Abstract
Objective:
The current state of the art for compartment modeling of dynamic PET data can be described as a two-stage approach. In Stage 1, individual estimates of kinetic parameters are obtained by fitting models using standard techniques, such as nonlinear least squares (NLS), to each individual’s data one subject at a time. Population-level effects, such as the difference between diagnostic groups, are analyzed in Stage 2 using standard statistical methods by treating the individual estimates as if they were observed data. While this approach is generally valid, it is possible to increase efficiency and precision of the analysis, allow more complex models to be fit, and also to permit parameter-specific investigation by fitting data across subjects simultaneously. We explore the application of nonlinear mixed-effects (NLME) models for estimation and inference in this setting.
Methods:
In the NLME framework, subjects are modeled simultaneously through the inclusion of random effects of subjects for each kinetic parameter; meanwhile, population parameters are estimated directly in a joint model.
Results:
Simulation results indicate that NLME outperforms the two-stage approach in estimating group-level effects and also has improved power to detect differences across groups. We applied our NLME approach to clinical PET data and found effects not detected by the two-stage approach.
Conclusion:
The proposed NLME approach is more accurate and correspondingly more powerful than the two-stage approach in compartment modeling of PET data. Significance: The NLME method can broaden the methodological scope of PET modeling because of its efficiency and stability.
Keywords: Compartment model, Nonlinear least squares, Model building
I. Introduction
DYNAMIC PET imaging has been widely used in studies of mental and neurological disorders. One very common application of PET imaging involves estimating the distribution of various macromolecules, often proteins, throughout the brain. In dynamic PET studies, a time activity curve (TAC) reflects the sequence of concentrations across time for any given voxel or region and is often used to estimate quantities related to the density of the target protein at each location.
The TAC, denoted as CT, is conceptualized as the convolution between two functions
| (1) |
where t is time, CP is the input function and H is the voxel- or region-specific impulse response function (IRF). The input function CP represents the concentration of the tracer in the arterial plasma over time, corrected to account for the radioactive metabolites of the tracer, and quantifies the amount of tracer molecules that are available to enter the brain at any given time. In practice, the input function requires blood data during the scan. The location-specific IRF may be interpreted as the hypothetical concentration of the tracer over time in the corresponding region if the input function were an instantaneous bolus spike. Because the IRF describes the physiological and pharmacological properties of the system, the analysis of the kinetic behavior of the tracer centers on estimating the IRF in Model (1).
The most widely used approach for tracer kinetic analysis is compartment modeling. Under the assumptions of this approach, the IRF has the form
| (2) |
where J is the total number of tissue compartments, and Lj and Rj are functions of the rate parameters k. The rate parameters are the key elements to be estimated because they completely characterize the kinetic behavior of the tracer based on the assumed model. Standard quantities of clinical importance, such as volume of distribution (VND & VT) and binding potential (BPND & BPP), can be expressed as functions of the rate parameters. For example, under the assumption of two-tissue compartment model which has four rate parameters (K1, k2, k3, k4), the forms of these measures are given by Innis et al. [1]:
A well established and almost universally applied method for compartment modeling of PET data can be characterized as a two-stage approach: in Stage 1, individual estimates of all kinetic parameters are obtained by fitting each individual’s data, one subject and one voxel/region at a time, using common techniques such as nonlinear least squares (NLS). Standard outcome measures of binding, including VND, VT, BPND, BPP, etc., can be calculated based on the estimated rate parameters. In Stage 2, the binding measures are compared across subjects, treating the individual estimates of the outcome measures as if they were the originally observed data; population-level effects, such as the difference between patients and controls, are examined using standard statistical methods. While this two-stage approach is statistically valid, it represents an inefficient use of the observed PET data, limiting the complexity of models that can be fit, the nature of comparisons that can be made, the precision of population-level estimates, and the power to detect population-level effects.
We advocate an alternative approach to the analysis of dynamic PET data that provides a flexible and efficient alternative to the two-stage approach. Specifically, we model all subjects simultaneously rather than one at a time by fitting nonlinear mixed-effects (NLME) models. NLME addresses the inherent instability of subject-level rate parameter estimates in the two-stage approach by jointly modeling all subjects, and produces improved individual estimates. This approach accounts for subject-to-subject variability directly by modeling each subject’s rate parameter as coming from a distribution of rate parameters, i.e., by considering a subject-level random effect. For instance, assuming one distribution for patients and another for controls, with an NLME approach, we are essentially analyzing the difference between these two distributions. Under the modeling framework of this approach, the difference is limited to a mean shift as we assume there is a shared variability in the random effects of both groups. In the NLME modeling framework, both individual rate parameters and the effects of some covariates on the rate parameters can be estimated in a single analysis, and taking this approach allows for more complex models than could be fit otherwise.
In addition to pursuing an NLME modeling approach for compartment modeling with PET data, we describe a model building procedure that is important when applying our approach to real data. Two main issues that need to be addressed are selecting the explanatory variables that should be included as fixed effects and identifying the parameters that should have an associated random effect with non-zero variance. We illustrate the general principles for model building through the careful analysis of our motivating clinical PET data.
Some previous work has used NLME models in the context of PET data. Most relevant to our contribution, Van Rij et al. [2] and Syvnen et al. [3] analyze PET data using NLME under the assumption of a two compartment model with a parameterization that is different from the model we use in this paper, and account for effects of potential covariates. Berges et al. [4] and Lim et al. [5] apply the NLME approach to PET data in the context of a direct PK-receptor occupancy relationship described by an Emax model, rather than in the context of a compartment model. Zamuner et al. [6] introduces an NLME approach accounting for intra- and inter-subject variability on the time-activity curve under the assumption of the Simplified Reference Tissue Model (SRTM); Kgedal et al. [7] extends this to a two tissue compartment model and simultaneously estimates the radioligand kinetics of two regions. Other papers have used NLME in the analysis of PET data, but in ways that are less relevant to our focus on improved compartment modeling. Veronese et al. [8] takes an NLME approach to model metabolite data across subjects in order to efficiently estimate the input function required for standard kinetic modeling. Bertoldo et al. [9] proposes an iterative two stage (ITS) method to model multiple ROIs, rather than multiple subjects, simultaneously, and assesses its performance by fitting a complex five-rate constant model to PET data.
In this paper we describe a general framework for modeling PET data using NLME with emphasis on model fitting and assessment. We present a model building procedure and establish criteria to select fixed and random effects. We conduct novel simulations to assess the improvement in estimation accuracy and power to detect group differences comparing NLME and the two-stage approach. In contrast to previous work, which used the commercial NONMEM software [10], our algorithm is implemented in R [11], a free and open source software, and code files that are used to generate the results in this paper are provided in the supplementary materials. The NLS model in the two-stage approach can be implemented using kinfitr package [12] in R.
The rest of the paper is organized as follows. In Section II, we present the NLME modeling framework for PET data and introduce criteria that can be used to select the fixed and random effects. The results of simulations comparing our proposed NLME method to the two-stage approach are given in Section III, with particular emphasis on power to detect differences across groups. In Section IV, we illustrate the model building procedure by using clinical PET data as an example. Finally, we summarize the main results in Section V and present a short discussion in Section VI.
II. Methodology
Intravascular activity may have a significant contribution to the total concentration of the tracer, so the whole blood concentration should be accounted for in the data analysis. Under the assumption of compartment models, Model (1) can be reformulated as
| (3) |
where CB is the time activity curve in the whole blood and Vb is the fractional blood volume of the tissue. As in (2), Hk here is the IRF which, under the assumptions of the compartment model, depends on a vector of rate parameters k.
We now describe how Model (3) can be cast in the general NLME framework. A general expression for the NLME model is given by
| (4) |
where yij is the jth observation of the ith subject, n is the number of subjects, ni is the number of measurements for subject i, and ϵij is the error for yij. In practice, the continuous-valued function CT in (3) is observed on a discrete grid of time points {tij}. Therefore, in the context of Model (3), the response yij is the TAC observations CT (tij); f is defined by the functions Hk, CP and CB; and the parameter vector θi, specific to subject i, consists of rate parameters ki and the fractional blood volume Vbi, i.e. . The exact form of f under the assumption of compartment models is given in the appendix.
Strictly speaking, CT(tij) j = 1, ⋯ , ni are derived from the decay counts observed over a given time interval across {tij}j=1, ⋯ , ni. By design, the time frames are gradually longer over time during the scan, because of the radioactive decay process and because of decreasing concentration of the tracer. The change in time frames over the scan has two practical implications. First, the discrete grid of time points on which TACs are observed is taken to be the midpoints of the frames, which can be irregular. Second, since data are observed over consecutive time frames of different lengths, the frame duration, the radioactive decay and the overall concentration affect the variability of the response. As a result, weighting schemes that account for these factors are necessary. In the simulation and real data analyses below, weights are set to be the duration of the time-frame corresponding to tij, as in Zanderigo et al. [13], although our methodology allows other weighting schemes to be used. Errors are assumed to be uncorrelated over time as they arise originally from decay count data, which are naturally independent, and then are reconstructed and registered separately for each time interval. Therefore, the errors of subject i
| (5) |
Wi is a diagonal matrix with diagonal elements {wij}l=1, ⋯ , ni, where {wij} are as fixed and known observation weights.
Within this modeling framework, we regard each subject’s kinetic parameter as following a particular probability distribution, often a Gaussian distribution, with mean and variance estimated from the data. For example, the value k4i for patient i is modeled as coming from a normal distribution with a mean and variance shared across all patients. This is a natural way to directly account for the (“biological”) variability in these parameters across subjects. Kinetic parameters can arise from distributions that have different means, e.g. a mean for the control group and another mean for a patient group. A major goal is thus to determine whether these two means, and thus the distributions of rate parameters in these two groups, are the same.
In Model (4), we expand the subject-level parameter vector θi as
| (6) |
In this representation, the first component accounts for deterministic factors (or “fixed effects”, depending on factors such as diagnosis, age, or sex) and the second component accounts for natural variability among subjects who have the same fixed effect specification. β is a matrix of fixed effect coefficients; xi is the “design matrix” which indicates the diagnosis, age, and other covariate values for each subject; and
| (7) |
is the vector of random effects for the ith subject, which represent the subject-specific deviations from the population averages. This expansion separates population averages from subject-specific deviations, and provides the mechanism through which group-level and subject-level kinetic behaviors can be understood. By modeling all subjects simultaneously, the properties of this distribution (including the variance Σ) can be estimated and used to stabilize subject-level estimates. This approach allows us to jointly model all subjects in a single analysis and to directly estimate and test for the significance of effects of covariates, such as diagnosis or age, on any of the individual rate parameters individually or jointly.
Taken together, the fixed and random effects are the parameters of interest in NLME representation of Model (1). These can be estimated either by maximum likelihood (ML) or by restricted maximum likelihood (REML) using the Lindstrom and Bates (LB) algorithm [14]. For our analyses, we use the implementation of this algorithm in the nlme package [15] in R. From the fitted model, estimates of the summarized measures, such as binding potentials (BPND and BPP) and total volume (VND and VT), can be computed at the population level using the fixed effects estimates. Furthermore, subject level predictions can also be obtained with the estimated random effects.
Key issues that arise when fitting NLME models for PET data include selecting covariates to include as fixed effects and determining which elements of the parameter vector θi should have associated random components. Because covariates can affect each of the rate parameters through the fixed effects specification, a global test can be used to assess the global significance of the covariate effect, for example, whether there exist non-zero differences comparing patients to controls for any of the rate parameters. Choices for determining the fixed effects include the likelihood ratio test (LRT) [16], alternative likelihood-based tests, such as Wald test [17] and score test [18], and information criterion statistics, such as Akaike information criterion (AIC) [19] and Bayesian information criterion (BIC) [20]. AIC and BIC can also be used to determine which effects should have associated random components. Alternatively, a non-standard likelihood ratio test can be applied on nested models to test whether one random effect component is zero, i.e., whether a parameter has significant between-subject variability. This non-standard LRT, proposed by Stram and Lee [21], addresses the issue that the testing value under the null hypothesis is on the boundary of the support of the parameter. Lastly, to obtain inferences directly for BPND, BPP, VND and VT the Delta method [22] is used to derive the standard errors based on the estimates and variance of rate parameter fixed effects.
III. Simulation
In this section we undertake a simulation exercise to understand the properties of NLME modeling for PET data and to compare the performance of the proposed methods to that of the two-stage approach.
Simulated datasets are designed to mimic our motivating data (PET data with [11C]WAY100635 tracer described in Section IV) in the following way. We begin by fitting Model (3) under a two-tissue compartment model assumption to the observed data with no covariates. From this model fit, we extract estimates of the fixed effects β, the random effect covariance Σ, and the error variance σ2, as shown in (6), (7) and (5), respectively, in Section II. These estimates are set to be the “truth” for the purposes of this simulation. To simulate new subject data, we sample observed input functions CP and whole blood time activity curves CB from subjects in the observed data with replacement. CP and CB in our data have the same forms as in Parsey et al. [23]. Subject-specific random effects are generated from a multivariate normal distribution with mean β and covariance Σ. The sampled functions and generated random effects are combined with estimated fixed effect parameters to produce simulated time activity curves CT according to Model (3). Simulated errors ϵi are generated from where σ2 = 0.01 and Wi is defined in the same way described in Section II. Each simulated dataset consists of 90 subjects, with half in each covariate group. Values of the β and Σ used to generate individual parameters are provided in the appendix.
A. Quality of fixed effect estimation
Our first objective is to assess how well NLME modeling estimates the “true” fixed effects. To do so, we generate 1000 datasets under the above design. For each of the simulated datasets, we apply both our proposed approach and two stage approach assuming there exists a group effect on all rate parameters but not blood volume. We arbitrarily choose one of the two groups to be the reference group, analogous to the control group in a medical study. In this simulation, every subject has a different set of rate parameters, drawn from a distribution that differs for the control group and the patient group. Also, every subject has a different blood volume Vbi and subjects in both groups have observations drawn from a common distribution. Below we compare the estimated values for both approaches to the “truth”, i.e., the values used to generate the data.
Figure 1 compares the estimates of both approaches for fixed effects related to each rate parameter as well as the summarized measures VT, BPND and BPP. The top row shows the relative estimation errors of fixed effects for subjects in the reference group, and the bottom row shows the absolute estimation errors of the difference between groups. As expected, the distribution of estimated values for the proposed NLME approach are generally narrower and include fewer outlying values than the corresponding distributions for the two-stage approach. As described in Section II, NLME improves and stabilizes the estimation of rate parameters at the subject level; this, in turn, leads to the observed improvement in estimation for population-level fixed effects.
Fig. 1.
Relative estimation errors of fixed effects in the reference group (top) and absolute estimation errors of the difference between groups (bottom) using both approaches.
B. Comparison of power for detecting group differences
The preceding simulation indicates that the proposed NLME approach is more accurate than two-stage approach for estimating group differences. We now explore how this difference affects the power to detect true differences in rate parameters or in binding measures when testing hypotheses.
We use the simulation design described above, with modifications that allow a careful comparison of power between approaches. Keeping the fixed effects in the reference group as they were, we initially set all group differences to zero. Then, we increase the group difference for a single rate parameter while keeping group differences for other rate parameters equal to zero. This process is repeated for each rate parameter in turn, and for each collection of true fixed effects we apply both approaches to 200 simulated datasets.
First, we compare methods on their ability to detect differences corresponding to individual rate parameters, i.e., to test , where and are rate parameters (ℓ ∈ {1, 2, 3, 4}) for controls and patients, respectively. Although all approaches are able to test effects on individual rate parameters, previous studies focus on testing effects on various outcome measures that are generally functions of the rate parameters. To evaluate this parameter-specific hypothesis test in simulations, we focus only on the rate parameter for which groups differ. Results for the NLME approach are obtained directly from the model fitting procedure, while for the two-stage approach we perform a t test on the individual rate parameter estimated from subject-specific NLS fits.
A better understanding about potential differences in individual rate parameters comparing groups, genders, or other covariates might lead new research to explore hypotheses about specific rate parameters. To date, though, tests for differences have focused only on summary measures like BP and little is known about differences in rate parameters. In the absence of an priori hypothesis about which rate parameters may be affected by which covariates, we focus here on applying a “global test” that is designed to find differences if any of the component rate parameters are affected, e.g. , , , , with the alternative hypothesis being that there is a difference in at least one of these. To test the global hypothesis using the standard two-stage approach, we use a multivariate analysis of variance (MANOVA) test, which is designed to test an effect on several dependent variables. In this case, once we have the individual rate parameters from Stage 1, we can use MANOVA to test whether there is a significant group effect on any of the four rate parameters. To conduct this test using NLME, we apply the derived LRT by fitting two different models: one in which the rate parameters do not depend on group and the other in which the rate parameters may depend on group. Then the two models are compared according to standard likelihood ratio testing.
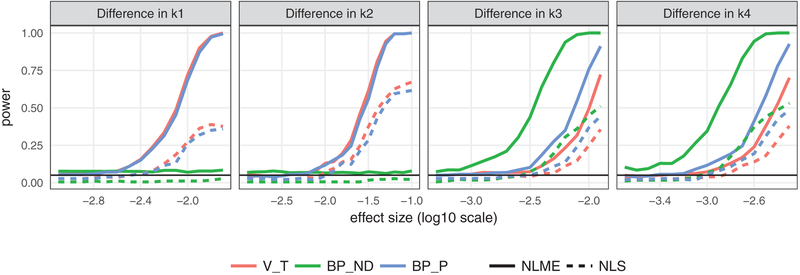
Figure 2 shows results for true differences in each of the four rate parameters, with power defined as the proportion of rejected null hypotheses across the 200 simulated datasets for each effect size. Unsurprisingly, the parameter-specific test of the form is more powerful than the global test in all cases. Importantly, for either test, the NLME approach is more powerful than the two-stage approach, often substantially so. This improvement in power derives from the better estimation of fixed effects observed in Section III-A.
Fig. 2.
Power curves of detecting the group mean difference on rate parameters using four different tests: parameter specific t test of group effect on each rate parameter using NLME model; LRT of overall group effect comparing nested NLME models; parameter specific t test of group effect on the rate parameters based on the two-stage approach; MANOVA test of overall group effect based on the two-stage approach. The black line in each plot represents the 0.05 nominal level.
Next, we compare methods on their ability to detect differences in the summary measures VT, BPND and BPP. The simulation design is as before, meaning that differences between groups exist in only one rate parameter at a time. The power to detect resulting differences in summary measures is shown in Figure 3. Because K1 and k2 do not affect BPND, group differences in these rate parameters are not detectable through this summary measure. However, VT and BPP are affected by such differences and NLME has much greater power than the two-stage approach to detect differences in those measures. Group differences in k3 and k4 affect all the summary measures, and as these group differences in rate parameters increases so does the power to detect differences in the summary measures. Again, NLME uniformly outperforms the two-stage approach.
Fig. 3.
Power curves of detecting the group mean difference on the summarized measures using six different tests: t test of group effect in VT based on NLME model; t test of group effect in BPND based on NLME model; t test of group effect in BPP based on NLME model; t test of group effect in VT based on two-stage approach; t test of group effect in BPND based on two-stage approach; t test of group effect in BPP based on two-stage approach. The black line in each plot represents the 0.05 nominal level.
IV. PET data analysis
Recent studies have shown that the serotonin 1A receptor (5-HT1A) plays a key role in major depressive disorder (MDD) [24] and bipolar disorder [25]. The [11CIWAY100635 tracer has been used widely to quantify 5-HT1A binding and the rate constant parameters when a compartment model is assumed [23].
Our data consist of TACs in the midbrain of 97 subjects who can be divided into three groups based on their prior medication history: MDD subjects who have not recently been on medication (NRM); antidepressant-exposed (AE) MDD subjects and MDD subjects who are on an adequate dose of antidepressant for at least 4 weeks [24]; and control subjects. Other covariates that may have effects on the rate parameters include age and gender. Metabolite corrected plasma data and whole blood data are available for all subjects.
In this section, we apply the NLME modeling approach described in Section II to the PET data under the assumption of a two-tissue compartment model. We use this data as an example to illustrate a model building procedure of NLME models on PET data. A related model building framework for NLME models can be found in Pinheiro et al. [26]. Because the primary interest lies in analyzing the group differences, our starting point is a model that includes this variable. Throughout, we will use global tests under the assumption that covariates may affect the four rate parameters but not the blood volume, and to start we assume that all rate parameters plus Vb have associated random effects.
A. Testing for random effects
The first question to address is whether all parameters exhibit subject-level variability, i.e., whether a particular parameter is identical for all the subjects with the same fixed effect specification or a parameter-specific random effect is needed. We fit separate models in which each the random effect for each of the parameters is omitted, and compare the results with the initial model using AIC and the LRT described in Section II. The initial model has the smallest AIC among all candidate models, and the p-values from the non-standard LRTs indicate that the random effect on each of the parameter is significant (largest p-value = 5.667 × 10−4). Thus, both criteria indicate that the model in which all the parameters have associated random components is superior, and we proceed to the selection of covariates for fixed effects.
Results are shown in Table I. Here we refer the model in which only the variance of random effect of kℓ is zero as Model ℓ and the model in which only the variance of random effect of Vb is zero as Model 5. For instance, Model 1 represents the model in which the variance of random effect of K1 is zero and the random effects of other parameters are allowed to have non-zero variances.
TABLE I.
AIC and LRT results for models with different number of random components.
| Model | AIC | log likelihood | test | p-value |
|---|---|---|---|---|
| Model 0 | −11617.98 | 5837.990 | ||
| Model 1 | −11003.95 | 5525.974 | 0 vs 1 | 6.972 × 10−133 |
| Model 2 | −11596.87 | 5822.436 | 0 vs 2 | 5.917 × 10−6 |
| Model 3 | −11606.99 | 5827.497 | 0 vs 3 | 5.667 × 10−4 |
| Model 4 | −11588.86 | 5818.430 | 0 vs 4 | 1.452 × 10−7 |
| Model 5 | −11579.03 | 5813.515 | 0 vs 5 | 1.435 × 10−9 |
B. Including covariate fixed effects
Covariates such as age and gender may affect rate parameters, and we now consider their addition to our model. We add these variables as fixed effects in a global way by including covariate effects on all rate parameters, and build our model using forward selection with a global hypothesis test. Both main effects and interactions between variables are considered. The results of our model building process are shown in Table II, and indicate that age and gender are significant predictors and none of the two-way interactions are significant. Therefore, we determine that the model with only the main effects of group, age and gender is our final model. Model fits of the final model using NLME and the two-stage approach are given in Table III. Estimates and standard errors of rate parameters and summary measures in each group are presented in Table A.1 in the appendix as a supplement. The estimated correlation matrix of the random effects for (K1, k2, k3, k4, Vb) is
TABLE II.
Results of LRT comparing nested models with difference combination of covariates
| Model | Fixed effect structure | Test | p-value |
|---|---|---|---|
| 1 | Group | ||
| 2 | Group + Gender | 1 vs 2 | 0.0293 |
| 3 | Group + Age | 1 vs 3 | 0.0013 |
| 4 | Group + Gender + Age | 3 vs 4 | 0.0154 |
| 5 | Group + Gender + Age + Gender * Age | 4 vs 5 | 0.4331 |
| 6 | Group + Gender + Age + Group * Gender | 4 vs 6 | 0.4100 |
| 7 | Group + Gender + Age + Group * Age | 4 vs 7 | 0.6280 |
TABLE III.
Results of the NLME and two-stage approaches
| Parameter | Variable | NLME | Two-stage approach | ||||
|---|---|---|---|---|---|---|---|
| Estimate (× 10−3) | Std.Error (× 10−3) | p-value | Estimate (×10−3) | Std.Error (×10−3) | p-value | ||
| K1 | AE - Control | −6.182 | 4.023 | 0.125 | −6.821 | 4.167 | 0.105 |
| NRM - Control | 5.415 | 4.179 | 0.195 | 3.827 | 4.235 | 0.369 | |
| Male - Female | −8.232 | 3.302 | 0.013 | −7.028 | 3.407 | 0.042 | |
| Age (1 year diff.) | −0.021 | 0.120 | 0.858 | −0.035 | 0.126 | 0.783 | |
| k2 | AE - Control | 7.863 | 7.956 | 0.323 | −1.502 | 11.690 | 0.898 |
| NRM - Control | 14.148 | 8.697 | 0.104 | 2.240 | 11.882 | 0.851 | |
| Male - Female | −14.756 | 6.544 | 0.024 | −11.014 | 9.557 | 0.252 | |
| Age (1 year diff.) | 0.115 | 0.213 | 0.589 | 0.015 | 0.352 | 0.965 | |
| k3 | AE - Control | −1.909 | 2.450 | 0.436 | −3.728 | 3.433 | 0.280 |
| NRM - Control | 2.180 | 2.679 | 0.416 | 0.697 | 3.489 | 0.842 | |
| Male - Female | −2.427 | 2.073 | 0.242 | −2.084 | 2.807 | 0.460 | |
| Age (1 year diff.) | −0.145 | 0.065 | 0.025 | −0.121 | 0.104 | 0.244 | |
| k4 | AE - Control | −0.022 | 0.968 | 0.982 | −0.130 | 1.331 | 0.922 |
| NRM - Control | −0.544 | 0.963 | 0.572 | −0.476 | 1.353 | 0.726 | |
| Male - Female | 0.501 | 0.777 | 0.520 | 0.536 | 1.088 | 0.624 | |
| Age (1 year diff.) | 0.031 | 0.026 | 0.236 | 0.063 | 0.040 | 0.123 | |
| VND | AE - Control | −46.093 | 25.715 | 0.076 | −41.397 | 26.081 | 0.116 |
| NRM - Control | 1.819 | 26.138 | 0.945 | 9.783 | 26.510 | 0.713 | |
| Male - Female | −29.146 | 21.024 | 0.169 | −27.244 | 21.323 | 0.205 | |
| Age (1 year diff.) | −0.396 | 0.775 | 0.611 | −0.218 | 0.786 | 0.782 | |
| VT | AE - Control | −199.169 | 94.978 | 0.039 | −4509.015 | 5484.074 | 0.413 |
| NRM - Control | 64.530 | 96.539 | 0.506 | −3983.753 | 5574.219 | 0.477 | |
| Male - Female | −165.526 | 77.651 | 0.036 | −4190.468 | 4483.598 | 0.352 | |
| Age (1 year diff.) | −4.699 | 2.864 | 0.104 | 62.156 | 165.370 | 0.708 | |
| BPND | AE - Control | −103.513 | 43.309 | 0.019 | −21594.627 | 27 285.339 | 0.431 |
| NRM - Control | 180.970 | 44.021 | <0.001 | −20 077.763 | 27 733.845 | 0.471 | |
| Male - Female | −193.907 | 35.408 | <0.001 | −20 251.287 | 22 307.596 | 0.366 | |
| Age (1 year diff.) | −11.674 | 1.306 | <0.001 | 320.045 | 822.780 | 0.698 | |
| BPP | AE - Control | −153.075 | 69.782 | 0.031 | −4467.618 | 5486.624 | 0.418 |
| NRM - Control | 62.711 | 70.929 | 0.379 | −3993.535 | 5576.812 | 0.476 | |
| Male - Female | −136.380 | 57.051 | 0.019 | −4163.224 | 4485.684 | 0.356 | |
| Age (1 year diff.) | −4.303 | 2.104 | 0.044 | 62.374 | 165.447 | 0.707 | |
Among the rate parameters, there is low correlation between k2 and k3 and between K1 and k4; Vb has a weak correlation with all the rate parameters. Additionally, in order to evaluate the effect of bias introduced by any covariates, we compared the individual estimates of rate parameters and summary measures using the NLME models with and without covariates including diagnostic group, age and gender. The similarity between the estimates indicates that any bias in the estimation is not greatly influenced by inclusion/exclusion of covariates.
C. Comparison with the two-stage approach
Next, we compare NLS estimates of the parameters obtained from the two-stage approach by fitting a two-tissue compartment model on each subject to those obtained from the NLME fit of the final model. Figure 4 plots these estimates for all the parameters, including the summary measures, and includes an identity line for reference. The approaches give similar estimates for K1, VND, VT and BPP, but the impact of assuming a random effects structure is clear for k2, k3, k4 and Vb: the NLME estimates have smaller variances. This “shrinkage” is expected from the NLME approach, and in fact is a reason why the approach is less vulnerable to individual outliers than NLS estimates. That is, it is difficult to obtain accurate and stable estimates for these rate parameters using NLS. In contrast, by simultaneously estimating rate parameters for all subjects and using the random effects distribution, NLME is able to balance subject- and population-level data to improve rate parameter estimation even at the level of individual subject.
Fig. 4.
Individual NLME estimates vs NLS estimates. The solid line on each panel is the identity line with intercept 0 and slope 1.(In VT, BPND and BPP panels, 2 points out of 97 are out of the figure axis ranges.)
The parameter estimates, standard errors and the p-values of the t tests, are given in Table III. According to the p-values of t tests associated with the comparisons of different covariates, our final NLME model identifies gender as a significant factor for K1, k2, VT, BPND and BPP; and age is a significant factor for k3, BPND and BPP. Based on parameter estimates, it is possible to draw several interesting conclusions. For example, adjusted for group and age, the mean of the K1 distribution for males is is significantly lower than the mean of the corresponding distribution of K1 values for females. Also, again adjusted for group and gender, as the age of a subject increases by 1 year the model predicts a slight decrease in mean k3.
Table IV shows the significance level of the overall effects of the covariates in both NLME and the two-stage approaches. Likelihood ratio tests are performed to assess the global effects on all rate parameters for NLME while the MANOVA F-tests are used for the two-stage approach. Both models identify age as a significant factor, but only the NLME approach detects a significant overall effect of gender. Neither approach suggests a significant overall effect of prior medication history group, although the p-value is somewhat smaller for NLME than for the two-stage approach.
TABLE IV.
p-values of overall effects in NLME and two-stage approaches
| NLME | Two-stage approach | |
|---|---|---|
| Group | 0.0798 | 0.1140 |
| Gender | 0.0154 | 0.1376 |
| Age | 0.0006 | 0.0006 |
V. Conclusion
We proposed a NLME approach for compartment modeling of PET data. The NLME approach addresses known shortcomings of the standard two-stage approach by fitting all subjects simultaneously and estimating covariate effects in a one-step model process. Our simulations indicate that the proposed NLME approach is more accurate and correspondingly more powerful in detecting group differences than the two-stage approach. In real data analyses, the NLME estimates of individual rate parameters often had narrower distributions than estimates derived from two-stage approach, an expected byproduct of the balancing subject and population data to estimate individual effects. We applied a model building procedure for the NLME approach to WAY tracer based on the two-tissue compartment model, and found effects not detected by a two-stage approach.
VI. Discussion
The instability of NLS for estimating rate parameters is a frequently encountered issue in practice. One potential solution is to set bounds on rate parameter estimates, but this is arbitrary and the bounds would have to set separately for each tracer. These bounds artificially reduce the range of rate parameters, and introduce a new problem of sensitivity to their specification. Instead, an NLME approach is based on a statistically principled modeling technique that naturally stabilizes individual rate parameter estimates and assesses covariate effects in a single step.
To demonstrate that the two-tissue compartment model that we fit on the target region can achieve meaningful parameter estimates, we fit a one-tissue compartment model to data of the white matter cerebellum, which is widely used as a reference region for [11C]WAY100635 tracer. A reference region is a region that is devoid of the target protein and is often used to isolate the effect of non-specific binding in a compartment modeling framework. The estimates obtained with the help of the reference region, which we refer as the “indirect” estimates, are compared to the “direct” estimates obtained by solely fitting the target region. The results indicate that the estimates match for meaningful parameters such as VND, BPP, BPND and for both NLME and NLS methods. For example, the NLME estimates of VND listed in Table A.1 (Control: 0.331 ± 0.033; AE: 0.285 ± 0.039; NRM: 0.333 ± 0.036) are compatible with the NLME estimate of VT from the 1TC fit of the reference region (0.339 ± 0.099), as well as the VT estimate (0.298 ± 0.112) as reported in [27].
Our work has focused on the two-tissue compartment model; extending this to a more complicated three-tissue compartment (3TC) model will introduce additional complexity but which will be important in some applications. Fitting a 3TC model is not generally possible when fitting one subject at a time, but the mixed effects framework may make these models more feasible by using information across subjects to improve estimation. Restrictions on the structure of variance-covariance matrix of the random effects might also be necessary as it reduces the number of the parameters and increases computational efficiency. Another direction we might take includes developing an NLME modeling approach to model multiple voxels/regions simultaneously. It is at least conceptually possible to extend the model to allow multiple voxels/regions. Modeling the correlation structure, however, becomes a rather complex problem because of the increased size of the variance-covariance matrix.
The availability of user-friendly software for NLME modeling of dynamic PET data is important for the wide adoption of these methods. Previous work has used NONMEM to implement NLME models; our models were fit using R, a free open-source platform.
Supplementary Material
VII. Acknowledgments
This research was supported in part by Grant 1R01EB024526 from the National Institute of Biomedical Imaging and Bioengineering.
Appendix I
Forms of nonlinear models
The exact forms of f in Model 4 are shown as follows. The input function CP and the whole blood function CB have the same forms as in Parsey et al. [23]. is the indicator function. f is based on an analytic convolution of the functions, while other approaches just involve in numerical convolutions.
A. One-tissue compartment (1TC) model
B. Two-tissue compartment (2TC) model
Appendix II
Parameter values used to simulate data
In Section III, the individual parameters are generated from the following multivariate normal distribution:
TABLE A.1.
Estimates and standard errors of parameters in each group
| Parameter | Variable | NLME | Two-stage approach | ||
|---|---|---|---|---|---|
| Estimate (×10−3) | Std.Error (×10−3) | Estimate (×10−3) | Std.Error (×10−3) | ||
| K1 | Control | 61.019 | 5.203 | 60.998 | 5.369 |
| AE | 54.836 | 6.089 | 54.177 | 6.333 | |
| NRM | 66.433 | 5.669 | 64.825 | 5.811 | |
| Female | 61.020 | 5.203 | 60.998 | 5.369 | |
| Male | 52.788 | 5.493 | 53.970 | 5.741 | |
| k2 | Control | 192.73 | 10.417 | 203.273 | 15.061 |
| AE | 200.585 | 11.662 | 201.771 | 17.767 | |
| NRM | 206.864 | 11.285 | 205.513 | 16.301 | |
| Female | 192.735 | 10.415 | 203.273 | 15.061 | |
| Male | 177.978 | 10.038 | 192.258 | 16.105 | |
| k3 | Control | 58.112 | 3.267 | 59.298 | 4.423 |
| AE | 56.201 | 3.555 | 55.569 | 5.217 | |
| NRM | 60.287 | 3.427 | 59.995 | 4.787 | |
| Female | 58.113 | 3.266 | 59.298 | 4.423 | |
| Male | 55.686 | 3.101 | 57.214 | 4.730 | |
| k4 | Control | 18.750 | 1.179 | 17.417 | 1.715 |
| AE | 18.728 | 1.402 | 17.287 | 2.023 | |
| NRM | 18.205 | 1.260 | 16.942 | 1.856 | |
| Female | 18.750 | 1.179 | 17.417 | 1.715 | |
| Male | 19.251 | 1.183 | 17.953 | 1.834 | |
| VND | Control | 331.485 | 33.132 | 317.583 | 33.603 |
| AE | 285.392 | 39.084 | 276.186 | 39.640 | |
| NRM | 333.304 | 35.861 | 327.365 | 36.371 | |
| Female | 331.485 | 33.132 | 317.583 | 33.603 | |
| Male | 302.339 | 35.429 | 290.339 | 35.933 | |
| VT | Control | 1341.069 | 122.372 | 4223.632 | 7065.783 |
| AE | 1141.900 | 144.356 | −285.383 | 8335.144 | |
| NRM | 1405.599 | 132.449 | 239.879 | 7647.676 | |
| Female | 1341.069 | 122.372 | 4223.632 | 7065.783 | |
| Male | 1175.543 | 130.855 | 33.164 | 7555.584 | |
| BPND | Control | 3104.599 | 55.800 | 17 548.266 | 35 154.939 |
| AE | 3001.085 | 65.824 | −4046.361 | 41 470.491 | |
| NRM | 3285.569 | 60.395 | −2529.497 | 38 050.077 | |
| Female | 3104.599 | 55.800 | 17 548.266 | 35 154.939 | |
| Male | 2910.691 | 59.668 | −2703.020 | 37 591.887 | |
| BPP | Control | 1009.584 | 89.908 | 3906.049 | 7069.069 |
| AE | 856.509 | 106.060 | −561.569 | 8339.021 | |
| NRM | 1072.295 | 97.312 | −87.486 | 7651.233 | |
| Female | 1009.584 | 89.908 | 3906.049 | 7069.069 | |
| Male | 873.204 | 96.141 | −257.175 | 7559.099 | |
Contributor Information
Yakuan Chen, AT&T Services, Inc., 200 South Laurel Avenue, Middletown, NJ 07080 USA (yc236k@att.com)..
Jeff Goldsmith, Department of Biostatistics, Columbia University Mailman School of Public Health, New York, NY 10032 USA (ajg2202@cumc.columbia.edu)..
R. Todd Ogden, Department of Biostatistics, Columbia University Mailman School of Public Health, New York, NY 10032 USA (to166@cumc.columbia.edu)..
References
- [1].Innis RB et al. , “Consensus nomenclature for in vivo imaging of reversibly binding radioligands,” J. Cereb. Blood Flow Metab, vol. 27, no. 9, pp. 1533–1539, 2007. [DOI] [PubMed] [Google Scholar]
- [2].Van Rij CM et al. , “Population plasma pharmacokinetics of 11C-flumazenil at tracer concentrations,” Br. J. Clin. Pharmacol, vol. 60, no. 5, pp. 477–485, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Syvänen S et al. , “(R)-[11C] Verapamil PET studies to assess changes in P-glycoprotein expression and functionality in rat blood-brain barrier after exposure to kainate-induced status epilepticus,” BMC Med. Imaging, vol. 11, no. 1, p. 1, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Berges A et al. , “Non linear mixed effects analysis in PET PK-receptor occupancy studies,” NeuroImage, vol. 76, pp. 155–166, 2013. [DOI] [PubMed] [Google Scholar]
- [5].Lim H-S, “Exploration of optimal dosing regimens of haloperidol, a D2 antagonist, via modeling and simulation analysis in a D2 receptor occupancy study,” Pharm. Res, pp. 1–11, 2013. [DOI] [PubMed] [Google Scholar]
- [6].Zamuner S et al. , “Estimate the time varying brain receptor occupancy in PET imaging experiments using non-linear fixed and mixed effect modeling approach,” Nucl. Med. Biol, vol. 29, no. 1, pp. 115–123, 2002. [DOI] [PubMed] [Google Scholar]
- [7].Kågedal M et al. , “Non-linear mixed effects modelling of positron emission tomography data for simultaneous estimation of radioligand kinetics and occupancy in healthy volunteers,” NeuroImage, vol. 61, no. 4, pp. 849–856, 2012. [DOI] [PubMed] [Google Scholar]
- [8].Veronese M et al. , “A non-linear mixed effect modelling approach for metabolite correction of the arterial input function in PET studies,” NeuroImage, vol. 66, pp. 611–622, 2013. [DOI] [PubMed] [Google Scholar]
- [9].Bertoldo A et al. , ““Population” approach improves parameter estimation of kinetic models from dynamic PET data,” IEEE Trans. Med. Imaging, vol. 23, no. 3, pp. 297–306, 2004. [DOI] [PubMed] [Google Scholar]
- [10].Beal SL et al. , NONMEM users guides, 1992. [Google Scholar]
- [11].R Core Team, R: A Language and Environment for Statistical Computing, R Foundation for Statistical Computing, Vienna, Austria, 2016. [Online]. Available: https://www.R-project.org/ [Google Scholar]
- [12].Matheson G, kinfitr: PET Kinetic Modelling using R, 2017. [Online]. Available: https://github.com/mathesong/kinfitr [Google Scholar]
- [13].Zanderigo F et al. , “Model-free quantification of dynamic pet data using nonparametric deconvolution,” Journal of Cerebral Blood Flow & Metabolism, vol. 35, pp. 1368–1379, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Lindstrom MJ and Bates DM, “Nonlinear mixed effects models for repeated measures data,” Biometrics, pp. 673–687, 1990. [PubMed] [Google Scholar]
- [15].Pinheiro J et al. , NLME: linear and nonlinear mixed effects models, 2016, r package version 3.1-128. [Online]. Available: http://CRAN.R-project.org/package=nlme [Google Scholar]
- [16].Neyman J and Pearson ES, “On the problem of the most efficient tests of statistical hypotheses,” in Breakthroughs in statistics. Springer, 1992, pp. 73–108. [Google Scholar]
- [17].Wald A, “Tests of statistical hypotheses concerning several parameters when the number of observations is large,” Trans Am Math Soc, vol. 54, no. 3, pp. 426–482, 1943. [Google Scholar]
- [18].Rao CR, “Large sample tests of statistical hypotheses concerning several parameters with applications to problems of estimation,” in Math. Proc. Cambridge Philos. Soc, vol. 44, no. 01 Cambridge Univ Press, 1948, pp. 50–57. [Google Scholar]
- [19].Akaike H, “Information theory and an extension of the maximum likelihood principle,” in Selected Papers of Hirotugu Akaike. Springer, 1998, pp. 199–213. [Google Scholar]
- [20].Schwarz G et al. , “Estimating the dimension of a model,” Ann. Stat, vol. 6, no. 2, pp. 461–464, 1978. [Google Scholar]
- [21].Stram DO and Lee JW, “Variance components testing in the longitudinal mixed effects model,” Biometrics, pp. 1171–1177, 1994. [PubMed] [Google Scholar]
- [22].Dorfman R, “A note on the delta-method for finding variance formulae,” The Biometric Bulletin, vol. 1, no. 129–137, p. 92, 1938. [Google Scholar]
- [23].Parsey RV et al. , “Validation and reproducibility of measurement of 5-HT1A receptor parameters with [carbonyl-11C] WAY-100635 in humans: comparison of arterial and reference tissue input functions,” J. Cereb. Blood Flow Metab, vol. 20, no. 7, pp. 1111–1133, 2000. [DOI] [PubMed] [Google Scholar]
- [24].Parsey RV et al. , “Higher serotonin 1A binding in a second major depression cohort: Modeling and reference region considerations,” Biol. Psychiatry, vol. 68, no. 2, pp. 170–178, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Sullivan GM et al. , “Positron emission tomography quantification of serotonin-1A receptor binding in medication-free bipolar depression,” Biol. Psychiatry, vol. 66, no. 3, pp. 223–230, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Pinheiro JC et al. , Model building for nonlinear mixed effects models. Citeseer, 1995. [Google Scholar]
- [27].Parsey RV et al. , “Regional heterogeneity of 5-HT1A receptors in human cerebellum as assessed by positron emission tomography,” Journal of Cerebral Blood Flow & Metabolism, vol. 25, no. 7, pp. 785–793, 2005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.