Abstract
Fecal contamination from sewage and agricultural runoff is a pervasive problem in Great Lakes watersheds. Most work examining fecal pollution loads relies on discrete samples of fecal indicators and modeling land use. In this study we made empirical measurements of human and ruminant-associated fecal indicator bacteria and combined these with hydrological measurements in eight watersheds ranging from predominantly forested to highly urbanized. Flow composited river samples were collected over low-flow (n=89) and rainfall or snowmelt runoff events (n=130). Approximately, 90% of samples had evidence of human fecal pollution, with highest loads from urban watersheds. Ruminant indicators were found in ~60–100% of runoff-event samples in agricultural watersheds, with concentrations and loads related to cattle density. Rain depth, season, agricultural tile drainage, and human or cattle density explained variability in daily flux of human or ruminant indicators. Mapping host-associated indicator loads to watershed discharge points sheds light on the type, level, and possible health risk from fecal pollution entering the Great Lakes and can inform Total Maximum Daily Load implementation and other management practices to target specific fecal pollution sources.
Keywords: watershed, TMDL, Human Bacteroides markers, Human Lachnospiraceae markers, ruminant markers, fecal pollution, Great Lakes pollution
1. Introduction
Coastal regions are increasingly stressed from human activities that contribute fecal pollution to nearshore waters. In the Laurentian Great Lakes, drinking water intakes can be vulnerable to pathogen contamination. This vulnerability was demonstrated in 1993, when approximately 400,000 people in Milwaukee, Wisconsin suffered gastrointestinal (GI) illness caused by Cryptosporidium in a public water supply obtained from Lake Michigan1. This outbreak is a stark reminder that even in developed countries, watershed discharges contain high pathogen loads and the only barrier preventing illness is treatment of the source water. Nearly 40 million people living in the United States and Canada rely on the Great Lakes for drinking water2. The Great Lakes also support a tourist industry dependent on healthy swimming beaches and a commercial/recreational fisheries industry worth well over 1 billion dollars annually3.
Elevated levels of fecal indicator bacteria (FIB) are the most frequent cause of water quality impairment in the United States4, including in the Great Lakes5. Human pathogens carried in fecal pollution are the actual concern, but it is difficult and expensive to monitor for pathogens. Therefore, FIB are commonly used by municipalities as a general method to monitor waterways for the broad array of fecal contamination sources that might be present, including sources that may or may not be associated with human pathogens. Wildlife and pets can contribute FIB to stormwater runoff, but human and agricultural FIB sources carry the highest risk of pathogen exposure and disease outcome6,7.
Runoff associated with rainfall/precipitation is a driver of degraded water quality in receiving waters8 and is linked to waterborne disease1,9,10. Stormwater contaminated with sewage can increase presence of human viruses in surface waters11–13 and carries with it other biological and chemical contaminants harmful to humans and aquatic wildlife. Although infrastructure and population vary widely between urban and rural watersheds, both inputs contribute to the nearshore pollution that travels from land to water14–17. Urban fecal pollution from leaking sewage infrastructure or sewage overflows may introduce human pathogens18–21 to waterways. Animals and septic systems from rural areas can impair drinking, irrigation, and recreational waters22,23.
Storms have been shown to increase human-associated fecal indicator bacteria by several orders of magnitude in Great Lakes tributaries during storm events8,24,25. Contamination plumes from river mouths into the Great Lakes can dilute quickly based on wind direction, currents, and surface temperature26,27. It is likely that this plume dilution causes reduction and die-off of associated bacteria in the nearshore. However, Newton et al. (2013) reported evidence of sewage and fecal contamination in Lake Michigan as far as 3 km offshore after heavy rains and as far as 8 km offshore after severe storms led to sewage overflows, illustrating the widespread impact of rainfall on nearshore waters.
Understanding fecal pollution sources and watershed load contributions from different types of land use is critical for devising management priorities and reducing the overall impact on the Great Lakes. Only a few studies have measured pollution loads using host associated fecal indicators, and these have been in single watersheds8,28–30. In this study, eight watersheds with a gradient of land use were examined for sources of human and ruminant fecal contamination impacting Great Lakes receiving waters during low-flow and runoff-event periods. DNA-based assays using host-associated indicators were combined with detailed hydrological measurements to perform a comprehensive study comparing fecal contamination loads discharging from watersheds ranging from forest to agricultural to urban land use.
2. Materials and methods
Study sites
Eight Great Lakes watersheds were selected that represent a varying degree of land use from high to low urban and agricultural intensity (Table S1; Table S2; Figure S1). Urban land cover comprises the majority of the Clinton and Rouge watersheds. The Milwaukee watershed contains mixed urban and agricultural land use. Maumee, Portage, Raisin, and Manitowoc watersheds are predominantly agricultural land use. The Menominee watershed has primarily forest and wetland land cover. Each watershed or river is abbreviated with the first name of the river. Land cover compositions were obtained from 2011 National Land Cover Database31. Percent of watershed area underlain by agricultural drainage tiles, which are essentially piping systems under fields intended to remove excess water, was computed using data from the 1995 National Resources Inventory32. Human and cattle population (dairy and non-dairy) densities (Table S2) were calculated based on the total population within a watershed divided by the total drainage area for that watershed31,33–35.
Sample Collection
A total of 219 samples were collected over a 24-month period from July 2011 through June 2013 at USGS streamgages that were furthest downstream along each river before the discharge point to the lake36. Flow-weighted composite samples were collected during low-flow periods (n = 89) and during periods of increased runoff (n = 130) due to rainfall and snowmelt (designated as “runoff-event periods”) and used to determine event-mean concentrations of host-associated indicator. With this flow-weighted sampling approach, subsample (60 mL) collection frequency increased as streamflow increased throughout the event hydrograph. Runoff-event sampling was initiated when water level increased above the low-flow level and lasted generally 1 – 7 days, when sampling was terminated. Low-flow samples were collected over approximately 24 – 36 h. Concurrent samples were separately collected from the same water line and analyzed for pathogens as reported by Lenaker et al. (2017) and these data were compared to host-associated indicators. Samples were kept refrigerated during collection and shipped overnight on ice for analysis.
Culture-based analysis
Immediately after arrival, samples were analyzed for Escherichia coli (E. coli) and enterococci using standard EPA methods38,39.
DNA extraction and quantitative PCR analysis
Extraction of DNA from filtered water samples was performed as previously described24, with extraction efficiencies determined to be 46%8. Inhibition was not observed in a subset of samples, consistent with previous studies that included highly contaminated stormwater samples8,25. Quantitative PCR was carried out using an Applied Biosystems StepOne Plus™ Real-Time PCR System Thermal Cycling Block (Applied Biosystems; Foster City, CA) with Taqman hydrolysis probe chemistry. Previously published primers and probe were used for all analyses: human Bacteroides (HB)8,24,25,28, which is an assay modified from Kildare et al. (2007) that substitues the forward primer with the HF183 primer41; human Lachnospiraceae (Lachno2)8,20,24,28; ruminant Bacteroides (BacR)42,43; and enterococci (ENT)44. Both HB and Lachno2 are highly correlated and have steady concentrations in sewage8. The HB assay targets the human associated HF183 bacterial cluster45 and mapping of family specific clone libraries using DNA from untreated sewage46 verified that the HF183F and the forward primer of the assay described by Kildare et al40 target the same organism. Both the HB and Lachno2 assay have been used as human-associated indicators at the Milwaukee site8,24,25,28. Standard curves and amplification conditions were carried out as described in Templar et al. (2016). Results were reported in copy number (CN) of respective markers per 100 mL sample volume. The limit of quantification was 225 CN/100 mL and the limit of detection was set at 40 cycles (1 to 224 CN/100 mL) for all assays. All field blanks (one per sample event per location) and no DNA template controls (one per qPCR plate set-up) were negative. See Supporting Information for standard curves, assay efficiencies and qPCR primers (Table S3).
Data Analysis
Concentrations of HB and Lachno2 were highly correlated in runoff-event and low-flow periods (Pearson’s r = 0.83 and 0.80, respectively; p < 0.001); therefore, only the HB was used as a proxy for human fecal pollution in some calculations because it is equivalent to multiple assays targeting the HF183 cluster. Additionally, Lachno2 has been found to give sporadic false-positives with animal markers47. Bacterial and viral data were log base 10-transformed for statistical analysis. Streamflow volume, measured by continuous flow gauges at or near the collection site, was computed by integrating instantaneous discharge values over the sampling period for low-flow periods, and throughout the runoff hydrograph during event periods. Because sampling was flow weighted, concentrations represented an average for that period. Mean concentrations for wet weather and low-flow events were reported as event mean concentrations. Event loads (units of CN) were computed by multiplying concentration by the associated streamflow volume as defined previously37. When multiple samples were collected during the same runoff-event period, loads were calculated for each sample concentration and corresponding volume and then summed to get a total runoff-event load. Watershed yields were computed by dividing the event load by the watershed drainage area (DA) as a normalization step for comparison among sites, resulting in units of CN/km2. Bacteria flux was computed by dividing the yield by the duration of the sample collection resulting in units of CN/km2/h. All concentrations have been published36 and streamflow data are available in the U.S. Geological Survey (USGS) National Water Information System (http://waterdata.usgs.gov/nwis). Sample volumes, sampling time periods, and detailed instructions to query the USGS National Water Information System for the specific data reported in this study have been previously published37.
One-way analysis of variance (ANOVA) was used to compare mean host-associated fecal indicator concentrations and mean event yields for each site and groupings of sites based on land use. Urban (Clinton, Rouge) and mixed (Milwaukee) land use watersheds were grouped into high percent urban use (range 30–92% of surface area); agricultural watersheds (Maumee, Portage, Raisin, and Manitowoc) were grouped into low percent urban use (range 7–11% of surface area); and Menominee forested watershed was considered a least-impacted reference category (3.8% urban land use). ANOVA was used to compare low-flow, rain-event, and snowmelt-event mean concentrations of fecal indicators for land use categories. When significant (α = 0.05) differences were found, post-hoc analysis with Tukey Honest Significant Difference resolved which groups were different. Quantum GIS48 was used to map watershed runoff-event yields to receiving waters, applying Jenk’s natural breaks classification method to cluster median yield values into similar categories.
Multiple regression analysis (Tobit regression for left-censored data assuming the Weibull distribution) for samples that were collected in periods without snow influence was used to explore variables that help explain fecal indicator flux49. Flux was used in this analysis to remove differences in sampling period. Regression models were selected based on variables that logically have potential to influence magnitude of bacteria markers including season (sine and cosine of decimal day of the year x 2 x π), rainfall depth, percent of tile drainage in the watershed, and population density for humans or cattle (Table S2). Resulting standardized coefficients were reported to provide information on the relative influence of each variable in the regressions. All regression coefficients reported were significant at α = 0.05. A logistic regression model was created based on virus results from these same samples, published in Lenaker et al (2017). Human virus data used in the model included the sum of adenovirus C, D, F, adenovirus A, norovirus groups I and II, and enterovirus to examine human virus occurrence in relation to human associated indicators. Bovine viruses in the summation included bovine polyomavirus, bovine rotavirus a, bovine enterovirus, and bovine viral diarrhea virus type 237. Data analysis was performed using Excel®, R version 3.1.150 open source programming language, and RStudio version 1.1.38351.
3. Results
3.1. Detection of human-associated and ruminant-associated fecal indicators in eight Great Lakes watersheds
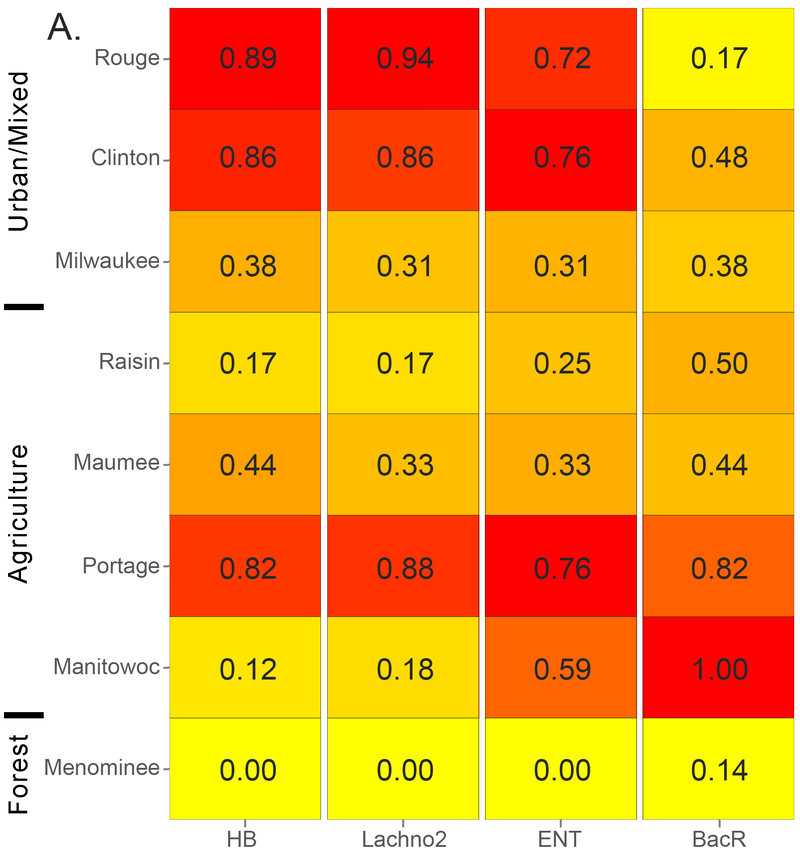
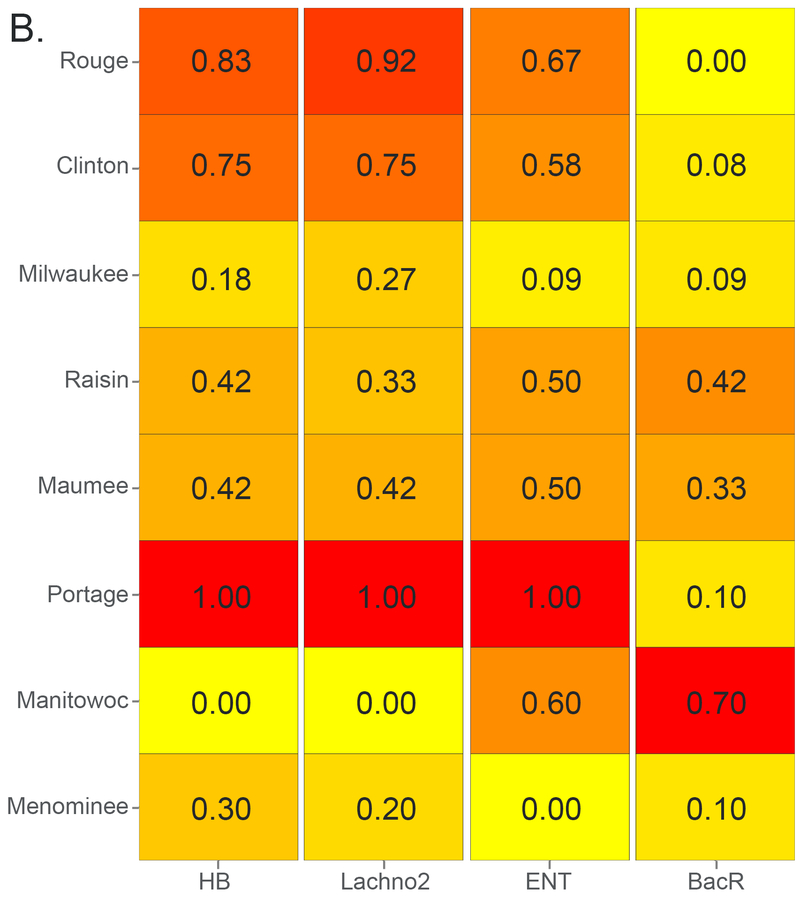
Overall, HB and Lachno2 markers were detected in 94% and 87% of 219 total samples, respectively, and concentrations ranged from just above the limit of quantification (225 CN/100 mL) to >105 CN/100 mL (Table S4). A total of 11 samples were below the limit of detection for both indicators. The watersheds with the greatest number of samples exceeding median concentrations (i.e., in the upper half of the overall dataset) for each indicator are shown in Figure 1. Over 85% of runoff-event samples in the two most urban watersheds (Rouge and Clinton) exceeded median concentrations of 1470 CN HB/100 mL and 1970 CN Lachno2/100 mL for human-associated indicators (Figure 1A), and more than 75% of the samples exceeded the median during low-flow periods (Figure 1B). Portage stood out from agricultural sites with significantly higher mean concentrations for human-associated indicators than the other agricultural sites (p < 0.05) and had unusually high frequency of samples over the median level in both runoff-event and in low-flow periods (Figure 1). There are combined sewage outfalls upstream and in close proximity to the Portage sampling location (see Woodville results at http://wwwapp.epa.ohio.gov/dsw/maps/cso/index.php).
Figure 1.
Heatmaps with frequency of sample concentrations over the median for each host-associated marker during (A) runoff-event periods and (B) low-flow periods for eight Great Lakes tributaries. Ordered top to bottom from most to least urban. HB = human Bacteroides; Lachno2 = human Lachnospiraceae; ENT = enterococci; BacR = ruminant Bacteroides.
Mean runoff-event concentrations of human markers were highest in the two urban (Rouge and Clinton) and one agricultural watershed (Portage), with moderate concentrations in the mixed land use watershed (Milwaukee) and one agricultural watershed (Maumee) (Table 1, Figure S2). The two remaining agricultural watersheds (Raisin and Manitowoc) had evidence of human fecal pollution, but with the lowest mean HB and Lachno2 concentrations. In urban watersheds, mean concentrations of human markers were ~10 to 30-fold higher during runoff-events compared with low-flow periods; however, in agricultural watersheds, mean concentrations of human markers during runoff-event periods were similar to low-flow periods and within the same order of magnitude.
Table 1.
Mean concentrations (in bold) and standard deviations (in parenthesis) of host-associated indicators during runoff events (n=130) and low-flow periods (n=89) for samples collected at eight Great Lakes watersheds from 2011–2013a.
| Lachno2 | HB | ENT | BacR | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Watershed | Basinb | Land Use Categoryc | Runoff Event | Low-flow Period | Runoff Event | Low-flow Period | Runoff Event | Low-flow Period | Runoff Event | Low-flow Period |
| Rouge River | DR | Urban/Mix | 113,000 (± 125,000) | 5,630 (± 5,610) | 53,900 (± 86,600) | 10,200 (± 12,400) | 28,000 (± 29,100) | 5,830 (± 6,050) | 298 (± 641) | 1 (± 0) |
| Clinton River | LSC | Urban/Mix | 74,000 (± 188,000) | 6,190 (± 6,860) | 61,600 (± 151,000) | 11,900 (± 15,700) | 22,900 (± 23,700) | 3,610 (± 2,630) | 4,140 (± 7,880) | 40 (± 138) |
| Milwaukee River | LM | Urban/Mix | 14,000 (± 36,300) | 457 (± 367) | 16,500 (± 42,600) | 685 (± 678) | 8,460 (± 10,100) | 970 (± 879) | 8,150 (± 23,000) | 2,500 (± 8,270) |
| Raisin River | LE | Agriculture | 1,090 (± 1,640) | 3,810 (± 9,550) | 1,160 (± 2,090) | 5,720 (± 11,500) | 4,030 (± 2,540) | 7,260 (± 10,500) | 6,120 (± 10,800) | 653 (± 1,540) |
| Maumee River | LE | Agriculture | 11,000 (± 26,100) | 2,420 (± 4,260) | 9,170 (± 23,200) | 4,720 (± 8,400) | 19,200 (± 50,800) | 4,200 (± 4,920) | 18,100 (± 57,300) | 2,160 (± 4,230) |
| Portage River | LE | Agriculture | 29,000 (± 36,500) | 12,000 (± 9,920) | 32,000 (± 40,700) | 43,100 (± 82,000) | 24,800 (± 31,800) | 22,600 (± 35,800) | 29,600 (± 65,200) | 37 (± 114) |
| Manitowoc River | LM | Agriculture | 794 (± 1,170) | 143 (± 183) | 444 (± 560) | 247 (± 67) | 12,700 (± 12,200) | 4,280 (± 3,600) | 113,000 (± 218,000) | 2,180 (± 5,050) |
| Menominee River | LM | Forested | 179 (±191) | 309 (± 324) | 286 (± 343) | 812 (± 1,100) | 412 (± 456) | 674 (± 374) | 182 (± 248) | 50 (± 151) |
Lachno2 = human Lachnospiraceae; HB = human Bacteroides; ENT = enterococci; BacR = ruminant Bacteroides. Units for concentration are CN/100 mL.
basin abbreviations: LM = Lake Michigan, LSC = Lake St. Clair, DR = Detroit River, LE = Lake Erie.
land use categories as defined in text.
Mean concentrations of the ruminant-associated fecal bacteria marker were highest in three agricultural watersheds (Manitowoc, Portage, and Maumee), but were also in moderate concentrations in the Clinton and Milwaukee during runoff-event periods (Table 1, Figure S2). In the Manitowoc watershed, dairy farming is a primary agricultural activity and BacR was detected over the median concentration of 510 CN BacR/100 mL in 100% and 70% of runoff-event and low-flow period samples, respectively (Figure 1). The ruminant-associated indicator was higher than the median concentration (of the entire dataset) in 82% of runoff-event samples from the Portage watershed; whereas the ruminant-associated indicators were over the median concentration in roughly 40% of the runoff-event period samples at other agriculturally impacted sites (Figure 1, Table 1).
In this study, we measured enterococci by qPCR for a general indicator of fecal pollution. The median event concentrations for all samples was 6400 CN ENT/100 mL, and the Clinton, Rouge and Portage had the most samples over the dataset median concentration, mirroring what was found for the human-associated indicators (Figure 1). Mean concentrations of ENT in samples collected at the forested site (Menominee) were significantly lower than all other sites during runoff-event periods (p < 0.01) (Table 1, Figure S2) and all sample results were below the median concentration for the dataset (Figure 1). Overall, the human-associated indicators were moderately correlated with ENT under runoff conditions (Spearman’s rank correlation coefficient: HB rho = 0.71, Lachno2 rho = 0.74, p < 0.001). Spearman’s results for individual sites are in (Table S5).
Enterococci and E. coli were also tested by standard plate-count methods. Overall, the plated indicators were highly correlated to each other (Pearson’s r = 0.83; p < 0.001). Table S6 provides the Federal recreational water quality standard exceedance percentages for enterococci and E. coli plate-counts. During events, exceedance of these standards in the Rouge and Clinton mirrored the high concentrations of human-associated marker levels (Figure 1 and Table S6); however low-flow FIB concentrations were unrelated to HB concentrations, indicating that FIB may not be associated with human sewage contamination in all hydrologic conditions. Additionally, other watersheds did not have a correspondence of patterns between cultured FIB and HB assays.
3.2. Influence of rainfall and snowmelt within watersheds with different land use
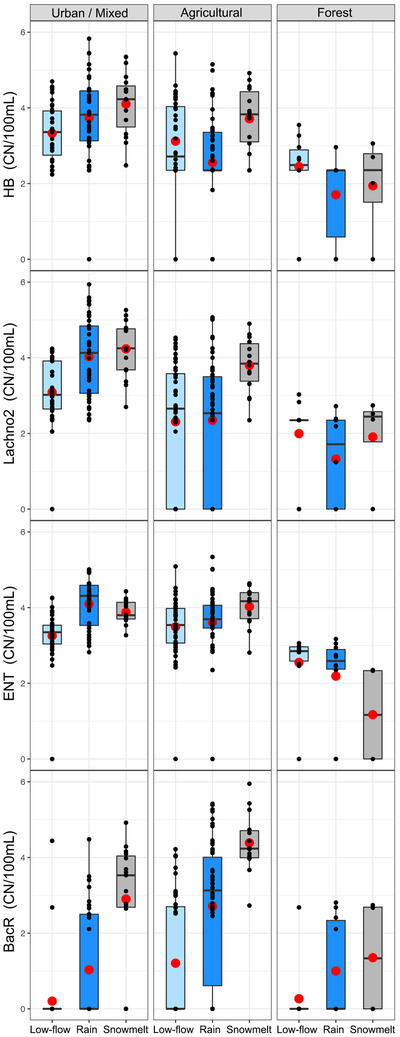
The impact of hydrologic conditions on human-associated, ruminant-associated, and general fecal indicator concentrations was examined by grouping watersheds into three land use categories (agricultural, forest, and urban/mixed; Figure 2). Mean concentrations of all fecal indicators increased significantly up to two orders of magnitude during snowmelt periods compared to low-flow periods in the urban/mixed land use category (p<0.02 for HB, Lachno2, BacR, and ENT). Concentrations of BacR in samples from sites with agricultural land use increased one order of magnitude in samples collected during rainfall-periods and three orders of magnitude in samples from snowmelt-event periods compared to low-flow periods (p < 0.001). In the forested category, mean concentrations of each fecal indicator type were similar in low-flow and both runoff-event conditions.
Figure 2.
Boxplots showing sample distribution for fecal indicator concentrations in low-flow (light-blue), rainfall-event (dark-blue) and snowmelt-event (gray) periods. Characterized by Urban/Mixed (urban land use 30–92%), Agriculture (urban land use 7–11%) and Forest (urban land use 3.8%). Y-axis is log-base10. The center line of each boxplot represents the median, the red dot represents the mean, the upper line represents the 75th percentile, the lower line represents the 25th percentile, the upper whisker represents the maximum, the lower whisker represents the minimum, black dots represent sample distribution. HB = human Bacteroides; Lachno2 = human Lachnospiraceae; ENT = enterococcus; BacR = ruminant Bacteroides.
3.3. Loads and yields of human and ruminant markers from eight watersheds
Human-associated indicator loads were compared among watersheds for runoff-event periods (Table 2). The ~16,000 km2 Maumee watershed (agricultural) had the greatest mean event load for Lachno2 and second greatest for HB. Similar mean loads were measured from the smaller Milwaukee, Clinton, and Rouge watersheds (Table 2). Despite the Maumee having a much smaller percent urban land use, it still had the largest urban acreage due to sheer size of the watershed (Table S2). The median load value reflects a “typical” event, perhaps more so than the mean. The smallest watershed (Rouge) had the greatest median load. When considering loads normalized to drainage area (yields), the mean and the median watershed yields of human fecal indicators from Milwaukee, Clinton, Rouge, and Portage were higher than the Maumee (Table 2).
Table 2.
Study Sites with Median Load and Yield and Mean Load and Yield Calculated for Runoff Eventsa
| Watershed | Drainage Area (km2) | (n) | Statistic | Load HB | Yield HB | Load Lachno2 | Yield Lachno2 | Load BacR | Yield BacR |
|---|---|---|---|---|---|---|---|---|---|
| Rouge River | 484 | 18 | Median Mean |
9.15E+14 1.62E+15 |
3.44E+15 3.84E+15 |
4.51E+10 1.67E+13 |
|||
| Clinton River | 1,901 | 21 | Median Mean |
7.23E+14 1.50E+16 |
3.80E+11 7.88E+12 |
1.04E+15 1.86E+16 |
5.49E+11 9.78E+12 |
2.20E+13 1.03E+15 |
1.16E+10 5.42E+11 |
| Milwaukee River | 2,258 | 13 | Median Mean |
1.77E+14 2.92E+16 |
7.83E+10 1.29E+13 |
2.49E+14 2.50E+16 |
1.10E+11 1.11E+13 |
4.74E+12 1.64E+16 |
2.10E+09 7.26E+12 |
| Raisin River | 2,699 | 12 | Median Mean |
4.51E+13 6.19E+14 |
1.67E+10 2.29E+11 |
6.39E+13 6.03E+14 |
2.37E+10 2.23E+11 |
4.59E+13 3.35E+15 |
1.70E+10 1.24E+12 |
| Maumee River | 16,395 | 18 | Median Mean |
8.15E+14 2.47E+16 |
4.97E+10 1.51E+12 |
2.80E+14 2.93E+16 |
1.71E+10 1.78E+12 |
1.93E+14 5.11E+16 |
1.18E+10 3.12E+12 |
| Portage River | 1,109 | 17 | Median Mean |
5.89E+14 6.21E+15 |
5.31E+11 5.60E+12 |
2.71E+14 6.39E+15 |
2.45E+11 5.77E+12 |
4.41E+14 6.05E+15 |
3.98E+11 5.45E+12 |
| Manitowoc River | 1,362 | 17 | Median Mean |
1.24E+13 4.93E+13 |
9.07E+09 3.62E+10 |
3.55E+13 1.16E+14 |
2.61E+10 8.54E+10 |
4.72E+14 1.67E+16 |
3.46E+11 1.23E+13 |
| Menominee River | 10,179 | 14 | Median Mean |
8.37E+13 2.05E+14 |
8.22E+09 2.02E+10 |
4.84E+13 2.03E+14 |
4.75E+09 2.00E+10 |
8.63E+11 1.31E+14 |
8.47E+07 1.29E+10 |
Color depth highlights drainage area size, going from the largest in dark blue (Maumee) to the smallest in very light blue (Rouge). Some watersheds share a color because of similar drainage area sizes. Units for load are CN (copy number) and yield are CN/km2; n = number of events for each watershed.
For the ruminant-associated indicator, the large Maumee watershed had relatively low cattle density (12.4 cattle/km2) but had the greatest BacR fecal indicator mean load of all of the study sites, likely due to its large area (Table 2). On the other hand, the smaller Manitowoc watershed with a high cattle density (143.2 cattle/km2) had the greatest mean yield (Table 2) of all sites.
3.4. Factors influencing the presence and magnitude of host-associated marker levels
Multiple regression analysis was used to explore factors that explain variability in fecal bacteria flux36. Season, rainfall depth, percent drain tile coverage in the watershed, and population density (human for HB and Lachno2, and cattle for BacR) were all found to be significant contributors for explaining variability of the flux of these host-associated markers (Table 3). Coefficients for these variables were not significantly different than each other except for one variable in one regression: the coefficient for population density was significantly greater than other variables for the Lachno2 regression (Table 3). Significant regressions were developed with human population density (a metric of urbanization) or cattle density for each respective host associated indicators for instances when the data were limited to the urban watersheds for human markers or agricultural watersheds for the ruminant marker; however, adding the percent drain tile coverage in the watershed (a metric of potential connections to waterways) allowed for a significant regression to explain human-associated marker variability with data from all eight watersheds.
Table 3.
Standardized Coefficients for Multiple Regressions used to Explain Variability of Host-Associated Fecal Indicator Bacteria Flux Measured during Periods without Snow at Eight Great Lakes Tributaries, 2011–2013.
| Standardized Coefficients | 95% Confidence intervals | |||
|---|---|---|---|---|
| Host-associated fecal indicator | 2.50% | 97.50% | ||
| Human Lachnospiraceae2 | Population density | 0.22 | 0.18 | 0.26 |
| Rainfall depth | 0.11 | 0.08 | 0.15 | |
| Cosine(decimal day of year) | 0.10 | 0.07 | 0.13 | |
| Sine(decimal day of year) | 0.08 | 0.05 | 0.11 | |
| Tile drainage % | 0.13 | 0.09 | 0.17 | |
| Human Bacteroides | Population density | 0.20 | 0.16 | 0.24 |
| Rainfall depth | 0.08 | 0.05 | 0.11 | |
| Cosine(decimal day of year) | 0.10 | 0.07 | 0.13 | |
| Sine(decimal day of year) | 0.06 | 0.03 | 0.10 | |
| Tile drainage % | 0.13 | 0.10 | 0.17 | |
| Ruminant Bacteroides | Cattle density | 0.08 | 0.03 | 0.13 |
| Rainfall depth | 0.21 | 0.15 | 0.27 | |
| Cosine(decimal day of year) | 0.12 | 0.07 | 0.17 | |
| Sine(decimal day of year) | 0.12 | 0.06 | 0.17 | |
| Tile drainage % | 0.08 | 0.03 | 0.13 | |
3.5. Correlation on bacterial indicator markers and documented viral load
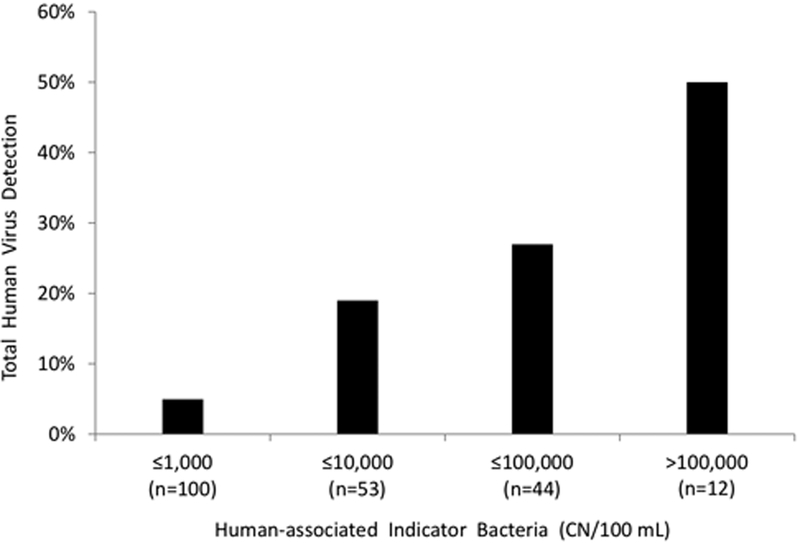
Using previously reported virus data for these sites37, we examined the relationship between viruses and host-associated indicators. In the 2017 Lenaker et al. study, human viruses were detected in 33 of 290 samples from runoff-event (n=189) and low-flow periods (n=101). Virus concentrations were generally low, with only 3 of the 33 samples above 100 genomic copies per L. Human viruses were detected more frequently in samples with high concentrations of human-associated indicator bacteria (Figure 3). Using logistic regression, combined human-associated indicators (HB plus Lachno2) had a relationship to human virus occurrence (five pooled human viruses), where the odds of observing human virus increased by 119% for every 10-fold increase in the human indicator abundance. For the ruminant markers, the odds of observing bovine viruses (a pool of four viruses) increased by 88% for every 10-fold increase in BacR abundance. Over all, bovine virus was detected in 77% of samples with ≥225 CN BacR/100 mL.
Figure 3.
Human-associated fecal indicator concentrations (the sum of HB and Lachno2) grouped into bins and plotted against the percentage of those samples with human virus detection. The human viruses pooled for detection included: adenovirus C, D, F, adenovirus A, norovirus groups I and II, and enterovirus.
4. Discussion
The Great Lakes hold approximately 20% of the Earth’s fresh surface water and functions largely as a closed system, with residence times of 62 years for Lake Michigan and 2.7 years for Lake Erie52. Pollution enters from the watersheds connected to these immense resources, with sewage discharges and overland runoff delivering pathogens, nutrients, and trace organic contaminants. While there are good inventories of impaired rivers in the United States due to FIB concentrations, a holistic assessment of watersheds draining to the Great Lakes is needed to understand how different land uses are related to the concentrations (intensity) and loads (total amounts) of sewage and agricultural pollutants entering the system on a regional basis.
4.1. Host-associated indicators quantify pollution sources
Most information on watershed pollution loads are derived from land use models and monitoring data (i.e. discrete samples) for general indicators53,54. This study used flow weighted, high frequency sampling to capture representative samples of actual loads of sewage and ruminant fecal pollution. These data demonstrated the large extent in which human fecal indicators were found in agricultural watersheds, and the occurrence of ruminant indicators in heavily urbanized watersheds with upstream agricultural land use (Clinton and Milwaukee). Understanding the levels of general fecal indicators that co-occur with these source specific indicators provide benchmarks to the magnitudes of fecal pollution for these sources across multiple watersheds.
In urban environments, some stormwater outfalls have the microbial signature of untreated sewage55. Thus, it is not surprising that human markers are detected in stormwater25,56 and that concentrations in receiving waters increase during runoff-event periods24. A difference between urban and agricultural watersheds was seen in the influence of rainfall and snowmelt on human marker concentration and could be due to the differences in the mechanisms by which sewage enters receiving waters; i.e. sewer pipes versus septic systems. It is possible that faulty septic systems need a build-up period over winter, without storm flushing of leachate, to increase marker concentrations to receiving waters, whereas the infrastructure of urban areas has many leaking sewer pipes that maintain high concentration of human indicators readily available for flushing. Septic tank density has been linked with human fecal pollution23 and in a primarily non-urban Ohio watershed, Peed et al. found human fecal markers strongly associated with wet weather and saw a positive correlation between human markers and septic tank density15.
In this study, sewage pollution was high in the agricultural Portage watershed, which may be reflective of the sampling location that was near a small town with a combined sewage/stormwater system and an outfall upstream of the sampling site. Septic systems near the stream could also account for contributions of human fecal pollution. Sewage releases to the river near the sampling location, or efficient delivery of leeched sewage from septic tanks could be the cause of the consistently high human-associated fecal signal at the Portage site.
In all weather conditions, HB and Lachno2 were closely correlated to each other and found at ratios consistent with what is found in wastewater treatment plant influent8,20,24, suggesting that overall the two human markers behave similarly in the environment. There is little information on the ecology of host-associated fecal indicators in the environment apart from microcosm studies57–59, which is needed to determine the window of time that these indicators would be detectable in the environment. There are many processes that influence the overall concentrations captured by sampling in downstream portions of a watershed. Microorganism survival models often assume a first-order decay60 and recent microcosm studies suggest that genetic markers are reduced by 1–3 orders of magnitude over a 7-day time frame58,61,62. This gives some confidence that genetic markers, which are generally fecal anaerobes, persist long enough to be detected up to a week’s time over the runoff-event. However, concentrations captured over the event may underestimate pollution signals compared to a single-day event and further refinement of estimates are necessary to take into account attenuation of signals over time for watershed comparisons.
4.2. Sewage sources create a human health risk
Quantitative microbial risk assessment modeling has shown that water containing a mix of human and non-human sources of enterococci have a smaller probability of producing gastrointestinal illness in humans compared with enterococci that is exclusively from human sources7,63. Similarly, a study in Singapore64 found predictions of human pathogens improved when human-associated fecal indicators were included as variables along with traditional enterococci and E. coli indicators. We found it difficult to generalize the relationship between human-associated markers and enterococci and E. coli plate-counts, which highlights the non-specific nature of FIB.
Human markers have been previously associated with increased occurrence of human viruses in Milwaukee watershed and nearshore20,25. In this study, the relationship between human markers and viruses was surprisingly consistent with previous work; we reported a 119% increase in the probability of detection of viruses with every 10-fold increase in human marker, and Newton et al. reported 154% increase in the likelihood of adenovirus detection with a 10-fold increase of human marker20. Boehm et al. reported that levels of 4200 CN/100 mL human-Bacteroides65, detected by a similar assay as the HB assay, correlated to a risk of gastroenteritis of 0.03; however, the Boehm study did not concurrently measure pathogens and human markers in samples, but employed published values for pathogen concentrations in sewage. In a subsequent study, estimates of HB levels of 7800 CN/100 mL HB were formulated using untreated sewage samples with both constituents measured in the same sample8.
4.3. Agriculture carries health risks
In the Midwestern region of the U.S., fall and spring are typical times for manure spreading and this likely influenced seasonal increases in BacR concentrations. Spring concentrations were generally higher than fall, and snowmelt had a greater effect on increases in BacR concentrations in most watersheds except for Manitowoc, an agricultural watershed that had the greatest BacR concentrations. Multiple variables are likely linked to high snowmelt concentrations and loads including winter manure spreading, saturated soil conditions during snowmelt periods, reduced UV light exposure due to snow and ice cover, persistence of indicators in colder temperatures, and differences in direct farm runoff vs. manure spreading. In a 2010 study, Soller et al. found that human health risks associated with the presence of fresh cattle feces were not substantially different from risks associated with human fecal sources63. However, according to a USEPA study (2010) that examined the human health risk associated with land-applied manure runoff, the risk was estimated at 25 to 150 times lower than risk from viruses carried by human fecal sources66. For many regions the sampling season should play an important role in assessing ruminant-associated health risk.
4.4. Watershed attributes explain flux variability
A single regression equation for each individual host-associated indicator that included flux data from all watersheds was sufficient to represent the relation with predictor variables for periods of rainfall. This result suggests that some generalizations can be made about influences on the delivery of fecal pollution sources that could be extrapolated to watersheds to inform modeling efforts for watershed assessments such as TMDLs when empirical data are not available. This would refine pollution source and load estimates to help watershed managers prioritize resource management efforts. Uncertainty in such estimates could be determined with targeted validation measurements for host associated indicators and potential model recalibration.
Standardized regression coefficients indicated that all predictor variables were relatively similar in importance of explaining variability of these host-associated markers. Three of the four predictor variables for each regression have relations to hydrology including rainfall, season, and tile drainage. Coefficients for the seasonal variables reflected a peak seasonal contribution in late winter and early spring for human and ruminant indicators. This is typically the time of year in the Great Lakes region when the ground is saturated or frozen and baseflow levels in streams are greatest, leading to efficient runoff mechanisms during rainfall periods67–69. In the mixed land use Milwaukee watershed, high frequency sampling demonstrated the greatest loads and concentrations in spring8. Tile drainage also increases efficiency of watershed hydraulics, rapidly draining water from areas that would typically have a much longer travel time before discharging to surface water. Efficiency in the watershed hydraulics can also lead to efficiency in contaminant transport, including microorganisms70. While the predictor variables used in these regressions were chosen based on likelihood of mechanistic linkage to contaminant transport, these are statistical relations that do not necessarily indicate causation, and validation would be necessary to provide confidence that management actions would be effective in reducing fecal pollution.
4.5. Management Implications
Managing pollution discharge to Areas of Concern in the Great Lakes is an important issue for the Great Lakes states, and remediation for effective load reductions must be weighed against other municipal needs so that managers might identify the best targets for financial investments. Variables that are driven by natural forces are difficult to change, but the impact of tiles, people, and cattle may be altered by thoughtful design of infrastructure investments or runoff management practices. TMDLs are based on modeling studies of general indicators, but closer examination of modeling estimates using source specific fecal indicator information could improve remediation efforts. While pathogen contamination is a major concern associated with fecal sources, nutrients and trace organic contaminants, like antibiotics and pharmaceuticals, are also present16 and therefore host-associated fecal indicators may serve as a proxy for other constituents that enter the Great Lakes. Further work is needed to understand how different pollutants of concern are coupled and identify where there are co-benefits for certain management strategies. Results from the current study provide unique information that may be used to assist in these complex watershed management tasks.
Supplementary Material
Acknowledgements
Support of this research was provided by the Graham Sustainability Institute Water Center at University of Michigan (grant number N017027), the Great Lakes Restoration Initiative (contract number DW-014–92453901), and National Institutes of Health (grant number R01AI091829). Thanks to colleagues in the Michigan, Ohio and Wisconsin U.S. Geological Survey Water Science Centers.
Footnotes
ASSOCIATED CONTENT
*S Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.est.8b01945.
Site locations and land use; qPCR primers and standard curves; plate count and qPCR results for all samples; Spearman correlation coefficients for ENT or Lachno2 for all sites; percentage of enterococci and E. coli over water quality standards for each site; watershed land use map; boxplots of fecal markers concentrations at each site in various weather conditions (PDF)
Any use of trade, product or firm names is for descriptive purposes only and does not imply endorsement by the U.S. Government.
The authors declare no competing financial interest.
Citations:
- (1).Mac Kenzie WR; Hoxie NJ; Proctor ME; Gradus MS; Blair KA; Peterson DE; Kazmierczak JJ; Addiss DG; Fox KR; Rose JB A Massive Outbreak in Milwaukee of Cryptosporidium Infection Transmitted through the Public Water Supply. N. Engl. J. Med 1994, 331 (3), 161–167. [DOI] [PubMed] [Google Scholar]
- (2).NOAA. About Our Great Lakes: Great Lakes Basin Facts https://www.glerl.noaa.gov/education/ourlakes/facts.html.
- (3).Swackhamer DL The Past, Present, and Future of the North American Great Lakes: What Lessons Do They Offer? J. Environ. Monit 2005, 7 (6), 540–544. [DOI] [PubMed] [Google Scholar]
- (4).U.S. EPA. National Summary of Impaired Waters and TMDL Information http://iaspub.epa.gov/waters10/attains_nation_cy.control?p_report_type=T#causes_303d.
- (5).U.S. EPA. National Water Quality Inventory: 2004 Report to Congress; United States Environmental Protection Agency, 2009. [Google Scholar]
- (6).Boehm AB; Ashbolt NJ; Colford JM; Dunbar LE; Fleming LE; Gold MA; Hansel JA; Hunter PR; Ichida AM; McGee CD; et al. A Sea Change Ahead for Recreational Water Quality Criteria. J. Water Health 2009, 7 (1), 9–20. [DOI] [PubMed] [Google Scholar]
- (7).Soller JA; Schoen ME; Varghese A; Ichida AM; Boehm AB; Eftim S; Ashbolt NJ; Ravenscroft JE Human Health Risk Implications of Multiple Sources of Faecal Indicator Bacteria in a Recreational Waterbody. Water Res. 2014, 66, 254–264. [DOI] [PubMed] [Google Scholar]
- (8).McLellan SL; Sauer EP; Corsi SR; Bootsma MJ; Boehm AB; Spencer SK; Borchardt MA Sewage Loading and Microbial Risk in Urban Waters of the Great Lakes. Elementa 2018, 6 (46). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Drayna P; McLellan SL; Simpson P; Li S-H; Gorelick MH Association between Rainfall and Pediatric Emergency Department Visits for Acute Gastrointestinal Illness. Environ. Health Perspect. 2010, 118 (10), 1439–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Curriero FC; Patz JA; Rose JB; Lele S The Association between Extreme Precipitation and Waterborne Disease Outbreaks in the United States, 1948–1994. Am. J. Public Health 2001, 91 (8), 1194–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Fong T-T; Lipp EK Enteric Viruses of Humans and Animals in Aquatic Environments: Health Risks, Detection, and Potential Water Quality Assessment Tools. Microbiol. Mol. Biol. Rev 2005, 69 (2), 357–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Corsi SR; Borchardt MA; Spencer SK; Hughes PE; Baldwin AK Human and Bovine Viruses in the Milwaukee River Watershed: Hydrologically Relevant Representation and Relations with Environmental Variables. Sci. Total Environ 2014, 490, 849–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Haile RW; Witte JS; Gold M; Cressey R; McGee C; Millikan RC; Glasser A; Harawa N; Ervin C; Harmon P; et al. The Health Effects of Swimming in Ocean Water Contaminated by Storm Drain Runoff. Epidemiology 1999, 10 (4), 355–363. [PubMed] [Google Scholar]
- (14).Lee D-Y; Lee H; Trevors JT; Weir SC; Thomas JL; Habash M Characterization of Sources and Loadings of Fecal Pollutants Using Microbial Source Tracking Assays in Urban and Rural Areas of the Grand River Watershed, Southwestern Ontario. Water Res. 2014, 53, 123–131. [DOI] [PubMed] [Google Scholar]
- (15).Peed LA; Nietch CT; Kelty CA; Meckes M; Mooney T; Sivaganesan M; Shanks OC Combining Land Use Information and Small Stream Sampling with PCR-Based Methods for Better Characterization of Diffuse Sources of Human Fecal Pollution. Environ. Sci. Technol 2011, 45 (13), 5652–5659. [DOI] [PubMed] [Google Scholar]
- (16).Baldwin AK; Corsi SR; De Cicco LA; Lenaker PL; Lutz MA; Sullivan DJ; Richards KD Organic Contaminants in Great Lakes Tributaries: Prevalence and Potential Aquatic Toxicity. Sci. Total Environ 2016, 554–555, 42–52. [DOI] [PubMed] [Google Scholar]
- (17).Newton RJ; Bootsma MJ; Morrison HG; Sogin ML; McLellan SL A Microbial Signature Approach to Identify Fecal Pollution in the Waters off an Urbanized Coast of Lake Michigan. Microb. Ecol 2013, 65 (4), 1011–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Bosch A Human Enteric Viruses in the Water Environment: A Minireview. Int. Microbiol 1998, 1 (3), 191–196. [PubMed] [Google Scholar]
- (19).Lipp EK; Farrah SA; Rose JB Assessment and Impact of Microbial Fecal Pollution and Human Enteric Pathogens in a Coastal Community. Mar. Pollut. Bull 2001, 42 (4), 286–293. [DOI] [PubMed] [Google Scholar]
- (20).Newton RJ; Vandewalle JL; Borchardt MA; Gorelick MH; McLellan SL Lachnospiraceae and Bacteroidales Alternative Fecal Indicators Reveal Chronic Human Sewage Contamination in an Urban Harbor. Appl. Environ. Microbiol 2011, 77 (19), 6972–6981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Sedmak G; Bina D; MacDonald J Assessment of an Enterovirus Sewage Surveillance System by Comparison of Clinical Isolates with Sewage Isolates from Milwaukee, Wisconsin, Collected August 1994 to December 2002. Appl. Environ. Microbiol 2003, 69 (12), 7181–7187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Jamieson R; Gordon R; Joy D; Lee H Assessing Microbial Pollution of Rural Surface Waters: A Review of Current Watershed Scale Modeling Approaches. Agric. Water Manag 2004, 70 (1), 1–17. [Google Scholar]
- (23).Verhougstraete MP; Martin SL; Kendall AD; Hyndman DW; Rose JB Linking Fecal Bacteria in Rivers to Landscape, Geochemical, and Hydrologic Factors and Sources at the Basin Scale. Proc. Natl. Acad. Sci 2015, 201415836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Templar HA; Dila DK; Bootsma MJ; Corsi SR; McLellan SL Quantification of Human-Associated Fecal Indicators Reveal Sewage from Urban Watersheds as a Source of Pollution to Lake Michigan. Water Res. 2016, 100, 556–567. [DOI] [PubMed] [Google Scholar]
- (25).Sauer EP; VandeWalle JL; Bootsma MJ; McLellan SL Detection of the Human Specific Bacteroides Genetic Marker Provides Evidence of Widespread Sewage Contamination of Stormwater in the Urban Environment. Water Res. 2011, 45 (14), 4081–4091. [DOI] [PubMed] [Google Scholar]
- (26).McLellan SL; Hollis EJ; Depas MM; Van Dyke M; Harris J; Scopel CO Distribution and Fate of Escherichia Coli in Lake Michigan Following Contamination with Urban Stormwater and Combined Sewer Overflows. J. Great Lakes Res 2007, 33 (3), 566–580. [Google Scholar]
- (27).Bravo HR; McLellan SL; Klump VJ; Hamidi SA; Talarczyk D Modeling the Fecal Coliform Footprint in a Lake Michigan Urban Coastal Area. Environ. Sci. Technol 2016, 95, 401–419. [Google Scholar]
- (28).Olds HT; Corsi SR; Dila DK; Halmo KM; Bootsma MJ; McLellan SL High Levels of Sewage Contamination Released from Urban Areas after Storm Events: A Quantitative Survey with Sewage Specific Bacterial Indicators. PLoS Med. 2018, 15 (7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Steele JA; Blackwood AD; Griffith JF; Noble RT; Schiff KC Quantification of Pathogens and Markers of Fecal Contamination during Storm Events along Popular Surfing Beaches in San Diego, California. Water Res. 2018, 136, 137–149. [DOI] [PubMed] [Google Scholar]
- (30).Jarde E; Communal PY; Jaffrezic A; Pourcher AM; Derrien M Development of Microbial and Chemical MST Tools to Identify the Origin of the Faecal Pollution in Bathing and Shellfish Harvesting Waters in France. 2010, 4. [DOI] [PubMed] [Google Scholar]
- (31).Jin S; Yang L; Danielson P; Homer C; Fry J; Xian G A Comprehensive Change Detection Method for Updating the National Land Cover Database to circa 2011. Remote Sens. Environ 2013, 132, 159–175. [Google Scholar]
- (32).USDA. Natural Resources Conservation Service and Statistical Laboratory; Ames, Iowa, 1995. [Google Scholar]
- (33).USDA. 2012 Census of agriculture. http://www.agcensus.usda.gov/Publications/2012/.
- (34).USDA-NRCS; USGS; USEPA. The Watershed Boundary Dataset (WBD) (Vector Digital Data).; Fort Worth, Texas, 2009. [Google Scholar]
- (35).USCB-Geography Division. 2010 State-Based Census Block TIGER/Line Shapefile; Michigan; Ohio; Wisconsin, 2010. [Google Scholar]
- (36).Corsi SR; Dila DK; Lenaker PL; Baldwin AK; Bootsma MJ; McLellan SL Regression models and associated data for describing variability of host specific bacteria fluxes in eight Great Lakes tributaries, 2011–2013: U.S. Geological Survey data release; 10.5066/F7VX0DRH. [DOI] [Google Scholar]
- (37).Lenaker PL; Corsi SR; Borchardt MA; Spencer SK; Baldwin AK; Lutz MA Hydrologic, Land Cover, and Seasonal Patterns of Waterborne Pathogens in Great Lakes Tributaries. Water Res. 2017, 113, 11–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).EPA US. Method 1603: Escherichia coli (E. coli) in Water by Membrane Filtration Using Modified Membrane-Thermotolerant Escherichia coli Agar (Modified MTEC), EPA-821-R-02–023 U.S. Environmental Protection Agency, 2002. [Google Scholar]
- (39).EPA US. Method 1600: Enterococci in Water by Membrane Filtration Using Membrane-Enterococus Indoxyl-B-D-Glucoside Agar (MEI), EPA-821-R-06–009; U.S. Environmental Protection Agency, 2006. [Google Scholar]
- (40).Kildare BJ; Leutenegger CM; McSwain BS; Bambic DG; Rajal VB; Wuertz S 16S RRNA-Based Assays for Quantitative Detection of Universal, Human-, Cow-, and Dog-Specific Fecal Bacteroidales: A Bayesian Approach. Water Res. 2007, 41 (16), 3701–3715. [DOI] [PubMed] [Google Scholar]
- (41).Bernhard AE; Field KG Identification of Nonpoint Sources of Fecal Pollution in Coastal Waters by Using Host-Specific 16S Ribosomal DNA Genetic Markers from Fecal Anaerobes. Appl. Environ. Microbiol 2000, 66 (4), 1587–1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Reischer GH; Kasper DC; Steinborn R; Mach RL; Farnleitner AH Quantitative PCR Method for Sensitive Detection of Ruminant Fecal Pollution in Freshwater and Evaluation of This Method in Alpine Karstic Regions. Appl. Environ. Microbiol 2006, 72 (8), 5610–5614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Reischer GH; Ebdon JE; Bauer JM; Schuster N; Ahmed W; Åström J; Blanch AR; Blöschl G; Byamukama D; Coakley T; et al. Performance Characteristics of QPCR Assays Targeting Human- and Ruminant-Associated Bacteroidetes for Microbial Source Tracking across Sixteen Countries on Six Continents. Environ. Sci. Technol 2013, 47 (15), 8548–8556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Ludwig W; Schleifer KH How Quantitative Is Quantitative PCR with Respect to Cell Counts? Syst. Appl. Microbiol 2000, 23 (4), 556–562. [DOI] [PubMed] [Google Scholar]
- (45).Harwood VJ; Staley C; Badgley BD; Borges K; Korajkic A Microbial Source Tracking Markers for Detection of Fecal Contamination in Environmental Waters: Relationships between Pathogens and Human Health Outcomes. FEMS Microbiol. Rev 2014, 38 (1), 1–40. [DOI] [PubMed] [Google Scholar]
- (46).Jeter SN; McDermott CM; Bower PA; Kinzelman JL; Bootsma MJ; Goetz GW; McLellan SL Bacteroidales Diversity in Ring-Billed Gulls (Laurns Delawarensis) Residing at Lake Michigan Beaches. Appl. Environ. Microbiol 2009, 75 (6), 1525–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Mayer RE; Reischer G; Ixenmaier SK; Derx J; Blaschke AP; Ebdon JE; Linke R; Egle L; Ahmed W; Blanch A; et al. Global Distribution of Human-Associated Fecal Genetic Markers in Reference Samples from Six Continents. Environ. Sci. Technol 2018, acs.est.7b04438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).QGIS Development Team. Quantum GIS Geographic Information System; Open Source Geospatial Foundation Project, 2016. [Google Scholar]
- (49).Therneau T A Package for Survival Analysis in S, version 2.38; Mayo Foundation, 2015. [Google Scholar]
- (50).R Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria, 2015. [Google Scholar]
- (51).Team RStudio. RStudio: Integrated Development for R. RStudio, Inc.: Boston, 2016. [Google Scholar]
- (52).Quinn FL Hydraulic Residence Times for the Laurentian Great Lakes. J. Great Lakes Res 1992, 18 (1), 22–28. [Google Scholar]
- (53).Gonzalez RA; Noble RT Comparisons of Statistical Models to Predict Fecal Indicator Bacteria Concentrations Enumerated ByqPCR- and Culture-Based Methods. Water Res. 2014, 48 (1), 296–305. [DOI] [PubMed] [Google Scholar]
- (54).Wolfand J; Bell CD; Boehm AB; Hogue T; Luthy RG Multiple Pathways to Bacterial Load Reduction by Stormwater Best Management Practices: Tradeoffs in Performance, Volume, and Treated Area. Environ. Sci. Technol 2018, 52, 6370–6379. [DOI] [PubMed] [Google Scholar]
- (55).Fisher JC; Newton RJ; Dila DK; McLellan SL Urban Microbial Ecology of a Freshwater Estuary of Lake Michigan. Elem. Sci. Anthr 2015, 3, 000064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Sercu B; Van De Werfhorst LC; Murray JLS; Holden PA Sewage Exfiltration as a Source of Storm Drain Contamination during Dry Weather in Urban Watersheds. Environ. Sci. Technol 2011, 45, 7151–7157. [DOI] [PubMed] [Google Scholar]
- (57).Dick LK; Stelzer EA; Bertke EE; Fong DL; Stoeckel DM Relative Decay of Bacteroidales Microbial Source Tracking Markers and Cultivated Escherichia coli in Freshwater Microcosms. Appl. Environ. Microbiol 2010, 76 (10), 3255–3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Bae S; Wuertz S Survival of Host-Associated Bacteroidales Cells and Their Relationship with Enterococcus Spp., Campylobacter Jejuni, Salmonella Enterica Serovar Typhimurium, and Adenovirus in Freshwater Microcosms as Measured by Propidium Mono. Appl. Environ. Microbiol 2012, 78 (4), 922–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Jeanneau L; Solecki O; Wéry N; Jardé E; Gourmelon M; Communal PY; Jadas-Hécart A; Caprais MP; Gruau G; Pourcher AM Relative Decay of Fecal Indicator Bacteria and Human-Associated Markers: A Microcosm Study Simulating Wastewater Input into Seawater and Freshwater. Environ. Sci. Technol 2012, 46 (4), 2375–2382. [DOI] [PubMed] [Google Scholar]
- (60).Benham B; Baffaut C; Zeckoski R; Mankin K; Pachepsky Y; Sadeghi A; Brannan K; Soupir M; Habersack M Modeling Bacteria Fate and Transport in Watersheds to Support TMDLs. Trans. ASABE 2006, 49 (4), 987–1002. [Google Scholar]
- (61).Korajkic A; McMinn BR; Shanks OC; Sivaganesan M; Fout GS; Ashbolt NJ Biotic Interactions and Sunlight Affect Persistence of Fecal Indicator Bacteria and Microbial Source Tracking Genetic Markers in the Upper Mississippi River. Appl. Environ. Microbiol 2014, 80 (13), 3952–3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Sassoubre LM; Yamahara KM; Boehm AB Temporal Stability of the Microbial Community in Sewage-Polluted Seawater Exposed to Natural Sunlight Cycles and Marine Microbiota. Appl. Environ. Microbiol 2015, 81 (6), 2107–2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Soller JA; Bartrand T; Ashbolt NJ; Ravenscroft J; Wade TJ Estimating the Primary Etiologic Agents in Recreational Freshwaters Impacted by Human Sources of Faecal Contamination. Water Res. 2010, 44 (16), 4736–4747. [DOI] [PubMed] [Google Scholar]
- (64).Liang L; Goh SG; Vergara GGRV; Fang HM; Rezaeinejad S; Chang SY; Bayen S; Lee WA; Sobsey MD; Rose JB; et al. Alternative Fecal Indicators and Their Empirical Relationships with Enteric Viruses, Salmonella Enterica, and Pseudomonas Aeruginosa in Surface Waters of a Tropical Urban Catchment. Appl. Environ. Microbiol 2015, 81 (3), 850–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Boehm AB; Soller JA; Shanks OC Human-Associated Fecal Quantitative Polymerase Chain Reaction Measurements and Simulated Risk of Gastrointestinal Illness in Recreational Waters Contaminated with Raw Sewage. Environ. Sci. Technol. Lett 2015, 2 (10), 270–275. [Google Scholar]
- (66).EPA US. Report on 2009 National Epidemiologic and Environmental Assessment of Recreational Water Epidemiology Studies; U.S. Environmental Protection Agency, 2010. [Google Scholar]
- (67).Hollinger SE; Isard SA A Soil Moisture Climatology of Illinois. J. Clim 1994, 7 (5), 822–833. [Google Scholar]
- (68).Sinha T; Cherkauer KA; Mishra V Impacts of Historic Climate Variability on Seasonal Soil Frost in the Midwestern United States. J. Hydrometeorol 2010, 11 (2), 229–252. [Google Scholar]
- (69).Wang D; Cai X Comparative Study of Climate and Human Impacts on Seasonal Baseflow in Urban and Agricultural Watersheds. Geophys. Res. Lett 2010, 37 (6), 1–6. [Google Scholar]
- (70).Jamieson RC; Gordon RJ; Sharples KE; Stratton GW; Madani A Movement and Persistence of Fecal Bacteria in Agricultural Soils and Subsurface Drainage Water: A Review. Can. Biosyst. Eng 2002, 44, 1.1–1.9. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.