Abstract
The discovery of recurrent mutations in subunits of the vacuolar-type H+-translocating ATPase (v-ATPase) in follicular lymphoma (FL) highlights a role for the amino acid– and energy-sensing pathway to mTOR in the pathogenesis of this disease. Here, through the use of complementary experimental approaches involving mammalian cells and Saccharomyces cerevisiae, we have demonstrated that mutations in the human v-ATPase subunit ATP6V1B2 (also known as Vma2 in yeast) activate autophagic flux and maintain mTOR/TOR in an active state. Engineered lymphoma cell lines and primary FL B cells carrying mutated ATP6V1B2 demonstrated a remarkable ability to survive low leucine concentrations. The treatment of primary FL B cells with inhibitors of autophagy uncovered an addiction for survival for FL B cells harboring ATP6V1B2 mutations. These data support the idea of mutational activation of autophagic flux by recurrent hotspot mutations in ATP6V1B2 as an adaptive mechanism in FL pathogenesis and as a possible new therapeutically targetable pathway.
Keywords: Cell Biology, Oncology
Keywords: Autophagy, Lymphomas, Molecular pathology
Introduction
Follicular lymphoma (FL) constitutes the most common indolent B cell lymphoma, with an incidence and prevalence of approximately 14,000 and approximately 100,000 cases, respectively, in the US (1). FL remains incurable with conventional therapies, and most patients receive multiple treatment regimens during the course of their illness. The development of targeted FL-directed therapies is in the early stages (2–8), and the identification of novel targets remains a research priority.
An expanding group of recurrently mutated genes has recently been shown to underlie the pathogenesis of FL (KMT2D [also known as MLL2], CREB-binding protein [CREBBP], TNF receptor superfamily member 14 [TNFRSF14], enhancer of zeste 2 polycomb repressive complex 2 subunit [EZH2], E1A-binding protein p300 [EP300], AT-rich interaction domain 1A [ARID1A], HIST1H1B-HIST1H1E, signal transducer and activator of transcription 6 [STAT6], Bruton tyrosine kinase [BTK], Ras-related GTP-binding C [RRAGC], those encoding components of the v-ATPase, and others), and some of these may afford novel opportunities for the development of targeted therapy (9–23). The rational design of such targeted therapies necessitates an in-depth understanding of the effects of mutations on the gene product and the consequences for cellular function.
The identification of mTOR-activating mutations in the small G protein RRAGC, a component of the amino acid signaling pathway to mTOR, has highlighted a potential opportunity to target this pathway in FL (24–26). Upstream of RRAGC in this pathway lies the multisubunit vacuolar-type H+-translocating ATPase (v-ATPase) (27–29), which has also been identified as a commonly mutated target in FL (18, 24, 30). However, a comprehensive functional analysis of the consequences of point mutations in the various v-ATPase subunits has not been reported, and it remains unclear why FL among all B cell non-Hodgkin lymphomas would uniquely select for such mutations.
In this study, we examined the effect of mutations in the ATP6V1B2 subunit of the v-ATPase on mammalian cell physiology. In addition, we took advantage of the conserved nature of the v-ATPase and generated a similar mutation in the yeast VMA2 gene (31), allowing us to carry out certain assays that are not available in more complex eukaryotes. Together, the results indicate that the mutant human ATP6V1B2 subunit (also known as the Vma2 subunit in yeasts) results in compromised v-ATPase activity, causing an upregulation of macroautophagy (referred to hereafter as autophagy) and autophagic flux. Even though there was an increase in autophagy, these cells continued to display activity for mTOR/TOR, a major negative regulator of autophagy. These data suggest that it is possible to uncouple the amino acid status of the cell (which is affected by autophagy and which regulates mTOR) and mTOR-dependent control of autophagy. In support of the physiological relevance of these findings, we show that one of the consequences of mutations in ATP6V1B2 is the ability of lymphoma cells to grow and survive under reduced leucine concentrations. This acquired ability to survive under nutrient stress is likely involved in the outgrowth of mutated FL cells and suggests opportunities for therapeutic interventions. Our findings highlight the potential for such interventions, as we demonstrated preferential sensitivity of ATP6V1B2 mutant primary FL B cells to inhibition of autophagic flux. In summary, our data provide insights into the role of macroautophagy and mutations in the v-ATPase in FL pathogenesis.
Results
The spectrum of ATP6V1B2 mutations in 144 FL and 14 transformed FL cells.
Recent reports of relatively frequent mutations in the v-ATPase subunit ATP6V1B2 in FL, and mTOR-activating mutations in RRAGC, along with findings of involvement of both proteins in a lysosomal amino acid–sensing pathway to mTOR, prompted us to study the underlying biology of these mutations (18, 24, 25, 27, 30). We first determined the frequency and nature of ATP6V1B2 mutations in 144 FL and 14 transformed FL (t-FL) cells using direct Sanger sequencing. We identified a total of 10% (16 of 158) of cases with nonsynonymous ATP6V1B2 mutations, 3 of which occurred in t-FL cases. The most common mutations in ATP6V1B2 were located in the previously reported amino acid hotspots p.Y371Y>Y/C (n = 5) and p.R400R>R/Q (n = 8). In addition, we detected the mutations p.D367E>D/E, p.R400R>R/W, and p.R471R>R/S (Figure 1A). We found that clonal mutations in RRAGC and ATP6V1B2 in FL did not occur together, suggesting that the corresponding proteins have overlapping functions in a shared pathway (see below) (24, 25).
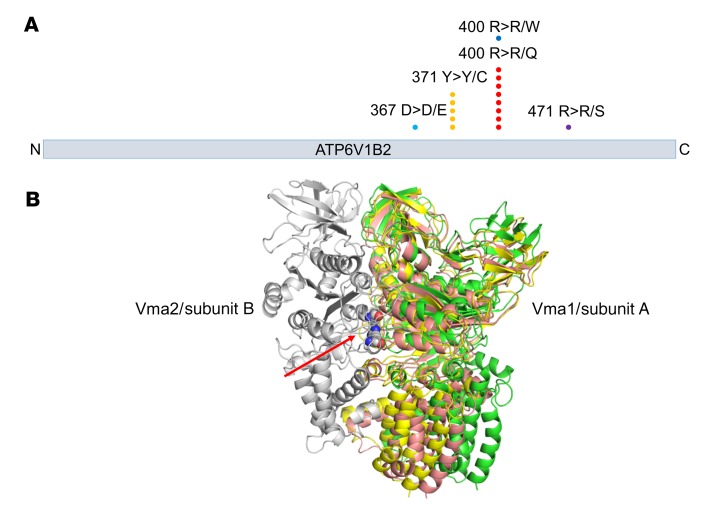
Figure 1. Graphical display and 3D modeling of FL-associated ATP6V1B2 (v-ATPase) mutations.
(A) ATP6V1B2 mutations at known hotspots (p.Y371Y>C and p.R400R>Q) and the mutations identified in this study are indicated. (B) 3D model of yeast v-ATPase based on electron microscopy data published by Zhao et al. (32). The location of yeast amino acid residues corresponding to the human ATP6V1B2 hotspot mutations p.Y371Y>C and p.R400R>Q are indicated by the red arrow. The mutations are located in a region of yeast Vma2/v-ATPase subunit B, which is involved in the ability of the complex to adopt different functional states (green: open; pink: loose; yellow: tight; all 3 states are superimposed in this figure).
FL-associated mutations in ATP6V1B2 are located at the dimer interface with ATP6V1A.
We modeled the location of the ATP6V1B2 hotspot mutations p.Y371Y>Y/C and p.R400R>R/Q on the published cryoelectron microscopy model of the yeast v-ATPase (32) (the human ATP6V1B2 protein has 77% sequence identity to its yeast counterpart). We found that both ATP6V1B2 hotspot mutations are located at the interface of the 2 subunits that correspond to the human/yeast v-ATPase subunits ATP6V1A/Vma1 and ATP6V1B2/Vma2 (Figure 1B). Zhao et al. recently reported that the v-ATPase in yeast exists in 3 states (open, loose, and tight) and that these states are linked to enzymatic activity, ATP-ADP binding, and signaling to the Vma3 subunit for proton translocation into the organelle lumen (32). The 3 conformations are thought to bind ATP, ADP, and phosphate, and no nucleotide, respectively. We found that yeast Vma2 residues Y352 and R381 (homologous to the FL-associated ATP6V1B2 hotspot mutations p.Y371Y>Y/C and p.R400R>R/Q) undergo significant conformational changes from one catalytic conformation to the other (Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/JCI98288DS1), coupled with changes in the interaction with the partner Vma1. This suggests that the Y371C and R400Q mutations may have an impact on the interconversion between the 3 conformational states, influencing the rate and efficiency of the ATPase and proton pumping activity (33).
FL-associated ATP6V1B2 mutations activate autophagic flux.
The v-ATPase is a key component of the cellular autophagic apparatus (34, 35). Upon assembly on lysosomal membranes, the v-ATPase pumps protons into the lysosomal lumen, and the resulting acidification activates lumenal proteases and peptidases, thus facilitating degradation of autophagy-derived and endocytic contents into free amino acids (36, 37). The proton gradient is also implicated in the inside-out active transport of amino acids from the lysosomal lumen into the cytoplasm as well as the activation of mTORC1 (27, 38).
We tested for possible effects of ATP6V1B2 mutations on autophagy using the steady-state levels of the well-studied autophagosomal marker LC3-II (39). Nascent LC3 is typically cleaved at the C-terminus and then conjugated to phosphatidylethanolamine (PE, termed LC3-II), allowing it to become associated with the membrane of the phagophore, and part of the LC3-II pool remains associated with the completed autophagosome.
Following transient transfection of human embryonic kidney 293 T cells (HEK293T cells) with either vector alone or vector containing cDNAs for WT or mutant HA-tagged ATP6V1B2, we detected highly elevated LC3-II levels in the mutant transfectants (Figure 2A). The LC3-II that remains associated with the autophagosome is located inside this compartment; subsequent fusion with the lysosome results in the degradation of this protein along with the autophagic cargo. For this reason, an increase in LC3-II may correspond to an induction of autophagy or to a block in the turnover of this protein in the lysosome (40). In order to distinguish between these two possibilities, we blocked lysosomal LC3-II degradation using the protease inhibitors pepstatin A and E-64D in parallel experiments and detected a further increase in LC3-II levels (Figure 2, A and C), indicative of elevated autophagic flux and normal fusion of autophagosomes with lysosomes in the mutant cells.
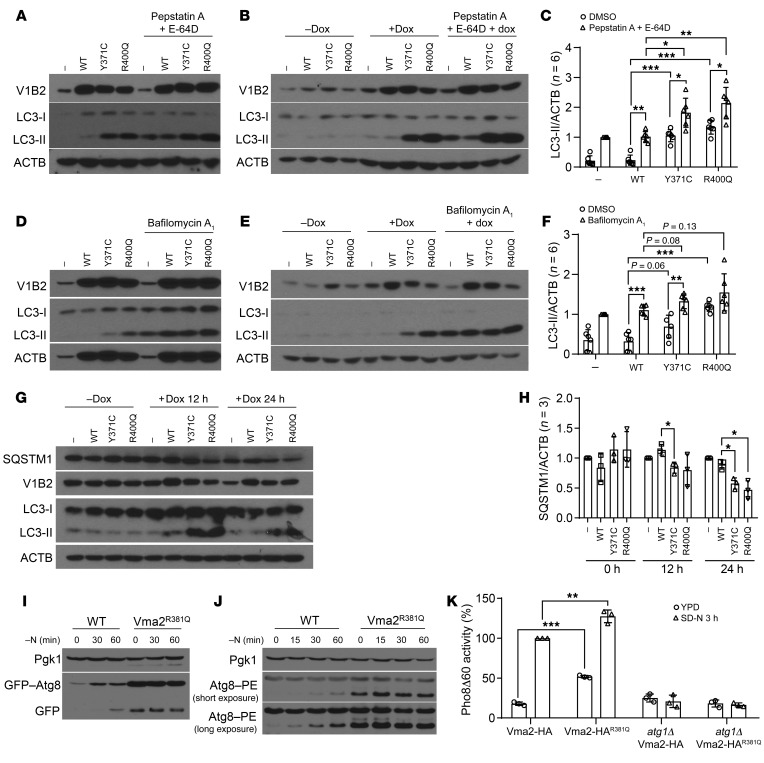
Figure 2. FL-associated ATP6V1B2 mutations activate autophagic flux.
(A) HEK293T cells transfected with empty vector, WT, or mutant (Y371C; R400Q) ATP6V1B2 treated or not with pepstatin A and E-64D. (B) Stable OCI-LY1 lymphoma cells carrying an inducible lentivirus (pCW57.1) expressing WT or mutated ATP6V1B2 (with or without doxycycline [Dox]) treated or not with pepstatin A and E-64D. (C) Densitometric quantification (LC3-II/ACTB) of 3 independent experiments per cell line from representative panels A and B. (A and B) Statistical comparisons: R400Q or Y371C versus WT (A, lanes 2–4; B, lanes 6–8); R400Q or Y371C versus WT plus pepstatin A and E-64D (A, lanes 6–8; B, lanes 10–12); R400Q or Y371C or WT with or without pepstatin A and E-64D. (C) *P < 0.05, **P < 0.01, and ***P < 0.001, by unpaired, 2-tailed t test with Bonferroni’s correction. Error bars indicate the SD. (D) HEK293T cells as in A treated or not with bafilomycin A1. (E) Stable OCI-LY1 lymphoma cells as in B (with or without doxycycline) treated or not with bafilomycin A1. (F) Densitometric quantification (LC3-II/ACTB) of 3 independent experiments per cell line from representative panels D and E. Statistical comparisons: R400Q or Y371C versus WT (D, lanes 2–4; E, lanes 6–8); R400Q or Y371C versus WT plus bafilomycin A1 (D, lanes 6–8; E, lanes 10–12); R400Q or Y371C or WT with or without bafilomycin A1. **P < 0.01 and ***P < 0.001, by unpaired, 2-tailed t test with Bonferroni’s correction. Error bars indicate the SD. (G) Stable HEK293T cells carrying inducible WT or mutated ATP6V1B2 induced with doxycycline for 12 or 24 hours. (H) Densitometric quantification (SQSTM1/ACTB) of 3 independent experiments from G (*P < 0.05, R400Q or Y371C versus WT comparisons within time points; unpaired, 2-tailed t test with Bonferroni’s correction). (I–K) The S. cerevisiae strain WLY176 was used to generate a Vma2R381Q-HA–knockin point mutation in the genome. Autophagy activity was measured with or without nitrogen (N) using (I) a GFP-Atg8 processing assay, (J) an Atg8 lipidation assay, and (K) a Pho8Δ60 activity assay. (K) **P < 0.01 and ***P < 0.001, by unpaired, 2-tailed t test. Error bars indicate the SD. SD-N, synthetic starvation medium with glucose, but lacking nitrogen.
We generated various stable doxycycline-inducible lymphoma cell lines expressing WT or mutant ATP6V1B2 and conducted experiments similar to the above-detailed HEK293T cell experiments. Upon doxycycline induction of ATP6V1B2 expression in OCI-LY1 cells, we again detected highly elevated LC3-II levels in the mutants. Like the findings with HEK293T cells, treatment with pepstatin A and E-64D resulted in an additional increase in the levels of LC3-II (Figure 2, B and C).
We also performed similar experiments with the v-ATPase inhibitor bafilomycin A1 and found that LC3-II accumulated to a higher level than that detected in the ATP6V1B2 mutants without inhibitor treatment (Figure 2, D–F).
Next, we followed the protein levels of SQSTM1 (also known as p62) for 12 and 24 hours after induction of WT and mutant ATP6V1B2 proteins (41). As can be seen in Figure 2, G and H, SQSTM1 levels declined in the ATP6V1B2 mutants, in agreement with induction of autophagic flux.
To further investigate the effect of the ATP6V1B2R400Q mutation on autophagy and autophagic flux, we proceeded to test its effects in a yeast model. Sequence alignment showed that the ATP6V1B2R400Q mutation corresponds to yeast Vma2R381Q, and we generated a Vma2R381Q-HA mutation in the genome. As a readout of autophagy activity, we measured GFP-Atg8 processing. Autophagy activation leads to the lipidation of Atg8 (akin to LC3-II in mammalian cells), which in turns leads to Atg8 binding to both sides of the phagophore. A portion of Atg8, or GFP-tagged Atg8, is delivered to the vacuole via autophagy; GFP is relatively resistant to vacuolar hydrolases, and the free form can be resolved and detected by Western blotting. Accordingly, this assay directly monitors the delivery of the autophagosome to the vacuole. The Vma2R381Q mutant showed increased delivery of GFP-Atg8 and subsequent processing under both growing and nitrogen starvation conditions, which was indicative of elevated autophagic flux (Figure 2I). Furthermore, the GFP-Atg8 chimera is under the control of the endogenous ATG8 promoter, and the ATG8 gene was substantially upregulated following autophagy induction. The increase in the amount of full-length GFP-Atg8 seen in the Vma2R381Q strain, even at time zero, is another indication of upregulated autophagy as a result of the v-ATPase mutation.
As a parallel method of monitoring autophagic induction, we examined the endogenous Atg8 protein through a lipidation assay. The attachment of the Atg8 C-terminus to PE results in a faster electrophoretic mobility relative to nonlipidated Atg8 (42). The cells expressing Vma2R381Q showed increased Atg8 lipidation under growing and nitrogen-starvation conditions (Figure 2J).
Finally, we measured autophagy activity using the quantitative Pho8Δ60 assay (43). In brief, Pho8Δ60 is an engineered form of the Pho8 phosphatase that can only be delivered to the vacuole (the yeast equivalent of lysosomes) nonselectively through autophagy. Subsequently, the Pho8Δ60 propeptide gets cleaved by vacuolar hydrolases, thereby activating its phosphatase activity. The Vma2R381Q mutant showed a significant increase in autophagy activity under both growing and nitrogen-starvation conditions (Figure 2K) that was dependent on autophagy related 1 (Atg1); deletion of ATG1 (the yeast homolog of unc-51 like autophagy activating kinase 1 [ULK1]) fully prevented the induction of autophagic flux by nitrogen starvation or Vma2R381Q (Figure 2K). Furthermore, we observed a similar increase in autophagy activity during nitrogen starvation in a strain carrying both a WT and R381Q-mutant version of the VMA2 gene (Vma2-HA WT/R381Q), a situation aimed at mimicking the heterozygous mutations occurring in FL tumors (Supplemental Figure 2, A and B). Together, these data show that the mutation R381Q in Vma2 increases autophagy activity, similar to the result seen with mutant ATP6V1B2 in mammalian cells.
FL-associated ATP6V1B2-mutant proteins activate mTOR.
mTOR is the major regulator of cellular metabolism and a negative regulator of autophagy (44–46). We hypothesized that mutations in the v-ATPase resulted in reduced degradation of lysosomal proteins and reduced release of amino acids from the lysosome, which would decrease mTOR activity (47, 48); this prediction would fit with the increase in autophagy activity.
HEK293T cells have been extensively used in the functional characterization of components of the v-ATPase/Ragulator/RRAGA/-B/-C/-D/mTOR pathway (27, 49, 50). In doxycycline-inducible stable HEK293T cells induced for 24, 48, or 72 hours, we followed the expression of ATP6V1B2, LC3-II, total RPS6KB (referred to hereafter as S6K), and phosphorylated S6K (p-S6K) at residue Thr389 over time. With prolonged ATP6V1B2 induction, we found substantially elevated LC3-II levels and increased S6K phosphorylation, indicative of elevated mTOR activity (Figure 3A). We conducted an extensive series of similar experiments in doxycycline-inducible stable lymphoma cells and detected equivalent changes (Figure 3,B–E; note that the induced expression of ATP6V1B2 p.R400Q in some of the lines is lower than that for WT ATP6V1B2 or the p.Y371C mutation). The induction of mTOR activity was not immediate after ATP6V1B2 induction, but rather was delayed.
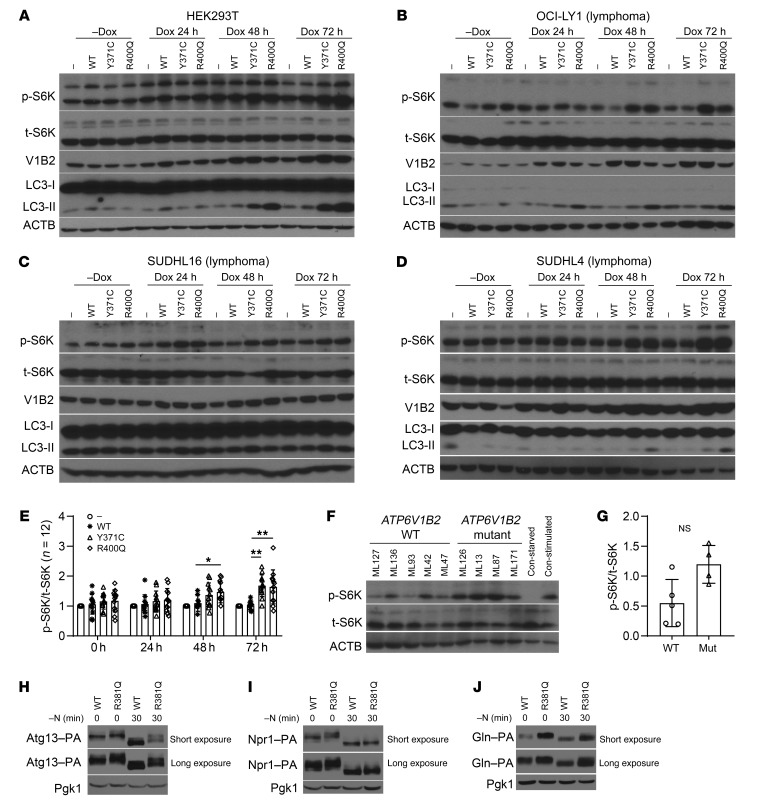
Figure 3. FL-associated ATP6V1B2 mutations activate both autophagic flux and mTOR.
(A) Stable HEK293T cells carrying the inducible lentivirus pCW57.1 with WT or mutated (Y371C; R400Q) ATP6V1B2. Cells were induced with doxycycline for 0, 24, 48, or 72 hours. (B–D) Stable OCI-LY1, SUDHL16, or SUDHL4 lymphoma cells carrying the inducible lentivirus pCW57.1 with WT or mutated (Y371C; R400Q) ATP6V1B2. Cells were induced with doxycycline for 0, 24, 48, or 72 hours. Delayed activation of mTOR, as evidenced by elevated p-S6K/total S6K (p-S6K/t-S6K) levels, was detected. (E) Combined densitometric quantification (p-S6K/t-S6K) of 3 independent experiments per cell line from A–D. *P < 0.05 and **P < 0.01, R400Q or Y371C versus WT comparisons within time points; unpaired 2-tailed t testing with Bonferroni’s correction. Error bars indicate the SD. (F and G) Phosphorylation of S6K in purified primary human FL B cells carrying WT or mutated ATP6V1B2. (F) Immunoblot results for lysates of purified FL B cells from 5 patients carrying WT ATP6V1B2 and from 4 patients carrying mutated ATP6V1B2, as well as for control lysates made from unstimulated or insulin- and serum-stimulated OCI-LY7 cells. One of two representative immunoblots is shown. Con-stimulated, stimulated control; Con-starved, starved control. (G) Densitometric quantification (p-S6K/t-S6K) of the blots shown in F. Error bars indicate the SD. (H–J) TOR activity measured by phosphorylation in S. cerevisiae in growing conditions and after a 30-minute nitrogen starvation (substrate gel migration assay; the higher the phosphorylation state, the slower the gel migration) for (H) Atg13, (I) Npr1, and (J) Gln3. Varying exposure durations are shown.
We also measured mTOR activity in the inducible HEK293T cell lines using various p-EIF4EBP1 epitopes as readouts (Supplemental Figure 3, A–D) and by monitoring S6K phosphorylation in primary purified FL B cells in fully supplemented medium carrying either WT (n = 5) or mutant (n = 4) ATP6V1B2 (Figure 3, F and G). Overall, the data again indicated elevated mTOR activity in ATP6V1B2-mutant cells.
As above, we proceeded with complementary experiments in yeast. To determine the effect of the Vma2R381Q mutation on TOR activity, phosphorylation levels of 3 TOR substrates, Atg13, Npr1, and Gln3 (51–53), were used as readouts (Figure 3, H–J). TOR phosphorylates these proteins in nutrient-rich conditions; however, during nitrogen starvation, TOR inactivation leads to dephosphorylation of these target proteins, which can be observed by a downward shift in their apparent molecular weight. During growing conditions (time zero), Atg13, nitrogen permease reactivator 1 (Npr1), and a nitrogen-responsive transcriptional regulator (Gln3) showed increased TOR phosphorylation in the Vma2R381Q cells, as evidenced by decreased mobility compared with the WT cells, indicating that TOR was hyperactive in the mutant strain (Figure 3, H–J). After 30 minutes of nitrogen starvation, all 3 substrates displayed increased mobility in extracts from the WT strain, corresponding to a decrease in TOR activity. In contrast, Atg13, Npr1, and Gln3 still showed increased phosphorylation levels in the Vma2R381Q mutant. This finding suggests that TOR is partially nonresponsive to nitrogen depletion in Vma2R381Q-mutant cells. Together, the findings in mammalian cells and yeast indicate an mTOR-activated phenotype for FL-associated ATP6V1B2-mutant proteins.
The activation of autophagic flux and intact SLC38A9 functions are necessary for mTOR activation by mutant ATP6V1B2.
Next, we decided to investigate the molecular mechanism responsible for the increase in autophagy activity in the presence of active mTOR/TOR. Considering the central role of ULK1 in autophagy initiation, we measured previously identified regulatory mTOR- and AMPK-mediated phosphorylation events in ULK1 (mTOR: p–757-ULK1; AMPK: p–317-ULK1) in ATP6V1B2 doxycycline-inducible HEK293T and LY1 lymphoma cells. As expected, glucose deprivation strongly increased p–317-ULK1, and leu deprivation reduced p–757-ULK1; however, neither site was substantially influenced by mutant ATP6V1B2 (Supplemental Figure 4, A and B).
Next, we examined the role of the stress-responsive activating transcription factor 4 (ATF4) (54). We found that ATF4 protein expression was elevated in doxycycline-inducible HEK293T and LY1 lymphoma cells expressing ATP6V1B2 p.R400Q and, to a lesser degree, ATP6V1B2 p.Y371C, indicative of metabolic stress (Supplemental Figure 5, A and B). We disrupted ATF4 in stable ATP6V1B2 WT or mutant inducible HEK293T cells using CRISPR-Cas9 targeting. While leucine starvation strongly induced ATF4 expression in the parental cells, its expression was completely lost after ATF4 disruption (Supplemental Figure 6A). However, LC3-II levels remained highly elevated in the ATP6V1B2-mutant strains lacking ATF4, arguing against a critical role for this transcription factor in autophagic flux initiation by mutant ATP6V1B2 (Supplemental Figure 6B).
The yeast homolog of ATF4, the amino acid starvation–responsive transcription factor Gcn4, has been implicated in autophagy regulation (55, 56). Accordingly, we measured the expression of several ATG genes in Saccharomyces cerevisiae carrying Vma2 WT or the Vma2R381Q-HA–knockin point mutation (55, 56). Under full-medium culture conditions, we detected substantially elevated expression of these ATG genes in the cells carrying Vma2R381Q-HA, further confirming the autophagy-inducing properties of the ATP6V1B2 mutants (Supplemental Figure 6C). Subsequently, we deleted GCN4 or the gene encoding its upstream regulatory kinase GCN2 (EIF2A/eIF2α kinase) in these yeast strains. Despite the deletion of one or both of these genes, the elevated expression of the ATG genes persisted, although there appeared to be a partial reduction in the double-knockout strain, arguing against the GCN2/GCN4 pathway as the critical mediator of autophagy induction in the ATP6V1B2 mutants (Supplemental Figure 6C).
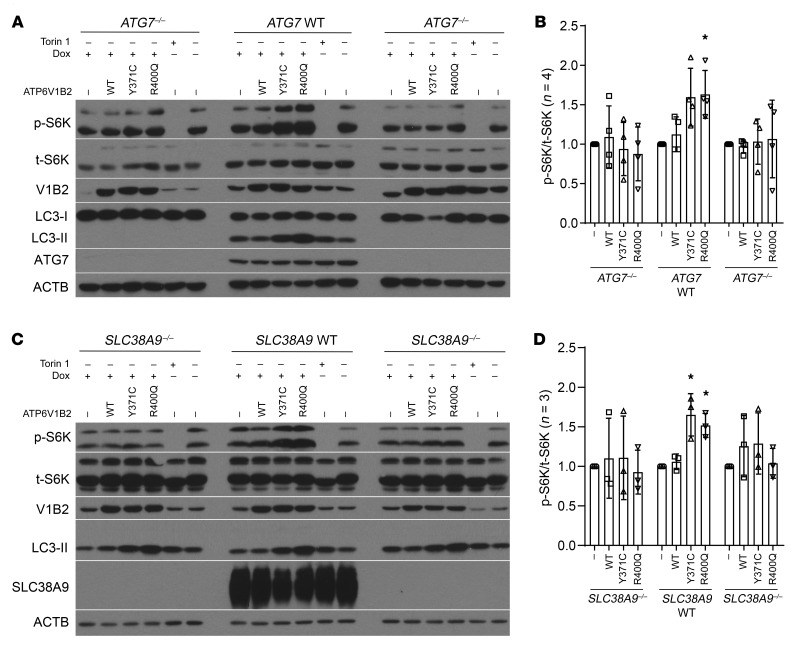
The second major phenotype resulting from the ATP6V1B2/Vma2 point mutations was the delayed activation of mTOR/TOR under nutrient stress (i.e., normally inactivating) conditions. To understand the relationship between activation of autophagic flux and mTOR activation, we generated HEK293T cells null for ATG7, a critical component of the autophagic machinery. We generated clonal HEK293T ATG7–/– cells that also carried inducible WT and mutant ATP6V1B2 expression cassettes and induced expression of ATP6V1B2 proteins using doxycycline. While the induction of LC3-II and mTOR activation was readily detectable in the ATG7 WT cells that had also gone through the CRISPR-Cas9 targeting process, we observed that both phenotypes were abolished in the ATG7–/– cells (Figure 4, A and B). This finding indicates that the activation of autophagic flux is the primary molecular consequence of mutations in ATP6V1B2 and that intact macroautophagy is required for the observed delayed mTOR activation.
Figure 4. An intact autophagy pathway and lysosomal amino acid sensing and transport are necessary for delayed activation of mTOR by mutant ATP6V1B2.
(A and B) ATG7–/– HEK293T cells were generated using CRISPR-Cas9 targeting, and independent single-cell clones were expanded. Control ATG7 WT cells were expanded from cells that were subjected to CRISPR-Cas9 targeting but remained intact for ATG7. All clones were subsequently infected with lentiviruses carrying doxycycline-inducible WT or mutant ATP6V1B2. Cells were induced with doxycycline for 72 hours, and cell lysates were prepared for immunoblotting using the indicated antibodies. (B) Results of quantification (p-S6K/t-S6K) of 4 independent experiments using data from all lanes labeled WT, Y371C, and R400Q. Statistical analysis compared the difference of the differences between measurements under ATG7 WT and ATG7–/– conditions (see Methods). P = 0.06 for Y371C and P = 0.04 for R400Q for ATG7–/–/ATG7 comparisons (standard regression-based Wald-type tests). Error bars indicate the SD. (C and D) SLC38A9–/– HEK293T cells were generated using CRISPR-Cas9 targeting, and independent single-cell clones were expanded. Control SLC38A9 WT cells were expanded from cells that were subjected to CRISPR-Cas9 targeting but remained intact for SLC38A9. All clones were infected with lentiviruses carrying doxycycline-inducible WT or mutant ATP6V1B2. Cells were induced with doxycycline for 72 hours, and cell lysates were prepared for immunoblotting using the indicated antibodies. (D) Results of quantification (p-S6K/t-S6K) of 3 independent experiments using data from all lanes labeled WT, Y371C, and R400Q. Statistical analysis compared the difference of the differences between measurements under SLC38A9 WT and SLC38A9–/– conditions (see Methods). P = 0.01 for Y371C and P = 0.01 for R400Q for SLC38A9–/– and SLC38A9 comparisons (standard regression-based Wald-type tests). (B and D) *P < 0.05, by standard regression-based Wald-type tests. Error bars indicate the SD.
The previously reported delayed reactivation of mTOR following starvation has been ascribed to the substantial upregulation of autophagic flux under starvation conditions (57). We reasoned that increased cargo delivery to lysosomes would activate mTOR through canonical pathways and mechanisms. Given the recently elucidated role of SLC38A9 in amino acid sensing and transport in lysosomes, we proceeded to generate HEK293T SLC38A9–/– cells (58–60). While the induction of LC3-II and mTOR was readily detectable in the SLC38A9 WT cells that had also gone through the CRISPR-Cas9 targeting process as expected, both phenotypes were abolished in the SLC38A9–/– cells (Figure 4, C and D). Combined, the data strongly suggest that increased protein delivery to lysosomes followed by elevated lysosomal amino acid concentrations and signaling through SLC38A9 ultimately results in mTOR activation in the setting of aberrantly persistent elevated autophagic flux conditions.
Mutant ATP6V1B2 confers a defect on the v-ATPase holoenzyme proton pump.
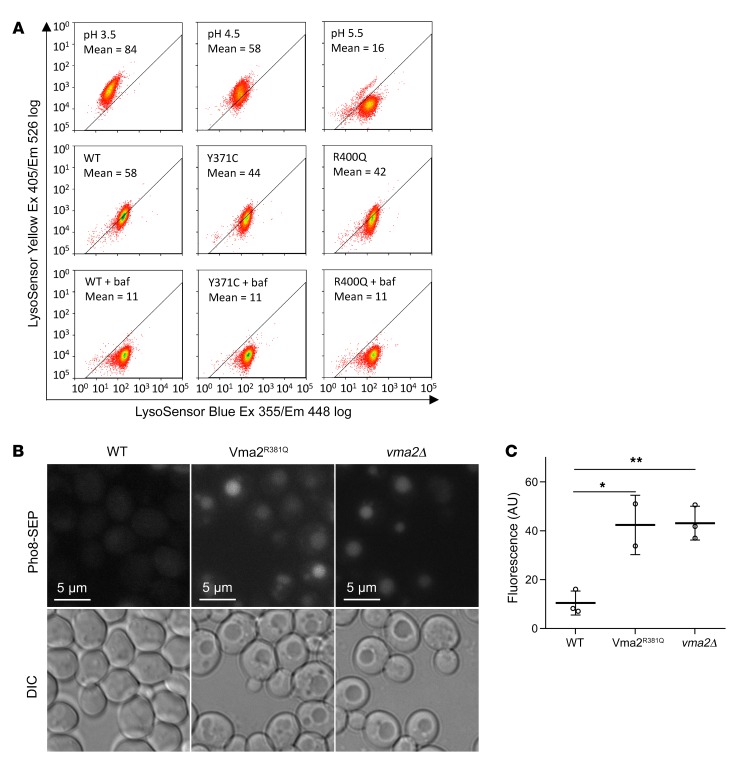
Given the central role of the v-ATPase as a proton pump, we proceeded with measurements of lysosomal pH in stable transduced HEK293T cells expressing WT or mutant ATP6V1B2. Cells were loaded with a pH-sensitive dual reporter dye conjugated to dextran and subsequently analyzed using FACS. As controls, cells in parallel were briefly incubated with buffers at a defined pH of 3.5, 4.5, or 5.5, respectively (Figure 5A, top row), or were treated with bafilomycin A1 to completely inhibit v-ATPase function (Figure 5A, bottom row). Whereas treatment with bafilomycin A1 resulted in an elevated pH in all cells as expected from its ability to completely inhibit the v-ATPase, we detected an acidic lysosomal pH of approximately 4.5 for the ATP6V1B2 WT and an acidic lysosomal pH ranging above 4.5 to 5.5 in the mutant cell lines; therefore, the mutants displayed a reduced ability for acidification (Figure 5A; middle row).
Figure 5. FL-associated ATP6V1B2 mutations reduce the ability of the v-ATPase to acidify lysosomes.
(A) Stable HEK293T cells were generated using the doxycycline-inducible lentivirus pCW57.1 carrying WT or mutated cDNAs encoding ATP6V1B2. Cells were induced with doxycycline for 48 hours and loaded afterwards with a pH indicator dye (dextran-conjugated LysoSensor Blue/Yellow) for 12 hours. In parallel, untransfected HEK293T cells, serving as pH controls, were treated with EMS buffer (see Methods) calibrated to pH 3.5, pH 4.5, or pH 5.5. The fluorescence intensity of cell suspensions was read at various wavelengths using flow cytometry. The MFI of the yellow dye fluorescence signal is a measure of lysosomal pH. Bafilomycin A1 (baf), which completely blocks lysosomal acidification, was used as a control for neutral pH. Ex, excitation; Em, emission. (B) In S. cerevisiae, the C-terminus of the protein Pho8 was tagged with a pH-sensitive SEP protein that increases fluorescence with increasing pH. Like vma2Δ (a knockout strain of the yeast homolog of human ATP6V1B2), Vma2R381Q cells demonstrated increased vacuolar pH compared with WT cells. Upper images show the fluorescence signal; lower images show the corresponding light microscopy. (C) Summary of the fluorescence intensity of vacuolar fluorescence measurements in S. cerevisiae for the data in B. *P < 0.05 and **P < 0.01, by t test with Bonferroni’s correction. Data represent the SD.
We extended this analysis to determine whether the R381Q mutation in Vma2 affects the ability of the v-ATPase to acidify the vacuolar lumen in yeast. To this end, the Pho8 C-terminus was tagged with a pH-sensitive super ecliptic pHluorin (SEP) protein that displays increased fluorescence with higher pH levels (61). Like the control vma2Δ strain, Vma2R381Q cells showed increased vacuolar pH compared with that of WT cells (Figure 5, B and C) (33).
Increased lysosomal localization of mutant ATP6V1B2 proteins and increased interaction with the Ragulator complex.
The v-ATPase is composed of 2 primary subcomplexes that can associate and dissociate in response to changing nutrient conditions (62, 63). We tested whether the 2 hotspot mutations in ATP6V1B2 (Figure 1B) affected interaction with ATP6V1A and could therefore have effects on the assembly of an intact v-ATPase. In transient cotransfection studies using HEK293T cells expressing HA-tagged WT and mutant ATP6V1B2 followed by HA-bead–based IPs, we detected no effect of the ATP6V1B2 mutants on the amount of coimmunoprecipitated ATP6V1A (Supplemental Figure 7).
Next, we used confocal microscopy to assess the amount of WT and mutant ATP6V1B2 localizing to lysosomes. Using fluorescence color changes as a readout for colocalization of ATP6V1B2 and lysosomal-associated membrane protein 1 (LAMP1), an endosomal and lysosomal membrane marker, we detected increased amounts of mutant ATP6V1B2 on endosomes and/or lysosomes when compared with the WT ATP6V1B2 staining (Supplemental Figure 8, A and B).
To study the interaction between ATP6B1V2 and the Ragulator (referred to hereafter as LAMTOR1 to LAMTOR5) proteins (49), we transiently cotransfected plasmids encoding HA-tagged WT or mutant ATP6V1B2, together with a mixture of plasmids that express FLAG-tagged LAMTOR1 to LAMTOR5, into HEK293T cells. After cell lysis, we performed reciprocal co-IPs using anti-HA– or anti-FLAG–conjugated beads. Upon immunoblotting of lysates and immunoprecipitates, we detected modestly increased interaction of the mutant ATP6V1B2 protein (anti-HA band) with the LAMTOR complex (compare lane 2 with lanes 3 and 4) (Supplemental Figure 8C).
Improved survival and growth of lymphoma cell lines carrying mutant ATP6V1B2 cultured under leucine-deprivation conditions.
We hypothesized that the dual activation of autophagic flux and mTOR by the FL-associated ATP6V1B2 mutant proteins may confer increased survival or progrowth properties on afflicted lymphoma cells. To test this hypothesis, we used lentivirus-based transient dual sequential transductions of ATP6V1B2 WT and mutant cDNAs into the SUDHL4 lymphoma cell line, and, 48 hours later, we incubated cells in full medium (100% leucine) or in medium with only 25% leucine. We followed cell counts and viability for 9 days after infection. For the cells grown in full medium, we noticed no significant differences; however, for the cells grown in 25% leucine, we measured a significantly greater number of viable mutant cells (Supplemental Figure 9B; P < 0.05 for both mutants). Data from 4 independent experiments are summarized in Supplemental Figure 9, A and B, and indicate a survival or growth advantage especially for the cells carrying the ATP6V1B2 R400Q mutation.
Next, we took advantage of the doxycycline-inducible stable cell lines described above. As with the experiments using SUDHL4 cells, we incubated cell lines expressing WT or mutant ATP6V1B2 after doxycycline induction for 48 hours in either full- or limited-leucine medium. Whereas OCI-LY1 cell lines carrying the ATP6V1B2 mutants did not grow as well under full-medium conditions (Supplemental Figure 9C), both mutant lines survived substantially and significantly better in low leucine concentrations (Supplemental Figure 9, D–F).
Improved survival and growth of primary FL B cells carrying mutant ATP6V1B2 cultured under leucine-deprivation conditions.
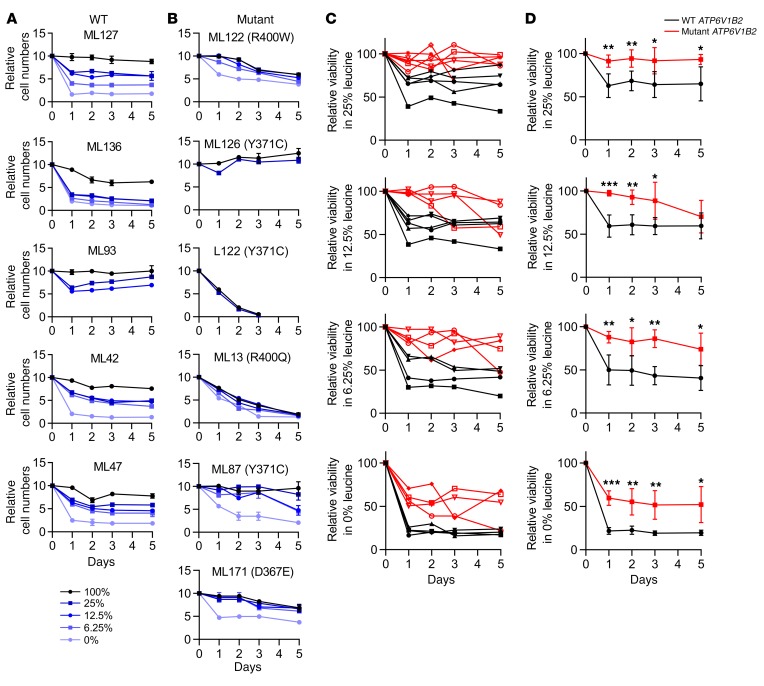
Next, we studied primary human FL B cells purified from cryopreserved lymphoma biopsy specimens. FL B cells carrying WT (n = 5) or mutant (n = 6) ATP6V1B2 were cultured in dialyzed serum–supplemented RPMI 1640 medium containing either 100% leucine or various reduced leucine concentrations depending on the availability of sufficient numbers of cells. FL B cells were cultured, and the cell numbers and fractions of living cells were determined for up to 5 days. FL B cells carrying WT ATP6V1B2 demonstrated substantial cell death induction in the first 24–48 hours of culture under reduced leucine concentrations (Figure 6A); in contrast, most FL B cells carrying mutated ATP6V1B2 largely survived under low leucine concentrations (Figure 6B). At all tested leucine concentrations, a substantially larger fraction of FL B cells carrying mutated ATP6V1B2 survived (Figure 6, C and D). Even at 0% leucine, more than 60% of FL B cells carrying mutated ATP6V1B2 survived, while only approximately 20% of cells carrying WT ATP6V1B2 remained alive.
Figure 6. FL-associated ATP6V1B2 mutations increase the viability of primary human FL B cells under leucine-starvation conditions.
Purified FL B cells carrying (A) WT (n = 5) or (B) mutant (n = 6) ATP6V1B2 were cultured in dialyzed serum–supplemented RPMI 1640 medium containing either 100% leucine or, depending on the availability of sufficient cells, reduced leucine concentrations (0%, 6.25%, 12.5%, and 25%). FL B cells were cultured, and the cell number and fraction of living cells were determined out to 5 days. (C and D) Summary of the results of the individual data displayed in A and B. Shown is the number of viable cells at various leucine concentrations, divided by the number of living cells at 100% leucine concentration. Significance levels are indicated by asterisks: *P < 0.05, **P < 0.01, and ***P < 0.001, compared with results for WT at the same time point (comparisons were done with a t test). Data represent the SD.
These data support the hypothesis that FL-associated ATP6V1B2 mutations facilitate lymphoma cell growth and survival under conditions of amino acid deprivation, probably through the generation of free leucine resulting from elevated autophagy.
Primary human FL B cells carrying mutant ATP6V1B2 are sensitive to inhibition of autophagic flux.
In preliminary experiments, we incubated purified FL B cells in fully supplemented RPMI 1640 medium and added the autophagy inhibitor bafilomycin A1 (35). We determined the fraction of cells that were alive, apoptotic, or dead using ANXA5 (also known as annexin V) and propidium iodide–based flow analyses. We found that FL B cells from 4 of 5 patients carrying mutated ATP6V1B2 showed sensitivity to bafilomycin A1, whereas cells from only 1 of 5 patients carrying WT ATP6V1B2 demonstrated sensitivity (Supplemental Figure 10). These data provided preliminary evidence that autophagic flux sustains survival of ATP6V1B2-mutated FL B cells.
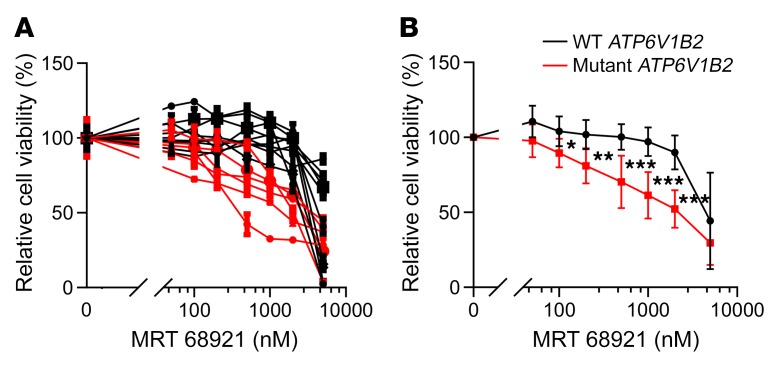
Two recent reports describe the discovery and functional studies of novel ULK1 kinase inhibitors (MRT68921 and SBI-0206965) (64, 65). We confirmed the ability of both molecules to inhibit phosphorylation of the direct ULK1 substrate ATG13 (Supplemental Figure 11) and, on the basis of potency, selected MRT68921 for further studies. Primary purified human FL B cells isolated from lymph node biopsies that carried WT or mutant ATP6V1B2 were treated with escalating doses of MRT68921 and the cell fraction that was alive after 48 hours was measured using ANXA5/annexin V–propidium iodide–based flow analyses. We found that FL B cells carrying mutant ATP6V1B2 were substantially and significantly more sensitive to ULK1 inhibitor–induced cell death than were ATP6V1B2 WT lymphoma cells (Figure 7, A and B).
Figure 7. FL-associated ATP6V1B2 mutations and dependence on autophagic flux for survival of primary FL B cells.
(A and B) Primary human purified FL B cells carrying WT or mutant ATP6V1B2 were cultured in serum-supplemented RPMI 1640 medium containing 100% leucine and treated with the ULK1 inhibitor MRT68921 for 72 hours at the indicated concentrations. Cell viability was measured using CellTiter-Glo and was normalized to the viability of cells cultured for 72 hours but left untreated. *P < 0.05, **P < 0.01, and ***P < 0.001 by t test, compared with results for WT at the same time point. Data represent the SD.
We also tested the ULK1 inhibitor MRT68921 in combination with mTOR inhibitors (rapamycin or torin 1) or chemotherapeutics (doxorubicin or vincristine) used clinically to treat patients with FL (Supplemental Figure 12, A–H). We found that ULK1 and mTOR inhibition was antagonistic, which was probably the result of further mTOR inhibitor–mediated elevations in autophagic flux (Supplemental Figure 12, E and F). In contrast, the combinations of ULK1 inhibitors with doxorubicin and especially with vincristine preferentially enhanced the killing of ATP6V1B2-mutant FL B cells, and in this context it is of interest that vinblastine, a drug with properties similar to those of vincristine, is known to inhibit autophagic flux (Supplemental Figure 12, G and H) (66).
In summary, these data uncover a survival addiction to activated autophagic flux in ATP6V1B2-mutant FL B cells, which we posit offers opportunities for the development of novel therapeutic approaches to treat FL.
Discussion
Whole-exome sequencing has begun to provide direction for investigating the genetic basis of FL (11–13, 15, 18, 30). For example, recurrent mutations in RRAGC and subunits of the v-ATPase in FL suggest a role for amino acid (via the RRAG proteins) and energy-sensing (through the v-ATPase) mechanisms that feed into the regulation of mTOR in the pathogenesis of FL. Whereas mutations in RRAGC activate mTOR in part through facilitated interactions with RPTOR (24, 25), the functional consequences of mutations in the v-ATPase subunits have remained unexplored. FL-associated mutations in the v-ATPase occur predominantly in ATP6AP1 and ATP6V1B2, the latter of which is a principle component of the multimeric V1 hexameric subunit of the v-ATPase.
In this study, we set out to address the mechanistic consequences of the heterozygous ATP6V1B2 hotspot mutations p.Y371Y>Y/C and p.R400R>R/Q, which occur at a frequency of 10% in FL. Given the established roles of v-ATPase as a major component of the autophagy machinery important for acidification of lysosomes and as a regulator of mTOR through mechanisms that have yet to be fully defined, we focused our initial studies on these pathways. Through a series of complementary experiments in mammalian cells as well as in S. cerevisiae yeast, we made the following observations: (a) the ATP6V1B2/Vma2 hotspot mutations activate autophagy and autophagic flux; (b) the ATP6V1B2/Vma2 hotspot mutations also activate mTOR/TOR, which led us to identify a gene mutation that activates both pathways simultaneously; (c) lymphoma cell lines and primary FL B lymphoma cells harboring ATP6V1B2 hotspot mutations survive and/or grow better in media containing a lower concentration of the amino acid leucine, demonstrating that these mutations counteract the effects of nutrient deprivation and providing a credible pathophysiologically selective advantage conferred by these mutations; and (d) FL B cells harboring mutant ATP6V1B2 are dependent on autophagic flux for survival.
The lysosomal acidification caused by activated v-ATPase enables activation of a set of lysosome-resident peptidases that facilitate the degradation of proteins delivered to the lysosome. Previous work has demonstrated that autophagosome-lysosome fusion can proceed in the absence of v-ATPase, but the resulting neutral lysosomal pH is unlikely to support the generation of sufficient free amino acids for active cell growth (35). We found in mammalian cells and yeast that mutated ATP6V1B2/Vma2 was hypomorphic and resulted in modestly impaired lysosomal acidification. Simultaneously, the amount of lysosomal v-ATPase was increased, indicating a compensatory adaptation. In addition, the impaired lysosomal acidification is associated with a substantial increase in autophagic flux, again indicating a compensatory adaptive mechanism. The substantial increase in autophagic flux constitutes one principle mechanism of FL-associated mutations in ATP6V1B2. These findings, combined with a recent report demonstrating an inhibitory role of EP300 in autophagy through PIK3C3 (also known as VPS34) acetylation and the frequent genetic inactivation of EP300 in FL, suggest that mutational activation of autophagy is prevalent in FL (13, 67).
Somewhat unexpectedly, we detected increased mTOR activity in recombinant lymphoma cells and primary FL B cells and toward various protein substrates in yeast. The activation of mTOR in lymphoma cells carrying inducible ATP6V1B2 transgenes was delayed, which is compatible with a model in which activation of autophagic flux precedes and ultimately results in the activation of mTOR. In support of this model, ablation of autophagic flux through ATG7 inactivation prevented ATP6V1B2 mutants from activating mTOR. Given the previous findings of an inside-out signaling pathway (lysosomal lumen to cytoplasm) for amino acids leading to mTOR activation (27) (68), we generated SLC38A9-knockout cells and confirmed the essential role for this amino acid sensor/transporter in mutant ATP6V1B2–induced mTOR activation.
In the course of these studies, we show for the first time to our knowledge an effect of ATP6V1B2 mutations on the survival or growth of lymphoma cell lines and primary FL B cells under experimentally reduced leucine concentrations. These data fully support the hypothesis that lymphoma cells carrying ATP6V1B2 mutations are preferentially able to generate leucine from internal stores, probably through the activation of autophagy. This interpretation is further supported by the fact that the majority of primary FL B cells carrying ATP6V1B2 mutations were preferentially sensitive to autophagy inhibitors. The latter finding may open up new avenues for the treatment of FL, possibly through intermittent inhibition of autophagy (69, 70), and inhibitors of ULK or PIK3C3 kinases that are in development may be useful in this setting (64, 65, 71).
In summary, our studies elucidate mechanisms of action of FL-associated mutations in components of the v-ATPase and demonstrate for the first time to our knowledge dual activation of autophagy and mTOR, findings that will provide a framework for future biological and therapeutic studies of v-ATPase in FL.
Methods
Patients’ characteristics and study source material
Results from the Sanger-based resequencing of ATP6V1B2 using template DNA isolated from 144 flow-sorted FL and 14 flow-sorted t-FL cells are reported here. The methods used for FACS of lymphoma specimens have been described previously (12). Specimens from 85 FL patients and 14 t-FL patients (cohort 1) were obtained from 2 separate lymphoma repositories at the University of Michigan. Furthermore, we performed Sanger sequencing to analyze flow-sorted cellular DNA derived from deidentified leftover clinical material collected between 1990 and 2005 from 59 FL patients (cohort 2), as previously described (72).
Exon resequencing of ATP6V1B2 in FL
Primers were designed to amplify and sequence all coding exons and adjacent intronic sequences of ATP6V1B2 using direct sequencing as described previously (12). Mutations were confirmed to be somatically acquired using unamplified FL cell–derived DNA and paired CD3 cell–derived DNA as templates isolated from flow-sorted cells.
ATP6V1B2 cDNA mutagenesis, retroviral and lentiviral vector generation, cell transfection, and cell transduction to generate stable cell lines
Cell lines.
The OCI-LY1 line was obtained from the Ontario Cancer Institute (Toronto, Ontario, Canada). The SUDHL4 cell line was obtained from K. Elenitoba-Johnson (University of Michigan, Ann Arbor, Michigan, USA). Cell line authentication was performed through sequence analysis of 6 to 8 gene mutations per line, based on the COSMIC cell line project (Catalog Of Somatic Mutations In Cancer; http://cancer.sanger.ac.uk/cell_lines). The SUDHL16 cell line was purchased from the DSMZ repository (https://www.dsmz.de/).
Reagents and mutagenesis.
A plasmid containing the ATP6V1B2 cDNA (CH817565) was purchased from Vigene and used as a template to generate mutant ATP6V1B2 cDNAs using the QuikChange Lightning Site-Directed Mutagenesis Kit (Stratagene, Agilent). Full-length WT and mutant HA–tagged ATP6V1B2 were constructed using PCR and cloned into the lentiviral FG9 vector (a gift from Colin Duckett, Ann Arbor, Michigan, USA) (73) or into the retroviral vector MSCV-IRES-PURO (MIP) (74). Full-length WT and mutant HA–tagged ATP6V1B2 were subcloned into the doxycycline-inducible plasmid pCW57.1 (Addgene, plasmid 41393; deposited by David Root, Broad Institute) using Gateway cloning (Life Technologies, Thermo Fisher Scientific ).
Transient transfection studies using HEK293T cells.
HEK293T cells were transfected in 10-cm dishes with 3 μg MIP plasmids encoding either WT or mutant forms of ATP6V1B2 or various other constructs as indicated using polyethylenimine (PEI) (Polyscience Inc., 23966).
Generation of stable transduced HEK293T cell lines.
HEK293T packaging cells were transfected with 5 μg lentiviral vector (either the vector alone or vector expressing WT or various mutant forms of HA-ATP6V1B2), together with 1 μg REV, RRE, and VSVG plasmids. Viral supernatant media were collected 48–72 hours after transfection and cleared by low-speed centrifugation to remove cells and debris.
SUDHL4 cells were double spin-inoculated at 30°C at 1360 g using 8 μg/ml polybrene for 2 hours on day 0 and day 1 with recombinant FG9 lentiviruses carrying WT or mutant cDNAs encoding HA-ATP6V1B2. The infection efficiency was confirmed on day 3 through FACS on the basis of GFP fluorescence and was greater than 90%. Cells were sorted on GFP high fluorescence.
Generation of stable doxycycline-inducible lymphoma cell lines.
The lentiviral expression vector pCW57.1 was used to generate stable doxycycline-inducible cell lines of OCI-LY1, SUDHL4, and SUDHL16. After viral transduction, cells were treated with 2 μg/ml puromycin for 3 to 5 days and dead cells removed using Histopaque-1077 gradient centrifugation (Sigma-Aldrich, RNBF2746) and subsequently cultured in regular medium; cultured pools were used for the experiments described herein. The inducible expression of ATP6V1B2 following doxycycline treatment (1000 ng/ml for 24 to 72 h) was verified by immunoblotting for ATP6V1B2 and HA.
Generation of stable doxycycline-inducible HEK293T cell lines.
Stable doxycycline-inducible HEK293T cell lines carrying WT or mutated HA-ATP6V1B2 cDNAs were generated using the pCW57.1 vector as outlined above. The inducible expression of ATP6V1B2 following doxycycline treatment (1000 ng/ml for 24 to 72 h) was verified by immunoblotting for ATP6V1B2 and HA.
Culture and survival analyses of various stable cell lines in full medium and during leucine deprivation
All experiments were performed at ambient O2 levels and 5% CO2. Prior to leucine starvation, SUDHL4 and SUDHL16 cells were cultured in RPMI 1640 medium supplemented with 10% FCS. OCI-LY1 cells were cultured in DMEM supplemented with 20% FBS and 50 μM β-mercaptoethanol. To control for the leucine concentration, we used leucine-free RPMI medium (Sigma-Aldrich, R1780; with re-added Arg and Lys), leucine-free DMEM medium (Sigma-Aldrich, D9443; with re-added Arg and Lys), and dialyzed FCS using dialysis tubing (Sigma-Aldrich, D7884).
Stable SUDHL4 cells expressing WT or mutated ATP6V1B2 were seeded at 5 × 105 cells/ml and cultured in full RMPI 1640 medium supplemented with 20% FBS. Parallel cell aliquots were cultured under otherwise identical conditions but using only 25% of the leucine concentration.
Doxycycline-inducible stable OCI-LY1 or SUDHL16 cell lines were induced with 1000 ng/ml doxycycline on day 0 and after 48 hours (day 2); cells were seeded at 5 × 105 cells/ml and cultured in full DMEM or RPMI medium supplemented with 10% FBS. Parallel cell aliquots were cultured under otherwise identical conditions but using reduced leucine concentrations. Cells were counted every other day using trypan blue staining and diluted back to a concentration of 5 × 105 cells/ml.
mTOR signaling and autophagy assays in inducible stable cell lines
Doxycycline-inducible stable OCI-LY1, SUDHL4, or SUDHL16 cell lines were induced with 1000 ng/ml doxycycline for 24, 48, or 72 hours. Cells were seeded at 5 × 105 cells/ml and cultured in full DMEM or RPMI medium supplemented with 10% FBS. The cell pellets were lysed in lysis buffer containing 1% NP-40, 150 mmol/l NaCl, 25 mmol/l Tris, pH 8.0, 20 mmol/l NaF, 2 mmol/l EGTA, and 2 mmol/l EDTA, supplemented with protease inhibitors (Sigma-Aldrich, P3840), phosphatase inhibitors (Sigma-Aldrich, P0044), sodium orthovanadate (Sigma-Aldrich, 450243), and PMSF (Thermo Fisher Scientific, 36978). The detergent-soluble fraction of cell lysates was cleared by centrifugation at 17,000 g for 10 minutes. Protein was fractionated through SDS-PAGE and prepared for immunoblotting using standard procedures. For analyses of autophagic flux, various cell lines, as indicated below, were treated with the v-ATPase inhibitor bafilomycin A1 at 100 nM for 4 hours or with the protease (hydrolase) inhibitors pepstatin A for 12 hours at 100 μg/ml and E-64D for 1 hour at 10 μg/ml. Cells were harvested and prepared for immunoblotting with the antibodies listed in Supplemental Table 1. Band intensities were analyzed by densitometry (ImageJ, version 2.0.0, NIH).
Generation of HEK293T cells lacking intact ATG7 or SLC38A9
We generated HEK293T ATG7–/– or SLC38A9–/– cells using pLENTIcrispr v2 (75) containing guides with the following sequences: ATG7 sense, GAAGCTGAACGAGTATCGGC; antisense, CACCGGAAGCTGAACGAGTATCGGC; and SLC38A9 sense, AACTGGATATTCATAGGTCC; antisense, GGACCTATGAATATCCAGTT. After lentiviral infection, single-cell cloning, and screening of clones by immunoblotting, 2 independent single-cell clones for ATG7 or SLC38A9 were selected for further study. As controls, we used single-cell clones that had gone through the entire gene-targeting process but remained WT for ATG7 or SLC38A9. To generate stable doxycycline-inducible, ATP6V1B2-overexpressing cells, we infected all single-cell clones (ATG7–/– or SLC38A9–/– and controls) with the doxycycline-inducible recombinant lentiviral pCW57.1 vectors followed by puromycin selection as outlined above.
ATF4 expression and ULK phosphorylation assays in ATP6V1B2-inducible stable cell lines
Doxycycline-inducible stable HEK293T or OCI-LY1 cell lines carrying WT or mutant ATP6V1B2 were induced with 1 μg/ml doxycycline for 48 hours and incubated in fresh doxycycline-free medium for 4 hours prior to harvesting. Stable HEK293T or OCI-LY1 cells carrying empty vector were incubated for 4 hours in full medium or glucose-free or leucine-free medium, respectively, and used as controls. Western blotting was performed to detect ATF4 expression and ULK phosphorylation.
Measurement of lysosomal pH in stable transfected HEK293T cells
HEK293T cells overexpressing WT or mutated HA-ATP6V1B2 were grown in 24-well plates. LysoSensor Yellow/Blue dextran (500 μg/ml; Thermo Fisher Scientific, L22460) was added and cells incubated for 16 hours, followed by a 1-hour chase in LysoSensor Yellow/Blue dextran-free medium. Subsequently, cells were trypsinized and washed, and cell samples were immediately analyzed by flow cytometry. The wavelength of the fluorescence emitted by LysoSensor Yellow/Blue Dextran is pH dependent; in an acidic environment, LysoSensor Yellow/Blue dextran emits in the yellow spectrum, and in a neutral or basic environment, emission is in the UV spectrum. We measured fluorescence emitted at 405 and 526 nm in response to excitation at 355 and 448 nm, respectively, and subtracted the signal from unstained cells (background fluorescence control). Parallel cell samples for pH calibration were generated using fluorescein-dextran–loaded cells and a 25-mM MES calibration buffer (adjusted to a pH of 3.5 to 6.0 in 0.5-pH-unit increments) containing 10 μM monensin, 10 μM nigericin, 5 mM NaCl, 115 mM KCl, and 1.2 mM MgSO4.
Localization of WT and mutant HA-ATP6V1B2 in stable transfected HEK293T cells using confocal microscopy
Cells were plated on presterilized coverslips. Cells were fixed and permeabilized with 4% paraformaldehyde for 10 minutes at room temperature and subsequently in 0.1% Triton X-100 for 5 minutes at 4°C. Samples were blocked with 4% BSA in PBS for 1 hour at room temperature. Staining was performed using primary antibodies diluted in 4% BSA in PBS. Species-specific conjugated secondary antibodies (Supplemental Table 1) were diluted 1:1000 in 4% BSA in PBS and applied for 1 hour at room temperature. Nuclei were counterstained with DAPI (Molecular Probes). The coverslips were mounted onto glass slides with ProLong Gold Antifade Reagent (Invitrogen, Thermo Fisher Scientific, 1811419). Images were taken using a Leica SP5 confocal imaging system. Colocalization assays were performed using ImageJ, version 2.0.0.
IP assays
Cells were pelleted, washed, and lysed on ice for 20 minutes in lysis buffer containing 0.3% CHAPS detergent (Dot Scientific, DSC41010), 100 mM NaCl, 25 mM Tris, pH 8.0 (Sigma-Aldrich, T6066), 20 mM NaF, 2 mM EGTA, and 2 mM EDTA (Sigma-Aldrich, ED2SS), supplemented with protease and phosphatase inhibitors, sodium orthovanadate, and PMSF. The detergent-soluble fraction of the cell lysates was cleared by centrifugation at 17,000 g for 10 minutes. For anti-HA or anti-FLAG IPs, anti-HA–loaded beads (Sigma-Aldrich, A2095) or anti-FLAG–loaded beads (Sigma-Aldrich, A2220) were blocked with 5% BSA in TBST (25 mM Tris, pH 7.5, 150 mM NaCl, 0.05% Tween 20) for 0.5 hours and then washed 3 times with lysis buffer. Next, 20 μl anti-HA– or anti-FLAG–conjugated beads were added to precleared cell lysates and incubated with rotation for 5 hours at 4°C. The beads were washed 4 times with lysis buffer, and protein was liberated through boiling in SDS-PAGE loading buffer. Protein was fractionated through SDS-PAGE and prepared for immunoblotting using standard procedures. For immunoblottings, the detergent was 1% NP-40 instead of CHAPS (see IP conditions above), and NaCl was at 150 mM.
FL cell column purification through negative selection
Cryopreserved single-cell suspensions from human FL biopsy samples were thawed, washed, and resuspended in degassed BSA-EDTA (1× PBS, 0.5% BSA, 1 mmol/l EDTA) buffer at a concentration of approximately 107 cells/60 μl. Cells were treated with 20 μl/107 cells of Miltenyi CD3 magnetic microbeads (catalog 130-050-101) and 20 μl/107 cells of Miltenyi CD14 magnetic microbeads (catalog 130-050-201) and incubated at 4°C for 25 minutes. Cells were washed with BSA-EDTA, resuspended in 500 μl BSA-EDTA and passed through Miltenyi LS columns (catalog 145387) loaded onto a QuadriMax magnet according to the manufacturer’s recommendations. The flow-through fraction containing CD3– and CD14– highly enriched FL B cells was centrifuged and resuspended in RPMI 1640 medium supplemented with 20% heat-inactivated FBS.
Drug treatment and apoptosis assays of FL cells
Purified FL cells were plated in RPMI 1640 medium supplemented with 20% heat-inactivated FBS at a density of 106 cells/ml and cultured at 37°C with 5% CO2. MRT 68921 (catalog 1190379), bafilomycin A1 (catalog 11038), rapamycin (catalog 13346), torin 1 (catalog 10997), doxorubicin (catalog 15007), dexamethasone (catalog 11015), vincristine (catalog 11764), and chloroquine (catalog 14194), all procured from Cayman Chemical, were diluted serially in medium, and DMSO alone was used as a vehicle control. After 72 hours of incubation, cell viability was measured with CellTiter-Glo (Promega, G7572) and quantified on a GloMax microplate reader (Promega, SA3030). For apoptosis readouts after 72 hours of incubation, cells were washed twice with HBSS. The ANXA5/annexin V–FITC reagent (SouthernBiotech, 10039-02) was 1:20 diluted in ANXA5/annexin V–binding buffer (10 mmol HEPES, 140 mmol NaCl, 2.5 mmol CaCl2, 0.1% BSA, pH 7.4), added to each sample, and incubated on ice in the dark for 15 minutes. Subsequently, 400 μl ANXA5/annexin V–binding buffer and 20 μl of 20 μg/ml propidium iodide solution were added, and the samples were immediately analyzed by flow cytometry.
Survival analyses of primary purified FL B cells in full medium and during leucine deprivation
Purified FL B cells were plated in RPMI 1640 medium supplemented with 20% heat-inactivated FBS at a density of 106 cells/ml. Parallel cell aliquots were cultured under otherwise identical conditions but using a reduced leucine concentration of 25%, 12.5%, 6.25%, or 0%. Cells were counted and viability assessed on days 1, 2, 3, and 5 using trypan blue staining.
Immunoblotting of lysates of cultured primary purified FL B cells
Purified FL cells were resuspended in RPMI 1640 medium supplemented with 20% heat-inactivated FBS at a density of 106 cells/ml and cultured at 37°C with 5% CO2 for 4 hours. Cells were washed once, pelleted, and rapidly frozen using dry ice thereafter. Cell pellets were lysed and prepared for immunoblotting as described above.
Computational methods
Several homology models of the ATP6V1B2 subunit were generated using servers on the Protein Model Portal (https://www.proteinmodelportal.org/), and structural alignment was performed using PyMOL software. Structures of the yeast v-ATPase were downloaded from the RCSB Protein Data Bank (PDB) (http://www.rcsb.org/; IDs: 3J9T, 3J9U, and 3J9V), and aligned using residues Y352 and R381 in PyMOL software.
Yeast methods
The S. cerevisiae strain WLY176 was used to generate a Vma2-HA R381Q point mutation in the genome as previously described (76, 77). The deletion strains gcn2Δ, gcn4Δ, and gcn2Δ gcn4Δ were also generated from WLY176 as previously described (78). The WLY176 Vma2-HA WT/WT and WT/R381Q strains were generated by AflII digestion of a pRS405 plasmid containing either Vma2-HA WT or R381Q. The pRS405 Vma2-HA plasmid was constructed through fast cloning (79). Site-directed mutagenesis of pRS405 Vma2-HA was performed as previously described (80). Pho8Δ60 and GFP-Atg8 Western blot analyses were performed as previously described (81, 82). A vacuolar fluorescence Pho8-SEP assay was developed according to a previously published protocol (61). For culturing, yeast strains were grown in YPD medium (1% [w/v] yeast extract, 2% [w/v] peptone, and 2% [w/v] glucose) to mid-log phase and then collected. Strains grown in YPD medium were shifted into minus-nitrogen medium (0.17% yeast nitrogen base without ammonium sulfate or amino acids and 2% [w/v] glucose) for the indicated time points and then collected. Fluorescence measurements were performed using ImageJ software. RNA extraction and quantitative PCR experiments were performed as described previously (83).
Statistics
Statistical comparisons were carried out using unpaired, 2-sample t tests for comparisons between 2 conditions or linear regression for tests involving more than 2 conditions. In the latter case, linear contrasts were constructed and tested against a null value as appropriate using standard regression–based Wald-type tests. For example, if measurements were made under mutant (mut) and WT cells in 2 environments, A and B, the contrast “A:mut – A:WT – (B:mut - B:WT)” would be compared with zero to assess whether the impact of the mutation differed in the 2 environments. Regression models included blot-fixed effects for all measurements obtained from the same blot. Bonferroni adjustments were used when a positive result from any one of several tests would be used to support a single hypothesis.
Study approval
The lymphoma repositories IRBMED nos. HUM00007985 and HUM00017055 were approved by the IRB (IRBMED) of the University of Michigan Medical School, and all patients provided written informed consent. Genomic research involving all specimens was approved through IRBMED no. HUM00005467 (genomic analysis of B cell non-Hodgkin’s lymphoma).
Author contributions
MSK, AEC, TP, and SNM enrolled patients and analyzed clinical data. FW, DG, ZXY, PK, LFP, KSC, and SM performed the laboratory research. DB and SW assisted with the structural modeling. DG and DJK performed and/or interpreted all yeast experiments. SM and DJK conceived the study and wrote the manuscript.
Supplementary Material
Acknowledgments
We are grateful for the services provided by the genomics and bioinformatics core of the University of Michigan’s Comprehensive Cancer Center. We are grateful to Kerby Shedden, Professor of Statistics at the University of Michigan for consultations. This work was supported by the National Cancer Institute (NCI), NIH (R01CA190384, to SNM), the Weatherhall Foundation (to MK), and the National Institute of General Medical Sciences (NIGMS) (R01GM053396, to DJK).
Version 1. 02/05/2019
In-Press Preview
Version 2. 03/04/2019
Electronic publication
Version 3. 03/06/2019
Typographical errors corrected in text and legend to figure 2.
Version 4. 04/01/2019
Print issue publication
Funding Statement
to SNM
to DK
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2019 American Society for Clinical Investigation
Reference information: J Clin Invest. 2019;129(4):1626–1640.https://doi.org/10.1172/JCI98288.
Contributor Information
Fangyang Wang, Email: fangyanw@med.umich.edu.
Damián Gatica, Email: gaticad@umich.edu.
Zhang Xiao Ying, Email: xyingmm@gmail.com.
Luke F. Peterson, Email: lukepete@med.umich.edu.
Denzil Bernard, Email: denzil@atomwise.com.
Kamlai Saiya-Cork, Email: ksaiya@med.umich.edu.
Shaomeng Wang, Email: shaomeng@umich.edu.
Mark S. Kaminski, Email: mkaminsk@med.umich.edu.
Alfred E. Chang, Email: aechang@med.umich.edu.
Tycel Phillips, Email: tycelp@med.umich.edu.
Daniel J. Klionsky, Email: klionsky@umich.edu.
Sami N. Malek, Email: smalek@med.umich.edu.
References
- 1.Morton LM, Wang SS, Devesa SS, Hartge P, Weisenburger DD, Linet MS. Lymphoma incidence patterns by WHO subtype in the United States, 1992-2001. Blood. 2006;107(1):265–276. doi: 10.1182/blood-2005-06-2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dave SS, et al. Prediction of survival in follicular lymphoma based on molecular features of tumor-infiltrating immune cells. N Engl J Med. 2004;351(21):2159–2169. doi: 10.1056/NEJMoa041869. [DOI] [PubMed] [Google Scholar]
- 3.Stevenson FK, Stevenson GT. Follicular lymphoma and the immune system: from pathogenesis to antibody therapy. Blood. 2012;119(16):3659–3667. doi: 10.1182/blood-2011-11-367730. [DOI] [PubMed] [Google Scholar]
- 4.Kridel R, Sehn LH, Gascoyne RD. Pathogenesis of follicular lymphoma. J Clin Invest. 2012;122(10):3424–3431. doi: 10.1172/JCI63186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kahl BS, Yang DT. Follicular lymphoma: evolving therapeutic strategies. Blood. 2016;127(17):2055–2063. doi: 10.1182/blood-2015-11-624288. [DOI] [PubMed] [Google Scholar]
- 6.Gopal AK, et al. PI3Kδ inhibition by idelalisib in patients with relapsed indolent lymphoma. N Engl J Med. 2014;370(11):1008–1018. doi: 10.1056/NEJMoa1314583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pon JR, Marra MA. Clinical impact of molecular features in diffuse large B-cell lymphoma and follicular lymphoma. Blood. 2016;127(2):181–186. doi: 10.1182/blood-2015-07-658401. [DOI] [PubMed] [Google Scholar]
- 8.Küppers R, Stevenson FK. Critical influences on the pathogenesis of follicular lymphoma. Blood. 2018;131(21):2297–2306. doi: 10.1182/blood-2017-11-764365. [DOI] [PubMed] [Google Scholar]
- 9.Yildiz M, et al. Activating STAT6 mutations in follicular lymphoma. Blood. 2015;125(4):668–679. doi: 10.1182/blood-2014-06-582650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pasqualucci L, et al. Genetics of follicular lymphoma transformation. Cell Rep. 2014;6(1):130–140. doi: 10.1016/j.celrep.2013.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Okosun J, et al. Integrated genomic analysis identifies recurrent mutations and evolution patterns driving the initiation and progression of follicular lymphoma. Nat Genet. 2014;46(2):176–181. doi: 10.1038/ng.2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li H, et al. Mutations in linker histone genes HIST1H1 B, C, D, and E; OCT2 (POU2F2); IRF8; and ARID1A underlying the pathogenesis of follicular lymphoma. Blood. 2014;123(10):1487–1498. doi: 10.1182/blood-2013-05-500264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pasqualucci L, et al. Inactivating mutations of acetyltransferase genes in B-cell lymphoma. Nature. 2011;471(7337):189–195. doi: 10.1038/nature09730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morin RD, et al. Somatic mutations altering EZH2 (Tyr641) in follicular and diffuse large B-cell lymphomas of germinal-center origin. Nat Genet. 2010;42(2):181–185. doi: 10.1038/ng.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morin RD, et al. Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature. 2011;476(7360):298–303. doi: 10.1038/nature10351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Green MR, et al. Hierarchy in somatic mutations arising during genomic evolution and progression of follicular lymphoma. Blood. 2013;121(9):1604–1611. doi: 10.1182/blood-2012-09-457283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ortega-Molina A, et al. The histone lysine methyltransferase KMT2D sustains a gene expression program that represses B cell lymphoma development. Nat Med. 2015;21(10):1199–1208. doi: 10.1038/nm.3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krysiak K, et al. Recurrent somatic mutations affecting B-cell receptor signaling pathway genes in follicular lymphoma. Blood. 2017;129(4):473–483. doi: 10.1182/blood-2016-07-729954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang J, et al. The CREBBP acetyltransferase is a haploinsufficient tumor suppressor in B-cell lymphoma. Cancer Discov. 2017;7(3):322–337. doi: 10.1158/2159-8290.CD-16-1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boice M, et al. Loss of the HVEM tumor suppressor in lymphoma and restoration by modified CAR-T cells. Cell. 2016;167(2):405–418.e13. doi: 10.1016/j.cell.2016.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang J, et al. Disruption of KMT2D perturbs germinal center B cell development and promotes lymphomagenesis. Nat Med. 2015;21(10):1190–1198. doi: 10.1038/nm.3940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiang Y, et al. CREBBP Inactivation Promotes the Development of HDAC3-Dependent Lymphomas. Cancer Discov. 2017;7(1):38–53. doi: 10.1158/2159-8290.CD-16-0975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McCabe MT, et al. EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature. 2012;492(7427):108–112. doi: 10.1038/nature11606. [DOI] [PubMed] [Google Scholar]
- 24.Okosun J, et al. Recurrent mTORC1-activating RRAGC mutations in follicular lymphoma. Nat Genet. 2016;48(2):183–188. doi: 10.1038/ng.3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ying ZX, et al. Recurrent mutations in the MTOR regulator RRAGC in follicular lymphoma. Clin Cancer Res. 2016;22(21):5383–5393. doi: 10.1158/1078-0432.CCR-16-0609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oricchio E, et al. Genetic and epigenetic inactivation of SESTRIN1 controls mTORC1 and response to EZH2 inhibition in follicular lymphoma. Sci Transl Med. 2017;9(396):eaak9969. doi: 10.1126/scitranslmed.aak9969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zoncu R, Bar-Peled L, Efeyan A, Wang S, Sancak Y, Sabatini DM. mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H(+)-ATPase. Science. 2011;334(6056):678–683. doi: 10.1126/science.1207056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cotter K, Stransky L, McGuire C, Forgac M. Recent insights into the structure, regulation, and function of the V-ATPases. Trends Biochem Sci. 2015;40(10):611–622. doi: 10.1016/j.tibs.2015.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maxson ME, Grinstein S. The vacuolar-type H(+)-ATPase at a glance - more than a proton pump. J Cell Sci. 2014;127(Pt 23):4987–4993. doi: 10.1242/jcs.158550. [DOI] [PubMed] [Google Scholar]
- 30.Green MR, et al. Mutations in early follicular lymphoma progenitors are associated with suppressed antigen presentation. Proc Natl Acad Sci U S A. 2015;112(10):E1116–E1125. doi: 10.1073/pnas.1501199112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Anraku Y, Hirata R, Wada Y, Ohya Y. Molecular genetics of the yeast vacuolar H(+)-ATPase. J Exp Biol. 1992;172:67–81. doi: 10.1242/jeb.172.1.67. [DOI] [PubMed] [Google Scholar]
- 32.Zhao J, Benlekbir S, Rubinstein JL. Electron cryomicroscopy observation of rotational states in a eukaryotic V-ATPase. Nature. 2015;521(7551):241–245. doi: 10.1038/nature14365. [DOI] [PubMed] [Google Scholar]
- 33.Liu Q, Kane PM, Newman PR, Forgac M. Site-directed mutagenesis of the yeast V-ATPase B subunit (Vma2p) J Biol Chem. 1996;271(4):2018–2022. doi: 10.1074/jbc.271.4.2018. [DOI] [PubMed] [Google Scholar]
- 34.Mijaljica D, Prescott M, Devenish RJ. V-ATPase engagement in autophagic processes. Autophagy. 2011;7(6):666–668. doi: 10.4161/auto.7.6.15812. [DOI] [PubMed] [Google Scholar]
- 35.Mauvezin C, Nagy P, Juhász G, Neufeld TP. Autophagosome-lysosome fusion is independent of V-ATPase-mediated acidification. Nat Commun. 2015;6:7007. doi: 10.1038/ncomms8007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Settembre C, Fraldi A, Medina DL, Ballabio A. Signals from the lysosome: a control centre for cellular clearance and energy metabolism. Nat Rev Mol Cell Biol. 2013;14(5):283–296. doi: 10.1038/nrm3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yamashiro CT, Kane PM, Wolczyk DF, Preston RA, Stevens TH. Role of vacuolar acidification in protein sorting and zymogen activation: a genetic analysis of the yeast vacuolar proton-translocating ATPase. Mol Cell Biol. 1990;10(7):3737–3749. doi: 10.1128/MCB.10.7.3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jewell JL, Russell RC, Guan KL. Amino acid signalling upstream of mTOR. Nat Rev Mol Cell Biol. 2013;14(3):133–139. doi: 10.1038/nrm3522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kabeya Y, et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19(21):5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tanida I, Minematsu-Ikeguchi N, Ueno T, Kominami E. Lysosomal turnover, but not a cellular level, of endogenous LC3 is a marker for autophagy. Autophagy. 2005;1(2):84–91. doi: 10.4161/auto.1.2.1697. [DOI] [PubMed] [Google Scholar]
- 41.Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell. 2010;140(3):313–326. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang WP, Scott SV, Kim J, Klionsky DJ. The itinerary of a vesicle component, Aut7p/Cvt5p, terminates in the yeast vacuole via the autophagy/Cvt pathways. J Biol Chem. 2000;275(8):5845–5851. doi: 10.1074/jbc.275.8.5845. [DOI] [PubMed] [Google Scholar]
- 43.Kanki T, et al. A genomic screen for yeast mutants defective in selective mitochondria autophagy. Mol Biol Cell. 2009;20(22):4730–4738. doi: 10.1091/mbc.e09-03-0225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Valvezan AJ, et al. mTORC1 couples nucleotide synthesis to nucleotide demand resulting in a targetable metabolic vulnerability. Cancer Cell. 2017;32(5):624–638.e5. doi: 10.1016/j.ccell.2017.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ben-Sahra I, Manning BD. mTORC1 signaling and the metabolic control of cell growth. Curr Opin Cell Biol. 2017;45:72–82. doi: 10.1016/j.ceb.2017.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Menon S, et al. Spatial control of the TSC complex integrates insulin and nutrient regulation of mTORC1 at the lysosome. Cell. 2014;156(4):771–785. doi: 10.1016/j.cell.2013.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wyant GA, et al. mTORC1 Activator SLC38A9 Is Required to Efflux Essential Amino Acids from Lysosomes and Use Protein as a Nutrient. Cell. 2017;171(3):642–654.e12. doi: 10.1016/j.cell.2017.09.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Abu-Remaileh M, et al. Lysosomal metabolomics reveals V-ATPase- and mTOR-dependent regulation of amino acid efflux from lysosomes. Science. 2017;358(6364):807–813. doi: 10.1126/science.aan6298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sancak Y, et al. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science. 2008;320(5882):1496–1501. doi: 10.1126/science.1157535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim E, Goraksha-Hicks P, Li L, Neufeld TP, Guan KL. Regulation of TORC1 by Rag GTPases in nutrient response. Nat Cell Biol. 2008;10(8):935–945. doi: 10.1038/ncb1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Beck T, Hall MN. The TOR signalling pathway controls nuclear localization of nutrient-regulated transcription factors. Nature. 1999;402(6762):689–692. doi: 10.1038/45287. [DOI] [PubMed] [Google Scholar]
- 52.Kamada Y, Funakoshi T, Shintani T, Nagano K, Ohsumi M, Ohsumi Y. Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J Cell Biol. 2000;150(6):1507–1513. doi: 10.1083/jcb.150.6.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schmidt A, Beck T, Koller A, Kunz J, Hall MN. The TOR nutrient signalling pathway phosphorylates NPR1 and inhibits turnover of the tryptophan permease. EMBO J. 1998;17(23):6924–6931. doi: 10.1093/emboj/17.23.6924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.B’chir W, et al. The eIF2α/ATF4 pathway is essential for stress-induced autophagy gene expression. Nucleic Acids Res. 2013;41(16):7683–7699. doi: 10.1093/nar/gkt563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bernard A, Jin M, Xu Z, Klionsky DJ. A large-scale analysis of autophagy-related gene expression identifies new regulators of autophagy. Autophagy. 2015;11(11):2114–2122. doi: 10.1080/15548627.2015.1099796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yao Z, Delorme-Axford E, Backues SK, Klionsky DJ. Atg41/Icy2 regulates autophagosome formation. Autophagy. 2015;11(12):2288–2299. doi: 10.1080/15548627.2015.1107692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yu L, et al. Termination of autophagy and reformation of lysosomes regulated by mTOR. Nature. 2010;465(7300):942–946. doi: 10.1038/nature09076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rebsamen M, et al. SLC38A9 is a component of the lysosomal amino acid sensing machinery that controls mTORC1. Nature. 2015;519(7544):477–481. doi: 10.1038/nature14107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jung J, Genau HM, Behrends C. Amino acid-dependent mTORC1 regulation by the lysosomal membrane protein SLC38A9. Mol Cell Biol. 2015;35(14):2479–2494. doi: 10.1128/MCB.00125-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang S, et al. Metabolism. Lysosomal amino acid transporter SLC38A9 signals arginine sufficiency to mTORC1. Science. 2015;347(6218):188–194. doi: 10.1126/science.1257132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hughes AL, Gottschling DE. An early age increase in vacuolar pH limits mitochondrial function and lifespan in yeast. Nature. 2012;492(7428):261–265. doi: 10.1038/nature11654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.McGuire CM, Forgac M. Glucose starvation increases V-ATPase assembly and activity in mammalian cells through AMP kinase and phosphatidylinositide 3-kinase/Akt signaling. J Biol Chem. 2018;293(23):9113–9123. doi: 10.1074/jbc.RA117.001327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kane PM. Disassembly and reassembly of the yeast vacuolar H(+)-ATPase in vivo. J Biol Chem. 1995;270(28):17025–17032. [PubMed] [Google Scholar]
- 64.Egan DF, et al. Small molecule inhibition of the autophagy kinase ULK1 and identification of ULK1 substrates. Mol Cell. 2015;59(2):285–297. doi: 10.1016/j.molcel.2015.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Petherick KJ, et al. Pharmacological inhibition of ULK1 kinase blocks mammalian target of rapamycin (mTOR)-dependent autophagy. J Biol Chem. 2015;290(48):28726. doi: 10.1074/jbc.A114.627778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kaizuka T, et al. An Autophagic Flux Probe that Releases an Internal Control. Mol Cell. 2016;64(4):835–849. doi: 10.1016/j.molcel.2016.09.037. [DOI] [PubMed] [Google Scholar]
- 67.Su H, et al. VPS34 acetylation controls its lipid kinase activity and the initiation of canonical and non-canonical autophagy. Mol Cell. 2017;67(6):907–921.e7. doi: 10.1016/j.molcel.2017.07.024. [DOI] [PubMed] [Google Scholar]
- 68.Milkereit R, Persaud A, Vanoaica L, Guetg A, Verrey F, Rotin D. LAPTM4b recruits the LAT1-4F2hc Leu transporter to lysosomes and promotes mTORC1 activation. Nat Commun. 2015;6:7250. doi: 10.1038/ncomms8250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rebecca VW, et al. A unified approach to targeting the lysosome’s degradative and growth signaling roles. Cancer Discov. 2017;7(11):1266–1283. doi: 10.1158/2159-8290.CD-17-0741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gayle S, et al. Identification of apilimod as a first-in-class PIKfyve kinase inhibitor for treatment of B-cell non-Hodgkin lymphoma. Blood. 2017;129(13):1768–1778. doi: 10.1182/blood-2016-09-736892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lazarus MB, Novotny CJ, Shokat KM. Structure of the human autophagy initiating kinase ULK1 in complex with potent inhibitors. ACS Chem Biol. 2015;10(1):257–261. doi: 10.1021/cb500835z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ross CW, Ouillette PD, Saddler CM, Shedden KA, Malek SN. Comprehensive analysis of copy number and allele status identifies multiple chromosome defects underlying follicular lymphoma pathogenesis. Clin Cancer Res. 2007;13(16):4777–4785. doi: 10.1158/1078-0432.CCR-07-0456. [DOI] [PubMed] [Google Scholar]
- 73.Galbán S, et al. Cytoprotective effects of IAPs revealed by a small molecule antagonist. Biochem J. 2009;417(3):765–771. doi: 10.1042/BJ20081677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Peterson LF, Wang Y, Lo MC, Yan M, Kanbe E, Zhang DE. The multi-functional cellular adhesion molecule CD44 is regulated by the 8;21 chromosomal translocation. Leukemia. 2007;21(9):2010–2019. doi: 10.1038/sj.leu.2404849. [DOI] [PubMed] [Google Scholar]
- 75.Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods. 2014;11(8):783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Longtine MS, et al. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14(10):953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 77.Toulmay A, Schneiter R. A two-step method for the introduction of single or multiple defined point mutations into the genome of Saccharomyces cerevisiae. Yeast. 2006;23(11):825–831. doi: 10.1002/yea.1397. [DOI] [PubMed] [Google Scholar]
- 78.Gueldener U, Heinisch J, Koehler GJ, Voss D, Hegemann JH. A second set of loxP marker cassettes for Cre-mediated multiple gene knockouts in budding yeast. Nucleic Acids Res. 2002;30(6):e23. doi: 10.1093/nar/30.6.e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Li C, Wen A, Shen B, Lu J, Huang Y, Chang Y. FastCloning: a highly simplified, purification-free, sequence- and ligation-independent PCR cloning method. BMC Biotechnol. 2011;11:92. doi: 10.1186/1472-6750-11-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Liu H, Naismith JH. An efficient one-step site-directed deletion, insertion, single and multiple-site plasmid mutagenesis protocol. BMC Biotechnol. 2008;8:91. doi: 10.1186/1472-6750-8-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Shintani T, Klionsky DJ. Cargo proteins facilitate the formation of transport vesicles in the cytoplasm to vacuole targeting pathway. J Biol Chem. 2004;279(29):29889–29894. doi: 10.1074/jbc.M404399200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Noda T, Klionsky DJ. The quantitative Pho8Delta60 assay of nonspecific autophagy. Meth Enzymol. 2008;451:33–42. doi: 10.1016/S0076-6879(08)03203-5. [DOI] [PubMed] [Google Scholar]
- 83.Hu G, et al. A conserved mechanism of TOR-dependent RCK-mediated mRNA degradation regulates autophagy. Nat Cell Biol. 2015;17(7):930–942. doi: 10.1038/ncb3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.