Abstract
Purpose
We have previously proposed temporal enhanced ultrasound (TeUS) as a new paradigm for tissue characterization. TeUS is based on analyzing a sequence of ultrasound data with deep learning and has been demonstrated to be successful for detection of cancer in ultrasound-guided prostate biopsy. Our aim is to enable the dissemination of this technology to the community for large-scale clinical validation.
Methods
In this paper, we present a unified software framework demonstrating near-real-time analysis of ultrasound data stream using a deep learning solution. The system integrates ultrasound imaging hardware, visualization and a deep learning back-end to build an accessible, flexible and robust platform. A client–server approach is used in order to run computationally expensive algorithms in parallel. We demonstrate the efficacy of the framework using two applications as case studies. First, we show that prostate cancer detection using near-real-time analysis of RF and B-mode TeUS data and deep learning is feasible. Second, we present real-time segmentation of ultrasound prostate data using an integrated deep learning solution.
Results
The system is evaluated for cancer detection accuracy on ultrasound data obtained from a large clinical study with 255 biopsy cores from 157 subjects. It is further assessed with an independent dataset with 21 biopsy targets from six subjects. In the first study, we achieve area under the curve, sensitivity, specificity and accuracy of 0.94, 0.77, 0.94 and 0.92, respectively, for the detection of prostate cancer. In the second study, we achieve an AUC of 0.85.
Conclusion
Our results suggest that TeUS-guided biopsy can be potentially effective for the detection of prostate cancer.
Keywords: Temporal enhanced ultrasound, Prostate cancer, 3D slicer, Real-time biopsy guidance
Introduction and background
According to statistics from the National Cancer Institute, approximately 11.6% of American men will be diagnosed with prostate cancer (PCa) at some point during their lifetime. In 2017, it is estimated that PCa will account for 161,360 newly diagnosed cancer cases and 26,730 deaths. Detection of early-stage PCa followed by treatment results in a 5-year survival rate of above 95% [9]. Definite diagnosis requires core needle biopsy of the prostate under TransRectal Ultra-Sound (TRUS) guidance. Conventional systematic biopsy has poor sensitivity (as low as 40%) [11,15,27] for differentiation between indolent and aggressive cancer. As a result, TRUS-guided biopsy frequently results in underdiagnosis of aggressive cancer and over-treatment of indolent prostate cancer [18]. There is an urgent need to develop patient-specific cancer diagnosis approaches based on TRUS, as this imaging modality is real time, inexpensive and relatively ubiquitous. Technologies for real-time tissue characterization in TRUS, that accurately identify PCa, will be key to this development.
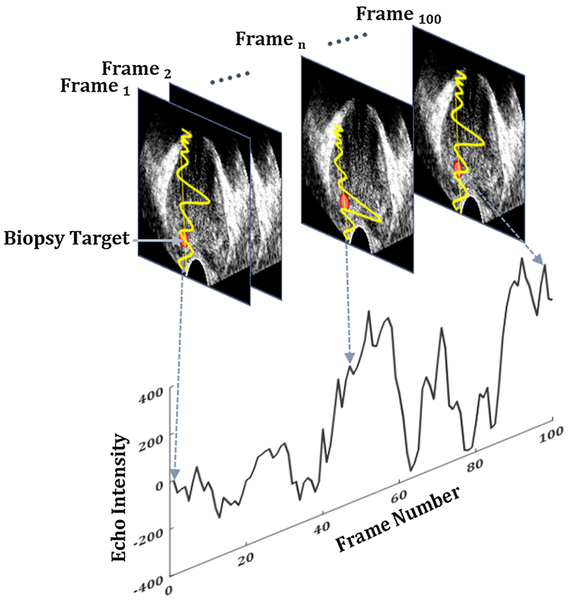
Conventional ultrasound (US)-based tissue typing techniques analyze the spectrum and texture of TRUS [20,26], measure mechanical properties of tissue in response to external excitation (elastography) [28] or determine the associated changes in blood flow (Doppler) [29]. To the best of our knowledge, these techniques to date have failed to successfully demonstrate distinguishing indolent from aggressive cancer in large-scale clinical trials. Using multi-parametric (mp)-MRI in fusion biopsy has resulted in the best clinical results to date; however, limited accessibility, difficulty in accurate MRI–TRUS registration and maintaining such registration for the duration of biopsy are key challenges. Recent studies from our group have shown promising results with temporal enhanced ultrasound (TeUS) imaging for tissue characterization in TRUS-guided prostate biopsies. TeUS (Fig. 1) is based on a machine learning framework and involves the analysis of a time series of US frames, captured following insonification of tissue over a short period of time, to extract tissue-specific information. Over the last decade, TeUS has been used for characterization of PCa in ex vivo [23,24] and in vivo studies [5,6,16,17,25], with reported areas under the receiver operating characteristic curve (AUC) of 0.76–0.93 [16,17]. Despite significant success of TeUS in research studies, its dissemination to clinical end-users demands multicenter studies to assess robustness and versatility.
Fig. 1.
TeUS: changes in backscattered time series of ultrasound, captured from a point in tissue (red dot), are analyzed using machine learning to characterize tissue
In this paper, we present a unified software framework demonstrating real-time analysis of ultrasound data stream using a deep learning solution. The system integrates cutting-edge machine learning software libraries with 3D Slicer [12] and the “Public software Library for UltraSound imaging research (PLUS)” [19] to build an accessible platform. To the best of our knowledge, this is the first system of its kind in the literature. We demonstrate the efficacy of the framework using two applications as case studies. First, we show that prostate cancer detection using near-real-time analysis of RF and B-mode TeUS data and deep learning is feasible. Second, we present real-time segmentation of prostate in ultrasound data using an integrated deep learning solution. This is the very first demonstration of automatic, real-time prostate segmentation in ultrasound in the literature. The proposed software system allows for depiction of live 2D US images augmented with patient-specific cancer likelihood maps that have been calculated from TeUS. We evaluate the performance of the proposed system with data obtained in two independent retrospective clinical studies. The first study includes data from 255 biopsy cores from 157 subjects obtained during mp-MRI–TRUS fusion biopsy and serves to establish a TeUS model for cancer likelihood map prediction. The second study further evaluates this model, with data obtained from six subjects during mp-MRI–TRUS fusion biopsy. We demonstrate that TeUS is able to accurately predict the presence of PCa in TRUS images prior to obtaining the biopsy core. This paper is the first step toward presenting an optimized platform that can enable the clinical validation, both within a single center, and across multiple centers that could get engaged in this research.
TeUS biopsy guidance system
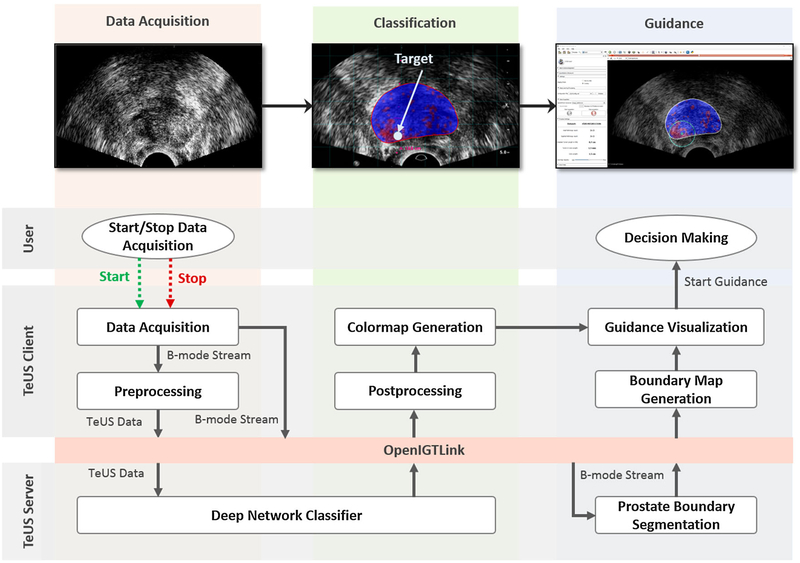
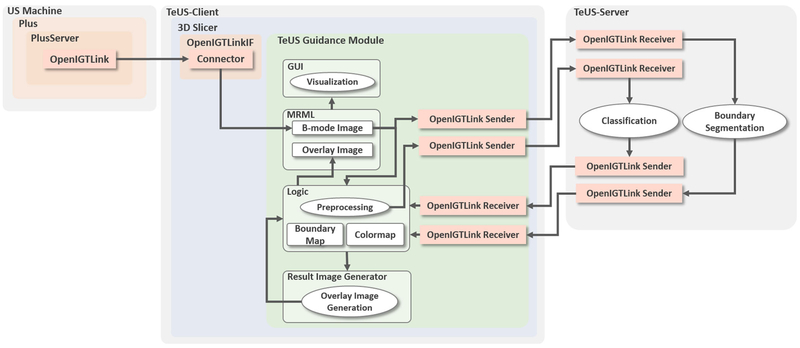
The overview of the components of the guidance system is given in Figs. 2 and 3. To allow for continuous localization of likely cancerous tissue in US data, the architecture incorporates state-of-the-art open-source software libraries for US data streaming, data visualization and deep learning. A client–server approach allows running computationally expensive algorithms simultaneously and in real time. The system has a three-tiered architecture as seen in Fig. 3: US-machine layer, TeUS-client, and TeUS-server. US-machine layer acquires data and streams it to the TeUS-client layer. TeUS-client is a 3D Slicer [12] extension responsible for US data management, preprocessing and visualization. Ultra-sound B-mode data, or B-mode and radio frequency (RF) data if both available, are streamed to the TeUS-server for tissue characterization and prostate localization and received back by the TeUS-client for real time displaying in 3D Slicer. The TeUS-server receives the US data through an OpenIGTLink network protocol [31], performs segmentation for continuous tracking and localization of the prostate and computes cancer likelihood maps that are transferred back to the TeUS-client through a second OpenIGTLink. The following subsections describe details of TeUS-client and TeUS-server.
Fig. 2.
Overview of the biopsy guidance system. The three steps in the guidance workflow are volume acquisition, classification and guidance. A client–server approach allows for simultaneous and real-time execution of computationally expensive algorithms including TeUS data classification and prostate boundary segmentation
Fig. 3.
The software system has a three-tiered architecture. Ovals represent processing elements, while arrows show the direction of data flow. In the US-machine layer, PLUS is responsible for US data acquisition and communicates with the TeUS-client via the OpenIGTLink protocol. The TeUS-client layer includes TeUS guidance, an extension module within the 3D Slicer framework. The TeUS-server layer is responsible for the simultaneous and real-time execution of computationally expensive algorithms and communicates with TeUS-client via the OpenIGTLink protocol
TeUS-client
The TeUS-client’s primary tasks are receiving streamed B-mode data and generating a TeUS sequence, preprocessing the data to divide it to smaller regions of interest (ROIs), and visualizing the cancer likelihood maps overlaid on US data. Data from ROIs are sent to the TeUS-server for analysis through the machine learning framework. TeUS-client receives the results as a colormap and segmented prostate boundary, and overlays this information on the US image. The TeUS-client includes a custom C++ loadable module, TeUS guidance module, created as an extension to 3D Slicer (Fig. 3).
Ultrasound data acquisition Once the TeUS guidance module is started, an instance of PLUS [19] is initiated on the US-machine layer by running the PlusServer application. Next, an OpenIGTLink receiver is created that continuously listens to the PlusServer to receive the US data (Fig. 3). Upon successful connection, the US data are displayed on the 3D Slicer window. Once the user issues a “Start” signal, data acquisition is initiated by buffering streamed B-mode images, followed by preprocessing. Data acquisition is halted by the user through a “Stop” signal.
Preprocessing Once the data acquisition begins, a independent preprocessing thread is initiated to avoid freezing 3D Slicer, while the TeUS ROIs are being generated. In this thread, each US image is divided into equally sized ROIs of 0.5 mm × 0.5 mm based on the scan conversion parameters. For the ith ROI, a sequence of TeUS data, , T = 100 frames, is generated by averaging all time series within that ROI and subtracting the mean value from the given time series.
Communication with the TeUS-server Following the completion of the preprocessing thread, the TeUS guidance module sends data from the extracted ROIs to the TeUS-server layer, using a standard OpenIGTLink protocol and by forking a sender thread. In addition to this thread, the TeUS-client layer also forks a receiver thread, which creates a new connection with the TeUS-server using the OpenIGTLink protocol. The receiver thread then waits for a message containing the resulting cancer likelihood colormap as well as the prostate localization information from the TeUS-server.
Receiving and visualizing the cancer likelihood colormap Upon the generation of results and receiving the output message from the TeUS-server, the TeUS-client’s receiver thread picks up the TeUS module’s execution. The guidance colormap and prostate boundary segmentation results are saved and 3D Slicer thread begins overlaying the information. During the guidance colormap visualization, the segmentation information is used to localize the prostate and mask out any colormap data falling outside the boundary. The boundary matrix is resized down to the colormap’s dimensions. Then, the guidance colormap is converted from single- channel float values (ranging from 0 to 1) to 3-channel RGB data (ranging from 0 to 255), with 0 being pure blue and 1 being pure red. (The green channel is always 0.) The boundary mask and processed colormap are multiplied together, masking out any data outside the prostate boundary.
TeUS-server
The integrated system encapsulates a machine learning framework where we specifically use two deep learning methods, implemented in Tensorflow [1]. These deep networks are responsible to identify target locations using TeUS data, and a state-of-the-art automatic prostate boundary segmentation technique is used to localize the prostate boundary during the prostate biopsy guidance. The TeUS-server is mainly responsible for simultaneous and real-time execution of computationally expensive deep learning models. The TeUS-server loads, prepares, and runs a Tensorflow graph [1] from a saved protocol buffers (.pb) file. Our Tensorflow graphs include the trained model parameters obtained from deep learning methods we will explain below. The TeUS-server also receives the extracted TeUS ROIs from the TeUS-client, buffers them into a Tensorflow Tensor object, and after running the Tensorflow graph, returns the result back to the TeUS-client. The TeUS-server is a stand-alone C++ application running on Linux and is built from within a clone of Tensorflow and is compiled using Bazel open-source toolbox.
Receiving the TeUS data As with the TeUS-client, the TeUS-server creates two socket-based objects: a sender and a receiver. Note that the TeUS-server’s receiver receives the TeUS ROIs data from the client’s sender, while the TeUS-server’s sender sends the output colormap to the client’s receiver. There is no need for multi-threading on the TeUS-server side because it needs to receive the frames in sequential order.
Running the deep neural networks Concurrent with the OpenIGTLink activity, the TeUS-server also performs a few steps to set up and run the Tensorflow graphs for PCa detection and prostate boundary segmentation. The TeUS-server initializes the Tensorflow graph, by loading the cancer classification and segmentation trained models from the protocol buffer files and initializes the buffer tensor, which will be filled with T = 100 frames. After receiving the T th frame, the TeUS-server runs the Tensorflow session, feeding the buffer tensor as input to the classification network graph. Simultaneously, prostate boundary segmentation graph is fed with the buffer tensor as the input to generate the segmentation results. Then, TeUS-server converts the returned tensors into a float vector, packs the float data from output vectors into a new OpenIGTLink “ImageMessage,” and sends them to the TeUS-client using the TeUS-server’s sender object.
Prostate cancer classification The deep networks are generated based on the methods that we presented in our earlier works and from a data set consisting of biopsy targets in mp-MRI–TRUS fusion biopsies with 255 biopsy cores from 157 subjects. (Here, we refer to these data as the first retrospective study.) We give a brief overview of these methods. For a detailed description of the models, the reader may refer to [4,8]. To generate the TeUS-based cancer detection models, we obtained TeUS data during fusion prostate biopsy. TeUS RF data were acquired on a Philips iU22 US imaging platform using an Endocavity curved probe (Philips C9–5ec) with the frequency of 6.6 MHz. All subjects provided informed consent to participate, and the study was approved by the institutional research ethics board. All biopsy targets were identified as suspicious for cancer in a preoperative mp-MRI examination where two radiologists assigned an overall MR suspicious level score of “low,” “moderate,” and “high” to each target [30]. Later, subjects underwent MRI–TRUS-guided biopsies using UroNav (Invivo Corp., FL) MR-US fusion system [21]. Prior to biopsy sampling from each target, T = 100 frames of TeUS data were obtained. We use histopathology information of each biopsy core as gold standard for generating labels for each target. In this dataset, 83 biopsy cores are cancerous with Gleason Score (GS) 3 + 3 or higher. These data are divided into mutually exclusive training, Dtrain, and test sets, Dtest. Training data are made up of 84 cores from patients with homogeneous tissue regions. For each biopsy target, we analyze an area of 2 mm × 10 mm around the target location, along the projected needle path. We divide this region into 80 equally sized regions of interest (ROIs) of 0.5 mm × 0.5 mm. Each ROI includes 27–55 RF samples based on the depth of imaging and the target location. We also augment the training data by creating ROIs using a sliding window of size 0.5 mm × 0.5 mm over the target region, which results in 1536 ROIs per target. Given the data augmentation strategy, we obtain a total number of 129,024 training samples including both TeUS radio frequency (RF) and TeUS B-mode data (see [8] for more details.). We further use the test data consisting of 171 cores to evaluate the trained model during the guidance system implementation, where 130 cores are labeled as benign and 31 cores are labeled as cancerous with GS ≥ 3 + 3.
An individual TeUS sequence of length T, xi, is composed of echo-intensity values for each time step, t, and is labeled as yi ϵ {0, 1}, where zero and one indicate benign and biopsy outcome, respectively. We aim to learn a mapping from xi to yi in using recurrent neural networks (RNNs) to model the temporal information in TeUS B-mode data. For this purpose, first, we used the unsupervised domain adaption method presented in [8] to find a common feature space between TeUS Radio Frequency (RF) and TeUS B-mode data. Our RNN architecture is built with long short-term memory (LSTM) cells [14], where each cell maintains a memory over time. We use two layers of LSTMs with T = 100 hidden units to capture temporal changes in data. Given an input TeUS B-mode sequence , our RNN computes a hidden vector sequence in the common feature space between the RF and B-mode data within a domain adaption step. Following this step, we use a fully connected layer to map the learned sequence to a posterior over binary classes of benign and cancer tissue in a supervised classification step. This final node generates a predicted label, , for a given TeUS B-mode ROI sequence, xi. The training criterion for the network is to minimize the loss function as the binary cross-entropy between y(i) and over all of the training samples where we use root-mean-square propagation (RMSprop) optimizer. As recommended, using a training–validation setting, we perform a grid search to find the optimum hyperparameter in our search space. Once the optimum hyperparameters are identified (the number of RNN hidden layers = 2, batch size = 64, learning rate = 0.0001, dropout rate = 0.4, and the regularization term =0.0001),= the entire training set, Dtrain, is used to learn the final classification model.
Prostate segmentation The segmentation deep networks are generated based on our earlier works [3]. The network is pre-trained on a dataset consisting of 4284 expert-labeled TRUS images of the prostate as explained in [3] and further fine-tuned using manually segmented B-mode images obtained during mp-MRI–TRUS fusion biopsy from train subjects. The method is based on residual neural networks and dilated convolution at deeper layers. The model takes an input image of the size of 224 × 224 pixels and generates a corresponding label map of the same size. For a detailed description of the models, the reader may refer to [3].
Results and discussion
Classification model validation
To evaluate the individual modules of the proposed system, we used clinical data from two retrospective studies to simulate the flow of information across different modules. Data are sent to the PlusServer which is subsequently transmitted to 3D slicer and the machine learning module for analysis and visualization.
To validate the accuracy of the classification model, we use Dtest. We use sensitivity, specificity, and accuracy in detecting cancerous tissue samples to report the validation results. We consider all cancerous cores as the positive class and other non-cancerous cores as the negative class. Sensitivity is defined as the ratio of cancerous cores that are correctly identified, while specificity is the ratio of non-cancerous cores that are correctly classified. Accuracy is the ratio of the correctly identified results over the total number of cores. We also report the overall performance of our approach using the AUC. Table 1 shows the model performance for classification of cores in Dtest for different MR suspicious levels. For samples of moderate MR suspicious level (70% of all cores), we achieve an AUC of 0.94 using the LSTM-RNN. In this group, our sensitivity, specificity, and accuracy are 0.75, 0.96, and 0.93, respectively. In comparison, only 26% of all of the cores identified in mp-MRI are cancerous after biopsy which means our approach can effectively complement mp-MRI during the guidance procedure to reduce the number of false positives for those targets with moderate MR suspicious level.
Table 1.
Model performance for classification of cores in the test data from the first retrospective study for different MR suspicious levels
| Method | Specificity | Sensitivity | Accuracy | AUC |
|---|---|---|---|---|
| All of the biopsy cores (N = 171) | 0.94 | 0.77 | 0.92 | 0.94 |
| Moderate MR suspicious level (N = 115) | 0.96 | 0.75 | 0.93 | 0.94 |
| High MR suspicious level (N = 20) | 0.85 | 0.98 | 0.95 | 0.95 |
N indicates number of cores in each group
System assessment
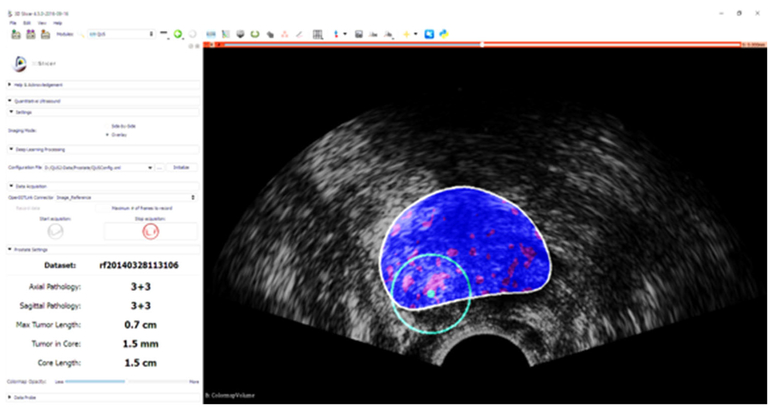
Figure 4 shows the guidance interface implemented as part of a 3D Slicer module running on the client machine. In order to evaluate the performance of the entire guidance system, other than the subjective evaluation of guidance visualization, the accuracy of the target detection and the run time are measured.
Fig. 4.
Guidance interface implemented as part of a 3D Slicer module: cancer likelihood map is overlaid on B-mode ultrasound images. Red indicates predicted labels as cancer, and blue indicates predicted benign regions. The boundary of the segmented prostate is shown with white, and the green circle is centered around the target location which is shown in green dot
Guidance accuracy To further assess the developed system, we performed a second independent MRI–TRUS fusion biopsy study. The study was approved by the institutional ethics review board, and all subjects provided informed consent to participate. Six subjects were enrolled in the study where they underwent preoperative mp-MRI examination prior to biopsy, to identify the suspicious lesions. From each MRI-identified target locations, two biopsies were taken, one in the axial imaging plane and one in the sagittal imaging plane. Only TeUS B-mode data were recorded for each target to minimize disruption to the clinical workflow. We use the histopathology labeling of the cores as the ground truth to assess the accuracy of the guidance system in detecting the cancerous lesions. This study resulted in 21 targeted biopsy cores with GS distribution as explained in Table 2. We achieve an AUC, sensitivity, specificity, and accuracy of 0.85, 0.93, 0.72, and 0.85, respectively, for the fusion biopsy targets. Our results show that the only miss-classified target is GS 3 + 3 with the tumor in core less than 0.4 cm.
Table 2.
Gleason score distribution in the second retrospective clinical study
| GS | Benign | GS 3+3 | GS 3+4 | GS 4+3 | GS 4+4 | GS 4+5 |
|---|---|---|---|---|---|---|
| Number of cores | 7 | 2 | 3 | 2 | 1 | 6 |
Run time The run time was measured for all parts of the workflow (classification, segmentation, and guidance visualization) using a timer log provided by the open-source Visualization Toolkit. The TeUS-client computer featured an 2.70 GHz Intel Core™ i7–6820 (8 CPUs) processor with 64 GB of RAM, Windows 10 Enterprise N, Visual Studio 2015, 3D Slicer 4.3.1, OpenMP 2.0, and Intel MKL 11.0. The TeUS-server was hosted by a computer running a Ubuntu 16.04 operating system and a 3.4 GHz Intel Core™ i7 CPU with 16 GB of RAM, equipped with GeForce GTX 980 Ti GPU with 2816 CUDA cores and 6 GB of memory.
Run-time results are summarized in Table 3. Averaged over all N = 21 trials in the second fusion biopsy study, the total time spent on TeUS-client for guidance visualization and post-processing is 0.40 ± 0.11 s. Since the classification and segmentation modules run simultaneously, the TeUS-server run time is constrained by the classification task run time of a batch of 100 ultrasound data frames, which is 1.66 ± 0.32 s. Within the context of prostate biopsy guidance workflow, this means an addition of only about 20-sec to a 20-min procedure. We consider this near-real-time performance sufficient for the requirements of this clinical procedure.
Table 3.
Run time of the steps of the prostate guidance system averaged over N = 21 trials with data from the second retrospective study (given as mean ± std)
| Host machine | Operation | Average time (s) |
|---|---|---|
| TeUS-Client | Guidance visualization | 0.40 (± 0.11) |
| TeUS-Server | Classification | 1.66 (± 0.32) |
| Segmentation | 0.12 (± 0.17) |
Discussion and comparison with other methods
The best result to date involving TeUS B-mode data (AUC = 0.7) is based on spectral analysis and deep belief network (DBN) as the underlying machine learning framework [5,7]. Comparing our LSTM-RNN approach with that method as the most related work, a two-way paired t-test shows statistically significant improvement in AUC (p < 0.05). Furthermore, using the RNN-based framework simplifies the real-time implementation of the guidance system. While analysis of a single RF ultrasound frame is not feasible in the context of our current clinical study, as we did not access to the transducer impulse response for calibration, previously, we have shown that analysis of TeUS significantly outperforms the analysis of a single RF frame [10,24]. The best results reported using a single RF frame analysis [13] involve 64 subjects with an AUC of 0.84, where they used prostate-specific antigen (PSA) as additional surrogate.
The performance of mp-MRI for detection of PCa has been established in a multicenter, 11-site study in the UK [2], consisting of 740 patients, to compare the accuracy of mp-MRI and systematic biopsy against template prostate mapping biopsy (TMP-biopsy) used as a reference test. The findings of the study were that for clinically significant cancer, mp-MRI was more sensitive (93%) than systematic biopsy (48%), but much less specific (41% for mp-MRI vs. 96% for systematic biopsy). In a prospective cohort study of 1003 men undergoing both targeted and standard biopsy [30], targeted fusion biopsy diagnosed 30% more high-risk cancers versus standard biopsy, with an area under the curve of 0.73 versus 0.59, respectively, based on whole-mount pathology. TeUS shows a balance of high specificity and sensitivity across all cancer grades, while enabling an ultrasound-only-based solution for guiding prostate biopsies.
To the best of our knowledge, this paper is also the first report on real-time automatic segmentation of prostate ultrasound images using deep learning. Prior work reported on prostate segmentation using deep learning within 3D Slicer [22] is only suitable for static MRI data, not dynamic ultrasound images which are streamed in real time from an ultrasound system.
Conclusion and future directions
In this work, we presented a software solution for a prostate biopsy guidance system based on the analysis of TeUS data. The solution allowed augmentation of the live standard 2D US image with a cancer likelihood map generated from a deep learning model trained using TeUS data. The system integrated open-source software libraries for ultrasound image research such as PLUS and 3D Slicer with the cutting-edge machine learning software libraries to achieve an accessible implementation.
From a clinical perspective, the motivation of the work is to enable real-time assessment of TeUS for prostate cancer detection. The immediate goal of the current work is to demonstrate the viability of this approach using retrospective data with known ground truth, so that we can optimize the system’s performance and understand how such system would fit within the standard clinical workflow. To reach our end goal, we have to demonstrate that the accuracy of the integrated system on clinical ultrasound devices that only provide B-mode data is comparable to those in benchtop prototypes in the laboratory. The system was validated using retrospective in vivo datasets including both TeUS RF and B-mode data of 255 biopsy target cores obtained in a large clinical study during mp-MRI–TRUS fusion biopsy. For the validation data, we achieve an AUC of 0.94, and sensitivity and specificity of 0.94, 0.77, respectively. The integrated system is then evaluated with an independent in vivo dataset obtained during mp-MRI–TRUS fusion biopsy from six subjects and includes only B-mode TeUS of 21 biopsy targets. We achieved an AUC of 0.85 for this dataset, and sensitivity and specificity of 0.93 and 0.72, respectively. The average sensitivity of the system considering both studies is 82% where 18% of cancerous cores (10 out of 55) were not identified. The promising results of this initial assessment indicate that our proposed TeUS-based system is capable of providing accurate guidance information for the prostate biopsy procedure. Future work should focus on prospective evaluation and feasibility assessment of the biopsy guidance system.
The development of a commercially viable prototype that can be used daily in a clinical environment requires many other steps, including the validation of the system in multicenter clinical studies, software certification, regulatory approval, and full integration with clinical ultrasound systems such as those manufactured today for fusion biopsy systems. However, the current solution is easily expandable to other guidance and decision-making systems that rely on real-time US data and machine learning. Moreover, the open-source nature of the implementation assures the dissemination and accessibility of the solution for the community. Our future direction is to further optimize the solution to reduce the number the needed host machines, by adding a Docker feature to run this distributed application on the same machine.
Acknowledgements
This work was supported in part by the Natural Sciences and Engineering Research Council of Canada (NSERC) and in part by the Canadian Institutes of Health Research (CIHR).
Footnotes
Conflict of interest The authors declare that they have no conflict of interest.
Ethical approval All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Declaration of Helsinki and its later amendments or comparable ethical standards.
Informed consent Informed consent was obtained from all individual participants included in the study.
References
- 1.Abadi M, Agarwal A, Barham P, Brevdo E, Chen Z, Citro C, Corrado GS, Davis A, Dean J, Devin M (2016) Tensorflow: large-scale machine learning on heterogeneous distributed systems. arXiv preprint arXiv:1603.04467
- 2.Ahmed HU, Bosaily AES, Brown LC, Gabe R, Kaplan R, Parmar MK, Collaco-Moraes Y, Ward K, Hindley RG, Freeman A (2017) Diagnostic accuracy of multi-parametric MRI and TRUS biopsy in prostate cancer (PROMIS): a paired validating confirmatory study. Lancet 389(10071):815–822 [DOI] [PubMed] [Google Scholar]
- 3.Anas EMA, Nouranian S, Mahdavi SS, Spadinger I, Morris WJ, Salcudean SE, Mousavi P, Abolmaesumi P (2017) Clinical target-volume delineation in prostate brachytherapy using residual neural networks In: International conference on medical image computing and computer-assisted intervention. Springer, pp 365–373 [Google Scholar]
- 4.Azizi S, Bayat S, Yan P, Tahmasebi A, Nir G, Kwak JT, Xu S, Wilson S, Iczkowski KA, Lucia MS, goldenberg L, Salcudean SE, Pinto P, Wood B, Abolmaesumi P, Mousavi P (2017) Detection and grading of prostate cancer using temporal enhanced ultrasound: combining deep neural networks and tissue mimicking simulations. Int J Comput Assist Radiol Surg 12(8):1293–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Azizi S, Imani F, Ghavidel S, Tahmasebi A, Wood B, Mousavi P, Abolmaesumi P (2016) Detection of prostate cancer using temporal sequences of ultrasound data: a large clinical feasibility study. IJCARS 11:947–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Azizi S, Imani F, Kwak JT, Tahmasebi A, Xu S, Yan P, Kruecker J, Turkbey B, Choyke P, Pinto P, Wood B, Mousavi P, Abolmaesumi P (2016) Classifying cancer grades using temporal ultrasound for transrectal prostate biopsy In: MICCAI. Springer, pp 653–661 [Google Scholar]
- 7.Azizi S, Imani F, Zhuang B, Tahmasebi A, Kwak JT, Xu S, Uniyal N, Turkbey B, Choyke P, Pinto P, Wood B, Mousavi P, Abolmaesumi P (2015) Ultrasound-based detection of prostate cancer using automatic feature selection with deep belief networks In: International conference on medical image computing and computer-assisted intervention. Springer, pp 70–77 [Google Scholar]
- 8.Azizi S, Mousavi P, Yan P, Tahmasebi A, Kwak JT, Xu S, Turkbey B, Choyke P, Pinto P, Wood B, Abolmaesumi P (2017) Transfer learning from RF to B-mode temporal enhanced ultrasound features for prostate cancer detection. Int J Computer Assist Radiol Surg 12:1111–1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barentsz JO, Richenberg J, Clements R (2012) ESUR prostate MR guidelines 2012. Eur Radiol 22(4):746–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Daoud MI, Mousavi P, Imani F, Rohling R, Abolmaesumi P (2013) Tissue classification using ultrasound-induced variations in acoustic backscattering features. IEEE TBME 60(2):310–320 [DOI] [PubMed] [Google Scholar]
- 11.Epstein JI, Feng Z, Trock BJ, Pierorazio PM (2012) Upgrading and downgrading of prostate cancer from biopsy to radical prostatectomy: incidence and predictive factors using the modified gleason grading system and factoring in tertiary grades. Eur Urol 61(5):1019–1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fedorov A, Beichel R, Kalpathy-Cramer J, Finet J, Fillion-Robin JC, Pujol S, Bauer C, Jennings D, Fennessy F, Sonka M (2012) 3D slicer as an image computing platform for the quantitative imaging network. Magn Reson Imaging 30(9):1323–1341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feleppa EJ, Rondeau MJ, Lee P, Porter CR (2009) Prostate-cancer imaging using machine-learning classifiers: potential value for guiding biopsies, targeting therapy, and monitoring treatment In: 2009 IEEE international ultrasonics symposium (IUS). IEEE, pp 527–529 [Google Scholar]
- 14.Gers FA, Schmidhuber J, Cummins F (2000) Learning to forget: continual prediction with LSTM. Neural Comput 12(10):2451–2471 [DOI] [PubMed] [Google Scholar]
- 15.Goossen T, Wijkstra H (2003) Transrectal ultrasound imaging and prostate cancer. Arch Ital Urol Androl 75(1):68–74 [PubMed] [Google Scholar]
- 16.Imani F, Abolmaesumi P, Gibson E, Khojaste A, Gaed M, Moussa M, Gomez JA, Romagnoli C, Leveridge M, Chang S (2015) Computer-aided prostate cancer detection using ultrasound RF time series: in vivo feasibility study. IEEE TMI 34(11):2248–2257 [DOI] [PubMed] [Google Scholar]
- 17.Imani F, Ramezani M, Nouranian S, Gibson E, Khojaste A, Gaed M, Moussa M, Gomez JA, Romagnoli C, Leveridge M (2015) Ultrasound-based characterization of prostate cancer using joint independent component analysis. IEEE TBME 62(7):1796–1804 [DOI] [PubMed] [Google Scholar]
- 18.Kuru TH, Roethke MC, Seidenader J, Simpfendörfer T, Boxler S, Alammar K, Rieker P, Popeneciu VI, Roth W, Pahernik S (2013) Critical evaluation of magnetic resonance imaging targeted, transrectal ultrasound guided transperineal fusion biopsy for detection of prostate cancer. J Urol 190(4):1380–1386 [DOI] [PubMed] [Google Scholar]
- 19.Lasso A, Heffter T, Rankin A, Pinter C, Ungi T, Fichtinger G (2014) PLUS: open-source toolkit for ultrasound-guided intervention systems. IEEE Trans Biomed Eng 61(10):2527–2537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Llobet R, Pérez-Cortés JC, Toselli AH, Juan A (2007) Computer-aided detection of prostate cancer. Int J Med Inform 76(7):547–556 [DOI] [PubMed] [Google Scholar]
- 21.Marks L, Young S, Natarajan S (2013) MRI-US fusion for guidance of targeted prostate biopsy. Curr Opin Urol 23(1):43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mehrtash A, Pesteie M, Hetherington J, Behringer PA, Kapur T, Wells III WM, Rohling R, Fedorov A, Abolmaesumi P (2017) Deepinfer: open-source deep learning deployment toolkit for image-guided therapy. In: Proceedings of SPIE–the International Society for Optical Engineering, vol 10135 NIH Public Access [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moradi M, Abolmaesumi P, Mousavi P (2010) Tissue typing using ultrasound RF time series: experiments with animal tissue samples. Med Phys 37(8):4401–4413 [DOI] [PubMed] [Google Scholar]
- 24.Moradi M, Abolmaesumi P, Siemens DR, Sauerbrei EE, Boag AH, Mousavi P (2009) Augmenting detection of prostate cancer in transrectal ultrasound images using SVM and RF time series. IEEE TBME 56(9):2214–2224 [DOI] [PubMed] [Google Scholar]
- 25.Moradi M, Mahdavi SS, Nir G, Jones EC, Goldenberg SL, Salcudean SE (2013) Ultrasound RF time series for tissue typing: first in vivo clinical results. In: SPIE medical imaging. International Society for Optics and Photonics, pp 86701I-1–86701I-8 [Google Scholar]
- 26.Oelze ML, O’Brien WD, Blue JP, Zachary JF (2004) Differentiation and characterization of rat mammary fibroadenomas and 4T1 mouse carcinomas using quantitative ultrasound imaging. IEEE TMI 23(6):764–771 [DOI] [PubMed] [Google Scholar]
- 27.Rapiti E, Schaffar R, Iselin C, Miralbell R, Pelte MF, Weber D, Zanetti R, Neyroud-Caspar I, Bouchardy C (2013) Importance and determinants of Gleason score undergrading on biopsy sample of prostate cancer in a population-based study. BMC Urol 13(1):19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Salcudean SE, French D, Bachmann S, Zahiri-Azar R (2006) Viscoelasticity modeling of the prostate region using vibroelastography In: MICCAI. Springer, pp 389–396 [DOI] [PubMed] [Google Scholar]
- 29.Sauvain JL, Sauvain E, Rohmer P, Louis D, Nader N, Papavero R, Bremon JM, Jehl J (2013) Value of transrectal power doppler sonography in the detection of low-risk prostate cancers. Diagn Interv Imaging 94(1):60–67 [DOI] [PubMed] [Google Scholar]
- 30.Siddiqui MM, Rais-Bahrami S, Turkbey B, George AK, Roth-wax J, Shakir N, Okoro C, Raskolnikov D, Parnes HL, Linehan WM (2015) Comparison of MR/ultrasound fusion-guided biopsy with ultrasound-guided biopsy for the diagnosis of prostate cancer. JAMA 313(4):390–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tokuda J, Fischer GS, Papademetris X, Yaniv Z, Ibanez L, Cheng P, Liu H, Blevins J, Arata J, Golby AJ (2009) OpenIGTLink: an open network protocol for image-guided therapy environment. Int J Med Robot Comput Assist Surg 5(4):423–434 [DOI] [PMC free article] [PubMed] [Google Scholar]