Introduction
Myelodysplastic syndromes (MDS) frequently progress to acute myeloid leukemia (AML), however, the cells leading to malignant transformation have not been directly elucidated. As progression of MDS to AML in humans provides a biological system to determine the cellular origins and mechanisms of neoplastic transformation, we studied highly fractionated stem cell populations in longitudinal samples of patients with MDS who progressed to AML. Targeted deep sequencing combined with single-cell sequencing of sorted cell populations revealed that stem cells at the MDS stage, including immunophenotypically and functionally defined pre-MDS stem cells (preMDS-SC), had a significantly higher subclonal complexity compared to blast cells and contained a large number of aging-related variants. Single-cell targeted re-sequencing of highly fractionated stem cells revealed a pattern of non-linear, parallel clonal evolution, with distinct subclones within pre-MDS and MDS stem cells contributing to generation of MDS blasts or progression to AML, respectively. Furthermore, phenotypically aberrant stem cell clones expanded during transformation and stem cell subclones that were not detectable in MDS blasts became dominant upon AML progression. These results reveal a crucial role of diverse stem cell compartments during MDS progression to AML, and have implications for current bulk cell-focused precision oncology approaches in MDS and possibly other cancers that evolve from pre-malignant conditions that may miss preexisting rare aberrant stem cells that drive disease progression and leukemic transformation.
Myelodysplastic syndromes (MDSs) are malignant, pre-leukemic, hematologic disorders with poor clinical outcome and median overall survival of less than 2 years in higher risk subtypes1,2. Delaying progression to secondary AML (sAML) is one of the key challenges in the clinical management of patients with MDS. The clonal origin of MDS and AML has been demonstrated to lie within the phenotypic and functionally defined stem cell compartment3–11. Previous seminal studies have investigated bulk tumor cells from patients with MDS, as well as fully transformed bulk cells (blasts) upon progression to sAML12–14. However, stem cell compartments, which represent a very small subset of total bone marrow cells cannot be effectively interrogated by bulk sequencing even when performed at significant depth. Clonal evolution at the stem cell level, which is crucial for MDS pathogenesis and progression to sAML, has not yet been directly examined.
To obtain direct insights into the pathogenesis of MDS and progression to sAML at the stem cell level, we utilized longitudinal, paired samples from 7 patients with MDS who had later progressed to sAML (Supplementary Table 1). For both MDS and paired sAML samples, we utilized multi-parameter fluorescence-activated cell sorting (FACS) to fractionate phenotypically defined malignant stem cells (MDS-SC, AML-SC), pre-malignant stem cells (preMDS-SC, preAML-SC), as well as blast populations (MDS blasts, AML blasts) (Fig. 1a; Supplementary Fig. 1, 2). Specifically, we isolated hematopoietic stem and progenitor cells (HSPC, Lin−CD34+CD38−) expressing at least one of the LSC markers (CD45RA, CD123, or IL1RAP) that were previously identified15–18, to enrich for malignant stem cells (MDS-SC, AML-SC) (Supplementary Fig. 1a). At the same time, we isolated HSPCs that were triple-negative for CD45RA, CD123, and IL1RAP to enrich for pre-malignant stem cells (preMDS-SC, preAML-SC) (Supplementary Fig. 1a). We observed significant expansion of the phenotypic malignant stem cell population within the total HSPC population during progression from MDS to sAML, increasing from 30.3% (MDS) to 66.9% (sAML) on average (p < 0.001; Supplementary Fig. 1b, c). Xenotransplantation of phenotypic MDS-SC led to predominantly myeloid engraftment (CD33+) compared to preMDS-SCs (73.2% versus 11.5%; Supplementary Fig. 3b, c), whereas phenotypic preMDS-SCs resulted in significantly higher lymphoid engraftment (CD19+) compared to MDS-SCs (82.4% versus 18.8%; Supplementary Fig. 3b, c). Similar findings were obtained upon xenotransplantation of sorted preAML-SC and AML-SC (Supplementary Fig. 3d-f). Moreover, consistent with previous reports19,20, we also observed significant lower clonogenicity (Supplementary Fig. 4a, b), and increased myeloid bias (Supplementary Fig. 4c, d) of sorted MDS-SCs and AML-SCs, compared to preMDS-SC and preAML-SC, respectively. These data indicate that CD45RA/CD123/IL1RAP expressing HSPCs are indeed enriched for malignant stem cells and CD45RA/CD123/IL1RAP triple-negative HSPCs are enriched for pre-malignant stem cells in MDS and AML.
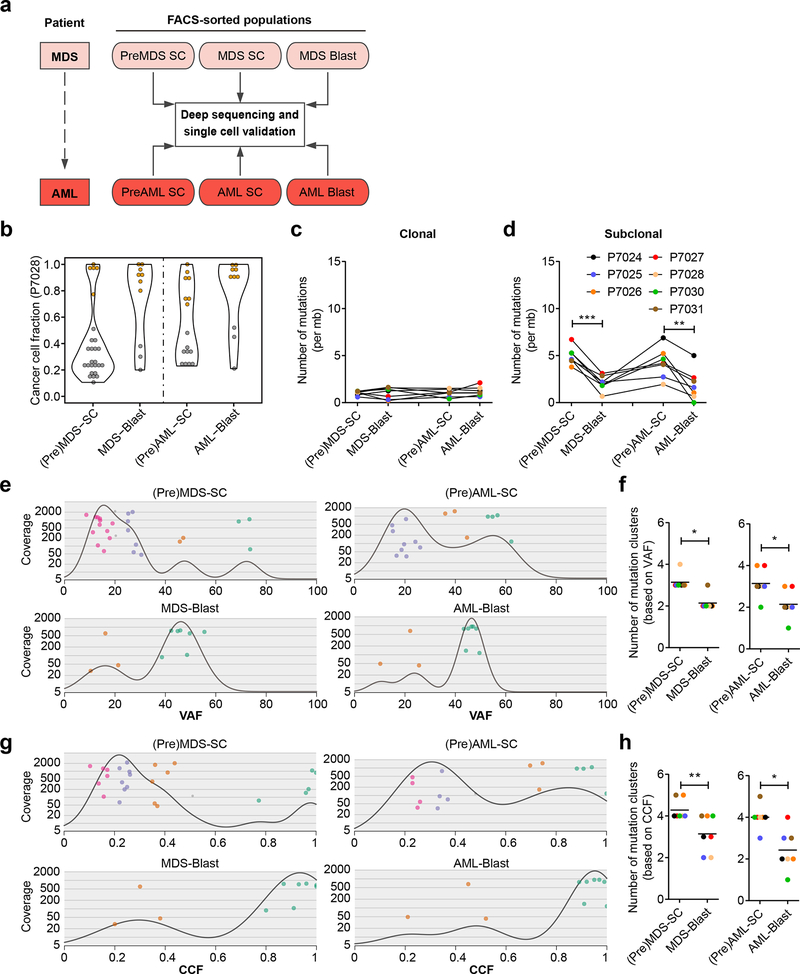
Fig. 1 |. Higher subclonal diversity at the stem cell level than in blasts in patients with MDS and sAML.
a, Schematics of experimental strategy of deep targeted sequencing and single cell validation of longitudinal, paired samples from patients with MDS who later progressed to secondary AML. Multi-parameter cell sorting was used to fractionate premalignant stem cells (PreMDS-SC, PreAML-SC), malignant stem cells (MDS-SC, AML-SC), and blast populations (MDS blasts, AML blasts). Non-hematopoietic cells (CD45-negative) were used as germline control for detection of somatic mutations and copy number changes. Selected mutations in each population were further examined with single cell sequencing. b, Representative distribution of CCFs in stem cells (preMDS-SC and MDS-SC; or preAML-SC and AML-SC) and blasts of patient P7028, showing that stem cells had more mutations at a lower frequency than blasts for both the MDS and sAML stages, respectively. Violin plot is showing frequency distribution (kernel density) of clonal mutations (orange) and subclonal mutations (grey). c, d, Burden of clonal (c) and subclonal (d) mutations in stem cell and blast populations at the MDS (p=0.0002) and AML (p=0.005) stages across patients (n=7). e, Clonal composition of stem cell and blast populations in MDS (upper left, lower left), and sAML (upper right, lower right), respectively, in patient P7028. Based on the VAFs, mutations covered by >30× are clustered as clones and denoted with the same color. Mutation was denoted with grey if the estimated possibility of the mutation to be clustered in the subclone was lower than 0.95. f, Number of mutation clusters, as estimated by VAFs of mutations, in stem cells and blasts at the MDS (left, p=0.013) and AML (right, p=0.021) stages across all patients studied (n=7). Black line represents the mean of clone numbers across the samples. g, h, Clonal composition of stem cell and blast populations at MDS (left, p=0.0047) and AML (right, p=0.02) estimated by CCFs of mutations (n=7). If not specified otherwise, data are mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001 (two-tailed paired Student’s t test).
To prospectively analyze clonal evolution at the stem cell level during the progression of MDS to AML, all seven cell populations (preMDS-SC, MDS-SC, MDS blasts; preAML-SC, AML-SC, AML blasts; non-hematopoietic germline control) from the same patient with MDS and sAML were subjected to targeted deep sequencing with a custom panel containing the most frequently altered genes in hematologic malignancies21, and other recent genes of interest involved in the development of MDS and AML (Fig. 1a, Supplementary Table 2). For each of the target genes, we included all of the exons, 5’ and 3’ UTRs, as well as the 1000bp up- and down-stream regions of the gene. Prior to sequencing, we performed whole genome amplification (WGA) of the sorted cell populations, which was shown to not distort the variant allele frequency (VAF) of mutations (Supplementary Fig. 5a,b). Targeted sequencing achieved consistent coverage across different cell populations in the same patient, and ranging from 300× to 1000× between patients (Supplementary Fig. 5c). To assess mutation patterns across different cell populations, we detected somatic mutations in each of the cell populations by comparing to the germline control (Fig. 1a and Supplementary Table 3), and validated the selected mutations by Sanger sequencing (Supplementary Fig. 5d, e). Interestingly, we found a significantly higher number of mutations, in both exonic as well as non-exonic regions, in stem cells compared to blasts in both MDS and sAML (Supplementary Fig. 5f-h).
Thereafter, we calculated the cancer cell fraction (CCF) within each cell population, considering VAF, purity, and ploidy as previously described22 (Supplementary Fig. 6a). Mutations were defined as “clonal” if the 95% confidence interval of the CCF was greater than 0.95, otherwise they were called “subclonal”22. We found that, while the frequencies of clonal mutations were similar across the cell populations (Fig. 1c and Supplementary Fig. 6), the frequency of subclonal mutations was significantly higher in stem cells than in blast cells in both MDS (4.9 ± 0.92 versus 2.1 ± 0.79 per Megabase; p < 0.001) and AML (4.2 ± 1.6 versus 1.9 ± 1.6 per megabase; p < 0.01) (Fig. 1d). These results indicated that, in both MDS and sAML, stem cells possess higher subclonal complexity compared to the blast cells. Previous studies have found associations of the intrinsic mutational processes in stem cells during life with various cancers, and the burden of mutations in tissue-specific stem cells is highly correlated with age23,24. In addition, as several DNA repair pathways are dependent on cell cycling, relative quiescence may render stem cells vulnerable to accumulation of DNA damage during aging25–27. Consistent with this idea, we found that mutation patterns in both MDS and sAML stem cells were associated with DNA repair pathways in addition to association with age-related signatures (Supplementary Fig. 7).
To compare the subclonal diversity of stem cells versus blasts, we inferred the clonal architectures of stem and blast cells with Sciclone28, using VAFs (Fig. 1e, f) as well as CCFs (Fig. 1g, h) of mutations. Interestingly, compared to blast cells, stem cells had a significantly higher total number of inferred mutation clusters (ranging from 2 to 4 versus 1 to 3; p < 0.05) at the MDS and sAML stages (Fig. 1e, f). Consistent findings were obtained through clonality analyses using CCFs, in that stem cells had a higher number of mutation clusters compared to the blasts (3 to 5 versus 1 to 4; p < 0.01) (Fig. 1g, h and Supplementary Fig. 8a-f). The difference was mainly attributable to a difference in number of non-dominant clones with lower CCFs (Fig. 1g and Supplementary Fig. 8a-f). Taken together, our results indicated that in both MDS and sAML, stem cell compartments have a higher subclonal diversity compared to blasts.
We next examined the patterns of clonal evolution during the progression from MDS to sAML of stem versus blast cell populations. Across all populations, pre-malignant stem cells, malignant stem cells, and blast cells, we identified shared mutations between MDS and sAML, that either had high (clonal) or low (subclonal) CCFs (Supplementary Fig. 9). Interestingly, our results also revealed substantially different patterns of clonal evolution between stem cell compartments and blast cells during MDS progression to sAML in several of the patients (Supplementary Fig. 9). In addition, we found a somewhat variable extent of clonal evolution of preMDS-SC and MDS-SC in individual patients. This may also reflect the phenotypic heterogeneity of putative disease stem cells29, which will be interesting to study in larger cohorts of patients.
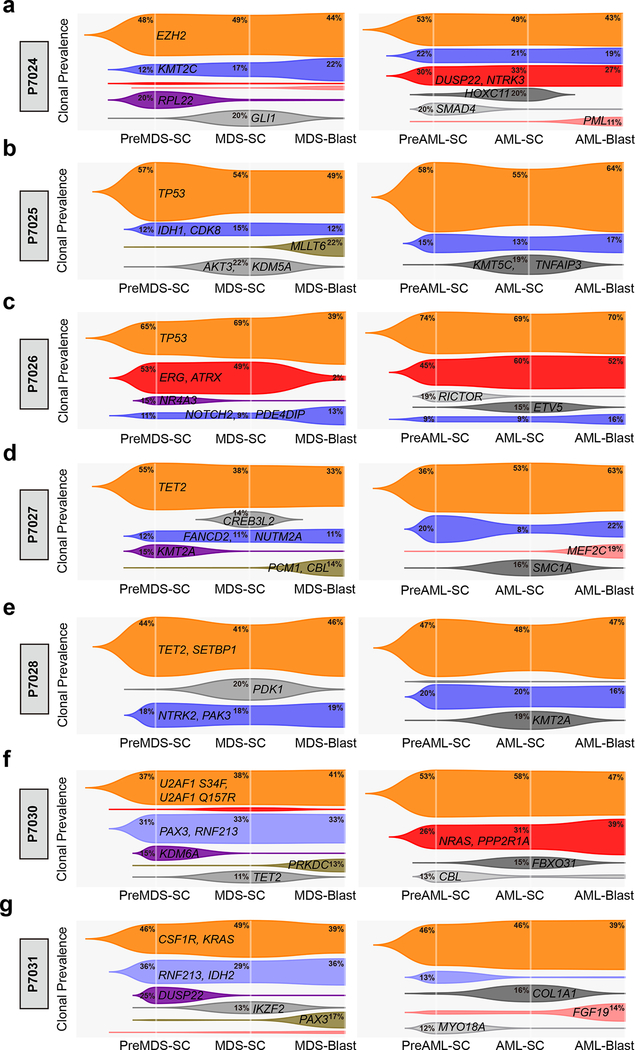
We next compared clonal evolution across all cell populations and during MDS to sAML progression longitudinally. In all the patients studied, we observed one dominant clone that was shared (denoted with orange) in stem cells and blast cells at both MDS and sAML stages (Fig. 2a-g). Within these dominant clones, we found mutations in genes (e.g. TET2, EZH2, TP53, SETBP1, U2AF1, CSF1R, and KRAS, etc) that are frequently observed in bulk cell sequencing studies of human MDS and AML30,31, as well as in elderly individuals with clonal hematopoiesis (CH) – albeit typically at a low subclonal size –32–34. Interestingly, both clonal shared mutations (e.g. TET2, EZH2, TP53, U2AF1, CSF1R, and KRAS), and subclonal shared mutations (e.g. KMT2C, NOTCH2 and FANCD2) were detectable in T cells (Supplementary Fig. 10), indicating that these shared mutations are acquired early during MDS disease initiation and that the presence of these mutations in stem cells is still compatible with T cell differentiation. This is in line with a recent study that found CH-associated mutations, including DNMT3A, TET2, TP53, and SF3B1 in virtually all hematopoietic populations, including HSCs, in elderly individuals35. Furthermore, two recent longitudinal studies of healthy individuals who eventually developed AML also detected mutations in some of the shared dominant genes (e.g. TET2, TP53, U2AF1, etc) in peripheral blood DNA many years before the actual diagnosis of AML, and the mutations were associated with increased risk of developing AML36,37.
Fig. 2 |. Schematic models of subclonal evolution of stem cell and blast populations during the progression from MDS to sAML.
a-e, Trajectory of individual clones in the different pre-malignant and malignant stem cell and blast populations at the MDS (left) and sAML (right) stages in individual patients. (a) Patient P7024, (b) patient P7025, (c) patient P7026, (d) patient P7027, (e) patient P7028, (f) patient P7030, and (g) patient P7031. Clonal prevalence was defined as the mean of VAFs of mutations (as shown) in the clone estimated by SciClone. Relative clonal prevalence within the same cell population is depicted on the Y-axis in the plots. Phylogenetic relationships of different cell populations were inferred by LICHeE and visualized by Timescape R package. Same clones in MDS and sAML are shown with the same color within each stem or blast population of the same patient, indicating the dynamics of clonal architecture in different cell populations, as well as longitudinal clonal evolution following progression from MDS to sAML. Clone is shown if the frequency is >1% in at least one of the three populations at MDS or sAML stages. And representative mutated genes in each clone are indicated.
In line with the results above (Supplementary Fig. 8), we consistently identified more subclones at the stem cell level compared to blasts in all patients, again revealing distinct subclonal architectures between stem and blast cell compartments. Interestingly, in patient P7026, one subclone (colored with red) was well detectable in pre-MDS-SC and MDS-SC, but had a frequency of only 2% in MDS blasts, and then expanded to be the dominant clone across all populations upon progression to sAML (Fig. 2c). Moreover, in patients P7024 and P7030, we observed large subclones at the AML stages (colored with red; Fig. 2a, f). Most interestingly, these subclones were undetectable in MDS blasts, but were inferred at frequencies of 2–3% in MDS stem cells (Fig. 2a, f). Taken together, these results suggested a potential model of non-linear clonal evolution at the stem cell level during initiation of MDS and progression to sAML: the mutational process would generate a dominant clone as well as distinct subclones at the stem cell level, and only one or few of these clones would become apparent at the bulk/blast level (Supplementary Fig. 11).
To definitively determine the relationship between different subclones in the same population as well as clonal dynamics across all the cell populations, we performed single cell targeted sequencing of sorted stem and blast populations (Supplementary Fig. 12) with selected mutations from each of the inferred subclones (Fig. 2). We calculated the CCFs of mutations using the single cell sequencing results, and found significant correlation between the CCFs estimated by Hiseq of sorted cell populations and CCFs determined by single cell sequencing in all patients (Supplementary Fig. 12d-h).
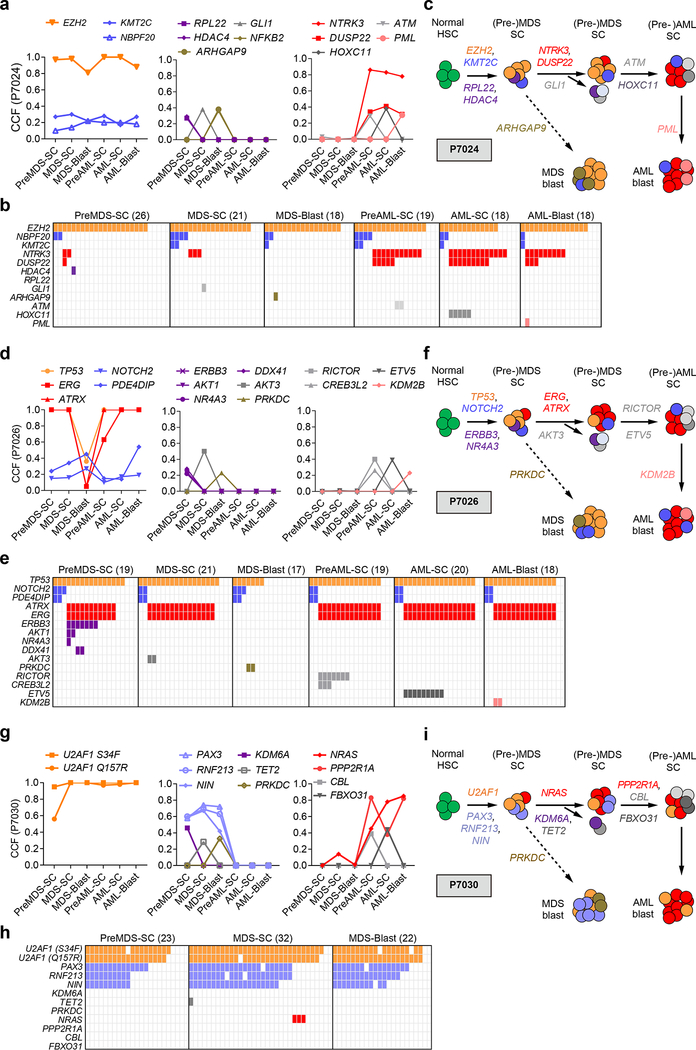
Targeted deep sequencing of sorted populations from patient P7024 had identified that clonal mutations in EZH2 and subclonal mutations (e.g. KMT2C) were shared across all stem cell and blast populations (Fig. 3a, left; Supplementary Fig. 13a). By single cell sequencing, we found that EZH2 mutations were indeed present in the majority of cells across different populations, whereas KMT2C mutations resided in a subclone within EZH2-mutated cells (Fig. 3b). Interestingly, mutations in HDAC4, GLI1, and RPL22 were present in small subclones of MDS stem cells only, and not responsible for MDS blast generation or progression to sAML (Fig. 3a-c). Co-mutations in NTRK3 and DUSP22 co-occurred in AML stem and blast cell populations within EZH2 mutated cells, but were not detectable in MDS blasts cells; strikingly, however, single cell sequencing demonstrated small subclones containing these mutations within preMDS-SC and MDS-SC stem cell compartments (Fig. 3b, c). In AML populations, we identified mutations of ATM and HOXC11 within the NTRK3 and DUSP22 mutated stem cells, whereas mutation of PML was only observed in a small subclone of NTRK3 and DUSP22 mutated blast cells (Fig. 3a-c). Taken together, the findings obtained by single cell sequencing lead to a patient-specific model of clonal evolution across different stem and blast populations in MDS and sAML (Fig. 3b, c). In this patient, mutations in EZH2 were acquired early in the founding clone at the MDS stage, and acquisition of additional mutations in NTRK3 and DUSP22 contributed to the progression to sAML (Fig. 3c), while MDS blasts were characterized by different co-mutations. Thus, sAML developed from a rare subclone contained within MDS stem cells, and not through further evolution of MDS blasts (Fig. 3c).
Fig. 3 |. Spatiotemporal subclonal evolution during the progression from MDS to sAML determined by single cell sequencing of sorted stem and blast cells.
a, CCFs of shared (left), MDS-specific (middle), AML specific (right) mutations across all cell populations in patient P7024. b, Single cell targeted sequencing of mutations across different cell populations of patient P7024. Each column represents the sequencing results of one single cell of the indicated cell population (preMDS-SC, MDS-SC, MDS-blasts, preAML-SC, AML-SC, AML-blasts), and the number of single cells tested in each population is shown in parentheses. The occurrence of a mutation in a single cell is indicated with the same color as in (a). c, Schematic model of clonal evolution in different stem and blast cell populations in patient P7024. Mutations in EZH2 were acquired early in the founding clone at the MDS stage, and acquisition of additional mutations in NTRK3 and DUSP22 contributed to the progression to sAML, while MDS blasts were characterized by different co-mutations. In this patient sAML developed from a rare subclone contained within MDS stem cells, and not through further evolution of MDS blasts. d, CCFs of shared (left), MDS-specific (middle), AML specific (right) mutations across all cell populations in patient P7026. e, Single cell targeted sequencing of mutations across different cell populations of patient P7026. f, Schematic model of clonal evolution in different stem and blast cell populations in patient P7026. Data again indicate that the dominant clone present in sAML stem and blast cells developed from a clone within the MDS stem cells that was nearly undetectable in MDS blast, indicating a crucial role of MDS stem cells in sAML initiation. g, CCFs of shared (left), MDS-specific (middle), AML-specific (right) mutations in different stem and blast populations at the MDS and sAML stage of patient P7030. h, Single cell targeted sequencing of mutations across different cell populations of patient P7030. i, Schematic model of clonal evolution in different stem and blast cell populations in patient P7030. Subclones of MDS stem cells with early founding mutations (i.e. U2AF1) remained present during MDS blast generation as well as AML progression, whereas other mutations, e.g. PAX3, RNF213, NIN and KDM6A, only occurred in MDS but not during progression to sAML. Progression to sAML originated from a subclone of MDS stem cells with NRAS mutation.
In patient P7026, we detected that a TP53 mutation was shared in the majority of single cells across all cell populations (Fig. 3d, e; Supplementary Fig. 13b). We also observed a less frequent, but stable subclone with co-mutations of NOTCH2 and PDE4DIP within the TP53-mutated cells (Fig. 2b and Fig. 3d, e). On the other hand, ERG and ATRX co-mutations were present in a more frequent (dominant) clone within preMDS-SC and MDS-SC (Fig. 3d, e), that was distinct from the subclone with NOTCH2 and PDE4DIP co-mutations. Interestingly, this subclone was nearly undetectable (VAF = 1.95%) in MDS blast bulk cell sequencing and undetectable in MDS blast single cell sequencing (Fig. 2b and Fig. 3d, e), but became dominant in all sAML stem and blast cell populations (Fig. 3d-f), again demonstrating that the subclones contributing to the generation of MDS blasts were different from those contributing to the progression to sAML (Fig. 3e, f). Single cell sequencing also identified two distinct subclones within the preMDS-SC subclone with ERBB3 mutation, one with co-mutations of AKT1 and NR4A3, and another one with mutation of DDX41 (Fig. 3e). However, none of these specific subclones persisted in MDS blasts or during sAML progression. Taken together, in this patient the dominant clone present in sAML stem and blast cells developed from a clone within the MDS stem cells that, however, was undetectable in MDS blasts (Fig. 3f). Mutations of ERG are relatively rare in MDS and AML; and mutations of ATRX are also infrequent and found in 0.2–0.8% of the MDS patients, but higher in the MDS subtype associated with α-thalassaemia38,39. It will be interesting to assess whether these mutations play functional roles in promoting the progression of MDS to sAML in future studies.
In patient P7030, we identified two clonal mutations in U2AF1 (Q157R and S34F) that were shared across all populations (Fig. 2f, Fig. 3g, h, and Supplementary Fig. 13d). We also identified a relatively large subclone within the U2AF1-muteted cells with mutations of PAX3, RNF213 and NIN that was shared in all the MDS populations, but did not appear at the sAML stages (Fig. 2f, Fig. 3g, h). A mutation in NRAS was only detectable in MDS-SC (VAF = 6.5%; Supplementary Fig. 13d) at the MDS stage (and not in MDS blasts), and resided in a subclone within U2AF1-muteted cells that was distinct from the PAX3-mutated subclone (Fig. 3h). Interestingly, this NRAS-mutated MDS-SC subclone then expanded at the sAML stage (Fig. 2f, Fig. 3g), accompanied by the acquisition of an additional mutation in PPP2R1A (VAF = 0% at MDS-SC; Supplementary Fig. 13d). In this patient, the progression to sAML originated from a small subclone of U2AF1-mutated MDS-SCs bearing the NRAS mutation (Fig. 3g-i). Similarly, in patient P7027, we observed that the AML progression was associated with a small subclone of MDS stem cells with RUNX1 mutation (Supplementary Fig. 14). Both NRAS and RUNX1 mutations are recurrent in patients with MDS and AML with markedly higher incidence in high-risk MDS and AML14,30,31, and NRAS mutations are rarely found at initial diagnosis14,40. Our results suggest that NRAS and RUNX1 mutations may pre-exists at least in some patients, and reside in rare stem cell subclones at a very early disease stage.
Interestingly, in comparison with the patients shown above, we observed slightly more stable clonal evolution at the level of both stem and blast cells in patients P7025 and P7028 (Fig. 2b, e and Supplementary Fig. 15a-d). While most of the clonal mutations were shared between MDS and sAML (e.g. TET2 and SETBP1 in P7028; TP53 in P7025), we again observed MDS- and AML-specific mutations, respectively, in particular within MDS-SC and AML-SC (Fig. 2b, e, and Supplementary Fig. 15a-d). In patient P7031, we identified clonal mutations on CSF1R and KRAS that were shared across all cell populations (Fig. 2g, Supplementary Fig. 15e, f). We also observed a larger subclone with mutations in RNF213, RUNX1, and IDH2 that were shared in all MDS populations as well as preAML-SC, but did not contribute to the generation of AML blasts (Fig. 2g, Supplementary Fig. 15e-g). A U2AF1 (Q157R) mutation was detected in MDS-SC and MDS-blast cells with CCFs of 0.26 and 0.17, respectively, and cells with this mutation expanded upon the progression to sAML with CCFs ranging from 0.51 to 0.61 (Supplementary Fig. 15e, f). Overall, compared to patients P7024, P7026, P7027, P7030 (Fig. 3c, f, i), results of P7025, P7028, P7031 revealed a model of slightly later branching of MDS stem cells towards progression to sAML (Supplementary Fig. 15b, d, g).
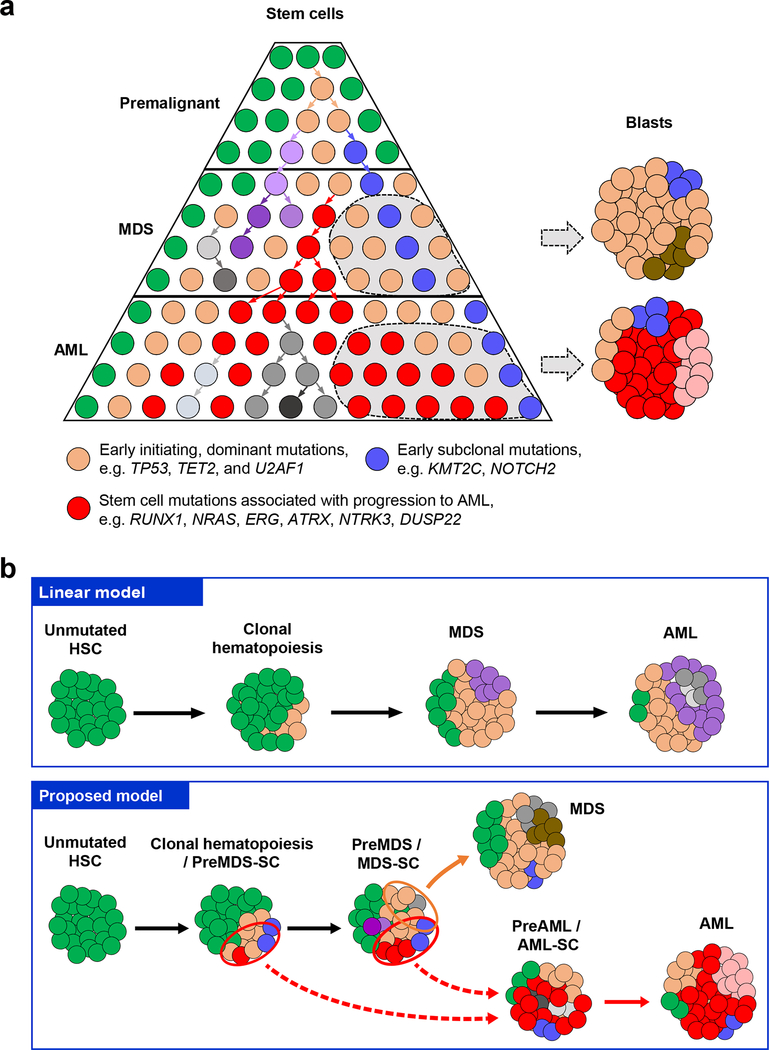
In summary, we chose a strategy of combining rigorous cell sorting with targeted deep sequencing of both stem and blast cells from patients with MDS who progressed to sAML, which resulted in a thus far unprecedented resolution at the stem cell level (effective depth equivalent to what could only be achieved by 250,000× to 5,000,000× deep bulk sequencing; as a result of ~0.01–0.2 % frequency of sorted stem cells, and average sequencing depth of ~500×). By ensemble as well as single cell sequencing of both stem cell and blast populations of MDS and matched sAML, we found that stem cells at the MDS stage have a significantly higher complexity of subclonal mutations compared to blast cells (Fig. 4a). Subclonal mutations mostly resided within the dominant clone with early mutations (e.g. TET2, TP53, and U2AF1), but can dramatically increase in size towards progression to sAML, suggesting that an upfront distinction at the MDS stage of “dominant” and “passenger” clones/mutations solely based on clone-size may not have disease pathogenetic or predictive relevance. Our findings reveal the crucial role of a diverse stem cell pool towards full transformation and MDS blast cell generation as well as progression to sAML in a non-linear and rather parallel manner (Fig. 4). These findings have implications for currently employed bulk cell-focused precision oncology approaches and provide a rationale to consider mutational examination of fractionated stem cell populations in patients with MDS, and possibly other cancers arising from premalignant conditions, in order to more comprehensively assess pharmacologically ‘actionable’ mutations relevant for later disease progression and development of AML.
Fig. 4 |. Proposed model of subclonal evolution of stem cells during the progression of MDS to sAML.
a, Our results suggest a model of non-linear clonal evolution arising from the stem cell level during development of MDS and progression to sAML. Accumulation of mutations in stem cell compartments gives rise to a highly diverse subclonal architecture (indicated by different colors) in MDS stem cells. Certain subclones (orange, e.g. with TP53, TET2, or U2AF1 mutations, ‘clonal hematopoiesis’) provide a shared basis for both MDS development (MDS blasts) as well as formation of preAML- and AML-stem cells. However, preMDS- or MDS-stem cells acquire different additional mutations which then drive MDS blast formation, or progression to sAML, respectively, in a non-linear and rather parallel manner in all patients studied. In four (P7024, P7026, P7027, and P7030) out of seven cases studied, we identified that the dominant clone at the sAML stage originated from a clone (red, e.g. with RUNX1, NRAS, or ERG and ATRX mutations) that was detectable in preMDS- and/or MDS-stem cells, but was undetectable in MDS blast cells. These results indicate that MDS stem cells leading to the generation of MDS blast can be different from those contributing to the progression to sAML, highlighting a crucial role of the entirety of the diverse MDS stem cell pool in sAML disease progression, which has implications for current bulk cell-focused diagnostic and therapeutic precision oncology approaches. b, Schematics of different models of MDS and sAML development and progression. In comparison to the linear model (top panel), which has been proposed based on bulk sequencing and suggests serial mutation accumulation during disease progression, our data support a model of parallel clonal evolution at the stem cell level during development of MDS and progression to sAML (bottom panel). 7 out of 7 cases showed a highly diverse pool of (Pre-)MDS stem cells as the basis of MDS and sAML development; in 4 out of 7 patients we found very early branching at the MDS stem cell level towards progression to AML stem cells leading to distinct clonal composition between MDS and AML bulk cells, 3 out 7 patients showed a pattern of slightly later branching (dashed red arrows) leading to more similar clonal composition between MDS and AML bulk cells compared to the early branching cases.
Materials and Methods
Multiparameter high-speed FACS of stem and blast cells from patient samples
Bone marrow samples from 7 patients with MDS and matched secondary AML (sAML) were obtained after written informed consent, from Montefiore Medical Center / Albert Einstein Cancer Center (IRB# 11-02-060E; for patients’ characteristics see Supplementary Table 1). All patients studied received treatment with hypomethylating agents between MDS and AML progression. Frozen BM aspirates were thawed in a water bath at 37°C and resuspended in IMDM supplemented with 2% FBS. After repeated washes with IMDM 2% FBS, cells were resuspended in MACS buffer (PBS supplemented with 0.5% BSA and 2mM EDTA, pH 7.2). Thereafter, CD34+ were immunomagnetically separated with Miltenyi MACS technology (130-046-702, Miltenyi Biotec) according to the manufacturer’s protocol. CD34+ enriched cells were stained for 30 minutes on ice with antibodies: PE-Cy5 (Tri-Color)–conjugated lineage markers (CD2, CD3, CD4, CD7, CD8, CD10, CD11b, CD14, CD19, CD20, CD56, Glycophorin A), APC-conjugated blast marker CD33, and hematopoietic stem and progenitor markers (Pacific blue CD34, PE-Cy7 CD38, FITC CD45RA, Alexa Fluor 700 CD123 and PE IL1RAP). A list of antibodies is provided in Supplementary Table 4. We used Lin−CD34+CD38−CD45RA−CD123−IL1RAP− to enrich for preMDS or preAML stem cells, and Lin−CD34+CD38−(CD45RA+ and/or CD123+ and/or IL1RAP+) to enrich for MDS or AML stem cells. Cell were also stained with PE CD45, APC CD33, and pacific orange CD4, to isolate blast cells (CD45+CD33+), T cells (CD45+CD4+) and non-hematopoietic cells (CD45−) as germline control for somatic variant calling. Inter-patient heterogeneity in the profile of surface markers for disease-relevant stem cells have been observed in patients with MDS and AML41,42, suggesting the need to utilize a combination of surface markers. In addition, the coexistence of residual normal HSC, numerous subclones of partially transformed pre-MDS-SC, as well as fully transformed MDS-SC, makes their distinction based on phenotypic markers challenging in individual patients. Isolation of cell populations based on phenotypic markers remains a relative enrichment strategy, which requires functional and genetic validation. Xenografting experiments with the respective populations (Supplementary Fig. 3) demonstrated functionality consistent with pre-MDS-SC versus MDS-SC properties. In addition, the fact that the here described sorting strategy was able to detect relevant mutations in pre-MDS-SC and MDS-SC indicates the validity of the strategy, at least in this cohort of patients. It will be interesting to further validate this sorting scheme for pre-MDS-SC in larger patient cohorts in the future.
Methylcellulose assay
To assess differentiation potential of phenotypic pre-malignant stem cells (preMDS/AML-SC) and malignant stem cells (MDS/AML-SCs), cells were FACS-sorted from additional patients with the same strategy (Supplementary Fig. 1a), and plated in HSC003 methylcellulose medium according to the manufacturer’s recommendation (R&D Systems, Minneapolis, MN). Colonies of different hematopoietic lineages were scored two weeks after plating using an Inverted Infinity and Phase Contrast Microscope (Fisher Scientific, Hampton, NH). In addition, to examine the expression of lineage makers, methylcellulose medium was dissolved in PBS to dissociate the colonies into single cell suspension. Cells were stained with antibodies against CD14, CD15 and CD235a on ice for 30 minutes, then analyzed on a BD FACSAria II system.
Xenotransplantation assays
Bone marrow samples from additional patients with MDS or AML (unpaired) were processed and stained for surfaces markers for pre-malignant stem cells (preMDS/AML-SC) and malignant stem cells (MDS/AML-SCs) as described above (Supplementary Fig. 1a). Thereafter, 30,000 to 100,000 sorted cells were washed with and resuspended in Hank’s Balanced Salt Solution (HBSS, Corning, NY), and transplanted into nonirradiated NOD,B6.SCID Il2rγ−/−KitW41/W41 (NBSGW) immunocompromised mice (aged 6–8 weeks) via retro-orbital injection43. All experiments conducted on mice were approved by the Institutional Animal Care and Use Committee at Albert Einstein College of Medicine (protocol #2016–0103). Engraftment analysis of patient-derived cells was performed >12 weeks after transplantation. Mouse bone marrow cells were incubated with ammonium chloride potassium (ACK) buffer for 1 min on ice, and then stained for surface markers for mouse leukocytes CD45.1, and markers for human leukocytes including CD45, CD19 and CD33. The stained cells were then analyzed on a BD FACSAria II system. While several studies have found some remaining lymphoid reconstitution of MDS/AML-SCs in irradiated recipient mice in a subset of patients44,45, many studies found exclusively myeloid output of MDS/AML-SCs8,15. The observed partially lymphoid engraftment in our study could be due to the nonirradiated NBSGW xenograft model we utilized43, as myeloid-biased engraftment of stem cells seems to be most pronounced in irradiation-conditioned transplantation assays46,47.
Whole genome amplification
Whole genome amplification (WGA) was performed with REPLI-g kit (Qiagen, MD), which utilizes the proofreading enzyme Phi 29 polymerase to achieve high-fidelity amplification of genomic DNA48,49. For sorted samples with yield cell number larger than 1000, cells were washed with PBS and then resuspended with 5μl of sterile PBS. REPLI-g mini kit (Qiagen, MD) was used for WGA according to the manufacturer’s protocol. For sorted samples with less than 1000 cells or single cell analysis, cells were sorted into 5μl PBS, thereafter, REPLI-g single cell kit (Qiagen, MD) was used for WGA according to the manufacturer’s protocol. For DNA samples, we used 1 to 10ng DNA as input, and REPLI-g mini kit (Qiagen) was used for the WGA. All the products of WGA were purified with Agencourt AMPure XP beads (Beckman Coulter, IN) to remove residual dNTP, primers and random products with size < 100bp.
Targeted sequencing with HiSeq 2500
From the same patient, seven cell populations (preMDS-SC, MDS-SC, MDS blasts; preAML-SC, AML-SC, AML blasts; non-hematopoietic germline control) were subjected to targeted sequencing of a 504-gene customized panel containing all the genes in the FoundationOne Heme panel21, as well as other genes of interest involved in the development of MDS and AML (full list of genes is provided in Supplementary Table 2). For each of the target genes, we included all the exons, 5’ and 3’ UTRs, as well as the 1000bp up- and down-stream regions of the gene. For targeted sequencing, 500ng of DNA was used as input for sequencing with an Illumina HiSeq 2500 system (Illumina, CA). In brief, DNA was fragmented by a Covaris ultrasonicator (Covaris, MA) with target size of ~200bp, followed by end repair and A-tailing with KAPA LTP library preparation kit for Illumina platforms (Kapa Biosystems, MA) according to the manufacturer’s instructions. Thereafter, we linked the DNA products with Illumina TrueSeq sequencing adapters, and performed size selection with dual-SPRI beads (Beckman Coulter, IN). Next, we performed 8 cycles of pre-capture LM-PCR with the adapter-ligated DNAs according to the user’s guide for NimbleGen SeqCap EZ Library (Roche NimbleGen, CA). Afterwards, LM-PCR products of different cell populations from the same patient were incubated together for 72 hours with NimbleGen SeqCap EZ probes (Roche NimbleGen). Hybridization products were then incubated with capture beads at 47°C for 45min, followed by washing and elution with PCR-grade water according to the manufacturer’s protocol. Captured DNAs were then amplified with 8 cycles of post-capture LM-PCR according to the user’s guide for NimbleGen SeqCap EZ Library (Roche NimbleGen). At last, DNA products were purified with Agencourt AMPure XP beads (Beckman Coulter, IN) and then subjected to massively parallel sequencing (100bp paired-end) on the HiSeq 2500 platform according to the manufacturer’s instructions.
Analysis of sequencing data
We assessed the quality of the raw sequencing data from HiSeq with FastQC v0.11.4 (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Reads contaminated with sequencing adapter and reads with low quality were removed by Trim Galore 0.4.1 using the default parameters (https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/). Thereafter, we performed genome alignment (hg19) using Bowtie2 v2.2.950. Alignment results were processed as described in GATK best practice for detection of somatic mutation recommended by the Broad institute51. Briefly, duplicated reads were marked with Picard toolkit (http://broadinstitute.github.io/picard/). Thereafter, indel realignment and base recalibration were performed for each of the individual samples with GATK v3.751. Moreover, we performed a second run of indel realignment with merged samples from the same human patient to remove false positive mutations caused by alignment artifacts. After pre-processing of the reads, sequencing coverage of each sample was calculated with DepthOfCoverage module of GATK. For detection of somatic mutations, we used Mutect2 of GATK v3.7 comparing each of the cell populations to the matched germline control with the default parameters52. Then we merged all the Mutect2 results passing the filter from the same human patient to generate a combined set of mutations for each of the patients. Moreover, FreeBayes v0.9.20 was used to perform joint variant calling with all the samples from the same human patient53, using the parameters of -m 1 -q 3 -F 0.05 -C 2 -U 3 --read-indel-limit 2 --min-coverage 20. We also excluded the variants from FreeBayes results with quality score <10. Thereafter, high-confidence mutations consistently detected by both Mutect2 and FreeBayes were used for downstream analysis. In addition, to address potential false negatives due to tumor cell contamination of germline controls, we also included somatic mutations reported in MDS or AML by more than 2 groups in the COSMIC database (http://cancer.sanger.ac.uk/cosmic). Thereafter, we excluded the mutations that were: 1) covered less than 20× in germline control or test cells; 2) supported by < 3 reads or 5% of the reads in test samples; 3) reported in dbSNP database (SNPs v147), 1000 genome phase 3 or ExAC database 1.0 with population frequency >0.5%. To further remove mutation artifacts caused by sequencing context with low complexity, we excluded mutations that were: 1) located within 10bp of an indel; 2) within 20bp of another SNV; 3) less than 5bp to microsatellite or simple repeats of the UCSC database (https://genome.ucsc.edu); 4) less than 5bp to homopolymer (> 5bp). Thereafter, mutations were annotated using hg19 database by SnpEff v4.1k54.
For analysis of mutation signatures, we combined the somatic mutations in each cell population from the five patients sequenced, and examined the pattern of mutation signatures with deconstructSigs 1.8 with the signatures defined previously55. Weight of each signature was normalized by number of times each trinucleotide context is observed in the targeted regions.
Clonal analysis
Variant allele frequency (VAF) for each mutation was calculated by the number of reads supporting the variant divided by total reads, using the FreeBayes output. Moreover, sample purity and local copy number variation (CNV) were estimated by FACETS v0.5.6 package of R v3.2.356, which utilizes the read counts of both heterozygous and homozygous single-nucleotide polymorphism (SNP) loci. In brief, for each of the samples, we first extracted the read counts of reference and alternative alleles of each SNP reported in dbSNP (Common SNPs v147) or 1000 genome SNP phase 3 database with population frequency larger than 5%. Thereafter, the read count information of the SNP loci covered by at least 20× in the targeted sequencing of each sample were subjected to FACETS as input to estimate the purity and CNV using the default parameters. Thereafter, cancer cell fraction (CCF) of each mutation was estimated using the VAF, purity and local CNV of the mutation as described before22. Mutations were defined as “clonal” if the 95% confidence interval of CCF overlapped with 0.95, otherwise were defined as “subclonal”. To investigate the clonal architecture, both VAFs and CCFs of mutations covered >30× were subjected to SciClone v1.1.0 allowing a maximum cluster number of 1028. When comparing the clonal architecture of different cell populations of the same patients, we first generated a combined list of mutations that covered at least 20× in all samples, then subjected the VAFs of mutations in different populations to SciClone analysis. We excluded the mutations in the cluster if the estimated possibility of the mutation to be clustered in the subclone was lower than 0.95. In addition, to examine the clonal relationship between different cell populations in the same samples, we performed phylogenetic reconstruction by LICHeE v1.0 using VAFs of the mutations and the prevalence of each subclone in the samples estimated by SciClone, with the standard parameters (-maxVAFAbsent 0.005 -minVAFPresent 0.005 -n 0) recommended by LICHeE’s instructions57. Thereafter, results of phylogenetic relationships determined by LICHeE were visualized by TimeScape v1.0.0 package58.
Single cell targeted sequencing
After staining of surface markers, single cells were directly deposited, using a MoFlo Astrios EQ system (Beckman Coulter, IN), into 96-well PCR plate containing 5μl of sterile PBS per well. Thereafter, WGA was performed using Repli-g single cell kit (Qiagen, MD) according to the manufacturer’s protocol. WGA products were purified with Agencourt AMPure XP beads (Beckman Coulter, IN). For targeted sequencing, we designed primers for each mutation target using Primer 3, with product sizes less than 200bp (Supplementary Table 5). Target specific primers were linked with Fluidigm forward (5’-ACACTGACGACATGGTTCTACA-3’) and reverse (5’-TACGGTAGCAGAGACTTGGTCT-3’) common sequence (CS) tag for downstream barcoding. To pre-amplify the DNA of target regions, we first performed specific target amplification (STA) of WGA products using FastStart™ Taq DNA Polymerase (Roche). In brief, all the CS-tagged primers for the same sample were pooled, and diluted to make a final concentration of 1μM for each primer. The amplification mix for each sample was prepared as follows: 0.5 μl of 10× reaction buffer with MgCl2, 0.5 μl MgCl2, DMSO, 10 mM nucleotide mix, 0.2 μl FastStart polymerase, 1 μl 1μM primer pool and 10 ng DNA. Afterwards, PCR amplification was performed as follows: 95°C for 10 min; 2 cycles of 95°C for 15s and 60°C for 4min; 10 cycles of 95°C for 15s and 72°C for 4min. As a negative control, we included a no template control (NTC) in the STA experiment. Thereafter, 10μl of each STA product diluted to 100ng/μl was transferred to half of a new 96-well plate (47 single cell samples plus one NTC per plate), and treated with ExoSAP-IT (Affymetrix, MA) for purification. For primer preparation, each primer pair was diluted to 1μM in the 96-well plate with Fluidigm Access Array loading reagent (Fluidigm, CA). Thereafter, plates of STA products and primer pairs were loaded onto 48.48 integrated fluidic circuits (IFC, Fluidigm, CA) in Biomark HD system (Fluidigm, CA). Each of the STA products were mixed with each primer pair, and PCR amplification was performed in the IFC array according to manufacturer’s protocol. Thereafter, PCR products of the same sample were pooled together, and sample barcoding PCR was performed with primers containing the barcode sequence (Fluidigm, CA) and Illumina sequencing adapter (Illumina, CA). We assessed the quality of the barcoded samples with a 2100 Bioanalyzer (Agilent, CA), then all samples were pooled at equal ratios and subjected to sequencing with the MiSeq (150bp paired-end) system according to the manufacturer’s protocol (Illumina, CA).
For analysis of the MiSeq data, we trimmed reads with CS tag and reads contaminated with sequencing adapter, and we also removed reads with low quality by Trim Galore using the default parameters. Thereafter, we performed genome alignment to hg10 with BWA-MEM v0.7.1559, and then variant calling with FreeBayes. We also manually confirmed each of the target mutation with Integrative Genomics Viewer (IGV), and mutation with > 20% supporting reads (covered at least 5×) were considered as positive.
T cell receptor sequencing
To assess diversity of the T cell receptor (TCR) repertoire, we extracted total RNAs of T cells isolated from the patient samples, as well as cord blood samples as healthy controls, using RNeasy Micro Kit (Qiagen) according to the manufacturer’s protocol. 50ng of total RNAs were used as input for first-strand cDNA synthesis with the supplied reagents of SMARTer Human TCR a/b Profiling Kit (Takara Bio USA, Mountain View, CA) according to the manufacturer’s protocol. Thereafter, first round of PCR (PCR 1) was performed with SMART Primer 1 to link the Illumina Read 2 sequence to the cDNA, and TCRα and TCRβ primers to specifically amplify the variable regions and constant regions of TCRα and TCRβ cDNA. PCR 1 reaction was performed for 21 cycles with in a preheated thermal cycler (C1000, Bio-Rad; Hercules, CA) according to manufacturer’s protocol. Afterwards, 1μl PCR1 product was subjected to second round PCR (PCR 2), which was performed with TCRα and TCRβ Human Primer 2 Reverse HT Index primers (D501) to link the Illumina Read 1 sequence and P5-i5 index sequences. In addition, for different samples, we used different TCR Primer 2 Forward HT Index primers for the linkage of Illumina P7-i7 index sequences. PCR 2 reaction was performed for 20 cycles with in a preheated thermal cycler according to the manufacturer’s protocol. Lastly, the products of PCR 2 were purified using Agencourt AMPure XP beads (Beckman Coulter) with a double size selection approach according to the manufacturer’s instructions. Quality and quantity of the purified products (sequencing-ready libraries) were assessed with a 2100 Bioanalyzer (Agilent) and Qubit 2.0 Fluorometer, respectively. Sequencing was performed on an Illumina MiSeq sequencer with paired-end, 300bp reads. For the analyses of the sequencing data, the first 30bp of read 2, which include the SMART primer sequence, was trimmed with Trim Galore. The trimmed data was then analyzed with LymAnalyzer 1.2.2 separately for TCRα and TCRβ genes60. We then calculated the frequency of each Vα or Vβ gene segment relative to the total sequences mapped to Vα or Vβ genes.
Statistical analysis
Data are presented as mean ± s.d. if not otherwise specified. Student’s t test was performed with GraphPad Prism 7.0, as indicated. Pearson correlation coefficient R and statistical significance p-values were calculated with built-in cor.test function of R, and data was visualized with the ggplot2 package of R.
Supplementary Material
Acknowledgments
We thank P. Schultes from the Department of Cell Biology for expert technical assistance. We thank Ariana Fiallo from the from Einstein Genomics Core Facility for technical assistance in single cell targeted sequencing, and Shahina Maqbool and Shijun Mi from Einstein Epigenomics Core Facility for assistance in the targeted sequencing with the HiSeq platform. We thank Victor Thiruthuvanathan from the Department of Cell Biology for the assistance in processing the patient samples. We also thank W. Li for advice regarding whole genome amplification, and F.C. Chan, C. Steidl, and H. Steidl for helpful discussion. This work was supported by NIH grants R01CA166429, R01CA217092 (to U.S.), R01HL139487, R01DK103961 (to A.V.), K01DK105134 (to B.W.), Translational Research Program (TRP) grants from the Leukemia & Lymphoma Society (to U.S., and to A.V., respectively); a research grant from the Taub Foundation for MDS Research (to U.S.); and a research grants from the Evans Foundation (to A.V.). J.C. was supported by The Einstein Training Program in Stem Cell Research from the Empire State Stem Cell Fund through New York State Department of Health Contract (C30292GG). U.S. is a Research Scholar of the Leukemia and Lymphoma Society and the Diane and Arthur B. Belfer Faculty Scholar in Cancer Research of the Albert Einstein College of Medicine. This work was supported through the Albert Einstein Cancer Center core support grant (P30CA013330).
Competing interests
U.S. has received research funding from GlaxoSmithKline, Bayer Healthcare, Aileron Therapeutics, Novartis, has received compensation for consultancy services and for serving on scientific advisory boards from GlaxoSmithKline, Bayer Healthcare, Celgene, Aileron Therapeutics, Stelexis Therapeutics, and Pieris Pharmaceuticals, and has equity ownership in and is serving on the board of directors of Stelexis Therapeutics. A.V. has received research funding from GlaxoSmithKline, Incyte, MedPacto, Novartis, Eli Lilly and Company, has received compensation as a scientific advisor to Novartis, Stelexis Therapeutics, Acceleron Pharma, and Celgene, and has equity ownership in Stelexis Therapeutics. B.W. has received research support from Novartis Pharmaceuticals.
Footnotes
Data and materials availability
The high-throughput DNA sequencing data have been deposited in the database of Genotypes and Phenotypes (dbGaP) under accession code (pending).
References
- 1.Greenberg PL, et al. Revised International Prognostic Scoring System for Myelodysplastic Syndromes. Blood 120, 2454–2465 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ades L, Itzykson R & Fenaux P Myelodysplastic syndromes. Lancet 383, 2239–2252 (2014). [DOI] [PubMed] [Google Scholar]
- 3.Fialkow PJ, et al. Clonal Development, Stem-Cell Differentiation, and Clinical Remissions in Acute Nonlymphocytic Leukemia. New Engl J Med 317, 468–473 (1987). [DOI] [PubMed] [Google Scholar]
- 4.Nilsson L, et al. Involvement and functional impairment of the CD34(+)CD38(−)Thy-1(+) hematopoietic stem cell pool in myelodysplastic syndromes with trisomy 8. Blood 100, 259–267 (2002). [DOI] [PubMed] [Google Scholar]
- 5.Steidl U, et al. Essential role of Jun family transcription factors in PU.1 knockdown-induced leukemic stem cells. Nat Genet 38, 1269–1277 (2006). [DOI] [PubMed] [Google Scholar]
- 6.Will B, et al. Stem and progenitor cells in myelodysplastic syndromes show aberrant stage-specific expansion and harbor genetic and epigenetic alterations. Blood 120, 2076–2086 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jan M, et al. Clonal evolution of preleukemic hematopoietic stem cells precedes human acute myeloid leukemia. Science translational medicine 4, 149ra118 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pang WW, et al. Hematopoietic stem cell and progenitor cell mechanisms in myelodysplastic syndromes. P Natl Acad Sci USA 110, 3011–3016 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Corces-Zimmerman MR, Hong WJ, Weissman IL, Medeiros BC & Majeti R Preleukemic mutations in human acute myeloid leukemia affect epigenetic regulators and persist in remission. Proceedings of the National Academy of Sciences of the United States of America 111, 2548–2553 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shlush LI, et al. Identification of pre-leukaemic haematopoietic stem cells in acute leukaemia. Nature 506, 328–333 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Will B, et al. Minimal PU.1 reduction induces a preleukemic state and promotes development of acute myeloid leukemia. Nat Med 21, 1172–1181 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Walter MJ, et al. Clonal architecture of secondary acute myeloid leukemia. N Engl J Med 366, 1090–1098 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Walter MJ, et al. Clonal diversity of recurrently mutated genes in myelodysplastic syndromes. Leukemia 27, 1275–1282 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Makishima H, et al. Dynamics of clonal evolution in myelodysplastic syndromes. Nat Genet 49, 204–212 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goardon N, et al. Coexistence of LMPP-like and GMP-like leukemia stem cells in acute myeloid leukemia. Cancer Cell 19, 138–152 (2011). [DOI] [PubMed] [Google Scholar]
- 16.Jordan C, et al. The interleukin-3 receptor alpha chain is a unique marker for human acute myelogenous leukemia stem cells. Leukemia 14, 1777 (2000). [DOI] [PubMed] [Google Scholar]
- 17.Barreyro L, et al. Overexpression of IL-1 receptor accessory protein in stem and progenitor cells and outcome correlation in AML and MDS. Blood 120, 1290–1298 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mitchell K, et al. IL1RAP potentiates multiple oncogenic signaling pathways in AML. J Exp Med 215, 1709–1727 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jan M, et al. Prospective separation of normal and leukemic stem cells based on differential expression of TIM3, a human acute myeloid leukemia stem cell marker. Proc Natl Acad Sci U S A 108, 5009–5014 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chung SS, et al. CD99 is a therapeutic target on disease stem cells in myeloid malignancies. Science translational medicine 9, eaaj2025 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He J, et al. Integrated genomic DNA/RNA profiling of hematologic malignancies in the clinical setting. Blood 127, 3004–3014 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McGranahan N, et al. Clonal status of actionable driver events and the timing of mutational processes in cancer evolution. Science translational medicine 7, 283ra254 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blokzijl F, et al. Tissue-specific mutation accumulation in human adult stem cells during life. Nature 538, 260–264 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Adams PD, Jasper H & Rudolph KL Aging-induced stem cell mutations as drivers for disease and cancer. Cell Stem Cell 16, 601–612 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rossi DJ, et al. Deficiencies in DNA damage repair limit the function of haematopoietic stem cells with age. Nature 447, 725–729 (2007). [DOI] [PubMed] [Google Scholar]
- 26.Mandal PK, Blanpain C & Rossi DJ DNA damage response in adult stem cells: pathways and consequences. Nat Rev Mol Cell Biol 12, 198–202 (2011). [DOI] [PubMed] [Google Scholar]
- 27.Mohrin M, et al. Hematopoietic stem cell quiescence promotes error-prone DNA repair and mutagenesis. Cell Stem Cell 7, 174–185 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miller CA, et al. SciClone: inferring clonal architecture and tracking the spatial and temporal patterns of tumor evolution. PLoS Comput Biol 10, e1003665 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yanagisawa B, Ghiaur G, Smith BD & Jones RJ Translating leukemia stem cells into the clinical setting: Harmonizing the heterogeneity. Experimental Hematology 44, 1130–1137 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Haferlach T, et al. Landscape of genetic lesions in 944 patients with myelodysplastic syndromes. Leukemia 28, 241–247 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cancer Genome Atlas Research, N., et al. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med 368, 2059–2074 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xie M, et al. Age-related mutations associated with clonal hematopoietic expansion and malignancies. Nat Med 20, 1472–1478 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Genovese G, et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N Engl J Med 371, 2477–2487 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jaiswal S, et al. Age-related clonal hematopoiesis associated with adverse outcomes. N Engl J Med 371, 2488–2498 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arends CM, et al. Hematopoietic lineage distribution and evolutionary dynamics of clonal hematopoiesis. Leukemia 32, 1908–1919 (2018). [DOI] [PubMed] [Google Scholar]
- 36.Abelson S, et al. Prediction of acute myeloid leukaemia risk in healthy individuals. Nature 559, 400–404 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Desai P, et al. Somatic mutations precede acute myeloid leukemia years before diagnosis. Nature medicine 24, 1015 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Herbaux C, et al. Incidence of ATRX mutations in myelodysplastic syndromes, the value of microcytosis. Am J Hematol 90, 737–738 (2015). [DOI] [PubMed] [Google Scholar]
- 39.Steensma DP, Higgs DR, Fisher CA & Gibbons RJ Acquired somatic ATRX mutations in myelodysplastic syndrome associated with alpha thalassemia (ATMDS) convey a more severe hematologic phenotype than germline ATRX mutations. Blood 103, 2019–2026 (2004). [DOI] [PubMed] [Google Scholar]
- 40.Bacher U, Haferlach T, Kern W, Haferlach C & Schnittger S A comparative study of molecular mutations in 381 patients with myelodysplastic syndrome and in 4130 patients with acute myeloid leukemia. Haematol-Hematol J 92, 744–752 (2007). [DOI] [PubMed] [Google Scholar]
- 41.Shastri A, Will B, Steidl U & Verma A Stem and progenitor cell alterations in myelodysplastic syndromes. Blood 129, 1586–1594 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thomas D & Majeti R Biology and relevance of human acute myeloid leukemia stem cells. Blood 129, 1577–1585 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McIntosh BE, et al. Nonirradiated NOD,B6.SCID Il2rgamma−/− Kit(W41/W41) (NBSGW) mice support multilineage engraftment of human hematopoietic cells. Stem Cell Reports 4, 171–180 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Woll PS, et al. Myelodysplastic Syndromes Are Propagated by Rare and Distinct Human Cancer Stem Cells In Vivo. Cancer Cell 25, 794–808 (2014). [DOI] [PubMed] [Google Scholar]
- 45.Terwijn M, et al. Leukemic stem cell frequency: a strong biomarker for clinical outcome in acute myeloid leukemia. Plos One 9, e107587 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang C, et al. Non-Lethal Ionizing Radiation Promotes Aging-Like Phenotypic Changes of Human Hematopoietic Stem and Progenitor Cells in Humanized Mice. Plos One 10, e0132041 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lu R, Czechowicz A, Seita J, Jiang D & Weissman IL Clonal level lineage commitment pathways of hematopoietic stem cells in vivo. bioRxiv, 262774 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hosono S, et al. Unbiased whole-genome amplification directly from clinical samples. Genome Res 13, 954–964 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.de Bourcy CF, et al. A quantitative comparison of single-cell whole genome amplification methods. Plos One 9, e105585 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Langmead B & Salzberg SL Fast gapped-read alignment with Bowtie 2. Nat Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McKenna A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20, 1297–1303 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cibulskis K, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol 31, 213–219 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Garrison E & Marth G Haplotype-based variant detection from short-read sequencing. arXiv preprint arXiv:1207.3907 (2012).
- 54.Cingolani P, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w(1118); iso-2; iso-3. Fly 6, 80–92 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rosenthal R, McGranahan N, Herrero J, Taylor BS & Swanton C DeconstructSigs: delineating mutational processes in single tumors distinguishes DNA repair deficiencies and patterns of carcinoma evolution. Genome Biol 17, 31 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shen R & Seshan VE FACETS: allele-specific copy number and clonal heterogeneity analysis tool for high-throughput DNA sequencing. Nucleic Acids Res 44, e131 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Popic V, et al. Fast and scalable inference of multi-sample cancer lineages. Genome Biol 16, 91 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Smith M timescape: Patient Clonal Timescapes. R package version 1.0.0(2017). [Google Scholar]
- 59.Li H Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv preprint arXiv:1303.3997 (2013).
- 60.Yu Y, Ceredig R & Seoighe C LymAnalyzer: a tool for comprehensive analysis of next generation sequencing data of T cell receptors and immunoglobulins. Nucleic Acids Res 44, e31 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.