Summary
Protein transport into the nucleus is mediated by transport receptors. Import of highly charged proteins, such as histone H1 and ribosomal proteins, requires a dimer of two transport receptors. In this study, we determined the cryo-EM structure of the Imp7:Impβ:H1.0 complex, showing that the two importins form a cradle that accommodates the linker histone. The H1.0 globular domain is bound to Impβ, whereas the acidic loops of Impβ and Imp7 chaperone the positively charged C-terminal tail. Although it remains disordered, the H1 tail serves as a zipper that closes and stabilizes the structure through transient non-specific interactions with importins. Moreover, we found that the GGxxF and FxFG motifs in the Imp7 C-terminal tail are essential for Imp7:Impβ dimerization and H1 import, resembling importin interaction with nucleoporins, which, in turn, promote complex disassembly. The architecture of many other complexes might be similarly defined by rapidly exchanging electrostatic interactions mediated by disordered regions.
Keywords: nuclear import, histone H1, Imp7, Impβ, transport receptors, karyopherins, cryo-EM, disordered proteins, IDP
Graphical Abstract
Highlights
-
•
Importin 7 and Importin β form a cradle to chaperone and transport histone H1
-
•
Transient and non-specific electrostatic interactions form and shape the complex
-
•
The H1 tail serves as a zipper that closes and stabilizes Imp7:Impβ:H1
-
•
FxFG motifs in nucleoporins facilitate RanGTP-dependent disassembly of the complex
Ivic et al. determined the cryo-EM structure of the Imp7:Impβ:H1 complex and show that transient and non-specific charged interactions form and shape the complex. They also found that the FxFG motif in Imp7 is essential for complex formation and that nucleoporins promote complex disassembly.
Introduction
In eukaryotic cells, proteins are guided by their targeting sequences into different compartments. Nuclear proteins contain nuclear localization or nuclear export signals that are recognized by transport receptors (Xu et al., 2010). These receptors mediate transport through large channels called nuclear pore complexes (NPCs) (Adams and Wente, 2013, Beck and Hurt, 2017). Although the exact transport mechanism remains unknown (Hayama et al., 2017, Timney et al., 2016), it has been shown that karyopherins bind highly flexible Phe-Gly (FG) repeats of nucleoporins, which are the building blocks of NPCs (Bayliss et al., 2000, Bayliss et al., 2002, Zahn et al., 2016). One-third of all nucleoporins contain multiple intrinsically disordered FG repeat regions that are oriented toward the center of the pore and promote active translocation through weak and transient interactions with the nuclear transport receptors (Hough et al., 2015, Milles et al., 2015, Raveh et al., 2016). On the nuclear side, the guanosine triphosphate (GTP)-bound form of the Ran protein (RanGTP) binds importins and releases the cargo (Görlich et al., 1996, Stewart, 2007).
Although most proteins are transported by a single β-karyopherin transport receptor, some require two independent receptors for their transport through the NPC. The import of linker histones requires the formation of a heterodimeric transport complex consisting of two members of importin β family, importin β (Impβ) and importin 7 (Imp7) (Jäkel et al., 1999). The heterodimeric Imp7:Impβ complex is also required for the import of the ribosomal proteins rpL4 and rpL6 and HIV integrase (Fassati et al., 2003, Jäkel et al., 2002), whereas a homodimer of the Crm1 export receptor is required for the export of HIV RNA (Booth et al., 2014). These data imply that dimerization of autonomous transport receptors is required for the transport of highly charged proteins that interact with nucleic acids and are prone to aggregation.
Histones are small basic proteins that pack DNA into nucleosomes, the fundamental structural units of chromatin. Although small enough to diffuse into the nucleus, histones are bound by specific chaperons and are actively imported by karyopherins (Baake et al., 2001, Keck and Pemberton, 2013, Mosammaparast et al., 2002, Mühlhäusser et al., 2001). Active transport of histones is required to ensure their rapid and efficient concentration in the nucleus during S phase and is essential for chromatin assembly (Mosammaparast et al., 2005). Linker histones bind to linker DNA between two nucleosome core particles and are essential for higher-order chromatin packing, gene expression, and genomic stability (Hergeth and Schneider, 2015, Torres et al., 2016, Fyodorov et al., 2018).
The formation of the Imp7:Impβ heterodimer and its interaction with its cargo are poorly understood. To address this, we determined a cryoelectron microscopy (cryo-EM) structure of Imp7:Impβ in a complex with the linker histone H1.0. The structure reveals why two importins are required for H1 import. The H1.0 globular domain is bound by Impβ only, whereas both Impβ and Imp7 bind and chaperone the highly positively charged and disordered C-terminal tail of H1.0. Cross-linking mass spectrometry reveals that H1 C-terminal tail residues come into close proximity with many different importin residues, indicating that the H1 tail remains disordered in the complex. Our data show that the transient electrostatic interactions between the H1.0 tail and importins stabilize the entire complex. Moreover, in the Imp7 C-terminal tail, we observed conserved GGxxF and FxFG motifs, which are commonly found in FG-nucleoporins. We show that these motifs are essential for complex formation and for the import of linker histones, indicating that Imp7 and nucleoporins use the same motif and interface to bind Impβ. Furthermore, we show that FxFG motifs in nucleoporins facilitate RanGTP-dependent complex disassembly when the complex has translocated through the pore. Our data show that the disordered H1 and Imp7 tails are required for formation of the complex shape and structure by rapidly exchanging non-specific interactions with importins.
Results
Cryo-EM Structure of the Imp7:Impβ:H1.0 Complex
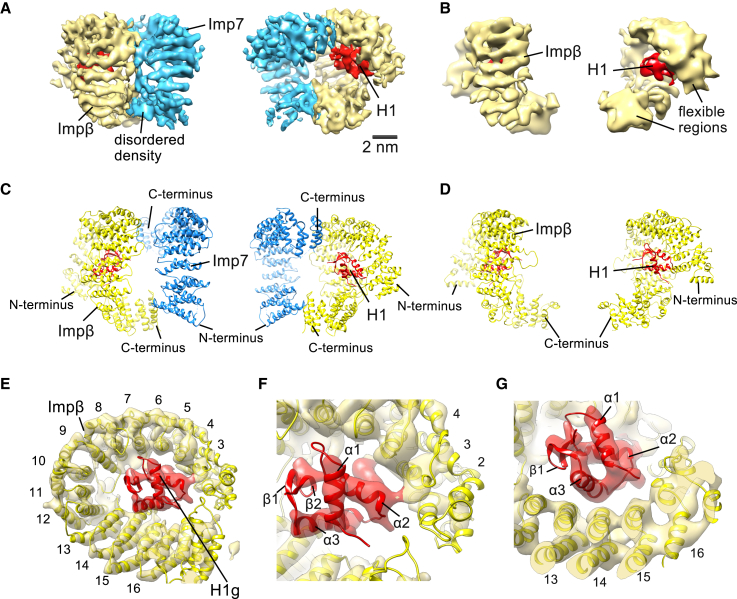
To prepare a homogeneous sample, we purified individual components from E. coli, reconstituted the Imp7:Impβ:H1.0 complex, and isolated it by size exclusion chromatography (Figures S1A–S1C; Ivic et al., 2017). The presence of H1.0 in the complex was confirmed by western blot analysis (Figure S1D). We used the random conical tilt reconstruction method (Radermacher, 1988) to generate an initial model of the Imp7:Impβ:H1.0 complex (Figures S1E–S1G). The random conical tilt model was further refined and used as a reference for subsequent structure determination using cryo-EM and single-particle analysis (Figures S1H and S1I). Extensive 3D classification yielded two major classes that were refined to a final resolution of 6.2 Å and 7.5 Å, respectively (Figures 1A, 1B, S2, S3A, and S3B; Table 1). In both maps, we could identify a curved density with rod-like features characteristic of the α-helical HEAT repeats as well as the winged helix domain of the linker histone (Figures 1A and 1B). The first map, refined to 6.2 Å, is larger, and the density contains two importins and histone H1 (Figure 1A). The smaller second map, at 7.5 Å, appears to have only one importin and histone H1 (Figure 1B). Because biochemical data (Bäuerle et al., 2002) show that Impβ interacts with the H1 globular domain, we conclude that the smaller structure represents Impβ and the H1 globular domain. This implies that the additional density of the larger structure is Imp7. We have also solved a 9.2 Å structure of Imp7:Impβ:H1.0 that was not cross-linked (Figure S3C). This structure has the same appearance as the cross-linked sample, showing that cross-linking does not affect the complex arrangement. According to the local resolution (Kucukelbir et al., 2014), the middle region of the complexes is better resolved than the peripheral regions, confirming the flexible nature of importin molecules (Cingolani et al., 2000, Conti et al., 2006; Figure S3D).
Figure 1.
Structures of Imp7:Impβ:H1.0 and Impβ:H1.0
(A) Cryo-EM map of the Imp7:Impβ:H1.0 complex resolved to 6.2 Å. Impβ is shown in yellow, Imp7 in blue, and H1 in red. Contour level, ∼0.04.
(B) Cryo-EM map of the Impβ:H1.0 complex resolved to 7.5 Å. Contour level, ∼0.04.
(C) Model of the Imp7:Impβ:H1.0 complex.
(D) Model of the Impβ:H1.0 complex.
(E) Model of Impβ (yellow) and the H1.0 globular domain (red) fitted into the cryo-EM map of Imp7:Impβ:H1.0 (transparent). For clarity, only Impβ is shown, and the Impβ HEAT repeats are numbered. Contour level, ∼0.04.
(F and G) Close-up view showing H1.0 (red) in the cradle of Impβ. H1.0 interacts with Impβ HEAT repeat 3 (F) and HEAT repeats 13 and 14 (G). Contour level, ∼0.04.
See also Figures S1–S4.
Table 1.
Cryo-EM Data Collection, Refinement, and Validation Statistics
| #1 Imp7:Impβ:H1.0 (EMDB-0366) (PDB: 6N88) | #2 Impβ:H1.0 (EMDB-0367) (PDB: 6N89) | #3 Imp7:Impβ (EMDB-0368) | |
|---|---|---|---|
| Data Collection and Processing | |||
| Magnification | 78,000 | 78,000 | 78,000 |
| Voltage (kV) | 300 | 300 | 300 |
| Electron exposure (e–/Å2) | 80 | 80 | 80 |
| Defocus range (μm) | −1 to −4 | −1 to −4 | −1 to −4 |
| Pixel size (Å) | 1.36 | 1.36 | 1.36 |
| Symmetry imposed | no | no | no |
| Initial particle images (no.) | 415,000 | 415,000 | 415,000 |
| Final particle images (no.) | 18 900 | 15 800 | 7 600 |
| Map resolution (Å) Fourier shell correlation (FSC) threshold | 6.2 | 7.5 | 10.4 |
| Map resolution range (Å) | 6−8 | 7.5−10 | 10−15 |
| Refinement | |||
| Initial model used (PDB code) | 3W5K | 3W5K | |
| Model resolution range (Å) | 6–8 | 7.5–10 | |
| Map sharpening B factor (Å2) | −150 | −200 | |
| Model Composition | |||
| Non-hydrogen Atoms | |||
| Protein residues | 1,561 | 947 | |
| Ligands | 0 | 0 | |
| Root-Mean-Square Deviations | |||
| Bond lengths (Å) | 0.01 | 0.01 | |
| Bond angles (°) | 2.23 | 1.71 | |
| Validation | |||
| MolProbity score | 2.56 | 2.05 | |
| Clashscore | 41.84 | 10.45 | |
| Poor rotamers (%) | 0.00 | 0.00 | |
| Ramachandran Plot | |||
| Favored (%) | 93.57 | 91.41 | |
| Allowed (%) | 5.47 | 7.10 | |
| Disallowed (%) | 0.96 | 1.48 | |
Based on the subunit assignment, we fitted the crystal structure of Impβ (Choi et al., 2014) and homology models of Imp7 and H1.0 into the cryo-EM densities (Figures S3E–S3H). Impβ consists of 19 HEAT repeats and Imp7 of 20 HEAT repeats, as shown by multiple secondary structure predictions and homology models (Figure S3I). Our biochemical assays and those of other groups show that the N-terminal part of Impβ interacts with the C-terminal end of Imp7 (Bäuerle et al., 2002, Jäkel et al., 1999; Figure S4A). Therefore, we positioned Imp7 in an anti-parallel manner with respect to Impβ (Figure 1C). The orientation of Imp7 in the structure was further confirmed through chemical cross-links identified by mass spectrometry, where lysine side chains were linked through succinimide esters (Herzog et al., 2012, Leitner et al., 2014; Figures S4B and S4C; see also Mendeley Data: https://doi.org/10.17632/dfjzx4sxrs.1). Although most cross-links involved disordered regions, we obtained multiple lysine-lysine cross-links between Imp7, Impβ, and H1 that are consistent with the orientation of the components in the structure (Figures S4B and S4C; see also Mendeley Data). The model for the H1.0 globular domain was fitted into the remaining α-helical density in a cradle formed by Impβ (Figure 1).
In both cryo-EM maps, the importins show a curved solenoid structure typical of the β-karyopherin family of proteins (Christie et al., 2016; Figure 1). In the Impβ:H1.0 structure, the middle HEAT repeat region (comprising HEAT repeats 9–15) of Impβ is structurally nearly identical to that of Impβ in the Imp7:Impβ:H1.0 complex. In contrast, the N- and C-terminal HEAT repeat regions show greater flexibility and are in a more open conformation in the Impβ:H1.0 complex (Figures S4D and S4E), indicating that binding of Imp7 to the Impβ complex rigidifies the structure.
Notably, six N-terminal HEAT repeats of Imp7 were visible only in a lower-resolution map obtained as an intermediate during the 3D classification (Figure S4F). In the final map, these HEAT repeats are poorly visible, indicating a high degree of flexibility. This is consistent with our data and that of others showing that the N-terminal part of Imp7 is dispensable for complex formation (Bäuerle et al., 2002, Jäkel et al., 1999).
Impβ Binds the H1.0 Globular Domain
The H1.0 globular domain (H1.0g) is located in the cradle of Impβ (HEAT repeats 3–16) in both structures (Figure 1). The N- and C-terminal regions of Impβ come into close proximity (HEAT repeats 2 and 17) and surround the H1.0 globular domain (Figures 1E–1G and S4E). Consistent with our structures, biochemical analysis shows that Impβ HEAT repeats 4–14 (residues 127–641) are required for H1 binding (Wohlwend et al., 2007). The cryo-EM structure reveals that the α2 helix of H1.0 interacts with Impβ HEAT repeat 3 and that the H1.0 α3 helix is in close proximity to Impβ HEAT repeats 13 and 14 (Figures 1F and 1G). Notably, the H1 binding site in Impβ overlaps with the RanGTP binding site (Cingolani et al., 1999, Vetter et al., 1999), which is consistent with RanGTP dissociating Impβ from the trimeric complex (Jäkel et al., 1999; Figure S4G).
The C-Terminal Tail of H1 Shapes the Imp7:Impβ:H1.0 Complex
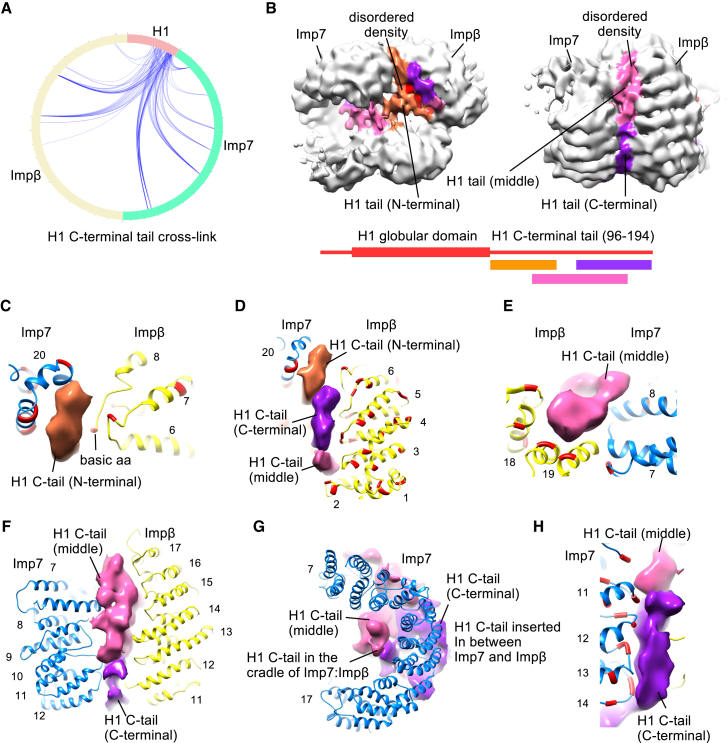
In the cryo-EM map, we observed an additional density between the two importins and in the cradle of Imp7 that does not belong to the structured parts of the complex (Figure 1A). This density is more pronounced at a lower contour level and is generated by one or more disordered regions: (1) an ∼100-amino acid (aa) basic H1 C-terminal tail with a net charge of +40 (residues 97–194), (2) an ∼90-aa loop of Imp7 connecting HEAT repeats 19 and 20 with a net charge of –39 (residues 872–955), and (3) the last 40 aa of the Imp7 C-terminal tail (Figures S3E and S3G). These disordered regions were assigned in the map with the help of cross-links detected by mass spectrometry. In cross-linking, aspartate and glutamate carboxy groups were activated to react directly with the primary amines of lysine side chains, also known as zero-length cross-links (Herzog et al., 2012, Leitner et al., 2014). In our structure, we mapped the Imp7 and Impβ residues that cross-link and show direct interaction with ∼30 lysines in the H1.0 C-terminal tail (Figures 2A and S5A; see also Mendeley Data). It is noteworthy that an individual lysine in H1.0 cross-links to multiple acidic residues in Imp7 and Impβ and that a single acidic residue in the importins cross-links to many lysines in H1.0 (Figure 2A). This indicates that, in different particles, different regions of H1 interact with importins. Our data show that the linker histone tail interactions do not require a defined binding site or specific residues. On the contrary, the H1.0 C-terminal tail remains disordered and can adopt multiple conformations, which explains why there is less defined density between HEAT repeats and in the cradle of importins. We observed that many Imp7 and Impβ residues preferentially cross-link to the N-terminal, middle, or C-terminal part of the H1.0 tail (Figure S5A). Using this information, we mapped the regions in disordered density that are enriched in the N-terminal, middle, and C-terminal part of the H1.0 tail (Figures 2B and S5B).
Figure 2.
A Disordered Region of H1 Shapes the Imp7:Impβ:H1.0 Complex
(A) Circular plot showing zero-length cross-links of the H1 C-terminal tail to Impβ and Imp7. Each Imp7 and Impβ residue cross-links to many lysines in the H1 C-terminal tail. The cross-links are shown in our Mendeley Data.
(B) The disordered density in the cradle and between two importins was segregated based on the cross-linking and mass spectrometry data. The density in proximity to the importin residues that cross-link predominantly to the N-terminal part of the H1 C-terminal tail is shown in orange, the middle part in pink, and the C-terminal part in violet. Contour level, ∼0.025.
(C) Close-up view of Imp7 HEAT repeat 20 and Impβ HEAT repeats 6 and 7. The N-terminal part of the H1 C-terminal tail inserts between the Impβ and Imp7 HEAT repeats and mediates interaction. The lysine-rich H1 tail interacts with the negatively charged residues (red) in the importins. Contour level, ∼0.025.
(D) Acidic residues (red) in the inner loops of Impβ N-terminal HEAT repeats 1–6 interact with the middle and C-terminal parts of the H1 C-terminal tail. Contour level, ∼0.025.
(E) Close-up view of Imp7 HEAT repeat 7 and Impβ HEAT repeats 18 and 19. The middle part of the H1 C-terminal tail inserts between the Impβ and Imp7 HEAT repeats and mediates interaction. The H1 tail interacts with the negatively charged residues (red) in the importins in the region. Contour level, ∼0.025.
(F) The middle and C-terminal parts of the H1 C-terminal tail insert between Impβ HEAT repeats 11–17 and Imp7 HEAT repeats 7–12. The disordered H1 tail holds the two importins together.
(G) The middle part of the H1 C-terminal tail is partially localized in the cradle formed by Impβ and Imp7. Contour level, ∼0.025.
(H) Acidic residues (red) in the inner loops of Imp7 HEAT repeats 11–14 interact with the middle and C-terminal parts of the H1 C-terminal tail. Contour level, ∼0.025.
See also Figure S5.
The HEAT repeats of Imp7 and Impβ are in close proximity at two locations (Figure S5C). The first such contact is between the inner helix of Imp7 HEAT repeat 20 and the loop connecting Impβ HEAT repeats 7 and 8 (Figures 2C and S5D). The N-terminal part of the H1 C-terminal tail is localized near this contact. The highly positively charged H1 tail inserts between Impβ HEAT repeat 6 and Imp7 HEAT repeat 20, both of which contain several acidic residues (Figure 2C). This indicates that the H1 tail mediates interaction between Imp7 and Impβ and stabilizes the contact between the two importins. The middle and C-terminal parts of the H1 tail protrude along Impβ HEAT repeats 1–6. The inner loops of these HEAT repeats are negatively charged and bind the positively charged H1 tail, showing that Impβ serves as a transport receptor and a chaperone for the H1 tail (Figure 2D).
The second area of contact between Imp7 and Impβ HEAT repeats comprises the inner helix of Impβ HEAT repeat 19 and the loop of Imp7 HEAT repeat 7 (Figures S5C and S5E). Near this interaction, we observed the density for the middle part of the H1 tail (Figure 2E). The H1 tail inserts between Impβ HEAT repeats 18–19 and Imp7 HEAT repeats 7–8, and it interacts with negatively charged amino acids in the region. The middle and C-terminal parts of the H1.0 tail extend along Impβ HEAT repeats 10–19 and Imp7 HEAT repeats 8–13, forming a large contact area between the two importins (Figures 2F and 2G). In this region, the inner loops of importins contain many negatively charged aa and bind the positively charged H1 C-terminal tail (Figure 2H). These nonspecific electrostatic interactions glue together two importins and define the structure and stability of the complex.
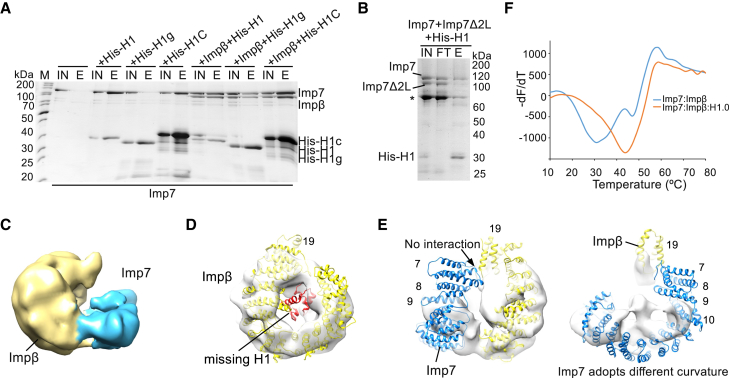
The results obtained from the cryo-EM structure and in previous studies (Bäuerle et al., 2002) agree with those of our biochemical assays. Imp7 binds the C-terminal tail of H1.0, whereas Impβ binds both the globular domain and the tail (Figure 3A). Imp7 also contains a large acidic loop between HEAT repeats 19 and 20, and this loop might chaperone the H1 tail located in the cradle that does not make direct contact with the structured part of the Imp7:Impβ complex (Figure 2G). To verify these findings, we replaced the acidic residues in the loop (aa 884–912 and aa 923–954) with a glycine-serine linker and generated the Imp7Δ2L mutant. Imp7Δ2L can still bind Impβ, indicating that the acidic loop is not essential for importin dimerization (Figure S5F). In a competitive pull-down assay, however, H1.0 interacted much more strongly with the full-length Imp7 than with the Imp7Δ2L mutant, suggesting that the large Imp7 acidic loop binds H1.0 (Figure 3B).
Figure 3.
The H1 C-Terminal Tail Stabilizes and Shapes Imp7:Impβ:H1.0
(A) SDS-PAGE analysis of a pull-down experiment. His-tagged wild-type full-length H1.0 or its domains was used as bait to investigate H1 interaction with Imp7 and Impβ. H1g, H1 globular domain; H1C, H1 C-terminal domain; IN, input; E, elution; M, protein marker.
(B) Results of a competition pull-down assay showing that Imp7 acidic loops contribute to H1 binding. His-H1.0 is bound to the nickel-nitrilotriacetic acid (Ni-NTA) resin. Equal amounts of Imp7 and the Imp7Δ2L were added to the assay. The Imp7Δ2L mutant lacks two acidic loops from the unstructured region between HEAT repeats 19 and 20. FT, flowthrough. A degradation product of Imp7Δ2L is labeled with an asterisk.
(C) Cryo-EM map of the Imp7:Impβ complex lacking density for H1.0 resolved to 10.4 Å. Impβ is shown in yellow, and Imp7 is shown in blue. Contour level, ∼0.035.
(D) Model of the Imp7:Impβ:H1.0 complex superimposed onto the Imp7:Impβ map. Impβ and H1.0 are shown in yellow and red, respectively. H1 is not bound by Impβ. Contour level, ∼0.035.
(E) Model of the Imp7:Impβ:H1.0 complex superimposed onto the Imp7:Impβ map. Imp7 and Impβ are shown in blue and yellow, respectively. The N-terminal part of Imp7 is not visible. Imp7 adopts a different curvature. Contour level, ∼0.035.
(F) TSA curves of the Imp7:Impβ:H1.0 and Imp7:Impβ complexes, showing that Imp7:Impβ:H1.0 is more stable than Imp7:Impβ.
See also Figure S5.
H1 Determines Complex Stability
Our data show that H1.0 brings together Imp7 and Impβ and imply that the substrate is essential for the complex to adopt its structure. This suggests that the Imp7:Impβ dimeric complex might have a different conformation in the absence of bound cargo. Notably, in our cryo-EM data, we found a class that had a weaker density for the H1.0 globular domain. Further classification revealed a subclass in which the H1.0 globular domain is not bound to Impβ (Figures 3C and S5G). As we used local angular searches for the classification, the overall orientation of the complex was preserved, enabling us to assign subunits. Although Impβ adopts a similar conformation, the density for Imp7 is weaker and incomplete in this class, indicating that Imp7 is loosely bound to Impβ in the absence of H1.0 (Figures 3C, 3D, and S5H).
In the Imp7:Impβ complex, we observed two major contacts. The first is formed near the contact point in the trimeric complex and involves Imp7 HEAT repeat 20 and Impβ HEAT repeats 7–8. The second contact is formed between Imp7 HEAT repeats 11–12 and Impβ HEAT repeats 12–13. At both contact points, the importins adopt curvature in Imp7:Impβ that differs from that in the Imp7:Impβ:H1 complex (Figure S5I). Moreover, the density for Imp7 HEAT repeats 1–9 and Impβ HEAT repeats 18 and 19 is missing in the Imp7:Impβ complex (Figure 3E). These HEAT repeats combine with the H1.0 tail to form the large interface between the two importins in Imp7:Impβ:H1.0 (Figures 2E, S5C, and S5E), which is missing in Imp7:Impβ. The remaining Imp7 HEAT repeats, 10–19, are visible in our map but have different curvature in the absence of H1.0, indicating that the H1 tail is required for the trimeric complex to adopt its structure (Figure 3E). In conclusion, our data show that the linker histone is required for the formation of the interface between Imp7 and Impβ, underlining the importance of the H1 C-terminal tail for the architecture of the entire complex.
Consistent with the cryo-EM structure and biochemical observations, a thermal shift assay (TSA) (Choudhary et al., 2017) showed that the Imp7:Impβ:H1.0 complex is more stable than the Imp7:Impβ complex (Figure 3F).
Taken together, our results show that the two importins interact through a large surface and form a cradle to bind and chaperone histone H1. Disordered regions of the substrate, i.e., the H1 C-terminal tail, mediate interactions with the transport receptors and are crucial for the shape and structure of the entire complex. Cross-linking and structural data show that the H1 C-terminal tail, although in a confined space, remains disordered in the complex and that the binding is achieved by many non-specific and transient electrostatic interactions.
The FxFG Motif in the Imp7 C Terminus Is Critical for Impβ Binding and H1 Import
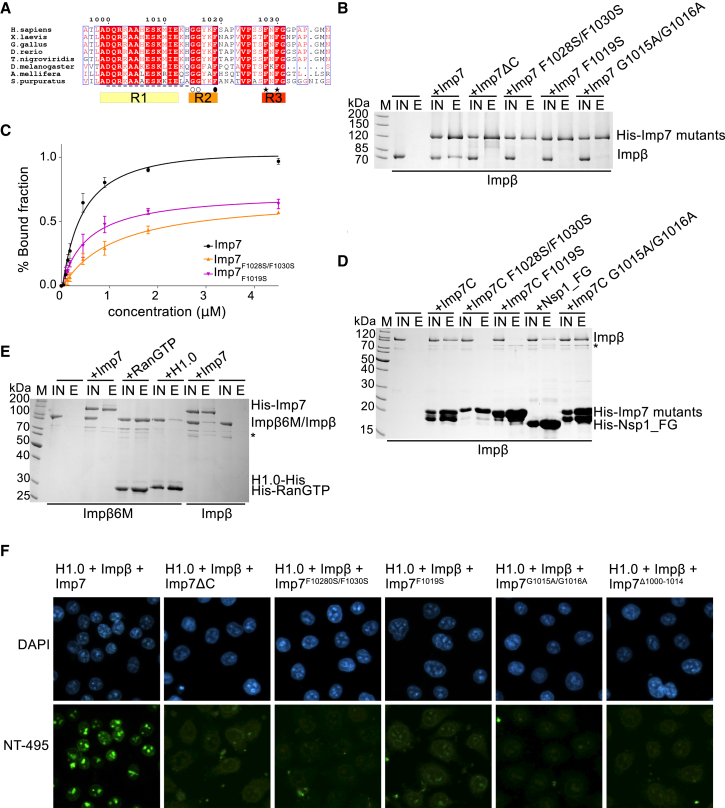
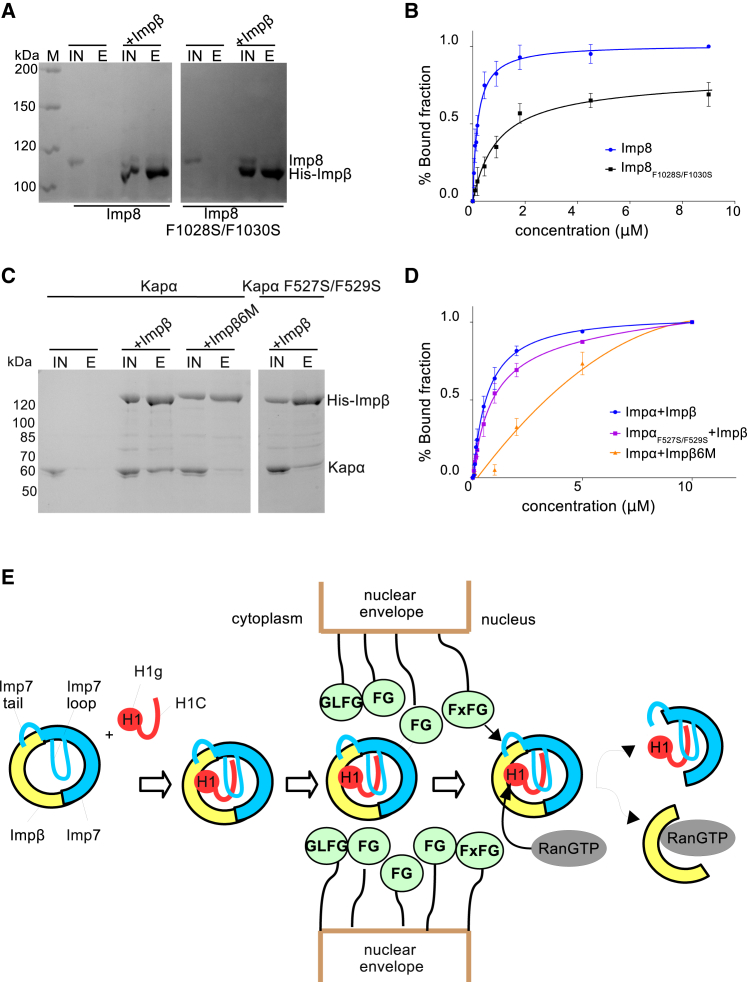
The third flexible region in the Imp7:Impβ:H1.0 complex is the C terminus of Imp7. The Imp7 C-terminal tail binds Impβ aa 203–362 and is essential for complex formation (Jäkel et al., 1999). We noticed three highly conserved regions in the Imp7 tail, which suggests their functional role (Figure 4A). The first conserved region (R1) is rich in charged residues and is followed by the GGxxF motif (R2) and finally by the FxFG motif (R3) (Figure 4A), which is also found in FG-nucleoporins. FG-nucleoporins mediate interactions with the transport receptors and are critical for establishing the nuclear pore diffusion barrier. We hypothesized that the Imp7:Impβ interaction resembled the well characterized interaction between importins and FG-nucleoporins. The crystal structures show that phenylalanines from the nucleoporin FxFG motifs bind hydrophobic grooves on the convex side of Impβ, with major binding pockets between HEAT repeats 5–6 and 6–7 (Bayliss et al., 2000, Bayliss et al., 2002, Liu and Stewart, 2005; Figure S6A). To test whether phenylalanine residues in the Imp7 C-terminal tail are important for Impβ binding, we mutated the FxFG motif in Imp7 and performed pull-down and gel shift assays with Impβ. Our results show that mutations of two phenylalanine residues in the FxFG motif (Imp7 F1028S and F1030S, R3 region) were sufficient to reduce the interaction with Impβ more than 7-fold (Figures 4B, 4C, S6C, and S6D). Even a single mutation of a phenylalanine in the R2 region, Imp7 F1019S, reduced Impβ binding, underlining the importance of these residues for heterodimerization of Imp7 and Impβ (Figures 4B, 4C, S6C, and S6D).
Figure 4.
The FG Motif in the Imp7 C-Terminal Tail Is Essential for Interaction with Impβ
(A) Sequence alignment of the disordered C-terminal tail of Imp7 (aa 996–1,038). Three conserved regions are labeled as R1, R2, and R3. F residues of the FxFG motif are indicated by black stars, conserved GG residues by white circles, and a following F residue by a black circle. The linker preceding the GG residues is indicated by a black dashed line. Conserved residues are highlighted in red. The alignment was generated with the ESPript server (http://espript.ibcp.fr).
(B) Results of a pull-down experiment showing that conserved phenylalanines in Imp7 are essential for interaction with Impβ. Shown is SDS-PAGE analysis of the IN and E fractions of the pull-down experiment with the His-tagged wild-type and mutant Imp7 bound to the resin.
(C) Quantification of three independent gel shift experiments showing Imp7 and Imp7 mutant binding to Impβ. The Kd for the Imp7/Impβ interaction was estimated from the binding curve. The Kd is 0.38 μM for wild-type Imp7, 2.89 μM for Imp7 F1028S and F1030S, and 1.22 μM for Imp7 F1019S.
(D) Results of a pull-down experiment showing that conserved phenylalanines in the Imp7 C-terminal tail (Imp7C, aa 1,000–1,038) are essential for interaction with Impβ. Shown is SDS-PAGE analysis of the IN and E fractions of the pull-down experiment with the His-tagged wild-type and mutant Imp7 C-terminal tail bound to the resin. Nsp1_FG (497–608) is a construct containing five FG repeats from nucleoporin Nsp1, which can bind Impβ. Degradation products of Impβ and Imp7 are indicated by an asterisk.
(E) Results of a pull-down experiment showing that the canonical FG-nucleoporin binding site in Impβ is essential for interaction with Imp7. SDS-PAGE analysis was performed of the IN and E fractions of the pull-down experiment with Impβ6M (Impβ L174S, T175A, I178D, E214A, F217A, and I218D) and resin-bound His-tagged Imp7, RanGTP, and H1.0. A degradation product of Impβ is indicated by an asterisk.
(F) Results of a nuclear import assay in permeabilized HeLa S3 cells, using NT-495-labeled H1.0 in the presence of an energy-regenerating system, Impβ, and wild-type or mutant Imp7. In the images in the top row, cell nuclei are stained blue with DAPI. In the images in the bottom row, cells that take up NT-495-labeled H1 appear green.
See also Figure S6.
The high degree of conservation of two glycine residues in the GGxxF motif (R2 region) in the Imp7 C-terminal tail suggests their functional role (Figure 4A). Glycine residues permit greater flexibility of the adjacent aa, which might be necessary for the bulky phenylalanine residues to bind to the hydrophobic pockets in Impβ. Mutation of the glycine residues G1015A and G1016A strongly reduced Imp7 interaction with Impβ (Figure 4B), indicating that these residues are essential for Impβ binding.
The results of our pull-down assays show that the isolated Imp7 C-terminal tail (Imp7C, residues 1,000–1,038) is sufficient to bind Impβ (Figure 4D). Consistent with the results of our assay using the full-length Imp7, mutations F1019S, F1028S, and F1030S in the Imp7C peptide abolished the interaction with Impβ (Figure 4D). The G1015A and G1016A mutations, however, did not affect binding of the Imp7 C-terminal tail to Impβ, showing that the isolated tail is sufficiently flexible to interact with Impβ and that the two glycine residues are not required for the interaction (Figure 4D). When the C-terminal tail is incorporated into the bulky structural core of Imp7, these glycines become crucial to providing sufficient flexibility for the phenylalanines to reach the binding pocket in Impβ (Figure S6B). Thus, the conserved phenylalanine residues in the Imp7 C-terminal tail are indispensable for Imp7:Impβ complex formation, whereas the two glycines preceding F1019 facilitate backbone motion for this region and enable the phenylalanine residues to have access to the Impβ binding site.
The Imp7:Impβ:H1.0 structure shows that C-terminal HEAT repeat 20 of Imp7 is located in close proximity of Impβ HEAT repeats 5–7, which are the primary interaction site for FG-nucleoporins (Bayliss et al., 2000; Figures 2C and S6B). To test whether the same pocket binds the Imp7 C-terminal tail, we mutated six aa that are crucial for FG-nucleoporin binding (L174S, T175A, I178D, E214A, F217A, and I218D) (Figure S6A). The resulting Impβ6M mutant did not bind full-length Imp7, nor did it bind the isolated Imp7 C-terminal tail (Figures 4E and S6E). Importantly, Impβ6M could bind H1.0, RanGTP, and Impα, indicating that the mutations did not affect the overall folding and interactions (Figures 4E and S6E). In summary, our data indicate that Imp7 interaction with Impβ resembles the interaction between Impβ and FG-nucleoporins.
To further confirm the importance of conserved motifs in the Imp7 C-terminal tail, we studied the efficiency of H1.0 nuclear import by using digitonin-permeabilized human HeLa cells. Consistent with previous results (Jäkel et al., 1999), our data show that both Impβ and Imp7 are required for H1.0 import (Figures 4F, S6F, and S6G). Imp7 with a deleted C-terminal tail as well as Imp7 F1019S, F1028S and F1030S mutant proteins were deficient in H1.0 import into the nucleus. In addition, the Imp7 G1015A and G1016A mutant and the mutant with a deleted R1 motif (aa 1,000–1,014) also showed a defect in nuclear transport efficiency (Figures 4F and S6G). These data show that the FG motif in the Imp7 C-terminal tail is required for Imp7:Impβ dimerization and for nuclear import of H1.
The FxFG Nucleoporins Cooperate with RanGTP to Promote Complex Disassembly
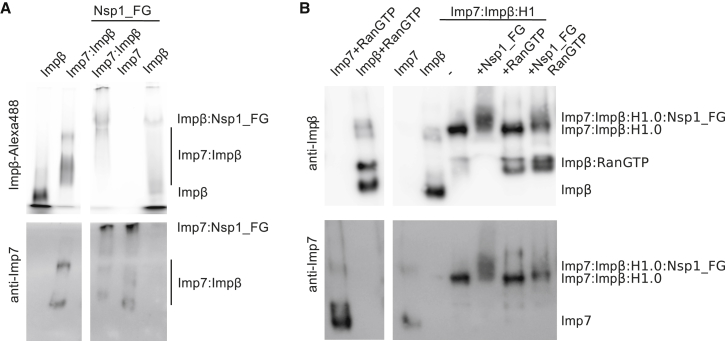
Our results suggested that a high local concentration of FxFG-nucleoporins on the nuclear side of the NPC might compete with Imp7 binding to Impβ and promote Imp7:Impβ dissociation. To test this possibility, we added an Nsp1 construct containing five FxFG repeats (Nsp1_FG) to the Imp7:Impβ complex (Figure S6H). We observed that the Nsp1_FG construct dissociated Imp7:Impβ, leading to the formation of Impβ:Nsp1 and Imp7:Nsp1 complexes (Figures 5A). The FxFG repeats in Nsp1 bound with higher affinity to Impβ, which out-competes Imp7 and leads to disassembly of the complex (Figure S6I). To mimic passage through NPC, we incubated the Imp7:Impβ:H1 complex with the Nsp1 FxFG repeats and RanGTP. Although the FxFG repeats by themselves were not sufficient to disassemble the trimeric complex, when combined with RanGTP, they promoted RanGTP-dependent disassembly of the complex (Figure 5B). Thus, our data indicate that FG-nucleoporins contribute to RanGTP-dependent disassembly of multimeric transport complexes.
Figure 5.
The FG Repeats Promote RanGTP-Dependent Disassembly of Impβ:Imp7:H1
(A) Results of a gel shift assay showing that the FG repeats from Nsp1 compete with Imp7 for the binding site in Impβ. FxFG repeats bind Imp7 (Imp7:Nsp1_FG) and Impβ (Impβ:Nsp1_FG), leading to disassembly of the Imp7:Impβ complex.
(B) Results of a gel shift assay showing that the FG repeats from Nsp1 promote RanGTP-dependent disassembly of the Imp7:Impβ:H1 complex. RanGTP binds to Impβ, leading to disassembly of the trimeric complex. FxFG repeats promote RanGTP-dependent complex disassembly.
See also Figure S6.
Conservation of the FG-Motif in Other Transport Factors
Similar to Imp7, RanBP8 (importin 8 [Imp8]) has been shown to form a heterodimer with Impβ (Görlich et al., 1997). Like Imp7, Imp8 contains conserved GGxxF and FxFG motifs at its C terminus (Figure S7A). Analogous to Imp7, mutations in the FxFG repeat in Imp8 reduced its interactions with Impβ more than 10-fold (Figures 6A, 6B, S7B, and S7C). By using ScanProsite (de Castro et al., 2006), we found FxF motifs and a preceding phenylalanine at the C-terminal end of Impα (importin α, Kapα) proteins (Figure S7D). Impα binds nuclear localization signal (NLS)-containing cargo molecules and acts as an adaptor for Impβ through the Impβ-binding domain (IBB) (Görlich et al., 1995). Consistent with previous data, we showed that Impα binds Impβ, but the binding to the Impβ6M mutant was reduced more than 5-fold (Figures 6C, 6D, and S7E). Moreover, mutations of the C-terminal FxF motif in Impα (F527S and F529S) reduced the interaction with Impβ 2-fold, showing that these residues in Impα contribute to the dimerization of the two importins and strengthen their interaction (Figures 6C, 6D, and S7E). This suggests that Impα C-terminal phenylalanine residues near the FxF motif also contribute to interaction with Impβ, similar to the Imp7 tail.
Figure 6.
The FG Motif Contributes to Dimerization of Several Importins
(A) Results of a pull-down experiment showing that conserved phenylalanines in the Imp8 C-terminal tail are required for the interaction with Impβ. Shown is SDS-PAGE analysis of IN and E fractions of the pull-down experiment, with wild-type or mutant Imp8 binding to His-tagged wild-type Impβ.
(B) Quantification of the results of three independent gel shift experiments, showing Imp8 and Imp8 mutant binding to Impβ. The Kd was estimated from the binding curve. The Kd is 0.19 μM for wild-type Imp8 and 1.72 μM for Imp8 F1028S and F1030S.
(C) Results of a pull-down experiment showing that conserved phenylalanines in the Impα C-terminal tail (Impα F527S/F529S) contribute to the interaction with Impβ. Likewise, the mutations in the FxFG binding pocket of Impβ, Impβ6M, destabilize the interaction with wild-type Impα. Shown is SDS-PAGE analysis of IN and E fractions of the pull-down experiment with His-tagged wild-type and mutant Impβ.
(D) Quantification of the results of three independent gel shift experiments showing Impα and Impα mutant binding to Impβ and the Impβ6M mutant. The Kd was estimated from the binding curve. The Kd is 0.62 μM for wild-type Impα and 0.92 μM for Impα F527S/F529S. The Kd for Impα binding to Impβ6M is 3.34 μM.
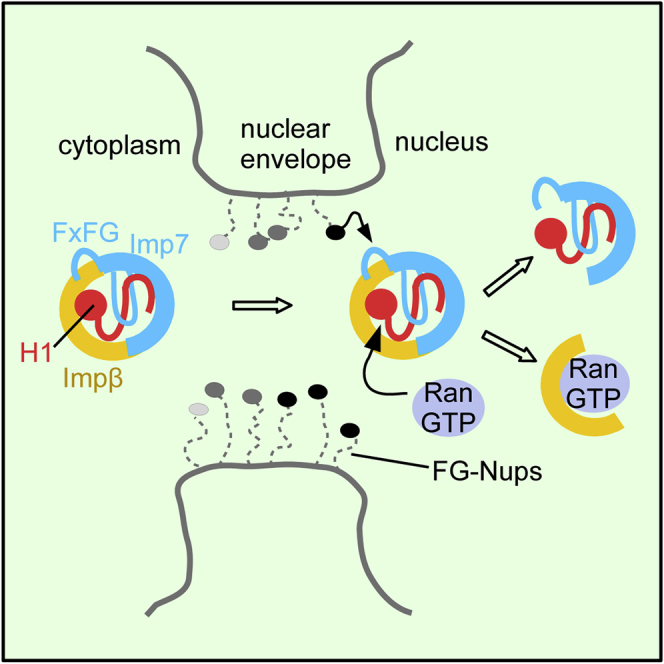
(E) In the first step, the FG motif in the Imp7 C-terminal tail binds to Impβ, mimicking FG-nucleoporin interactions. The affinity of this complex for H1 in the cytoplasm is higher than that of individual importins because the complex can efficiently accommodate its charged C-terminal tail in the cradle and between the HEAT repeat motifs. At the same time, the disordered C-terminal tail of H1 stabilizes and shapes the complex structure. The Imp7:Impβ:H1 complex translocates through the NPC mainly via transient FG-nucleoporin:importin interactions. At the nuclear side of the NPC, Impβ dissociates from H1 by RanGTP binding and by competing FxFG-nucleoporin interactions. H1 is then transferred to nuclear chaperones.
See also Figure S7.
Our data show that dimerization through the FG motif is a more general mechanism that involves many transport receptors and might also be used by other proteins.
Discussion
Proteins smaller than 40 kDa commonly diffuse through the nuclear pore. However, when small proteins are highly charged, they require import receptors to pass through the NPC. These difficult cargoes, such as ribosomal proteins or histones, need to be shielded and protected from aggregation or premature unspecific interactions (Jäkel et al., 2002, Mosammaparast et al., 2002, Mosammaparast et al., 2005). Some of these proteins are transported with the help of specific chaperones and single transport receptors, whereas others require two transport receptors (Jäkel et al., 2002, Booth et al., 2014). In this study, we addressed the question of how such a combined transport and/or chaperone system assembled and transported these complicated cargoes through the NPC. Our work has revealed the first structure of two import receptors, Imp7 and Impβ, in a complex with the linker histone H1.0 as a cargo.
In the Imp7:Impβ:H1.0 complex structure, two importins are positioned side by side with a large interaction surface. The interface is established by HEAT repeats and by the intrinsically flexible C-terminal tail of H1, as reflected by the unstructured density located between the importins and in the concave cavity of the complex. The acidic residues in loops connecting HEAT repeats of the importins interact with the H1 C-terminal tail and protect it from aggregation, providing further evidence of the role of importins as chaperones for the linker histone tail. Similar patches of acidic aa are found in the other histone chaperones and are important for interaction with histones (Warren and Shechter, 2017). Imp7 has a long loop with the highest content of acidic residues among all human importins, making it suitable for transporting highly positively charged proteins such as histones and ribosomal proteins. A similar acidic loop is present in Imp8; however, the cargo of the dimeric Imp8:Impβ complex remains unknown (Görlich et al., 1997). Long loops with a high content of acidic residues are also found in Imp4, Imp9, and transportin-1, which import core histones and ribosomal proteins. This suggests that other importins transporting highly charged proteins use the same mechanism.
Although the H1 C-terminal tail shapes and stabilizes the complex, our data indicate that it remains disordered in the confined space of the complex. H1 binding is achieved by multiple nonspecific electrostatic interactions with acidic residues of two importins. A similar high-affinity fuzzy interaction was recently described for H1 and its chaperone prothymosin-α (Borgia et al., 2018). In the H1:prothymosin-α complex, interactions are completely disordered. In the Imp7:Impβ:H1.0 complex, however, fuzzy interactions are essential for the architecture of the complex. In the absence of H1, the Imp7:Impβ complex is highly flexible and is fully assembled with the help of the H1 C-terminal tail, which serves as a zipper that closes and stabilizes the structure.
Another disordered region, the Imp7 C-terminal tail, is required for the initial interaction between Imp7 and Impβ. The complex formation is triggered by insertion of the Imp7 C-terminal tail into the Impβ FG-nucleoporin binding surface located on its convex side. The Imp7:Impβ complex is, however, highly flexible and is only fully assembled with the help of the H1 C-terminal tail.
Our data suggest that a high concentration of FG motifs within the NPC might disassemble the Imp7:Impβ complex. H1 stabilizes the trimeric complex, which allows the transient and weakly interacting FG motif in Imp7 to rebind rapidly to Impβ. When the cargo is bound, the complex is more stable and can translocate through the NPC. On the nuclear side of the NPC, a high local concentration of FxFG repeats, nuclear RanGTP, and histone chaperones jointly promote complex dissociation and H1 transfer to the nuclear chaperones (Figure 6E).
X-ray structures of the scaffold Nups (Andersen et al., 2013, Bilokapic and Schwartz, 2012, Flemming et al., 2012, Stuwe et al., 2014) have shown that the NPC components share structural properties with the members of the karyopherin family. We have shown that a subset of transport factors contains an FxFG motif, a hallmark of FG-nucleoporins, which are intrinsically disordered proteins that undergo fuzzy interactions with transport factors themselves (Hough et al., 2015, Milles et al., 2015, Raveh et al., 2016, Sparks et al., 2018). This points to an additional evolutionary connection between the stationary (NPC) and soluble (transport receptor) parts of the nucleocytoplasmic transport machinery.
In addition to nucleoporins, FG motifs are present in transmembrane proteins of the inner nuclear membrane and nuclear envelope (Zuleger et al., 2011). It has been speculated that the role of these motifs is to facilitate the translocation of proteins into the inner nuclear membrane by interacting with FG-nucleoporins of the nuclear pore complex (Zuleger et al., 2011). The results of our study suggests a possible general mechanism by which FG-containing proteins can recruit transport receptors or other proteins by mimicking importin:FG-nucleoporin interactions.
Our data reveal that the architecture and organization of the Imp7:Impβ:H1.0 complex are mediated by nonspecific charged interactions of the disordered region. Many other complexes might be structured in a similar way; that is, through transient electrostatic interactions of disordered regions.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Goat polyclonal anti-Importin 7 | Abcam | RRID:AB_302077; Lot: GR238864-11 |
| Rabbit polyclonal anti-NTF97/Importin beta | Abcam | RRID:AB_944512; Lot: GR3206575-1 |
| Rabbit Anti-Goat IgG (H+L) Horseradish Peroxidase Conjugate | Bio-Rad | Cat. # 172-1034; RRID:AB_11125144 |
| Goat Anti-Rabbit IgG (H+L) Horseradish Peroxidase Conjugate | Bio-Rad | Cat. # 170-6515; RRID:AB_11125142 |
| Bacterial and Virus Strains | ||
| E. coli XL1-Blue | NEB | C2992 |
| E. coli Rosetta(DE3) | Novagen/Merck | Cat. # 70954-3 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Ni Sepharose 6 Fast Flow | GE Healthcare | Cat. # 17-5318-01 |
| Glutathione Sepharose 4 Fast Flow | GE Healthcare | Cat. # 17-5132-01 |
| Superdex 200 Increase column 10/30 | GE Healthcare | Cat. # 28-9909-44 |
| HiTrap Q Sepharose FF | GE Healthcare | Cat. # 17-5156-01 |
| HiTrap SP FF | GE Healthcare | Cat. # 17-5054-01 |
| PD-10 column | GE Healthcare | Cat. # 17-0851-01 |
| Thrombin | Sigma-Aldrich | Cat. # T7513-50UN |
| Trypsin | Promega | Cat. # V5111 |
| Lysyl endopeptidase | FUJIFILM Wako | Cat. # 125-05061 |
| R3.5/1 2nm carbon grids | Quantifoil | N/A |
| R2/1 | Quantifoil | N/A |
| (d0/d12) Bis[sulfosuccinimidyl] suberate (BS3) | Thermo Fisher Scientific | Cat# 21580 |
| Critical Commercial Assays | ||
| Monolith Protein Labeling Kit BLUE-NHS (Amine Reactive) | NanoTemper Technologies | Cat. # MO-L003 |
| Alexa Fluor 488 Protein Labeling Kit | Thermo Fisher Scientific | Cat. # A10235 |
| Alexa Fluor 647 Microscale Protein Labeling Kit | Thermo Fisher Scientific | Cat. # A30009 |
| Deposited Data | ||
| EM map and Atomic coordinates, Imp7:Impβ:H1.0 | Protein Data Bank | PDB: 6N88 |
| EMD-0366 | ||
| EM map and Atomic coordinates, Impβ:H1.0 | Protein Data Bank | PDB: 6N89 |
| EMD-0367 | ||
| EM map, Imp7:Impβ | Protein Data Bank | EMD-0368 |
| Oligonucleotides | ||
| Primers for Imp7C (aas 1000–1038) | metabion | N/A |
| 676F: 5′-GATCAGAGGCGGGCAGCAC-3′ | ||
| 382R: 5′-GGATCCGGGCCCCTGGAA-3′ | ||
| Primers for Imp7 ΔC (aas 1–999) | metabion | N/A |
| pETDuet_avr_f: 5′-CCTAGGCTGCTGCCACC-3′ | ||
| 678R: 5′-TTATGCCAGTGTAGCGATGTCTTG-3′ | ||
| Primers for Imp7 F1028S F1030S | metabion | N/A |
| 677F: 5′-CAAGTACGTCTAATTCCGGCAACCCG-3′ | ||
| 677R: 5′-GCACGACTGGGGCGTTGAAT-3′ | ||
| Primers for Imp7 F1019S | metabion | N/A |
| 823F: 5′-GGATTTGTAGCCTCCGTG-3′ | ||
| 823R: 5′-AACGCCCCAGTCGTG-3′ | ||
| Primers for Imp7 GG1015AA | metabion | N/A |
| 824F: 5′-GCATACAAATTCAACGCC-3′ | ||
| 824R: 5′-ATGATCGAGAAACACGCT-3′ | ||
| Primers for Imp7 Δ aas 1000–1014 | metabion | N/A |
| 826F: 5′-GGAGGCTACAAATTCAACG-3′ | ||
| 826R: 5′-CATCGCTACACTGGCA-3′ | ||
| Primers for Imp7 Δ aas 884–912 replaced with SGGSGG | metabion | N/A |
| 679F: 5′-CAGGAGGCCAGGAGTACCTGGAGATTTTAG-3′ | ||
| 679R: 5′-GCCTCCTGAGTTCTCTTGCTCTGCATGG-3′ | ||
| Primers for Imp7 Δ aas 923–954 replaced with SGGSGG | metabion | N/A |
| 680F: 5′-CAGGAGGCCCTATTGATGAATATCAGATATTT-3′ | ||
| 681R: 5′-GCCTCCTGAGGCCTGTTTGGCTAAAAT-3′ | ||
| Primers for Impβ I178D L174S and T175A (forImpβ6M) | metabion | N/A |
| 774F: 5′-GCTGCCATAGACCAGGGGA-3′ | ||
| 774R: 5′-CGAAATCTCATTGGATTTATCTTG-3′ | ||
| Primers for Impβ E214A F217A I218D (Impβ6M) | metabion | N/A |
| 775F: 5′-CTGATATGCAGGTGGTCTGTG-3′ | ||
| 775R: 5′-CGTGCCTTGCAGACTCTTTATC-3′ | ||
| Primers for ScNsp1 aa 497-608, FF5 (BamHI/XhoI) | metabion | N/A |
| 780F: 5′-CATGGATCCACTAAATCGAATGAAAAAAAG-3′ | ||
| 780R: 5′-CTTCTACGGGGAAGTCATAACTCGAG-3′ | ||
| Primers for HRV-3C site removal | metabion | N/A |
| 688F: 5′-TTCCAGGGGCCCGGATC-3′ | ||
| 687R: 5′-CCGCGGACCAATCTGTTCT-3′ | ||
| Primers for Imp8 F1028S F1030S | metabion | N/A |
| 1219F: 5′-CTCCGCA TCT AAT TCC GGGACTGTG-3′ | ||
| 1219R: 5′-AGGACTCCTTTGTTTTCAAAGGTG-3′ | ||
| Primers for Kapα F527S F529S | metabion |
N/A |
| 1237F: 5′-GCTCCTGGGACCTCTAACTCCTAG-3′ | ||
| 1237R: 5′-CCCATCCTGAACTTGGAAAGTG-3′ | ||
| Recombinant DNA | ||
| Plasmid pETDuet-1 + 6His-SUMO-Impβ | This paper | 918 |
| Plasmid pETDuet-1 + 6His-SUMO-Impβ6M | This paper | 922 |
| Plasmid pETDuet-1 + 6His-7Arg-SUMO-Imp7 | This paper | 924 |
| Plasmid pETDuet-1 + 6His-7Arg-SUMO-Imp7Δ2L (Δ aas 884–912 and 923-954, replaced with SGGSGG linkers) | This paper | 934 |
| Plasmid pETDuet-1 + 6His-7Arg-SUMO-Imp7ΔC (aas 1–999) | This paper | 931 |
| Plasmid pETDuet-1 + 6His-7Arg-SUMO-Imp7 F1028S F1030S | This paper | 930 |
| Plasmid pETDuet-1 + 6His-7Arg-SUMO-Imp7 F1019S | This paper | 936 |
| Plasmid pETDuet-1 + 6His-7Arg-SUMO-Imp7 G1015A G1016A | This paper | 938 |
| Plasmid pETDuet-1 + 6His-7Arg-SUMO-Imp7Δaa1000–1014 | This paper | 942 |
| Plasmid pETDuet-1 + 6His-7Arg-SUMO-Imp7C (aas 1000–1038) | This paper | 927 |
| Plasmid pETDuet-1 + 6His-7Arg-SUMO-Imp7C F1028S F1030S (aas 1000–1038) | This paper | 929 |
| Plasmid pETDuet-1 + 6His-7Arg-SUMO-Imp7C F1019S (aas 1000–1038) | This paper | 937 |
| Plasmid pETDuet-1 + 6His-7Arg-SUMO-Imp7C G1015A G1016A (aas 1000–1038) | This paper | 939 |
| Plasmid pGEX-4T-1 + H1.0-6His | This paper | 945 |
| Plasmid pETDuet-1 + 6His-SUMO-H1.0g (aas 1–95) | This paper | 947 |
| Plasmid pETDuet-1 + 6His-SUMO-H1.0g (aas 96–194) | This paper | 950 |
| Plasmid pETDuet-1 + 6His-SUMO-Sc_Nsp1 (FF5, aas 497–608) | This paper; Bayliss et al., 2000 | 952 |
| Plasmid pGEX-4T-3 + Imp8 | Volpon et al., 2016 | 1138 |
| Plasmid pGEX-4T-3 + Imp8 F1028S/F1030S | This paper | 1140 |
| Plasmid pGEX-4T-3 + Impα | Volpon et al., 2016 | 1153 |
| Plasmid pGEX-4T-3 + Impα F527S/F529S | This paper | 1155 |
| Plasmid pETDuet-1 + 6His-SUMO-Imp8 | This paper | 1141 |
| Plasmid pETDuet-1 + 6His-SUMO-Imp8 F1148S/F1150S | This paper | 1143 |
| Plasmid pETDuet-1 + Impα | This paper | 1159 |
| Plasmid pETDuet-1 + Impα F527S F529S | This paper | 1161 |
| Software and Algorithms | ||
| EMAN2 | Tang et al., 2007 | N/A |
| XMIPP | Scheres et al., 2008 | N/A |
| Unblur | Grant and Grigorieff, 2015 | N/A |
| CTFFIND4 | Rohou and Grigorieff, 2015 | N/A |
| Relion 2.0 | Scheres, 2012 | N/A |
| HHpred | Söding et al., 2005 | N/A |
| MODELER | Webb and Sali, 2016 | N/A |
| UCSF Chimera | Pettersen et al., 2004 | N/A |
| COOT | Emsley et al., 2010 | https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/ |
| Phenix | Adams et al., 2010 | https://www.phenix-online.org |
| Prism5 | GraphPad | https://www.graphpad.com/ |
| xQuest | Walzthoeni et al., 2012 | N/A |
Contact for Reagent and Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Mario Halic (mario.halic@stjude.org).
Method Details
Cloning Procedures
The Homo sapiens gene encoding Impβ was amplified from cMarathon DNA by PCR and cloned into the modified pETDuet vector containing a 6xHis-SUMO tag followed by an HRV-3C recognition site. For technical reasons, we used the Xenopus laevis gene encoding Imp7 and not the corresponding H. sapiens gene (the two genes have 92.5% identity and 97.2% similarity). The X. laevis gene encoding Imp7 was cloned into a modified pETDuet vector containing a 6xHis-6xArg-SUMO tag followed by a HRV-3C recognition site (the original plasmid containing the gene for Imp7 was a gift from K. Ribbeck). The H. sapiens gene encoding histone H1.0 was cloned from genomic DNA into an in-house–modified version of the pGEX-4T-1 plasmid to yield a GST tag followed by an HRV-3C recognition site at the N terminus and a 6xHis tag at the C terminus of H1.0. pGEX plasmids containing genes encoding the H. sapiens Imp8 and Kapα proteins were a gift from Yuh Min Chook. Imp8 and Kapα were additionally pre-cloned into a pETDuet vector containing a 6xHis-6xArg-SUMO tag followed by an HRV-3C recognition site by using BamHI/NotI and BamHI/XhoI restriction enzymes, respectively. All mutant constructs were made by inverse PCR (iPCR) using plasmids that contained the gene for the full-length protein (Ulrich et al., 2012). For the pull-down assays, the HRV-3C protease recognition site was removed from Imp7 and its mutants by using iPCR to prevent any unspecific cleavage. The Saccharomyces cerevisiae Nsp1 fragment (aa 497–608, Nsp1 FF5 containing five FxFG repeats; Bayliss et al., 2000) was cloned from genomic DNA into an in-house–modified pETDuet vector containing 6xHis-SUMO tag at the N terminus followed by an HRV-3C recognition site.
Protein expression and purification
Each of the proteins was individually expressed and purified. Transformed E. coli cells (Rosetta strain) were grown in LB medium (with ethanol, 1% final concentration) at 37°C until the OD600nm reached 0.5–0.6. The temperature was then lowered to 18°C and after 30 min, cells were induced with 0.4 mM IPTG. After an overnight induction, cells were collected by centrifugation (4000 rpm for 10 min at 4°C). Pelleted cells that contained His-tagged proteins were resuspended in lysis buffer (50 mM sodium phosphate pH 8.0, 500 mM NaCl, 20 mM imidazole, 10% glycerol (v/v), 3 mM β-mercaptoethanol and 0.1 mM PMSF). After being lysed with a French press, the disrupted cells were centrifuged for 20 min at 17,000 rpm, 4°C. The cleared supernatant was mixed and incubated for 20 min at 4°C with Ni Sepharose 6 Fast Flow resin (GE Healthcare) pre-equilibrated in lysis buffer. The resin with bound His-tagged protein was washed thoroughly in batches with the lysis buffer and then transferred to a disposable column (Pierce). The column was washed with 4 bed volumes of wash buffer (50 mM sodium phosphate pH 8.0, 500 mM NaCl, 40 mM imidazole, 10% glycerol (v/v), 3 mM β-mercaptoethanol and 0.1 mM PMSF) and the protein was eluted with 5 bed volumes of elution buffer (50 mM sodium phosphate pH 8.0, 50 mM NaCl, 300 mM imidazole, 10% glycerol (v/v), 3 mM β-mercaptoethanol and 0.1 mM PMSF).
The presence of the 6xArg-tag on Imp7 enabled its further purification on a cation exchange column (Bilokapic and Schwartz, 2012). After being eluted from the Ni-NTA resin, Imp7 was diluted to a salt concentration of approximately 100 mM and then quickly loaded onto a HiTrap SP HP column. The purest fractions were collected, the salt concentration was adjusted to 300 mM and HRV-3C protease was added at a 1:100 ratio. After an overnight incubation at 4°C, the sample salt concentration was increased to 600 mM NaCl. The sample was concentrated and loaded onto a size-exclusion chromatography Superdex 200 Increase 10/30 column (GE Healthcare), pre-equilibrated in buffer containing 15 mM HEPES/NaOH pH 7.5, 600 mM NaCl and 1 mM DTT. The cleanest fractions were pooled and concentrated.
Impβ, first purified using affinity chromatography (Ni-NTA), was loaded onto a HiTrap Q Sepharose FF (GE Healthcare) ion exchange column. The purest fractions were pooled and dialyzed over-night at 4°C with HRV-3C protease (1:100 ratio) in a buffer containing 15 mM HEPES/NaOH pH 7.5, 150 mM NaCl and 1 mM DTT. The protein was concentrated and loaded onto a size-exclusion chromatography Superdex 200 Increase 10/30 column (GE Healthcare) pre-equilibrated in buffer containing 15 mM HEPES/NaOH pH 7.5, 150 mM NaCl and 1 mM DTT. The cleanest fractions were pooled and concentrated.
H1.0 does not elute from the Ni-NTA resin if the buffer has a high imidazole concentration and a low salt concentrations (50 mM NaCl) (Ivic et al., 2017). Therefore, this step was used as an additional washing step that prevented subsequent protein precipitation. The elution fraction (50 mM sodium phosphate pH 8.0, 500 mM NaCl, 300 mM imidazole, 10% glycerol (v/v), 3 mM β-mercaptoethanol and 0.1 mM PMSF) was diluted to a salt concentration of 200 mM NaCl and quickly loaded onto a HiTrap SP FF ion exchange column. The fractions containing the protein were pooled and diluted to a 300 mM NaCl concentration. HRV-3C protease was added at a 1:50 ratio and the sample was incubated overnight at 4°C. To remove the GST-tag, the sample was diluted and again loaded onto a HiTrap SP HP ion exchange. Clean protein fractions were pooled and concentrated using Amicon Ultra-15 centrifugal filters.
Clarified cell lysates containing GST-Imp8 or GST-Kapα proteins in PBS buffer with PMSF were mixed with Glutathione Sepharose 4 Fast Flow resin (GE Healthcare) and incubated for 30 min at 4°C. The resin with bound GST-tagged proteins was washed in batches three times with 5 bed volumes of PBS buffer and then transferred to a disposable column (Pierce). The proteins were eluted with 3 bed volumes of buffer containing 50 mM Tris pH 8.0 and 20 mM reduced glutathione. Immediately after elution proteins were dialyzed into buffer containing 15 mM HEPES pH 7.5, 300 mM NaCl and 1 mM DTT. The GST tag was removed by diluting the proteins to a concentration of around 1 mg/mL in cleavage buffer (50 mM Tris pH 8.0, 150 mM NaCl and 10 mM CaCl2), adding 1 U of thrombin per 100 μg of protein, and incubating the mixture at room temperature for 1–2 h.
Complex assembly and cross-linking
Purified Imp7, Impβ and H1.0 were mixed in 1:1:1.2 ratio and incubated for 30 min on ice. The complex was isolated on a size-exclusion Superdex 200 Increase 10/30 column (GE Healthcare) equilibrated in 15 mM HEPES/NaOH pH 7.5, 150 mM NaCl and 1 mM DTT. Single peak fractions were analyzed on SDS-PAGE and collected. The complex (0.25 mg/mL) was cross-linked by adding 2.3% glutaraldehyde to a final concentration of 0.1% and incubating the mixture for 2 min at 37°C. The reaction was stopped by adding 1 M Tris/HCl pH 8.0 (150 mM final concentration). Cross-links were checked by 7% SDS-PAGE and then desalted on a disposable PD-10 column (GE Healthcare). To remove any higher order cross-links and aggregates, the sample was run again on a size-exclusion Superdex 200 Increase 10/30 column and concentrated.
Electron microscopy sample preparation
For negative stain analysis the cross-linked sample was diluted in size-exclusion buffer to around 0.2 mg/mL. A 3.5 μL aliquot of the sample was loaded onto a freshly glow discharged thin carbon-coated grid (Quantifoil R3.5/1 with a 2 nm carbon coating), incubated for 45 s at room temperature and washed with six drops of size-exclusion buffer. The grid was then stained with a drop of 2% uranyl-acetate for 15 s and dried on a filter paper.
For cryo-EM grids, the cross-linked complex was diluted to around 0.4 mg/ml. Four microliters of the sample were loaded on a previously glow discharged holey carbon (Quantifoil R2/1) grid. After 1 s of blotting time, grids were plunge frozen in liquid ethane by using a Leica EM GP-Automatic Plunge Freezer. The temperature in the chamber was kept at +15°C and the humidity at 95%.
EM data collection and processing
Image acquisition of negative stain data was performed with a Titan HALO transmission electron microscope (FEI) operating at 300kV. Random conical tilt data were collected at 0° and 40°. Particle pairs (2721) were picked manually using the EMAN2 e2RCTboxer tool (Tang et al., 2007). Untilted particles were aligned and classified using EMAN2. The random conical tilt reconstruction was calculated from different classes and subsequently refined with the single particles using EMAN2.
The cryo-EM data (∼1250 micrographs) were collected on a Titan Krios with a Gatan Summit K2 electron detector (NeCEN, Leiden, the Netherlands) and analyzed as described (Bilokapic et al., 2018a, Bilokapic et al., 2018b). The image pixel size was 1.36 Å per pixel on the object scale. Data were collected in a defocus range of 10,000 – 40,000 Å with a total exposure of 80 e/Å2. Sixty frames were collected and aligned with the Unblur software package by using a dose filter (Grant and Grigorieff, 2015). Several thousand particles were manually picked and carefully cleaned in XMIPP (Scheres et al., 2008) to remove inconsistent particles. The resulting useful particles were then used for semi-automatic and automatic particle picking in XMIPP. The contrast transfer function parameters were determined using CTFFIND4 (Rohou and Grigorieff, 2015). The 2D class averages were generated with the RELION software package (Scheres, 2012). Inconsistent class averages were removed from further data analysis. The 3D refinements and classifications were subsequently performed in RELION. Imp7:Impβ classification was performed with local angular searches, keeping the complex orientation consistent with that for Imp7:Impβ:H1. All final refinements were done in RELION using the auto-refine option. The initial reference was filtered to 60 Å in RELION. Particles were split into two datasets and refined independently, and the resolution was determined using the 0.143 cut-off (with the RELION auto-refine option). Local resolution was determined with RELION 2.0. All maps were filtered to local resolution using RELION 2.0 with a B-factor determined by RELION.
The templates for modeling Imp7 and H1.0 were obtained from homologous proteins through the HHpred homology detection software. Secondary structure predictions for Imp7 HEAT-repeat elements are highly similar for all obtained templates (Figure S3I). We used 1WA5 (chain C), which had the best score and the longest target length, as the template for the MODELER software to obtain a structural model for Imp7. The same approach was used to obtain the H1.0 structural model. Based on the target length, we used 5NL0 as a template to obtain the H1.0 structural model.
Molecular models were built manually, first by inserting individual HEAT repeats, and then by building loops where possible using Coot (Emsley et al., 2010). Rigid-body real-space refinement was performed in Phenix (Adams et al., 2010). The UCSF Chimera software package was used for rigid-body fitting of models (Pettersen et al., 2004). Visualization of all cryo-EM maps was performed with Chimera.
Pull-down assay
For pull-down assays, 10 μg of purified protein was mixed with 10 μg of His-tagged protein and incubated at room temperature. The sample was then diluted with pull-down buffer (50 mM sodium phosphate pH 8.0, 150 mM NaCl, 20 mM imidazole) to a final volume of 200 μL. The mixture (180 μL) was added to 60 μL of 50% Ni Sepharose 6 Fast Flow slurry equilibrated in pull-down buffer and incubated in a thermoblock for 30 min at 25°C with shaking at 900 rpm. After incubation, the supernatant was separated by centrifugation, at 4000 rpm for 3 min and the resin was washed three times with 200 μL of the pull-down buffer. Proteins bound to the resin were eluted with 30 μL of buffer containing 50 mM sodium phosphate pH 8.0, 150 mM NaCl and 400 mM imidazole. Aliquots of 15 μL of the initial mixture and eluate were analyzed by SDS-PAGE. Each pull-down experiment was repeated three times.
Nuclear import assay
Purified human H1.0 was labeled with the NT-495 fluorescent dye by using a MO-L003 Monolith Protein Labeling Kit BLUE-NHS (Amine Reactive), NanoTemper Technologies. The nuclear import assay was performed according to the published protocol (Cassany and Gerace, 2009) with some minor adjustments. HeLa S3 cells were grown to 80% confluence in 8 well μ-Slides (ibidi GmbH) or 16 well plates in DMEM medium with 10% (v/v) fetal bovine serum and 1% (w/v) penicillin/streptomycin. The medium was then removed and the cells were washed three times with 200 μL of cold transport buffer (TB) containing 20 mM HEPES/NaOH pH 7.4, 110 mM potassium acetate, 2 mM magnesium acetate, 1 mM EGTA, 2 mM DTT, 0.1 mM PMSF and 1 μg/mL each of leupeptin, pepstatin, and aprotinine. Cells were permeabilized with 0.005% digitonin for 5 min then again washed three times with TB. Next, 1 mL of fresh, cold TB or WGA was added and the cells were incubated for 15 min at room temperature. To remove all of the cytosol, cells were washed a further five times with 1 mL of TB. After washing, 50 μL of the import reaction mix (0.4 μM ND-495 H1.0, 0.3 μM Impβ, 0.3 μM Imp7, 1 μM NTF2, 1 mM ATP, 0.1 mM GTP, 1 mg/mL CP, 15 U/mL CPK and if used 0.8 mg/mL WGA) was added to the cells and incubated for 30 min at room temperature. As negative controls, reaction mixtures without importin receptors, energy suppliers, and both were used. After incubation, cells were again washed three times with 200 μL TB, stained with DAPI (300 nM) and then inspected under the fluorescent microscope. The experiment was repeated three times. Fluorescence quantification was measured using ImageJ software as described previously (Walker et al., 2009). All visible cells from an acquired image (50–150 cells) were scored into the following categories: N > C (more H1.0 in the nucleus than in the cytoplasm), N = C (an equal distribution of H1.0 between nucleus and cytoplasm), and N < C (more H1.0 in the cytoplasm than in the nucleus).
Native gel-shift assays
Impβ and H1.0 were labeled with fluorescent Alexa488 and Alexa647 dyes by using the Alexa Fluor 488 Protein Labeling Kit and the Alexa Fluor 647 Microscale Protein Labeling Kit (Thermo Fisher Scientific), respectively. The Imp7:Impβ:H1.0 and Imp7:Impβ complexes were isolated by size-exclusion chromatography (Superdex 200 Increase 10/30 column [GE Healthcare]). After isolation, 0.5–1 μg of the Imp7:Impβ complex was mixed with increasing amounts of Nsp1_FG (10, 30 and 60 μg) in buffer containing 50 mM NaCl and 15 mM HEPES pH 7.5 (the final volume of the reaction was 15 μL). Imp7:Impβ:H1.0 (0.5–1 μg) was mixed with Nsp1_FG, RanGTP, or both and incubated for 2 h at 37°C. Native loading dye was then added to the samples, and they were loaded onto a 4%–20% Mini-PROTEAN-TGX Gel (Bio-Rad). The running buffer contained 25 mM Tris and 192 mM glycine, pH 8.2, and the gels were run for 2–3 h at 4°C and 120 V. For visualization, gels containing fluorescently labeled samples were scanned using the Typhoon imaging system (GE Healthcare). To visualize non-labeled proteins, native gels were then electroblotted onto PVDF membranes (GE Healthcare). Membranes were blocked with 5% milk in PBS buffer with 0.1% v/v Tween (PBS-T) overnight at 4°C. Immunoblotting was performed with the goat polyclonal antibody anti-Importin 7 (ab15840, Abcam) and rabbit polyclonal antibody anti-NTF97/Importin beta (ab45938, Abcam). All antibodies were used at dilutions recommended by the suppliers. Immunoblots were detected using horseradish peroxidase (HRP)-conjugated secondary antibodies (Bio-Rad) and enhanced chemiluminescence (Thermo Scientific). The resulting western blot images were scanned using an Amersham Imager 600 (GE Healthcare).
For affinity measurements, Impβ and Impβ6M labeled with fluorescent Alexa 488 dye were used. Reaction mixtures (15 μL in volume) were assembled on ice, with increasing concentrations of non-labeled protein, whereas the concentration of Impβ/Impβ6M was unchanged. The reactions were incubated for 2 h on ice and then loaded onto 4%–20% gradient acrylamide gels. The gels were run for 2 h at 4°C and 120 V and scanned using the Typhoon imaging system (GE Healthcare). To quantify the decrease in the Impβ/Impβ6M fluorescent signal, Quantity one 1-D Analysis Software (Bio-Rad) was used. The data obtained were fitted using non-linear regression in GraphPad Prism software. Affinity measurements were performed in triplicate.
Chemical cross-linking and mass spectrometry
The Imp7:Impβ:H1.0 complex was cross-linked by using an equimolar mixture of isotopically labeled (d0/d12) BS3 (bis[sulfosuccinimidyl] suberate) (Creative Molecules) for 30 min at 30°C. The reaction was quenched by adding ammonium bicarbonate to a final concentration of 100 mM for 10 min. Another sample of the complex was cross-linked by using 4-(4,6-dimethoxy-1,3,5-triazin-2-yl)-4-methylmorpholinium chloride (DMTMM) (Sigma Aldrich), a reagent that couples carboxylic acids to the primary amine of lysines. Cross-linking was performed using 40mM DMTMM for 6 min at 35°C. The reaction was stopped by buffer exchange using a desalting column (Thermo Scientific) equilibrated with PBS, followed by the addition of ammonium bicarbonate. Cross-linked samples were denatured by adding 2 sample volumes of 8 M urea, reduced with 5 mM TCEP (Thermo Scientific), and alkylated by adding 10 mM iodoacetamide (Sigma-Aldrich) and incubating them for 40 min at room temperature in the dark. A digestion was performed with lysyl endopeptidase (Wako) (1:50 w/w) for 2 h, and this was followed by a second digestion with trypsin (Promega) (1:50 ratio) at 35°C overnight. Proteolysis was stopped by adding 1% (v/v) trifluoroacetic acid (TFA). Cross-linked peptides were purified by reversed-phase chromatography using C18 cartridges (Sep-Pak, Waters) and enriched on a Superdex Peptide PC 3.2/30 column (300 × 3.2 mm). Fractions of the cross-linked peptides were analyzed by liquid chromatography coupled to tandem mass spectrometry using an LTQ Orbitrap Elite instrument (Thermo Scientific) (Herzog et al., 2012). Cross-linked peptides were identified using xQuest (Walzthoeni et al., 2012). The results from BS3 cross-linking were filtered with an MS1 tolerance window of −4 to 4 ppm, score ≥ 22, and were manually validated. Identifications of zero-length cross-links were filtered using the following parameters: MS1 tolerance window of −3 to 3 ppm, min delta score ≤ 0.85 and score ≥ 22 followed by manual validation.
Data and Software Availability
Data resources
The accession numbers for the Imp7:Impβ:H1.0 and Impβ:H1.0 atomic coordinates have been deposited in the PDB: 6N88 and 6N89. The Imp7:Impβ:H1.0, Imp7:Impβ and Impβ:H1.0 cryo-EM maps have been deposited in the EMDB: EMD-0366, EMD-0367 and EMD-0368, respectively. The original data are deposited to https://doi.org/10.17632/dfjzx4sxrs.1.
Acknowledgments
We thank Elena Conti for providing access to the cryo-EM facility at the Max Planck Institute of Biochemistry, where the negative stain and initial cryo-EM data were collected, and we especially thank Mike Strauss for support with data acquisition. We acknowledge support from and use of the resources of Instruct-ERIC. Instruct provided data collection at the cryo-EM facilities at CEITEC (Brno, Czech Republic) and NeCEN (Leiden, the Netherlands). We thank Martin Turk for making and inspecting the initial negative stain grids of individual importins and the complex, and we thank Sigrun Jaklin and Carlos Rojas Cordova for help with experiments. We would like to thank Ilaria Ugolini, Thomas Becker, Marija Luic, and Joseph Bartho for comments on the manuscript. We also thank Keith A. Laycock, Ph.D., ELS, for scientific editing of the manuscript. N.I. has received funding from the European Union Seventh Framework Program (FP7 2007-2013) under grant agreement 291823 Marie Curie FP7-PEOPLE-2011-COFUND (The New International Fellowship Mobility Programme for Experienced Researchers in Croatia – NEWFELPRO project under grant agreement 26). This work was supported by ERC-smallRNAhet-309584 to M.H.
Author Contributions
S.B. initiated and designed the project. N.I. planned and performed the biochemical experiments. M.P., V.S.-M., and F.H. performed cross-linking mass-spectrometry. N.I., S.B., and M.H. performed electron microscopy and analyzed the data. N.I. built the model. S.B. advised on all aspects of the project. and N.I., S.B., and M.H. wrote the paper.
Declaration of Interests
The authors declare no competing financial interests.
Published: February 26, 2019
Footnotes
Supplemental Information can be found with this article online at https://doi.org/10.1016/j.molcel.2019.01.032.
Contributor Information
Silvija Bilokapic, Email: silvija.bilokapic@stjude.org.
Mario Halic, Email: mario.halic@stjude.org.
Supplemental Information
References
- Adams R.L., Wente S.R. Uncovering nuclear pore complexity with innovation. Cell. 2013;152:1218–1221. doi: 10.1016/j.cell.2013.02.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams P.D., Afonine P.V., Bunkóczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.-W., Kapral G.J., Grosse-Kunstleve R.W. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen K.R., Onischenko E., Tang J.H., Kumar P., Chen J.Z., Ulrich A., Liphardt J.T., Weis K., Schwartz T.U. Scaffold nucleoporins Nup188 and Nup192 share structural and functional properties with nuclear transport receptors. eLife. 2013;2:e00745. doi: 10.7554/eLife.00745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baake M., Bäuerle M., Doenecke D., Albig W. Core histones and linker histones are imported into the nucleus by different pathways. Eur. J. Cell Biol. 2001;80:669–677. doi: 10.1078/0171-9335-00208. [DOI] [PubMed] [Google Scholar]
- Bäuerle M., Doenecke D., Albig W. The requirement of H1 histones for a heterodimeric nuclear import receptor. J. Biol. Chem. 2002;277:32480–32489. doi: 10.1074/jbc.M202765200. [DOI] [PubMed] [Google Scholar]
- Bayliss R., Littlewood T., Stewart M. Structural basis for the interaction between FxFG nucleoporin repeats and importin-beta in nuclear trafficking. Cell. 2000;102:99–108. doi: 10.1016/s0092-8674(00)00014-3. [DOI] [PubMed] [Google Scholar]
- Bayliss R., Littlewood T., Strawn L.A., Wente S.R., Stewart M. GLFG and FxFG nucleoporins bind to overlapping sites on importin-beta. J. Biol. Chem. 2002;277:50597–50606. doi: 10.1074/jbc.M209037200. [DOI] [PubMed] [Google Scholar]
- Beck M., Hurt E. The nuclear pore complex: understanding its function through structural insight. Nat. Rev. Mol. Cell Biol. 2017;18:73–89. doi: 10.1038/nrm.2016.147. [DOI] [PubMed] [Google Scholar]
- Bilokapic S., Schwartz T.U. Molecular basis for Nup37 and ELY5/ELYS recruitment to the nuclear pore complex. Proc. Natl. Acad. Sci. USA. 2012;109:15241–15246. doi: 10.1073/pnas.1205151109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilokapic S., Strauss M., Halic M. Histone octamer rearranges to adapt to DNA unwrapping. Nat. Struct. Mol. Biol. 2018;25:101–108. doi: 10.1038/s41594-017-0005-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilokapic S., Strauss M., Halic M. Structural rearrangements of the histone octamer translocate DNA. Nat. Commun. 2018;9:1330. doi: 10.1038/s41467-018-03677-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth D.S., Cheng Y., Frankel A.D. The export receptor Crm1 forms a dimer to promote nuclear export of HIV RNA. eLife. 2014;3:e04121. doi: 10.7554/eLife.04121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borgia A., Borgia M.B., Bugge K., Kissling V.M., Heidarsson P.O., Fernandes C.B., Sottini A., Soranno A., Buholzer K.J., Nettels D. Extreme disorder in an ultrahigh-affinity protein complex. Nature. 2018;555:61–66. doi: 10.1038/nature25762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassany A., Gerace L. Reconstitution of nuclear import in permeabilized cells. Methods Mol. Biol. 2009;464:181–205. doi: 10.1007/978-1-60327-461-6_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi S., Yamashita E., Yasuhara N., Song J., Son S.-Y., Won Y.H., Hong H.R., Shin Y.S., Sekimoto T., Park I.Y. Structural basis for the selective nuclear import of the C2H2 zinc-finger protein Snail by importin β. Acta Crystallogr. D Biol. Crystallogr. 2014;70:1050–1060. doi: 10.1107/S1399004714000972. [DOI] [PubMed] [Google Scholar]
- Choudhary D., Kumar A., Magliery T.J., Sotomayor M. Using thermal scanning assays to test protein-protein interactions of inner-ear cadherins. PLoS ONE. 2017;12:e0189546. doi: 10.1371/journal.pone.0189546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie M., Chang C.-W., Róna G., Smith K.M., Stewart A.G., Takeda A.A.S., Fontes M.R.M., Stewart M., Vértessy B.G., Forwood J.K., Kobe B. Structural Biology and Regulation of Protein Import into the Nucleus. J. Mol. Biol. 2016;428(10 Pt A):2060–2090. doi: 10.1016/j.jmb.2015.10.023. [DOI] [PubMed] [Google Scholar]
- Cingolani G., Petosa C., Weis K., Müller C.W. Structure of importin-beta bound to the IBB domain of importin-alpha. Nature. 1999;399:221–229. doi: 10.1038/20367. [DOI] [PubMed] [Google Scholar]
- Cingolani G., Lashuel H.A., Gerace L., Müller C.W. Nuclear import factors importin alpha and importin beta undergo mutually induced conformational changes upon association. FEBS Lett. 2000;484:291–298. doi: 10.1016/s0014-5793(00)02154-2. [DOI] [PubMed] [Google Scholar]
- Conti E., Müller C.W., Stewart M. Karyopherin flexibility in nucleocytoplasmic transport. Curr. Opin. Struct. Biol. 2006;16:237–244. doi: 10.1016/j.sbi.2006.03.010. [DOI] [PubMed] [Google Scholar]
- de Castro E., Sigrist C.J.A., Gattiker A., Bulliard V., Langendijk-Genevaux P.S., Gasteiger E., Bairoch A., Hulo N. ScanProsite: detection of PROSITE signature matches and ProRule-associated functional and structural residues in proteins. Nucleic Acids Res. 2006;34:W362-5. doi: 10.1093/nar/gkl124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P., Lohkamp B., Scott W.G., Cowtan K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fassati A., Görlich D., Harrison I., Zaytseva L., Mingot J.-M. Nuclear import of HIV-1 intracellular reverse transcription complexes is mediated by importin 7. EMBO J. 2003;22:3675–3685. doi: 10.1093/emboj/cdg357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flemming D., Devos D.P., Schwarz J., Amlacher S., Lutzmann M., Hurt E. Analysis of the yeast nucleoporin Nup188 reveals a conserved S-like structure with similarity to karyopherins. J. Struct. Biol. 2012;177:99–105. doi: 10.1016/j.jsb.2011.11.008. [DOI] [PubMed] [Google Scholar]
- Fyodorov D.V., Zhou B.R., Skoultchi A.I., Bai Y. Emerging roles of linker histones in regulating chromatin structure and function. Nat. Rev. Mol. Cell Biol. 2018;19:192–206. doi: 10.1038/nrm.2017.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Görlich D., Vogel F., Mills A.D., Hartmann E., Laskey R.A. Distinct functions for the two importin subunits in nuclear protein import. Nature. 1995;377:246–248. doi: 10.1038/377246a0. [DOI] [PubMed] [Google Scholar]
- Görlich D., Panté N., Kutay U., Aebi U., Bischoff F.R. Identification of different roles for RanGDP and RanGTP in nuclear protein import. EMBO J. 1996;15:5584–5594. [PMC free article] [PubMed] [Google Scholar]
- Görlich D., Dabrowski M., Bischoff F.R., Kutay U., Bork P., Hartmann E., Prehn S., Izaurralde E. A novel class of RanGTP binding proteins. J. Cell Biol. 1997;138:65–80. doi: 10.1083/jcb.138.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant T., Grigorieff N. Measuring the optimal exposure for single particle cryo-EM using a 2.6 Å reconstruction of rotavirus VP6. eLife. 2015;4:e06980. doi: 10.7554/eLife.06980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayama R., Rout M.P., Fernandez-Martinez J. The nuclear pore complex core scaffold and permeability barrier: variations of a common theme. Curr. Opin. Cell Biol. 2017;46:110–118. doi: 10.1016/j.ceb.2017.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hergeth S.P., Schneider R. The H1 linker histones: multifunctional proteins beyond the nucleosomal core particle. EMBO Rep. 2015;16:1439–1453. doi: 10.15252/embr.201540749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzog F., Kahraman A., Boehringer D., Mak R., Bracher A., Walzthoeni T., Leitner A., Beck M., Hartl F.-U., Ban N. Structural probing of a protein phosphatase 2A network by chemical cross-linking and mass spectrometry. Science. 2012;337:1348–1352. doi: 10.1126/science.1221483. [DOI] [PubMed] [Google Scholar]
- Hough L.E., Dutta K., Sparks S., Temel D.B., Kamal A., Tetenbaum-Novatt J., Rout M.P., Cowburn D. The molecular mechanism of nuclear transport revealed by atomic-scale measurements. eLife. 2015;4:e10027. doi: 10.7554/eLife.10027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivic N., Bilokapic S., Halic M. Preparative two-step purification of recombinant H1.0 linker histone and its domains. PLoS ONE. 2017;12:e0189040. doi: 10.1371/journal.pone.0189040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jäkel S., Albig W., Kutay U., Bischoff F.R., Schwamborn K., Doenecke D., Görlich D. The importin beta/importin 7 heterodimer is a functional nuclear import receptor for histone H1. EMBO J. 1999;18:2411–2423. doi: 10.1093/emboj/18.9.2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jäkel S., Mingot J.-M., Schwarzmaier P., Hartmann E., Görlich D. Importins fulfil a dual function as nuclear import receptors and cytoplasmic chaperones for exposed basic domains. EMBO J. 2002;21:377–386. doi: 10.1093/emboj/21.3.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keck K.M., Pemberton L.F. Histone chaperones link histone nuclear import and chromatin assembly. Biochim. Biophys. Acta. 2013;1819:277–289. [PubMed] [Google Scholar]
- Kucukelbir A., Sigworth F.J., Tagare H.D. Quantifying the local resolution of cryo-EM density maps. Nat. Methods. 2014;11:63–65. doi: 10.1038/nmeth.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitner A., Joachimiak L.A., Unverdorben P., Walzthoeni T., Frydman J., Förster F., Aebersold R. Chemical cross-linking/mass spectrometry targeting acidic residues in proteins and protein complexes. Proc. Natl. Acad. Sci. USA. 2014;111:9455–9460. doi: 10.1073/pnas.1320298111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S.M., Stewart M. Structural basis for the high-affinity binding of nucleoporin Nup1p to the Saccharomyces cerevisiae importin-beta homologue, Kap95p. J. Mol. Biol. 2005;349:515–525. doi: 10.1016/j.jmb.2005.04.003. [DOI] [PubMed] [Google Scholar]
- Milles S., Mercadante D., Aramburu I.V., Jensen M.R., Banterle N., Koehler C., Tyagi S., Clarke J., Shammas S.L., Blackledge M. Plasticity of an ultrafast interaction between nucleoporins and nuclear transport receptors. Cell. 2015;163:734–745. doi: 10.1016/j.cell.2015.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosammaparast N., Guo Y., Shabanowitz J., Hunt D.F., Pemberton L.F. Pathways mediating the nuclear import of histones H3 and H4 in yeast. J. Biol. Chem. 2002;277:862–868. doi: 10.1074/jbc.M106845200. [DOI] [PubMed] [Google Scholar]
- Mosammaparast N., Del Rosario B.C., Pemberton L.F. Modulation of histone deposition by the karyopherin kap114. Mol. Cell. Biol. 2005;25:1764–1778. doi: 10.1128/MCB.25.5.1764-1778.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mühlhäusser P., Müller E.C., Otto A., Kutay U. Multiple pathways contribute to nuclear import of core histones. EMBO Rep. 2001;2:690–696. doi: 10.1093/embo-reports/kve168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Radermacher M. Three-dimensional reconstruction of single particles from random and nonrandom tilt series. J. Electron Microsc. Tech. 1988;9:359–394. doi: 10.1002/jemt.1060090405. [DOI] [PubMed] [Google Scholar]
- Raveh B., Karp J.M., Sparks S., Dutta K., Rout M.P., Sali A., Cowburn D. Slide-and-exchange mechanism for rapid and selective transport through the nuclear pore complex. Proc. Natl. Acad. Sci. USA. 2016;113:E2489–E2497. doi: 10.1073/pnas.1522663113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohou A., Grigorieff N. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 2015;192:216–221. doi: 10.1016/j.jsb.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres S.H.W. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres S.H.W., Núñez-Ramírez R., Sorzano C.O.S., Carazo J.M., Marabini R. Image processing for electron microscopy single-particle analysis using XMIPP. Nat. Protoc. 2008;3:977–990. doi: 10.1038/nprot.2008.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Söding J., Biegert A., Lupas A.N. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33:W244-8. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparks S., Temel D.B., Rout M.P., Cowburn D. Deciphering the “Fuzzy” Interaction of FG Nucleoporins and Transport Factors Using Small-Angle Neutron Scattering. Structure. 2018;26:477–484.e4. doi: 10.1016/j.str.2018.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart M. Molecular mechanism of the nuclear protein import cycle. Nat. Rev. Mol. Cell Biol. 2007;8:195–208. doi: 10.1038/nrm2114. [DOI] [PubMed] [Google Scholar]
- Stuwe T., Lin D.H., Collins L.N., Hurt E., Hoelz A. Evidence for an evolutionary relationship between the large adaptor nucleoporin Nup192 and karyopherins. Proc. Natl. Acad. Sci. USA. 2014;111:2530–2535. doi: 10.1073/pnas.1311081111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang G., Peng L., Baldwin P.R., Mann D.S., Jiang W., Rees I., Ludtke S.J. EMAN2: an extensible image processing suite for electron microscopy. J. Struct. Biol. 2007;157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- Timney B.L., Raveh B., Mironska R., Trivedi J.M., Kim S.J., Russel D., Wente S.R., Sali A., Rout M.P. Simple rules for passive diffusion through the nuclear pore complex. J. Cell Biol. 2016;215:57–76. doi: 10.1083/jcb.201601004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres C.M., Biran A., Burney M.J., Patel H., Henser-Brownhill T., Cohen A.S., Li Y., Ben-Hamo R., Nye E., Spencer-Dene B. The linker histone H1.0 generates epigenetic and functional intratumor heterogeneity. Science. 2016;353:aaf1644. doi: 10.1126/science.aaf1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulrich A., Andersen K.R., Schwartz T.U. Exponential megapriming PCR (EMP) cloning--seamless DNA insertion into any target plasmid without sequence constraints. PLoS ONE. 2012;7:e53360. doi: 10.1371/journal.pone.0053360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vetter I.R., Arndt A., Kutay U., Görlich D., Wittinghofer A. Structural view of the Ran-Importin beta interaction at 2.3 A resolution. Cell. 1999;97:635–646. doi: 10.1016/s0092-8674(00)80774-6. [DOI] [PubMed] [Google Scholar]
- Volpon L., Culjkovic-Kraljacic B., Osborne M.J., Ramteke A., Sun Q., Niesman A., Chook Y.M., Borden K.L. Importin 8 mediates m7G cap-sensitive nuclear import of the eukaryotic translation initiation factor eIF4E. Proc. Natl. Acad. Sci. USA. 2016;113:5263–5268. doi: 10.1073/pnas.1524291113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker P., Doenecke D., Kahle K. Importin 13 mediates nuclear import of histone fold-containing chromatin accessibility complex heterodimers. J. Biol. Chem. 2009;284:11652–11662. doi: 10.1074/jbc.M806820200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walzthoeni T., Claassen M., Leitner A., Herzog F., Bohn S., Förster F., Beck M., Aebersold R. False discovery rate estimation for cross-linked peptides identified by mass spectrometry. Nat. Methods. 2012;9:901–903. doi: 10.1038/nmeth.2103. [DOI] [PubMed] [Google Scholar]
- Warren C., Shechter D. Fly Fishing for Histones: Catch and Release by Histone Chaperone Intrinsically Disordered Regions and Acidic Stretches. J. Mol. Biol. 2017;429:2401–2426. doi: 10.1016/j.jmb.2017.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb B., Sali A. Comparative Protein Structure Modeling Using MODELLER. Curr. Protoc. Bioinformatics. 2016;54:5.6.1–5.6.37. doi: 10.1002/cpbi.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohlwend D., Strasser A., Dickmanns A., Doenecke D., Ficner R. Thermodynamic analysis of H1 nuclear import: receptor tuning of importinbeta/importin7. J. Biol. Chem. 2007;282:10707–10719. doi: 10.1074/jbc.M610409200. [DOI] [PubMed] [Google Scholar]
- Xu D., Farmer A., Chook Y.M. Recognition of nuclear targeting signals by Karyopherin-β proteins. Curr. Opin. Struct. Biol. 2010;20:782–790. doi: 10.1016/j.sbi.2010.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahn R., Osmanović D., Ehret S., Araya Callis C., Frey S., Stewart M., You C., Görlich D., Hoogenboom B.W., Richter R.P. A physical model describing the interaction of nuclear transport receptors with FG nucleoporin domain assemblies. eLife. 2016;5:e14119. doi: 10.7554/eLife.14119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuleger N., Kelly D.A., Richardson A.C., Kerr A.R.W., Goldberg M.W., Goryachev A.B., Schirmer E.C. System analysis shows distinct mechanisms and common principles of nuclear envelope protein dynamics. J. Cell Biol. 2011;193:109–123. doi: 10.1083/jcb.201009068. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data resources
The accession numbers for the Imp7:Impβ:H1.0 and Impβ:H1.0 atomic coordinates have been deposited in the PDB: 6N88 and 6N89. The Imp7:Impβ:H1.0, Imp7:Impβ and Impβ:H1.0 cryo-EM maps have been deposited in the EMDB: EMD-0366, EMD-0367 and EMD-0368, respectively. The original data are deposited to https://doi.org/10.17632/dfjzx4sxrs.1.