Abstract
Matrix-assisted laser desorption/ionization (MALDI) mass spectrometry imaging (MSI) has emerged as a label-free analytical tool for fast biomolecule profiling on tissue sections. Among various functional molecules, mapping neurotransmitters and related metabolites is of tremendous significance, as these compounds are critical to signaling in the central nervous system. Here, we demonstrated the use of both derivatization and reaction-free approaches that greatly reduced signal complexity and thus enabled complementary signaling molecule visualization on crab brain sections via MALDI-LTQ-Orbitrap XL platform. Pyrylium salt served as a primary amine derivatization reagent and produced prominent signal enhancement of multiple neurotransmitters, including dopamine, serotonin, γ-aminobutyric acid, and histamine that were not detected in underivatized tissues. Molecules with other functional groups, such as acetylcholine and phosphocholine, were directly imaged after matrix application. The identities of discovered neurotransmitters were verified by standards using LC-MS/MS. This study broadens our understanding of metabolic signaling in the crustacean nervous system and highlights potential of multifaceted MS techniques for unambiguous neurotransmitter characterization.
Keywords: MALDI imaging, LC-MS/MS, neurotransmitter, amine metabolites, crustacean, on-tissue derivatization
Graphical Abstract:
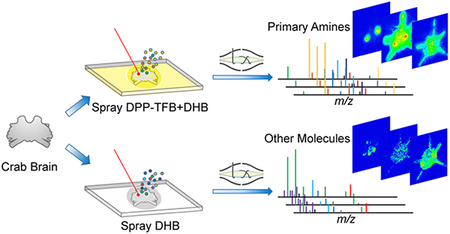
Neurotransmitters and related low molecular weight metabolites (MW < 1000 Da) are distributed among the nervous system and possess diverse physiochemical properties. Specifically, neurotransmitters are released from synaptic vesicles in neuronal axons and then bind to receptors on a dendrite of another neuron. The synthesis and degradation of neurotransmitters are regulated by sophisticated metabolic pathways where numerous small molecules are associated. These molecules have been extensively studied due to their essential roles involved in neuronal development and cellular communication. The changes in expression profiles of neurotransmitters and metabolites are relevant to neural malfunction. For instance, dopamine depletion is related to extrapyramidal disorders,1 and progressive serotonergic neuron loss occurs in Parkinsonism.2 Therefore, deeper understanding toward neurotransmitters and regulatory metabolites in situ distribution offer insights into neurological functions, neuro-modulation processes as well as potential treatments to nervous system diseases.
For a long time, the crustacean nervous system model has been extensively applied in neurological studies owing to the well-studied and relatively simple anatomical structures. The distribution of multiple neurotransmitters in the crustacean nervous system has been uncovered by a constellation of approaches. For example, monoamines are visible after fluorescence histochemical treatment.3,4 Later on, the utilization of immunocytochemical techniques permitted more reliable and precise localization of compound of interest.5 Although these techniques require specialized tools and skills, their widespread use with proven findings demonstrated their unique leverage in neuroscience. However, research and discovery in the modern era are in high demand of advanced techniques for simultaneous molecular visualization of multiple compounds on tissues. Thus, the development of a user-friendly and unequivocal imaging method is essential to better understanding of neurotransmitter metabolism and regulatory pathways.
Mass spectrometry has been widely adapted for the study of biomolecules in diverse disciplines owing to its high sensitivity, selectivity, and analysis speed. While liquid chromatography (LC) mass spectrometry (MS) is the gold standard for metabolomics research, tissue undergoes extensive sample preparation where tissue-specific morphology and anatomical context is lost. Alternatively, matrix-assisted laser desorption/ionization mass spectrometry imaging (MALDI MSI) facilitates molecule visualization with simple-step matrix pretreatment without sacrificing sensitivity. Unlike immunology imaging techniques that require target-specific antibodies, MALDI MSI allows untargeted profiling in the tissue area. Numerous m/z distribution patterns can be acquired in a single MSI experiment to reveal biomolecular in situ localization. Since the first report of MALDI MSI by Caprioli et al.6 in 1997, MSI has undergone prosperous development and generated fruitful applications in mapping a wide variety of biomolecules, including proteins,7–9 peptides,10–12 metabolites,13–18 and lipids.19–24 Utilizing this technique, we have successfully visualized several amino acids, nucleotides, and organic acids in the crustacean nervous system in a previous study25 that improved our understanding of crustacean metabolism. Nevertheless, a rapid and highly sensitive method is still lacking to map neurotransmitters inside the crustacean central nervous system.
While matrix compounds facilitate molecule ionization, the in situ MSI of low mass compounds via MALDI presents enormous challenges. One of the major difficulties comes from matrix background signal interference. Typically, matrices are sprayed onto tissue section before MS analysis. As commonly used matrices are small molecules, neurotransmitter and metabolite signals are largely suppressed by abundant matrix. Another obstacle is caused by compounds of interest themselves. Low molecular weight molecules spanning broad dynamic range of concentrations consist of diverse functional groups that can contribute to distinct ionization efficiencies in MALDI experiments. Extensive efforts have been devoted to address these issues. For instance, low-background and background-free matrices have been utilized for small molecule imaging.26–28 Other useful approaches include selectively derivatizing typical functional groups via on tissue chemical reaction for signal enhancement. For example, 4-hydroxy-3-methoxycinnamaldehyde29 and pyrylium salts efficiently react with primary amines,30–32 1,1′-thiocarbonyldiimidazole was used for 3-methoxysalicylamine derivatization,33 and carboxyl groups can be derivatized by amine.34 Those derivatization approaches benefit compound detection by reducing spectral complexity and increasing compound molecular weight to keep molecules away from intensive matrix signal region. The addition of acryl groups to small molecule neurotransmitters tremendously increases compound ionization efficiency and promotes energy transfer from matrix to analytes and in return enhance signal intensity.
Herein, we applied derivatization reagent 2,4-diphenylpyranylium tetrafluoroborate (DPP-TFB) and, for the first time, simultaneously detected major primary amine-containing neurotransmitters and metabolites with enhanced sensitivity, in the crustacean brain. Additionally, other types of metabolites were imaged directly without derivatization using improved sample preparation methodology, resulting in better identi-fication rate. In addition to accurate mass matching using high resolution accurate mass (HRAM) MALDI Orbitrap instrument, neurotransmitter identities were further confirmed by MS/MS information and retention times obtained from standard compounds via LC-MS/MS. The integration of multifaceted mass spectrometric approaches allowed simultaneous localization via imaging and identification via LC-MS/MS of neurotransmitters in the crustacean brain. The localization information on neurotransmitters provides deeper insights into crustacean neurobiological and physiological studies and demonstrates a facile approach for future neuroscience research.
RESULTS AND DISCUSSION
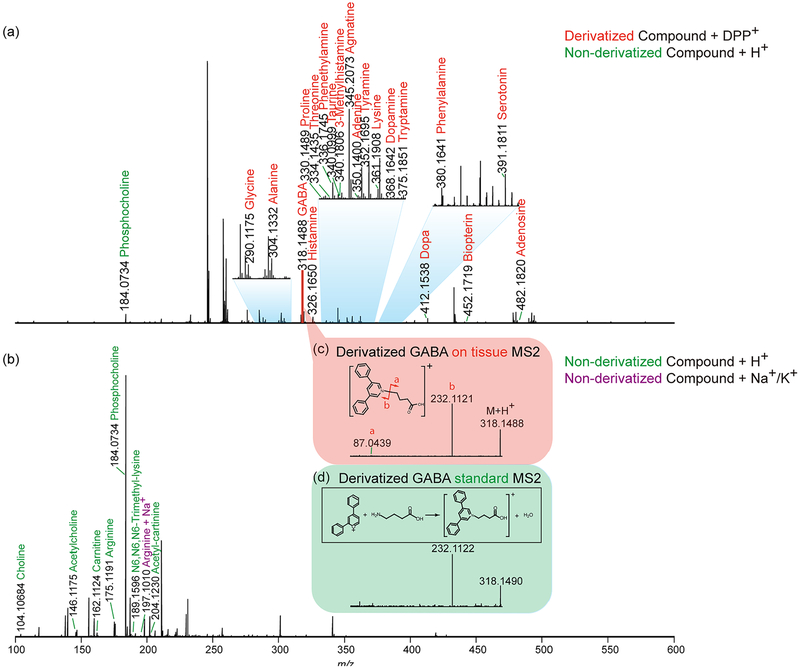
To investigate performance of labeling and label-free MSI approaches, a pilot experiment was performed by acquiring MS1 profiles after a laser was randomly fired 12 times across crab brain sections. The HRAM feature of this Orbitrap platform allows both spectral differentiation and confident compound identification. The accurate mass detection permits reliable low-mass molecule characterization using both approaches with small ppm error (most compounds were detected at mass error < 2 ppm, Supporting Information Table S1). The MS1 spectrum obtained from the pyrylium labeled tissue is shown in Figure 1a. Due to the derivatization reaction selectivity, primary amines were detectable with enhanced signals. Several amino acids, as well as metabolites with amines, were displayed in the spectrum, such as phenylalanine and dopa. Moreover, several representative neurotransmitters like histamine, GABA, serotonin, and dopamine were detected, while they remained invisible with direct matrix application approach without derivatization (Figure 1b). The derivatization step has efficiently boosted signals of amines with the addition of the C17H11 group to the original molecular structure. With a molecular mass increase of 215.0855, the low-mass molecules are free from interference of the intensive matrix peaks as well as phosphocholine, an abundant lipid the brain tissue. Another advantage enabled by derivatization in situ MS2 fragmentation. For example, the on-tissue labeled GABA precursor ion was selected for further HCD fragmentation, and the generated MS2 spectrum (Figure 1c) contained an intense peak b originated from derivatization reagent as well as peak a belonging to GABA structure. The MS2 spectrum matched well with fragments from pyrylium reacted GABA standard (Figure 1d), further supporting confident identification of GABA, an inhibitory neuro-transmitter in the crustacean brain. Without derivatization, GABA could not be detected at the MS1 level, making impossible to perform on-tissue MS2 fragmentation with MALDI source. Via the replacement of primary amine group with conjugated aromatic group in neurotransmitters, laser energy transfer efficiency may be enhanced that could lead to improved full MS scan signals for these neurotransmitters. The conjugation structure could better stabilize attached proton, producing enhanced precursor signals and lead to high intensity fragments generated in MS2 scans. It is also noticeable that, in the spectrum from derivatized tissue (Figure 1a), the sodium and potassium adduct peaks were barely detectable whereas they were commonly observed in tissues samples only sprayed with matrix (Figure 1b). This is another benefit of derivatization as it generates less-complex spectra. This proof-of-principle experiment demonstrates the usefulness of the on-tissue derivatization reaction for neuro-transmitter and monoamine detection. However, the non derivatization method should not be abandoned, as it complements molecular identification. As shown in Figure 1b, acetylcholine (one of major neurotransmitters) and carnitine (a critical metabolite) exhibited reasonable intensity in the spectrum, while after derivatization they could not be detected, most likely due to signal suppression from excess derivatization reagent. Furthermore, this matrix coating process could be performed directly after sample collection and tissue section to best preserve physiological states, while the derivatization reaction required overnight reaction where degradation could occur. To maximize information collected from brain tissue and gain an in-depth understanding about the crustacean nervous system, the utilization of complementary derivatization and nonderivatization approaches is essential. Unlike immunohistochemical approaches that require specific antibodies to visualize certain molecules, MS-based methods demonstrate their unique advantages via a less time-consuming and more cost-effective manner. Within a few hours, the MALDI MSI approach enabled simultaneous recording of m/z values of more than hundreds of compounds along with corresponding abundances and location information. The ease of use for the MSI workflow also minimizes errors during sample handling thus ensures authenticity of the collected information. While immuno-based fluorescence imaging provides better spatial resolution, recent advancements in MALDI MSI have enabled cellular and subcellular resolution.35
Figure 1.
Distinct and complementary identification of low-MW molecules provided by derivatization and reaction-free approaches after on-tissue profiling with the MALDI-LTQ-Orbitrap XL platform. (a) Detected primary amines after DPP-TFB derivatization by accurate mass matching with 5 ppm error window. (b) Detected metabolites with DHB as matrix without derivatization by accurate mass matching with 5 ppm error window. Derivatized GABA (c) on-tissue and (d) standard MS2.
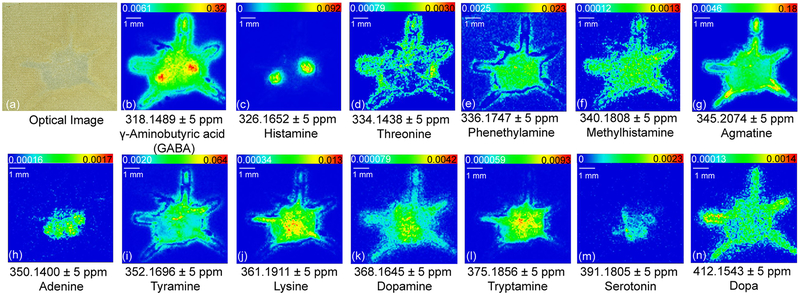
By employing on-tissue derivatization, the primary amine-containing molecules demonstrate distinct localization patterns in the crustacean central nervous system (Figure 2). Previously researchers have localized both serotonin and dopamine immunoreactivity in female mud crab brain.36 To the best of our knowledge, our study represents the first study on crustacean brain that allows simultaneous visualization of multiple primary amine neurotransmitters and metabolites. The crab brain anatomy (Supporting Information Figure S1) facilitates better understanding of tissue-specific in situ distribution. Some neurotransmitters are specifically localized in certain regions on the brain section; e.g., histamine (Figure 2c) is only distributed in the olfactory lobes (ON) and serotonin (Figure 2m) exhibits localization in ON, median antenna I neuropil (MAN), anterior medial protocerebral neuropil (AMPN), and posterior medial protocerebral neuropil (PMPN). In comparison, some neurotransmitters exhibit more widespread distribution throughout the brain where abundance level varies. For example, GABA (Figure 2b) exhibits distribution throughout the whole brain region with the ON showing the most abundant signal. Additionally, dopamine (Figure 2k) exhibits the most intense signals in the MAN, AMPN, and PMPN regions, while tryptamine (Figure 2l) is more abundant in MAN and PMPN. The revealed distribution patterns of these compounds together with corresponding molecular identity enable deeper understanding about crustacean neurochemical signaling and region-specific functions. For instance, GABA, histamine, serotonin, and dopamine all have distributions in the ON region. This observation suggests that ON is a highly active brain region where many neurotransmitters are involved in neuromodulation and signal transmission. While neurotransmitters are featured with shared distribution area, localization differences may be explained by distinct roles signaling molecules play in the central nervous system. It is also interesting to note that dopa (Figure 2n), the precursor of dopamine, has more widespread distribution in the crab brain but the localization pattern does not perfectly overlap with dopamine. This slight discrepancy might be resulted from region-specific enzymatic activities in neurotransmitter synthesis pathway across the nervous system. Different metabolites also display various in situ localization patterns. Metabolites with ubiquitous distribution include phenethylamine (Figure 2e), methylhistamine (Figure 2f), and lysine (Figure 2j), indicating their involvement in a wide range of metabolic activities. In contrast, the localization of threonine (Figure 2d), agmatine (Figure 2g), adenine (Figure 2h), and tyramine (Figure 2i) all differ from each other, indicating their distinct roles in the nervous system. Overall, this simple on-tissue derivatization method enabled localization of multiple primary amines in a single MALDI MSI experiment with enhanced throughput and chemical specificity.
Figure 2.
Selected ion images showing in situ distribution of biogenic amines after DPP-TFB derivatization. Major neurotransmitters are illustrated, including GABA, histamine, serotonin, and dopamine.
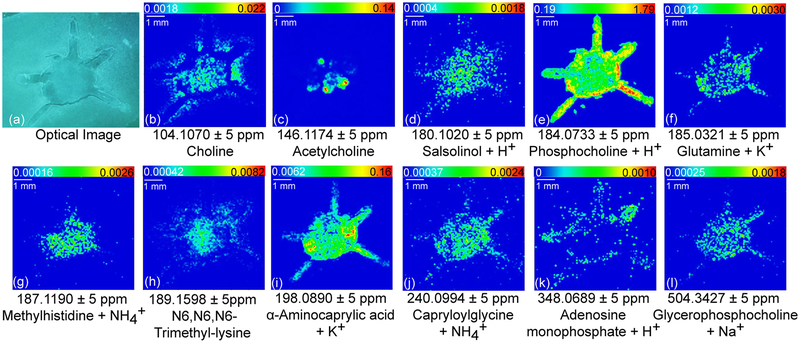
Molecular imaging could also be achieved by directly applying DHB matrix without performing derivatization, as shown in Figure 3. A drier matrix application method37 was utilized to reduce molecule delocalization and produce fine matrix crystals. Using a robotic nozzle, DHB was sprayed preprogrammed moving patterns via relatively high moving velocity and low spray flow. This approach guaranteed prompt solvent evaporation during matrix application thus minimized molecular diffusion. Matrix application step is extremely important to reduce compound diffusion probabilities because crustacean brains are of higher water content than typical model system, such as mouse brain. Sample handling another determinant of high-quality images in MALDI experiments. In our study, tissue sectioning was performed the same day of brain collection to best prevent molecular degradation. Through these improved sample preparation processes, we successfully localized a key neurotransmitter acetylcholine (Figure 3c) not reported in a previous study25 of the crab central nervous system. Unlike classical approaches indirectly revealing acetylcholine distribution by mapping acetylcholine metabolism-related enzymes,38,39 MALDI MSI offers fast and straightforward visualization of acetylcholine (Figure 3c). Although acetylcholine metabolite choline (Figure 3b) has ubiquitous distribution among the whole brain, acetylcholine displays more specific localization pattern. By comparing acetylcholine with typical crab brain anatomy (Supporting Information Figure S1), the most acetylcholine-rich region is found to be AMPN. In addition to neuro-transmitter, the direct matrix deposition method allows putative identification and visualization of many metabolites with functional groups other than amines. Rich distribution information can be extracted, including lipids (Figure 3e, l), nucleotide metabolites (Figure 3k), organic acid (Figure 3i), isoquinoline (Figure 3d), and amino acid metabolites (Figure 3h, j). The location of these molecules together derivatized-amines offer more comprehensive understanding about the nervous system. Unlike chemical derivatization method that is free from adduct ions, applying matrix directly generates adduct ion bound metabolites, such as glutamine + K+ (Figure 3f) and methylhistidine + NH4+ (Figure 3g). interference of adduct ions (e.g., NH4+, Na+, and K+) and matrix signals can complicate low-MW MS spectra. Therefore, some compounds of interest remain buried and undetectable in this direct matrix application approach.
Figure 3.
Selected ion images showing spatial distributions of neurotransmitters and metabolites revealed directly with DHB matrix.
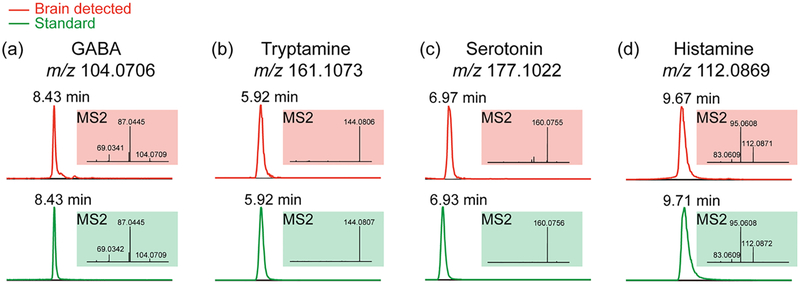
LC-MS/MS experiments were performed to further confirm putatively identified neurotransmitters. It is naturally difficult to conduct on-tissue MS2 for each neurotransmitter as relatively high abundance level is required to generate high quality MS2 spectra. To verify identities of molecules, utilized LC-MS/MS to achieve confident identification with both retention time in the chromatography domain and structural fragments in the MS2 domain. By comparing these features with those of standard compounds, one can easily identify unknown molecules and more confidently characterize the analytes of interest. To reduce sample complexity, crab brain extract was cleaned using an MCX SPE after extraction.40 Figure 4 illustrates representative spectra acquired from crab brain extract and the comparison with corresponding standard compound. MS2 information was acquired to differentiate isomers that might coelute with compound of interest. As different molecules are composed of distinct structures, MS2 fragmentation patterns reveal structural variations among molecules. The detected MS2 fragments from crab brain extract matched well with those generated from standards. For instance, GABA MS2 fragments (shown in Figure 4a) include the loss of NH3 (m/z 87.0445) and the loss of both NH3 and H2O (m/z 69.0342). Tryptamine (Figure 4b) and serotonin (Figure 4c) MS2 spectra show the major fragmentation products after losing NH3 (m/z 144.0807 for tryptamine and m/z 160.0756 for serotonin, respectively). Figure 4d demonstrates histamine product (m/z 95.0608) after loss of NH3 and product (m/z 83.0609) most likely due to loss of CH2=NH. Excellent match was found between target compound retention time in the crab brain extract and that of standard, as well as their resulting MS2 fragmentation spectra, indicating the effectiveness of this 16 min LC-MS/MS method. Given dopamine’s extremely low abundance, we were only able to detect an extracted ion chromatogram of m/z 154.0863 while MS2 fragments suffered from abundant noise interference. The well-matched retention time suggested identity of detected ion at m/z 154.0863 to be dopamine (Supporting Information Figure S2). Overall, LC-MS/MS facilitates the neurotransmitter identification in this complex biological sample.
Figure 4.
Neurotransmitters verified via hydrophilic interaction liquid chromatography (HILIC) LC-MS/MS. The top panel shows detected chromatogram peaks and corresponding MS2 spectra in crab brain extract. The bottom panel illustrates retention times and MS2 fragments from corresponding standard compounds. (a) GABA. (b) Tryptamine. (c) Serotonin. (d) Histamine.
In conclusion, our study established and demonstrated a viable HRAM platform to visualize in situ localization of neurotransmitters as well as to achieve their confident identifications in the central nervous system of rock crab, Cancer irroratus, a classical model organism. The employment of on-tissue chemical derivatization and direct matrix application was advantageous as the combined approach generated comprehensive coverage of small signaling molecules. HILIC LC-MS/MS enabled further separation and confirmation of detected molecules in MALDI MSI experiments, demonstrating the necessity of utilizing multifaceted mass spectrometric techniques in the investigation of complex biological systems. The novel findings of neurotransmitters and their metabolites enabled by HRAM MSI are anticipated to advance related neuroscience areas and offer effective approaches to probe a plethora of neurochemical signaling molecules.
METHODS
Chemicals and Materials.
2,5-Dihydroxybenzoic acid (DHB) was from Acros Organics (Morris Plains, NJ). α-Cyano-4-hydroxycinnamic acid (CHCA) and DPP-TFB were from Sigma-Aldrich (St. Louis, MO). Optima grade methanol (MeOH), acetonitrile (ACN), formic acid (FA), water, and microscope glass slides (25 mm × 75 mm × 1 mm) were purchased from Fisher Scientific (Fair Lawn, NJ). Mixed-Mode Cation eXchange (MCX) solid phase extraction (SPE) 1 cm3 cartridges were obtained from Waters Corporation (Milford, MA). Standard compounds were purchased from Sigma-Aldrich unless specified otherwise.
Animal Tissue Collection.
Rock crabs Cancer irroratus were purchased from Ocean Resources Inc. (Sedgwick, ME) and maintained in man-made saline water. All animal experiments were performed under institutional guidelines (University of Wisconsin—Madison IACUC). Before animal dissection, the crab was anesthetized in ice for ~15 min. For MALDI MSI experiment, the isolated brain was rinsed by deionized water for removal of salt then embedded in 100 mg/mL gelatin in water and snap-frozen in dry ice. For LC-MS/MS experiment, brain tissue was transferred into 100 μL acidified MeOH (MeOH/H2O/HAc = 90/9/1, v/v/v). All samples were stored at −80 °C and processed the same day of collection to minimize degradation and best preserve fast-degrading neuro-transmitters.
MALDI MSI Sample Preparation.
The crab brain was sectioned by a cryostat (Thermo Scientific Microm HM 525) at −20 °C into 16 μm thickness and thaw-mounted onto microscope glass slides. The tissue slides were dried for 30 min at room temperature inside a desiccator. TM-Sprayer (HTX Technologies, Carrboro, NC) was utilized for on-tissue derivatization reagent and matrix application. For on tissue derivatization, the nozzle temperature was set to 80 °C and moving velocity to 1100 mm/min. Then 1.33 mg/mL DPP-TFB (9.6 mg compound dissolved in solvent with 5.4 mL MeOH, 1.8 mL H2O and 3.5 μL triethylamine) was homogeneously sprayed for 30 passes (6 s dry time between each pass) with 3 mm tracking space and flow rate of 0.08 mL/min. The tissue slide after derivatization was kept in a desiccator at room temperature for 24 h before spraying the DHB matrix. Here, 40 mg/mL DHB matrix (MeOH/H2O/FA = 49.95/49.95/0.1, v/v/v) was sprayed on the tissue slide following the method previously described.37 Briefly, nozzle moving velocity was set to 1250 mm/min with temperature at 80 °C, and DHB was sprayed for 24 passes with 0.05 mg/mL flow rate and 3 mm tracking space. For experiments without on-tissue derivatization, tissue slides were sprayed with DHB matrix directly after sectioning. Tissue slides were dried at room temperature in the desiccator before instrumental analysis.
MALDI MSI Data Acquisition and Data Analysis.
All MALDI MSI experiments were performed on the MALDI-LTQ-Orbitrap XL platform (Thermo Scientific, Bremen, Germany) with a 60 Hz 337 nm N2 laser. Mass resolution was set to 60 K (at m/z 400) for full MS scan with microscan of 1, raster step of 75 μm and laser energy of 15 μJ. Mass range was set to m/z 270−1300 for derivatized tissue and m/z 50−600 for nonderivatized tissue. On-tissue profiling data were analyzed and searched against small molecule theoretical masses using home-built Python program with 5 ppm error window. The MALDI MSI images were generated using Thermo ImageQuest 1.1.0.
MALDI Spot Analysis of Derivatized GABA Standard.
A volume of 10 μL of GABA solution (3.2 mg/mL in water) was mixed with 90 μL of DPP-TFB (concentration same as on-tissue derivatization) and reacted at room temperature for 30 min. Reaction was quenched with 1 μL of acetic acid. Then 1 μL of mixture was added to 9 μL of CHCA (3.5 mg/mL in 0.1% FA H2O/EtOH/ACN = 3/13/84, v/v/v). Then 1 μL of solution was spotted on MALDI plate for MS2 analysis.
Tissue Processing for LC-MS/MS.
Crab brain was probe sonicated in acidified MeOH, and the extract was further separated by using a 3K MWCO filter and cleaned up using SPE as stated previously.40 Briefly, small molecules were extracted in 100 μL of acidified MeOH by using a sonic dismembrator (Fisher Scientific, Pittsburgh, PA) for three cycles with 8 s pulse on and 15 s pulse off. The supernatant was mixed with 85 μL of Optima water and then separated by using a 3K MWCO filter. The flow-through was dried down, resuspended in 1 mL 2% FA water, and loaded into a MCX SPE cartridge preconditioned with MeOH then water. Basic compounds were eluted by 5% ammonia in MeOH after washing with FA water and MeOH. The collected basic elution was dried down and stored at −20 °C until further handling.
LC-MS/MS Analysis.
Chromatographic separation was performed at 40 °C using a Waters Acquity UPLC BEH Amide column (2.1 mm × 150 mm, 1.7 μm) on a Dionex UltiMate 3000 LC system coupled to a Thermo Q Exactive Orbitrap mass spectrometer. The binary gradient consists of mobile phase A (0.1% FA H2O with 2.5 mM ammonium formate) and mobile phase B (95/5 ACN/H2O with 0.1% FA and 2.5 mM ammonium format). The flow rate was 0.4 mL/min with gradient set as 0−3 min, 100% B; 3−13 min, 100−50% B; 13−13.1 min 50−100% with 2.9 min equilibrium at the end. Full MS and parallel reaction monitoring (PRM) mode were utilized during mass spectrometry data acquisition. Full MS scans (m/z 70−1000) were performed with mass resolution of 70 K, automatic gain control (AGC) target 1e6, and maximum injection time 100 ms. In PRM mode, targeted ions were isolated with 0.5 m/z and MS2 spectra were acquired with isolation window 0.5 m/z, normalized collision energy 30%, resolution 17.5 K, AGC target 1e5, and maximum injection time 100 ms.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Cameron O. Scarlett and Molly Pellitteri Hahn from Analytical Instrumentation Center in the School of Pharmacy at UW-Madison for instrument maintenance and helpful suggestions. Q.C. gratefully acknowledges a former Li Lab member Dr. Junhua Wang for scientific discussions and a current Li Lab member Jillian Johnson for providing helpful suggestions.
Funding
This work was supported by the National Institutes of Health Grants R56MH110215 and R01 DK071801. The MALDI Orbitrap instrument and Q Exactive Orbitrap instrument were purchased through the support of an NIH shared instrument grant S10RR029531. L.L. thanks a Vilas Distinguished Achievement Professorship and the Charles Melbourne Johnson Distinguished Chair Professorship with funding provided by the Wisconsin Alumni Research Foundation and University of Wisconsin-Madison School of Pharmacy.
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acschemneuro.8b00730.
Neurotransmitters and metabolites identified from crab brain tissue via HRAM MALDI-LTQ-Orbitrap XL platform; illustration of a crab brain from the dorsal view; identification of dopamine by matching ion m/z 154.0863 retention time with dopamine standard (PDF)
The authors declare no competing financial interest.
REFERENCES
- (1).Kurian MA, Gissen P, Smith M, Heales SJR, and Clayton PT (2011) The monoamine neurotransmitter disorders: an expanding range of neurological syndromes. Lancet Neurol 10, 721–733. [DOI] [PubMed] [Google Scholar]
- (2).Politis M, and Niccolini F (2015) Serotonin in Parkinson’s disease. Behav. Brain Res 277, 136–145. [DOI] [PubMed] [Google Scholar]
- (3).Falck B, Hillarp N-Å, Thieme G, and Torp A. a. (1962) Fluorescence of catechol amines and related compounds condensed with formaldehyde. J. Histochem. Cytochem 10, 348–354. [DOI] [PubMed] [Google Scholar]
- (4).Felten DL, Felten SY, Sladek JR Jr, Notter MD, Carlson SL, Bellinger DL, and Wiegand SJ (1990) Fluorescence histochemical techniques for catecholamines as tools in neurobiology. J. Microsc 157, 271–283. [DOI] [PubMed] [Google Scholar]
- (5).Fingerman M, Nagabhushanam R, Sarojini R, and Reddy PS (1994) Biogenic Amines in Crustaceans: Identification, Localization, and Roles. J. Crustacean Biol 14, 413–437. [Google Scholar]
- (6).Caprioli RM, Farmer TB, and Gile J (1997) Molecular Imaging of Biological Samples: Localization of Peptides and Proteins Using MALDI-TOF MS. Anal. Chem 69, 4751–4760. [DOI] [PubMed] [Google Scholar]
- (7).Chaurand P, Schwartz SA, and Caprioli RM (2002) Imaging mass spectrometry: a new tool to investigate the spatial organization of peptides and proteins in mammalian tissue sections. Curr. Opin. Chem. Biol 6, 676–681. [DOI] [PubMed] [Google Scholar]
- (8).van Remoortere A, van Zeijl RJM, van den Oever N, Franck J, Longuespée R, Wisztorski M, Salzet M, Deelder AM, Fournier I, and McDonnell LA (2010) MALDI imaging and profiling MS of higher mass proteins from tissue. J. Am. Soc. Mass Spectrom 21, 1922–1929. [DOI] [PubMed] [Google Scholar]
- (9).Grey AC, Chaurand P, Caprioli RM, and Schey KL (2009) MALDI Imaging Mass Spectrometry of Integral Membrane Proteins from Ocular Lens and Retinal Tissue. J. Proteome Res 8, 3278–3283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Caprioli RM, Farmer TB, and Gile J (1997) Molecular imaging of biological samples: localization of peptides and proteins using MALDI-TOF MS. Anal. Chem 69, 4751–4760. [DOI] [PubMed] [Google Scholar]
- (11).Altelaar AM, Taban IM, McDonnell LA, Verhaert PD, de Lange RP, Adan RA, Mooi WJ, Heeren RM, and Piersma SR (2007) High-resolution MALDI imaging mass spectrometry allows localization of peptide distributions at cellular length scales in pituitary tissue sections. Int. J. Mass Spectrom 260, 203–211. [Google Scholar]
- (12).OuYang C, Chen B, and Li L (2015) High throughput in situ DDA analysis of neuropeptides by coupling novel multiplex mass spectrometric imaging (MSI) with gas-phase fractionation. J. Am. Soc. Mass Spectrom 26, 1992–2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Lee YJ, Perdian DC, Song Z, Yeung ES, and Nikolau BJ (2012) Use of mass spectrometry for imaging metabolites in plants. Plant J 70, 81–95. [DOI] [PubMed] [Google Scholar]
- (14).Esquenazi E, Coates C, Simmons L, Gonzalez D, Gerwick WH, and Dorrestein PC (2008) Visualizing the spatial distribution of secondary metabolites produced by marine cyanobacteria and sponges via MALDI-TOF imaging. Mol. BioSyst 4, 562–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Ye H, Gemperline E, Venkateshwaran M, Chen R, Delaux P-M, Howes-Podoll M, Ané J-M, and Li L (2013) MALDI mass spectrometry-assisted molecular imaging of metabolites during nitrogen fixation in the Medicago truncatula−Sinorhizobium meliloti symbiosis. Plant J 75, 130–145. [DOI] [PubMed] [Google Scholar]
- (16).He J, Sun C, Li T, Luo Z, Huang L, Song X, Li X, and Abliz Z (2018) A Sensitive and Wide Coverage Ambient Mass Spectrometry Imaging Method for Functional Metabolites Based Molecular Histology. Advanced science (Weinheim, Baden-Wurttemberg, Germany) 5, 1800250–1800250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Guo S, Wang Y, Zhou D, Xu Y, Chen T, and Li Z (2018) Association of alteration of nucleosides and nucleotides with gastric cancer microenvironment. Int. J. Mass Spectrom 434, 37–42. [Google Scholar]
- (18).Sun C, Li T, Song X, Huang L, Zang Q, Xu J, Bi N, Jiao G, Hao Y, Chen Y, Zhang R, Luo Z, Li X, Wang L, Wang Z, Song Y, He J, and Abliz Z (2019) Spatially resolved metabolomics to discover tumor-associated metabolic alterations. Proc. Natl. Acad. Sci. U. S. A 116, 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Murphy RC, Hankin JA, and Barkley RM (2009) Imaging of lipid species by MALDI mass spectrometry. J. Lipid Res 50, S317–S322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Jackson SN, Ugarov M, Egan T, Post JD, Langlais D, Albert Schultz J., and Woods AS (2007) MALDI-ion mobility-TOFMS imaging of lipids in rat brain tissue. J. Mass Spectrom 42, 1093–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Anderson DMG, Ablonczy Z, Koutalos Y, Spraggins J, Crouch RK, Caprioli RM, and Schey KL (2014) High Resolution MALDI Imaging Mass Spectrometry of Retinal Tissue Lipids. J. Am. Soc. Mass Spectrom 25, 1394–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Guo S, Zhou D, Zhang M, Li T, Liu Y, Xu Y, Chen T, and Li Z (2017) Monitoring changes of docosahexaenoic acid-containing lipids during the recovery process of traumatic brain injury in rat using mass spectrometry imaging. Sci. Rep 7, 5054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Li G, Cao Q, Liu Y, DeLaney K, Tian Z, Moskovets E, and Li L (2019) Characterizing and Alleviating Ion Suppression Effects in Atmospheric Pressure Matrix-Assisted Laser Desorption/Ionization (AP-MALDI). Rapid Commun. Mass Spectrom 33, 327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Zhao C, Xie P, Yong T, Wang H, Chung ACK, and Cai Z (2018) MALDI-MS Imaging Reveals Asymmetric Spatial Distribution of Lipid Metabolites from Bisphenol S-Induced Nephrotoxicity. Anal. Chem 90, 3196–3204. [DOI] [PubMed] [Google Scholar]
- (25).Ye H, Wang J, Greer T, Strupat K, and Li L (2013) Visualizing Neurotransmitters and Metabolites in the Central Nervous System by High Resolution and High Accuracy Mass Spectrometric Imaging. ACS Chem. Neurosci 4, 1049–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Wang J, Sun J, Wang J, Liu H, Xue J, and Nie Z (2017) Hexagonal boron nitride nanosheets as a multifunctional background-free matrix to detect small molecules and complicated samples by MALDI mass spectrometry. Chem. Commun 53, 8114–8117. [DOI] [PubMed] [Google Scholar]
- (27).Liu H, Zhou Y, Wang J, Xiong C, Xue J, Zhan L, and Nie Z (2018) N-Phenyl-2-naphthylamine as a Novel MALDI Matrix for Analysis and in Situ Imaging of Small Molecules. Anal. Chem 90, 729–736. [DOI] [PubMed] [Google Scholar]
- (28).Lin Z, Wu J, Dong Y, Xie P, Zhang Y, and Cai Z (2018) Nitrogen and Sulfur Co-doped Carbon-Dot-Assisted Laser Desorption/Ionization Time-of-Flight Mass Spectrometry Imaging for Profiling Bisphenol S Distribution in Mouse Tissues. Anal. Chem 90, 10872–10880. [DOI] [PubMed] [Google Scholar]
- (29).Manier ML, Spraggins JM, Reyzer ML, Norris JL, and Caprioli RM (2014) A derivatization and validation strategy for determining the spatial localization of endogenous amine metabolites in tissues using MALDI imaging mass spectrometry. J. Mass Spectrom 49, 665–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Shariatgorji M, Nilsson A, Goodwin RJA, Källback P, Schintu N, Zhang X, Crossman AR, Bezard E, Svenningsson P, and Andren PE (2014) Direct Targeted Quantitative Molecular Imaging of Neurotransmitters in Brain Tissue Sections. Neuron 84, 697–707. [DOI] [PubMed] [Google Scholar]
- (31).Shariatgorji M, Nilsson A, Källback P, Karlsson O, Zhang X, Svenningsson P, and Andren PE (2015) Pyrylium Salts as Reactive Matrices for MALDI-MS Imaging of Biologically Active Primary Amines. J. Am. Soc. Mass Spectrom 26, 934–939. [DOI] [PubMed] [Google Scholar]
- (32).Shariatgorji M, Strittmatter N, Nilsson A, Källback P,Alvarsson A, Zhang X, Vallianatou T, Svenningsson P, Goodwin RJA, and Andren PE (2016) Simultaneous imaging of multiple neurotransmitters and neuroactive substances in the brain by desorption electrospray ionization mass spectrometry. NeuroImage 136, 129–138. [DOI] [PubMed] [Google Scholar]
- (33).Chacon A, Zagol-Ikapitte I, Amarnath V, Reyzer ML, Oates JA, Caprioli RM, and Boutaud O (2011) On-tissue chemical derivatization of 3-methoxysalicylamine for MALDI-imaging mass spectrometry. J. Mass Spectrom 46, 840–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Wu Q, Comi TJ, Li B, Rubakhin SS, and Sweedler JV (2016) On-Tissue Derivatization via Electrospray Deposition for Matrix-Assisted Laser Desorption/Ionization Mass Spectrometry Imaging of Endogenous Fatty Acids in Rat Brain Tissues. Anal. Chem 88, 5988–5995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Duenas ME, Feenstra AD, Korte AR, Hinners P, and Lee YJ (2018) Cellular and Subcellular Level Localization of Maize Lipids and Metabolites Using High-Spatial Resolution MALDI Mass Spectrometry Imaging, In Maize: Methods and Protocols (Lagrimini LM, Ed.), pp 217–231, Springer New York, New York. [DOI] [PubMed] [Google Scholar]
- (36).Khornchatri K, Kornthong N, Saetan J, Tinikul Y, Chotwiwatthanakun C, Cummins SF, Hanna PJ, and Sobhon P (2015) Distribution of serotonin and dopamine in the central nervous system of the female mud crab, Scylla olivacea (Herbst). Acta Histochem 117, 196–204. [DOI] [PubMed] [Google Scholar]
- (37).Gemperline E, Rawson S, and Li L (2014) Optimization and Comparison of Multiple MALDI Matrix Application Methods for Small Molecule Mass Spectrometric Imaging. Anal. Chem 86, 10030–10035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Jacobowitz DM, and Palkovits M (1974) Topographic atlas of catecholamine and acetylcholinesterase-containing neurons in the rat brain. I. Forebrain (telencephalon, diencephalon). J. Comp. Neurol 157, 13–28. [DOI] [PubMed] [Google Scholar]
- (39).Armstrong DM, Saper CB, Levey AI, Wainer BH, and Terry RD (1983) Distribution of cholinergic neurons in rat brain: demonstrated by the immunocytochemical localization of choline acetyltransferase. J. Comp. Neurol 216, 53–68. [DOI] [PubMed] [Google Scholar]
- (40).Cao Q, Ouyang C, Zhong X, and Li L (2018) Profiling of small molecule metabolites and neurotransmitters in crustacean hemolymph and neuronal tissues using reversed-phase LC-MS/MS. Electrophoresis 39, 1241–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.