Abstract
Background:
Prostaglandin-endoperoxide synthase 2 (PTGS2, cyclooxygenase-2, COX-2)-prostaglandin E2 (PGE2) pathway promotes tumour progression. Considering evidence suggesting increased PGE2 synthesis by BRAF mutation in tumour cells, we hypothesised that the association of tumour PTGS2 (COX-2) expression with colorectal cancer mortality might be stronger in BRAF-mutated tumours than in BRAF-wild-type tumours.
Methods:
Using 1,708 patients, including 1,200 stage I-IV colorectal carcinoma cases in the Nurses’ Health Study (NHS) and the Health Professionals Follow-up Study (HPFS) and 508 stage III colon cancer cases in a National Cancer Institute-sponsored randomised controlled trial of adjuvant therapy (CALGB/Alliance 89803), we evaluated tumour PTGS2 (COX-2) expression status using immunohistochemistry. We examined the prognostic association of PTGS2 (COX-2) expression in strata of BRAF mutation status by multivariable Cox proportional hazards regression models to adjust for potential confounders, including disease stage, tumour differentiation, microsatellite instability status, and KRAS and PIK3CA mutations.
Results:
In NHS and HPFS, the association of PTGS2 (COX-2) expression with colorectal cancer-specific survival differed by BRAF mutation status (Pinteraction = 0.0005); compared with PTGS2 (COX-2)-negative/low carcinomas, the multivariable-adjusted hazard ratios for PTGS2 (COX-2)-high carcinomas were 2.44 (95% confidence interval, 1.39–4.28) in BRAF-mutated cases and 0.82 (95% confidence interval, 0.65–1.04) in BRAF-wild-type cases. Differential prognostic associations of PTGS2 (COX-2) expression in strata of BRAF mutation status were similarly observed in CALGB/Alliance 89803 trial (Pinteraction = 0.03).
Conclusions:
The association of tumour PTGS2 (COX-2) expression with colorectal cancer mortality is stronger in BRAF-mutated tumours than in BRAF-wild-type tumours, supporting interactive roles of PTGS2 (COX-2) expression and BRAF mutation statuses in prognostication of patients with colorectal cancer; ClinicalTrials.gov Identifier, NCT00003835.
Keywords: adenocarcinoma, clinical outcome, colorectal neoplasm, immunity, immunology, inflammation, inflammatory mediator, molecular pathological epidemiology, precision medicine, prostaglandin, PTGS, RAF
1. Introduction
Prostaglandin-endoperoxide synthase 2 (PTGS2, cyclooxygenase-2, COX-2) regulates the synthesis of prostaglandin E2 (PGE2), which provokes chronic inflammation and plays important roles in the development of colorectal cancer.[1–4] Epidemiological studies have shown that regular use of the PTGS (COX) inhibitor aspirin is associated with lower colorectal cancer incidence and mortality.[5–10] Evidence has reinforced the theory that the PTGS2 (COX-2)-PGE2 pathway plays a critical role in suppression of anti-tumour immunity in the tumour microenvironment.[11–16] Our incomplete knowledge of the interactions between the immune system and cancer proves that there is a significant need for transdisciplinary integrated analyses of cancer and immunity.[17–19]
Colon and rectal cancers consist of heterogeneous diseases with tumour cells possessing varying sets of genetic and epigenetic alterations,[20] influenced by host-tumour interactions.[21] A mutation in BRAF is present in approximately 10% to 15% of colorectal cancers.[22–24] BRAF mutation in colorectal cancer is associated with high-level CpG island methylator phenotype (CIMP) which is associated with microsatellite instability (MSI).[25] Considering the association between BRAF mutation and worse clinical outcome in colorectal cancer patients,[26,27] further developments of effective treatment strategies are required for BRAF-mutated colorectal cancer patients.[23] Emerging evidence indicates that upregulation of the RAF-MAPK pathway by BRAF mutation may activate PTGS2 (COX-2) in tumour cells to increase the production of PGE2.[28,29] Therefore, we hypothesised that the association of tumour PTGS2 (COX-2) expression with colorectal cancer mortality might be stronger in BRAF-mutated tumours than in BRAF-wild-type tumours.
To test this hypothesis, we utilised molecular pathological epidemiology databases of 1,708 patients, including 1,200 stage I-IV colorectal cancer cases in two large U.S. prospective cohort studies and 508 stage III colon cancer cases in a randomised controlled trial of adjuvant therapy.
2. Methods
2.1. Study population
We utilised the database on colorectal cancer cases within two prospective cohort studies in the U.S.: the Nurses’ Health Study (NHS, 121,701 women aged 30–55 years followed since 1976) and the Health Professionals Follow-up Study (HPFS, 51,529 men aged 40–75 years followed since 1986).[6] Every two years, study participants have been sent follow-up questionnaires to collect information on lifestyle factors and medical history, including physician-confirmed diseases. The National Death Index was used to confirm deaths of study participants and to identify unreported lethal colorectal cancer cases. Participating physicians reviewed medical records to confirm diagnoses of colorectal cancer, record tumour characteristics [e.g., size, location, and the American Joint Committee on Cancer tumour, node, and metastases (TNM) classification], and record causes of deaths for participants who died. Formalin-fixed paraffin-embedded (FFPE) tissue blocks were collected from hospitals where participants diagnosed with colorectal cancer underwent tumour resection. For this analysis, we included 1,200 patients with available data on tumour PTGS2 (COX-2) expression and BRAF mutation status. We included both colon and rectal cancers based on the colorectal continuum model.[30] Patients were followed until death or the end of follow-up (January 1, 2014 for the HPFS; June 30, 2014 for the NHS), whichever came first. Written informed consent was obtained from all study participants. This study was approved by the institutional review boards at Harvard T.H. Chan School of Public Health and Brigham and Women’s Hospital (Boston, MA, USA).
As a validation set, we used 508 stage III colon cancer patients with available data on tumour PTGS2 (COX-2) expression and BRAF mutation status within Cancer and Leukemia Group B (CALGB) 89803 trial. CALGB is now part of the Alliance for Clinical Trials in Oncology. CALGB/Alliance 89803 (ClinicalTrials.gov NCT000038350) is a National Cancer Institute-sponsored adjuvant therapy trial for stage III colon cancer, comparing weekly 5-fluorouracil and leucovorin (FU/LV) and weekly irinotecan, 5-fluorouracil, and leucovorin (IFL).[26] Between April 1999 and April 2001, 1,264 patients were enrolled in the treatment trial. The details of this study have been described elsewhere.[26] Data collection and statistical analyses were conducted by the Alliance Statistics and Data Center at Duke University Medical Center and Mayo Clinic. Data quality was ensured by review of data by the Alliance Statistics and Data Center. All analyses were based on the study database frozen on November 9, 2009. Written informed consent was obtained from all patients. This study was approved by the institutional review board at each institution.
In NHS, HPFS, and CALGB/Alliance 89803 trial, a single pathologist (S.O.), who was unaware of other data, conducted a centralised review of hematoxylin and eosin-stained tissue sections of all colorectal cancer cases. Tumour differentiation was categorised as well to moderate or poor (> 50% vs. ≤ 50% glandular area, respectively).
2.2. Immunohistochemistry for PTGS2 (COX-2) expression
We constructed tissue microarrays that included up to four cores from colorectal cancer blocks and up to two cores from normal tissue blocks from the NHS and HPFS cohorts.[5] Immunohistochemistry for PTGS2 (COX-2) was performed using an anti-PTGS2 (COX-2) antibody (dilution 1:300; Cayman Chemical, Ann Arbor, MI, USA).[5] We used whole tissue sections for immunohistochemical analysis in the CALGB/Alliance 89803 set. Tumour PTGS2 (COX-2) expression level, compared with adjacent normal colonic epithelium, was evaluated by a single pathologist (S.O.) and categorised as negative/low or high. A selected sample of 124 tumours was examined by a second pathologist (T.M.); concordance between the two observers was 0.85 (κ = 0.69).[7]
2.3. Analyses of microsatellite Instability (MSI) and KRAS, BRAF, and PIK3CA mutations
DNA was extracted from archival FFPE tissue blocks using QIAamp DNA FFPE Tissue Kit (Qiagen, Hilden, Germany). MSI status[25,26] and mutation statuses for KRAS,[26,30] BRAF,[26,30] and PIK3CA[26,30] were determined.
2.4. Statistical analysis
Descriptive statistics of patient clinical features were presented according to dichotomised BRAF status for categorical variables, or mean and standard deviation for continuous variables. Our primary hypothesis testing focused on the assessment of a statistical interaction (using the Wald test on the cross-product) between tumour PTGS2 (COX-2) expression (negative/low vs. high) and BRAF mutation (mutant vs. wild-type) in the Cox proportional hazards regression model for colorectal cancer-specific survival. We also estimated the hazard ratios (HRs) for PTGS2 (COX-2)-high vs. PTGS2 (COX-2)-negative/low cases in strata of BRAF mutation status using a re-parameterization of the interaction term in a single regression model. All other analyses, including evaluation of individual HR estimates, represent secondary analyses. We used the two-sided α level of 0.005 for our primary hypothesis testing on new discovery.[31] To account for the multiple hypothesis testing in secondary analyses, we interpreted the results of our secondary analyses conservatively. All statistical analyses were performed using SAS software (version 9.4; SAS Institute, Cary, NC, USA), and all P values were two-sided. The authors had access to the study data, and had reviewed and approved the final manuscript.
In NHS and HPFS, survival time was defined as the period from colorectal cancer diagnosis to death or the end of follow-up. For analyses of colorectal cancer-specific survival, participants who died from other causes were censored at the time of death. In CALGB/Alliance 89803, the definitions of survival time were: (i) colorectal cancer-specific survival, defined as the time from study enrollment to death from the primary colon cancer; (ii) recurrence-free survival, defined as the time from study enrollment to tumour recurrence or occurrence of the new primary colon tumour; (iii) disease-free survival, defined as the time from study enrollment to tumour recurrence, occurrence of the new primary colon tumour, or death from any cause; and (iv) overall survival, defined as the time from study enrollment to death from any cause.[26] For recurrence-free survival, patients who died without known tumour recurrence were censored at the last documented evaluation by a treating provider.
In NHS and HPFS, the multivariable Cox proportional hazards regression models initially included sex, age at diagnosis (continuous), year of diagnosis (continuous), family history of colorectal cancer in any first-degree relative (present vs. absent), tumour location (proximal colon vs. distal colon vs. rectum), tumour differentiation (well/moderate vs. poor), disease stage (I-II vs. III-IV), MSI status (high vs. non-high), KRAS (mutant vs. wild-type), and PIK3CA (mutant vs. wild-type). In CALGB/Alliance 89803, the multivariable Cox model initially included sex, age at diagnosis (continuous), year of diagnosis (continuous), family history of colorectal cancer in any first-degree relative (present vs. absent), tumour location (proximal colon vs. distal colon), tumour differentiation (well/moderate vs. poor), pT stage (T1 vs. T2 vs. T3 vs. T4), pN stage (N1 vs. N2), treatment arm (FU/LV vs. IFL), Eastern Cooperative Oncology Group (ECOG) performance status (0 vs. 1–2), obstruction or perforation (present vs. absent), MSI status (high vs. non-high), KRAS (mutant vs. wild-type), and PIK3CA (mutant vs. wild-type). A backward elimination was conducted with a threshold P = 0.05 to select variables for the final models. Cases with missing data were included in the majority category of a given categorical covariate to avoid excluding patients with missing data (Supplementary Table S1). For cases with missing information on PIK3CA mutation in CALGB/Alliance 89803, since the missing percentage is higher (15%), we assigned a separate missing indicator variable. We confirmed that excluding cases with missing information in any of the covariates did not substantially alter results (data not shown). The assumption of proportional hazards was satisfied using the assessment of a time-varying covariate; i.e., the cross-product of tumour PTGS2 (COX-2) expression and survival time in strata of BRAF mutation status (P > 0.12). The Kaplan-Meier method was used to describe the distribution of colorectal cancer-specific survival, and the log-rank test was used to compare survival probabilities across PTGS2 (COX-2) expression status.
3. Results
We included 1,200 patients with colorectal cancer in NHS and HPFS (Table 1). During the median follow-up time of 15.8 years (interquartile range, 12.0 to 19.0 years) for all censored patients, there were 745 all-cause deaths, including 352 colorectal cancer-specific deaths.
Table 1.
Characteristics of colorectal cancer cases according to BRAF mutation status in the Nurses’ Health Study (NHS), Health Professionals Follow-up Study (HPFS), and CALGB/Alliance 89803 trial
| NHS |
HPFS |
CALGB/Alliance 89803
trial |
|||||||
|---|---|---|---|---|---|---|---|---|---|
|
BRAF mutation
status |
BRAF mutation
status |
BRAF mutation
status |
|||||||
| Characteristica | All cases (N = 676) | Wild-type (N = 542) | Mutant (N = 134) | All cases (N = 524) | Wild-type (N = 483) | Mutant (N = 41) | All cases (N = 508) | Wild-type (N = 433) | Mutant (N = 75) |
| Mean age ± SD (years) | 66.4 ± 8.2 | 65.8 ± 8.3 | 68.9 ± 7.3 | 70.7 ± 8.8 | 70.6 ± 8.7 | 71.3 ± 9.6 | 59.9 ± 11.5 | 58.7 ± 11.6 | 66.5 ± 8.0 |
| Sex | |||||||||
| Male | - | - | - | 524 | 483 | 41 | 276 (54%) | 247 (57%) | 29 (39%) |
| Female | 676 | 542 | 134 | - | - | - | 232 (46%) | 186 (43%) | 46 (61%) |
| Year of diagnosis | |||||||||
| 1995 or before | 249 (37%) | 211 (39%) | 38 (28%) | 212 (40%) | 198 (41%) | 14 (34%) | - | - | - |
| 1996–2000 | 240 (36%) | 191 (35%) | 49 (37%) | 162 (31%) | 147 (30%) | 15 (37%) | 345 (68%) | 290 (67%) | 55 (73%) |
| 2001–2008 | 187 (28%) | 140 (26%) | 47 (35%) | 150 (29%) | 138 (29%) | 12 (29%) | 163 (32%) | 143 (33%) | 20 (27%) |
| Family history of colorectal cancer in first-degree relative(s) | |||||||||
| Absent | 534 (79%) | 429 (80%) | 105 (78%) | 421 (80%) | 388 (80%) | 33 (80%) | 421 (84%) | 363 (85%) | 58 (77%) |
| Present | 138 (21%) | 109 (20%) | 29 (22%) | 102 (20%) | 94 (20%) | 8 (20%) | 82 (16%) | 65 (15%) | 17 (23%) |
| Tumour location | |||||||||
| Cecum | 99 (15%) | 82 (15%) | 17 (13%) | 109 (21%) | 101 (21%) | 8 (20%) | 127 (25%) | 104 (24%) | 23 (31%) |
| Ascending to transverse | 233 (35%) | 139 (26%) | 94 (71%) | 125 (24%) | 100 (21%) | 25 (61%) | 162 (32%) | 117 (27%) | 45 (61%) |
| Descending to sigmoid | 201 (30%) | 184 (34%) | 17 (13%) | 169 (32%) | 163 (34%) | 6 (15%) | 214 (43%) | 208 (48%) | 6 (8.1%) |
| Rectum | 141 (21%) | 136 (25%) | 5 (3.8%) | 119 (23%) | 117 (24%) | 2 (4.9%) | - | - | - |
| Tumour differentiation | |||||||||
| Well to moderate | 592 (88%) | 498 (92%) | 94 (70%) | 486 (93%) | 458 (95%) | 28 (68%) | 381 (76%) | 342 (79%) | 39 (53%) |
| Poor | 81 (12%) | 41 (7.6%) | 40 (30%) | 35 (6.7%) | 22 (4.6%) | 13 (32%) | 123 (24%) | 89 (21%) | 34 (47%) |
| pT stage (depth of tumour invasion) | |||||||||
| pT1 (submucosa) | 71 (11%) | 61 (12%) | 10 (7.6%) | 55 (12%) | 54 (13%) | 1 (2.7%) | 12 (2.4%) | 12 (2.8%) | 0 (0%) |
| pT2 (muscularis propria) | 112 (18%) | 97 (19%) | 15 (11%) | 113 (24%) | 106 (25%) | 7 (19%) | 45 (9.0%) | 37 (8.6%) | 8 (11%) |
| pT3 (subserosa) | 410 (64%) | 313 (62%) | 97 (73%) | 281 (60%) | 256 (60%) | 25 (68%) | 412 (82%) | 354 (83%) | 58 (78%) |
| pT4 (serosa or other organs) | 43 (6.8%) | 33 (6.6%) | 10 (7.6%) | 18 (3.9%) | 14 (3.3%) | 4 (11%) | 33 (6.6%) | 25 (5.8%) | 8 (11%) |
| pN stage (number of positive lymph nodes) | |||||||||
| pN0 (0) | 377 (62%) | 298 (61%) | 79 (63%) | 293 (65%) | 269 (65%) | 24 (67%) | - | - | - |
| pN1 (1–3) | 140 (23%) | 115 (24%) | 25 (20%) | 104 (23%) | 96 (23%) | 8 (22%) | 319 (63%) | 278 (65%) | 41 (55%) |
| pN2 (≥ 4) | 94 (15%) | 72 (15%) | 22 (17%) | 55 (12%) | 51 (12%) | 4 (11%) | 186 (37%) | 153 (35%) | 33 (45%) |
| AJCC disease stage | |||||||||
| I | 147 (23%) | 126 (25%) | 21 (16%) | 128 (28%) | 121 (29%) | 7 (18%) | - | - | - |
| II | 209 (33%) | 155 (31%) | 54 (41%) | 144 (31%) | 128 (30%) | 16 (40%) | - | - | - |
| III | 186 (29%) | 156 (31%) | 30 (23%) | 129 (28%) | 120 (29%) | 9 (23%) | 508 | 433 | 75 |
| IV | 97 (15%) | 71 (14%) | 26 (20%) | 59 (13%) | 51 (12%) | 8 (20%) | - | - | - |
| MSI status | |||||||||
| MSI-high | 126 (19%) | 52 (10%) | 74 (56%) | 58 (11%) | 36 (7.6%) | 22 (54%) | 86 (17%) | 53 (12%) | 33 (44%) |
| Non-MSI-high | 534 (81%) | 476 (90%) | 58 (44%) | 457 (89%) | 438 (92%) | 19 (46%) | 421 (83%) | 379 (88%) | 42 (56%) |
| KRAS mutation | |||||||||
| Wild-type | 424 (64%) | 296 (55%) | 128 (96%) | 285 (55%) | 246 (52%) | 39 (95%) | 322 (64%) | 248 (58%) | 74 (99%) |
| Mutant | 243 (36%) | 238 (45%) | 5 (3.8%) | 232 (45%) | 230 (48%) | 2 (4.9%) | 180 (36%) | 179 (42%) | 1 (1.3%) |
| PIK3CA mutation | |||||||||
| Wild-type | 524 (86%) | 414 (85%) | 110 (88%) | 403 (83%) | 373 (83%) | 30 (75%) | 378 (88%) | 318 (87%) | 60 (92%) |
| Mutant | 87 (14%) | 72 (15%) | 15 (12%) | 85 (17%) | 75 (17%) | 10 (25%) | 54 (13%) | 49 (13%) | 5 (7.6%) |
| PTGS2 (COX-2) expression | |||||||||
| Negative/low | 260 (38%) | 186 (34%) | 74 (55%) | 202 (39%) | 180 (37%) | 22 (54%) | 337 (66%) | 290 (67%) | 47 (63%) |
| High | 416 (62%) | 356 (66%) | 60 (45%) | 322 (61%) | 303 (63%) | 19 (46%) | 171 (34%) | 143 (33%) | 28 (37%) |
| Performance status (ECOG)b | |||||||||
| 0 | - | - | - | - | - | - | 384 (76%) | 333 (77%) | 51 (69%) |
| 1–2 | - | - | - | - | - | - | 120 (24%) | 97 (23%) | 23 (31%) |
| Treatment arm | |||||||||
| FU/LV | - | - | - | - | - | - | 266 (52%) | 233 (54%) | 33 (44%) |
| IFL | - | - | - | - | - | - | 242 (48%) | 200 (46%) | 42 (56%) |
| Clinical bowel perforation or obstruction | |||||||||
| Absent | - | - | - | - | - | - | 384 (76%) | 328 (76%) | 56 (75%) |
| Present | - | - | - | - | - | - | 124 (24%) | 105 (24%) | 19 (25%) |
Percentage indicates the proportion of patients with a specific clinical, pathologic, or molecular characteristic among all patients or in strata of BRAF mutation status within each cohort.
Definition of performance status (ECOG): 0, fully active, able to carry on all pre-disease performance without restriction; 1, restricted in physically strenuous activity but ambulatory and able to carry out work of a light or sedentary nature; 2, ambulatory and capable of all selfcare but unable to carry out any work activities (up and about more than 50% of waking hours).
Abbreviations: AJCC, American Joint Committee on Cancer; ECOG, Eastern Cooperative Oncology Group; FU/LV, 5-fluorouracil and leucovorin; HPFS, Health Professionals Follow-up Study; IFL, irinotecan, 5-fluorouracil and leucovorin; MSI, microsatellite instability; NHS, Nurses’ Health Study; SD, standard deviation.
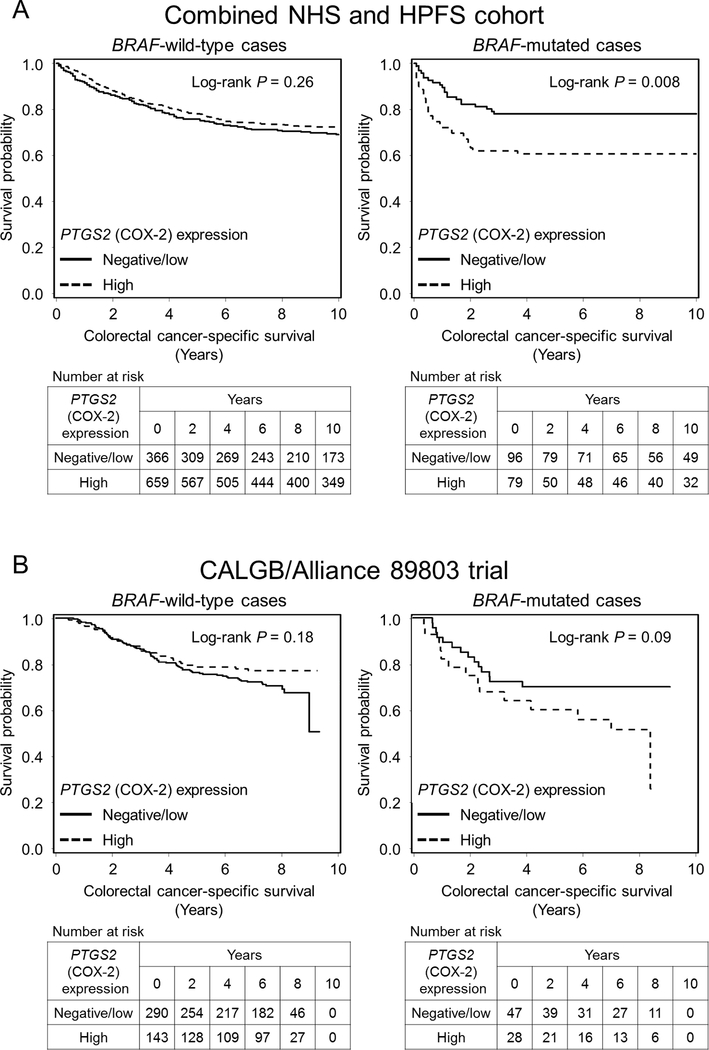
In the combined NHS and HPFS cohort, we examined the prognostic association of tumour PTGS2 (COX-2) expression status in strata of BRAF mutation status. In Kaplan-Meier survival analyses, tumour PTGS2 (COX-2) expression was associated with shorter colorectal cancer-specific survival in BRAF-mutated cases, but not in BRAF-wild-type cases (Figure 1A). In our primary hypothesis testing using Cox regression analysis, we observed a statistically significant interaction between tumour PTGS2 (COX-2) expression and BRAF mutation status in colorectal cancer-specific survival analysis (Pinteraction = 0.0005; Table 2 and Supplementary Table S2). After adjustment for potentially prognostic factors, high tumour PTGS2 (COX-2) expression was significantly associated with shorter colorectal cancer-specific survival in BRAF-mutated tumours [multivariable HR, 2.44; 95% confidence interval (CI), 1.39–4.28], but not in BRAF-wild-type tumours (multivariable HR, 0.82; 95% CI, 0.65–1.04). These interactive associations between PTGS2 (COX-2) expression and BRAF mutation status in colorectal cancer survival were observed in both the NHS and HPFS cohorts when examined separately, although statistical power was limited for cohort-specific analyses (Table 2 and Supplementary Figure S1).
Figure 1.
Kaplan-Meier analysis of colorectal cancer-specific survival according to tumour PTGS2 (COX-2) expression status in strata of BRAF mutation status. A, Combined Nurses’ Health Study (NHS) and Health Professionals Follow-up Study (HPFS) cohort; B, CALGB/Alliance 89803 trial. P values were calculated by the log-rank test (two-sided).
HPFS, Health Professionals Follow-up Study; NHS, Nurses’ Health Study.
Table 2.
Tumour PTGS2 (COX-2) expression and colorectal cancer survival according to BRAF mutation status in the Nurses’ Health Study (NHS) and Health Professionals Follow-up Study (HPFS)
| Colorectal cancer-specific
survival |
Overall survival |
||||||
|---|---|---|---|---|---|---|---|
| No. of cases | No. of events | Univariable HR (95% CI) | Multivariable HR (95% CI)a | No. of events | Univariable HR (95% CI) | Multivariable HR (95% CI)a | |
| Combined NHS and HPFS cohort | |||||||
| All cases | |||||||
| PTGS2 (COX-2) expression | |||||||
| Negative/low | 462 | 134 | 1 (referent) | 1 (referent) | 288 | 1 (referent) | 1 (referent) |
| High | 738 | 218 | 1.01 (0.81–1.25) | 0.97 (0.77–1.21) | 457 | 0.94 (0.81–1.09) | 0.98 (0.84–1.14) |
| BRAF-wild-type | |||||||
| PTGS2 (COX-2) expression | |||||||
| Negative/low | 366 | 113 | 1 (referent) | 1 (referent) | 231 | 1 (referent) | 1 (referent) |
| High | 659 | 187 | 0.89 (0.71–1.13) | 0.82 (0.65–1.04) | 401 | 0.89 (0.76–1.05) | 0.91 (0.77–1.07) |
| BRAF-mutant | |||||||
| PTGS2 (COX-2) expression | |||||||
| Negative/low | 96 | 21 | 1 (referent) | 1 (referent) | 57 | 1 (referent) | 1 (referent) |
| High | 79 | 31 | 2.16 (1.24–3.76) | 2.44 (1.39–4.28) | 56 | 1.42 (0.98–2.05) | 1.45 (1.00–2.11) |
| Pinteractionb | 0.004 | 0.0005 | 0.02 | 0.02 | |||
| NHS | |||||||
| All cases | |||||||
| PTGS2 (COX-2) expression | |||||||
| Negative/low | 260 | 79 | 1 (referent) | 1 (referent) | 157 | 1 (referent) | 1 (referent) |
| High | 416 | 130 | 1.03 (0.78–1.36) | 0.97 (0.72–1.30) | 242 | 0.93 (0.76–1.14) | 0.97 (0.79–1.19) |
| BRAF-wild-type | |||||||
| PTGS2 (COX-2) expression | |||||||
| Negative/low | 186 | 64 | 1 (referent) | 1 (referent) | 118 | 1 (referent) | 1 (referent) |
| High | 356 | 107 | 0.85 (0.63–1.16) | 0.78 (0.57–1.07) | 199 | 0.82 (0.65–1.03) | 0.84 (0.67–1.06) |
| BRAF-mutant | |||||||
| PTGS2 (COX-2) expression | |||||||
| Negative/low | 74 | 15 | 1 (referent) | 1 (referent) | 39 | 1 (referent) | 1 (referent) |
| High | 60 | 23 | 2.23 (1.16–4.28) | 2.42 (1.24–4.72) | 43 | 1.70 (1.10–2.63) | 1.55 (1.00–2.40) |
| Pinteractionb | 0.009 | 0.003 | 0.004 | 0.02 | |||
| HPFS | |||||||
| All cases | |||||||
| PTGS2 (COX-2) expression | |||||||
| Negative/low | 202 | 55 | 1 (referent) | 1 (referent) | 131 | 1 (referent) | 1 (referent) |
| High | 322 | 88 | 0.98 (0.70–1.37) | 0.96 (0.68–1.35) | 215 | 0.94 (0.76–1.17) | 1.02 (0.82–1.28) |
| BRAF-wild-type | |||||||
| PTGS2 (COX-2) expression | |||||||
| Negative/low | 180 | 49 | 1 (referent) | 1 (referent) | 113 | 1 (referent) | 1 (referent) |
| High | 303 | 80 | 0.94 (0.66–1.34) | 0.88 (0.61–1.26) | 202 | 0.98 (0.78–1.23) | 1.00 (0.80–1.27) |
| BRAF-mutant | |||||||
| PTGS2 (COX-2) expression | |||||||
| Negative/low | 22 | 6 | 1 (referent) | 1 (referent) | 18 | 1 (referent) | 1 (referent) |
| High | 19 | 8 | 1.92 (0.66–5.54) | 2.20 (0.71–6.81) | 13 | 0.77 (0.38–1.58) | 1.22 (0.58–2.57) |
| Pinteractionb | 0.21 | 0.13 | 0.54 | 0.62 | |||
The initial multivariable Cox regression model initially included sex, age, year of diagnosis, family history of colorectal cancer, tumour location, tumour differentiation, disease stage, microsatellite instability, and KRAS, BRAF (except for BRAF-stratified analyses), and PIK3CA mutations. A backward elimination with a threshold P of 0.05 was used to select variables for the final models. The variables which remained in the final models are shown in Supplementary Table S2.
Pinteraction (two-sided) was calculated by the Wald test on the cross-product term of PTGS2 (COX-2) expression (negative/low vs. high) and BRAF mutation (wild-type vs. mutant) in the Cox regression model.
Abbreviations: CI, confidence interval; HR, hazard ratio; HPFS, Health Professionals Follow-up Study; NHS, Nurses’ Health Study.
In analyses limited to patients with stage I-III colorectal cancer, a similar differential prognostic association of tumour PTGS2 (COX-2) expression by BRAF mutation status was observed, although statistical power was limited (Supplementary Table S3).
We validated our findings using an independent cohort of 508 patients with stage III colon cancer in CALGB/Alliance 89803 (Table 1). The median age was 59.9 years, 46% were women, and 76% were performance status (ECOG) 0. During the median follow-up time of 7.6 years (interquartile range, 7.1 to 8.0 years) for all censored patients, there were 159 all-cause deaths, including 140 colon cancer-specific deaths. The multivariable HR for colorectal cancer-specific survival for PTGS2 (COX-2)-high cases compared to PTGS2 (COX-2)-negative/low cases was higher in the BRAF-mutated group (multivariable HR, 1.85; 95% CI, 0.88–3.88) than in the BRAF-wild-type group (multivariable HR, 0.74; 95% CI, 0.49–1.12; Pinteraction = 0.03; Table 3 and Supplementary Table S4). Similar differential survival association were observed for recurrence-free survival (Pinteraction = 0.005; Figure 1B) and disease-free survival (Pinteraction = 0.006).
Table 3.
Tumour PTGS2 (COX-2) expression and colorectal cancer survival according to BRAF mutation status in CALGB/Alliance 89803 trial
| Colorectal cancer-specific
survival |
Recurrence-free
survival |
Disease-free survival |
Overall survival |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. of cases | No. of events | HR (95% CI) |
No. of events | HR (95% CI) |
No. of events | HR (95% CI) |
No. of events | HR (95% CI) |
|||||
| Univariable | Multivariablea | Univariable | Multivariablea | Univariable | Multivariablea | Univariable | Multivariablea | ||||||
| All cases | |||||||||||||
| PTGS2 (COX-2) expression | |||||||||||||
| Negative/low | 337 | 95 | 1 (referent) | 1 (referent) | 128 | 1 (referent) | 1 (referent) | 141 | 1 (referent) | 1 (referent) | 108 | 1 (referent) | 1 (referent) |
| High | 171 | 45 | 0.93 (0.65–1.33) | 0.88 (0.61–1.25) | 54 | 0.82 (0.60–1.13) | 0.78 (0.57–1.07) | 60 | 0.82 (0.61–1.11) | 0.79 (0.58–1.07) | 51 | 0.93 (0.67–1.30) | 0.87 (0.62–1.22) |
| BRAF-wild-type | |||||||||||||
| PTGS2 (COX-2) expression | |||||||||||||
| Negative/low | 290 | 81 | 1 (referent) | 1 (referent) | 113 | 1 (referent) | 1 (referent) | 123 | 1 (referent) | 1 (referent) | 91 | 1 (referent) | 1 (referent) |
| High | 143 | 31 | 0.76 (0.50–1.14) | 0.74 (0.49–1.12) | 39 | 0.66 (0.46–0.95) | 0.64 (0.44–0.92) | 44 | 0.68 (0.48–0.96) | 0.67 (0.48–0.95) | 36 | 0.78 (0.53–1.15) | 0.79 (0.53–1.16) |
| BRAF-mutant | |||||||||||||
| PTGS2 (COX-2) expression | |||||||||||||
| Negative/low | 47 | 14 | 1 (referent) | 1 (referent) | 15 | 1 (referent) | 1 (referent) | 18 | 1 (referent) | 1 (referent) | 17 | 1 (referent) | 1 (referent) |
| High | 28 | 14 | 1.93 (0.92–4.04) | 1.85 (0.88–3.88) | 15 | 2.11 (1.03–4.32) | 2.04 (0.98–4.21) | 16 | 1.90 (0.97–3.72) | 1.96 (0.99–3.87) | 15 | 1.71 (0.85–3.42) | 1.43 (0.71–2.89) |
| Pinteractionb | 0.03 | 0.03 | 0.005 | 0.005 | 0.008 | 0.006 | 0.05 | 0.15 | |||||
The initial multivariable Cox regression model initially included sex, age, year of diagnosis, family history of colorectal cancer, tumour location, tumour differentiation, pT stage, pN stage, treatment arm, performance status, perforation or obstruction, microsatellite instability, and KRAS, BRAF (except for BRAF-stratified analyses), and PIK3CA mutations. A backward elimination with a threshold P of 0.05 was used to select variables for the final models. The variables which remained in the final models are shown in Supplementary Table S4.
Pinteraction (two-sided) was calculated by the Wald test on the cross-product term of PTGS2 (COX-2) expression (negative/low vs. high) and BRAF mutation (wild-type vs. mutant) in the Cox regression model.
Abbreviations: CI, confidence interval; HR, hazard ratio.
4. Discussion
To test our hypothesis that the association of tumour PTGS2 (COX-2) expression with colorectal cancer mortality might be stronger in BRAF-mutated tumours than in BRAF-wild-type tumours, we conducted this study utilising the two U.S. prospective cohort studies and the randomised controlled trial. We observed a differential prognostic association of tumour PTGS2 (COX-2) expression in strata of BRAF mutation status.
The PTGS2 (COX-2)-PGE2 pathway plays key roles in tumour progression in a variety of tumour types, including colorectal cancer.[1,2] Evidence indicates that PGE2 overproduction may enable tumour cells to evade host immune surveillance mechanisms through accumulation of myeloid-derived suppressor cells, suppression of dendritic cells, and evasion of the T cell-mediated anti-tumour immune response.[12–14,32] Considering that the immunomodulatory effect by PGE2 inhibition can synergise with immune checkpoint blockade therapies targeting PDCD1 (programmed cell death 1, PD-1) or CD274 (PDCD1 ligand 1, PD-L1) in various cancer types,[12,33,34] a better understanding of the roles of tumour PTGS2 (COX-2) expression in the context of tumour-immune interactions would have considerable clinical implications.[35]
Gain-of-function BRAF mutation leads to accelerated production and activity of a number of critical cellular substrates involved in cell proliferation and survival through phosphorylation of the MAPK kinases.[23,24] Studies indicate that BRAF mutation has been associated with high-level CIMP and worse clinical outcomes in colorectal cancer.[22–27,36,37] Evidence suggests that BRAF mutation may increase microRNA MIR21 (miR-21) expression level through the activation of the MAPK and STAT3 signalling pathways.[28,29,38] Given that MIR21 increases local levels of PGE2 by suppressing PGE2 degradation,[29,38,39] the prognostic association of PTGS2 (COX-2) expression might be especially pronounced in BRAF-mutated colorectal cancer. Overall, increased synthesis of PGE2 resulting from BRAF mutation and heightened PTGS2 (COX-2) activity may serve as one possible pathway through which the survival of colorectal cancer patients with this combination is affected.
We acknowledge limitations in our study. Data on cancer recurrence were unavailable in NHS and HPFS. However, colorectal cancer-specific survival can be considered a reasonable cancer-specific outcome in a population-based study with long-term follow-up, because median survival for recurrent (metastatic) colorectal cancer was approximately 10 to 20 months during the time period of this study. Moreover, we found the association of tumour PTGS2 (COX-2) expression with recurrence-free survival and disease-free survival stratified by BRAF mutation status remained consistent in the validation set of CALGB/Alliance 89803. Data on cancer treatment were also limited in the NHS and HPFS cohorts. However, the decision to undergo chemotherapy and the specific regimen utilised would be unlikely to differ substantially according to tumour PTGS2 (COX-2) expression in resected specimens, as these data were not available to treating physicians. We also recognise another limitation that the current study is an observational cohort study, not an intervention trial such as a randomized controlled trial using aspirin and/or BRAF inhibitor. Therefore, we cannot conclude that inhibiting PTGS2 (COX-2) in BRAF-mutated colorectal cancer is an effective therapeutic strategy. In the current study, we certainly observed that the association of tumour PTGS2 (COX-2) expression with colorectal cancer mortality is stronger in BRAF-mutated tumours than in BRAF-wild-type tumours, and further research is warranted to investigate the therapeutic roles of BRAF and PTGS2 (COX-2) inhibitors in patients with this malignancy.
A major strength of this study is utilisation of a molecular pathological epidemiology database of rectal and colon cancer cases from the two large U.S. prospective cohort studies,[40] which integrates clinicopathologic features, long-term survival data, and tumour molecular features. This population-based colorectal cancer database enabled us to rigorously examine the interactive prognostic association of tumour PTGS2 (COX-2) expression and BRAF mutation status while controlling for potential confounders. Use of the randomised controlled trial as a validation set was another significant strength of this study. The colorectal cancer patient data in our study were derived from a large number of hospitals from diverse locations within the U.S., which adds greatly to the generalisability of our findings.
In conclusion, we found a stronger association of tumour PTGS2 (COX-2) expression with colorectal cancer mortality in BRAF-mutated tumours than in BRAF-wild-type tumours. Our population-based data suggest the potential of tumour PTGS2 (COX-2) expression status as a prognostic biomarker in patients with BRAF-mutated colorectal cancer.
Supplementary Material
Acknowledgments:
We would like to thank the participants and staff of the Nurses’ Health Study and the Health Professionals Follow-up Study for their valuable contributions as well as the following state cancer registries for their help: AL, AZ, AR, CA, CO, CT, DE, FL, GA, ID, IL, IN, IA, KY, LA, ME, MD, MA, MI, NE, NH, NJ, NY, NC, ND, OH, OK, OR, PA, RI, SC, TN, TX, VA, WA, WY.
We would like to thank the CALGB/Alliance Pathology Coordinating Office at the Ohio State University for banking and preparing the materials for the study.
The authors assume full responsibility for analyses and interpretation of these data.
Funding: Research reported in this publication was supported by U.S. National Cancer Institute of the National Institutes of Health (NIH) under Award Numbers U10CA180821, U10CA180882, and U24CA196171 (to the Alliance for Clinical Trials in Oncology), and U10CA180791, U10CA180867, P01 CA87969 (to M.J. Stampfer), UM1 CA186107 (to M.J. Stampfer), P01 CA55075 (to W.C. Willett), UM1 CA167552 (to W.C. Willett), U01 CA167552 (to L.A. Mucci, W.C. Willett), P50 CA127003 (to C.S.F.), R01 CA118553 (to C.S.F.), R01 CA169141 (to C.S.F.), R01 CA137178 (to A.T.C.), K24 DK098311 (to A.T.C.), R01 CA205406 (to K.N.), R35 CA197735 (to S.O.), R01 CA151993 (to S.O.), K07 CA190673 (to R.N.), and K07 CA188126 (to X.Z.). This work was also supported by Nodal Award (2016-02) from the Dana-Farber Harvard Cancer Center (to S.O.); by grants from the Project P Fund, The Friends of the Dana-Farber Cancer Institute, Bennett Family Fund, the Entertainment Industry Foundation through National Colorectal Cancer Research Alliance, and by the Stand Up to Cancer Colorectal Cancer Dream Team Translational Research Grant (SU2C-AACR-DT22-17 to C.S.F. and M.G.), administered by the American Association for Cancer Research, a scientific partner of SU2C. CALGB/Alliance 89803 was supported in part by Pfizer. K.K. was supported by grants from Overseas Research Fellowship from Japan Society for the Promotion of Science (JP2017-775). T.H. was supported by a fellowship grant from the Mitsukoshi Health and Welfare Foundation. L.L. was supported by a scholarship grant from Chinese Scholarship Council and a fellowship grant from Huazhong University of Science and Technology. A.T.C. is a Stuart and Suzanne Steele MGH Research Scholar. The content is solely the responsibility of the authors and does not necessarily represent the official views of NIH. The funding sources had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, and approval of the manuscript; and decision to submit the manuscript for publication.
Abbreviations:
- CALGB
Cancer and Leukemia Group B (now part of Alliance for Clinical Trials in Oncology)
- CI
confidence interval
- CIMP
CpG island methylator phenotype
- ECOG
Eastern Cooperative Oncology Group
- FFPE
formalin-fixed paraffin-embedded
- FU/LV
5-fluorouracil and leucovorin
- HPFS
Health Professionals Follow-up Study
- HR
hazard ratio
- IFL
irinotecan, 5-fluorouracil, and leucovorin
- MSI
microsatellite instability
- NHS
Nurses’ Health Study
- PGE2
prostaglandin E2
Footnotes
Conflict of interest statement
A.T.C. previously served as a consultant for Bayer Healthcare, and Pfizer Inc. for areas unrelated to this research. K.N. has participated in an advisory board for Bayer. No other conflict of interest exists. The other authors declare that they have no conflicts of interest.
Use of standardised official symbols: We use HUGO (Human Genome Organisation)-approved official symbols (or root symbols) for genes, gene products, and gene families, including BRAF, CD274, KRAS, MAPK, MIR21, NFKB, PDCD1, PIK3CA, PTGS2, RAF, STAT3, and WNT; all of which are described at www.genenames.org. The official symbols are italicised to differentiate from non-italicised colloquial names that are used along with the official symbols. This format enables readers to familiarise the official symbols for genes and gene products together with common colloquial names.
ClinicalTrials.gov Identifier: NCT00003835
References
- 1.Wang D, Fu L, Sun H, Guo L, DuBois RN. Prostaglandin E2 Promotes Colorectal Cancer Stem Cell Expansion and Metastasis in Mice. Gastroenterology 2015;149:1884–1895 e4. doi: 10.1053/j.gastro.2015.07.064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu Y, Sun H, Hu M, Zhang Y, Chen S, Tighe S, et al. The Role of Cyclooxygenase-2 in Colorectal Carcinogenesis. Clin Colorectal Cancer 2017;16:165–172. doi: 10.1016/j.clcc.2016.09.012 [DOI] [PubMed] [Google Scholar]
- 3.Benelli R, Vene R, Ferrari N. Prostaglandin-endoperoxide synthase 2 (cyclooxygenase-2), a complex target for colorectal cancer prevention and therapy. Transl Res 2018;196:42–61. doi: 10.1016/j.trsl.2018.01.003 [DOI] [PubMed] [Google Scholar]
- 4.Morgillo F, Dallio M, Della Corte CM, Gravina AG, Viscardi G, Loguercio C, et al. Carcinogenesis as a Result of Multiple Inflammatory and Oxidative Hits: a Comprehensive Review from Tumor Microenvironment to Gut Microbiota. Neoplasia 2018;20:721–733. doi: 10.1016/j.neo.2018.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chan AT, Ogino S, Fuchs CS. Aspirin and the risk of colorectal cancer in relation to the expression of COX-2. N Engl J Med 2007;356:2131–42. doi: 10.1056/NEJMoa067208 [DOI] [PubMed] [Google Scholar]
- 6.Liao X, Lochhead P, Nishihara R, Morikawa T, Kuchiba A, Yamauchi M, et al. Aspirin use, tumor PIK3CA mutation, and colorectal-cancer survival. N Engl J Med 2012;367:1596–606. doi: 10.1056/NEJMoa1207756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nishihara R, Lochhead P, Kuchiba A, Jung S, Yamauchi M, Liao X, et al. Aspirin use and risk of colorectal cancer according to BRAF mutation status. JAMA 2013;309:2563–71. doi: 10.1001/jama.2013.6599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hua X, Phipps AI, Burnett-Hartman AN, Adams SV, Hardikar S, Cohen SA, et al. Timing of Aspirin and Other Nonsteroidal Anti-Inflammatory Drug Use Among Patients With Colorectal Cancer in Relation to Tumor Markers and Survival . J Clin Oncol 2017;35:2806–2813. doi: 10.1200/JCO.2017.72.3569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frouws MA, Reimers MS, Swets M, Bastiaannet E, Prinse B, van Eijk R, et al. The Influence of BRAF and KRAS Mutation Status on the Association between Aspirin Use and Survival after Colon Cancer Diagnosis. PLoS One 2017;12:e0170775. doi: 10.1371/journal.pone.0170775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gray RT, Cantwell MM, Coleman HG, Loughrey MB, Bankhead P, McQuaid S, et al. Evaluation of PTGS2 Expression, PIK3CA Mutation, Aspirin Use and Colon Cancer Survival in a Population-Based Cohort Study. Clin Transl Gastroenterol 2017;8:e91. doi: 10.1038/ctg.2017.18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Prima V, Kaliberova LN, Kaliberov S, Curiel DT, Kusmartsev S. COX2/mPGES1/PGE2 pathway regulates PD-L1 expression in tumor-associated macrophages and myeloid-derived suppressor cells. Proc Natl Acad Sci U S A 2017;114:1117–1122. doi: 10.1073/pnas.1612920114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zelenay S, van der Veen AG, Bottcher JP, Snelgrove KJ, Rogers N, Acton SE, et al. Cyclooxygenase-Dependent Tumor Growth through Evasion of Immunity. Cell 2015;162:1257–70. doi: 10.1016/j.cell.2015.08.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Q, Zhu B, Li Y. Resolution of Cancer-Promoting Inflammation: A New Approach for Anticancer Therapy. Front Immunol 2017;8:71. doi: 10.3389/fimmu.2017.00071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gobel C, Breitenbuecher F, Kalkavan H, Hahnel PS, Kasper S, Hoffarth S, et al. Functional expression cloning identifies COX-2 as a suppressor of antigen-specific cancer immunity. Cell Death Dis 2014;5:e1568. doi: 10.1038/cddis.2014.531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zelenay S, Reis ESC. Reducing prostaglandin E2 production to raise cancer immunogenicity. Oncoimmunology 2016;5:e1123370. doi: 10.1080/2162402X.2015.1123370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Renner K, Singer K, Koehl GE, Geissler EK, Peter K, Siska PJ, et al. Metabolic Hallmarks of Tumor and Immune Cells in the Tumor Microenvironment. Front Immunol 2017;8:248. doi: 10.3389/fimmu.2017.00248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ogino S, Nowak JA, Hamada T, Phipps AI, Peters U, Milner DA Jr., et al. Integrative analysis of exogenous, endogenous, tumour and immune factors for precision medicine. Gut 2018;67:1168–1180. doi: 10.1136/gutjnl-2017-315537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ogino S, Giannakis M. Immunoscore for (colorectal) cancer precision medicine. Lancet 2018;391:2084–2086. doi: 10.1016/S0140-6736(18)30953-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pages F, Mlecnik B, Marliot F, Bindea G, Ou FS, Bifulco C, et al. International validation of the consensus Immunoscore for the classification of colon cancer: a prognostic and accuracy study. Lancet 2018;391:2128–2139. doi: 10.1016/S0140-6736(18)30789-X [DOI] [PubMed] [Google Scholar]
- 20.Strickler JH, Loree JM, Ahronian LG, Parikh AR, Niedzwiecki D, Pereira AAL, et al. Genomic Landscape of Cell-Free DNA in Patients with Colorectal Cancer. Cancer Discov 2018;8:164–173. doi: 10.1158/2159-8290.CD-17-1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dienstmann R, Vermeulen L, Guinney J, Kopetz S, Tejpar S, Tabernero J. Consensus molecular subtypes and the evolution of precision medicine in colorectal cancer. Nat Rev Cancer 2017;17:79–92. doi: 10.1038/nrc.2016.126 [DOI] [PubMed] [Google Scholar]
- 22.Van Cutsem E, Cervantes A, Adam R, Sobrero A, Van Krieken JH, Aderka D, et al. ESMO consensus guidelines for the management of patients with metastatic colorectal cancer. Ann Oncol 2016;27:1386–422. doi: 10.1093/annonc/mdw235 [DOI] [PubMed] [Google Scholar]
- 23.Seligmann JF, Fisher D, Smith CG, Richman SD, Elliott F, Brown S, et al. Investigating the poor outcomes of BRAF-mutant advanced colorectal cancer: analysis from 2530 patients in randomised clinical trials. Ann Oncol 2017;28:562–568. doi: 10.1093/annonc/mdw645 [DOI] [PubMed] [Google Scholar]
- 24.Sanz-Garcia E, Argiles G, Elez E, Tabernero J. BRAF mutant colorectal cancer: prognosis, treatment, and new perspectives. Ann Oncol 2017;28:2648–2657. doi: 10.1093/annonc/mdx401 [DOI] [PubMed] [Google Scholar]
- 25.Ogino S, Nosho K, Kirkner GJ, Kawasaki T, Meyerhardt JA, Loda M, et al. CpG island methylator phenotype, microsatellite instability, BRAF mutation and clinical outcome in colon cancer. Gut 2009;58:90–6. doi: 10.1136/gut.2008.155473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ogino S, Shima K, Meyerhardt JA, McCleary NJ, Ng K, Hollis D, et al. Predictive and prognostic roles of BRAF mutation in stage III colon cancer: results from intergroup trial CALGB 89803. Clin Cancer Res 2012;18:890–900. doi: 10.1158/1078-0432.CCR-11-2246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goey KKH, Sorbye H, Glimelius B, Adams RA, Andre T, Arnold D, et al. Consensus statement on essential patient characteristics in systemic treatment trials for metastatic colorectal cancer: Supported by the ARCAD Group. Eur J Cancer 2018;100:35–45. doi: 10.1016/j.ejca.2018.05.010 [DOI] [PubMed] [Google Scholar]
- 28.Becker TM, Boyd SC, Mijatov B, Gowrishankar K, Snoyman S, Pupo GM, et al. Mutant B-RAF-Mcl-1 survival signaling depends on the STAT3 transcription factor. Oncogene 2014;33:1158–66. doi: 10.1038/onc.2013.45 [DOI] [PubMed] [Google Scholar]
- 29.Yu H, Lee H, Herrmann A, Buettner R, Jove R. Revisiting STAT3 signalling in cancer: new and unexpected biological functions. Nat Rev Cancer 2014;14:736–46. doi: 10.1038/nrc3818 [DOI] [PubMed] [Google Scholar]
- 30.Yamauchi M, Morikawa T, Kuchiba A, Imamura Y, Qian ZR, Nishihara R, et al. Assessment of colorectal cancer molecular features along bowel subsites challenges the conception of distinct dichotomy of proximal versus distal colorectum. Gut 2012;61:847–54. doi: 10.1136/gutjnl-2011-300865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Benjamin DJ, Berger JO, Johannesson M, Nosek BA, Wagenmakers EJ, Berk R, et al. Redefine statistical significance. Nature Human Behaviour 2018;2:6–10. doi: 10.1038/s41562-017-0189-z [DOI] [PubMed] [Google Scholar]
- 32.Wu AA, Drake V, Huang HS, Chiu S, Zheng L. Reprogramming the tumor microenvironment: tumor-induced immunosuppressive factors paralyze T cells. Oncoimmunology 2015;4:e1016700. doi: 10.1080/2162402X.2015.1016700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang D, DuBois RN. The Role of Prostaglandin E(2) in Tumor-Associated Immunosuppression. Trends Mol Med 2016;22:1–3. doi: 10.1016/j.molmed.2015.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hamada T, Cao Y, Qian ZR, Masugi Y, Nowak JA, Yang J, et al. Aspirin Use and Colorectal Cancer Survival According to Tumor CD274 (Programmed Cell Death 1 Ligand 1) Expression Status. J Clin Oncol 2017;35:1836–1844. doi: 10.1200/JCO.2016.70.7547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Emile JF, Julie C, Le Malicot K, Lepage C, Tabernero J, Mini E, et al. Prospective validation of a lymphocyte infiltration prognostic test in stage III colon cancer patients treated with adjuvant FOLFOX. Eur J Cancer 2017;82:16–24. doi: 10.1016/j.ejca.2017.04.025 [DOI] [PubMed] [Google Scholar]
- 36.Blaker H, Alwers E, Arnold A, Herpel E, Tagscherer KE, Roth W, et al. The Association Between Mutations in BRAF and Colorectal Cancer-Specific Survival Depends on Microsatellite Status and Tumor Stage. Clin Gastroenterol Hepatol 2018. doi: 10.1016/j.cgh.2018.04.015 [DOI] [PubMed] [Google Scholar]
- 37.Taieb J, Le Malicot K, Shi Q, Penault-Llorca F, Bouche O, Tabernero J, et al. Prognostic Value of BRAF and KRAS Mutations in MSI and MSS Stage III Colon Cancer. J Natl Cancer Inst 2017;109. doi: 10.1093/jnci/djw272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Iliopoulos D, Jaeger SA, Hirsch HA, Bulyk ML, Struhl K. STAT3 activation of miR-21 and miR-181b-1 via PTEN and CYLD are part of the epigenetic switch linking inflammation to cancer. Mol Cell 2010;39:493–506. doi: 10.1016/j.molcel.2010.07.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shi C, Yang Y, Xia Y, Okugawa Y, Yang J, Liang Y, et al. Novel evidence for an oncogenic role of microRNA-21 in colitis-associated colorectal cancer. Gut 2016;65:1470–81. doi: 10.1136/gutjnl-2014-308455 [DOI] [PubMed] [Google Scholar]
- 40.Hamada T, Liu L, Nowak JA, Mima K, Cao Y, Ng K, et al. Vitamin D status after colorectal cancer diagnosis and patient survival according to immune response to tumour. Eur J Cancer 2018;103:98–107. doi: 10.1016/j.ejca.2018.07.130 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.