Targeted nanocarriers deliver HIV-1 latency-reversing agents to CD4+ T cells in the lymph nodes.
Abstract
A proposed strategy to cure HIV uses latency-reversing agents (LRAs) to reactivate latent proviruses for purging HIV reservoirs. A variety of LRAs have been identified, but none has yet proven effective in reducing the reservoir size in vivo. Nanocarriers could address some major challenges by improving drug solubility and safety, providing sustained drug release, and simultaneously delivering multiple drugs to target tissues and cells. Here, we formulated hybrid nanocarriers that incorporate physicochemically diverse LRAs and target lymphatic CD4+ T cells. We identified one LRA combination that displayed synergistic latency reversal and low cytotoxicity in a cell model of HIV and in CD4+ T cells from virologically suppressed patients. Furthermore, our targeted nanocarriers selectively activated CD4+ T cells in nonhuman primate peripheral blood mononuclear cells as well as in murine lymph nodes, and substantially reduced local toxicity. This nanocarrier platform may enable new solutions for delivering anti-HIV agents for an HIV cure.
INTRODUCTION
Highly active antiretroviral therapy (HAART) has revolutionized the treatment of HIV-1 and transformed it into a chronic disease but does not cure the infection. Long-term HIV infection is maintained by several factors including limited accessibility of antiretroviral drugs (ARVs) to certain anatomical sites where viral replication may occur (1–3), and latent infection of resting cells where integrated provirus is invisible to drug treatment as well as the immune system (4, 5). The latent cell pool decays slowly despite suppressive HAART (6, 7), requiring patients to take lifelong ARV regimens, associated with short- and long-term side effects (8).
Strategies to accelerate the decay of the HIV reservoir pool are being explored, but none has yet proven effective in reducing reservoir size in vivo. One strategy, which has been studied extensively, including in clinical studies, is to induce HIV reactivation using latency-reversing agents (LRAs) while continuing suppressive HAART (9). Upon reversal of the latent state, the reactivated cells can be eliminated by host immune responses or other killing strategies (10). This “shock and kill” approach is still controversial based on recent safety issues and limited success in reducing reservoir sizes in clinical settings (11). There are several hypotheses to explain the failures in clinical trials: First, these studies were restricted to individual LRAs from a single class (11, 12), but recent evidence shows that combinations of multiple mechanistic classes may be needed to effectively overcome latency in vivo (13–15). Second, some LRAs are associated with high toxicity (16, 17) or nonspecific T cell activation that is unnecessary for latency reversal and causes off-target toxicity (14). Last, LRAs might not achieve sufficient concentrations in the lymphatic tissues including lymph nodes (LNs) and gut-associated lymphoid tissue (GALT), where vast numbers of latently infected CD4+ T cells reside (1, 18). Systemic administration of LRAs may work well on circulating CD4+ T cells, but those cells make up less than 2% of the body’s CD4+ T cells (19, 20).
Nanocarrier drug delivery systems provide a promising approach for overcoming these three challenges to using LRAs (21–23). Specifically, nanocarrier drug delivery systems allow LRAs to be delivered specifically to target cells such as CD4+ T cells within organs of interest, thus achieving high concentrations at relevant sites while minimizing off-target effects by keeping concentrations low at other sites. However, few studies have reported on the use of nanocarriers for HIV cure, and those studies used single LRAs (24–27). Of these studies, none has investigated CD4-targeting or lymphatic tissue–targeting functions in vivo. Here, we developed lipid-coated poly(lactic-co-glycolic acid) (PLGA) nanoparticles (termed “LCNPs”) to address the current barriers to LRA treatment. Through simple chemical synthesis and bioconjugation processes, we successfully loaded a variety of LRAs into our LCNPs and formulated them to target CD4+ T cells in the LNs. We demonstrate that these LCNPs can co-deliver multiple LRAs with varying release kinetics for synergistic latency reversal, and successfully traffic to LNs after subcutaneous administration. Our CD4-targeted LCNPs incorporate a promising LRA candidate, ingenol-3-angelate (Ing3A), inducing long-acting and specific CD4+ T cell activation in the complex lymphatic environment while substantially reducing local toxicity compared to the free drug. Our approach is unique and potentially transformative because we investigate agents of multiple mechanistic classes and specifically target them to the cell types and tissues sustaining latency.
RESULTS
Development of LCNPs loaded with mechanistically diverse LRAs
Combinations of LRAs have recently shown synergistically enhanced potency for latency reversal as compared to single LRAs in multiple ex vivo assays (13). However, the varied physicochemical properties of different LRAs make combined formulation difficult to achieve and administer. In addition, the low potency but high toxicity of some LRAs limits their tolerability when used in monotherapy. To improve LRA potency while minimizing toxicity, we co-formulated and targeted the drugs to CD4+ T cells using lipid-polymer hybrid nanoparticles (termed LCNPs) surface-modified with targeting antibodies and with LRA incorporated into both the lipid bilayer and the polymer core (Fig. 1).
Fig. 1. Strategies for loading LRAs into LCNPs.
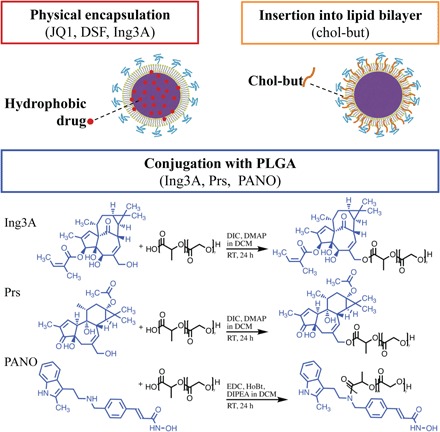
Hydrophobic LRAs were physically encapsulated into the PLGA core. Chol-but, as the prodrug of butyric acid, was inserted into the lipid bilayer. LRAs with hydroxyl or amine groups were conjugated to the PLGA followed by LCNP synthesis. Prs, prostratin; PANO, panobinostat; DIC, N,N′-diisopropylcarbodiimide; DMAP, 4-(dimethylamino)pyridine; RT, room temperature; EDC, 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide; HoBt, 1-hydroxybenzotriazole; DIPEA, N,N-diisopropylethylamine.
For ease of formulation, we first attempted to physically encapsulate all LRAs individually into LCNPs by co-dissolving each compound with PLGA in ethyl acetate, followed by a modified single-emulsion evaporation method as described previously (23). An initial 5 weight % (wt %) (ratio of LRA to PLGA and lipids) input of hydrophobic drugs JQ1, disulfiram (DSF), and Ing3A resulted in drug loadings of 1.70 ± 0.08 wt %, 2.54 ± 0.60 wt %, and 1.00 ± 0.04 wt %, respectively (Table 1). However, prostratin and romidepsin were encapsulated into LCNPs at less than 0.02 wt % (table S1). Physical encapsulation of panobinostat was also impractical because of its poor solubility in ethyl acetate and dichloromethane (DCM). We used DCM as an alternative solvent for romidepsin, which increased its drug loading to 0.2 wt % but failed to improve the loading of either prostratin or panobinostat. We were unable to encapsulate butyric acid, a short fatty acid that has shown potential in HIV latency reversal (28, 29), into the PLGA core. Instead, we inserted its prodrug cholesteryl butyrate (chol-but) into the LCNP lipid bilayer (30), achieving a loading of 2.72 wt % (Fig. 1).
Table 1. Physicochemical properties of LCNP-formulated LRAs.
BET, bromodomain and extra-terminal proteins; NF-κβ, nuclear factor κ–light chain enhancer of activated B cells; PKC, protein kinase C; HDAC, histone deacetylase; EE, encapsulation efficiency; n.d., not determined.
| LRAs | JQ1 | DSF | Ing3A | Ing3A-PLGA | Prs-PLGA | PANO-PLGA | Chol-but |
| Molecular target | BET | NF-κβ | PKC | PKC | PKC | HDAC | HDAC |
| Formulation* strategy | Single emulsion | Steglich esterification | EDC | Lipid insertion | |||
| Size (d, nm)† | 187.2 ± 4.0 | 183.7 ± 1.2 | 182.4 ± 1.7 | 164.6 ± 0.7 | 176.2 ± 1.3 | 169.9 ± 3.1 | 219.3 ± 2.6 |
| Polydispersity index† | 0.09 ± 0.01 | 0.07 ± 0.02 | 0.05 ± 0.02 | 0.07 ± 0.03 | 0.09 ± 0.01 | 0.07 ± 0.01 | 0.06 ± 0.02 |
| ζ Potential (mV)† | 12.0 ± 3.8 | 15.7 ± 1.5 | 13.8 ± 5.9 | 15.2 ± 9.6 | 8.4 ± 8.0 | 9.41 ± 2.5 | −10.4 ± 1.5 |
| Drug input (wt %) | 5 | 5 | 5 | 1.7 | 1.2 | 0.4 | 4.3 |
| Drug loading (wt %)† | 1.70 ± 0.08 | 2.54 ± 0.60 | 1.00 ± 0.04 | 1.62 ± 0.05 | 1.08 ± 0.03 | 0.31 ± 0.03 | 2.72 ± 0.03 |
| EE (%)†‡ | 35.8 ± 1.6 | 53.3 ± 1.3 | 21.1 ± 0.8 | 93.3 ± 2.5 | 92.6 ± 2.2 | 70.6 ± 6.3 | 63.2 ± 7.7 |
| Colloidal stability§ | ≥10 days | ≥10 days | ≥10 days | ≥10 days | ≥10 days | ≥10 days | ≥10 days |
| Time to release 50%|| | <1 hour | 14 hours | 1.6 days | 25.7 days | 24.0 days | 1.4 days | n.d. |
*Formulation was by physical encapsulation in the PLGA core (single emulsion), conjugation with PLGA (Steglich esterification and EDC chemistry), or insertion into lipid bilayer surface coating.
†Data represent means ± SD from three independently formulated batches.
‡EE is the ratio of the actual loading (wt %) to the drug input (wt %) expressed as a percentage.
§Colloidal stability was measured in cell culture media (fig. S3).
||Drug release curves were fit by nonlinear regression to estimate the time when 50% of the drug was released from the LCNP formulation (Fig. 2A and table S3).
For Ing3A, prostratin, and panobinostat, we also evaluated chemical conjugation into LCNPs (Fig. 1). The ester or amide bond formed between PLGA and LRA is hydrolyzed to release free LRAs under physiological conditions by host esterases or the esterase-like activity of human serum albumin (31). Chemical conjugation was expected to decrease the release rate of LRAs compared to physical encapsulation. Drugs were conjugated in a 1:1 molar ratio with a low–molecular weight PLGA to increase LRA content. However, we found that the LCNPs using PLGA of molecular weight less than 20 kDa led to poor particle size distribution and low stability (table S2). Therefore, to maximize LCNP drug loading while minimizing polydispersity, we selected a 24- to 38-kDa carboxyl-terminated PLGA. We used proton nuclear magnetic resonance (H-NMR) and high-performance liquid chromatography (HPLC) to verify the conjugation chemistry and efficiency. Both methods supported successful conjugation of each LRA with PLGA (figs. S1 and S2), with drug loadings of 1.62 ± 0.05 wt % (Ing3A), 1.08 ± 0.03 wt % (prostratin), and 0.31 ± 0.03 wt % (panobinostat).
All LRA-loaded LCNPs showed well-distributed sizes with diameters of 160 to 190 nm and polydispersity index (PDI) of <0.1. Chol-but–inserted LCNPs (cbLCNPs) showed an increased size of ~220 nm in diameter and significant changes in ζ potential going from positive to negative, as would be expected because of the replacement of the positively charged lipid 1,2-dioleoyl-3-trimethylammonium-propane (DOTAP) (Table 1). All LRA-loaded LCNPs were colloidally stable in physiological conditions for more than 10 days in cell culture media supplemented with 10% fetal bovine serum (FBS) (fig. S3).
Efficacy of LRA-loaded LCNPs in an in vitro model of latent HIV-1 infection
LRA action requires release from LCNPs and subsequent binding to intracellular molecular targets. Previously, we and others have shown that physical encapsulation of drugs into LCNPs results in an initial burst release upon mixing with cell culture medium (23). We hypothesized that the physically encapsulated LRAs would show similar burst release, while chemically conjugated LRAs would be released slowly. To test this hypothesis, we measured release kinetics of LRAs from LCNPs in cell culture media (Fig. 2A). As expected, all physically encapsulated LRAs (JQ1, DSF, and Ing3A) showed burst release from LCNPs, resulting in 50% of the drug released between 1 hour and 1.6 days as measured by nonlinear regression (Table 1, Fig. 2A, and table S3). In contrast, chemically conjugated LRAs showed slower release kinetics. For Ing3A and prostratin, which were conjugated by Steglich esterification, it took more than 1 month to reach 50% release. Because Ing3A was both physically encapsulated and chemically conjugated, combining these separate formulations would provide both slow and fast release kinetics. For panobinostat conjugated by amide bond using 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide (EDC) chemistry, 50% of the drug was released over 1.4 days.
Fig. 2. In vitro dose-response and time-dependent HIV-1 latency reversal correlates with LRA release kinetics from LCNPs.
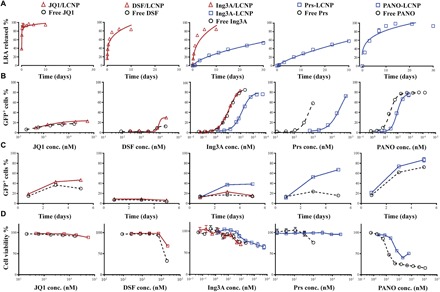
(A) Release kinetics of physically encapsulated LRAs (red triangle) and chemically conjugated LRAs (blue square) from LCNPs in cell culture media at 37°C. Data were fit by nonlinear regression, detailed in table S3. (B) Dose-response curve for latent HIV reactivation (indicated as a percentage of GFP+ cells) on J-Lat A1 cells incubated with single LRA formulations for 20 hours. Data were fit in the four-parameter log-logistic model, detailed in table S4. (C) Time-dependent curve for latent HIV reactivation of J-Lat A1 cells incubated with single LRA formulations over 5 days. (D) Cell viability of J-Lat A1 cells after incubation with single free LRAs or LCNP-formulated LRAs at different concentrations for 20 hours. Cell viability was measured by monitoring metabolic activity with the CellTiter-Blue Assay. Each experiment was performed once with n = 3 wells of each treatment. Data represent means ± SD. LRA/LCNP, LRA was physically encapsulated into LCNPs (red curve); LRA-LCNP, LRA was chemically conjugated to the PLGA (blue curve).
Next, we compared the HIV-1 latency reactivation potency of these LCNP-formulated LRAs with free LRAs on the J-Lat Tat-GFP (A1) cell line model, which expresses green fluorescent protein (GFP) upon reactivation of latent HIV-1 integrated into the cell genome (32, 33). The percentage of GFP+ cells measured by flow cytometry was used to assess the potency of LRAs (Fig. 2B and fig. S4). Physically encapsulated JQ1, DSF, and Ing3A showed similar potency to the free drug used at the same concentrations after 20 hours of treatment. In contrast, all chemically conjugated LRAs (Ing3A, prostratin, and panobinostat) showed lower potency than the free drug, likely due to their slower release from LCNPs (table S3). To investigate whether the slow drug release kinetics could lead to delayed latency activation at later time points, we extended cell incubation with LRA formulations to 5 days and observed that the conjugated LRAs (Ing3A-LCNP, Prs-LCNP, and PANO-LCNP) showed accumulated induction of GFP expression over time (Fig. 2C). In contrast, the free LRAs (DSF, Ing3A, and prostratin) and the physically encapsulated drugs (DSF/LCNP and Ing3A/LCNPs) exhibited constant activity over 5 days. Exceptions are physically encapsulated JQ1 and free panobinostat, which also showed accumulated effects. All the LCNP-formulated LRAs showed equal or lower cytotoxicity compared to the free drug (Fig. 2D), meaning that LCNPs could potentially deliver higher LRA doses required for efficacy while avoiding the high toxicity. cbLCNPs induced only ~5% GFP expression in this cell model at the highest concentration we could achieve (fig. S5A). However, because chol-but reduces nonspecific cell binding (34), we kept it in our formulations for the subsequent targeting and animal studies.
Synergistic induction of HIV-1 mRNA levels by Ing3A-LCNP and JQ1/LCNP in CD4+ T cells from virally suppressed individuals
Mechanistically distinct LRAs delivered in combination have shown synergistic interactions in ex vivo latent HIV-1 reactivation (13, 14). To identify the best LRA formulations to evaluate for HIV-1 latency reversal in human clinical samples, we first measured viral reactivation and cytotoxicity for every pairwise combination of LCNP-formulated LRA using J-Lat A1 cells. To better compare therapeutic index in future planned nonhuman primate (NHP) studies, our criterion for selecting LRA combinations was that both the free and LCNP-formulated drugs were nontoxic. Hence, we balanced trade-off between toxicity, potency, and synergy for selecting LRA combinations. LRA concentrations were chosen to achieve similar induction of GFP expression (~20% GFP+ cells) when dosed individually, and this concentration was used to measure efficacy of the drug combination (Fig. 3, A and B). Most binary combinations showed higher GFP induction than the corresponding single drugs. We used the Bliss independence model to select LRAs with synergistic effects as measured by Δfaxy > 0 (details in Materials and Methods). JQ1 in combination with any of the other four LRAs, and DSF in combination with Ing3A or prostratin, displayed synergy with Δfaxy above 0.1 (Fig. 3C). Ing3A-LCNP and PANO-LCNP were the most potent, as indicated by the lower dose necessary to achieve equivalent efficacy of ~20% GFP+ cells as well as their median effective dose (ED50) (Figs. 2B and 3A and table S4). However, panobinostat demonstrated high cytotoxicity both individually and in combination with JQ1 (Figs. 2D and 3D). A similar relationship between efficacy and cytotoxicity was observed for DSF. DSF combined with prostratin in LCNPs led to the highest measured synergy and also high cell viability (Fig. 3, C and D). However, this LRA combination required use at 10-fold higher total dose (~18,000 nM) compared to the combination of JQ1/LCNP and Ing3A-LCNP (~1500 nM) (Fig. 3A). The free drug combination of DSF and prostratin also showed low viability (Fig. 3D). Last, the combination of Ing3A and JQ1 was chosen as it showed equivalent and synergistic activity at a lower dose with notably better viability (Fig. 3, A to D).
Fig. 3. LCNP-formulated Ing3A and JQ1 enhance latent HIV reactivation, reduce cytotoxicity from J-Lat A1 cells, and synergistically increase HIV-1 mRNA expression in CD4+ T cells from infected individuals on suppressive HAART.
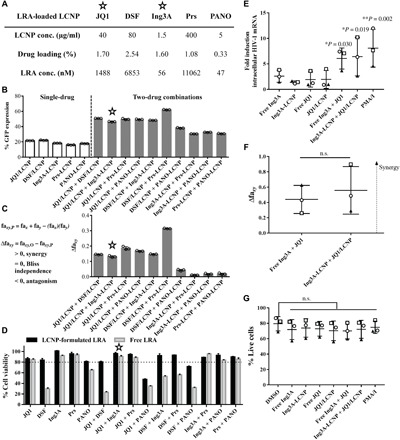
(A) Concentrations of single and combination LCNP-formulated LRAs. LRA concentrations were calculated as total LRA in LCNPs. (B) In vitro latent HIV reactivation using single or combination LCNP-formulated LRAs on J-Lat A1 cells for 20 hours. (C) Calculation of synergy for LCNP-formulated LRA combinations using the Bliss independence model. Data are presented as the difference between the observed and predicted percentage of GFP+ cells. fax or fay, percentage of GFP expression by drug x or y; faxy,O, observed percentage of GFP expression by a combination of drug x and y; faxy,P, predicted percentage of GFP expression by a combination of drug x and y using the equation detailed in Materials and Methods. (D) Cell viability of J-Lat A1 cells after incubation with single or combination LRA formulations for 20 hours. Free or LCNP-formulated LRAs were dosed at the concentrations that achieved similar latent HIV reactivation (JQ1, 1488 nM; DSF, 14,840 nM; Ing3A, 3.5 nM; Prs, 251 nM; and PANO, 13.2 nM). The combination of JQ1 and Ing3A (✫) was chosen for high potency, synergy, and low cytotoxicity. The experiment (A to D) was performed once with n = 3 wells of each treatment. Data represent means ± SD. (E) Intracellular HIV-1 mRNA levels in CD4+ T cells isolated from peripheral blood of infected individuals and treated with free Ing3A, Ing3A-LCNP, free JQ1, JQ1/LCNP, or their binary combinations. Data are presented as fold induction relative to DMSO control. Statistical analysis was performed using paired one-way analysis of variance (ANOVA) with Bonferroni’s test comparing each group with the DMSO control. *P < 0.05, **P < 0.005. (F) Calculation of synergy for Ing3A and JQ1 combinations using the Bliss independence model. Data are presented as the difference between the observed and predicted fractional effect by the LRAs compared to the PMA/I positive control (see Materials and Methods for details). Statistical analysis was performed using paired Student’s t test. n.s., not significant. (G) Percentage of live cells after treatments, measured by live/dead staining following the fluorescence-activated cell sorting analysis. The experiments (E to G) were performed using peripheral blood from three different individuals, represented as a circle, square, or triangle. Each data point represents the mean fold induction of three replicate LRA treatments of 1 × 106 CD4+ T cells per individual. Error bars represent means ± SD from three individuals.
CD4+ T cells isolated from HIV-1–infected individuals under suppressive ART were used to validate the activity of LCNP-formulated Ing3A and JQ1. Cells from three donors were used to test the induction of intracellular HIV-1 mRNA after treatment with Ing3A and JQ1 alone as well as their combination as free drugs and formulated in LCNPs. The single drugs in free or LCNP formulations increased intracellular HIV by one- to fourfold over dimethyl sulfoxide (DMSO) control treatment, but the differences were not statistically significant (Fig. 3E). Combining Ing3A and JQ1 significantly increased intracellular HIV-1 mRNA expression 6.1-fold (free Ing3A + JQ1) and 6.4-fold (LCNP-formulated Ing3A + JQ1), which was similar to the positive control of phorbol 12-myristate 13-acetate plus ionomycin (PMA/I; 8.1-fold induction). Because Ing3A and JQ1 act through different mechanisms, we compared the experimentally observed synergistic effect to the predicted effect by the Bliss independence model. We found that the combined effects from both free and LCNP-formulated Ing3A and JQ1 significantly exceeded the Bliss model prediction, resulting in a calculated Δfaxy values of 0.44 and 0.56 (Fig. 3F), respectively, validating that the combination synergistically induces intracellular HIV-1 mRNA from clinical samples. None of these LRA formulations caused significant cytotoxicity in comparison to the DMSO control (Fig. 3G).
CD4-targeted LCNPs selectively activate CD4+ T cells from pigtail macaque PBMCs
We have previously shown that conjugating an anti-CD4 monoclonal antibody (mAb) to our optimized cbLCNPs led to high specificity for CD4+ T cells (34). Here, we conjugated an anti-CD4 mAb to our Ing3A-cbLCNPs (Fig. 4A). We chose conjugated Ing3A as the model drug because our screening showed that it is the most potent LRA and the chemically conjugated formulation provides sustained release, which is especially beneficial for targeting. To facilitate a higher level of passive targeting to the draining LNs (3, 35), we synthesized smaller LCNPs by optimizing the single-emulsion process with a different lipid-to-PLGA ratio (table S5). After conjugation with anti-CD4 mAb, the diameter of LCNPs remained less than 150 nm (Fig. 4A). We confirmed the activity of these smaller Ing3A-LCNPs in J-Lat A1 cells and observed slightly higher latency reversal compared to the larger particle (fig. S6A). To evaluate reactivation of the targeted formulations, we used primary cells obtained from pigtail macaque peripheral blood mononuclear cells (PBMCs) and compared CD69 expression between CD4+ (CD3+CD14−CD8−) and CD8+ (CD3+CD14−CD8+) T cells (fig. S7). CD69 is a marker for T cell activation and positively correlates with HIV-1 latency reversal (25, 36). PBMCs were treated with free drug, bare LCNPs, anti-CD4 LCNPs, and isotype LCNPs (Iso-LCNPs). The targeted Ing3A-CD4-cbLCNPs showed twofold increased CD69 expression in CD4+ T cells compared with CD8+ T cells (Fig. 4B), demonstrating the ability of our targeted LCNPs to specifically activate CD4+ T cells.
Fig. 4. CD4-targeted LCNPs selectively activate CD4+ T cells from macaque PBMCs and accumulate in the draining LNs after subcutaneous injection to mouse left flank.
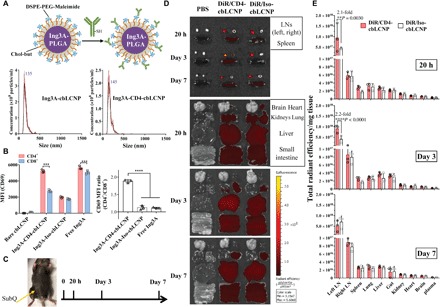
(A) Ing3A-LCNP (<150 nm) conjugated with anti-CD4 mAb, resulting in size distribution measured by NanoSight. DSPE, 1,2-distearoyl-sn-glycero-3-phosphoethanolamine; PEG, polyethylene glycol. (B) CD69 median fluorescence intensity (MFI) of CD4+ (CD14−CD3+CD8−) and CD8+ (CD14−CD3+CD8+) cells (left) and their respective MFI ratio (right) from PBMCs isolated from pigtail macaque blood and treated with free Ing3A, bare LCNPs, CD4-targeted LCNPs, and Iso-LCNPs for 20 hours. The experiment was performed once with blood samples from three pigtail macaques (n = 3). (C) Schedule of injection and tissue harvest. C57BL/6J mice were injected with phosphate-buffered saline (PBS), DiR-loaded CD4-cbLCNPs, and Iso-cbLCNPs subcutaneously at the left flank and sacrificed at 20 hours, 3 days, and 7 days for analysis. (D) Representative fluorescent images of inguinal LNs, spleen (top), and other major organs at 20 hours, 3 days, and 7 days. (E) Region of interest quantification of tissue fluorescence normalized by tissue mass. Statistical significance was calculated using paired two-way ANOVA with Bonferroni’s test. **P < 0.005, ***P < 0.005, ****P < 0.0001. Data represent means ± SD; n = 3 mice per group. (Photo credit: Shijie Cao, Department of Bioengineering, University of Washington)
CD4-LCNP biodistribution in mice over 7 days
Most of the latent HIV reservoir is harbored in lymphatic tissues such as LNs, making it inaccessible to many anti-HIV-1 agents (2, 3, 21). Nanocarriers have been developed to target ARVs to anatomical HIV reservoirs including LNs, the central nervous system, and GALTs (21, 23). However, none of those nanocarriers have been used to deliver LRAs to lymphatic tissues. We hypothesized that our nanocarriers could traffic to draining LNs after subcutaneous administration and enter the lymphatic system to deliver LRAs to resident CD4+ T cells. To test this hypothesis, we labeled our LCNPs with DiR dye, subcutaneously injected them into the left flank of C57BL/6J mice, and tracked the LCNPs by imaging the draining and nondraining inguinal LNs, as well as other major tissues and organs (spleen, lung, liver, gut, kidney, heart, brain, and plasma). First, we surprisingly found that LCNPs with DOTAP in the lipid bilayer (dtLCNPs) showed little accumulation to LNs after subcutaneous injection (fig. S8A). In contrast, negatively charged cbLCNPs with chol-but in the lipid bilayer led to a marked increase in LN accumulation (fig. S8B). We assumed that the difference was due to nonspecific cell binding of neutrally to positively charged dtLCNPs at the injection site that hindered LN trafficking. Next, we found that the smaller-size optimized cbLCNPs (<150 nm) enhanced LN accumulation compared to the larger particle (fig. S8C). This was consistent with other reported studies that bigger nanoparticles might be more easily trapped at the injection site instead of efficiently draining to the LN (3, 35). Because of their desirable trafficking properties, as well as the increased targeting specificity reported previously (34), we moved forward with smaller CD4-targeted cbLCNPs for all animal studies.
We selected 20 hours, 3 days, and 7 days as time points for investigating the biodistribution of CD4-cbLCNPs in mice (Fig. 4, C to E). For all three time points, both targeted and nontargeted cbLCNPs exhibited preferential accumulation in the left draining inguinal LN compared to other organs. The second highest fluorescent signal among all major organs was observed in the right inguinal LN. Comparing the targeted and nontargeted cbLCNPs, CD4-cbLCNPs showed 2.1- and 2.2-fold increased left LN accumulation over the Iso-cbLCNPs at 20 hours and 3 days, respectively, suggesting that CD4 targeting also led to higher LN accumulation and retention.
CD4-LCNPs selectively activate lymphatic CD4+ T cells and protect tissues from drug toxicity over 7 days
Having shown that our LCNPs target and reactivate CD4+ T cells ex vivo and accumulate in LNs in vivo, we investigated in vivo lymphatic CD4+ T cell activation using our targeted Ing3A-cbLCNPs. We performed this in vivo study following similar procedures as in the biodistribution study but instead injected mice with free Ing3A, Ing3A-CD4-cbLCNPs, or Ing3A-Iso-cbLCNPs containing equal amounts of drug. We also injected mice with 20-fold lower free Ing3A, because our previous in vitro and ex vivo studies (Figs. 2A and 3E) show that this concentration demonstrated similar or equivalent efficacy to our Ing3A-LCNPs (due to the slow release nature of the LCNPs). We isolated cells from the left and right inguinal LNs and evaluated CD4+ and CD8+ T cell activation at all time points by measuring CD69 expression (fig. S9). At 20 hours, free Ing3A induced the highest activation of both CD4+ and CD8+ T cells from left LN, without any significant difference between the two cell populations as expected (Fig. 5A). In contrast, our targeted Ing3A-CD4-cbLCNPs selectively activated CD4+ T cells, while CD8+ T cells were not activated. Surprisingly, the induction in the right LN was as high as that in the left LN, although the left LN showed higher LCNP accumulation in the biodistribution study. Ing3A-CD4-cbLCNP induced less CD69 expression than free Ing3A, which was expected because of the slower drug release from these chemically conjugated LCNP formulations. We did not observe statistically significant activation of T cells from any other treatment groups. At days 3 and 7 after administration, treatment groups with free Ing3A showed ~10-fold reduction in T cell activation from the levels seen at 20 hours down to levels indistinguishable from the untreated controls. In contrast, the Ing3A-CD4-cbLCNP–treated mice resulted in sustained CD69 expression on CD4+ T cells in both LNs out to day 3 and, to a lesser extent, day 7. These results are the first investigation of Ing3A delivered subcutaneously in vivo for T cell activation.
Fig. 5. CD4-targeted LCNPs selectively activate CD4+ T cells in inguinal LNs after subcutaneous injection to mouse left flank and protect local tissues from toxicity.
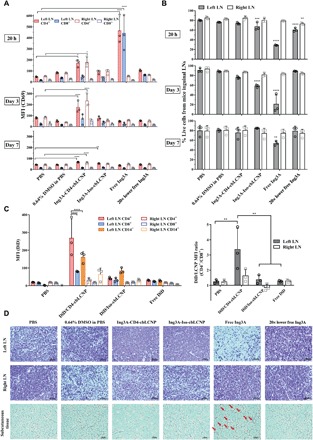
(A) MFI of CD69 expression on CD4+ (CD14−CD3+CD8−) and CD8+ (CD14−CD3+CD8+) cells from mouse left and right inguinal LNs. Mice were injected with 0.3 ml of PBS, 0.64% DMSO, Ing3A-CD4-cbLCNP (0.48 mg/kg; Ing3A dose per body weight), Ing3A-Iso-cbLCNP (0.48 mg/kg), or free Ing3A (0.48 or 0.024 mg/kg) and were sacrificed after 20 hours, 3 days, and 7 days for analysis. (B) Percentage of live cells from all cell populations in the left or right inguinal LNs after treatment. (C) Left: MFI of DiD fluorescent signal from CD4+ (CD14−CD3+CD8−), CD8+ (CD14−CD3+CD8+), and CD14+ cells from mouse left or right inguinal LNs. Mice were injected subcutaneously at the left flank with 0.3 ml of PBS, DiD/CD4-cbLCNP, DiD/Iso-cbLCNP, or equivalent amount of free DiD (50 μg/kg; DiD dose per body weight) in PBS and were sacrificed after 20 hours for analysis. Right: MFI ratio of DiD fluorescent signal between CD4+ and CD8+ T cells. Statistical significance was calculated using paired two-way ANOVA with Bonferroni’s test comparing each treatment group with PBS. *P < 0.05, **P < 0.005, ***P < 0.0005, ****P < 0.0001. Data represent means ± SD; n = 3 mice per group. (D) Hematoxylin and eosin staining of tissue slides (left and right inguinal LNs and subcutaneous tissues near the injection site) from mice at day 3. From free Ing3A treatment group, condensed cell nucleus in the left LN that indicated cell death and immune cell infiltration into the adipose tissue near the injection site (red arrows) were observed.
To further investigate the targeting effect of our LCNP formulations, we measured the cell-specific biodistribution of DiD fluorescently labeled LCNP formulations in primary cells isolated from the left and right inguinal LNs at 20 hours after subcutaneous dosing (Fig. 5C). We observed that mice treated with CD4-cbLCNPs showed threefold higher signal in CD4+ compared to CD8+ cells from the left draining LN, indicating preferential targeting to CD4+ T cells (Fig. 5C) and consistent with our CD69 activation studies (Fig. 5A). In addition, the CD14+ monocytes from those LNs also showed significant DiD signals, indicating uptake or binding by monocytes. In the right nondraining LN, we were unable to measure cell-specific biodistribution above the phosphate-buffered saline (PBS) control (Fig. 5C), which is consistent with our biodistribution studies showing a ~10-fold lower accumulation of DiR/LCNP in the right LN (Fig. 4, D and E). Last, the control Iso-LCNPs showed no significant uptake by either CD4+ or CD8+ T cells but was taken up or bound by monocytes, possibly via Fc receptor (FcR) binding.
From our efficacy studies, we noticed that free Ing3A induced severe inflammation-like toxicity in the tissues near the injection sites at all time points (fig. S10). To test for toxicity at a cellular level, we isolated cells from the inguinal LNs and found that most cells (>70%) in the left LN were dead after treatment with free Ing3A at the 20-hour and 3-day time points, while less than 30% of cells were dead from Ing3A-CD4-cbLCNP treatments (Fig. 5B). Our Ing3A-CD4-cbLCNP did not show significant toxicity, although they had better CD4-specific activation at day 3. In addition, while our targeted LCNPs showed preferential accumulation to CD4+ T cells in the draining LNs, the CD4+ T cells retained high viability (fig. S11). To further understand the injection site toxicity that resulted from free Ing3A, we performed histological examination of the LNs and tissues at the injection site (Fig. 5D). The free Ing3A treatment group was the only group with obvious morphological changes in the draining LNs and subcutaneous tissues. Free Ing3A caused cell apoptosis as indicated by condensed cell nucleus from the left inguinal LN near the injection site but no effect on the right LN. In addition, we observed inflammatory cell infiltration into the adipose tissue near the injection site from the mice treated with free Ing3A. These data suggest that, although free Ing3A exhibits high T cell activation efficacy, its activation is not CD4 specific and causes severe cell apoptosis in the draining LNs as well as inflammatory-like toxicity in vivo near the injection site. In contrast, our Ing3A-CD4-cbLCNPs induce specific CD4+ T cell activation for at least 7 days without toxicity.
DISCUSSION
The major challenges for the current shock and kill approach to HIV cure include insufficient potency of single LRAs, low LRA concentration in the lymphatic tissues that harbor most latent reservoirs, and toxicity. Nanocarriers have shown great promise in delivering combination therapeutics to specific sites or cells and therefore could address these challenges. However, nanocarriers have not been tested in a rigorous fashion as a strategy to deliver LRAs for HIV cure. In a few reported studies, LRAs were evaluated in vitro (24, 26, 27) or in vivo through systemic administration without specific targeting (25). In addition, no studies have investigated the delivery of combination LRAs, which may be necessary to sufficiently stimulate HIV reactivation (13). In this study, we focused on using targeted nanocarriers to deliver mechanistically distinct LRAs in combination and to specifically act on CD4+ T cells in the LNs.
Our approach shows many advantages over conventional drug delivery systems and existing nanocarrier platforms to deliver LRAs. One of the biggest advantages is the ability of nanocarriers to passively traffic to LNs and actively target CD4+ T cells. We optimized our LCNPs to achieve a high level of LN accumulation by altering lipid composition and reducing their size (table S5). Our CD4-targeted Ing3A-LCNPs selectively bind to and activate CD4+ T cells from both NHP PBMCs ex vivo and mouse LNs in vivo. Together, the high accumulation and long-term retention of targeted LCNPs in the LNs, along with sustained release of conjugated Ing3A, result in up to 7 days of specific CD4+ T cell activation in the LNs. Notably, significantly more CD4-targeted LCNPs accumulated in draining LNs compared to control Iso-LCNPs at 20 hours and 3 days after administration, demonstrating that the targeted LCNPs preferentially traffic to and remain in the LNs. As we observed particle binding to or uptake by monocytes in vivo, it will be needed to determine whether some entry of LCNP-formulated Ing3A into monocytes can cause any side effects. Further studies are also necessary to investigate why we saw similar levels of CD4+ T cell activation in the left and right LNs despite the observed lower biodistribution of LCNPs in the right LN, opposite the injection site. To achieve CD4+ T cell activation and eventually reservoir eradication throughout the body, it will be necessary to assess T cell activation in other LNs and lymphatic tissues such as the GALT. The impact of multiple subcutaneous injection sites on drug efficacy, as well as alternative administration routes on systemic activity, also warrants further investigation.
The CD4-targeting effect also allows LRAs to only activate lymphatic CD4+ T cells without affecting other cells such as CD8+ T cells. This avoids inducing aberrant autoimmune responses and focuses drug activity on HIV-1 latency reversal. We found that free Ing3A caused severe cell apoptosis and inflammation-like toxicity near the injection sites in mice (Fig. 5, B and D, and fig. S10), although it caused little cytotoxicity in cell line and primary cell studies (Figs. 2D and 3D and fig. S7D). In contrast, Ing3A in our targeted LCNP formulation did not cause severe inflammation in local tissues or cell apoptosis in the draining LNs. In addition to the targeting effects, we also hypothesize that the lack of toxicity is due to the sustained release of Ing3A from LCNPs, which results in consistent low-level exposure over time in contrast to exposure to brief, high levels of free drug exposure. Ing3A is approved by the U.S. Food and Drug Administration for topical treatment of actinic keratosis (37) but has not been studied for HIV cure research until recently and then only in an ex vivo model of latency reversal (13–15). Our approach is the first to deliver Ing3A to mice subcutaneously and to demonstrate its T cell activation and toxicity profiles in vivo.
Some other advantages of our LCNPs include the capability to incorporate multiple physicochemically diverse LRAs and to improve drug solubility. LRAs vary in their solubility and bioavailability, and it may therefore be difficult to achieve the expected efficacy when dosed in combination with conventional delivery approaches. Our LCNPs deliver multiple LRAs with fixed dose ratios to the target sites that may be more easily translated from in vitro to in vivo studies. Furthermore, our LCNPs can be dosed at concentrations up to 10 mg/ml in saline, which could significantly increase the solubility of hydrophobic LRAs.
In summary, we have developed hybrid nanocarriers that can deliver multiple types of LRA and determined an LCNP-formulated JQ1 and Ing3A combination that induces robust and synergistic HIV-1 latent reactivation with low toxicity. Our targeted formulations provide long-term and specific activation of CD4+ T cells in LNs while reducing local drug toxicity in vivo. These nanocarriers will be tested in an NHP model for reactivation of latent simian immunodeficiency virus (SIV). This robust platform for targeting CD4+ T cells in the LNs has potential for the delivery of other types of anti-HIV agents, vaccines, immunomodulating agents, and gene-modifying oligonucleotide drugs for many biomedical applications.
MATERIALS AND METHODS
Study design
The objective of the study was to develop a nanocarrier-based drug delivery system that permits the loading of multiple diverse anti-HIV agents and specific targeting to HIV reservoir cells and tissues in vivo to address the major clinical hurdles in the shock and kill strategy. We first developed several strategies to load different LRAs into our hybrid nanocarriers, characterized the performance of every single formulation, and determined the best drug combination in an HIV latency cell line model. We further confirmed their latency reversal in CD4+ T cells from virologically suppressed patient, a more clinically relevant model. Next, we evaluated CD4+ T cell–specific activation in NHP PBMCs ex vivo. We used C57BL/6J mice to evaluate the in vivo targeting properties of our optimized nanocarriers, especially the ability to target and activate CD4+ T cells in LNs and their toxicity profile after subcutaneous administration. Sample sizes were chosen to detect statistical significance. Statistical analysis is detailed in the last section.
Materials
PLGA B6013-2 (75:25 L:G; ester-terminated, inherent viscosity range: 0.55 to 0.75 dl/g in chloroform) was purchased from Lactel. All Resomer PLGA polymers were purchased from Sigma-Aldrich. All lipids for the nanoparticle synthesis were purchased from Avanti Polar Lipids. Chol-but was purchased from Santa Cruz Biotechnology. LRAs were purchased from Cayman Chemical (Ing3A), LC Laboratories (prostratin and panobinostat), Active Biochem (romidepsin), or Sigma-Aldrich (DSF and JQ1). The EasySep Human CD4+ T Cell Isolation Kit was purchased from STEMCELL Technologies. Anti-human antibodies and anti-mouse antibodies including phycoerythrin (PE) anti-CD14, peridinin chlorophyll protein (PerCP)/Cy5.5 anti-CD4, fluorescein isothiocyanate (FITC) anti-CD8, allophycocyanin (APC) anti-CD69, and their isotype control antibodies were purchased from BD Biosciences. Rhesus recombinant anti-CD4 antibody and rhesus recombinant immunoglobulin G1 (IgG1) isotype control antibody were purchased from the National Institutes of Health (NIH) Nonhuman Primate Reagent Resource. Anti-mouse CD4 antibody and its isotype IgG2b control antibody were purchased from BioXCell. RPMI 1640 cell culture media containing 2 mM l-glutamine and 25 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) supplemented with heat-inactivated FBS (10%, v/v) and penicillin-streptomycin (100 U/ml) were purchased from Thermo Fisher Scientific and used for most of the in vivo and ex vivo studies. All other chemicals were purchased from Sigma-Aldrich and Thermo Fisher Scientific unless otherwise specified.
PLGA-drug conjugation and LCNP fabrication
Ing3A and prostratin were conjugated to carboxyl-ended PLGA 503H using the Steglich esterification. Briefly, PLGA (1.29 × 10−5 mol) and Ing3A or prostratin (3.87 × 10−5 mol) were dissolved in 6 ml of DCM. N,N′-diisopropylcarbodiimide (1.16 × 10−4 mol) and 4-(dimethylamino)pyridine (3.87 × 10−5 mol) were added, and the mixture was incubated at room temperature (RT) for 24 hours. After that, 160 ml of cold methanol was added to precipitate polymer-drug conjugates. The conjugates were then collected by centrifugation and dissolved in DCM for another washing. Final products were dissolved in DCM and lyophilized for future use. Panobinostat was conjugated to the same PLGA using the EDC chemistry. The process was similar to Ing3A or prostratin conjugation, except that EDC, 1-hydroxybenzotriazole hydrate (HoBt), and N,N-diisopropylethylamine (DIPEA) (all at 3.87 × 10−5 mol) were used as the catalyst.
These PLGA-LRA conjugates as well as PLGA and each LRA were analyzed by H-NMR (AV500, Bruker) after dissolving in deuterated DMSO. They were also verified by HPLC (Shimadzu) with a Phenomenex Kinetex C18 column (250 × 4.6 mm), eluting with water-acetonitrile [0.1% trifluoroacetic acid (TFA)] gradients (fig. S2A).
LCNPs were synthesized using a modified single-emulsion evaporation method as described previously (23). In brief, the lipid mixture [either dioleoylphosphatidylcholine (DOPC), DOTAP, and 1,2-distearoyl-sn-glycero-3-phosphoethanolamine (DSPE)–polyethylene glycol (PEG)–maleimide (Mal) or DOPC, chol-but, and DSPE-PEG-Mal at a 4:4:1 molar ratio] in chloroform was dried under nitrogen and then suspended in Milli-Q water. PLGA (10 mg/ml in ethyl acetate) was added dropwise to the lipid suspension while vortexing. The mixture was then homogenized using a probe sonicator (500 W, Ultrasonic Processor GEX500) three times for 30 s each. The residual ethyl acetate in the mixture was removed by rotary evaporation (Rotavapor R-210, BÜCHI Labortechnik) for 10 min. LCNPs were then washed by high-speed centrifugation at 14,000 rpm for 10 min at 4°C. To physically encapsulate LRAs into LCNPs, different LRAs were co-dissolved with PLGA B6013-2 in ethyl acetate before LCNP synthesis. PLGA-LRA conjugates were dissolved in ethyl acetate at 10 mg/ml before LCNP synthesis to make LRA-conjugated LCNPs. For DiR or DiD fluorescent LCNPs, 1% (w/w) (DiR or DiD to PLGA) DiR or DiD dissolved in ethyl acetate was added to PLGA/ethyl acetate solution right before LCNP synthesis. Anti-CD4 mAbs were conjugated to synthesize LCNPs via maleimide-functionalized DSPE-PEG in the lipid bilayer as described previously (23, 34). In brief, the mAb was thiolated using Traut’s reagents and then incubated with LCNPs in PBS (containing 5 mM EDTA) for 2 hours. Final mAb-conjugated LCNPs were centrifuged at 10,000 rpm for 5 min in a 50-ml falcon tube to remove unreacted antibodies.
Characterization of LRA-loaded LCNPs and their in vitro release kinetics
The size, PDI, and ζ potential of all LCNP formulations were measured by dynamic light scattering using Zetasizer Nano ZS90 (Malvern Instruments), and the size was also measured by NanoSight NS300 (Malvern Instruments). Nanoparticles were suspended in Milli-Q water for size and PDI measurement or in NaCl buffer (10 mM, pH 7.4) for ζ-potential measurement. Drug loading was measured by dissolving preweighted lyophilized LRA-loaded LCNPs in DMSO and analyzing the samples using HPLC. The separation was made on a C18 column. The gradient mobile phase described above was used to measure LRA content in conjugated LCNP formulations. For other LRAs, the mobile phase was isopropanol-acetonitrile (40:60, v/v) for chol-but (38), water-acetonitrile (30:70, v/v; with <0.045% TFA) for JQ1 and DSF, or water-acetonitrile (60:40, v/v; with <0.045% TFA) for prostratin and romidepsin. The drug loadings (DLs) were calculated using Eq. 1
| (1) |
The encapsulation efficiencies were calculated using Eq. 2
| (2) |
The drug release studies were all performed in RPMI 1640 cell culture media as described above. After synthesis and centrifugation, LRA-loaded LCNPs were resuspended in cell culture media and divided into individual microcentrifuge tubes for each time point per replicate. At each time point, replicate tubes were centrifuged at 14,000 rpm for 10 min. Supernatants were taken out and mixed with acetonitrile or DMSO at a ratio of 1:1 (v/v) and centrifuged to remove precipitated proteins. The LRA contents in the supernatant were measured by HPLC. The values were normalized to the total amount of LRA present. Release kinetics data were fitted in Eq. 3
| (3) |
where D(t) is the percentage of released drug and t is the time (days).
Efficacy and cytotoxicity of LRA-loaded LCNPs in J-Lat A1 cells
J-Lat Tat-GFP (A1) cells were obtained from the NIH AIDS Research and Reference Reagent Program (32, 33). Cells were maintained in RPMI 1640 media as described above and incubated at 37°C in a humidified 5% CO2 air environment. Cells were seeded at a concentration of 1 × 106 per well in 24-well culture plates and then treated with free LRAs or LCNP-formulated LRAs at different concentrations for 20 hours at 37°C. Subsequently, cells were washed with PBS and incubated with LIVE/DEAD violet dead cell stain for 30 min at RT. Cells were washed again and fixed in 2% paraformaldehyde (PFA) for flow cytometry analysis. The dose-response curves for each LRA formulation were fitted in the four-parameter log-logistic model, or Eq. 4, using the DRC package in R (39).
| (4) |
where x is the drug concentration and f(x) is the percentage of GFP+ cells.
Cytotoxicity was measured simultaneously with the efficacy study using CellTiter-Blue Cell Viability Assay (Promega) following the manufacturer’s recommended procedures. After cells were centrifuged and resuspended in PBS, 100 μl of cells from each well was incubated with a CellTiter-Blue reagent (20 μl) in a 96-well culture plate for 4 hours, and fluorescence was recorded at excitation/emission wavelengths of 560/590 nm using a fluorescent plate reader (Infinite 200 PRO, TECAN, Männedorf, Switzerland).
LRA combination effects and cytotoxicity were assessed in J-Lat A1 cells using the same methods. Cells were incubated with single LRAs at the concentrations that achieved approximately 20% GFP+ cell induction, as well as every pair of those LRAs, for 20 hours, followed by flow cytometry analysis and CellTiter-Blue assay as described above. The Bliss independence model was used to analyze combined effects of LRAs. The model was defined by Eq. 5
| (5) |
where faxy,P is the predicted fraction (here, the percentage of GFP+ A1 cells after treatment) affected by a combination of two LRAs, given the experimentally observed fraction affected by single-drug fax or fay individually. Then, the experimentally observed fraction affected by a combination of two LRAs (faxy,O) was compared with faxy,P using Eq. 6
| (6) |
If Δfaxy > 0 with statistical significance, then the two drugs display synergy. If Δfaxy = 0, then the two drugs show independent action. If Δfaxy < 0 with statistical significance, then the two drugs display antagonism.
Measurement of intracellular HIV-1 mRNA levels in CD4+ T cells from infected individuals
We measured intracellular HIV mRNA levels from CD4+ T cells isolated from three HIV+ PBMC donors from the HIV Vaccine Trials Network clinical cohort (institutional review board number: FH IR file #5567) in three independent experiments. CD4+ T cells were isolated by negative selection using the EasySep Human CD4+ T Cell Isolation Kit (STEMCELL Technologies) and plated in a 24-well plate at 1 × 106 cells/ml in 1 ml of RPMI 1640 cell culture media as described above with 10 μM T20 [fusion inhibitor; from AIDS, National Institute of Allergy and Infectious Diseases (NIAID)] to prevent new infections. We exposed the cells to six experimental conditions, all performed in triplicate: (i) Ing3A-LCNP (3 μg/ml) containing Ing3A (50 ng/ml), (ii) JQ1/LCNP (60 μg/ml) containing JQ1 (1020 ng/ml), (iii) both Ing3A-LCNP and JQ1/LCNP in the same concentrations as the single drug nanoparticle conditions, (iv) free Ing3A (10 ng/ml), (v) free JQ1 (4000 ng/ml), and (vi) both free Ing3A and free JQ1 in the same concentrations as the single free drug conditions. As a positive control for cell stimulation and production of HIV mRNA, we stimulated the cells with PMA (50 ng/ml) and ionomycin (1 μM). For a negative control, we added DMSO in an equal volume to the PMA/I. The cells were incubated with these treatments for 20 to 21 hours at 37°C and 5% CO2.
After incubation, the cells were harvested and an aliquot was taken for live/dead staining by flow cytometry. RNA was extracted from the remaining cells using the Qiagen RNeasy Mini RNA Extraction Kit (Qiagen). Complementary DNA (cDNA) was generated using the qScript cDNA Synthesis Kit (QuantaBio) starting with an equivalent input mass of RNA from each experimental replicate. After cDNA synthesis, the samples were diluted 1:1 with molecular-grade water. For the droplet digital polymerase chain reaction (ddPCR), we combined 5 μl of diluted cDNA, 11 μl of ddPCR supermix (no deoxyuridine triphosphate; Bio-Rad), 1.1 μl of HIV mRNA primer/probe (Integrated DNA Technologies), and 4.9 μl of water. Droplet generation, thermal cycling, and droplet reading were performed according to the Bio-Rad ddPCR supermix protocol. All ddPCRs were performed in duplicate, and wells with fewer than 10,000 droplets were excluded from the analysis.
Primer and probe sequences for detecting HIV-1 mRNAs (40) were as follows: forward, 5′-CAGATGCTGCATATAAGCAGCTG-3′ (9501–9523); reverse, 5′-TTTTTTTTTTTTTTTTTTTTTTTTGAAGCAC-3′ (9629–poly A); and probe, 5′-FAM-CCTGTACTGGGTCTCTCTGG-MGB-3′ (9531–9550).
The Bliss independence model, from Eqs. 5 and 6 above, was used to analyze the combined effects of JQ1 and Ing3A. However, here, fa is defined as the percentage of intracellular HIV-1 mRNA induction compared to the PMA/I positive control group.
Ex vivo activation study from pigtail macaque PBMCs
Pigtail macaque blood was obtained from the tissue distribution program at the Washington National Primate Research Center, University of Washington. All macaques were confirmed to be serologically negative for simian type D retrovirus, SIV, and simian T cell lymphotropic virus before sample collection. PBMCs were isolated from the blood using lymphocyte separation medium (Mediatech Inc.) and maintained in the same RPMI 1640 cell culture media for the study. PBMCs were seeded at a concentration of 1 × 106 per well in 24-well culture plates and then treated with free Ing3A, Ing3A-cbLCNP, Ing3A-CD4-cbLCNP, or Ing3A-Iso-cbLCNP for 20 hours at 37°C. Subsequently, cells were washed and incubated with FITC anti-human CD8, PE anti-human CD14, PerCP/Cy5.5 anti-human CD3 antibodies, and APC anti-human CD69 for 30 min at 4°C. The FITC IgG1, PE IgG2a, PerCP IgG1, and APC IgG1 control antibodies were used as the isotype control for gating cell populations. After incubation, cells were washed with PBS and then treated with LIVE/DEAD violet dead cell stain for another 30 min at RT, followed by 2% PFA fixation and flow cytometry analysis.
Biodistribution of CD4-targeted LCNPs in mice
Animal studies were approved by the Institutional Animal Care and Use Committee (IACUC) at the University of Washington (protocol #4260-01). All animals were obtained and cared for in accordance with the IACUC guidelines. Eight- to 12-week-old male C57BL/6J mice (The Jackson Laboratory) were subcutaneously administered with DiR-loaded LCNP formulations to the left flank (Fig. 4C). In our pilot animal studies, LCNPs with different lipid compositions (DOTAP versus chol-but) and sizes (100 nm versus 200 nm) as well as equivalent free DiR dye were administered into mice, and the biodistribution in different organs was compared at different time points. In our scaled-up studies, selected CD4-cbLCNPs or control Iso-cbLCNPs were administered into mice. At 20 hours, 3 days, and 7 days after administration, mice were sacrificed by cardiac exsanguination under isoflurane for terminal blood collection, followed by cervical dislocation. Major organs including inguinal LNs, spleen, brain, heart, kidneys, lung, liver, and small intestine were harvested, and the fluorescence signal was quantified by the Xenogen IVIS Spectrum Imaging System using excitation/emission wavelengths of 745/800 nm. Regions of interest were drawn across each organ for quantification, and total radiant efficiencies were normalized by tissue mass. Plasma (100 μl) from the blood was put in a 96-well plate for imaging, and the signals were quantified the same way as other tissues.
In vivo CD4+ T cell binding, activation, and toxicity study
Eight- to 12-week-old male C57BL/6J mice were injected subcutaneously with 0.3 ml of PBS, free Ing3A, Ing3A-CD4-cbLCNPs, or Ing3A-Iso-LCNPs at 0.48 mg/kg (Ing3A dose per body weight). Twenty-fold lower free Ing3A (0.024 mg/kg) was also injected because that concentration indicated similar potency compared to LCNP-formulated Ing3A in vitro. In addition, free Ing3A formulation at 0.48 mg/kg contained 0.64% DMSO in PBS buffer; thus, a 0.64% DMSO in PBS control was included to demonstrate the negligible effects by DMSO. After 20 hours, 3 days, and 7 days, mice were sacrificed, and left and right inguinal LNs were harvested. Lymphocytes were then collected from those LNs. In brief, the LNs were incubated in the digestion media [RPMI 1640 cell culture media containing collagenase D (1.5 mg/ml) and deoxyribonuclease I (40 μg/ml)] at 37°C in the shaker. After 30-min incubation, 5 mM EDTA was added to the solution and incubated for another 5 min. Tissues and solutions were then smashed and passed through a 70-μm cell strainer. Collected cells were then washed and stained with FITC anti-mouse CD8, PE anti-mouse CD14, PerCP/Cy5.5 anti-mouse CD3, and APC anti-mouse CD69 or their isotype antibodies for 30 min at 4°C. After incubation, cells were washed with PBS and then treated with LIVE/DEAD violet dead cell stain for another 30 min at RT, followed by 2% PFA fixation and flow cytometry analysis.
For in vivo cell-specific biodistribution studies, mice were injected with 0.3 ml of PBS, DiD/CD4-cbLCNP, or DiD/Iso-cbLCNP or equivalent free DiD (50 μg/kg; DiD dose per body weight). After 20 hours, mice were sacrificed, and left and right inguinal LNs were harvested. Cells were isolated from those LNs as described above and stained with fluorescent antibody (FITC anti-mouse CD8, PE anti-mouse CD14, and PerCP/Cy5.5 anti-mouse CD3 antibodies) and LIVE/DEAD violet dead cell stain, followed by 2% PFA fixation and flow cytometry analysis.
For toxicity studies, at 3 days after administration, inguinal LNs and biopsy of subcutaneous tissue near the injection sites were collected and postfixed in 4% PFA for 24 hours. Tissues were then processed and sectioned for hematoxylin and eosin staining, performed by the Pathology Research Services Laboratory at the University of Washington. Tissue slides were then examined under a Nikon Eclipse Ti microscope equipped with a Nikon Digital Sight DS-Fi2 camera.
Statistical analysis
Significant differences between control and treatment groups were determined by various statistical analyses. Student’s t test was used for two groups comparison. One-way analysis of variance (ANOVA) with Bonferroni’s test was used for multiple groups comparison. Two-way ANOVA with Bonferroni’s test was used when there were subgroups in each group, for example, different LNs or cells in those LNs from different treatment groups. Data represent means ± SD in each figure and table as indicated. Statistical analyses were performed using GraphPad Prism 7.0 software (GraphPad Software). *P < 0.05, **P < 0.005, ***P < 0.0005, and ****P < 0.0001.
Supplementary Material
Acknowledgments
The following reagent was obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH: J-Lat Tat-GFP cells (A1) from E. Verdin (32, 33). T20 (fusion inhibitor) was from AIDS, NIAID. Funding: This work was supported by the NIH (R33AI094412, R01AI116292-04, and UM1AI126623-03), amfAR (The Foundation for AIDS Research; 109541-61-RGRL), and CFAR award 2037020 (Prime P30 AI064518-13). S.D.S. was partially supported by the Mary Gates Endowment. Author contributions: S.C. designed and carried out experiments, performed data analysis, and wrote the manuscript. S.D.S. performed the experiments and data analysis, and contributed to manuscript writing. C.N.L. and S.M.H. acquired donor samples, performed ex vivo experiments from donor samples, analyzed data, and contributed to manuscript writing. Y.J. acquired NHP samples and performed cell isolation. C.Y. assisted in nanoparticle fabrication. P.R. performed data analysis. K.R.J., J.T.S., and F.H. contributed to project planning. K.A.W. provided project planning, experimental design, and data analysis and contributed to manuscript writing. All authors provided active and valuable feedback on the manuscript. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/5/3/eaav6322/DC1
Fig. S1. H-NMR analysis of PLGA, LRA, and PLGA-LRA conjugates.
Fig. S2. HPLC analysis of PLGA-LRA conjugates.
Fig. S3. Colloidal stability of LRA-loaded LCNPs.
Fig. S4. Flow cytometry dot plots showing the entire gating strategy used to identify GFP+ cell populations of J-Lat A1 cells.
Fig. S5. In vitro dose-response HIV-1 latency reversal and cytotoxicity by free butyric acid or its prodrug inserted into LCNPs.
Fig. S6. In vitro dose-response HIV-1 latency reversal and cytotoxicity by smaller Ing3A-cbLCNP compared to previous Ing3A-LCNP formulation.
Fig. S7. Comparison of CD69 expression between CD8+ and CD4+ T cells from NHP PBMCs after treating with Ing3A formulations.
Fig. S8. Pilot mouse studies comparing biodistribution of different LCNP formulations.
Fig. S9. Flow cytometry dot plots showing the entire gating strategy applied in Fig. 5.
Fig. S10. Representative images of mouse subcutaneous tissues at 3 days after administration of different Ing3A formulations.
Fig. S11. Targeted LCNP-formulated Ing3A is nontoxic to CD4+ and CD8+ T cells in mouse LNs after subcutaneous dosing.
Table S1. Physicochemical properties of LCNP-formulated LRAs with unsatisfactory low drug loading.
Table S2. Physicochemical properties of LCNPs made of various PLGAs.
Table S3. Parameters from fitting to LRA release kinetics.
Table S4. Parameters from fitting to LRA dose-response curve.
Table S5. Synthesis optimization for smaller LCNPs.
REFERENCES AND NOTES
- 1.Lorenzo-Redondo R., Fryer H. R., Bedford T., Kim E.-Y., Archer J., Kosakovsky Pond S. L., Chung Y.-S., Penugonda S., Chipman J. G., Fletcher C. V., Schacker T. W., Malim M. H., Rambaut A., Haase A. T., McLean A. R., Wolinsky S. M., Persistent HIV-1 replication maintains the tissue reservoir during therapy. Nature 530, 51–56 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fletcher C. V., Staskus K., Wietgrefe S. W., Rothenberger M., Reilly C., Chipman J. G., Beilman G. J., Khoruts A., Thorkelson A., Schmidt T. E., Anderson J., Perkey K., Stevenson M., Perelson A. S., Douek D. C., Haase A. T., Schacker T. W., Persistent HIV-1 replication is associated with lower antiretroviral drug concentrations in lymphatic tissues. Proc. Natl. Acad. Sci. U.S.A. 111, 2307–2312 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Freeling J. P., Ho R. J. Y., Anti-HIV drug particles may overcome lymphatic drug insufficiency and associated HIV persistence. Proc. Natl. Acad. Sci. U.S.A. 111, E2512–E2513 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eisele E., Siliciano R. F., Redefining the viral reservoirs that prevent HIV-1 eradication. Immunity 37, 377–388 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barton K., Winckelmann A., Palmer S., HIV-1 reservoirs during suppressive therapy. Trends Microbiol. 24, 345–355 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Siliciano J. D., Kajdas J., Finzi D., Quinn T. C., Chadwick K., Margolick J. B., Kovacs C., Gange S. J., Siliciano R. F., Long-term follow-up studies confirm the stability of the latent reservoir for HIV-1 in resting CD4+ T cells. Nat. Med. 9, 727–728 (2003). [DOI] [PubMed] [Google Scholar]
- 7.Crooks A. M., Bateson R., Cope A. B., Dahl N. P., Griggs M. K., Kuruc J. A. D., Gay C. L., Eron J. J., Margolis D. M., Bosch R. J., Archin N. M., Precise quantitation of the latent HIV-1 reservoir: Implications for eradication strategies. J. Infect. Dis. 212, 1361–1365 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carr A., Cooper D. A., Adverse effects of antiretroviral therapy. Lancet 356, 1423–1430 (2000). [DOI] [PubMed] [Google Scholar]
- 9.Margolis D. M., Garcia J. V., Hazuda D. J., Haynes B. F., Latency reversal and viral clearance to cure HIV-1. Science 353, aaf6517 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barouch D. H., Deeks S. G., Immunologic strategies for HIV-1 remission and eradication. Science 345, 169–174 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rasmussen T. A., Lewin S. R., Shocking HIV out of hiding: Where are we with clinical trials of latency reversing agents? Curr. Opin. HIV AIDS 11, 394–401 (2016). [DOI] [PubMed] [Google Scholar]
- 12.Delagrèverie H. M., Delaugerre C., Lewin S. R., Deeks S. G., Li J. Z., Ongoing clinical trials of human immunodeficiency virus latency-reversing and immunomodulatory agents. Open Forum Infect. Dis. 3, ofw189 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Laird G. M., Korin Bullen C., Rosenbloom D. I. S., Martin A. R., Hill A. L., Durand C. M., Siliciano J. D., Siliciano R. F., Ex vivo analysis identifies effective HIV-1 latency-reversing drug combinations. J. Clin. Invest. 125, 1901–1912 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Darcis G., Kula A., Bouchat S., Fujinaga K., Corazza F., Ait-Ammar A., Delacourt N., Melard A., Kabeya K., Vanhulle C., Van Driessche B., Gatot J.-S., Cherrier T., Pianowski L. F., Gama L., Schwartz C., Vila J., Burny A., Clumeck N., Moutschen M., De Wit S., Matija Peterlin B., Rouzioux C., Rohr O., Van Lint C., An in-depth comparison of latency-reversing agent combinations in various in vitro and ex vivo HIV-1 latency models identified bryostatin-1+JQ1 and ingenol-B+JQ1 to potently reactivate viral gene expression. PLOS Pathog. 11, e1005063 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiang G., Mendes E. A., Kaiser P., Wong D. P., Tang Y., Cai I., Fenton A., Melcher G. P., Hildreth J. E. K., Thompson G. R., Wong J. K., Dandekar S., Synergistic reactivation of latent HIV expression by ingenol-3-angelate, PEP005, targeted NF-kB signaling in combination with JQ1 induced p-TEFb activation. PLOS Pathog. 11, e1005066 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gray L. R., On H., Roberts E., Lu H. K., Moso M. A., Raison J. A., Papaioannou C., Cheng W.-J., Ellett A. M., Jacobson J. C., Purcell D. F. J., Wesselingh S. L., Gorry P. R., Lewin S. R., Churchill M. J., Toxicity and in vitro activity of HIV-1 latency-reversing agents in primary CNS cells. J. Neurovirol. 22, 455–463 (2016). [DOI] [PubMed] [Google Scholar]
- 17.Wong V. C., Fong L. E., Adams N. M., Xue Q., Dey S. S., Miller-Jensen K., Quantitative evaluation and optimization of co-drugging to improve anti-HIV latency therapy. Cell. Mol. Bioeng. 7, 320–333 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Estes J. D., Kityo C., Ssali F., Swainson L., Makamdop K. N., Del Prete G. Q., Deeks S. G., Luciw P. A., Chipman J. G., Beilman G. J., Hoskuldsson T., Khoruts A., Anderson J., Deleage C., Jasurda J., Schmidt T. E., Hafertepe M., Callisto S. P., Pearson H., Reimann T., Schuster J., Schoephoerster J., Southern P., Perkey K., Shang L., Wietgrefe S. W., Fletcher C. V., Lifson J. D., Douek D. C., McCune J. M., Haase A. T., Schacker T. W., Defining total-body AIDS-virus burden with implications for curative strategies. Nat. Med. 23, 1271–1276 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Di Mascio M., Paik C. H., Carrasquillo J. A., Maeng J.-S., Jang B.-S., Soo Shin I., Srinivasula S., Byrum R., Neria A., Kopp W., Catalfamo M., Nishimura Y., Reimann K., Martin M., Clifford Lane H., Noninvasive in vivo imaging of CD4 cells in simian-human immunodeficiency virus (SHIV)–infected nonhuman primates. Blood 114, 328–337 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Westermann J., Pabst R., Distribution of lymphocyte subsets and natural killer cellsin the human body. Clin. Investig. 70, 539–544 (1992). [DOI] [PubMed] [Google Scholar]
- 21.Cao S., Woodrow K. A., Nanotechnology approaches to eradicating HIV reservoirs. Eur. J. Pharm. Biopharm. (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Freeling J. P., Koehn J., Shu C., Sun J., Ho R. J. Y., Anti-HIV drug-combination nanoparticles enhance plasma drug exposure duration as well as triple-drug combination levels in cells within lymph nodes and blood in primates. AIDS Res. Hum. Retroviruses 31, 107–114 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cao S., Jiang Y., Zhang H., Kondza N., Woodrow K. A., Core-shell nanoparticles for targeted and combination antiretroviral activity in gut-homing T cells. Nanomedicine 14, 2143–2153 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kovochich M., Marsden M. D., Zack J. A., Activation of latent HIV using drug-loaded nanoparticles. PLOS ONE 6, e18270 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Buehler D. C., Marsden M. D., Shen S., Toso D. B., Wu X., Loo J. A., Hong Zhou Z., Kickhoefer V. A., Wender P. A., Zack J. A., Rome L. H., Bioengineered vaults: Self-assembling protein shell–lipophilic core nanoparticles for drug delivery. ACS Nano 8, 7723–7732 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tang X., Liang Y., Liu X., Zhou S., Liu L., Zhang F., Xie C., Cai S., Wei J., Zhu Y., Hou W., PLGA-PEG nanoparticles coated with anti-CD45RO and loaded with HDAC plus protease inhibitors activate latent HIV and inhibit viral spread. Nanoscale Res. Lett. 10, 413 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jayant R. D., Atluri V. S., Agudelo M., Sagar V., Kaushik A., Nair M., Sustained-release nanoART formulation for the treatment of neuroAIDS. Int. J. Nanomedicine 10, 1077–1093 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Das B., Dobrowolski C., Shahir A.-M., Feng Z., Yu X., Sha J., Bissada N. F., Weinberg A., Karn J., Ye F., Short chain fatty acids potently induce latent HIV-1 in T-cells by activating P-TEFb and multiple histone modifications. Virology 474, 65–81 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Park J., Kim M., Kang S. G., Jannasch A. H., Cooper B., Patterson J., Kim C. H., Short-chain fatty acids induce both effector and regulatory T cells by suppression of histone deacetylases and regulation of the mTOR-S6K pathway. Mucosal Immunol. 8, 80–93 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brioschi A., Zara G., Calderoni S., Gasco M., Mauro A., Cholesterylbutyrate solid lipid nanoparticles as a butyric acid prodrug. Molecules 13, 230–254 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang F., Bian C., Zhu L., Zhao G., Huang Z., Huang M., Effect of human serum albumin on drug metabolism: Structural evidence of esterase activity of human serum albumin. J. Struct. Biol. 157, 348–355 (2007). [DOI] [PubMed] [Google Scholar]
- 32.Jordan A., Bisgrove D., Verdin E., HIV reproducibly establishes a latent infection after acute infection of T cells in vitro. EMBO J. 22, 1868–1877 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jordan A., Defechereux P., Verdin E., The site of HIV-1 integration in the human genome determines basal transcriptional activity and response to Tat transactivation. EMBO J. 20, 1726–1738 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cao S., Jiang Y., Levy C. N., Hughes S. M., Zhang H., Hladik F., Woodrow K. A., Optimization and comparison of CD4-targeting lipid–polymer hybrid nanoparticles using different binding ligands. J. Biomed. Mater. Res. A 106, 1177–1188 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jiang H., Wang Q., Sun X., Lymph node targeting strategies to improve vaccination efficacy. J. Control. Release 267, 47–56 (2017). [DOI] [PubMed] [Google Scholar]
- 36.Beans E. J., Fournogerakis D., Gauntlett C., Heumann L. V., Kramer R., Marsden M. D., Murray D., Chun T.-W., Zack J. A., Wender P. A., Highly potent, synthetically accessible prostratin analogs induce latent HIV expression in vitro and ex vivo. Proc. Natl. Acad. Sci. U.S.A. 110, 11698–11703 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gupta A. K., Paquet M., Ingenol mebutate: A promising treatment for actinic keratoses and nonmelanoma skin cancers. J. Cutan. Med. Surg. 17, 173–179 (2013). [DOI] [PubMed] [Google Scholar]
- 38.Duncan I. W., Culbreth P. H., Burtis C. A., Determination of free, total, and esterified cholesterol by high-performance liquid chromatography. J. Chromatogr. B Biomed. Sci. Appl. 162, 281–292 (1979). [DOI] [PubMed] [Google Scholar]
- 39.Ritz C., Baty F., Streibig J. C., Gerhard D., Dose–response analysis using R. PLOS ONE 10, e0146021 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bullen C. K., Laird G. M., Durand C. M., Siliciano J. D., Siliciano R. F., New ex vivo approaches distinguish effective and ineffective single agents for reversing HIV-1 latency in vivo. Nat. Med. 20, 425–429 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/5/3/eaav6322/DC1
Fig. S1. H-NMR analysis of PLGA, LRA, and PLGA-LRA conjugates.
Fig. S2. HPLC analysis of PLGA-LRA conjugates.
Fig. S3. Colloidal stability of LRA-loaded LCNPs.
Fig. S4. Flow cytometry dot plots showing the entire gating strategy used to identify GFP+ cell populations of J-Lat A1 cells.
Fig. S5. In vitro dose-response HIV-1 latency reversal and cytotoxicity by free butyric acid or its prodrug inserted into LCNPs.
Fig. S6. In vitro dose-response HIV-1 latency reversal and cytotoxicity by smaller Ing3A-cbLCNP compared to previous Ing3A-LCNP formulation.
Fig. S7. Comparison of CD69 expression between CD8+ and CD4+ T cells from NHP PBMCs after treating with Ing3A formulations.
Fig. S8. Pilot mouse studies comparing biodistribution of different LCNP formulations.
Fig. S9. Flow cytometry dot plots showing the entire gating strategy applied in Fig. 5.
Fig. S10. Representative images of mouse subcutaneous tissues at 3 days after administration of different Ing3A formulations.
Fig. S11. Targeted LCNP-formulated Ing3A is nontoxic to CD4+ and CD8+ T cells in mouse LNs after subcutaneous dosing.
Table S1. Physicochemical properties of LCNP-formulated LRAs with unsatisfactory low drug loading.
Table S2. Physicochemical properties of LCNPs made of various PLGAs.
Table S3. Parameters from fitting to LRA release kinetics.
Table S4. Parameters from fitting to LRA dose-response curve.
Table S5. Synthesis optimization for smaller LCNPs.


