Abstract
Hyperactivated AKT/mTOR signaling is a hallmark of pancreatic neuroendocrine tumors (PNETs). Drugs targeting this pathway are used clinically, but tumor resistance invariably develops. A better understanding of factors regulating AKT/mTOR signaling and PNET pathogenesis is needed to improve current therapies. We discovered that RABL6A, a new oncogenic driver of PNET proliferation, is required for AKT activity. Silencing RABL6A caused PNET cell-cycle arrest that coincided with selective loss of AKT-S473 (not T308) phosphorylation and AKT/mTOR inactivation. Restoration of AKT phosphorylation rescued the G1 phase block triggered by RABL6A silencing. Mechanistically, loss of AKT-S473 phosphorylation in RABL6A-depleted cells was the result of increased protein phosphatase 2A (PP2A) activity. Inhibition of PP2A restored phosphorylation of AKT-S473 in RABL6A-depleted cells, whereas PP2A reactivation using a specific small-molecule activator of PP2A (SMAP) abolished that phosphorylation. Moreover, SMAP treatment effectively killed PNET cells in a RABL6A-dependent manner and suppressed PNET growth in vivo. The present work identifies RABL6A as a new inhibitor of the PP2A tumor suppressor and an essential activator of AKT in PNET cells. Our findings offer what we believe is a novel strategy of PP2A reactivation for treatment of PNETs as well as other human cancers driven by RABL6A overexpression and PP2A inactivation.
Keywords: Cell Biology, Oncology
Keywords: Cancer, Drug therapy, Phosphoprotein phosphatases
Introduction
Neuroendocrine tumors (NETs) are malignancies that are clinically challenging to manage and treat whose incidence is steadily increasing (1, 2). They are slow growing, biologically diverse tumors that arise from neuroendocrine cells throughout the body (3). NETs in the pancreas (hereafter designated PNETs) are rare, islet-derived neoplasms that nonetheless represent the second most common cancer of the pancreas (4). Most patients with PNETs are asymptomatic or have nonspecific symptoms, delaying diagnosis until tumors have advanced and are often metastatic (1, 3, 4). Surgical resection is the first line therapy for primary PNETs, and hepatic cytoreduction may also play a role in the treatment of metastatic tumors (4–6). Somatostatin analogs and peptide receptor radionuclide therapy (PRRT) are important modalities for treating metastatic tumors, as are targeted therapies with the mammalian target of rapamycin (mTOR) inhibitor, everolimus, and the receptor tyrosine kinase inhibitor sunitinib. Chemotherapy is also used for disease control in advanced PNET patients. However, none of these approaches are curative and they have minimal impact on long-term survival, necessitating better therapeutic options for these patients (4, 5, 7–9).
The clinical use of everolimus in patients with advanced PNETs (10–13) was based on evidence that hyperactivation of PI3-kinase (PI3K)/AKT/mTOR signaling is central to PNET progression and associated with worse patient outcomes (14–18). Tumors harbor mutations in pathway genes (e.g., PIK3CA, TSC2, and PTEN) (14, 18) and dysregulation of the proteins is common, including downregulation of the PTEN and TSC2 tumor suppressors and increased phosphorylation of AKT and mTORC1 kinase targets/effectors (S6K, S6, and EIF4EBP1) (15–17). Everolimus therapy significantly increases median progression-free survival and, although not statistically significant, is associated with a 6-month improvement in median overall survival (to 44 months) compared with placebo (11, 12). However, sustained inhibition of mTOR with everolimus removes negative feedback control of AKT, paradoxically causing unwanted AKT activation and drug resistance in tumors (19, 20). This has encouraged combination therapies targeting both mTOR and AKT (10). Given the importance of PI3K/AKT/mTOR signaling in PNET pathogenesis, a greater understanding of factors controlling the pathway is needed to improve current therapies or offer new alternatives, particularly when drug resistance develops.
RABL6A is a novel oncogenic driver of PNETs whose role in cancer cell signaling is beginning to unfold. It is a large, RAB-like GTPase (also called c9orf86, RBEL1, or Parf [partner of ARF]) that physically and functionally interacts with the Alternative Reading Frame (ARF)–Mdm2-p53 tumor suppressor pathway (21–23). Elevated RABL6A expression is seen in various human cancers, including breast tumors and pancreatic ductal adenocarcinomas where its overexpression is associated with poor survival (24–26). We recently showed that the RABL6A gene is amplified and its protein highly expressed in patient PNETs (23). Moreover, RABL6A is required for PNET cell survival and cell-cycle progression, in part through its inactivation of the retinoblastoma (RB1) tumor suppressor (23). It was apparent, however, that additional, currently undefined pathways besides RB1 contribute to and are required for RABL6A-mediated PNET survival and proliferation.
This study sought to identify other clinically relevant pathways regulated by RABL6A in PNETs. We compared gene expression profiles from patient tumors and RABL6A-depleted cells and found that RABL6A expression correlated with AKT pathway activation. Analyses of PNET cells showed that RABL6A promotes S473 phosphorylation of AKT, which is required for AKT activation and G1-to-S phase progression. RABL6A controls AKT phosphorylation through inhibition of protein phosphatase 2A (PP2A), a powerful tumor suppressor and known regulator of AKT (27–31). Importantly, reactivation of endogenous PP2A with a small-molecule activator of PP2A (SMAP) killed PNET cells in a RABL6A-dependent manner and blocked tumor growth in vivo. These discoveries illustrate the biological importance of a novel RABL6A-PP2A-AKT pathway in driving PNET pathogenesis, suggesting new targets (PP2A and RABL6A) for PNET therapy.
Results
RABL6A is required for AKT-mTOR activation in PNETs.
We recently showed that RABL6A is required for PNET cell proliferation and promotes G1-to-S phase progression via inactivation of the RB1 tumor suppressor (23). Specifically, PNET cells with RABL6A knock down failed to proliferate and accumulated hypophosphorylated RB1, the growth inhibitory form of the protein. Inactivation of RB1 partially rescued the knockdown phenotype by promoting G1-to-S phase transition; however, it was unable to override the mitotic block and eventual cell death caused by RABL6A loss. This revealed that RABL6A controls additional pathways besides RB1 to promote PNET cell survival and proliferation (Figure 1A).
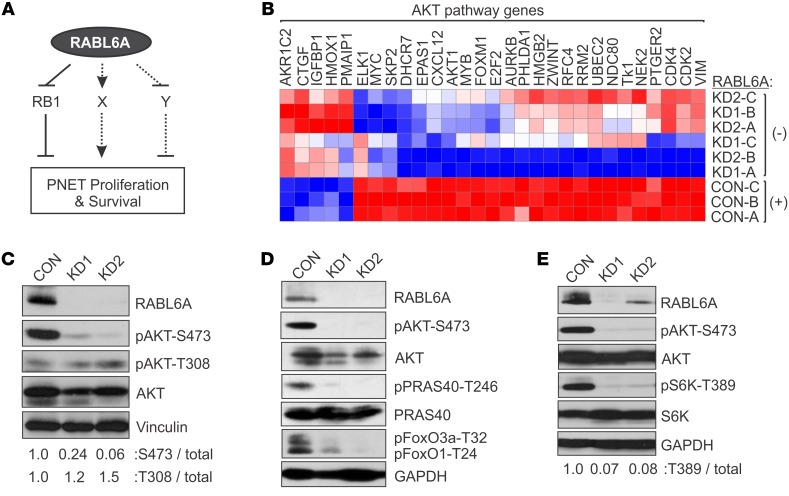
Figure 1. RABL6A depletion impairs AKT-S473 phosphorylation and AKT-mTOR signaling in PNETs.
(A) Schematic showing that RABL6A promotes PNET cell proliferation and survival through multiple mechanisms, including inhibition of RB1 signaling as well as regulation of other undefined (X, Y) pathways. Arrow, activating event; perpendicular bar, inhibitory event. (B) BON-1 cells expressing vector control (CON: with RABL6A) or shRNAs targeting RABL6A (KD1 and KD2: without RABL6A) were examined by microarray analyses. Heat map shows that RABL6A depletion significantly alters the expression of genes involved in AKT signaling; data from 3 experiments, designated A–C. Genes were categorized by IPA software and displayed 2-fold or greater changes in expression (P < 0.05). Red, relatively increased expression; blue, relatively decreased expression. (C) Representative Western blots showing that RABL6A knockdown (KD1, KD2) in BON-1 cells specifically reduces the activating phosphorylation of AKT at S473, not T308. Vinculin served as loading control. Relative phosphorylation of each residue relative to total AKT was quantified by ImageJ. (D) Representative Western blots showing that inactivation of AKT in RABL6A knockdown BON-1 cells coincides with reduced phosphorylation of AKT substrates, PRAS40-T246 and FoxO1-T24/FoxO3a-T32, with GAPDH as loading control. (E) Representative Western blots showing that inactivation of AKT in RABL6A knockdown BON-1 cells coincides with reduced activation of mTORC1, as measured by decreased S6K phosphorylation at T389. GAPDH was the loading control. S6K-T389 phosphorylation relative to total S6K was quantified by ImageJ. Experiments in C–E represent at least 3 independent experimental repeats.
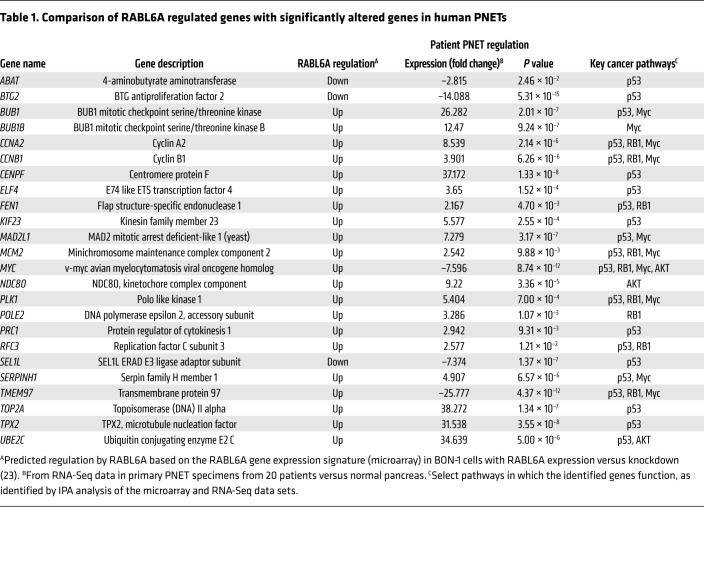
To identify other relevant pathways controlled by RABL6A in PNETs, we compared the gene signature of RABL6A expressing PNET cells with that of patient PNETs. Microarray data from BON-1 PNET cells that express or lack RABL6A (23) were used to define the RABL6A signature, while RNA-Seq data from 20 primary human PNETs (compared with normal human pancreas) were examined to identify the most significantly altered genes in patient tumors. The comparison shown in Table 1 revealed remarkable concordance between the 2 data sets, in keeping with RABL6A amplification (23) and increased mRNA expression (Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/JCI123049DS1) in human PNETs. Prominent cancer pathways affected in patient tumors and similarly regulated by RABL6A included p53, RB1, and AKT (Table 1). Having previously examined the importance of p53 and RB1 in RABL6A function in PNETs (23), we focused on the AKT pathway, a major driver of PNET pathogenesis (14–18). Ingenuity Pathway Analysis (IPA) of the gene expression profiles from RABL6A knockdown (KD) versus control BON-1 cells suggested that loss of RABL6A expression impaired AKT pathway activation (Figure 1B).
Table 1. Comparison of RABL6A regulated genes with significantly altered genes in human PNETs.
Activation of AKT requires phosphorylation at 2 key residues: T308 by phosphoinositide-dependent kinase-1 (PDK1) and S473 by mTORC2 (32, 33). To determine if RABL6A controls AKT activation and signaling, we knocked down endogenous RABL6A in BON-1 cells with 2 different shRNAs (KD1 or KD2) (23, 26) and assessed AKT phosphorylation by Western blotting. In these cells, RABL6A loss induced an initial cell-cycle arrest (mainly in G1 with a moderate G2/M block) that is sustained for 3–5 days followed by apoptosis at later time points (23). Downregulation of RABL6A greatly reduced S473 but not T308 phosphorylation of AKT (Figure 1C). This coincided with a failure of AKT to phosphorylate its substrates, PRAS40 and FoxO1/FoxO3a (Figure 1D), and reduced downstream signaling by mTORC1 kinase as measured by decreased S6K phosphorylation (Figure 1E). Thus, RABL6A is required for AKT-mTOR activation and signaling in PNET cells.
RABL6A promotes G1-to-S phase progression through AKT activation and sensitizes PNET cells to AKT inhibitors.
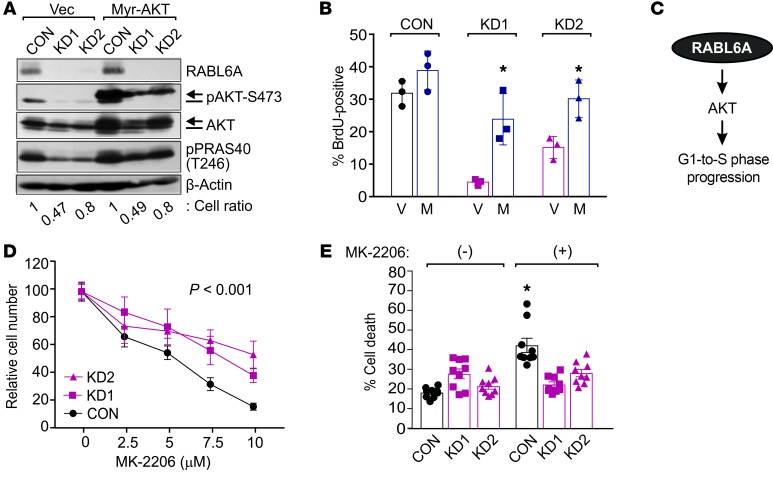
Given the importance of AKT to tumor cell proliferation and survival (32, 33), we speculated that its inactivation may contribute to the cell proliferation defects caused by RABL6A silencing in PNET cells. To test that idea, we expressed myristoylated AKT (Myr-AKT) in BON-1 cells to determine if constitutively activated AKT could rescue the RABL6A knockdown phenotype. Immunoblotting verified Myr-AKT expression and constitutive phosphorylation at S473, even in RABL6A-deficient cells where endogenous AKT-S473 phosphorylation was absent (Figure 2A). Activated Myr-AKT effectively restored phosphorylation of PRAS40-T246 in the RABL6A-deficient cells (Figure 2A). BrdU incorporation analyses showed that Myr-AKT promoted a significant increase in DNA synthesis in RABL6A knockdown cells (Figure 2B), although increased AKT activity was not able to restore subsequent cell division in RABL6A-depleted cells (see unchanged relative cell numbers, Figure 2A). These results demonstrate that AKT reactivation is sufficient to push cells lacking RABL6A from G1 phase into S phase, identifying a critical role for RABL6A-AKT signaling in PNET cell proliferation (Figure 2C).
Figure 2. RABL6A-AKT signaling is required for PNET cell cycle progression and response to AKT inhibitors.
(A) BON-1 cells expressing vector (Vec) or constitutively activated myristoylated AKT (Myr-AKT) were infected with control (CON) or RABL6A shRNAs (KD1, KD2). Representative Western blots of phosphorylated AKT-S473 detect endogenous AKT (bar, lower band) and Myr-AKT (arrow, upper band only in lanes 4-6). Heightened activity of Myr-AKT was assessed by levels of PRAS40-T246 phosphorylation. β-actin served as the loading control. Cell ratios (i.e., relative cell numbers normalized to CON cells) are indicated for each sample. (B) The percentage of BrdU-positive cells was quantified in BON-1 control and RABL6A knockdown cells expressing vector (V) versus Myr-AKT (M). Data were quantified from 3 independent experiments; *P < 0.02 for V versus M comparison, 2-way ANOVA. (C) Schematic of RABL6A promoting G1-to-S phase progression and PNET cell proliferation by activating AKT signaling. (D) Dose response curves, shown as relative cell number, in BON-1 control and RABL6A knockdown cells treated for 5 days with increasing concentrations of the AKT inhibitor, MK-2206. Data represent the mean ± SEM for triplicate samples from 3 separate experiments, in which results were normalized to values for untreated cells within each group. *P < 0.001 for KD1 or KD2 compared with CON, 2-way ANOVA and adjusted for multiple comparisons using the Bonferroni method. Overall differences between the curves were assessed by generalized linear regressions. (E) Percentage of cell death in BON-1 control and RABL6A knockdown cells following treatment with MK-2206 (10 μM) for 3 days. Data represent the mean ± SEM from 3 independent experiments. *P < 0.001 for (+) versus (–) comparison, 2-way ANOVA and adjusted for multiple comparisons using the Bonferroni method.
The regulation of AKT-mTOR by RABL6A predicts that levels of RABL6A may affect the response of PNET cells to drugs targeting the pathway. Indeed, control BON-1 cells expressing RABL6A were more sensitive to the AKT inhibitor MK-2206 than the RABL6A knockdown cells that displayed reduced AKT signaling (Figure 2D). The reduced efficacy of MK-2206 in RABL6A knockdown cells, where AKT is already inhibited, was associated with significantly less drug-induced cell death compared with control cells (Figure 2E). Similarly, RABL6A knockdown cells showed a diminished response to the mTORC1 inhibitor rapamycin in cell proliferation assays (Supplemental Figure 2).
RABL6A regulation of AKT-S473 phosphorylation is independent of mTORC2 and Nek2 kinases.
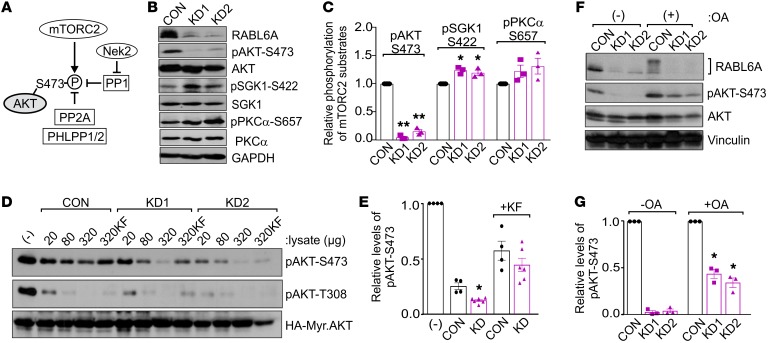
Phosphorylation of AKT-S473 is mediated by the mTORC2 kinase (34, 35) (Figure 3A). To determine if RABL6A affects mTORC2 kinase formation and activity, we examined endogenous mTORC2 complexes in control and RABL6A knockdown cells by IP Western analyses. Cells were first treated with a cell-permeable cross-linking reagent to enable detection of the endogenous complexes (36). No decrease in either the expression or interaction between the essential mTORC2 kinase subunits (mTOR, rictor, and GβL) was observed following RABL6A depletion (Supplemental Figure 3). Consistent with that finding, no decrease in the phosphorylation of other mTORC2 substrates, SGK1 and pKCα, was seen in RABL6A knockdown cells (Figure 3, B and C). In fact, phosphorylation of SGK1 was modestly increased (and PKCα trended similarly) in the same cells where AKT-S473 phosphorylation was dramatically reduced by RABL6A loss (Figure 3C), consistent with evidence that decreased AKT activity can promote mTORC2 activity through a TSC1-TSC2–dependent mechanism (37). Our data showed that mTORC2 remains active in RABL6A-depleted cells, suggesting reduced AKT-S473 phosphorylation may instead be caused by enhanced phosphatase activity.
Figure 3. RABL6A increases AKT-S473 phosphorylation through inhibition of PP2A.
(A) Schematic of the kinases and phosphatases that regulate AKT-S473 phosphorylation. P, phosphorylation; arrows, activating events; perpendicular bars, inhibiting events. (B) Western blots of BON-1 control (CON) and RABL6A knockdown (KD1 and KD2) cells showing selective loss of pAKT-S473 in RABL6A-depleted cells versus moderately increased phosphorylation of other mTORC2 substrates, SGK1 and PKCα. GAPDH was the loading control. (C) Relative phosphorylation of mTORC2 substrates in BON-1 CON, KD1, and KD2 cells was quantified by ImageJ. Data represent the mean ± SD from 3 independent experiments (*P < 0.05 and **P < 0.001 compared with CON, 2-way ANOVA and adjusted for multiple comparisons using the Bonferroni method). (D) Western blots of pAKT-S473 and pAKT-T308 following in vitro phosphatase assays using phosphorylated HA-tagged Myr-AKT as substrate to which the indicated amounts (μg) of BON-1 CON, KD1, or KD2 lysates were added. As controls, buffer (–) or the general phosphatase inhibitor potassium fluoride (KF), was added to substrate prior to the phosphatase reaction. (E) Relative levels of pAKT-S473, normalized to total HA-Myr-Akt, were quantified by ImageJ analysis of blots from 3 or more experiments in which 320 μg cell lysate was tested. *P < 0.005 KD (KD1 and KD2) versus CON, 2-way ANOVA adjusted for multiple comparisons using the Bonferroni test. (F) Western blots of pAKT-S473, AKT, and RABL6A following okadaic acid (OA) treatment (100 nM, 20 hours) in BON-1 CON and KD cells showing significant restoration of pAKT-S473 by PP2A inhibition. Vinculin was loading control. (G) Relative phosphorylation of AKT-S473 was quantified from 3 experiments. *P < 0.005 compared with untreated counterparts, 2-way ANOVA adjusted for multiple comparisons using the Bonferroni method. Western blots in B, D, and F are representative of 3 or more experiments.
Dephosphorylation of AKT-S473 has been associated with several different phosphatases including PP1, PP2A, and the Pleckstrin homology (PH) domain leucine-rich repeat protein phosphatases PHLPP1 and PHLPP2 (27, 38–45) (Figure 3A). PP1 activity toward AKT-S473 is inhibited by NIMA-related expressed kinase 2, Nek2 (46), an oncogenic kinase that we previously found is downregulated at the mRNA level by RABL6A knockdown (23). Immunoblots verified reduced Nek2 protein expression in RABL6A-depleted cells (Supplemental Figure 4A). Therefore, we expressed HA-tagged Nek2 in cells to test if it could override the effects of RABL6A loss on AKT-S473 phosphorylation. High levels of exogenous HA-Nek2 increased basal AKT-S473 phosphorylation in the cells (Supplemental Figure 4B) but failed to restore AKT-S473 phosphorylation in the face of RABL6A loss (Supplemental Figure 4C). Likewise, Nek2 overexpression was unable to stimulate reentry of RABL6A knockdown cells into the cell cycle, as indicated by the persistently low percentage of cells in S phase. Thus, RABL6A controls AKT-S473 phosphorylation through mechanisms that are independent of mTORC2 and Nek2-PP1.
RABL6A regulates AKT-S473 phosphorylation through protein phosphatase 2A (PP2A).
Compared with PP1, the PP2A and PHLPP1/2 (Figure 3A) phosphatases are more strongly linked to AKT dephosphorylation at S473 (38–42). To broadly assess the involvement of phosphatases in RABL6A-mediated AKT regulation, we performed in vitro phosphatase assays using phosphorylated HA-tagged Myr-AKT as substrate. Phospho-Myr-AKT was isolated from HEK 293 cells using HA antibody–coated agarose and incubated with increasing amounts of control or RABL6A knockdown lysates from BON-1 cells. Effects of the lysates on S473 and T308 phosphorylation were measured by Western blotting. Relative to control cell lysates that contain normal levels of RABL6A, the RABL6A-depleted lysates displayed greater phosphatase activity toward AKT-S473, particularly when 320 μg of lysate was used (Figure 3, D and E). By comparison, control and RABL6A-depleted lysates displayed equal activity toward T308 (Figure 3D), as expected given the specific effects of RABL6A toward S473 phosphorylation.
To test the involvement of PP2A, control and RABL6A knockdown cells were treated with okadaic acid. Okadaic acid is a serine/threonine protein phosphatase inhibitor that specifically blocks PP1 and PP2A activity although it is more potent against the latter, completely inhibiting PP2A at 1 nM compared with PP1 at 1 μM (47, 48). Notably, PHLPP-mediated dephosphorylation of AKT-S473 is insensitive to okadaic acid (38). In our assays, cells were treated with 100 nM okadaic acid, which yields low nanomolar intracellular concentrations of the drug that are selective toward PP2A (49). Western blotting of drug-treated cells relative to untreated controls showed that okadaic acid restored a significant amount of AKT-S473 phosphorylation in cells lacking RABL6A (Figure 3, F and G). Moreover, okadaic acid exposure to control cells caused a marked upward shift in endogenous RABL6A mobility on protein gels (Figure 3F, lane 4). These data support 2 conclusions. First, RABL6A is a likely phosphoprotein and new target (possibly a direct substrate) of PP2A. Second, and more importantly, PP2A is a key downstream target of RABL6A whose activity toward AKT-S473 is normally inhibited by RABL6A.
PP2A activation reduces AKT-S473 phosphorylation, downregulates RABL6A levels, and induces PNET cell death in a RABL6A-dependent manner.
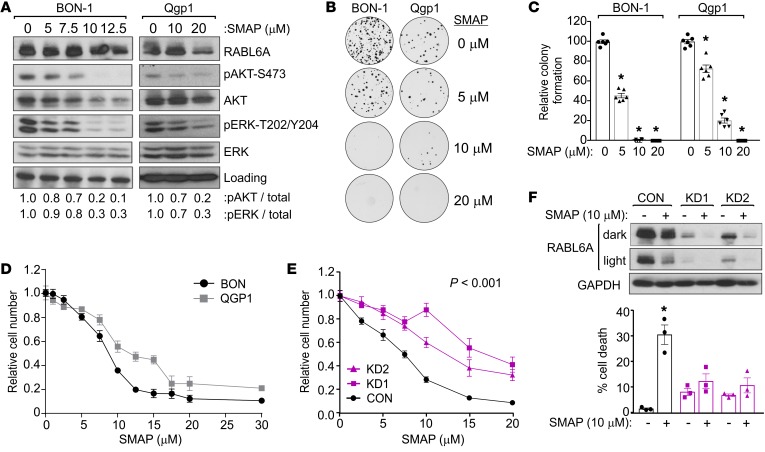
Recent papers published by Michael Ohlmeyer’s group and us describe specific SMAPs (50, 51). These tricyclic compounds were originally derived from neuroleptic drugs that were reengineered to neutralize their CNS effects, resulting in a novel class of specific PP2A-activating compounds (51, 52). Through extensive testing, SMAPs have shown remarkable efficacy and specificity in stimulating PP2A phosphatase activity toward oncogenic targets (such as phosphorylated ERK1/2) and halting the growth of lung and prostate cancer cells, both in vitro and in vivo, in a PP2A-dependent manner (53–56). We transiently treated parental BON-1 cells, as well as Qgp1 cells (the only other PNET cell line available at the time of this study), overnight with increasing doses of SMAP. SMAP effectively reduced AKT-S473 phosphorylation in both cell lines in a dose-dependent manner with an equivalent effect on ERK1/2 phosphorylation (Figure 4A). This correlated with dramatic reduction in the long-term growth of BON-1 and Qgp1 cells, with essentially no colonies detected in cells exposed to 10–20 μM SMAP (Figure 4, B and C). Quantitative proliferation assays were conducted to determine the IC50 of SMAP, showing a slightly lower IC50 of approximately 8 μM in BON-1 cells compared with an IC50 of approximately 15 μM in Qgp1 cells (Figure 4D). The IC50 for SMAP in PNET cells are similar to those defined for other tumor cell types (53, 54).
Figure 4. PP2A reactivation with SMAP reduces AKT-S473 phosphorylation, downregulates RABL6A, and induces PNET cell death in a RABL6A-dependent manner.
(A) Western blots (repeated 3 or more times) of lysates from BON-1 and Qgp1 cells treated 20 hours with SMAP showing effective reduction of pAKT-S473 and pERK-T202/Y204. Loading, vinculin (BON-1), and GAPDH (Qgp1). (B) Representative images from clonogenic assays of BON-1 and Qgp1 cells treated with 0 (vehicle), 5, 10, or 20 μM SMAP for 3 weeks. (C) Quantification of clonogenic assay results, normalized to 100% for vehicle-treated control cells, from 2 separate experiments performed in triplicate. *P < 0.001, 2-way ANOVA and adjusted for multiple comparisons using the Bonferroni method. Identical results were obtained in 4 additional repeats per cell line for cells plated at different densities. (D) Representative experiment (repeated twice) showing dose response curves for BON-1 and Qgp1 cells following exposure for 3 days to indicated concentrations of SMAP. Data represent the mean ± SD for triplicate samples and were normalized to values from untreated cells. (E) Relative cell number (assayed by Cell-Quant) of BON-1 cells expressing CON, KD1, or KD2 shRNAs after exposure for 3 days to increasing concentrations of SMAP. Data (from 3 experiments done in triplicate) were normalized to untreated controls. *P < 0.001 for KD1 and KD2 compared with CON, 2-way ANOVA and adjusted for multiple comparisons using the Bonferroni method. Overall differences between curves were assessed using generalized linear regression. (F) BON-1 CON, KD1, and KD2 cells were treated with 10 μM SMAP for 3 days. Dark and light exposures of Western blots show PP2A activation decreased RABL6A levels and altered its migration on gels. *P < 0.05 for untreated versus treated samples, Student’s t test. Error bars in E and F indicate SEM for data from 3 independent experiments.
To determine if SMAP inhibited PNET cell growth in a RABL6A-dependent manner, BON-1 control and RABL6A knockdown cells were treated with increasing doses of SMAP and relative cell numbers were quantified. As shown in Figure 4E, loss of RABL6A significantly reduced the cellular response to SMAP treatment. Thus, RABL6A expression sensitizes PNET cells to the effects of PP2A activation by SMAP, demonstrating that SMAP activity in PNET cells is at least partially dependent on RABL6A. Analyses of cell viability, as measured by trypan blue staining in the same populations treated for several days with 10 μM SMAP, revealed that RABL6A is required for SMAP-induced cell death (Figure 4F). Notably, Western blot analyses of RABL6A in those cells showed that SMAP treatment led to a marked decrease in RABL6A expression, not only in control cells but also in RABL6A knockdown cells (Figure 4F, top). While the effect of SMAP on RABL6A expression was less pronounced in experiments involving shorter drug exposure times (as in Figure 4A), the reduction in RABL6A levels caused by SMAP was also associated with loss of slower migrating RABL6A forms on protein gels.
SMAP effectively suppresses PNET progression in vivo.
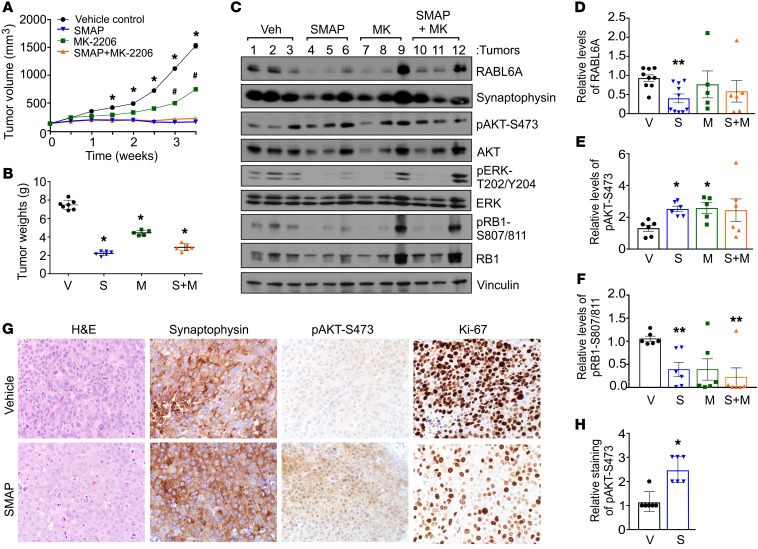
Patient-derived xenograft (PDX) models for nonfunctional PNETs are lacking, therefore BON-1 xenograft tumor studies were performed to determine the efficacy of SMAP treatment on PNET tumor progression in vivo. Tumors were grown to approximately 200 mm3 in size, at which time mice were treated by oral gavage with vehicle control, SMAP, the AKT inhibitor MK-2206, or the combination of SMAP plus MK-2206. SMAP treatment caused complete cessation of tumor growth that was superior to single-agent MK-2206 treatment (Figure 5, A and B). There was no benefit of combination therapy with SMAP plus MK-2206 (Figure 5, A and B), in agreement with in vitro drug studies that showed no synergistic effect of their combination (negative data not shown). Treatment with all drugs was well tolerated as indicated by the absence of behavioral abnormalities or changes in body weight during the treatment period (Supplemental Figure 5).
Figure 5. Therapeutic reactivation of PP2A suppresses PNET growth in vivo.
(A) BON-1 cells (5 × 106) were injected subcutaneously into NOD.SCID mice. Once tumors reached an average volume of 200 mm3, drug treatments were initiated. Tumor volumes were measured over a 4-week period in which mice were treated by oral gavage with vehicle control, SMAP (5 mg/kg, twice a day), MK-2206 (30 mg/kg, 3 times a week), and a combination of SMAP plus MK-2206. SEM for at least n = 5 mice per group; *P < 0.03 for vehicle versus SMAP or SMAP+MK-2206; #P < 0.05 for vehicle versus MK-2206; 2-way ANOVA and adjusted for multiple comparisons using Bonferroni method. (B) Comparison of tumor weights from vehicle (V), SMAP (S), MK-2206 (M), and SMAP plus MK-2206 (S+M) groups after the final treatment. Error bars, SEM; *P < 0.001, 2-way ANOVA and adjusted for multiple comparisons using the Bonferroni method. (C) Representative Western blot analyses of the indicated proteins in lysates of xenografted BON-1 tumors harvested from the treated mice. (D–F) Quantification of relative levels of RABL6A, pAKT-S473, and pRB1-S807/811 in xenograft tumors, respectively, obtained by ImageJ analysis of Western blots (as shown in C). Mean ± SEM; *P < 0.05; **P < 0.01; 2-way ANOVA and adjusted for multiple comparisons using the Bonferroni method. (G) Representative H&E and IHC staining for the indicated proteins in BON-1 xenograft tumors from vehicle control and SMAP-treated mice. Images were taken at ×400. (H) Quantification of pAKT-S473 staining (assessed as weak = 1, moderate = 2, strong = 3) in tumors from vehicle-treated (V) and SMAP-treated (S) mice. Mean ± SEM; *P < 0.001, Student’s t test.
The molecular and biological effects of SMAP treatment were evaluated by Western blotting (Figure 5, C–F) and immunohistochemical (IHC) staining (Figure 5, G and H) of the tumors. High synaptophysin expression in the tumors by both approaches verified their NET identity (Figure 5, C and G). Consistent with the downregulation of endogenous RABL6A expression by SMAP in cultured PNET cells, reduced levels of RABL6A were detected by immunoblotting in the majority of SMAP-treated tumors (Figure 5, C and D). Unexpectedly, AKT-S473 phosphorylation in most tumors was increased (not decreased) by treatment with SMAP as well as MK-2206 (Figure 5, C, E, G, and H). This unwanted effect of SMAP likely reflects loss of negative feedback loops controlling AKT due to sustained exposure to each drug, as seen with everolimus (10, 19, 20).
To determine how SMAP suppressed tumor growth in vivo, expression of apoptotic and proliferation markers was evaluated. No change in cleaved caspase-3 levels was caused by drug treatment (not shown), suggesting SMAP’s antitumor effects were unrelated to apoptosis. Interestingly, while there was only a slight (but significant) decrease in the percentage of Ki-67+ cells in SMAP-treated tumors (Figure 5G; 99.6% versus 93.2%, P < 0.05, Student’s t test), exposure to SMAP caused a dramatic reduction in Ki-67 levels per cell (Figure 5G). It is known that G1 phase cells express the lowest level of Ki-67 and that residual Ki-67 staining persists in arrested, nonproliferating cells (57, 58). In agreement with that observation, SMAP treatment alone or in combination with MK-2206 significantly decreased RB1-S807/811 phosphorylation (Figure 5, C and F). Reduced RB1 phosphorylation at those cyclin-dependent kinase 4/6–specific (CDK4/6-specific) sites correlates with its activation and G1 phase cell-cycle arrest. Thus, our data suggest that SMAP-mediated PP2A activation effectively halts PNET growth in vivo by inhibiting tumor cell proliferation, likely via a G1 phase blockade that may depend on RABL6A downregulation and RB1 activation.
Discussion
This work identifies a RABL6A-PP2A-AKT pathway that drives PNET tumorigenesis, providing new insights into critical upstream regulators that promote aberrant AKT-mTOR signaling in PNETs (Figure 6) and possibly other cancers. We found that RABL6A is a novel inhibitor of PP2A that is required for AKT signaling and G1-to-S phase progression in PNET cells. RABL6A maintains AKT phosphorylation at S473 by impeding PP2A phosphatase activity. Importantly, an orally bioavailable SMAP effectively suppressed PNET cell growth in vitro and in vivo. Pharmacological restoration of PP2A tumor suppressive phosphatase activity is an emerging strategy for cancer therapy (29, 30, 50), one that would deliver a new treatment option to clinicians and patients for managing advanced PNETs.
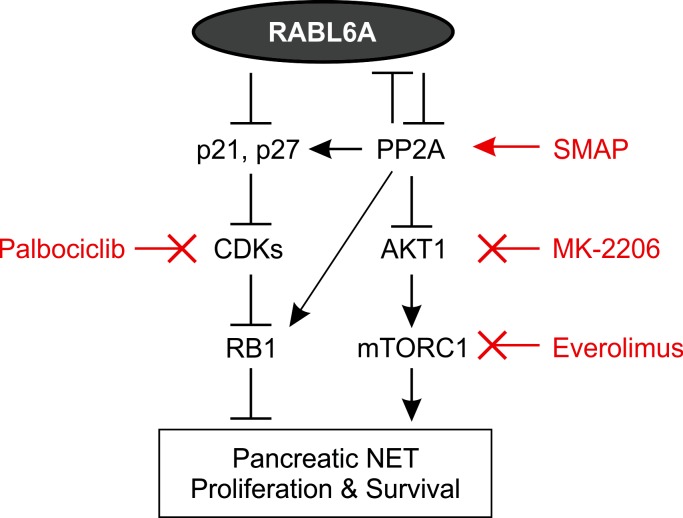
Figure 6. RABL6A oncogenic signaling in PNETs.
Schematic showing that RABL6A inhibits the PP2A tumor suppressor, thereby activating AKT-mTOR signaling to drive PNET proliferation and survival. In turn, PP2A can inhibit expression of RABL6A, possibly via dephosphorylation and destabilization of the protein. Earlier work showed that RABL6A can promote PNET pathogenesis by inhibiting RB1, which was associated with downregulation of CDK inhibitors (p21 and p27) and increased RB1 phosphorylation (23). PP2A-mediated dephosphorylation of p27 and RB1 increases their activities, representing additional mechanisms by which PP2A may antagonize RABL6A. Although not shown, AKT also inhibits RB1 signaling through several mechanisms. The pathways depicted here, as well as drug response assays performed in this study, predict that high RABL6A expression in patient PNETs will sensitize tumors to clinically relevant drugs that inhibit CDKs (Palbociclib), activate PP2A (SMAP), inhibit AKT (MK-2206), and inhibit mTORC1 (Everolimus). Arrows, activating events. Perpendicular bars, inhibitory events.
The rationale for reactivating PP2A in anticancer therapy is compelling. First and foremost, PP2A is a powerful tumor suppressor that accounts for over 50% of serine/threonine phosphatase activity in mammalian cells (31, 50). As such, PP2A controls a wide diversity of substrates and exerts its antitumorigenic effects by inhibiting numerous oncogenic proteins (such as AKT, Ras, Erk, Myc, JAK2, and STAT5) and activating various tumor suppressor proteins (such as RB1, PTPN6, and GSK-3β) (29, 31, 40). PP2A is inactivated in tumors through various mechanisms including somatic mutation of PP2A subunits, posttranslational modifications of the catalytic C subunit, and increased expression of endogenous inhibitors such as CIP2A and SET (29, 30, 59). Moreover, the transforming activity of certain viral oncoproteins, such as the small T antigen of SV40, depends on their ability to bind PP2A and disrupt its function (59–63). Thus, reactivation of PP2A in tumors using small molecules like SMAPs should elicit broad-acting anticancer effects with one therapeutic agent, increasing the likelihood that PP2A activation in combination with traditional chemotherapeutics and/or other pathway targeted drugs may eliminate cancers before resistance can develop.
The mechanism(s) by which RABL6A controls PP2A activity is currently not known. The complexity of PP2A itself is immense, with over 80 possible holoenzymes that can be formed from 2 structural (A) isoforms α and β, 2 catalytic (C) isoforms α and β, and more than 20 regulatory (B) subunits (31). In PNET cells, altered RABL6A levels had no effect on expression of the A or C subunits of PP2A (Supplemental Figure 6, A–C). RABL6A may instead control certain PP2A regulatory B subunits, particularly those (e.g., B56γ and B56β) that regulate AKT-S473 phosphorylation (40–42) and are downregulated in various cancers (29, 30). Among the PP2A inhibitors, increased expression of CIP2A (cancerous inhibitor of PP2A) has been observed in a wide variety of human cancers (reviewed in 29, 30, 50). We found that RABL6A loss caused significant downregulation of CIP2A expression (Supplemental Figure 6, A and D), consistent with elevated PP2A activity in those cells. It is also conceivable that RABL6A regulates PP2A through direct association. A large number of other RAB GTPases (namely RABs 1A, 2A, 8A, 9A, 14, 20, and 31) were recently found by mass spectrometry to interact with PP2A, with analyses of RAB9 showing that it disrupts PP2A holoenzymes via competitive binding to the A subunit (64). RABL6A could impair PP2A assembly and function through a similar mechanism.
Our studies uncovered a negative feedback loop between RABL6A and PP2A (Figure 6). RABL6A expression inhibited PP2A activity toward AKT, whereas activation of endogenous PP2A with SMAP downregulated RABL6A expression in both cultured PNET cells and tumors. The observation that SMAP activity was diminished in cells lacking RABL6A shows that its anticancer effects depend, in part, on inhibition of RABL6A. It remains to be determined if PP2A directly regulates RABL6A. Selective manipulation of PP2A activity with either okadaic acid (inhibitor) or SMAP (activator) had reciprocal effects on the migration of RABL6A on denaturing protein gels. We suspect those effects on RABL6A migration reflect altered phosphorylation and that dephosphorylated RABL6A is unstable. According to phosphorylation site databases, RABL6A is phosphorylated at numerous sites (Supplemental Table 1). Ongoing studies will test the possibility that PP2A antagonizes RABL6A activity by dephosphorylating and destabilizing the protein, thereby reducing RABL6A expression.
It is likely that RABL6A influences the activity of other cellular phosphatases since AKT-S473 phosphorylation levels were only partly restored by okadaic acid–mediated inhibition of PP2A. Two isoforms of the PHLPP phosphatase exist and inactivate different AKT isoforms through direct binding and dephosphorylation of S473 (39). While PHLPP1 acts on AKT2 and AKT3, PHLPP2 regulates S473 phosphorylation and activity of AKT1 and AKT3 (39). It will be important to determine which isoform(s) of AKT is affected by RABL6A and if PHLPP isoforms contribute to that regulation, particularly since PHLPP1 and PHLPP2 are tumor suppressors whose genetic deletion in certain cancers, such as prostate cancer, rivals that of another tumor suppressive phosphatase, PTEN (65).
This study suggests that drugs targeting RABL6A or PP2A could provide beneficial new therapies for PNET patients. There are currently no known inhibitors of RABL6A. SMAPs are attractive therapeutics since preclinical studies show they are well-tolerated oral drugs with significant in vivo activity toward PNETs (this study) as well as Ras-driven lung cancer and prostate adenocarcinoma (53, 54). Other PP2A activating drugs function as indirect activators of PP2A by targeting PP2A’s endogenous inhibitors, and typically have non–PP2A-related activity. One example, FTY720 (fingolimod), an FDA-approved drug for multiple sclerosis, increases PP2A activity through SET inhibition but also has immunosuppressive activities that may be undesirable for cancer patients (29, 30). As such, nonimmunosuppressive derivatives of FTY720 ([S]-FTY720-OMe, [S]-FTY720-regioisomer, and OSU-2S) may hold more promise as anticancer agents. Indeed, OSU-2S displayed potent antitumor activity in preclinical in vitro and in vivo studies against various hematologic malignancies and hepatocellular carcinoma (66–68).
As we look to improve and expand treatment options for patients with advanced PNETs, one significant clinical consideration is our finding that sustained reactivation of PP2A in tumors caused an unwanted increase in AKT activation. A similar effect is observed with prolonged everolimus therapy due to loss of negative feedback control and induction of tyrosine kinase receptor signaling (10, 19, 20). It is therefore encouraging that the PNETs still responded well to SMAP treatment (complete stoppage of tumor growth), implying that other cancer pathways regulated by PP2A override the oncogenic effects of AKT. One of those other pathways may be ERK signaling; it is promoted by RABL6A (26, 69), was inhibited in SMAP-treated PNETs (Figure 5C), and contributes to SMAP’s anticancer effects in Ras-driven tumors (53). As shown in Figure 6, PP2A also promotes RB1 tumor suppressive activity through dephosphorylation of RB1 (70) and dephosphorylation/stabilization of the CDK inhibitor, p27 (71, 72). We previously showed RB1 is inhibited by RABL6A in PNETs (23), and here found that PP2A reactivation in tumors caused marked RABL6A loss and RB1 activation. Additional studies will be needed to define the relative importance of RABL6A downregulation and/or RB1 activation in mediating the antitumor effects of SMAP, and to determine the degree to which RB1 hypo-phosphorylation depends on PP2A-induced RABL6A loss versus PP2A activation.
In summary, PP2A reactivation and RABL6A inhibition may be promising strategies for PNET treatment. The fact that RABL6A expression dictated PNET cell response to drugs targeting AKT, mTOR, and PP2A also suggests that tumor levels of RABL6A may be a predictive biomarker for therapies against each of those pathways. More research in patient NETs, a newly developed PNET cell line (73), and other types of NETs, such as bronchial NETs for which cell lines are available, will establish how generalizable aberrant RABL6A-PP2A-AKT signaling is to NET pathogenesis, and if pathway status predicts tumor responsiveness to targeted therapies.
Methods
Cell culture and reagents.
Two human PNET cell lines have been established and are available, and both were used for these studies. BON-1 cells (originally developed from a lymph node metastasis of a PNET and authenticated by Courtney Townsend, University of Texas Medical Branch, Galveston, TX) (74) were maintained in Dulbecco’s modified Eagle’s medium (DMEM)/F12 containing 10% fetal bovine serum (FBS), 4 mM glutamine, and 100 μg/ml penicillin-streptomycin. Qgp1 cells were purchased from the Japanese Collection of Research Bioresources (JCRB0183) and maintained in RPMI 1640 medium containing 10% FBS, 4 mM glutamine, and 100 μg/ml penicillin-streptomycin. RABL6A gene and protein expression in BON-1 and Qgp1 cells relative to nontransformed controls is not known since NET control cell lines are lacking. However, karyotype and quantitative PCR analyses suggest there are additional copies of the RABL6 gene in both PNET cell lines (Supplemental Figure 7). Moreover, both cell lines express elevated levels of RABL6A protein similar to primary PNET and small bowel NET cultures derived from patient tumors (Supplemental Figure 8). Human embryonic kidney (HEK) 293T cells were grown in DMEM with the same supplements listed above. Cells were routinely tested for mycoplasma contamination and found to be negative.
Cells were treated with the indicated concentrations of okadaic acid (PP2A inhibitor, Cell Signaling [catalog 5934], dissolved in DMSO and stored at –20°C), MK-2206 (AKT inhibitor, Selleck Chemicals, reconstituted in DMSO and stored at –20°C), or SMAP compound (PP2A activator, dissolved in DMSO and stored at room temperature, provided by Goutham Narla, The University of Michigan, Ann Arbor, MI).
RNA interference, virus production, and infection.
Human RABL6A shRNAs in the pLKO.1 lentiviral vector (Open Biosystems) have been described (23, 26). Lentiviruses encoding RABL6A shRNAs were produced in HEK 293T cells and infected into target cells (BON-1, Qgp-1) as described (23, 26). Cells were harvested 3 to 5 days after infection depending on the assay. For combined overexpression of HA-tagged myristoylated AKT (HA-Myr.AKT) and silencing of RABL6A, cells were sequentially infected with pMSCV-HA-Myr.AKT-IRES-GFP viruses followed by RABL6A shRNA or CON viruses. Lentiviruses coexpressing HA-tagged Nek2 kinase and GFP were similarly produced in HEK 293T cells and infected into BON-1 cells, and GFP-positive cells were selected by sterile cell sorting (FACS Aria II, Becton Dickinson). Cells infected with RABL6A shRNA viruses were selected with 1 μg/ml puromycin for 2 days and maintained in 0.5 μg/ml puromycin for the remainder of the assay.
Cell proliferation and survival assays.
Cell number and viability were directly measured by trypan blue staining (1:1 vol/vol) and manual counting using a hemocytometer following infection with RABL6A shRNA viruses and/or treatment with drugs. Where indicated, relative cell numbers were quantified using alamarBlue (Thermo Fisher Scientific) or Cell-Quant (Genecopoeia) assays in a 96-well format according to manufacturer’s instructions. To measure DNA synthesis, infected cells were plated onto coverslips and the next day incubated with 10 μM bromodeoxyuridine (BrdU) for 5 hours. BrdU incorporation was detected by immunofluorescent staining with anti–BrdU antibodies and 4′,6-diamidino-2-phenylindole (DAPI) to detect nuclei (75). Confocal microscopy (Zeiss LSM 710) was performed to capture fluorescence images and quantify BrdU positivity (100 or more cells per sample counted) from 3 or more separate experiments.
RNA expression analyses.
Microarray analysis of total RNA from 3 separately generated sets of BON-1 cells expressing RABL6A shRNAs or control vector has been described (23). Differentially expressed genes between the RABL6A knockdowns and vector were analyzed by IPA software (Ingenuity Systems). Significant gene set enrichment was defined as reaching a P value less than 0.01 after a Benjamini-Hochberg multiple testing correction. The “Upstream Analysis” component of the IPA Core Analysis was used to specifically assess for enrichment of genes involved in AKT pathways.
RNA-Seq analysis of human PNET specimens was performed on primary tumors collected from 20 patients during operative procedures at the University of Iowa Hospitals and Clinics. RNA was isolated from tissues stored in RNAlater (Thermo Fisher Scientific) using the RNeasy Plus kit (Qiagen), subjected to DNA digestion, and resuspended in water prior to RNA quality assessment using the Agilent 2100 Bioanalyzer (Agilent Technologies). Samples with an RNA integrity number (RIN) greater than 6 were sequenced using the Illumina TruSeq protocol (Illumina) at the University of Iowa Institute of Human Genetics Genomics Division, exactly as described (76). RNA-Seq reads were mapped with the STAR aligner (77). StringTie was used for assignment of reads to transcripts, quantification of expression, and normalization (78). BallGown was used to determine differential gene expression (79). RNA-Seq data will be deposited in the NCBI dbGaP database (accession no. phs001772.v1.p1, web address https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs001772.v1.p1). Statistically significant gene expression changes were indicated by an adjusted P value less than 0.05.
Antibodies.
For Western blotting, antibodies specific for AKT (no. 4691), p-AKT-S473 (no. 4060), p-AKT-T308 (no. 9275), ERK (no. 4695), p-ERK-T202/Y204 (no. 9106), PRAS40 (no. 2997), p-PRAS40-T246 (no. 2610), p70 S6 kinase (no. 2708), p-p70 S6 kinase-T389 (no. 9206), PKCα (no. 2056), cleaved caspase-3 (no. 9661), mTOR (no. 2983), rictor (no. 9476), GβL (no. 3274), p-RB1-S807/811 (no. 9308S), RB1 (no. 9309S), PP2A A subunit (no. 2041), and p-FoxO1-T24/p-FoxO3a-T32 (no. 9464) were obtained from Cell Signaling Technology. An SGK1 antibody was obtained from Millipore (no. 07-315). GAPDH (no. Ab8245), Synaptophysin (no. Ab32127) and PP2A C subunit (no. Ab106262) antibodies were purchased from Abcam. Vinculin (no. V9131) and HRP-coupled secondary antibodies (nos. NA931, NA934, and NA935) were obtained from MilliporeSigma. The 3F10 rat monoclonal antibody against HA (no. 11867423001) was obtained from Roche. Antibodies against p-SGK1-S422 (no. SC-16745), p-PKCα-S657 (no. SC-12356), Nek2 (no. SC-55601), and CIP2A (no. SC-80659) were purchased from Santa Cruz. A rabbit polyclonal antibody against RABL6A was produced in-house (21). For immunohistochemistry, antibodies against p-AKT-S473 (no. 4060, Cell Signaling Technology), synaptophysin (DAK-SYNAP clone, Dako-Agilent), Ki-67 (no. 16667, Abcam), and RABL6A (monoclonal generated in-house, ref. 26) were used.
Western blotting.
Cellular proteins were isolated from cells by lysis in SDS-PAGE loading buffer and identical cell equivalents electrophoresed through polyacrylamide gels. Whole-cell extracts and tumors were lysed for 30 minutes on ice with RIPA buffer (50 mM Tris, pH 8.0, 150 mM NaCl, 1% Triton X-100, 0.1% SDS, 0.5% sodium deoxycholate) supplemented with 1 mM NaF, protease, and phosphatase inhibitor cocktails (Sigma, P-8340 and P-0044) and 30 μM phenylmethylsulfonyl fluoride (PMSF). Protein concentrations of cell extracts were determined by Precision Red protein assay reagent (Cytoskeleton, ADV02) and equal quantities of protein were loaded for separation of proteins. Proteins were transferred onto PVDF membranes (Millipore), which were blocked with 5% nonfat milk or 3% BSA in TBST (Tris-buffered saline containing Tween-20) depending on the antibody used for protein detection. Western blotting conditions for primary antibodies can be found in Supplemental Table 2. After incubation with primary antibodies, membranes were washed in TBST and incubated for 40 minutes at room temperature with HRP-conjugated secondary antibodies. Proteins were visualized with enhanced chemiluminescence (ECL, Amersham) as described (80). Densitometry quantification was performed using ImageJ (NIH) software.
Analysis of mTORC2 kinase complexes.
To detect endogenous cellular mTORC2 complexes, BON-1 cells expressing control or RABL6A shRNAs were washed in phosphate buffered saline (PBS) and incubated with 1.25 mM DSP (dithiobis[succinimidylpropionate], Thermo Fisher Scientific, no. 22586, suspended in DMSO at 25 mM and diluted in PBS), a cell-permeable cross-linking reagent, for 30 minutes at room temperature (RT). The cross-linking reaction was halted by incubation with 20 mM Tris, pH 7.6, for 15 minutes at RT. Cells were rinsed in PBS and lysed directly in ice-cold NP-40 buffer (50 mM Tris, pH 8.0, 120 mM NaCl, 1 mM EDTA, 0.5% Nonidet P-40, and 0.1 mM Na3VO4) supplemented with 1 mM NaF, protease inhibitor cocktail (Sigma-Aldrich, P8340) and phosphatase inhibitor cocktail (Sigma-Aldrich, P0044). Samples were sonicated for 5 seconds and lysates clarified by centrifugation at 21,000g for 10 minutes at 4°C. mTORC2 complexes were immunoprecipitated by overnight incubation at 4°C using Rictor antibodies (Cell Signaling, no. 9476) and protein A/G-Sepharose versus an isotype-matched IgG control. Immune complexes were washed 4 times in NP-40 buffer, separated by SDS-PAGE, transferred to PVDF membranes, and analyzed by immunoblotting (see Supplemental Table 2 for antibodies).
In vitro phosphatase assay.
An in vitro phosphatase assay was performed using phosphorylated HA-Myr-AKT as the substrate. HEK 293T cells expressing HA-tagged Myr-AKT were lysed in RIPA buffer lacking phosphatase inhibitors and the protein purified by binding to anti-HA affinity matrix (Sigma-Aldrich) at 4°C overnight. The resin was washed 3 times in ice-cold RIPA buffer, 2 times in ice-cold, hypotonic phosphatase reaction buffer (50 mM Tris, pH 7.5, 10 mM MgCl2, 20 mM dithiothreitol, 5 mM EDTA), and finally resuspended in phosphatase reaction buffer containing 30% glycerol and stored at 4°C. BON-1 cells expressing CON or RABL6A shRNA constructs were harvested, washed once in PBS, suspended in phosphatase reaction buffer containing protease inhibitors (protease inhibitor cocktail [MilliporeSigma, P8340] and 30 μM phenylmethylsulfonyl fluoride) for 10 minutes on ice, and briefly sonicated (one 5-second pulse). Lysates were clarified by centrifugation at 21,000g for 5 minutes at 4°C, and protein concentrations determined by Precision Red (Cytoskeleton, ADV02). Varying amounts of each cell lysate were incubated with 5 μl (1:1 slurry) of purified HA-Myr-AKT substrate for 20 minutes at 30°C, with all reaction volumes made equivalent by addition of buffer. As negative controls, an equivalent volume of buffer (no lysate) was added to the substrate or 50 mM KF (phosphatase inhibitor) was added to the cell lysate prior to performing the phosphatase reaction. Reactions were stopped by the addition of SDS-PAGE buffer and boiling for 10 minutes. Phosphatase activity within each lysate was measured by Western blotting for levels of phosphorylated AKT-S473 and AKT-T308 relative to total levels of HA-Myr-AKT (using HA antibodies) in each reaction.
Drug response assays.
Chemotherapeutic drugs MK-2206 (AKT inhibitor) and rapamycin (mTORC1 inhibitor) were purchased from Selleck Chemicals and stocks stored at –20°C once in solution. Rapamycin was dissolved in sterile ddH2O whereas MK-2206 and SMAP were dissolved in dimethyl sulfoxide (DMSO). SMAP solution was stored at RT and used for up to 1 month. BON-1 and Qgp1 cells were seeded at a density of 2000 cells per well in 96-well flat-bottomed culture dishes. After overnight incubation, an equal volume of 2 times the concentrated drug was added to each well and cells were exposed to the drugs for 3 to 5 days. Each condition was performed in triplicate. Samples were assayed for relative cell number using alamarBlue (Thermo Fisher Scientific, DAL1025) or Cell-Quant (Genecopoeia, A014) cell proliferation assays. Results were quantified using a fluorescence microplate reader by measuring absorbance at wavelengths of 540/570 nm for alamarBlue and 480/530 nm for Cell-Quant.
To measure effects of MK-2206, SMAP, and okadaic acid on RABL6A levels and AKT-S473 phosphorylation, cells plated in 6-well dishes were treated with vehicle control or the indicated drug concentrations for 16–20 hours at 37°C, 5% CO2. Cells were harvested, lysed in RIPA buffer, and subjected to Western blotting. In some assays, cells were exposed for longer periods of time (up to 3 days) to the drugs, and cells were examined by trypan blue staining to assess cell viability and by Western blotting to evaluate protein status.
Clonogenic assay.
Cells were plated at low density (250, 500, and 1000 cells per well) in 6-well plates. After 24 hours, cells were treated in triplicate with vehicle control or increasing concentrations of SMAP and incubated for 3 weeks. Drug-containing medium was refreshed every 72 hours. Cells were then fixed and stained with 1% crystal violet solution (Sigma-Aldrich). Quantification was performed using a GelCount colony counter (Oxford Optronix Ltd).
Mouse tumor xenografts and treatment studies.
Mice were housed in the University of Iowa Animal Care barrier facility in rooms with a 12-hour light-dark cycle and free access to water and food. For tumor studies, the right flanks of 8-week-old immunodeficient female mice (homozygous NOD.CB17-Prkdc SCID/J strain, model 001303, Jackson Laboratory) were injected subcutaneously with BON-1 cells (5 × 106) suspended in 0.2 ml of 1:1 PBS/Matrigel (BD Biosciences). When tumor volumes reached an average of 200 mm3, mice were randomized into the different treatment groups and tumor volume was assessed by caliper measurement 2 times a week throughout the study. Tumor volume was determined using the formula: [width (2) × length]/2. Mice were treated by oral gavage with vehicle control, MK-2206 (30 mg/kg, 3 times per week), SMAP (5 mg/kg, twice daily), and SMAP plus MK-2206. Mouse body weights were recorded 2 times per week. SMAP was prepared in a N,N-Dimethylacetamide (DMA)/Kolliphor HS-15 (Solutol)/diH20 solution. Animals were observed for signs of toxicity (weight loss, ruffled fur, immobility, and abdominal rigidity). Tumor tissues, harvested 2 hours after the final dose of treatment, were weighed and split for processing by fixation in 10% neutral buffered formalin (for IHC analyses) or flash frozen in LN2 (for Western blotting).
Histopathological analysis.
Formalin (10% neutral buffered formalin) fixed tumors were routinely processed, paraffin embedded, and sectioned (~4 μm) onto glass slides. Slides were stained with H&E (81) and reviewed by a pathologist. For immunostaining of phosphorylated AKT-S473 (Cell Signaling Technology, antibody 4060) and Ki-67 (Abcam, antibody 16667), we used Dako Rabbit Envision (Dako-Agilent) and chromogen was DAB with Harris hematoxylin counterstain. The synaptophysin immunostain was run on a Dako Autostainer Link 48 after HpH antigen retrieval using a mouse monoclonal antibody (DAK-SYNAP clone from Dako-Agilent; 1:100; 15-minute incubation) followed by detection using the polymer-based EnVision FLEX+ detection system. Synaptophysin and phosphorylated AKT-S473 were scored as weak = 1, moderate = 2, or strong = 3. Ki-67 was quantified by the number of positively stained cells (regardless of staining intensity) in approximately 500 tumor cells per tumor sample.
Statistics.
Western blot data were imaged by scanning densitometry and quantified by ImageJ software (NIH). Values for phosphoproteins were normalized to expression of the total protein while values for all other proteins were normalized to the loading control. Differences in levels of protein phosphorylation or expression were expressed as fold change relative to untreated counterparts or vector controls. Quantified data were presented as the mean ± SD or SEM, as indicated. All P values, unless otherwise specified, were obtained by 2-way ANOVA and adjusted for multiple comparisons using the Bonferroni method. Overall differences between curves were assessed using generalized linear regressions. An adjusted P value less than 0.05 was considered statistically significant.
Study approval.
All patients were consented for genetic studies and entered into a neuroendocrine tumor registry approved by the University of Iowa Hospitals and Clinics institutional review board (IRB 199911057). All mouse studies were conducted according to protocols approved by the University of Iowa Institutional Animal Care and Use Committee (protocol 5121590, molecular mechanisms and biomarkers of neuroendocrine tumors). All efforts were made to minimize animal suffering.
Author contributions
SU, GN, BWD, FWQ, and DEQ conceived and designed the studies. SU, CAK, JLK, BL, JAR, AMS, RMS, JH, NT, MRL, TAB, BJB, ATS, TG, AFT, GKZ, AMB, BWD, and DEQ conducted experiments and analyzed the data. FZ, TMO, JD, JRH, DKM, ALP, RAM, AMB, SS, GN, BWD, FWQ, and DEQ provided observations and scientific interpretations. All authors reviewed, discussed, and approved the manuscript.
Supplementary Material
Acknowledgments
We thank David Gordon for advice on combination drug therapies and we are grateful for assistance from personnel in the core facilities within the University of Iowa College of Medicine and Holden Comprehensive Cancer Center (Flow Cytometry, Central Microscopy and Genomics Core). We especially thank the Iowa Neuroendocrine Tumor (NET) Specialized Program of Research Excellence (SPORE) group for critical feedback as this work was being conducted and Suzanne Kieffer for her tireless and exceptional management of the Iowa NET Registry Database (which is housed within the Clinical and Translational Science Award Research Electronic Data Capture). This work was supported by grants from the NIH including NCI NET SPORE grant P50 CA174521 (Project 2, DEQ and BWD; Project 3, JRH, AMB, and TAB; Developmental Research Project, DEQ and JRH), grant 2T32 GM0677954 (to JAK), 5T32 CA078586 (to RMS), and an NCI Core Grant P30 CA086862 (University of Iowa Holden Comprehensive Cancer Center).
Version 1. 02/05/2019
In-Press Preview
Version 2. 03/04/2019
Electronic publication
Version 3. 04/01/2019
Print issue publication
Footnotes
Conflict of interest: The Icahn School of Medicine has filed patents covering composition of matter on the small molecules disclosed herein for the treatment of human cancer and other diseases and for methods of use for using these small-molecule PP2A activators (patent no. 9,937,186 B2). RAPPTA Therapeutics LLC has optioned this intellectual property for the clinical and commercial development of this series of small-molecule PP2A activators. GN has an ownership interest in RAPPTA Therapeutics LLC.
Copyright: © 2019 American Society for Clinical Investigation
Reference information: J Clin Invest. 2019;129(4):1641–1653.https://doi.org/10.1172/JCI123049.
Contributor Information
Shaikamjad Umesalma, Email: Umesalma-shaikamjad@uiowa.edu.
Courtney A. Kaemmer, Email: courtney-waite@uiowa.edu.
Jordan L. Kohlmeyer, Email: jordan-kohlmeyer@uiowa.edu.
Angela M. Schab, Email: angela-schab@uiowa.edu.
Jacqueline A. Reilly, Email: jacqueline.erin.reilly@gmail.com.
Ryan M. Sheehy, Email: RSheehy@kcumb.edu.
Jussara Hagen, Email: jussara-hagen@uiowa.edu.
Nitija Tiwari, Email: nitija-tiwari@uiowa.edu.
Fenghuang Zhan, Email: fenghuang-zhan@uiowa.edu.
Mariah R. Leidinger, Email: mariah-leidinger@uiowa.edu.
Ronald A. Merrill, Email: ronald-merrill@uiowa.edu.
David K. Meyerholz, Email: david-meyerholz@uiowa.edu.
Abbey L. Perl, Email: alp80@case.edu.
Bart J. Brown, Email: bartley-brown@uiowa.edu.
Terry A. Braun, Email: terry-braun@uiowa.edu.
Aaron T. Scott, Email: aaron-scott@uiowa.edu.
Timothy Ginader, Email: timothy-ginader@uiowa.edu.
Agshin F. Taghiyev, Email: agshin-taghiyev@uiowa.edu.
Gideon K. Zamba, Email: gideon-zamba@uiowa.edu.
James R. Howe, Email: james-howe@uiowa.edu.
Stefan Strack, Email: stefan-strack@uiowa.edu.
Andrew M. Bellizzi, Email: andrew-bellizzi@uiowa.edu.
Goutham Narla, Email: gnarla@med.umich.edu.
Benjamin W. Darbro, Email: benjamin-darbro@uiowa.edu.
Frederick W. Quelle, Email: Frederick-Quelle@uiowa.edu.
Dawn E. Quelle, Email: dawn-quelle@uiowa.edu.
References
- 1.Vinik AI, et al. NANETS consensus guidelines for the diagnosis of neuroendocrine tumor. Pancreas. 2010;39(6):713–734. doi: 10.1097/MPA.0b013e3181ebaffd. [DOI] [PubMed] [Google Scholar]
- 2.Yao JC, et al. One hundred years after “carcinoid”: epidemiology of and prognostic factors for neuroendocrine tumors in 35,825 cases in the United States. J Clin Oncol. 2008;26(18):3063–3072. doi: 10.1200/JCO.2007.15.4377. [DOI] [PubMed] [Google Scholar]
- 3.Frilling A, Sotiropoulos GC, Li J, Kornasiewicz O, Plöckinger U. Multimodal management of neuroendocrine liver metastases. HPB (Oxford) 2010;12(6):361–379. doi: 10.1111/j.1477-2574.2010.00175.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ehehalt F, Saeger HD, Schmidt CM, Grützmann R. Neuroendocrine tumors of the pancreas. Oncologist. 2009;14(5):456–467. doi: 10.1634/theoncologist.2008-0259. [DOI] [PubMed] [Google Scholar]
- 5.Strosberg JR, et al. The North American Neuroendocrine Tumor Society Consensus Guidelines for Surveillance and Medical Management of Midgut Neuroendocrine Tumors. Pancreas. 2017;46(6):707–714. doi: 10.1097/MPA.0000000000000850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Howe JR, et al. The surgical management of small bowel neuroendocrine tumors: consensus guidelines of the North American Neuroendocrine Tumor Society. Pancreas. 2017;46(6):715–731. doi: 10.1097/MPA.0000000000000846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bilimoria KY, et al. Clinicopathologic features and treatment trends of pancreatic neuroendocrine tumors: analysis of 9,821 patients. J Gastrointest Surg. 2007;11(11):1460–7; discussion 1467. doi: 10.1007/s11605-007-0263-3. [DOI] [PubMed] [Google Scholar]
- 8.Sherman SK, Howe JR. Translational research in endocrine surgery. Surg Oncol Clin N Am. 2013;22(4):857–884. doi: 10.1016/j.soc.2013.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Falconi M, et al. ENETS consensus guidelines for the management of patients with digestive neuroendocrine neoplasms of the digestive system: well-differentiated pancreatic non-functioning tumors. Neuroendocrinology. 2012;95(2):120–134. doi: 10.1159/000335587. [DOI] [PubMed] [Google Scholar]
- 10.Wolin EM. PI3K/Akt/mTOR pathway inhibitors in the therapy of pancreatic neuroendocrine tumors. Cancer Lett. 2013;335(1):1–8. doi: 10.1016/j.canlet.2013.02.016. [DOI] [PubMed] [Google Scholar]
- 11.Yao JC, et al. Everolimus for advanced pancreatic neuroendocrine tumors. N Engl J Med. 2011;364(6):514–523. doi: 10.1056/NEJMoa1009290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yao JC, et al. Everolimus (Eve) for the treatment of advanced pancreatic neuroendocrine tumors (PNET): final overall survival (OS) results of a randomized, double-blind, placebo (PBO)-controlled, multicenter phase III trial (RADIANT-3) Ann Oncol. 2014;25(suppl 4):iv394 [Google Scholar]
- 13.Pusceddu S, et al. Everolimus treatment for neuroendocrine tumors: latest results and clinical potential. Ther Adv Med Oncol. 2017;9(3):183–188. doi: 10.1177/1758834016683905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiao Y, et al. DAXX/ATRX, MEN1, and mTOR pathway genes are frequently altered in pancreatic neuroendocrine tumors. Science. 2011;331(6021):1199–1203. doi: 10.1126/science.1200609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Missiaglia E, et al. Pancreatic endocrine tumors: expression profiling evidences a role for AKT-mTOR pathway. J Clin Oncol. 2010;28(2):245–255. doi: 10.1200/JCO.2008.21.5988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qian ZR, et al. Prognostic significance of MTOR pathway component expression in neuroendocrine tumors. J Clin Oncol. 2013;31(27):3418–3425. doi: 10.1200/JCO.2012.46.6946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fernandes I, et al. Prognostic significance of AKT/mTOR signaling in advanced neuroendocrine tumors treated with somatostatin analogs. Onco Targets Ther. 2012;5:409–416. doi: 10.2147/OTT.S36330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Scarpa A, et al. Whole-genome landscape of pancreatic neuroendocrine tumours. Nature. 2017;543(7643):65–71. doi: 10.1038/nature21063. [DOI] [PubMed] [Google Scholar]
- 19.Sun SY, et al. Activation of Akt and eIF4E survival pathways by rapamycin-mediated mammalian target of rapamycin inhibition. Cancer Res. 2005;65(16):7052–7058. doi: 10.1158/0008-5472.CAN-05-0917. [DOI] [PubMed] [Google Scholar]
- 20.O’Reilly KE, et al. mTOR inhibition induces upstream receptor tyrosine kinase signaling and activates Akt. Cancer Res. 2006;66(3):1500–1508. doi: 10.1158/0008-5472.CAN-05-2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tompkins V, Hagen J, Zediak VP, Quelle DE. Identification of novel ARF binding proteins by two-hybrid screening. Cell Cycle. 2006;5(6):641–646. [PubMed] [Google Scholar]
- 22.Lui K, et al. Negative regulation of p53 by Ras superfamily protein RBEL1A. J Cell Sci. 2013;126(Pt 11):2436–2445. doi: 10.1242/jcs.118117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hagen J, et al. RABL6A promotes G1-S phase progression and pancreatic neuroendocrine tumor cell proliferation in an Rb1-dependent manner. Cancer Res. 2014;74(22):6661–6670. doi: 10.1158/0008-5472.CAN-13-3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Montalbano J, Jin W, Sheikh MS, Huang Y. RBEL1 is a novel gene that encodes a nucleocytoplasmic Ras superfamily GTP-binding protein and is overexpressed in breast cancer. J Biol Chem. 2007;282(52):37640–37649. doi: 10.1074/jbc.M704760200. [DOI] [PubMed] [Google Scholar]
- 25.Li YY, Fu S, Wang XP, Wang HY, Zeng MS, Shao JY. Down-regulation of c9orf86 in human breast cancer cells inhibits cell proliferation, invasion and tumor growth and correlates with survival of breast cancer patients. PLoS ONE. 2013;8(8):e71764. doi: 10.1371/journal.pone.0071764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Muniz VP, et al. RABL6A promotes oxaliplatin resistance in tumor cells and is a new marker of survival for resected pancreatic ductal adenocarcinoma patients. Genes Cancer. 2013;4(7-8):273–284. doi: 10.1177/1947601913501074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hahn WC, Weinberg RA. Rules for making human tumor cells. N Engl J Med. 2002;347(20):1593–1603. doi: 10.1056/NEJMra021902. [DOI] [PubMed] [Google Scholar]
- 28.Jackson JB, Pallas DC. Circumventing cellular control of PP2A by methylation promotes transformation in an Akt-dependent manner. Neoplasia. 2012;14(7):585–599. doi: 10.1593/neo.12768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Perrotti D, Neviani P. Protein phosphatase 2A: a target for anticancer therapy. Lancet Oncol. 2013;14(6):e229–e238. doi: 10.1016/S1470-2045(12)70558-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sangodkar J, Farrington CC, McClinch K, Galsky MD, Kastrinsky DB, Narla G. All roads lead to PP2A: exploiting the therapeutic potential of this phosphatase. FEBS J. 2016;283(6):1004–1024. doi: 10.1111/febs.13573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eichhorn PJ, Creyghton MP, Bernards R. Protein phosphatase 2A regulatory subunits and cancer. Biochim Biophys Acta. 2009;1795(1):1–15. doi: 10.1016/j.bbcan.2008.05.005. [DOI] [PubMed] [Google Scholar]
- 32.Calvo E, Bolós V, Grande E. Multiple roles and therapeutic implications of Akt signaling in cancer. Onco Targets Ther. 2009;2:135–150. doi: 10.2147/ott.s4943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Manning BD, Toker A. AKT/PKB signaling: navigating the network. Cell. 2017;169(3):381–405. doi: 10.1016/j.cell.2017.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005;307(5712):1098–1101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- 35.Hresko RC, Mueckler M. mTOR.RICTOR is the Ser473 kinase for Akt/protein kinase B in 3T3-L1 adipocytes. J Biol Chem. 2005;280(49):40406–40416. doi: 10.1074/jbc.M508361200. [DOI] [PubMed] [Google Scholar]
- 36.Sarbassov DD, et al. Prolonged rapamycin treatment inhibits mTORC2 assembly and Akt/PKB. Mol Cell. 2006;22(2):159–168. doi: 10.1016/j.molcel.2006.03.029. [DOI] [PubMed] [Google Scholar]
- 37.Huang J, Dibble CC, Matsuzaki M, Manning BD. The TSC1-TSC2 complex is required for proper activation of mTOR complex 2. Mol Cell Biol. 2008;28(12):4104–4115. doi: 10.1128/MCB.00289-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gao T, Furnari F, Newton AC. PHLPP: a phosphatase that directly dephosphorylates Akt, promotes apoptosis, and suppresses tumor growth. Mol Cell. 2005;18(1):13–24. doi: 10.1016/j.molcel.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 39.Brognard J, Sierecki E, Gao T, Newton AC. PHLPP and a second isoform, PHLPP2, differentially attenuate the amplitude of Akt signaling by regulating distinct Akt isoforms. Mol Cell. 2007;25(6):917–931. doi: 10.1016/j.molcel.2007.02.017. [DOI] [PubMed] [Google Scholar]
- 40.Sablina AA, Hector M, Colpaert N, Hahn WC. Identification of PP2A complexes and pathways involved in cell transformation. Cancer Res. 2010;70(24):10474–10484. doi: 10.1158/0008-5472.CAN-10-2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rocher G, Letourneux C, Lenormand P, Porteu F. Inhibition of B56-containing protein phosphatase 2As by the early response gene IEX-1 leads to control of Akt activity. J Biol Chem. 2007;282(8):5468–5477. doi: 10.1074/jbc.M609712200. [DOI] [PubMed] [Google Scholar]
- 42.Van Kanegan MJ, Adams DG, Wadzinski BE, Strack S. Distinct protein phosphatase 2A heterotrimers modulate growth factor signaling to extracellular signal-regulated kinases and Akt. J Biol Chem. 2005;280(43):36029–36036. doi: 10.1074/jbc.M506986200. [DOI] [PubMed] [Google Scholar]
- 43.Vereshchagina N, Ramel MC, Bitoun E, Wilson C. The protein phosphatase PP2A-B’ subunit Widerborst is a negative regulator of cytoplasmic activated Akt and lipid metabolism in Drosophila. J Cell Sci. 2008;121(Pt 20):3383–3392. doi: 10.1242/jcs.035220. [DOI] [PubMed] [Google Scholar]
- 44.Strack S, Cribbs JT, Gomez L. Critical role for protein phosphatase 2A heterotrimers in mammalian cell survival. J Biol Chem. 2004;279(46):47732–47739. doi: 10.1074/jbc.M408015200. [DOI] [PubMed] [Google Scholar]
- 45.Thayyullathil F, et al. Protein phosphatase 1-dependent dephosphorylation of Akt is the prime signaling event in sphingosine-induced apoptosis in Jurkat cells. J Cell Biochem. 2011;112(4):1138–1153. doi: 10.1002/jcb.23033. [DOI] [PubMed] [Google Scholar]
- 46.Zhou W, et al. NEK2 induces drug resistance mainly through activation of efflux drug pumps and is associated with poor prognosis in myeloma and other cancers. Cancer Cell. 2013;23(1):48–62. doi: 10.1016/j.ccr.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Peirce MJ, Cox SE, Munday MR, Peachell PT. Preliminary characterization of the role of protein serine/threonine phosphatases in the regulation of human lung mast cell function. Br J Pharmacol. 1997;120(2):239–246. doi: 10.1038/sj.bjp.0700915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cohen P, Klumpp S, Schelling DL. An improved procedure for identifying and quantitating protein phosphatases in mammalian tissues. FEBS Lett. 1989;250(2):596–600. doi: 10.1016/0014-5793(89)80803-8. [DOI] [PubMed] [Google Scholar]
- 49.Cohen P, Holmes CF, Tsukitani Y. Okadaic acid: a new probe for the study of cellular regulation. Trends Biochem Sci. 1990;15(3):98–102. doi: 10.1016/0968-0004(90)90192-E. [DOI] [PubMed] [Google Scholar]
- 50.Shenolikar S. A SMAP in the face for cancer. J Clin Invest. 2017;127(6):2048–2050. doi: 10.1172/JCI94763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kastrinsky DB, et al. Reengineered tricyclic anti-cancer agents. Bioorg Med Chem. 2015;23(19):6528–6534. doi: 10.1016/j.bmc.2015.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gutierrez A, et al. Phenothiazines induce PP2A-mediated apoptosis in T cell acute lymphoblastic leukemia. J Clin Invest. 2014;124(2):644–655. doi: 10.1172/JCI65093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sangodkar J, et al. Activation of tumor suppressor protein PP2A inhibits KRAS-driven tumor growth. J Clin Invest. 2017;127(6):2081–2090. doi: 10.1172/JCI89548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McClinch K, et al. Small-molecule activators of protein phosphatase 2a for the treatment of castration-resistant prostate cancer. Cancer Res. 2018;78(8):2065–2080. doi: 10.1158/0008-5472.CAN-17-0123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wiredja DD, et al. Phosphoproteomics profiling of nonsmall cell lung cancer cells treated with a novel phosphatase activator. Proteomics. 2017;17(22):e1870214. doi: 10.1002/pmic.201700214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kauko O, et al. PP2A inhibition is a druggable MEK inhibitor resistance mechanism in KRAS-mutant lung cancer cells. Sci Transl Med. 2018;10(450):eaaq1093. doi: 10.1126/scitranslmed.aaq1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.van Oijen MG, Medema RH, Slootweg PJ, Rijksen G. Positivity of the proliferation marker Ki-67 in noncycling cells. Am J Clin Pathol. 1998;110(1):24–31. doi: 10.1093/ajcp/110.1.24. [DOI] [PubMed] [Google Scholar]
- 58.Sobecki M, et al. Cell-cycle regulation accounts for variability in Ki-67 expression levels. Cancer Res. 2017;77(10):2722–2734. doi: 10.1158/0008-5472.CAN-16-0707. [DOI] [PubMed] [Google Scholar]
- 59.Westermarck J, Hahn WC. Multiple pathways regulated by the tumor suppressor PP2A in transformation. Trends Mol Med. 2008;14(4):152–160. doi: 10.1016/j.molmed.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 60.Sablina AA, Hahn WC. SV40 small T antigen and PP2A phosphatase in cell transformation. Cancer Metastasis Rev. 2008;27(2):137–146. doi: 10.1007/s10555-008-9116-0. [DOI] [PubMed] [Google Scholar]
- 61.Chen W, Possemato R, Campbell KT, Plattner CA, Pallas DC, Hahn WC. Identification of specific PP2A complexes involved in human cell transformation. Cancer Cell. 2004;5(2):127–136. doi: 10.1016/S1535-6108(04)00026-1. [DOI] [PubMed] [Google Scholar]
- 62.Moreno CS, et al. Signaling and transcriptional changes critical for transformation of human cells by simian virus 40 small tumor antigen or protein phosphatase 2A B56gamma knockdown. Cancer Res. 2004;64(19):6978–6988. doi: 10.1158/0008-5472.CAN-04-1150. [DOI] [PubMed] [Google Scholar]
- 63.Yeh E, et al. A signalling pathway controlling c-Myc degradation that impacts oncogenic transformation of human cells. Nat Cell Biol. 2004;6(4):308–318. doi: 10.1038/ncb1110. [DOI] [PubMed] [Google Scholar]
- 64.Sacco F, et al. A subset of RAB proteins modulates PP2A phosphatase activity. Sci Rep. 2016;6:32857. doi: 10.1038/srep32857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Newton AC, Trotman LC. Turning off AKT: PHLPP as a drug target. Annu Rev Pharmacol Toxicol. 2014;54:537–558. doi: 10.1146/annurev-pharmtox-011112-140338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Omar HA, et al. Antitumor effects of OSU-2S, a nonimmunosuppressive analogue of FTY720, in hepatocellular carcinoma. Hepatology. 2011;53(6):1943–1958. doi: 10.1002/hep.24293. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 67.Neviani P, et al. PP2A-activating drugs selectively eradicate TKI-resistant chronic myeloid leukemic stem cells. J Clin Invest. 2013;123(10):4144–4157. doi: 10.1172/JCI68951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mani R, et al. ROR1-targeted delivery of OSU-2S, a nonimmunosuppressive FTY720 derivative, exerts potent cytotoxicity in mantle-cell lymphoma in vitro and in vivo. Exp Hematol. 2015;43(9):770–4.e2. doi: 10.1016/j.exphem.2015.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Montalbano J, Lui K, Sheikh MS, Huang Y. Identification and characterization of RBEL1 subfamily of GTPases in the Ras superfamily involved in cell growth regulation. J Biol Chem. 2009;284(27):18129–18142. doi: 10.1074/jbc.M109.009597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kurimchak A, Graña X. PP2A: more than a reset switch to activate pRB proteins during the cell cycle and in response to signaling cues. Cell Cycle. 2015;14(1):18–30. doi: 10.4161/15384101.2014.985069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kim SW, Kim HJ, Chun YJ, Kim MY. Ceramide produces apoptosis through induction of p27(kip1) by protein phosphatase 2A-dependent Akt dephosphorylation in PC-3 prostate cancer cells. J Toxicol Environ Health Part A. 2010;73(21-22):1465–1476. doi: 10.1080/15287394.2010.511553. [DOI] [PubMed] [Google Scholar]
- 72.Lee TY, et al. The B56gamma3 regulatory subunit of protein phosphatase 2A (PP2A) regulates S phase-specific nuclear accumulation of PP2A and the G1 to S transition. J Biol Chem. 2010;285(28):21567–21580. doi: 10.1074/jbc.M109.094953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Benten D, et al. Establishment of the first well-differentiated human pancreatic neuroendocrine tumor model. Mol Cancer Res. 2018;16(3):496–507. doi: 10.1158/1541-7786.MCR-17-0163. [DOI] [PubMed] [Google Scholar]
- 74.Townsend CM, Ishizuka J, Thompson JC. Studies of growth regulation in a neuroendocrine cell line. Acta Oncol. 1993;32(2):125–130. doi: 10.3109/02841869309083900. [DOI] [PubMed] [Google Scholar]
- 75.Korgaonkar C, Zhao L, Modestou M, Quelle DE. ARF function does not require p53 stabilization or Mdm2 relocalization. Mol Cell Biol. 2002;22(1):196–206. doi: 10.1128/MCB.22.1.196-206.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Keck KJ, et al. Changes in gene expression in small bowel neuroendocrine tumors associated with progression to metastases. Surgery. 2018;163(1):232–239. doi: 10.1016/j.surg.2017.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pertea M, Pertea GM, Antonescu CM, Chang TC, Mendell JT, Salzberg SL. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol. 2015;33(3):290–295. doi: 10.1038/nbt.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pertea M, Kim D, Pertea GM, Leek JT, Salzberg SL. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat Protoc. 2016;11(9):1650–1667. doi: 10.1038/nprot.2016.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.di Tommaso A, et al. Residues in the alternative reading frame tumor suppressor that influence its stability and p53-independent activities. Exp Cell Res. 2009;315(7):1326–1335. doi: 10.1016/j.yexcr.2009.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Meyerholz DK, Griffin MA, Castilow EM, Varga SM. Comparison of histochemical methods for murine eosinophil detection in an RSV vaccine-enhanced inflammation model. Toxicol Pathol. 2009;37(2):249–255. doi: 10.1177/0192623308329342. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.