pain data. Results of linear SVM indicated an accuracy of 71.67%, with a sensitivity of 73.81% and a specificity of 69.52%. This classifier may not have performed well since the EDA data set from Visit 1 came from participants with notable pain score variability (0 to 9), including scores typically not considered clinically relevant pain (0 to 3) [15] and may explain why considering all data from Visit 1 as pain did not lead to high accuracy. Such pain score variability may have been the result of clinical interventions (e.g., effects of residual surgical anesthesia).
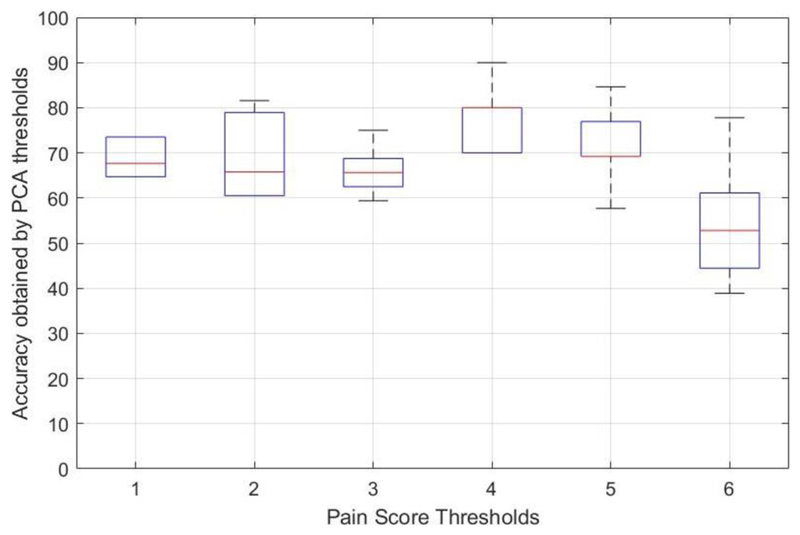
To improve accuracy, in our second analysis, EDA data were further categorized along with accompanying pain scores using varying pain score thresholds. Figure 2 indicates performance of linear SVM across a range of pain score thresholds where pain scores equal or larger than the threshold were assigned as pain and those lower than the threshold were assigned as no-pain. Table 2 indicates the averaged performance measurements over all PCA thresholds with pain score threshold. The best average classification accuracy (77.66%, with a sensitivity of 81.33%, and specificity of 74%) was achieved with a pain score threshold of 4. These observations demonstrate that our classification method is sensitive to pain thresholding for correct identification of both pain and no-pain conditions. Of note, a pain score threshold of 4 is a commonly used pain score criterion in clinical settings to categorize between mild and moderate pain [14].
FIGURE 2.
Linear SVM performance by various pain score thresholds
TABLE 2:
Averaged performance measurements over all PCA thresholds
| Pain score thresholds | Average Accuracy | Average Sensitivity | Average Specificity |
|---|---|---|---|
| 1 | 68.53 % | 70.58 % | 66.47 % |
| 2 | 68.69 % | 69.47 % | 67.89 % |
| 3 | 65.94 % | 67.50 % | 64.37 % |
| 4 | 77.66 % | 81.33 % | 74.00 % |
| 5 | 71.92 % | 78.46 % | 65.38 % |
| 6 | 53.88 % | 54.44 % | 53.33 % |
Prior to our present efforts, researchers have not evaluated the capability of EDA signals alone to distinguish between pain and no pain conditions using TSD for feature extraction followed by machine learning applications. Moreover, this is the first application of this methodology in the pediatric population. Children pose additional challenges in pain assessment given developmental and cognitive limitations that may further compromise standard assessment methods (self-report). To our knowledge, only Gruss et al. [15] have previously examined acute pain detection using a fusion of EDA, electromyography, and electroencephalography through machine learning applications with a significant accuracy rate of 90.94%, but the contributions of individual modalities could not be established, and the study was conducted with adults. In contrast, in our study, using only one modality (EDA signals) in children to discriminate between clinically moderate to severe pain vs. no pain, we accomplished 77.66% recognition accuracy applying TSD for feature extraction followed by machine learning analytic methods.
IV. CONCLUSION
The primary contributions of this paper are twofold. First, we demonstrate the utility of TSD as a novel feature extraction method for use with EDA data. Second, we present promising preliminary evidence for an accurate machine learning classification algorithm to discriminate clinically moderate to severe pain vs. no pain in children using EDA patterns alone. In summary, our results reveal that EDA signals alone can be used for pain assessment with a significant accuracy rate.
Our preliminary results for distinguishing between clinically moderate-to-severe pain and no-pain conditions using only EDA signals is promising. Using a single wearable EDA sensor and without requiring collection of additional physiological signals that might require significant hardware setup, our current method could translate to quick and effective pain identification in clinical settings and in an often difficult to assess patient population (children).
Acknowledgments
This work was supported by National Institutes of Health, National Institute of Nursing Research grant R01 NR013500.
Contributor Information
Busra T. Susam, Electrical and Computer Engineering Department, University of Pittsburgh, PA 15213, USA (bts38@pitt.edu).
Murat Akcakaya, Electrical and Computer Engineering Department, University of Pittsburgh, PA 15213, USA..
Hooman Nezamfar, the Department of Electrical and Computer Engineering, Northeastern University, Boston, MA, USA..
Damaris Diaz, Rady Children’s Hospital and Department of Pediatrics, University of California San Diego, La Jolla, CA 92093, USA..
Xiaojing Xu, Department of Electrical and Computer Engineering University of California, San Diego, La Jolla, CA 92093, USA (xix068@eng.ucsd.edu)..
Virginia R. de Sa, Department of Cognitive Science University of California San Diego, La Jolla, CA 92093, USA (desa@ucsd.edu).
Kenneth D. Craig, Department of Psychology University of British Columbia, Vancouver, BC, Canada (kcraig@psych.ubc.ca).
Jeannie S. Huang, Rady Children’s Hospital and Department of Pediatrics, University of California San Diego, La Jolla, CA 92093, USA (jshuang@ucsd.edu).
Matthew S. Goodwin, Dept. of Health Sciences, Northeastern University Boston, MA. (m.goodwin@northeastern.edu).
References
- [1].Williams Amanda C. de C., and Craig Kenneth D.. “Updating the definition of pain.” Pain 157.11 (2016): 2420–2423. [DOI] [PubMed] [Google Scholar]
- [2].Merskey Harold. “Pain terms: a list with definitions and notes on usage. Recommended by the IASP Subcommittee on Taxonomy.” Pain 6 (1979): 249–252. [PubMed] [Google Scholar]
- [3].Loggia Marco L., Juneau Mylène, and Bushnell M. Catherine. “Autonomic responses to heat pain: Heart rate, skin conductance, and their relation to verbal ratings and stimulus intensity.” PAIN® 152.3 (2011): 592–598. [DOI] [PubMed] [Google Scholar]
- [4].Eriksson Mats, et al. “Skin conductance compared to a combined behavioural and physiological pain measure in newborn infants.” Acta paediatrica 97.1 (2008): 27–30. [DOI] [PubMed] [Google Scholar]
- [5].Choo Eugene K., et al. “Skin conductance fluctuations correlate poorly with postoperative self-report pain measures in school-aged children.” Anesthesiology: The Journal of the American Society of Anesthesiologists 113.1 (2010): 175–182. [DOI] [PubMed] [Google Scholar]
- [6].Schestatsky Pedro, et al. “Skin autonomic reactivity to thermoalgesic stimuli.” Clinical Autonomic Research 17.6 (2007): 349–355. [DOI] [PubMed] [Google Scholar]
- [7].Bradley Margaret M., Silakowski Tammy, and Lang Peter J.. “Fear of pain and defensive activation.” PAIN® 137.1 (2008): 156–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Braithwaite JJ, Watson DG, Jones R, Rowe M A guide for analyzing electrodermal activity (EDA) & skin conductance responses (SCRs) for psychological experiments 2015 Technical Report 2nd version: Selective Attention & Awareness Laboratory (SAAL), Behavioural Brain Sciences Centre, University of Birmingham, UK. [Google Scholar]
- [9].Sikka K, Ahmed A, Diaz D, Goodwin MS, Craig KD, Bartlett MS, & Huang JS (2015). Automated assessment of children’s postoperative pain using computer vision. Pediatrics, 136, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Fletcher RR et al. , “iCalm: Wearable sensor and network architecture for wirelessly communicating and logging autonomic activity”, IEEE Trans. Inf. Technol. Biomed vol. 14, no. 2, pp. 215–223, Mar. 2010. [DOI] [PubMed] [Google Scholar]
- [11].Dawson Michael E., Schell Anne M., and Filion Diane L.. “The electrodermal system.” Handbook of psychophysiology 2 (2007): 200–223. [Google Scholar]
- [12].Sips Mike, et al. “A visual analytics approach to multiscale exploration of environmental time series.” IEEE Transactions on Visualization and Computer Graphics 18.12 (2012): 2899–2907. [DOI] [PubMed] [Google Scholar]
- [13].Akers Walt. “Visual resource monitoring for complex multi project environments.” International Journal of System of Systems Engineering 6.1–2 (2015): 112–126. [Google Scholar]
- [14].Hoffman Deborah L., et al. “How do changes in pain severity levels correspond to changes in health status and function in patients with painful diabetic peripheral neuropathy?” PAIN® 149.2 (2010): 194–201. [DOI] [PubMed] [Google Scholar]
- [15].Gruss Sascha, et al. “Pain intensity recognition rates via biopotential feature patterns with support vector machines [DOI] [PMC free article] [PubMed]