Abstract
Although clear cell renal cell carcinoma (ccRCC) has been shown to result in widespread aberrant cytosine methylation and loss of 5-hydroxymethylcytosine (5hmC), the prognostic impact and therapeutic targeting of this epigenetic aberrancy has not been fully explored. Analysis of 576 primary ccRCC samples demonstrated that loss of 5hmC was strongly associated with aggressive clinicopathologic features and was an independent adverse prognostic factor. Loss of 5hmC also predicted reduced progression-free survival after resection of nonmetastatic disease. The loss of 5hmC in ccRCC was not due to mutational or transcriptional inactivation of ten eleven translocation (TET) enzymes, but to their functional inactivation by l-2-hydroxyglutarate (L2HG), which was overexpressed due to the deletion and underexpression of L2HG dehydrogenase (L2HGDH). Ascorbic acid (AA) reduced methylation and restored genome-wide 5hmC levels via TET activation. Fluorescence quenching of the recombinant TET-2 protein was unaffected by L2HG in the presence of AA. Pharmacologic AA treatment led to reduced growth of ccRCC in vitro and reduced tumor growth in vivo, with increased intratumoral 5hmC. These data demonstrate that reduced 5hmC is associated with reduced survival in ccRCC and provide a preclinical rationale for exploring the therapeutic potential of high-dose AA in ccRCC.
Keywords: Oncology
Keywords: Cancer
Introduction
Metastatic renal cell cancer is a generally incurable malignancy that needs newer molecular and therapeutic insights. We and others have previously demonstrated that clear cell renal cell carcinoma (ccRCC) is an epigenetically aberrant solid tumor, characterized by widespread DNA cytosine hypermethylation (1–4). We observed that aberrant methylation was particularly enriched in kidney-specific enhancer regions (H3K4Me1-positive regions) associated with silencing of important genes, such as the TGF-β regulator SMAD6. Similar findings were also seen in the recent The Cancer Genome Atlas (TCGA) analysis of ccRCC (2). These data demonstrated widespread aberrant hypermethylation in ccRCC, though the etiology of increased cytosine methylation has not been fully elucidated.
Ten eleven translocations (TETs) are dioxygenase enzymes that convert 5-methylcytosine (5mC) to 5-hydroxymethylcytosine (5hmC). 5hmC is further oxidized by TETs to 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC), followed by conversion to cytosine by the base excision repair (BER) pathway, demonstrating the role of TET enzymes in passive demethylation. Loss of function of the TET-2 enzyme can occur with inactivating mutations in myeloid malignancies (5, 6) and result in increased cytosine methylation and decreased hydroxymethylation (7, 8). Functional inactivation of TET enzymes can also occur in the presence of oncometabolite 2-hydroxyglutarate (2HG), which is produced by gain-of-function mutations in isocitrate dehydrogenase (IDH) enzymes in brain tumors and myeloid malignancies (8, 9). A recent report demonstrated that the L isoform of 2HG is elevated in ccRCC samples and correlates with reduced 5hmC, as measured by the dot blot assay (10). Both functional and mutational inactivation of TET enzymes can thus cause hypermethylation, and this has not been comprehensively examined in the context of ccRCC.
Recent work in embryonic stem cells has demonstrated that ascorbic acid (AA) is a cofactor for the TET enzyme and can facilitate TET-mediated DNA demethylation (11, 12). We hypothesized that targeting the aberrant DNA methylation of ccRCC by enhancing the activity of the TET enzymes with AA would result in epigenetic reprogramming and have a therapeutic potential. We also aimed to determine the difference in the hydroxymethyl cytosine fraction of the genome between low-grade and high-grade ccRCC and to determine whether it has a prognostic impact, which would further add to the rationale of exploring the therapeutic potential of AA in ccRCC.
Results
Loss of 5hmC is strongly associated with advanced and higher grade ccRCC.
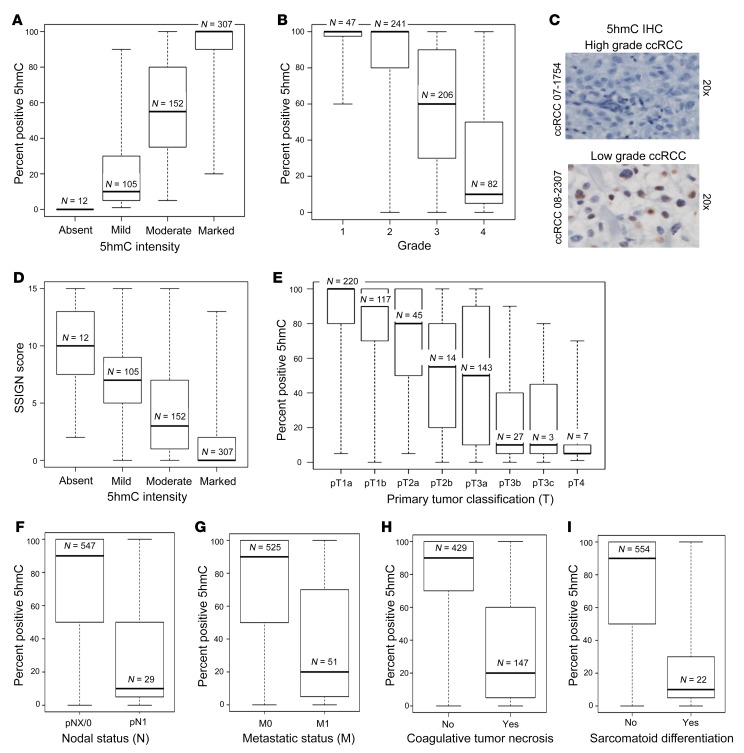
As increased cytosine methylation (5mC) has been reported in ccRCC (1, 2), we wanted to determine whether changes in 5hmC are also seen in these tumors and correlate with any clinicopathologic characteristics. Immunohistochemical evaluation of 5hmC was conducted on a large cohort (n = 576) of ccRCC patients. The percentage of tumor cells positive for 5hmC correlated well with the intensity of the stain (Figure 1A).
Figure 1. Loss of 5hmC is strongly associated with features of tumor aggressiveness in ccRCC.
(A) Correlation between median percentage positive for 5hmC and 5hmC intensity in IHC (P < 0.001). (B) Higher grade ccRCC is associated with loss of 5hmC (P < 0.001). (C) Representative photographs of low-grade and high-grade ccRCC with 5hmC IHC. (D) Loss of 5hmC correlates with higher SSIGN score, which predicts increased risk of progression of ccRCC after nephrectomy (P < 0.001). (E) Increased tumor size in ccRCC is associated with loss of 5hmC (P < 0.001). (F) Nodal metastasis in ccRCC is associated with loss of 5hmC (P < 0.001). (G) Presence of systemic metastatic disease in ccRCC is associated with loss of 5hmC (P < 0.001). (H) Presence of coagulative tumor necrosis is associated with loss of 5hmC (P < 0.001). (I) Presence of sarcomatoid differentiation is associated with loss of 5hmC (P < 0.001). Box plots have horizontal lines at the 25th percentile, the median, and the 75th percentile. The vertical lines extend to the minimum and maximum values. Associations of 5hmC expression with the clinical and pathologic features studied were evaluated using Spearman’s rank correlation coefficients, Kruskal-Wallis tests, and Wilcoxon’s rank sum tests.
Pathologically higher grade ccRCC tumors had a striking loss of 5hmC compared with lower grade tumors (Figure 1, B and C). Median percentage positive for 5hmC for grades 1, 2, 3, and 4 tumors were 100%, 100%, 60%, and 10%, respectively (P < 0.001) (Figure 1B). Loss of 5hmC was also associated with a higher primary tumor classification and nodal and systemic metastasis (Figure 1, D–F, P < 0.001). Tumor size negatively correlated with percentage positive for 5hmC (correlation coefficient = –0.52, P < 0.001), and median sizes for tumors with absent, mild, moderate, and marked 5hmC intensity were 11.1, 9.4, 6.2, and 3.6 cm, respectively (P < 0.001). The percentages of absent, mild, moderate, and marked 5hmC intensity tumors that were grade 4 were 50%, 45%, 12%, and 4%, respectively (P < 0.001). Tumors with additional signs of aggressiveness, such as coagulative tumor necrosis and sarcomatoid differentiation, were also associated with significantly lower percentages of positive 5hmC (Figure 1, G and H, and Supplemental Tables 1–3, showing associations of percentage positive for 5hmC and 5hmC intensity with clinical and pathologic features; supplemental material available online with this article; https://doi.org/10.1172/JCI98747DS1). Taken together, these data indicate that a loss of 5hmC is associated with a clinicopathological advanced phenotype of ccRCC and prompted us to investigate the prognostic value of loss of 5hmC in a univariable and multivariable setting.
Loss of 5hmC is an independent adverse prognostic factor in ccRCC and predicts shortened time to metastatic disease after surgical resection for localized (M0) disease.
In our cohort of ccRCC cases, 185 patients out of a total of 576 died at a median of 2.7 years following surgery (IQR, 1.1–5.1). The median duration of follow-up for the 391 patients who were still alive at last follow-up was 7.2 years (IQR, 6.2–8.7). Eight patients who died from unknown causes were excluded from the analyses of cancer-specific survival (CSS); of the remaining 568 patients, 112 died from RCC at a median of 2.1 years following surgery (IQR, 0.9–3.5).
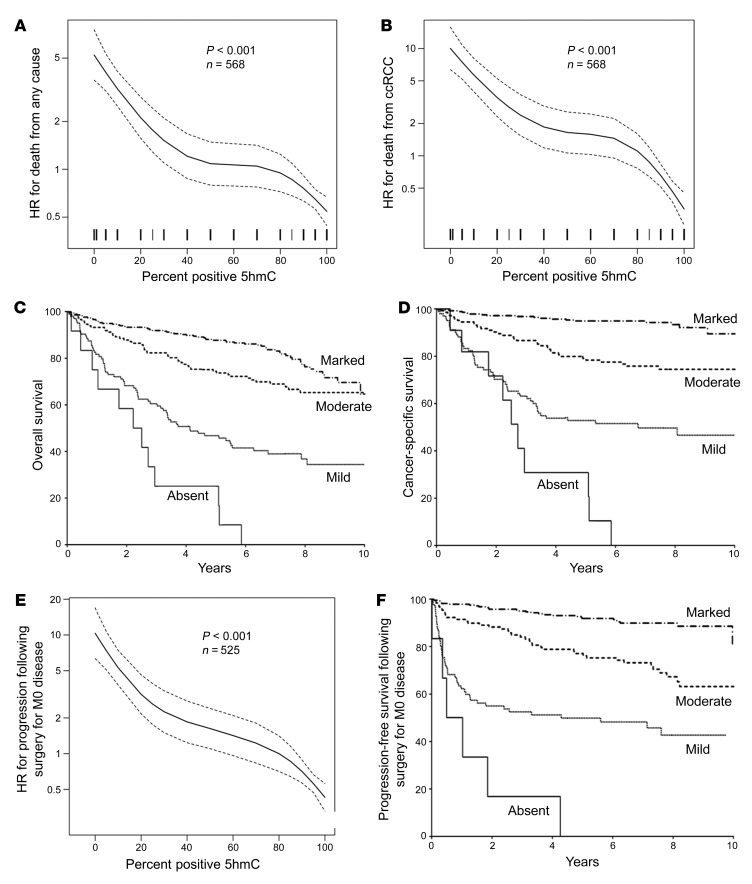
We found that loss of 5hmC was strongly associated with reduced CSS in both univariable and multivariable analysis. Associations of 5hmC expression with time to death from any cause and time to death from RCC are summarized in Supplemental Table 4. The percentage positive for 5hmC was inversely related to death from any cause (univariable HR for a 10% increase, 0.82; 95% CI, 0.79–0.85, P < 0.001, Figure 2A) and death from RCC (univariable HR for a 10% increase, 0.74; 95% CI, 0.70–0.78; P < 0.001, Figure 2B; multivariable HR, 0.93; 95% CI, 0.87–0.98; P = 0.013). Patients with absent, mild, and moderate 5hmC tumor-staining intensity had a univariable HR of death from any cause of 11.60 (P < 0.001), 4.44 (P < 0.001), and 1.69 (P = 0.007), respectively, compared with marked intensity. The median overall survival (OS) in the absent, mild, and moderate 5hmC intensity cohorts occurred at 2.4, 4.1, and 10.5 years, respectively. Median OS in the marked group has not been reached (Figure 2C). Patients with absent, mild, and moderate 5hmC tumor-staining intensity had a univariable HR of death from RCC of 27.27 (P < 0.001), 11.15 (P < 0.001), and 4.06 (P < 0.001), respectively, compared with marked intensity. The median CSS in the absent and mild intensity cohorts occurred at 2.7 and 6.8 years, respectively. Median CSS in the moderate and marked 5hmC intensity group has not been reached. Ten-year CSS in the marked 5hmC intensity group was 90% (Figure 2D).
Figure 2. Loss of 5hmC is an independent prognostic factor in ccRCC and predicts shortened time to metastatic disease after surgical resection for M0 disease.
(A) Univariable association of percentage positive for 5hmC with death from any cause (HR, 0.82; 95% CI, 0.79–0.85; P < 0.001; n = 568 patients). Dotted lines represent 95% CI. (B) Univariable association of percentage positive for 5hmC with death from RCC (HR, 0.74; 95% CI, 0.70–0.78; P < 0.001; n = 568 patients). Dotted lines represent 95% CI. (C) Univariable association of 5hmC intensity with OS. Median OS in the absent, mild, and moderate 5hmC intensity cohorts occurred at 2.4, 4.1, and 10.5 years, respectively. Median OS in the marked group has not been reached. (D) Univariable association of 5hmC intensity with CSS. Median CSS in the absent and mild intensity cohorts occurred at 2.7 and 6.8 years, respectively. Median CSS in the moderate and marked 5hmC intensity group has not been reached. Ten-year CSS in the marked 5hmC intensity group was 90%. (E) Univariable association of percentage positive for 5hmC with progression following surgery for M0 disease (HR, 0.76; 95% CI, 0.72–0.80; P < 0.001). Dotted lines represent 95% CI. (F) Univariable association of 5hmC intensity with PFS among M0 patients. Median PFS in the absent and mild intensity cohort occurred at 0.8 and 4.3 years, respectively. Median CSS in the moderate and marked 5hmC intensity group has not been reached. Ten-year PFS in the marked 5hmC intensity group was 81%.
Loss of 5hmC was also associated with progression following surgery for nonmetastatic (M0) disease in a large cohort of cases (n = 525). Associations of 5hmC with time to progression among M0 patients are summarized in Supplemental Table 4. There were 6 (1%), 86 (16%), 134 (26%), and 299 (57%) tumors with absent, mild, moderate, and marked 5hmC intensity, respectively. Five patients who died from unknown causes without experiencing distant metastases were excluded from the analyses of progression-free survival (PFS). Of the remaining 520 patients, 117 experienced progression at a median of 1.3 years following surgery (IQR, 0.3–3.6). The percentage positive for 5hmC was found to be inversely related to progression following surgery for M0 disease (univariable HR for a 10% increase, 0.76; 95% CI, 0.72–0.80; P < 0.001, Figure 2E; multivariable HR, 0.91; 95% CI, 0.86–0.97; P = 0.002). Patients with absent, mild, and moderate 5hmC tumor-staining intensity had a univariable HR of progression following surgery for M0 patients of 27.07 (P < 0.001), 8.44 (P < 0.001), and 3.23 (P < 0.001), respectively, compared with marked intensity. The median PFS in the absent and mild intensity cohorts occurred at 0.8 and 4.3 years, respectively. Median PFS in the moderate and marked 5hmC intensity groups has not been reached. Ten-year PFS in the marked 5hmC intensity group was 81% (Figure 2F). The relationship of absent 5hmC and decreased PFS was also validated in multivariable analysis, with an HR for progression of 4.69 (95% CI, 1.84–11.96; P = 0.001).
L2HGDH deletions and underexpression are seen in ccRCC and are strongly associated with hypermethylation and adverse prognosis.
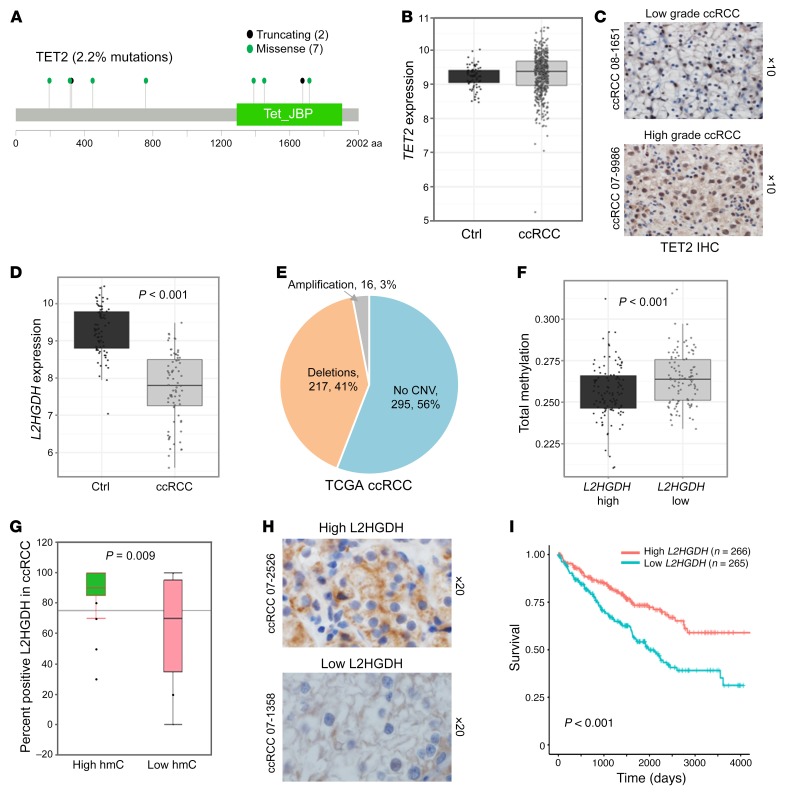
We then aimed to determine the reason for the loss of 5hmC in high-grade ccRCC. TET-2 mutations are common in hematologic malignances and are associated with hypermethylation. Analysis of a TCGA cohort (n = 418) revealed that TET-2 is mutated (heterozygous) in only 2.2% of ccRCC tumors (Figure 3A). No decrease in TET-2 expression was seen between ccRCC samples and matched controls obtained from TCGA (Figure 3B). TET-2 IHC revealed no differences in expression patterns between high-grade and low-grade ccRCC (representative photographs in Figure 3C). Although TET-2 expression is intact, its activity has been previously shown to be inhibited by the accumulation of oncometabolite l-2-hydroxyglutarate (L2HG) in ccRCC (10). Reduced expression of the enzyme L2HG dehydrogenase (L2HGDH) has been reported to be partly responsible for the accumulation of L2HG in ccRCC (10). Consistent with that observation, we observed that L2HGDH expression was significantly lower in ccRCC compared with matched normal kidney tissue (TCGA data, P < 0.001) (Figure 3D). Copy number data from TCGA showed that deletions at the L2HGDH locus were seen in 41% of samples (Figure 3E). Integration of methylation data with L2HGDH expression determined that ccRCC tumors with lower L2HGDH were significantly associated with higher cytosine methylation (Figure 3F, P < 0.001). L2HGDH IHC from 20 high-5hmC ccRCC and 20 low-5hmC ccRCC samples from our cohort suggested that lower 5hmC levels in ccRCC are associated with lower L2HGDH levels (Figure 3G, P = 0.009); representative photographs are shown in Figure 3H. Furthermore, lower L2HGDH expression was associated with worse survival in the cohort of 533 ccRCC patients in the TCGA (Figure 3I and Supplemental Figure 6, P < 0.001).
Figure 3. L2HGDH deletions and underexpression are seen in ccRCC and are significantly associated with adverse prognosis.
(A) Analysis of TCGA data set revealed that TET2 is mutated (heterozygous) only in 2.2% of ccRCC tumors. The mutations seen are predominantly missense (green) or nonsense (black) and are predicted to result in truncated proteins. (B) TCGA analysis revealed no significant difference in TET2 expression between ccRCC and normal kidney. (C) TET2 IHC revealed no difference in expression patterns between high-grade and low-grade ccRCC. Parts A, B and C, taken together, indicate that neither the mutation rate nor the expression of TET2 explains the loss of 5hmC in higher grade ccRCC. (D) L2HGDH gene-expression comparison between matched normal kidney and ccRCC tumor from TCGA (n = 72). The mean normalized log2 expression for the normal group is 9.28 and that of the tumor group is 7.75 (t test, P < 0.001). (E) Copy number variation (CNV) data for 528 patients (TCGA) with ccRCC are as follows: deletion, 217 (41%); no copy number variation, 295 (56%); amplification, 16 (3%). (F) TCGA analysis of methylation status of ccRCC tumors based on low L2HGDH and high L2HGDH. (G) L2HGDH IHC (n = 40; 20 with high 5hmC and 20 with low 5hmC) demonstrating that loss of L2HGDH is associated with loss of 5hmC (t test, P = 0.009). Box plots have horizontal lines at the 25th percentile, the median, and the 75th percentile. Vertical lines extend to the 10th and 90th percentiles. (H) Representative photographs showing loss of L2HDH in lower 5hmC ccRCC. (I) Lower L2HGDH expression is associated with shorter survival (P < 0.001). n = 533 ccRCC patients, grouped based on L2HGDH expression less than or greater than median. Survival analysis performed with log-rank test. The median survival (in days) for the low L2HGDH group is 1980. Median not reached for the high L2HGDH group.
AA treatment leads to increased TET activity, loss of methylation, and gain of hydroxymethyl cytosine levels in ccRCC cells.
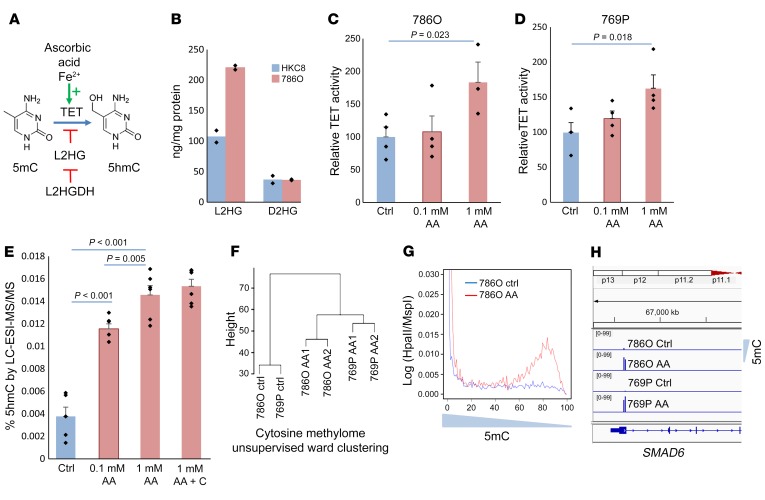
Since lower L2HGDH and, consequently, higher expression of L2HG can functionally inhibit TET enzymes, we then evaluated pharmacologic activation of TET in ccRCC. AA is an essential cofactor for TET enzymes (binds to the catalytic domain and aids in conversion of Fe3+ to Fe2+ that is used by TET enzymes for conversion of methyl to hydroxymethyl cytosines) (Figure 4A) (13, 14). We evaluated the effects of AA on 2 ccRCC cell lines that had heterozygous deletions affecting the L2HGDH locus (TCGA data). We also determined that L2HG levels were raised in 786-O cells when compared with kidney tubular control (Figure 4B). Next, we determined that exposure of ccRCC cell lines to pH-neutralized AA led to an increase in TET enzymatic activity (Figure 4, C and D). To determine the consequence of this AA-induced increased TET activity on DNA hydroxymethylation, mass spectrometry (liquid chromatography/electrospray ionization/tandem mass spectrometry [LC-ESI-MS/MS]) was performed and showed an increase in 5hmC after AA treatment (Figure 4E). Furthermore, as previously reported in embryonic stem cells, we found that the effect of AA on DNA methylation in ccRCC cell line 786-O was dependent on TET enzymes. Knockdown of TET1/TET2 and TET1/TET2/TET3 resulted in no change in 5hmC and no decrease in 5mC upon AA treatment (Supplemental Figure 1). We validated global changes in methylation by the HpaII tiny fragment enrichment by ligation–mediated PCR (HELP) assay, which relies on differential restriction digestion of methylated CpGs, followed by high-throughput sequencing analysis (15). Unsupervised clustering demonstrated that AA treatment led to changes in cytosine methylation patterns with epigenetic dissimilarity between control and AA-treated ccRCC cells (Figure 4F). Qualitatively, AA treatment led to loss of methylation (Figure 4G) and affected loci that had been previously shown to be hypermethylated in RCC (SMAD6 promoter is demethylated after AA treatment [Figure 4H] with increased expression [Supplemental Figure 3]). Given that the conventional HELP assay does not differentiate 5hmC from 5mC, we performed oxidative bisulphite sequencing, which revealed an acquisition of hydroxymethylcytosine with AA treatment in gene loci encoding KMT2C, DOCK8, PRKG1, CWH43, ZHX1, AKT3, MXI1, and others (Supplemental Figure 2, oxidative bisulphite sequencing [OxBS] table). Lower expression of several of these putative tumor suppressors was found to be associated with significantly worse prognosis in TCGA ccRCC data sets (Supplemental Figure 2).
Figure 4. AA leads to increased TET activity and 5hmC levels in ccRCC cells.
(A) Schematic showing the role of AA as an essential cofactor for TET enzymatic activity. (B) Intracellular L2HG levels measured by MS in ccRCC cell line 786-O are much higher than in the immortalized normal kidney cell line HKC8 (n = 2). (C and D) TET activity was measured in vitro with AA-treated RCC cells (769-P and 786-O) and was increased after treatment. t test, P values as indicated. Data are shown as mean ± SEM with individual data points overlaid (n = 2). Exposure time was 4 hours, mimicking bioavailability curves with i.v. AA, followed by 24-hour incubation with fresh media prior to harvesting the cells for nuclear extraction and TET activity analysis. We adjusted for multiple comparisons by dividing the significance level by the number of comparisons performed via Bonferroni’s correction. Hypotheses were deemed significant if P values were lower than 0.025 (0.05/2 to account for multiple comparisons). (E) 5hmC was measured by LC-ESI-MS/MS and was significantly increased after AA treatment of RCC cells 769-P. Addition of catalase did not change the percentage of 5hmC. t test, P values as indicated. Data are shown as mean ± SEM with individual data points overlaid (n = 2). We adjusted for multiple comparisons by dividing the significance level by the number of comparisons performed via Bonferroni’s correction. Hypotheses were deemed significant if P values were lower than 0.0125 (0.05/4 to account for multiple variations). (F) Unsupervised clustering based on genome-wide methylation analysis conducted by HELP assay. Ward clustering shows global methylation changes are induced by AA treatment. (G) Histograms based on methylation (log [HpaII/MspI]) show increased hypomethylation after AA treatment. (H) Smad6 promoter becomes demethylated after AA treatment in both 786-O and 769-P ccRCC cells.
Fluorescence quenching of recombinant TET-2 is unaffected by L2HG in the presence of AA.
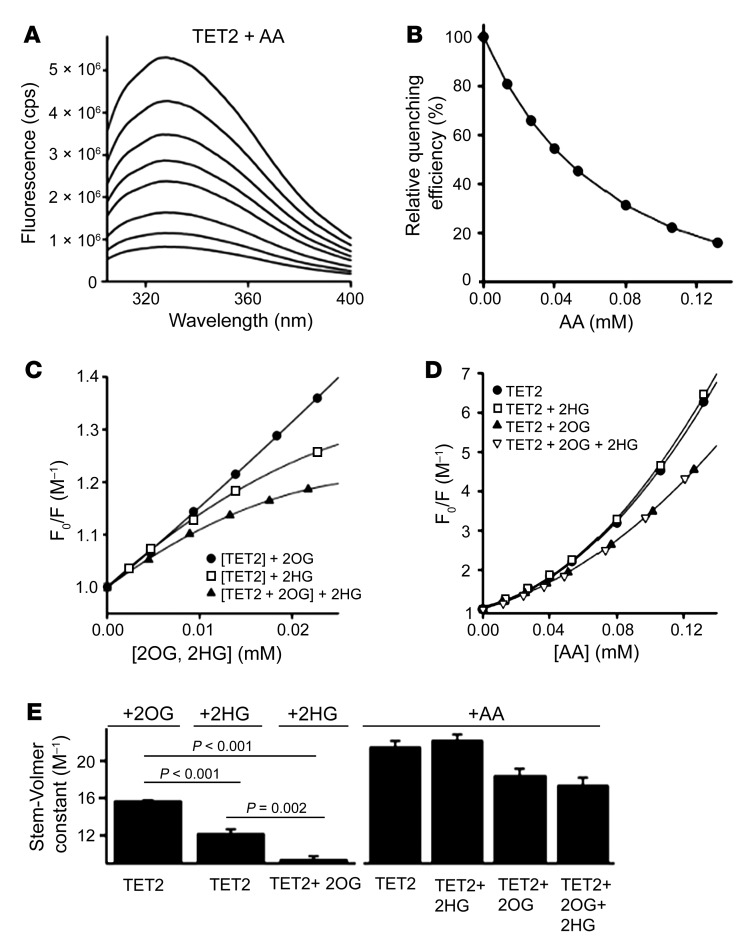
Next, we evaluated the interaction between the recombinant TET-2 protein with AA and its cofactor 2OG as well as its competitive inhibitor L2HG. We determined that there was significant quenching of fluorescence when AA was added to a solution containing 0.5 μM TET-2 (Figure 5A). Maximal quenching was obtained with a dose of 132 μM, for which a greater than 90% reduction of the fluorescence emission signal was observed (Figure 5B). This AA concentration would be very difficult to achieve even in the plasma with the maximum tolerated oral dose. However, the relation between intracellular AA concentration in vivo in kidney cancer cells and the plasma AA concentration is unknown. We then studied the interaction of TET-2 with 2OG and L2HG (Figure 5, C and E). The quenching efficiency of 2OG (15.66 ± 0.12 M–1) was found to be higher than for L2HG (12.15 ± 0.51 M–1), yielding a P value of less than 0.001. Furthermore, 2OG seems to prevent L2HG from interacting efficiently, as L2HG quenching is less efficient (9.36 ± 0.40 M–1) in the presence of 23 μM 2OG than without (P < 0.001). This indicates that the substrate specificity of TET-2 for 2OG is higher than L2HG.
Figure 5. Fluorescence quenching of recombinant TET-2 protein is unaffected by L2HG in the presence of AA.
(A) Fluorescence spectra of 0.5 μM TET-2 are shown after excitation at 280 nm with increasing amounts of AA (from top to bottom). (B) The relative fluorescence intensity at 328 nm is shown as a function of AA. A concentration of 132 μM AA reduces the fluorescence signal by 90 %. (C) Quenching effect on 0.5 μM TET-2 for 2OG (black circles), L2HG (white circles), and L2HG in the presence of 23 μM 2OG (triangles). Quenching efficiency of 2OG is higher than that of L2HG (P < 0.001). L2HG quenching is less efficient with 2OG than without (paired t test, P = 0.002). The substrate specificity of TET-2 is 2OG over L2HG. (D) The fluorescence quenching effect of AA on 0.5 μM TET-2 is shown. Symbols are as follows: black circles, TET-2; white squares, TET-2 in the presence of 100 μM L2HG; triangles, TET-2 in the presence of 23 μM 2OG; and inverted triangles, quenching of TET-2 in the presence of 23 μM L2HG and 23 μM 2OG. The overlapping curves indicate that TET-2 is unaffected by L2HG in the presence of AA. (E) Comparison of the Stern-Volmer constants ± SEM obtained from the linear range of the data in parts C and D. P values are as indicated in the figure (paired t test). In the equation y = m × x + c, m indicates the Stern-Volmer constant. The (y, x) points are obtained from C and D. For the Stern-Volmer constants derived from C, we adjusted for multiple comparisons by dividing the significance level by the number of comparisons performed via Bonferroni’s correction. Hypotheses were deemed significant if P values were less than 0.017 (0.05/3 to account for multiple comparisons).
We then performed the quenching of TET-2 with AA, in the presence and absence of L2HG and 2OG (Figure 5, D and E). We observed that the addition of AA leads to abrogation of decreased quenching that is seen in the presence of L2HG. There was an overlap of the quenching curves of TET-2 plus AA (21.46 ± 0.70 M–1) and TET-2 plus L2HG plus AA (22.17 ± 0.68 M–1) as well as an overlap of quenching curves of TET-2 plus 2OG plus AA (18.37 ± 0.80 M–1) and TET-2 plus 2OG plus L2HG plus AA (17.34 ± 0.87 M–1), suggesting that fluorescence quenching of TET-2 is unaffected by L2HG in the presence of AA. Figure 5, A and B, shows quenching of the fluorescence signal emitted by the TET protein consequent to structural changes induced by the binding of the protein with AA. These data reveal that the maximal effect of AA on the TET protein occurs with a concentration of approximately 132 μM (reduction of the fluorescence signal by >90%). Figure 5C shows that the quenching efficiency of 2OG is higher than that of L2HG (P < 0.001), suggesting that the substrate specificity of TET2 is 2OG over L2HG. Figure 5D shows that the fluorescence quenching of the TET enzyme with AA is largely unaffected by the presence of the oncometabolite 2HG, which suggests that in the presence of AA, the TET enzyme may be unaffected by L2HG.
High-dose AA treatment inhibits growth of ccRCC cells via non–free radical mechanisms.
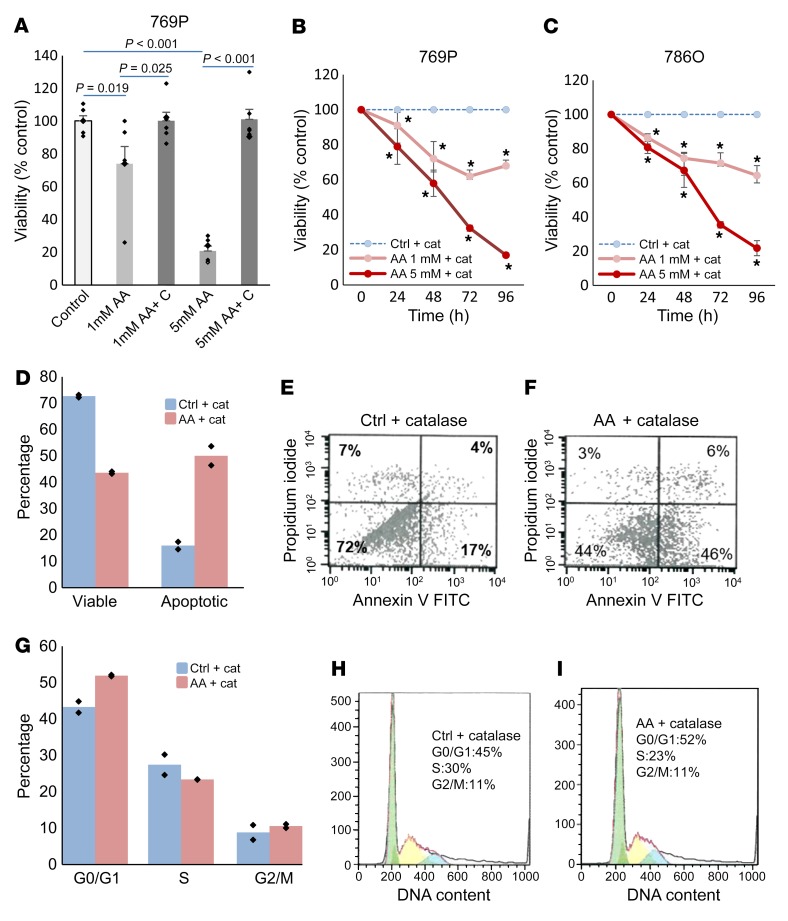
Next, we wanted to determine whether treatment with AA could lead to inhibitory effects in ccRCC. AA, when added to culture media, generates hydrogen peroxide (H2O2), which can cause cytotoxicity (16–18). To evaluate the functional impact of epigenetic effects of AA in vitro, ccRCC-derived cell lines were treated with short-term exposure to high-dose AA (mimicking the bioavailability curves of i.v. AA doses currently being used in early phase trials and roughly accounting for differences in plasma concentrations and those of tumor microenvironment) with or without catalase treatment to counter the H2O2 generated by AA. We observed acute loss in viability with high-dose AA that was reversed in the presence of catalase cotreatment, suggesting that the acute cytotoxicity with short-term exposure of high-dose AA (millimolar concentration) is primarily mediated by H2O2, as has been demonstrated previously (16, 17) (Figure 6A and Supplemental Figure 4). However, treatment of high-dose AA with catalase did result in reduced viability, which was detected after longer time points in ccRCC cells, demonstrating that AA could exert antitumor effects through non-H2O2 mechanisms with a longer-term exposure (Figure 6, B and C).
Figure 6. High-dose AA treatment leads to inhibition of ccRCC growth in vitro via non–free radical mechanisms.
(A) ccRCC (769-P) cells were treated with AA for 4 hours with and without catalase treatment (to abrogate H2O2), followed by incubation in fresh media for 24 hours. The acute cytotoxicity of AA was reversed by catalase treatment. t test, P values as indicated. Data are shown as mean ± SEM with individual data points overlaid (n = 2). We adjusted for multiple comparisons by dividing the significance level by the number of comparisons performed via Bonferroni’s correction. Hypotheses were deemed significant if P values were lower than 0.0125 (0.05/4 to account for multiple variations). Results were similar with 786-O (Supplemental Figure 5). (B and C) ccRCC cells were treated with AA and catalase and followed for longer time points. AA led to dose-dependent and progressive loss of viability despite catalase control, suggesting non–free radical mechanism of proliferation inhibition. t test. Data are shown as mean ± SEM (n = 2). We adjusted for multiple comparisons by dividing the significance level by the number of comparisons performed at each time point via Bonferroni’s correction. Hypotheses were deemed significant if P values were lower than 0.025 (0.05/2 to account for multiple comparisons). *P < 0.05. (D–F) ccRCC cells (786-O) were treated with catalase and AA (5 mM) and assessed for apoptosis by FACS. AA treatment led to significant increase in apoptosis after 48 hours of treatment. AA exposure for D1 (24 hours) and D3 (48–72 hours) with apoptosis assay at 96 hours. Representative flow figures are shown. n = 2. Data are shown as means with individual data points overlaid in D. (G–I) ccRCC cells (786-O) were treated with catalase and AA (5 mM) and assessed for cell cycle by FACS. AA treatment led to significant increase in G0/G1 arrest after 48 hours of treatment. AA exposure for D1 (24 hours) and D3 (48–72 hours) with apoptosis assay at 96 hours. Representative flow figures are shown. n = 2. Data are shown as means with individual data points overlaid in G.
To determine the mechanism of loss of viability, we treated ccRCC cells with AA and catalase and assessed for apoptosis and cell-cycle dynamics. We observed that high-dose AA treatment at the 96-hour time point led to increased apoptosis in ccRCC cells (Figure 6, D–F). ccRCC cells were also found to be significantly arrested in the G0/G1 stages of the cell cycle at 96 hours after high-dose AA treatment (Figure 6, G–I).
High-dose AA treatment inhibits ccRCC tumor growth in vivo.
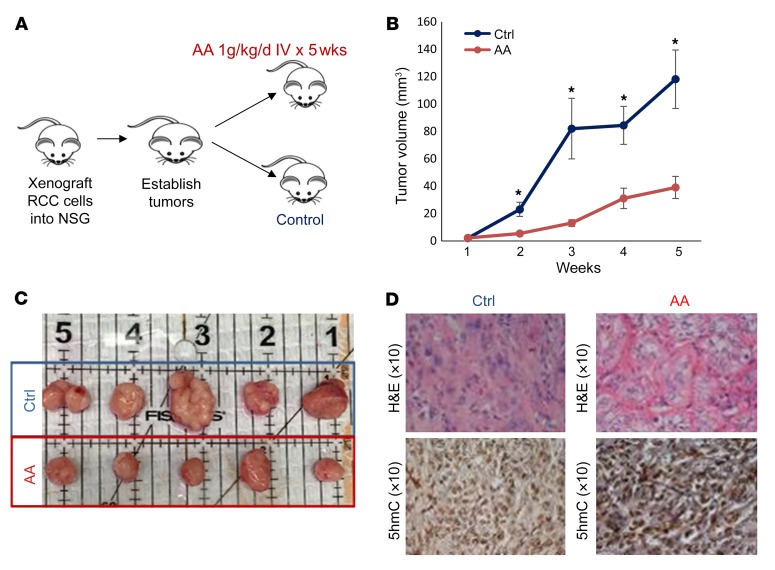
Next, we established xenografts from ccRCC cells in immunodeficient NSG mice and then treated them for 5 weeks with 1 g/kg/5 d/wk of AA given i.v. or with vehicle control (n = 10 in each cohort) (Figure 7A). Tumor measurements revealed a significantly reduced rate of ccRCC growth with AA treatment when compared with vehicle controls (Figure 7, B and C). Histological examination revealed that ccRCC tumor cells in the control (vehicle treated) xenograft group showed a higher grade (poor differentiation) based on increased prominent hyperchromatic nucleoli, nuclear pleomorphism, multilobation, and multinucleate giant cells when compared with the i.v. AA-treated group (Figure 7D and Supplemental Figure 7).
Figure 7. High-dose AA treatment leads to inhibition of ccRCC tumor growth in vivo.
(A) ccRCC cells (786-O) were xenografted into immunodeficient NSG mice. After tumors were established, treatment was initiated with i.v. AA (1 mg/kg/d) or vehicle and tumor measurements were conducted. (B and C) AA treatment led to significantly delayed tumor growth. t test, P < 0.05. Data are shown as mean ± SEM. n = 10 in each cohort. (D) Representative images of histologic examination showing the H&E and 5hmC IHC comparison of ccRCC xenograft control group versus i.v. AA-treated group. Tumor cells in the control group showed a higher grade (poorly differentiated) based on increased prominent hyperchromatic nucleoli, nuclear pleomorphism, multilobation, and multinucleate giant cells when compared with the i.v. AA-treated group. Tumor cells in the i.v. AA-treated group showed a higher intensity and increased staining of nuclei with 5hmC when compared with the control group.
Tumor cells in the control group showed a lesser intensity and decreased staining of nuclei with 5hmC when compared with the i.v. AA-treated group (Figure 7D and Supplemental Figure 7), consistent with the in vitro effects observed with AA treatment.
Discussion
We and others have previously demonstrated that ccRCC is an epigenetically aberrant solid tumor characterized by widespread DNA cytosine hypermethylation, particularly enriched in kidney-specific enhancer regions (H3K4Me1-positive regions), corresponding to important tumor-suppressor genes (1, 2). In this study, we report that loss of 5hmC is an independent adverse prognostic factor in ccRCC and predicts shortened time to metastatic disease after surgical resection for localized (M0) disease. Patients with tumors that retain high 5hmC levels (suggesting intact TET function) have a 10-year PFS of 81% after surgical resection for localized ccRCC. In contrast, those with “absent” and “mild” staining have a median PFS of 0.8 months and 4.3 years, respectively. The reason for the difference in outcomes seen in our study with 5hmC, as compared with a previous study (19), appears to be the sample size and the duration of follow-up. Our study has a sample size of 576 patient, whereas the previous study had a sample size of 70 ccRCC patients. In our study, 185 patients out of a total of 576 died at a median of 2.7 years following surgery (IQR, 1.1–5.1), and the median duration of follow-up for the 391 patients who were still alive at last follow-up was 7.2 years (IQR, 6.2–8.7). In the previous study, in the limited number of patients that had progression of disease in the observation period, although there was no statistically significant association between tumor 5hmC levels and mortality (P = 0.39), the HR was 0.48 (19). In summary, the increased sample size and duration of observation in our study enhanced the power to detect a statistically significant difference in the prognostic outcomes based on 5hmC in ccRCC. In addition to demonstrating that loss of 5hmC is an independent adverse prognostic factor in ccRCC, we have shown that a grading of 5hmC based on intensity (absent, mild, moderate, and marked) or based on percentage of positive tumor cells can be used as a strong tool to predict outcomes and could potentially be integrated in prognostic models and therapeutic decisions as well as clinical trial designs in the future.
From a mechanistic standpoint, we show that the increased 5mC or the loss of 5hmC in ccRCC is not explained by either inactivating mutations or underexpression of TET2 enzyme, but by functional inactivation of the TETs, at least partly due to a low expression of L2HGDH. This results in accumulation of L2HG — an oncometabolite that is known to inhibit the activity of TET enzymes by competitive inhibition of 2-oxoglutarate, a necessary cofactor (10). The expression of L2HGDH in kidney-tumor samples correlates well with the 5hmC levels. Patients with lower L2HGDH expression also have higher cytosine methylation content and a worse prognosis compared with those with higher expression. Interestingly, von Hippel–Lindau (VHL) knockdown, HIF stabilization, and activation of a hypoxia gene-expression pattern (which is seen in a majority of ccRCC) has been shown to be associated with an increase in L2HG levels via enhanced reductive carboxylation and without a change in L2HGDH mRNA expression (20). Taken together, the data suggest that in ccRCC, the reasons for an abnormal accumulation of L2HG (and consequent inhibition of 2OG-dependent enzymes, including TET) are both a reduction in the degradation of L2HG (due to deletions/underexpression of L2HGDH) and an increase in the production of L2HG (via enhanced reductive carboxylation).
We also report that AA enhances TET activity, resulting in epigenetic reprogramming with a genome-wide increase in hydroxymethylation in ccRCC cell lines (including 786-O, shown to have increased L2HG) and the demethylation of important tumor-suppressor genes, including SMAD6 and KMT2C. The epigenetic experiments were conducted mimicking the bioavailability curves of i.v. AA and considering potential differences between plasma concentrations and that of the tumor microenvironment. The rationale for mimicking the bioavailability of i.v. AA is that oral AA has been previously investigated in randomized trials and found to be ineffective as an anticancer agent (21, 22). However, recent studies indicate that AA at concentrations achieved only by the i.v. route (which are more than 100-fold higher than the oral route due to tight gastrointestinal regulation) has anticancer activity (16–18, 23, 24), and early phase trials have demonstrated that i.v. AA doses of up to 1–1.25 g/kg are not only well tolerated with chemotherapy, but also reduce the toxicity of chemotherapy and may have antitumor activity (25–30). With the short in vitro exposure time, epigenetic effects were more profound with doses achieved through the i.v. route.
We then aimed to determine the concentration of AA needed for its maximal effect on the TET enzymes. We realized this would be difficult to measure with conventional experiments for several reasons that make it hard to estimate accurate intracellular concentration of AA with a given extracellular concentration — Michaelis constant (Km) shifts of ascorbate transporters, decreased/mutated enzyme/transporter copy numbers, accelerated intracellular/extracellular oxidation, etc. Furthermore, Km shifts and copy numbers of TET enzymes could also play a role in the intracellular AA concentration needed for maximal effect on total TET activity. To eliminate these factors, we studied the quenching of fluorescence of the recombinant TET-2 protein with AA, which depends on the conformational changes of the protein induced by its binding with AA. We found that maximal (>90%) quenching was obtained with 132 μM AA, a concentration that would be very difficult to achieve even in the plasma with the maximum tolerated oral dose. Next, we studied the dynamics of fluorescence quenching of the recombinant TET-2 protein with the oncometabolite L2HG and the cosubstrate 2OG in the presence and absence of AA. We found that the quenching efficiency of 2OG is higher than that of L2HG, indicating that the substrate specificity of the TET-2 protein is 2OG over L2HG. We then found that the quenching of TET-2 protein with AA was unaffected by the presence of L2HG (with or without the presence of 2OG). Further studies need to be done to firmly establish the functional significance of these findings, specifically whether AA can partly or fully reverse the oncogenic effects of the metabolite L2HG in ccRCC.
We demonstrated that the H2O2 generated when AA is added to culture media does not play a role in the DNA demethylation and increase in hydroxymethylation in ccRCC with AA treatment and that this effect is TET enzyme dependent (both via catalase control and TET-knockdown experiments). This is an important consideration, since hydrogen peroxide is responsible for the acute cytotoxicity of cancer cells with short-term exposure to AA, as demonstrated in this study and others (16, 17, 31) with the complete reversal of short-term exposure cytotoxicity with catalase. However, despite the multiple in vitro studies demonstrating AA-induced H2O2-mediated cancer cytotoxicity and in vivo data demonstrating that H2O2 is produced in the extracellular fluid with high-dose parenteral AA, it remains a matter of debate whether H2O2 plays a significant role in AA-induced cytotoxicity in patients. For AA to generate H2O2, it requires the presence of free catalytic metal ions, which are available in plenty in vitro, but with questionable availability in the tumor microenvironment. We further demonstrate that high-dose AA can have proliferation-inhibition effects that occur at later time points with chronic high-dose AA exposure even with catalase neutralization, suggesting that AA could exert antitumor effects through non-H2O2, potentially epigenetic mechanisms. Through high-resolution genome-wide mapping of 5mC and 5hmC (using the OxBS and HELP techniques), we were able to demonstrate the epigenetic reprogramming of several putative tumor-suppressor genes in ccRCC. We further demonstrated that single-agent i.v. AA administration caused marked proliferation inhibition in a ccRCC cell line xenograft model and that the AA-treated xenografts revealed better differentiation as well as increased intratumoral 5hmC when compared with the vehicle-treated xenografts. It is possible that both mechanisms (enhancement of TET activity with epigenetic reprogramming as well as H2O2) may have anticancer effects in ccRCC (30).
The pharmacokinetic modeling data suggesting that supraphysiological plasma ascorbate concentrations are required to achieve effective delivery of ascorbate to tumor tissue (32), the data showing that the epigenetic effects of AA in vitro are more profound with doses achieved with the i.v. route with short exposure time, the potential additional benefit of H2O2 generation with the i.v. route, the xenograft data demonstrating ccRCC tumor proliferation inhibition with i.v. AA, the results to date in solid tumors demonstrating excellent tolerance of the i.v. schedule, and the lack of effect in cancer patients with oral AA together provide a strong rationale for future clinical trials in ccRCC of the i.v. schedule (30)
Methods
Cell lines.
ccRCC cell lines 786-O and 769-P were purchased from ATCC. Cell line authentication was done at ATCC. Cells were cultured in RPMI 1640 media supplemented with 10% v/v FBS and 1% v/v penicillin/streptomycin.
IHC (5hmC).
Tissue sectioning and IHC staining were performed at the Pathology Research Core, Mayo Clinic, using the Leica BOND RX stainer (Leica). Formalin-fixed, paraffin-embedded tissues were sectioned at 5 microns, and IHC staining was performed on site. Slides were retrieved for 20 minutes using Epitope Retrieval 1 (citrate, Leica) and incubated in protein block (Rodent Block M, Biocare) for 30 minutes. The 5hmc primary antibody (Active Motif, catalog 39769) was diluted to 1:1500 in Background Reducing Diluent (Dako) and incubated for 15 minutes.
The detection system used was the Polymer Refine Detection System (Leica). Immunostaining visualization was achieved by incubating slides for 10 minutes in DAB and DAB buffer (1:19 mixture) from the BOND Polymer Refine Detection System. For this, slides were rinsed between steps with 1× BOND Wash Buffer (Leica). Slides were counterstained for 5 minutes using Schmidt hematoxylin and molecular biology grade water (1:1 mixture), followed by several rinses in 1× BOND Wash Buffer and distilled water; this did not use the hematoxylin provided with the Refine kit. Once the immunochemistry process was completed, slides were removed from the stainer and rinsed in tap water for 5 minutes. Slides were dehydrated in increasing concentrations of ethyl alcohol and cleared in 3 changes of xylene prior to permanent coverslipping in xylene-based medium. IHC with L2HGDH (Sigma-Aldrich, catalog HPA065409) and TET2 (Abcam, catalog ab94580) was performed similarly.
Patient selection.
The Mayo Clinic Nephrectomy Registry was queried to identify 631 adults treated with radical or partial nephrectomy for sporadic, unilateral, noncystic ccRCC between 2007 and 2010. Of these, 576 (91%) had 5hmC expression available for analysis.
Statistical methods for clinicopathologic correlation with 5hmC.
The clinical and pathologic features studied were summarized with medians and IQRs or frequency counts and percentages and included age at surgery; sex; symptoms at diagnosis; Eastern Cooperative Oncology Group (ECOG) performance status; Charlson score; tumor size, 2010 primary tumor, regional lymph node (LN), and distant metastases classifications; WHO/International Society for Urological Pathology (ISUP) grade; coagulative tumor necrosis; sarcomatoid differentiation; SSIGN score; and progression score (33, 34). Patients with a palpable flank or abdominal mass, discomfort, gross hematuria, acute onset varicocele, or constitutional symptoms, including rash, sweats, weight loss, fatigue, early satiety, and anorexia, were considered symptomatic. Associations of 5hmC expression with the clinical and pathologic features studied were evaluated using Spearman’s rank correlation coefficients, Kruskal-Wallis tests, and Wilcoxon’s rank sum tests. OS, CSS, and PFS following surgery were calculated using the Kaplan-Meier method. Progression was defined as distant metastases or death from RCC based on the death certificate in the absence of documented distant metastases. Associations of 5hmC expression with time to death from any cause, time to death from RCC, and time to progression were evaluated using Cox proportional hazards regression models and summarized with HRs and 95% CIs. Statistical analyses were performed using SAS version 9.4 (SAS Institute) and R version 3.2.3 (R Foundation for Statistical Computing). All tests were 2 sided, and P values of less than 0.05 were considered statistically significant.
Measurement of L2HG by MS.
The enantiomers L2HG and d-2-hydroxyglutarate were measured as previously described (10, 35–37). Briefly, the cellular extracts were derivatized with (+)-Di-O-acetyl-L-tartaric anhydride (DATAN), a chiral derivatizing agent, followed by LC-MS/MS. Deuterated stable-isotope, d,l-[3,3,4,4-2H4]-2-hydroxyglutarate, was used as an internal standard, and the results were normalized to protein content of the cell extracts.
Nuclear protein extraction and in vitro TET enzymatic activity analysis.
Cells were treated with different concentrations of pH neutralized AA, with or without catalase. Exposure time to 1 mM AA was 2 hours (taking into account the plasma concentration curve with i.v. AA and potential differences between plasma concentrations and tumor microenvironment concentrations), and this was followed by the cells being washed and incubated in fresh media for 24 hours. Nuclear protein was then isolated from cells using the EpiQuik Nuclear Extraction Kit (EpiGentek Group Inc.), according to the manufacturer’s instructions. TET enzymatic activity was measured by using the ELISA-based Epigenase 5mC Hydroxylase TET Activity/Inhibition Assay Kit (Fluorometric) according to the manufacturer’s instructions. This technique relies on the conversion of methylated products at the bottom of the wells to hydroxymethylated products by the TET enzyme present in the nuclear extract. Thus, the amount of hydroxymethylated products formed is a measure of the TET activity of the nuclear extract harvested from the cells being tested. Incubation time of nuclear lysates was 90 minutes. Six micrograms of nuclear lysate was used per well for measurement of TET activity.
Measurement of 5hmC levels by MS.
DNA hydrolysis was performed as previously described (38). Briefly, 1 μg of genomic DNA was first denatured by heating at 100°C. Five units of Nuclease P1 (Sigma-Aldrich, catalog N8630) were added and the mixture incubated at 45°C for 1 hour. A 1/10 volume of 1 M ammonium bicarbonate and 0.002 units of venom phosphodiesterase 1 (Sigma-Aldrich, catalog P3243) were added to the mixture and the incubation continued for 2 hours at 37°C. Next, 0.5 units of alkaline phosphatase (Invitrogen, catalog 18009-027) were added, and the mixture was incubated for 1 hour at 37°C. Quantification was performed using a LC-ESI-MS/MS system in the multiple reaction monitoring mode, as described (38), with some modifications. Before injection into the Zorbax Eclipse Plus C18 2.1 mm × 150 mm column (1.8 μm particle size) (Agilent, catalog 959759-902), the reactions were diluted 10-fold to dilute out the salts and the enzymes. Samples were analyzed on an Agilent 1290 series LC instrument in tandem with the Agilent 6490 triple quadrupole mass spectrometer.
Genome-wide DNA methylation analysis using the HELP assay.
Intact DNA of high molecular weight was corroborated by electrophoresis on 1% agarose gel in all cases. One microgram of genomic DNA was digested overnight with either HpaII or MspI (NEB). The following day, the reactions were extracted once with phenol chloroform and resuspended in 11 μl of 10 mM Tris-HCl pH 8.0. The digested DNA was used to set up an overnight ligation of the JHpaII adapter using T4 DNA ligase. The adapter-ligated DNA was used to carry out the PCR amplification of the HpaII- and MspI-digested DNA as previously described (1). Both amplified fractions were submitted to Roche-NimbleGen Inc. for labeling and hybridization onto a human hg18 custom-designed oligonucleotide array (50 mers) covering 1.3 million HpaII amplifiable fragments (HAF). HELP microarray data were deposited in the NCBI’s Gene Expression Omnibus database (GEO GSE126823). All microarray hybridization was subjected to extensive quality control. Uniformity of hybridization was evaluated using a modified version of a previously published algorithm adapted for the NimbleGen platform, and any hybridization with strong regional artifacts was discarded. Bioinformatic analysis was done as previously reported (15, 39).
Whole-genome methylation and hydroxymethylation analysis by oxidative bisulfite sequencing.
One microgram genomic DNA from ccRCC cells (control and AA treated) was sonicated to 100–400 bp by Bioruptor, and 0.5% (w/w) sequencing spike-in control DNA was added thereafter and purified by AMPure XP beads (Beckman Coulter). Spike-in controls were added to the adapted library. Half of the library was subjected to oxidation reaction following the manufacturer’s protocol (Cambridge Epigenetix). Both oxidized and nonoxidized samples were then treated with bisulphite conversion reagent. The final PCR was performed according to the manufacturer’s guide using 10 cycles of amplification and was purified and sequenced at the Einstein Epigenomics Facility (40). Bismark was used to map the sample reads and make methylation calls. At every base location, for the estimation of 5-hmC, both bisulfite sequencing (BS) and OxBS samples were analyzed, and an estimate for the percentage of 5-hmC methylation was calculated by the difference between BS and OxBS conversions.
Since 5-hmC is a less frequent modification, for further stringency in measuring the difference in ratios, we used Fisher’s exact test of proportions, using the number of converted and nonconverted reads in the BS and OxBS samples and selected sites that had a P value of less than 0.05. 5-mC sites were calculated by the ratio of nonconverted bases to total bases in the OxBS sample, with a biologically influenced threshold of 50%. To compare methylation between cancer and control samples, we used Fisher’s test and adjusted for multiple comparisons through the Benjamini-Hochberg procedure. Data data were deposited in the NCBI’s Gene Expression Omnibus database (GEO GSE 124610).
For total number of reads analyzed, we combined runs from MISEQ and HISEQ across the BS and OxBS protocols, as follows: CO BS: 1542651 nReads (Miseq); 115456119 nReads (Hiseq); CO OxBS: 1598798 nReads (Miseq); 97154526 nReads (Hiseq); VITC BS: 1366239 nReads (Miseq); 146589506 nReads (Hiseq); VITC OxBS: 1431420 nReads (Miseq); and 65557437 nReads (Hiseq).
For the number of distinct loci found to acquire hmc, we used the following rule set: [ctrl.hmc < vitc.hmc & vitc.hmc > THRESH & ctrl.mc > vitc.mc], where THRESH is set to 0.25. With AA treatment, 827 distinct loci were found to gain hmC.
Cell viability.
Cells were incubated at varying concentrations and time periods with L-AA (Sigma-Aldrich, catalog 50-81-7) with or without catalase at 100 μg/ml (Sigma-Aldrich, catalog 9001-05-2). Viability was assessed by addition of Cell Titer Blue (Promega) and measured via the Fluostar Omega Microplate Reader (BMG Labtech). We found that antioxidant drugs interfere with cell viability measurements by assays that rely on the reducing property of viable cells. They directly reduce the reagent substrate to the reduced fluorescent form, giving spurious results. The protocol modification to counter this interference has been described and was used in this study (41, 42).
Flow cytometry for apoptosis and cell-cycle analysis.
The 786-O cells were treated with high-dose AA (+catalase), and apoptosis and cell-cycle dynamics were studied at the 96-hour time point. Cells were washed with annexin buffer solution and stained with both propidium iodide (PI) and FITC–annexin V (Life Technologies), and assayed on a BD FACSCalibur flow cytometer. Apoptosis results were analyzed with BD CellQuest software. For cell-cycle analysis, cells were fixed with 70% cold ethanol and subsequently analyzed after PI staining. Analysis was performed using FlowJo software.
Fluorescence spectroscopy.
All fluorescence measurements were performed at 20°C on a Horiba Jobin Yvon Fluorolog 3 spectrofluorometer equipped with a Wavelength electronics model LFI-3751 temperature controller. Protein fluorescence emission spectra of TET2 were averaged 3 times between 305 and 400 nm, with excitation at 280 nm. The step width was 1 nm and the integration time 1 second. All protein solutions contained 0.5 μM TET2 in PBS buffer. PBS-buffered stock solutions of 2OG, L2HG, and AA were added stepwise to the protein solutions prior to the measurement of the fluorescence spectra.
Quenching constants were obtained using the Stern-Volmer equation (43), as follows: F0/F = 1 + KD × [Q], where KD is the quenching constant and [Q] is a defined concentration of AA. F0 and F represent fluorescence intensities (at 328 nm) in the absence and presence of AA, respectively.
RT-PCR.
Total RNA was isolated from ccRCC cells using the RNeasy Mini Kit (QIAGEN) according to the manufacturer’s protocol. cDNA was generated using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems) with random primers. Quantitative reverse-transcriptase PCR (qRT-PCR) was performed using Fast SYBR Green Master Mix (Applied Biosystems, catalog 4385612) and primers specific to SMAD6/TET1/TET2/TET3. GAPDH was used as an internal loading control.
In vivo studies with AA.
ccRCC cells (786-O) were xenografted into immunodeficient NSG mice, obtained from The Jackson Laboratory. After tumors were established, treatment was initiated with tail-vein injections with i.v. AA at 1 g/kg/d or vehicle over 5 weeks (5 days dosing/wk), and tumor measurements were conducted.
IHC was performed on 5 μm thick paraffin sections following the standardized protocol (44). Briefly, slides were deparaffinized in 3 changes of xylene for 5 minutes each, followed by rehydration through a graded series of alcohol. Antigen retrieval was performed in citrate buffer (pH 6.0) for 10 minutes, followed by subsequent cooling for 20 minutes and blocking of endogenous peroxidase with 30% hydrogen peroxide. Slides were next incubated with the primary antibody 5-hMC (Active Motif, catalog 39769, 1:750) at 4°C, overnight, washed 3 times with Tris-buffered saline (5 minutes each), and incubated with biotinylated anti-rabbit secondary antibody (1:200) for 1 hour at room temperature. After treating the slides with HRP-conjugated ABC complex (Vectastain, Vector Laboratories) for 1 hour at room temperature, color was developed with DAB (Vector Laboratories), and slides were counterstained with hematoxylin, mounted with DPX, and examined under an Olympus DP73 microscope for imaging, analysis, and interpretation. Sections from a reactive LN served as a positive control, while those without the addition of a primary antibody served as negative controls.
Statistics.
For clinical data, associations of 5hmC expression with the clinical and pathologic features studied were evaluated using Spearman’s rank correlation coefficients, Kruskal-Wallis tests, and Wilcoxon’s rank-sum tests. OS, CSS, and PFS following surgery were calculated using the Kaplan-Meier method. Associations of 5hmC expression with time to death from any cause, time to death from RCC, and time to progression were evaluated using the Cox’s proportional hazards regression model. All tests were 2 sided, and P values of less than 0.05 were considered statistically significant. For data obtained from ccRCC TCGA in Figure 3 (parts D and F) as well as L2HGDH IHC correlation with 5hmC, 2-sided Student’s t test was used. For in vitro experiments, 1-sided Student’s t test was used for statistical analyses in Figure 4 and Figure 6. Paired t test was used for statistical analyses in Figure 5. For the in vivo xenograft study, 2-sided Student’s t test was used for statistical analyses. Statistical significance was defined as P < 0.05.
Study approval.
The study of the correlation between 5hmC and clinicopathologic features in ccRCC patients was approved by the Mayo Clinic IRB (17-009764). The ccRCC xenograft study was approved by the Albert Einstein College of Medicine IRB and IACUC.
Author contributions
NS and AV developed the study concepts and hypothesis and designed the study. NS, TDB, JC, CL, YZ, SAB, AT, VM, DR, SGM, GC, ER, BA, LFW, XW, LG, SAB, BL, RHT, and AV acquired data. NS, AV, and TDB analyzed and interpreted data. JC and CL performed scoring and statistical analysis of 5hmC IHC (clinical cases). KP, AV, and NS were responsible for bioinformatics. AT, VM, and NS performed fluorescence quenching. YZ and TDB performed xenograph studies. NS and AV prepared the manuscript. NS, AV, US, TW, LP, BG, MS, JMG, and KS edited and reviewed the manuscript.
Supplementary Material
Acknowledgments
We thank the Mayo Clinic Pathology Research Core team as well as E. Nieves (Biochemistry and Developmental & Molecular Biology, Albert Einstein College of Medicine). Funding was provided by a 2017 American Society of Hematology Research Training Award for Fellows (to NS).
Version 1. 01/31/2019
In-Press Preview
Version 2. 03/04/2019
Electronic publication
Version 3. 04/01/2019
Print issue publication
Footnotes
Conflict of interest: NS, AV, TW, JC have applied for a patent for the use of 5hmC as a biomarker for cancer prognostication and management (application no. 62/673,582). AV has received research funding from GSK, Incyte, MedPacto, Novartis, Curis, and Eli Lilly, has received compensation as a scientific advisor to Novartis, Stelexis Therapeutics, Acceleron, and Celgene, and has equity ownership in Stelexis Therapeutics.
Copyright: © 2019 American Society for Clinical Investigation
Reference information: J Clin Invest. 2019;129(4):1612–1625.https://doi.org/10.1172/JCI98747.
Contributor Information
Niraj Shenoy, Email: niraj.shenoy@einstein.yu.edu.
Tushar D. Bhagat, Email: tushar.bhagat@phd.einstein.yu.edu.
Christine Lohse, Email: Lohse.Christine@mayo.edu.
Sanchari Bhattacharyya, Email: sanchari.bhattacharyya@einstein.yu.edu.
Alexander Tischer, Email: tischer.alexander@mayo.edu.
Venkata Machha, Email: machha.venkata@mayo.edu.
Shanisha Gordon-Mitchell, Email: shanisha.gordon@einstein.yu.edu.
Li-Fan Wong, Email: lw54776n@pace.edu.
LouAnn Gross, Email: Gross.LouAnn@mayo.edu.
Stephen A. Boorjian, Email: boorjian.stephen@mayo.edu.
Ulrich Steidl, Email: ulrich.steidl@einstein.yu.edu.
Xiaosheng Wu, Email: wu.xiaosheng@mayo.edu.
Kith Pradhan, Email: kith.pradhan@einstein.yu.edu.
Benjamin Gartrell, Email: bgartrel@montefiore.org.
Beamon Agarwal, Email: beagarwa@gmail.com.
Lance Pagliaro, Email: Pagliaro.Lance@mayo.edu.
Masako Suzuki, Email: masako.suzuki@einstein.yu.edu.
John M. Greally, Email: jgreally@aecom.yu.edu.
Dinesh Rakheja, Email: dinesh.rakheja@utsouthwestern.edu.
R. Houston Thompson, Email: Thompson.Robert@mayo.edu.
Katalin Susztak, Email: ksusztak@mail.med.upenn.edu.
Thomas Witzig, Email: witzig.thomas@mayo.edu.
Yiyu Zou, Email: yiyu.zou@einstein.yu.edu.
Amit Verma, Email: amit.verma@einstein.yu.edu.
References
- 1.Hu CY, et al. Kidney cancer is characterized by aberrant methylation of tissue-specific enhancers that are prognostic for overall survival. Clin Cancer Res. 2014;20(16):4349–4360. doi: 10.1158/1078-0432.CCR-14-0494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cancer Genome Atlas Research Network Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature. 2013;499(7456):43–49. doi: 10.1038/nature12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shenoy N, et al. Role of DNA methylation in renal cell carcinoma. J Hematol Oncol. 2015;8:88. doi: 10.1186/s13045-015-0180-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shenoy N, Pagliaro L. Sequential pathogenesis of metastatic VHL mutant clear cell renal cell carcinoma: putting it together with a translational perspective. Ann Oncol. 2016;27(9):1685–1695. doi: 10.1093/annonc/mdw241. [DOI] [PubMed] [Google Scholar]
- 5.Delhommeau F, et al. Mutation in TET2 in myeloid cancers. N Engl J Med. 2009;360(22):2289–2301. doi: 10.1056/NEJMoa0810069. [DOI] [PubMed] [Google Scholar]
- 6.Gaidzik VI, et al. TET2 mutations in acute myeloid leukemia (AML): results from a comprehensive genetic and clinical analysis of the AML study group. J Clin Oncol. 2012;30(12):1350–1357. doi: 10.1200/JCO.2011.39.2886. [DOI] [PubMed] [Google Scholar]
- 7.Rasmussen KD, et al. Loss of TET2 in hematopoietic cells leads to DNA hypermethylation of active enhancers and induction of leukemogenesis. Genes Dev. 2015;29(9):910–922. doi: 10.1101/gad.260174.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kroeze LI, et al. Characterization of acute myeloid leukemia based on levels of global hydroxymethylation. Blood. 2014;124(7):1110–1118. doi: 10.1182/blood-2013-08-518514. [DOI] [PubMed] [Google Scholar]
- 9.Dang L, Jin S, Su SM. IDH mutations in glioma and acute myeloid leukemia. Trends Mol Med. 2010;16(9):387–397. doi: 10.1016/j.molmed.2010.07.002. [DOI] [PubMed] [Google Scholar]
- 10.Shim EH, et al. L-2-Hydroxyglutarate: an epigenetic modifier and putative oncometabolite in renal cancer. Cancer Discov. 2014;4(11):1290–1298. doi: 10.1158/2159-8290.CD-13-0696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blaschke K, et al. Vitamin C induces Tet-dependent DNA demethylation and a blastocyst-like state in ES cells. Nature. 2013;500(7461):222–226. doi: 10.1038/nature12362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yin R, et al. Ascorbic acid enhances Tet-mediated 5-methylcytosine oxidation and promotes DNA demethylation in mammals. J Am Chem Soc. 2013;135(28):10396–10403. doi: 10.1021/ja4028346. [DOI] [PubMed] [Google Scholar]
- 13.Cimmino L, et al. Restoration of TET2 function blocks aberrant self-renewal and leukemia progression. Cell. 2017;170(6):1079–1095.e20. doi: 10.1016/j.cell.2017.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Agathocleous M, et al. Ascorbate regulates haematopoietic stem cell function and leukaemogenesis. Nature. 2017;549(7673):476–481. doi: 10.1038/nature23876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bhattacharyya S, et al. Genome-wide hydroxymethylation tested using the HELP-GT assay shows redistribution in cancer. Nucleic Acids Res. 2013;41(16):e157. doi: 10.1093/nar/gkt601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen Q, et al. Pharmacologic ascorbic acid concentrations selectively kill cancer cells: action as a pro-drug to deliver hydrogen peroxide to tissues. Proc Natl Acad Sci U S A. 2005;102(38):13604–13609. doi: 10.1073/pnas.0506390102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen Q, et al. Ascorbate in pharmacologic concentrations selectively generates ascorbate radical and hydrogen peroxide in extracellular fluid in vivo. Proc Natl Acad Sci U S A. 2007;104(21):8749–8754. doi: 10.1073/pnas.0702854104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen Q, et al. Pharmacologic doses of ascorbate act as a prooxidant and decrease growth of aggressive tumor xenografts in mice. Proc Natl Acad Sci U S A. 2008;105(32):11105–11109. doi: 10.1073/pnas.0804226105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Munari E, et al. Global 5-hydroxymethylcytosine levels are profoundly reduced in multiple genitourinary malignancies. PLoS ONE. 2016;11(1):e0146302. doi: 10.1371/journal.pone.0146302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oldham WM, Clish CB, Yang Y, Loscalzo J. Hypoxia-mediated increases in L-2-hydroxyglutarate coordinate the metabolic response to reductive stress. Cell Metab. 2015;22(2):291–303. doi: 10.1016/j.cmet.2015.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Creagan ET, et al. Failure of high-dose vitamin C (ascorbic acid) therapy to benefit patients with advanced cancer. A controlled trial. N Engl J Med. 1979;301(13):687–690. doi: 10.1056/NEJM197909273011303. [DOI] [PubMed] [Google Scholar]
- 22.Moertel CG, Fleming TR, Creagan ET, Rubin J, O’Connell MJ, Ames MM. High-dose vitamin C versus placebo in the treatment of patients with advanced cancer who have had no prior chemotherapy. A randomized double-blind comparison. N Engl J Med. 1985;312(3):137–141. doi: 10.1056/NEJM198501173120301. [DOI] [PubMed] [Google Scholar]
- 23.Du J, Cullen JJ, Buettner GR. Ascorbic acid: chemistry, biology and the treatment of cancer. Biochim Biophys Acta. 2012;1826(2):443–457. doi: 10.1016/j.bbcan.2012.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Verrax J, Calderon PB. Pharmacologic concentrations of ascorbate are achieved by parenteral administration and exhibit antitumoral effects. Free Radic Biol Med. 2009;47(1):32–40. doi: 10.1016/j.freeradbiomed.2009.02.016. [DOI] [PubMed] [Google Scholar]
- 25.Hoffer LJ, et al. Phase I clinical trial of i.v. ascorbic acid in advanced malignancy. Ann Oncol. 2008;19(11):1969–1974. doi: 10.1093/annonc/mdn377. [DOI] [PubMed] [Google Scholar]
- 26.Monti DA, et al. Phase I evaluation of intravenous ascorbic acid in combination with gemcitabine and erlotinib in patients with metastatic pancreatic cancer. PLoS ONE. 2012;7(1):e29794. doi: 10.1371/journal.pone.0029794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stephenson CM, Levin RD, Spector T, Lis CG. Phase I clinical trial to evaluate the safety, tolerability, and pharmacokinetics of high-dose intravenous ascorbic acid in patients with advanced cancer. Cancer Chemother Pharmacol. 2013;72(1):139–146. doi: 10.1007/s00280-013-2179-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Crump M, et al. Randomized comparison of gemcitabine, dexamethasone, and cisplatin versus dexamethasone, cytarabine, and cisplatin chemotherapy before autologous stem-cell transplantation for relapsed and refractory aggressive lymphomas: NCIC-CTG LY.12. J Clin Oncol. 2014;32(31):3490–3496. doi: 10.1200/JCO.2013.53.9593. [DOI] [PubMed] [Google Scholar]
- 29.Ma Y, Chapman J, Levine M, Polireddy K, Drisko J, Chen Q. High-dose parenteral ascorbate enhanced chemosensitivity of ovarian cancer and reduced toxicity of chemotherapy. Sci Transl Med. 2014;6(222):222ra18. doi: 10.1126/scitranslmed.3007154. [DOI] [PubMed] [Google Scholar]
- 30.Shenoy N, Creagan E, Witzig T, Levine M. Ascorbic acid in cancer treatment: let the phoenix fly. Cancer Cell. 2018;34(5):700–706. doi: 10.1016/j.ccell.2018.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shenoy N, et al. Upregulation of TET activity with ascorbic acid induces epigenetic modulation of lymphoma cells. Blood Cancer J. 2017;7(7):e587. doi: 10.1038/bcj.2017.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kuiper C, Vissers MC, Hicks KO. Pharmacokinetic modeling of ascorbate diffusion through normal and tumor tissue. Free Radic Biol Med. 2014;77:340–352. doi: 10.1016/j.freeradbiomed.2014.09.023. [DOI] [PubMed] [Google Scholar]
- 33.Frank I, Blute ML, Cheville JC, Lohse CM, Weaver AL, Zincke H. An outcome prediction model for patients with clear cell renal cell carcinoma treated with radical nephrectomy based on tumor stage, size, grade and necrosis: the SSIGN score. J Urol. 2002;168(6):2395–2400. doi: 10.1016/S0022-5347(05)64153-5. [DOI] [PubMed] [Google Scholar]
- 34.Leibovich BC, et al. Prediction of progression after radical nephrectomy for patients with clear cell renal cell carcinoma: a stratification tool for prospective clinical trials. Cancer. 2003;97(7):1663–1671. doi: 10.1002/cncr.11234. [DOI] [PubMed] [Google Scholar]
- 35.Jones PM, Boriack R, Struys EA, Rakheja D. Measurement of oncometabolites D-2-hydroxyglutaric acid and L-2-hydroxyglutaric acid. Methods Mol Biol. 2017;1633:219–234. doi: 10.1007/978-1-4939-7142-8_14. [DOI] [PubMed] [Google Scholar]
- 36.Rakheja D, Mitui M, Boriack RL, DeBerardinis RJ. Isocitrate dehydrogenase 1/2 mutational analyses and 2-hydroxyglutarate measurements in Wilms tumors. Pediatr Blood Cancer. 2011;56(3):379–383. doi: 10.1002/pbc.22697. [DOI] [PubMed] [Google Scholar]
- 37.Rogers RE, Deberardinis RJ, Klesse LJ, Boriack RL, Margraf LR, Rakheja D. Wilms tumor in a child with L-2-hydroxyglutaric aciduria. Pediatr Dev Pathol. 2010;13(5):408–411. doi: 10.2350/09-12-0768-CR.1. [DOI] [PubMed] [Google Scholar]
- 38.Figueroa ME, et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell. 2010;18(6):553–567. doi: 10.1016/j.ccr.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Suzuki M, Jing Q, Lia D, Pascual M, McLellan A, Greally JM. Optimized design and data analysis of tag-based cytosine methylation assays. Genome Biol. 2010;11(4):R36. doi: 10.1186/gb-2010-11-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Booth MJ. Quantitative sequencing of 5-methylcytosine and 5-hydroxymethylcytosine at single-base resolution. Science. 2012;336(6083):934–937. doi: 10.1126/science.1220671. [DOI] [PubMed] [Google Scholar]
- 41.Shenoy N, Stenson M, Lawson J, Abeykoon J, Patnaik M, Wu X. Drugs with anti-oxidant properties can interfere with cell viability measurements by assays that rely on the reducing property of viable cells. Lab Invest. 2017;94:494–497. doi: 10.1038/labinvest.2017.18. [DOI] [PubMed] [Google Scholar]
- 42. Shenoy N, Wu X, Witzig T. Protocol Modification To Determine The Cytotoxic Potential Of Drugs Using Cell Viability Assays That Rely On The Reducing Property Of Viable Cells. Nature Protocol Exchange Web site. https://www.nature.com/protocolexchange/protocols/5735 Published March 20, 2017. Accessed January 30, 2019.
- 43. Lakowicz, JR. Principles of Fluorescence Spectroscopy. 3rd ed. New York, NY: Springer; 2006. [Google Scholar]
- 44.Agarwal B, Das P, Naresh KN, Borges AM. Angiogenic ability of metastatic squamous carcinoma in the cervical lymph nodes from unknown primary tumours. J Clin Pathol. 2011;64(9):765–770. doi: 10.1136/jclinpath-2011-200039. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.