Abstract
Haploinsufficiency occurs when loss of one copy of a diploid gene (hemizygosity) causes a phenotype. It is relatively rare, in that most genes can produce sufficient mRNA and protein from a single copy to prevent any loss of normal activity and function. Reproduction is a complex process relying on migration of GnRH neurons from the olfactory placode to the hypothalamus during development. We have studied 3 different homeodomain genes Otx2, Vax1, and Six3 and found that the deletion of one allele for any of these genes in mice produces subfertility or infertility in one or both sexes, despite the presence of one intact allele. All 3 heterozygous mice have reduced numbers of GnRH neurons, but the mechanisms of subfertility differ significantly. This review compares the subfertility phenotypes and their mechanisms.
Keywords: Heterozygous, Homeodomain, Infertility
Introduction
Normosmic idiopathic hypogonadotropic hypogonadism (IHH) and its anosmic counterpart, Kallmann syndrome, are 2 rare genetic disorders leading to various degrees of subfertility, including complete infertility and absent puberty [1]. This subfertility is often due to a reduction in the number of GnRH expressing neurons or impairment of the rhythmic release of GnRH. Over the last decade, numerous genes have been identified as responsible for these 2 conditions; however, more than 50% of IHH cases still have unknown origins [2, 3]. Genetic mutations known to cause IHH are frequently either autosomal recessive or dominant. Of note, it is becoming increasingly clear that a number of unidentified genetic causes of IHH are the result of compound heterozygosity [3]. Compound heterozygosity is particularly hard to detect, as it requires identification of mutations in 2 distinct genes. Despite the difficulty in detecting polygenic IHH, haploinsufficiencies adversely affecting fertility have been reported in both rodents and humans [3–8].
In all mammals, including humans, GnRH neurons have a unique cellular origin in the nasal placode. In mice, GnRH neurons are first detected on embryonic day 11 (e11) in the olfactory placode, concurrent with the onset of GnRH expression (Fig. 1a). They then migrate from the vomeronasal organ (VNO) across the nasal septum into the developing basal forebrain, following the terminal nerve and the olfactory nerve (Fig. 1a) [9–12]. The terminal nerve (cranial nerve zero or nervus terminalis) extends from the nasal submucosa, is located medially and in very close proximity to the olfactory tract, and projects to important limbic structures (e.g., amygdala, hypothalamic nuclei) [13, 14]. The olfactory nerve (cranial nerve I) contains axons from the olfactory neuroepithelium, travels up through the cribriform plate, and extends into the brain to innervate the olfactory bulb [14]. At e14, a population of ~800 GnRH neurons [15] migrates, as a continuum, from the developing vomeronasal epithelium to the preoptic/hypothalamic area. As the neurons migrate and mature, they increase their expression of GnRH [16, 17]. In adulthood, the same number of GnRH cell bodies is found scattered in the preoptic area, among the fibers of the diagonal band of Broca, and in the medial septum. GnRH fibers extend their axons not only to the medial eminence, but also throughout the hypothalamus and midbrain [10, 18]. Defects in this migration causing abnormal GnRH neuron location in the brain and incomplete maturation can result in infertility and IHH [19, 20].
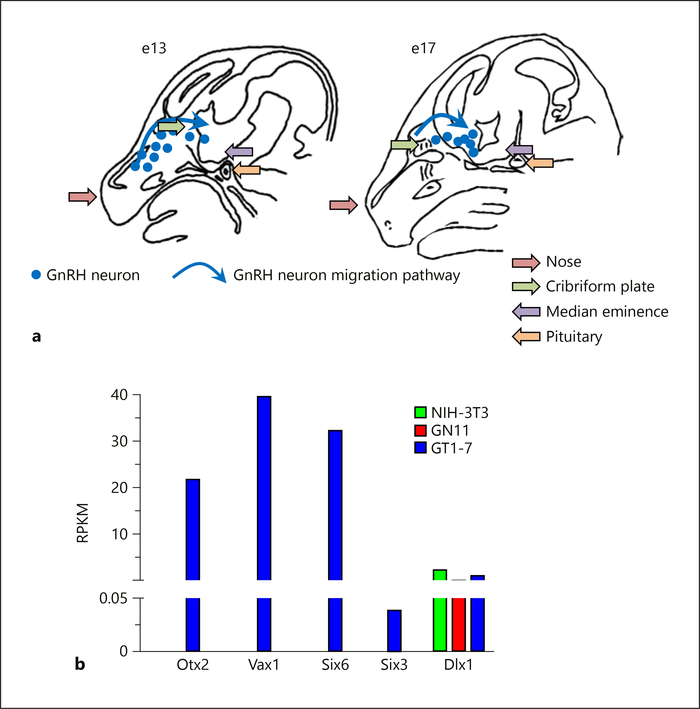
Fig. 1.
Developmental GnRH neuron migration and homeodomain gene expression. GnRH neuron maturation depends on internal and external factors to the GnRH neurons, allowing developmental migration and maturation. Correct GnRH neuron migration and increased Gnrh1 expression is required for fertility. a Schematic of a sagittal mouse head illustrating developmental GnRH neuron migration. GnRH neurons arise in the olfactory placode around embryonic day 11 (e11) in the mouse. From there they migrate through the cribriform plate into the brain. Once within the brain, GnRH neurons follow a more ventral trajectory to finally localize scattered throughout the anterior hypothalamus. On completion of their migration, GnRH neurons send projections to the median eminence, where GnRH is released in a pulsatile fashion into the hypophyseal portal system. GnRH neurons require ~4 days to complete this migration. Blue dots illustrate the location of GnRH neurons at e13 and e17, and the blue arrows indicate the migration path. b During GnRH neuron development, a complex gene expression pattern is required for increased Gnrh1 expression in parallel with expression of receptors and other factors allowing GnRH neuron pathfinding. We used RNA-Seq comparison of homeodomain transcription factor gene expression in immortalized mature non-migratory GnRH neurons (GT1–7), immature migratory GnRH neurons (GN11), and fibroblasts (NIH-3T3) to identify novel transcription factors potentially involved in GnRH neuron maturation. RNA-Seq data are shown as the average of 2 biological replicates, and expressed as RPKM (reads per kilobase million). These data support quantitative RTPCR analyses previously published for these genes [23–26].
We have identified 3 homeodomain genes, which when heterozygous in mice, impair fertility. These genes first came to our attention as they were highly expressed in the mature immortalized mouse GnRH neuronal cell line, GT1–7 [21] as compared to the immature GnRH neuronal cell line, GN11 [22], and NIH-3T3 fibroblasts (Fig. 1b). Using RNA-seq comparing transcript levels in GT1–7, GN11, and NIH-3T3 cells to screen for candidates that may act in GnRH neuron maturation, we found a set of homeodomain transcription factors strongly expressed in GT1–7 cells (Fig. 1b). We have focused on Otx2, Vax1, Six3, and Six6. All of these homeodomain transcription factors directly bind to and regulate the Gnrh1 gene at conserved ATTA sites in the proximal promoter and/or the conserved enhancer [23–28]. Although Otx2, Vax1, and Six3 homozygous deletions (null mice) are all neonatal or perinatal lethal, the heterozygous (Het) mice are viable, overall healthy, and born in Mendelian ratios. This review will address the contrasting mechanisms and sex-specificity of the subfertility of the mice with heterozygous mutations in the Otx2, Vax1, and Six3 genes.
Subfertility in Otx2 Heterozygous Mice
Otx2, the vertebrate homologue of Drosophila orthodenticle, is a transcription factor that has been shown to be critical for normal brain and eye development [29–32]. During embryogenesis, Otx2 is expressed in both the developing GnRH neurons [33] and presumptive pituitary at e12.5 [30] indicating its role in development of the HPG axis. This hypothesis is supported by the identification of several heterozygous OTX2 loss-of-function mutations in patients with combined pituitary hormone deficiency [34–36]. Several germline and conditional knockout mice have been generated; they have emphasized a role for Otx2 in head formation, postnatal survival, and growth [29, 37–39]. However, as Otx2-null mice are embryonic lethal due to a failure to develop the forebrain, midbrain, and anterior hindbrain, analysis of the development and maintenance of the HPG axis in these mice has not been possible.
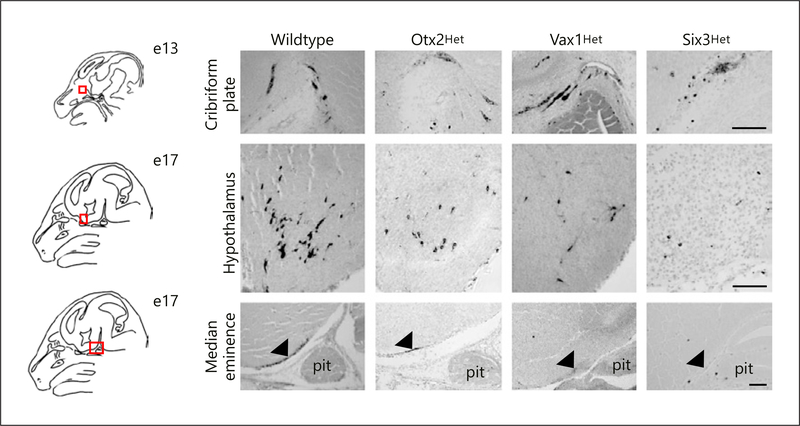
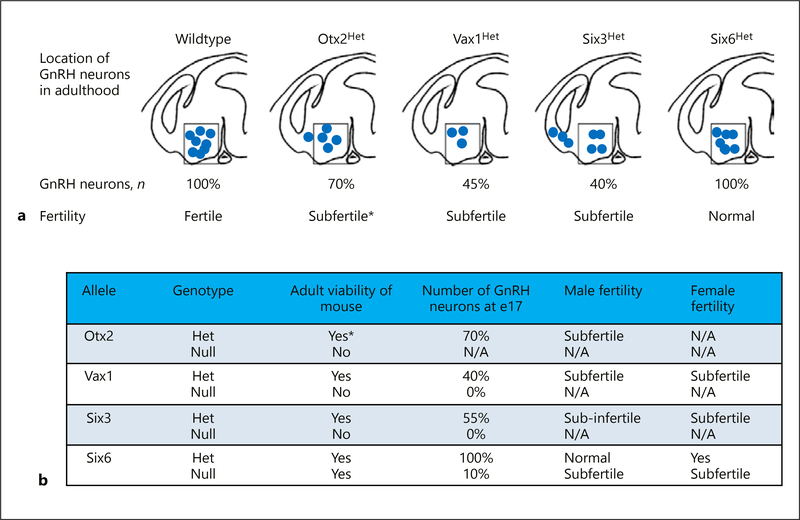
We studied the role of Otx2 in GnRH neurons in heterozygotes Otx2 (Otx2Het) mice [18, 37] to investigate Otx2 in GnRH neuron differentiation and migration in vivo. Male Otx2Het mice [5] exhibit a progressive loss of fertility. To determine the origin of subfertility, we investigated the number and location of GnRH neurons during development in Otx2Het mice. At e13.5, Otx2Het embryos of both sexes have a > 50% reduction in the total number of GnRH neurons in the head (nasal area, cribriform plate and brain combined; Fig. 2). Furthermore, by e17.5, when the majority of GnRH neurons are normally in the hypothalamus (Fig. 2; Wildtype), in the Otx2Het mouse, GnRH neurons are still present in the nose and crossing the cribriform plate to a greater extent than controls. At e17.5, mutant males have a ~30% reduction in total GnRH neurons (Fig. 3). These data show that Otx2 is important for development and progression of migration of GnRH neurons, and for GnRH expression in mature neurons [5]. The male Otx2Het mice display compromised fertility, and, while the loss of Otx2 does not affect expression of pituitary gonadotropin genes, it produces a significant fall in luteinizing hormone (LH) serum levels [5]. In contrast, the female Otx2Het mice did not survive to adulthood in our studies. Thus, correct gene dosage of Otx2 is critical for normal development of the GnRH neurons and expression of GnRH in adult, male mice. Diaczok et al. [28] established that deletion of Otx2 specifically from GnRH neurons results in hypogonadotropic hypogonadism in mice, adding in vivo data to previously published reports demonstrating the important role Otx2 plays as a transcriptional regulator of GnRH expression [26, 27, 40].
Fig. 2.
Reduced number of GnRH-expressing neurons in the hypothalamus of 17 day-old mouse embryos heterozygous for Otx2, Vax1, or Six3. To determine how haploinsufficiency of Otx2, Vax1, and Six3 impacted GnRH neuron development, we performed immunohistochemistry (IHC) for GnRH in mouse embryo heads from control, Otx2Het, Vax1Het, and Six3Het mice at e13 and e17. The red squares on the schematic illustrate the area imaged. GnRH IHC at e13 shows normal location and numbers of GnRH neurons at the cribriform plate in Otx2Het, Vax1Het, and Six3Het embryos as compared to wildtype. In contrast, at e17, Otx2Het, Vax1Het, and Six3Het embryos have fewer detectable hypothalamic GnRH neurons, causing a reduction of GnRH terminals at the median eminence (black arrow). Follow-up studies found that reduced release of GnRH caused subfertility in these heterozygote mouse lines. Scale bar represents 100 μm. pit, pituitary.
Fig. 3.
Summary of role of Otx2, Vax1, Six3, and Six6 in GnRH neuron development and fertility. a Sagittal illustration of GnRH neuron location after termination of migration in the adult brains of control and heterozygote animals and the correlation with fertility. Box indicates normal location of neurons that are most likely to project to the median eminence allowing GnRH release into the hypophysial portal system promoting LH and FSH release from the pituitary to promote gonadal function and fertility. b Summary of findings in the Otx2, Vax1, Six3, and Six6 heterozygote (Het) and knock-out (null) mice. * Studies only performed in males due to inability to generate adult Otx2Het females. N/A, not applicable.
Vax1 Heterozygous Mice Have Altered Hypothalamic Kiss1 and GnRH Expression Causing Subfertility
Ventral Anterior Homeobox (Vax1) is expressed in the eye, olfactory placode, and ventral hypothalamus and is known to have a role in neuronal fate determination [41–43]. It directs the formation of the ventral and rostral forebrain and Vax1-null mice have altered migration of olfactory placode neurons into the forebrain [44] and reduced neuronal proliferation [45]. The developmental defects in Vax1-null mice are in part caused by abnormal Sonic hedgehog signal transduction [45, 46]. Vax1-null mice are neonatal lethal due to severe holoprosencephaly and cleft palate [42, 44, 47]. Despite this severe phenotype, one report of a human child with a homozygous mutation in VAX1 phenocopies the findings in the Vax1null mouse model [48]. Interestingly, in humans, heterozygote mutations of VAX1 have been associated with cleft lip/palate [49–51], supporting a dosage effect of Vax1 [7].
While Vax1-null mice are neonatal lethal, male and female Vax1Het mice are subfertile indicating dosage sensitivity of the Vax1 allele on GnRH neuron development [7]. Vax1Het females produced normal numbers of superovulated oocytes, but corpora lutea were reduced, along with a slight increase in basal LH and estrogen. The cause of the subfertility originated in the hypothalamus where Kiss1 and Gnrh1 mRNAs were altered, along with a substantial reduction in GnRH neuron number (Fig. 2, 3). Although the pituitary responded normally to a GnRH challenge, diestrus females had reduced circulating LHβ and FSHβ. Furthermore, Vax1Het males had reduced Gnrh1 mRNA and neurons, while the pituitary had normal transcript levels and responses to GnRH. Interestingly, Vax1Het males had an 88% reduction in motile sperm. Thus, our data showed that Vax1Het subfertility originates in the hypothalamus making it a candidate for haploinsufficient IHH. In addition to the critical role of Vax1 in establishing normal Kiss1 levels and a correct GnRH neuronal population, the Vax1Het male sub-fertility might be caused by a combined role of Vax1 in the brain and the testis. Indeed, we detected Vax1 expression in the testis, but not in the mature sperm [7]. Thus, Vax1 haploinsufficiency might impact testis/sperm function and contribute to Vax1Het sub-fertility, although at this point the mechanism remains unknown.
To determine when Vax1 was required for GnRH neuron development, we counted GnRH expressing neurons in the developing wildtype, Vax1Het, and Vax1-null embryo. We found that Vax1Het and Vax1-null embryos have normal numbers of GnRH expressing neurons at e13 (Fig. 2), but Vax1Het embryos have a ~50% reduction in GnRH neurons at e17 (Fig. 2, 3), whereas no GnRH staining was observed in the Vax1-null embryo at this age (Fig. 3b). To identify the role of Vax1 specifically in GnRH neuron development, we generated Vax1flox mice. Lineage tracing in Vax1flox:GnRHcre:RosaLacZ mice identified Vax1 as essential for maintaining expression of GnRH, since the neurons survive but fail to express GnRH. The absence of GnRH staining in adult Vax1flox:GnRHcre mice results in delayed puberty, hypogonadism, and infertility [23, 52].
Defects in Olfaction Cause Male Infertility in Six3 Heterozygous Male Mice
Mammalian Six proteins are vertebrate homologues of Drosophila optix [53], and Six3 and Six6 are close homologs sharing partially overlapping expression patterns [53]. Six3 and Six6 are expressed early in development and strongly in the postnatal suprachiasmatic nucleus (SCN) [54]. However, despite their initial overlapping pattern, they become segregated in the postnatal brain [55], with Six6 confined to the adult hypothalamus, eye, and pituitary. Six6-null mice survive but have a hypoplastic pituitary and variable retinal hypoplasia, often with no optic chiasm or optic nerve [56, 57], traits that parallel defects associated with human chromosomal deletions that include the human SIX6 locus [56, 58]. Homozygous Six6-null mice survive but are subfertile (Fig. 3b) [25] and lack the structures of the SCN and circadian rhythms [57], while Six6 heterozygous mice are fertile [25]. In contrast, Six3-null mice die at birth, lacking most head structures anterior to the midbrain including the SCN, though the rest of the body appears normal [59].
Mating behavior in males and females is dependent on olfactory cues processed through both the main olfactory epithelium (MOE) and the VNO [60–63]. Signaling through the MOE is critical for male mating behavior. The development of olfactory neurons is closely linked to the development of GnRH neurons [9, 64–68], which originate in the primordial nasal placode, and migrate along olfactory nerves into the forebrain [11, 66, 69], and both are compromised in Kallmann syndrome (anosmic IHH) [2, 70–74]. We found that dosage of Six3 affects the development of MOE but not the VNO [8]. Anomalous MOE development in Six3Het mice leads to hyposmia, specifically disrupting male mounting behavior by impairing detection of volatile female estrus pheromones. Six3 is highly expressed in the MOE, main olfactory bulb (MOB), and hypothalamus; all regions essential in the proper migration of the GnRH neurons. Six3Het mice have compromised development of the MOE and MOB, resulting in 45% loss of GnRH neurons due to improper olfactory axon targeting (Fig. 2, 3). This leads to female subfertility but does not impact male hormone levels or sperm, indicating male infertility is exclusively linked to abnormal olfaction. Olfactory marker protein (OMP) specifically localizes in the primary neurons of the olfactory system. Remarkably, a total loss of OMP in MOE and MOB was observed in the Six3Het mice. In Six3-null e13.5 embryos [8], recognizable olfactory structures were not detectable. Instead, a bundle of neurons and axonal fibers with GnRH and OMP was detected in the region where the MOE normally exists. In contrast, OMP staining was preserved in the Six3Het VNO, explaining the normal non-volatile olfaction. Thus, we conclude that Six3 is haploinsufficient for MOE development, GnRH neuron migration, and fertility, and represents a novel candidate gene for Kallmann IHH. Remarkably, conditional deletion of Six3 using GnRHcre, instead caused a 30% increase in the number of GnRH neurons as detected by immunohistochemistry [8].
Conclusions and Perspectives
Heterozygous deletion of Otx2, Vax1, or Six3 is sufficient to produce subfertility. All 3 homeodomain transcription factors bind to and regulate the Gnrh1 gene directly through ATTA and related elements, but studies of their in vivo phenotypes are hampered by perinatal lethality of the null mice. We have studied the reproductive phenotypes of the heterozygous mice and found that all 3 have reduced numbers of GnRH neurons and are subfertile. However, there is specificity to the subfertility by sex and by mechanisms of action. Our findings confirm the importance of considering haploinsufficiency as a contributor to human disease and IHH.
Acknowledgments
Funding Sources
This work was supported by the National Institutes of Health grants R01 HD082567 and R01 HD072754 (to P.L.M.) and by the National Institute of Child Health and Human Development/National Institutes of Health P50 HD012303 as part of the National Centers for Translational Research in Reproduction and Infertility (P.L.M.). P.L.M. was partially supported by NIH grants P30 DK063491, P42 ES101337, and P30 CA023100. H.M.H. was partially supported by NIH grant K99/R00 HD084759. E.C.P. was partially supported by NIH grants R25 GM083275 and F31 HD098652.
Footnotes
Disclosure Statement
The authors have no conflicts of interest to declare.
Statement of Ethics
Animal experiments conform to internationally accepted standards and have been approved by the appropriate institutional review body.
References
- 1.Strauss JF, Barbieri RL. Yen and Jaffe’s Reproductive Endocrinology. 5th ed. Philadelphia (PA): Elsevier Saunders; 2004. [Google Scholar]
- 2.Bianco SD, Kaiser UB. The genetic and molecular basis of idiopathic hypogonadotropic hypogonadism. Nat Rev Endocrinol. 2009. October; 5(10): 569–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pitteloud N, Quinton R, Pearce S, Raivio T, Acierno J, Dwyer A, et al. Digenic mutations account for variable phenotypes in idiopathic hypogonadotropic hypogonadism. J Clin Invest. 2007. February; 117(2): 457–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Walters KA, Allan CM, Jimenez M, Lim PR, Davey RA, Zajac JD, et al. Female mice haploinsufficient for an inactivated androgen receptor (AR) exhibit age-dependent defects that resemble the AR null phenotype of dysfunctional late follicle development, ovulation, and fertility. Endocrinology. 2007. August; 148(8): 3674–84. [DOI] [PubMed] [Google Scholar]
- 5.Larder R, Kimura I, Meadows J, Clark DD, Mayo S, Mellon PL. Gene dosage of Otx2 is important for fertility in male mice. Mol Cell Endocrinol. 2013. September; 377(1–2): 16–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim HG, Herrick SR, Lemyre E, Kishikawa S, Salisz JA, Seminara S, et al. Hypogonadotropic hypogonadism and cleft lip and palate caused by a balanced translocation producing haploinsufficiency for FGFR1. J Med Genet. 2005. August; 42(8): 666–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hoffmann HM, Tamrazian A, Xie H, PérezMillán MI, Kauffman AS, Mellon PL. Heterozygous deletion of ventral anterior homeobox (vax1) causes subfertility in mice. Endocrinology. 2014. October; 155(10): 4043–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pandolfi EC, Hoffmann HM, Schoeller EL, Gorman MR, Mellon PL. Haploinsufficiency of SIX3 abolishes male reproductive behavior through disrupted olfactory development, and impairs female fertility through disrupted GnRH neuron migration. Mol Neurobiol. 2018. November; 55(11): 8709–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schwanzel-Fukuda M, Pfaff DW. Origin of luteinizing hormone-releasing hormone neurons. Nature. 1989. March; 338(6211): 161–4. [DOI] [PubMed] [Google Scholar]
- 10.Schwanzel-Fukuda M, Jorgenson KL, Bergen HT, Weesner GD, Pfaff DW. Biology of normal luteinizing hormone-releasing hormone neurons during and after their migration from olfactory placode. Endocr Rev. 1992. November; 13(4): 623–34. [DOI] [PubMed] [Google Scholar]
- 11.Tobet SA, Schwarting GA. Minireview: recent progress in gonadotropin-releasing hormone neuronal migration. Endocrinology. 2006. March; 147(3): 1159–65. [DOI] [PubMed] [Google Scholar]
- 12.Taroc EZ, Prasad A, Lin JM, Forni PE. The terminal nerve plays a prominent role in GnRH-1 neuronal migration independent from proper olfactory and vomeronasal connections to the olfactory bulbs. Biol Open. 2017. Oct; 6(10): 1552–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vilensky JA. The neglected cranial nerve: nervus terminalis (cranial nerve N). Clin Anat. 2014. January; 27(1): 46–53. [DOI] [PubMed] [Google Scholar]
- 14.Sonne J, Lopez-Ojeda W. Neuroanatomy, Cranial Nerve. Treasure Island (FL): StatPearls; 2018. [Google Scholar]
- 15.Wray S, Grant P, Gainer H. Evidence that cells expressing luteinizing hormone-releasing hormone mRNA in the mouse are derived from progenitor cells in the olfactory placode. Proc Natl Acad Sci USA. 1989. October; 86(20): 8132–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Simonian SX, Herbison AE. Regulation of gonadotropin-releasing hormone (GnRH) gene expression during GnRH neuron migration in the mouse. Neuroendocrinology. 2001. March; 73(3): 149–56. [DOI] [PubMed] [Google Scholar]
- 17.Gore AC, Roberts JL, Gibson MJ. Mechanisms for the regulation of gonadotropin-releasing hormone gene expression in the developing mouse. Endocrinology. 1999. May; 140(5): 2280–7. [DOI] [PubMed] [Google Scholar]
- 18.Yoon H, Enquist LW, Dulac C. Olfactory inputs to hypothalamic neurons controlling reproduction and fertility. Cell. 2005. November; 123(4): 669–82. [DOI] [PubMed] [Google Scholar]
- 19.Wierman ME, Kiseljak-Vassiliades K, Tobet S. Gonadotropin-releasing hormone (GnRH) neuron migration: initiation, maintenance and cessation as critical steps to ensure normal reproductive function. Front Neuroendocrinol. 2011. January; 32(1): 43–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seminara SB, Hayes FJ, Crowley WF Jr. Gonadotropin-releasing hormone deficiency in the human (idiopathic hypogonadotropic hypogonadism and Kallmann’s syndrome): pathophysiological and genetic considerations. Endocr Rev. 1998. October; 19(5): 521–39. [DOI] [PubMed] [Google Scholar]
- 21.Mellon PL, Windle JJ, Goldsmith PC, Padula CA, Roberts JL, Weiner RI. Immortalization of hypothalamic GnRH neurons by genetically targeted tumorigenesis. Neuron. 1990. July; 5(1): 1–10. [DOI] [PubMed] [Google Scholar]
- 22.Radovick S, Wray S, Lee E, Nicols DK, Nakayama Y, Weintraub BD, et al. Migratory arrest of gonadotropin-releasing hormone neurons in transgenic mice. Proc Natl Acad Sci USA. 1991. April; 88(8): 3402–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoffmann HM, Trang C, Gong P, Kimura I, Pandolfi EC, Mellon PL. Deletion of Vax1 from GnRH neurons abolishes GnRH expression and leads to hypogonadism and infertility. J Neurosci. 2016; 36: 3506–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hoffmann HM, Gong P, Tamrazian A, Mellon PL. Transcriptional interaction between cFOS and the homeodomain-binding transcription factor VAX1 on the GnRH promoter controls Gnrh1 expression levels in a GnRH neuron maturation specific manner. Mol Cell Endocrinol. 2018. February 5; 461: 143–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Larder R, Clark DD, Miller NL, Mellon PL. Hypothalamic dysregulation and infertility in mice lacking the homeodomain protein Six6. J Neurosci. 2011. January; 31(2): 426–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Larder R, Mellon PL. Otx2 induction of the gonadotropin-releasing hormone promoter is modulated by direct interactions with Grg co-repressors. J Biol Chem. 2009. June; 284(25): 16966–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim HH, Wolfe A, Cohen RN, Eames SC, Johnson AL, Wieland CN, et al. In vivo identification of a 107 bp promoter element mediating neuron-specific expression of mouse GnRH. Mol Endocrinol. 2007; 21: 457–71. [DOI] [PubMed] [Google Scholar]
- 28.Diaczok D, DiVall S, Matsuo I, Wondisford FE, Wolfe AM, Radovick S. Deletion of Otx2 in GnRH neurons results in a mouse model of hypogonadotropic hypogonadism. Mol Endocrinol. 2011. May; 25(5): 833–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Acampora D, Mazan S, Lallemand Y, Avantaggiato V, Maury M, Simeone A, et al. Forebrain and midbrain regions are deleted in Otx2−/− mutants due to a defective anterior neuroectoderm specification during gastrulation. Development. 1995. October; 121(10): 3279–90. [DOI] [PubMed] [Google Scholar]
- 30.Simeone A, Acampora D, Mallamaci A, Stornaiuolo A, D’Apice MR, Nigro V, et al. A vertebrate gene related to orthodenticle contains a homeodomain of the bicoid class and demarcates anterior neuroectoderm in the gastrulating mouse embryo. EMBO J. 1993. July; 12(7): 2735–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Frantz GD, Weimann JM, Levin ME, McConnell SK. Otx1 and Otx2 define layers and regions in developing cerebral cortex and cerebellum. J Neurosci. 1994. October; 14(10): 5725–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Puelles E, Annino A, Tuorto F, Usiello A, Acampora D, Czerny T, et al. Otx2 regulates the extent, identity and fate of neuronal progenitor domains in the ventral midbrain. Development. 2004. May; 131(9): 2037–48. [DOI] [PubMed] [Google Scholar]
- 33.Mallamaci A, Di Blas E, Briata P, Boncinelli E, Corte G. OTX2 homeoprotein in the developing central nervous system and migratory cells of the olfactory area. Mech Dev. 1996. August; 58(1–2): 165–78. [DOI] [PubMed] [Google Scholar]
- 34.Dateki S, Kosaka K, Hasegawa K, Tanaka H, Azuma N, Yokoya S, et al. Heterozygous orthodenticle homeobox 2 mutations are associated with variable pituitary phenotype. J Clin Endocrinol Metab. 2010. February; 95(2): 756–64. [DOI] [PubMed] [Google Scholar]
- 35.Dateki S, Fukami M, Sato N, Muroya K, Adachi M, Ogata T. OTX2 mutation in a patient with anophthalmia, short stature, and partial growth hormone deficiency: functional studies using the IRBP, HESX1, and POU1F1 promoters. J Clin Endocrinol Metab. 2008. October; 93(10): 3697–702. [DOI] [PubMed] [Google Scholar]
- 36.Diaczok D, Romero C, Zunich J, Marshall I, Radovick S. A novel dominant negative mutation of OTX2 associated with combined pituitary hormone deficiency. J Clin Endocrinol Metab. 2008. November; 93(11): 4351–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ang SL, Jin O, Rhinn M, Daigle N, Stevenson L, Rossant J. A targeted mouse Otx2 mutation leads to severe defects in gastrulation and formation of axial mesoderm and to deletion of rostral brain. Development. 1996. January; 122(1): 243–52. [DOI] [PubMed] [Google Scholar]
- 38.Matsuo I, Kuratani S, Kimura C, Takeda N, Aizawa S. Mouse Otx2 functions in the formation and patterning of rostral head. Genes Dev. 1995. November; 9(21): 2646–58. [DOI] [PubMed] [Google Scholar]
- 39.Fossat N, Chatelain G, Brun G, Lamonerie T. Temporal and spatial delineation of mouse Otx2 functions by conditional self-knockout. EMBO Rep. 2006. August; 7(8): 824–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kelley CG, Lavorgna G, Clark ME, Boncinelli E, Mellon PL. The Otx2 homeoprotein regulates expression from the gonadotropin-releasing hormone proximal promoter. Mol Endocrinol. 2000. August; 14(8): 1246–56. [DOI] [PubMed] [Google Scholar]
- 41.Taglialatela P, Soria JM, Caironi V, Moiana A, Bertuzzi S. Compromised generation of GABAergic interneurons in the brains of Vax1−/− mice. Development. 2004. September; 131(17): 4239–49. [DOI] [PubMed] [Google Scholar]
- 42.Soria JM, Taglialatela P, Gil-Perotin S, Galli R, Gritti A, Verdugo JM, et al. Defective postnatal neurogenesis and disorganization of the rostral migratory stream in absence of the Vax1 homeobox gene. J Neurosci. 2004. December; 24(49): 11171–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bertuzzi S, Hindges R, Mui SH, O’Leary DD, Lemke G. The homeodomain protein vax1 is required for axon guidance and major tract formation in the developing forebrain. Genes Dev. 1999. December; 13(23): 3092–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hallonet M, Hollemann T, Pieler T, Gruss P. Vax1, a novel homeobox-containing gene, directs development of the basal forebrain and visual system. Genes Dev. 1999. December; 13(23): 3106–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Geoghegan F, Xavier GM, Birjandi AA, Seppala M, Cobourne MT. Vax1 Plays an Indirect Role in the Etiology of Murine Cleft Palate. J Dent Res. 2017. December; 96(13): 1555–62. [DOI] [PubMed] [Google Scholar]
- 46.Zhao L, Saitsu H, Sun X, Shiota K, Ishibashi M. Sonic hedgehog is involved in formation of the ventral optic cup by limiting Bmp4 expression to the dorsal domain. Mech Dev. 2010. Jan-Feb; 127(1–2): 62–72. [DOI] [PubMed] [Google Scholar]
- 47.Bharti K, Gasper M, Bertuzzi S, Arnheiter H. Lack of the ventral anterior homeodomain transcription factor VAX1 leads to induction of a second pituitary. Development. 2011. March; 138(5): 873–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Slavotinek AM, Chao R, Vacik T, Yahyavi M, Abouzeid H, Bardakjian T, et al. VAX1 mutation associated with microphthalmia, corpus callosum agenesis, and orofacial clefting: the first description of a VAX1 phenotype in humans. Hum Mutat. 2012. February; 33(2): 364–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang BH, Shi JY, Lin YS, Shi B, Jia ZL. VAX1 gene associated non-syndromic cleft lip with or without palate in Western Han Chinese. Arch Oral Biol. 2018. November; 95: 40–3. [DOI] [PubMed] [Google Scholar]
- 50.Butali A, Suzuki S, Cooper ME, Mansilla AM, Cuenco K, Leslie EJ, et al. Replication of genome wide association identified candidate genes confirm the role of common and rare variants in PAX7 and VAX1 in the etiology of nonsyndromic CL(P). Am J Med Genet A. 2013. May; 161A(5): 965–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.de Aquino SN, Messetti AC, Bagordakis E, Martelli-Júnior H, Swerts MS, Graner E, et al. Polymorphisms in FGF12, VCL, CX43 and VAX1 in Brazilian patients with nonsyndromic cleft lip with or without cleft palate. BMC Med Genet. 2013. May; 14(1): 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hoffmann HM, Mellon PL. A small population of hypothalamic neurons govern fertility: the critical role of VAX1 in GnRH neuron development and fertility maintenance. Neurosci Commun (Houst). 2016; 2: 2. [PMC free article] [PubMed] [Google Scholar]
- 53.Jean D, Bernier G, Gruss P. Six6 (Optx2) is a novel murine Six3-related homeobox gene that demarcates the presumptive pituitary/ hypothalamic axis and the ventral optic stalk. Mech Dev. 1999. June; 84(1–2): 31–40. [DOI] [PubMed] [Google Scholar]
- 54.VanDunk C, Hunter LA, Gray PA. Development, maturation, and necessity of transcription factors in the mouse suprachiasmatic nucleus. J Neurosci. 2011. April; 31(17): 6457–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Conte I, Morcillo J, Bovolenta P. Comparative analysis of Six 3 and Six 6 distribution in the developing and adult mouse brain. Dev Dyn. 2005. November; 234(3): 718–25. [DOI] [PubMed] [Google Scholar]
- 56.Li X, Perissi V, Liu F, Rose DW, Rosenfeld MG. Tissue-specific regulation of retinal and pituitary precursor cell proliferation. Science. 2002. August; 297(5584): 1180–3. [DOI] [PubMed] [Google Scholar]
- 57.Clark DD, Gorman MR, Hatori M, Meadows JD, Panda S, Mellon PL. Aberrant development of the suprachiasmatic nucleus and circadian rhythms in mice lacking the homeodomain protein Six6. J Biol Rhythms. 2013. February; 28(1): 15–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gallardo ME, Lopez-Rios J, Fernaud-Espinosa I, Granadino B, Sanz R, Ramos C, et al. Genomic cloning and characterization of the human homeobox gene SIX6 reveals a cluster of SIX genes in chromosome 14 and associates SIX6 hemizygosity with bilateral anophthalmia and pituitary anomalies. Genomics. 1999. October; 61(1): 82–91. [DOI] [PubMed] [Google Scholar]
- 59.Lagutin OV, Zhu CC, Kobayashi D, Topczewski J, Shimamura K, Puelles L, et al. Six3 repression of Wnt signaling in the anterior neuroectoderm is essential for vertebrate forebrain development. Genes Dev. 2003. February; 17(3): 368–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Brown RE. Mammalian Social Odors: A Critical Review In: Rosenblat JS, Hinde RA, Beer C, Busnell MC, editors. Advances in the Study of Behavior. Volume 10 New York, New York: Academic Press; 1979. pp. 107–61. [Google Scholar]
- 61.Brennan PA, Zufall F. Pheromonal communication in vertebrates. Nature. 2006. November; 444(7117): 308–15. [DOI] [PubMed] [Google Scholar]
- 62.Thompson ML, Edwards DA. Olfactory bulb ablation and hormonally induced mating in spayed female mice. Physiol Behav. 1972. June; 8(6): 1141–6. [DOI] [PubMed] [Google Scholar]
- 63.Brennan PA, Keverne EB. Neural mechanisms of mammalian olfactory learning. Prog Neurobiol. 1997. March; 51(4): 457–81. [DOI] [PubMed] [Google Scholar]
- 64.Wray S From nose to brain: development of gonadotrophin-releasing hormone-1 neurones. J Neuroendocrinol. 2010. July; 22(7): 743–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Forni PE, Wray S. GnRH, anosmia and hypogonadotropic hypogonadism—where are we? Front Neuroendocrinol. 2015. January; 36: 165–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wray S Development of gonadotropin-releasing hormone-1 neurons. Front Neuroendocrinol. 2002. July; 23(3): 292–316. [DOI] [PubMed] [Google Scholar]
- 67.Rattazzi L, Cariboni A, Poojara R, Shoenfeld Y, D’Acquisto F. Impaired sense of smell and altered olfactory system in RAG-1(-∕-) immunodeficient mice. Front Neurosci. 2015. September; 9: 318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Origin Schwanzel-Fukuda M. and migration of luteinizing hormone-releasing hormone neurons in mammals. Microsc Res Tech. 1999. January; 44(1): 2–10. [DOI] [PubMed] [Google Scholar]
- 69.Wray S Development of luteinizing hormone releasing hormone neurones. J Neuroendocrinol. 2001. January; 13(1): 3–11. [DOI] [PubMed] [Google Scholar]
- 70.Tsai PS, Gill JC. Mechanisms of disease: insights into X-linked and autosomal-dominant Kallmann syndrome. Nat Clin Pract Endocrinol Metab. 2006. Mar; 2(3): 160–71. [DOI] [PubMed] [Google Scholar]
- 71.Kramer PR, Wray S. Novel gene expressed in nasal region influences outgrowth of olfactory axons and migration of luteinizing hormone-releasing hormone (LHRH) neurons. Genes Dev. 2000. July; 14(14): 1824–34. [PMC free article] [PubMed] [Google Scholar]
- 72.Xu N, Kim HG, Bhagavath B, Cho SG, Lee JH, Ha K, et al. Nasal embryonic LHRH factor (NELF) mutations in patients with normosmic hypogonadotropic hypogonadism and Kallmann syndrome. Fertil Steril. 2011. April; 95(5): 1613–20.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Silveira LG, Noel SD, Silveira-Neto AP, Abreu AP, Brito VN, Santos MG, et al. Mutations of the KISS1 gene in disorders of puberty. J Clin Endocrinol Metab. 2010. May; 95(5): 2276–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bless E, Raitcheva D, Henion TR, Tobet S, Schwarting GA. Lactosamine modulates the rate of migration of GnRH neurons during mouse development. Eur J Neurosci. 2006. August; 24(3): 654–60. [DOI] [PubMed] [Google Scholar]