Abstract
A promoter can be regulated by various cis-acting elements so that delineation of the regulatory modes among them may help understand developmental, environmental and genetic mechanisms in gene activity. Here we report that the human dopamine transporter gene SLC6A3 carries a 5’ distal 5 kb super enhancer (5KSE) which upregulated the promoter by 5-fold. Interestingly, 5KSE is able to prevent 3’ downstream variable number tandem repeats (VNTRs) from silencing the promotor. This new enhancer consists of a 5’ VNTR and three repetitive sub-elements those are conserved in primates. Two of 5KSE’s sub-elements, E-9.7 and E-8.7, upregulate the promoter but only the later could continue doing so in the presence of VNTRs. Finally, E-8.7 is activated by novel dopaminergic transcription factors including SRP54 and Nfe2l1. Together, these results reveal a multimodal regulatory mechanism in SLC6A3.
Keywords: epistasis, genetic marker, haplotype, primate-specific, transactivation
Introduction
Besides insulators which play an intervening role, cis-acting elements within a gene can be grouped as activating (enhancer) and silencing (repressor) in terms of direct effects on promoter activity[1–3]. It is unknown whether enhancers and silencers interact with each other in transcriptional regulation. Since these cis-elements usually carry polymorphisms[4,5], dissection of potential cis-interplays may provide effective association markers and identify underlying genetic risk factors.
The dopamine transporter (DAT) is a principal regulator of dopamine transmission in the brain[6–8]. Regulated transcription of the human DAT gene (SLC6A3: OMIM# 126455, cytogenetic location: 5p15.33, genomic coordinates (GRCh38): 5:1,392,789–1,445,427) determines spatial, temporal and quantitative aspects of DAT expression in the brain and therefore affects many dopamine-related function including reward, memory, movement, cognition and behavioral organization. Abnormal regulation may also cause pathological states implicated in a spectrum of neuropsychiatric disorders such as substance use disorders, schizophrenia, bipolar disorder, Parkinson’s disease, and ADHD[9]. It is urgent to uncover cis-acting elements in SLC6A3 for two main reasons. The first reason is that identification of these elements may guide us to clone the transcription factors (TFs) for signaling pathway dissection; another reason is that DNA sequence polymorphisms in these functional elements may serve as effective markers for genetic association study.
SLC6A3 spans 70 kb in the human chromosome 5 but only a little is known about its cis-acting activity. The best known are two downstream variable number tandem repeats (VNTRs), one in the 3’ untranslated region (3’VNTR)[10] and another, in Intron 8 (In8VNTR)[11]. 3’VNTR has 7–11 repeats of 40 bp and In8VNTR has 5–8 repeats of 30 bp and both have been shown to display inhibitory activity and used as genetic markers in association studies[12,13]. It has been reported that the TF Hey1 binds to 3’VNTR[14] and there is no information about what proteins recognize Int8VNTR.
The human promoter remains underexploited[15]. This 5’ promoter has a 16 kb DNA sequence with hundreds of polymorphisms among various populations (1,252 subjects used by the 1000 Genomes Project) [4,5] but unfortunately no specific cis-acting elements have been identified for the human promoter yet, especially in the 5’ distal regions. We and others have found a 121 bp cis-acting element located in Intron 1 as an inhibitory activity[16,17] and like Int8VNTR, no proteins are known to mediate the inhibition except an lncRNA molecule[18]. A few TFs are known to regulate the human core promoter, such as SP1/3, PITX3 and NURR1, but the target sequences are largely unknown[19,20,15,21]. Such paucity of transcription information in this critical brain gene prevents us from a better understanding of how SLC6A3 contributes to the regulation of brain neurochemistry and genetic etiology of related neuropsychiatric disorders.
In this study, we discovered a 5 kb super enhancer (5KSE) located in a distal promoter region that antagonizes the downstream VNTRs and further cloned TFs that regulated 5KSE.
Methods
Plasmid construction
F plasmid-based luciferase reporters of SLC6A3 promoters, 18A, 18ALL, 18B and 18BLL, have been described before[13]. Deletion of a 5 kb (4,899 bp) NheI fragment from 18A, 18ALL, 18B and 18BLL resulted in 10A, 10ALL, 10B and 10BLL.
Cloning of a 352 bp fragment (which was located at −8.7 kb and termed as E-8.7 whose sequence is listed below) from the 5 kb of haplotype A into the NheI site of 10A or 10ALL resulted in 10A_E-8.7 or 10ALL_E-8.7. Similarly, cloning of E-10.3 and E-9.7 resulted in 10A_E-10.3 or 10A_E-9.7. The PfuUltra High-fidelity DNA Polymerase (Agilent Technologies Inc, Santa Clara, CA, USA)-based PCR used the primers 2.6F aaaaaaagctagcgtaagttaaaaatacacatagt and 2.6R aaaaaaagctagcgagggggtcttactatgtgt for E-10.3; 3.2F ttttttgctagctgggcacagtggctcct and 3.2R ttttttgctagctttttgttttgtttttttatgaga for E-9.7; 4.2F ttttttgctagcggctggctaggtgcagatt and 4.2R ttttttgctagctttttcttttcttttctttgagat (synthesized by IDT, Skokie, IL, USA; NheI site: underlined) for E-8.7 to amplify the fragments, followed by NheI-digestion, cloning back to 10A or 10ALL and verification for correct orientation and PCR fidelity by DNA re-sequencing.
ggctggcta ggtgcagatt aactccctta aatgtaagaa aagtgaatgg gggccGgacg cagtggctca tgcctgtaat cccaacactt tgggaggctg aggcatgtag atcacctaag gttaggagtt ccagaccagt gtggccatca cagtgaaacc ctgtctctac taagaatatg aaaattagct gggtgtggtg gtgcacgcct gtaatcccag gtactctgga ggctgaggca agagaattgc ctgaacccag gaagcagaag ttgcagagag ctgagatcgc accactgcac tccagcctgg gcaacagagt gagactccat ctcaaagaaa agaaaagaaa aag (bold, E-8.7; underline, PCR primers; upper case, an rs1478435 allele)
Human SRP54 cDNA clone from Origene Inc (SC118182 for variant 1, NM_003136) was switched from vector pCMV6-XL5 into pcDNA3.1+ by NotI site for pcDNA3.1+SRP54 where insert orientation was verified by NheI digestion. Human Nfe2l1 (2,342 bp), Slitrk2 (2,558 bp) or Skp1 (527 bp) cDNA was cloned by PfuUltra High-fidelity DNA Polymerase-based PCR and ligated into the indicated sites in pcDNA3.1+, resulting pcDNA3.1+Nfe2l1, pcDNA3.1+Slitrk2 and pcDNA3.1+Skp1. Nfe2l1 PCR used the primers tttttgctagcttcagcaatgctttctctgaag and tttttgcggccgccttcttccccaggctcactt, Slitrk2 PCR used the primers aaaaagctagcccgaaggtgcctaaagatgctga and aaaaagcggccgcccttcacagctgactgattgca, Skp1 PCR used the primers tttttgctagcgtctccttaacaccgaacacca and tttttgcggccgccaggcacaacatttcacttctctt all for NheI/NotI sites. PCR templates used were cDNA synthesized by using mixed human RNAs isolated from human SK-N-AS, IMR-32 and SH-SH5Y cells combined[13]. PCR fidelity was verified by DNA re-sequencing.
Cell Culture
SK-N-AS and SN4741 were cultured in Dulbecco’s modified eagle medium (DMEM, ATCC, VA, USA) containing 10% Fetal Bovine Serum (FBS, Atlanta biologicals, GA, USA) in a 75 cm2 Falcon flask (Thermo Fisher Scientific, Waltham, MA, USA) at 37°C in humidified air with 5% CO2. SN4741 cells were cultured at 32oC for maintenance and at 37oC for gene expression after transfection.
Cloning of transcription factors (TFs) via Yeast One Hybrid System (Y1H)
The 352 bp E-8.7 fragment of haplotype A was used as bait after cloning to pB300/pASZ_Y1H as the vector[22], and a human adult brain cDNA library was constructed into pP6, a pGADGH derivative plasmid, both used in Y1H in search for proteins that bound to E-8.7 (Hybrigenics Services, Paris, France). The bait construct was checked by sequencing the entire insert and transformed into the yeast strain Y1H300 (MAT a, Gal4Δ, ade2–101, trp1–901, leu2–3,112, his3Δ200) to integrate the DNA bait into the yeast genome. Screening was performed against the random-primed cDNA library. Clones were screened using a mating approach with YHGX13 (Y187 ade2–101::loxP-kanMX-loxP, matα) and the E-8.7-containing yeast (mata) strain as previously described[23]. His+ colonies were selected on a medium lacking adenine, leucine and histidine. The prey fragments of the positive clones were amplified by PCR and sequenced at their 5’ and 3’ junctions. The resulting sequences were used to identify the corresponding interacting proteins in the GenBank database (NCBI).
Cell transfection for luciferase (Luc) activity assay and RNA extraction
For Luc activity assay, SN4741 cells in each well of a 24 well plate were transfected with 400 ng cDNA plasmid DNA of pcDNA3.1+ or a cDNA clone, using Superfect Transfection Reagent (Qiagen, Germantown, MD, USA) following the manufacturers’ procedures. After overnight incubation, 400 ng reporter plasmid DNA of the vector pGL3eBleo1, 18A, 10A, 10A_E-10.3, 10A_E-9.7, 10A_E-8.7, 10ALL_E-9.7, or 10ALL_E-8.7 Luc plasmid was further introduced by transfection. Twenty four hrs later, the cells were harvested and lysed with firefly luciferase buffer; Luc activity was analyzed using Luciferase Assay System (Promega, Madison, WI, USA) according to the manufacturer’s instructions, and read out in a Synergy HT luminometer (BioTek, Winooski, VT, USA). Cell density was estimated by protein quantification by using a BCA (Bicinchoninic acid) kit. Luc activity was a result of normalizing the readout by protein amount.
For effects of SRP54 or Nfe2l1 overexpression on endogenous SLC6A3 expression, SK-N-AS or BE(2)-M17 cells were cultured in a 24-well plate for a confluency level of about 80% next day. For each well, 400 ng plasmid DNA was mixed with 20µL DMEM, then 2.4 µL SuperFect Transfection Reagent was added into DNA solution. The tube was vortexed for 10s, and then was incubated at room temperature (RT) for 10 min. After that 190µL fresh complete medium was added into DNA-Superfect complex. The liquid in each well was aspirated before adding 200µl of the complex to each well. DNA-Superfect complex solution was changed with 400µL fresh complete medium in 2–3hrs. Cells were incubated overnight before RNA purification.
Quantitative reverse transcription PCR (qRT-PCR)
Cells of each well in a 24 well plate were extracted for total RNA in TRIZOL (Invitrogen, CA, USA) following the TRIZOL User Guide, and RNA was quantified by NanoDrop Lite (Thermo Fisher Scientific, Waltham, MA, USA). 0.6 µg RNA was subjected to cDNA synthesis in a volume of 10 µL, by using the Verso cDNA synthesis kit (Thermo Fisher Scientific, Waltham, MA, USA). All PCR reactions were performed using SsoAdvanced Universal SYBR green supermix (BIO-RAD, Hercules, California USA). Primers were selected based on an amplification coefficient (AC) of near “2” and single peak in melting curve. Fold differences in mRNA expression were calculated as per AC(ΔCTtest - ΔCTcontrol) where GAPDH was used as an internal control.
Immunofluorescent staining of cells, human postmortem midbrain tissue and mouse brain
For cell staining, SN-K-AS or SN4741 cells were cultured on coverslip until 80% confluence, fixed in 4% paraformaldehyde (PFA) in PBS for 15 min, and washed three times for 5 min each time with 0.01 M PBS. The cells were incubated with 3% bovine serum albumin (BSA) at 37°C for 30 min. After incubated at 4°C with rabbit anti-NFE2L1 (sc721) 1:50 dilution or rabbit antiSRP54 (AB154796) at 1:100 dilution antibodies in 0.03% Triton X-100 PBS for 24 hrs, the cells were washed three times for 5 min with 0.01 M PBS and then incubated with Alexa Fluor 568 goat anti-rabbit IgG (1:500, Life technologies, NY, USA) at RT for 2 hrs in darkness. Slides were mounted with VECTASHIELD anti-fade mounting medium containing DAPI stain (Vector Laboratories, Burlingame, CA, USA) for labeling cell nuclei. Images were captured in confocal laser scanning microscopy (Leica TCS SP8, IL, USA).
For frozen human nigral midbrain tissue which was from healthy controls as described before[24], 10 µm sections were collected on Superfrost Plus Gold slides (Fisher Scientific USA), fixed in 4% PFA for 30 min and rinsed 3 times with PBS and then incubated with 3% BSA at 37°C for 30 min. After incubated at 4°C with the same primary rabbit antibodies as for the cells, together with chicken anti-tyrosine hydroxylase (TH; 1:1000; Abcam, Cambridge, MA, USA) antibodies, the sections were washed three times for 5 min with 0.01 M PBS and then incubated with Alexa Fluor 568 goat anti-rabbit IgG (1:500, Life technologies) and Alexa Fluor 488 goat anti-chicken IgY (1:500, Abcam) at RT for 2 hrs in darkness. Slides were mounted with VECTASHIELD anti-fade mounting medium containing DAPI stain. For the control experiments, the same protocols were applied, but the SRP54 or Nfe2l1 primary antibodies were substituted by normal rabbit IgG. No positive immunolabeling was observed in any sections in which the primary antibody was substituted. Use of de-identified human postmortem tissue was approved by McLean Hospital IRB Committee.
For mouse midbrain, C57BL/6 adult male brains were fixed in 4% PFA in PBS for 20 hrs, and cryoprotected in 30% (g/v) sucrose overnight prior to embedding in OCT. Frozen sections were coronally collected at 40 µm-thickness. Midbrain sections containing the substantia nigra and the ventral tegmental area were kept at 4°C in 0.01 M PBS and processed for floating immunofluorescence staining and image capture through the same procedures as for the on-slide human postmortem sections.
Chromatin Immunoprecipitation
One million SN4741 or SK-N-AS cells were used for each ChIP reaction. For mouse tissue, 4 mg of VTA tissue was used for each ChIP. Cells were crosslinked using 1% formaldehyde at RT for 10 min; tissue was homogenized with a motor pellet-pestle before the crosslinking. Crosslinked sample was lysed in lysis buffers (1% SDS, 10mM EDTA-pH 8, 50 mM Tris-HCl pH 8). The DNA was then sheared with Diagenode Bioruptor Sonicator (Denville, NJ, USA) for 12 min. The lysate was precleared with Protein G magnetic beads (GenScript, Piscataway, NJ, USA) at 4°C for 1 hr. A total of 4 µg of rabbit anti-NFE2L1 (sc721) or anti-Srp54 (AB154796) was added and incubated at 4°C overnight with gentle rotation. Next day, 80 µL of protein G magnetic beads were added to each sample and incubated at 4°C for 2 hrs. The beads were eluted and elute was incubated with 200 mM NaCl at 65°C for 5 hrs to reverse the cross-linking, followed by treatment with Proteinase K (Cat no: 19131, Qiagen) at 45°C for 2 hrs to digest proteins. The treated DNA was purified by using QIAquick PCR Purification Kit (Cat: 28104, Qiagen). Protein-DNA binding activity was examined by PCR using primers 5’-tgtggccatcacagtgaaa-3’ and 5’-cttcctgggttcaggcaa-3’, PCR products resolved by 1.5%-agarose electrophoresis and visualized via ethidium bromide staining and UV illumination.
Experimental Design and Statistical Analysis
All numeric data were analyzed statistically using algorithms implemented in GraphPad Prism 5 software. Student’s t tests were used for comparing two conditions and ANOVA was used with Bonferroni post hoc tests for multiple comparisons. All data are expressed as means ± SEM. The statistical significance level was defined as p<0.05: * for p<0.05, ** for p<0.01 and *** for p<0.001.
Results
Identification of 5KSE in SLC6A3
During a fresh search for cis-acting elements in the human SLC6A3, we found that deletion of a 5 kb (4,899 bp, from −8,329 bp to −13,227 bp, assuming “+1” at transcription start site) NheI fragment from the 18 kb promoter of haplotype A decreased the promoter activity by almost 5-fold in two cultured models of dopamine neurons (Figure 1). When we increased the deletion size from 5 kb to 12 kb (−3.5kb to −15.6kb), no additional effects were found (data not shown). These data suggest that this 5 kb is a super enhancer (5KSE) of SLC6A3. DNA sequence analysis of 5KSE suggested that 5KSE carried four repetitive elements: the 5’VNTR (rs70957367, from - 11.28 kb to −10.86 kb), and three sub-elements named E-10.3 (348 bp), E-9.7 (301 bp) and E-8.7 (299 bp) localized from −10.62 kb to −10.33 kb, from −10.05 kb to −9.75 kb and from −9.13 kb to - 8.78 kb. 5’VNTR had no extensive homology to the three other sub-elements except 53.3% identity to E-8.7. By contrast, there were extensive sequence identities (75.6% - 80.7%) among the three sub-elements (75.6% between E-10.3 and E-8.7; 80.7% between E-9.7 and E-8.7).
Figure 1. Identification of a 5 kb fragment as a super enhancer (■, 5KSE) in the 5’ promoter regions of SLC6A3.
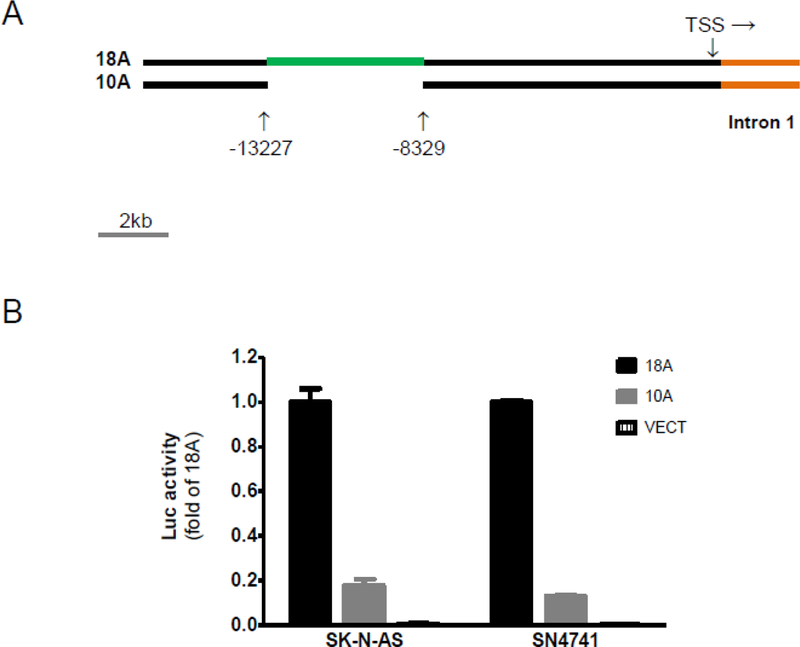
(A) 18 kb (haplotypes A) promoter regions and a deletion construct (10A ) generated and studied in this study. (B) Luc reporting of promoter activities on 18A, and 10A in two cultured models for dopamine neurons (***, P<0.0001 by ANOVA Tukey tests for significance; n=3). Vect, pGL3eBelo1.
Intragenic antagonism between 5KSE and downstream VNTRs
Previous studies have shown that the two downstream variable number tandem repeats (VNTRs), one located in Intron 8 (Int8VNTR) and another in the last exon (3’VNTR), displayed silencing activity on promoter activity. Int8VNTR has two common alleles, 5 repeats (S) and 6 repeats (L); 3’VNTR has two common alleles, 9 repeats (S) and 10 repeats (L). When the two L alleles were combined into LL, the most common haplotype of the two 3’ VNTRs, and LL was inserted into the 3’ end of the luciferase gene in the 18A or 10A plasmids, LL decreased the SLC6A3 promoter activity significantly with similar extents in both constructs (Figure 2A and B), suggesting that LL inhibited the promoter independently of 5KSE. In the presence of LL, addition of 5KSE into 10ALL increased the promoter by 5.2-fold in 18ALL. Together, an intragenic transcriptional antagonism was revealed between 5KSE and 3’ VNTRs, in terms of regulating the SLC6A3 promoter (Figure 2C).
Figure 2. SLC6A3 promoter activity of haplotype A silenced by Int8–3’VNTRs in the presence (A) or absence (B) of 5KSE in SK-N-AS.
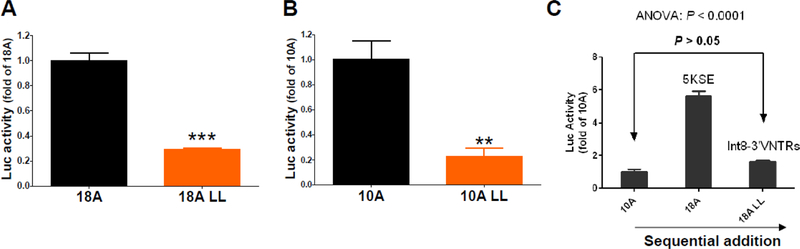
LL, combination of 6-repeat allele of Int8VNTR and 10-repeat allele of 3’VNTR; VNTRs were inserted at an FseI site immediately after luc+ in pGL3eBelo1. (C) Cis-acting mode: upstream 5KSE and downstream Int8–3’VNTRs antagonized each other in regulating 10 kb SLC6A3 promoter activity; **, p<0.01; ***, p<0.0001 by t-tests (n=4–5).
E-8.7 is a key sub-enhancer of 5KSE
In order to better understand 5KSE, we conducted a database search (via NCBI Blast), aiming to identify in other species homology to 5KSE. As a result, three sub-elements were found to share homologies with primate genomes only (Figure 3A). Among the three, E-10.3 had bit lower conservation (83%−86% identity among the top 200 hits) than the other two sub-elements (86%-91%). No extensive homologies were found in the rest of the human genome or in non-primate species, suggesting that 5KSE be a primate-specific enhancer.
Figure 3. Identification of two repetitive enhancers within 5KSE in SK-N-AS.
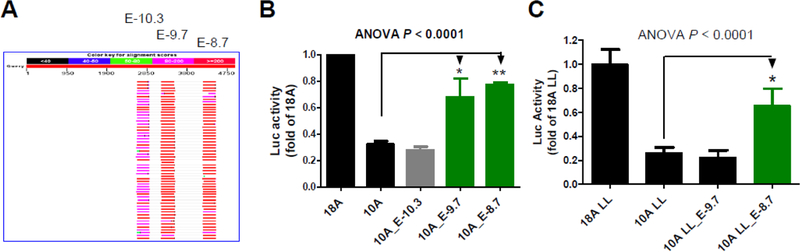
(A) Conservation of E-10.3, E-9.7 & E-8.7 of 5KSE in primates only. (B) Enhancing activity of E-9.7 and E-8.7 PCR-cloned into NheI site of 10A (n=3). (C) E-8.7 (not E-9.7) antagonized Int8– 3’VNTRs (n=4).
These primate homologies implied potential functionality. To verify the possibility, we cloned these three sub-elements, one at a time, back to the NheI site in the 10A plasmid, substituting 5KSE. Adding back of E-10.3 had no effect on the promoter activity. However, adding back of E-9.7 and E-8.7 each restored at least a half of 5KSE’s enhancing activity (2.1-fold and 2.4-fold, Figure 3B). That meant that E-9.7 and E-8.7 together restored 5-fold, the full enhancing activity of 5KSE.
Next, we examined whether E-9.7 and/or E-8.7 could antagonize the silencing activity of 3’VNTRs. In the presence of LL, E-9.7 was unable to restore any of 5KSE’s enhancing activity but E-8.7 was able to increase the promoter activity by 2.9-fold (Figure 3C). That is, E-8.7 contributed to 5KSE’s antagonizing activity.
TF activation of E-8.7
In order to better understand the cis-activating activity of E-8.7, we searched for proteins that mediated the transactivation. E-8.7 was used as bait and a human adult cDNA library used as the prey in the search. One hundred twenty nine million interactions (12-fold the complexity of the library) were analyzed and 214 clones processed. Potential clones were selected based on inframe infusion, repeated show-ups, and known expression in dopamine neurons. Four candidate cDNA clones were identified, including SRP54, Nfe2l1, Slitrk2 and Skp1.
Figure 4A shows that SRP54 and Nfe2l1 displayed the largest E-8.7-dependent upregulations of the SLC6A3 promoter; Slitrk2 and Skp1 showed smaller extents of the upregulation. None of the screened cDNA clones showed any downregulations. When SRP54 and Nfe2l1 combined, a 2.26-fold of upregulation was observed (Figure 4B), explaining E-8.7’s activating capacity (Figure 3B). Endogenously, overexpression of both proteins each upregulated SLC6A3 in two cell lines (Figure 4C).
Figure 4. Identification of TFs for E-8.7.
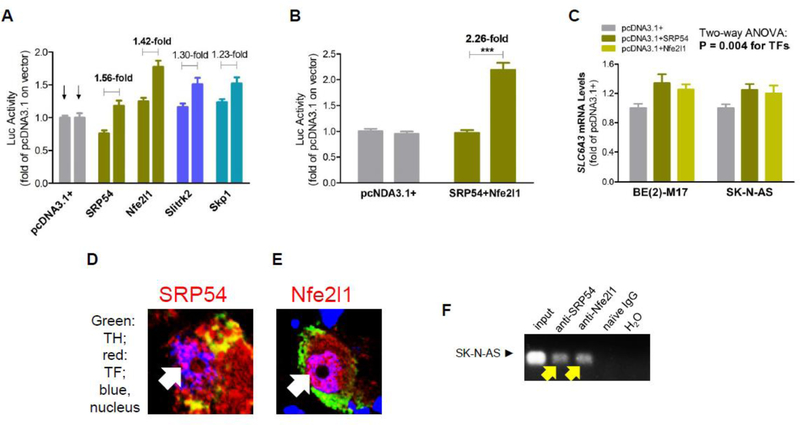
(A) Overexpression of individual cDNA clones regulated the SLC6A3 promoter; (B) More significant regulation by combined cDNA clones in human SK-N-AS cells: 1st bar, plasmid 10A (18A without 5KSE); 2nd bar, 10A_E-8.7; bracket, clones for combinations; 2-way ANOVA: P<0.05 (*) or 0.0001 (***) (n=4 for both A and B). (C) Overexpression of TFs upregulated endogenous SLC6A3 mRNA levels in two cell lines (n=6). Nfe2l1 and SRP54 showed nuclear expression (white arrow) in human postmortem midbrain (D,E), and binding (yellow arrow) to E-8.7 in SK-N-AS by ChIP-PCR (F).
In cultured cells, SK-N-AS of human origin and SN4741 of murine origin, expression of both SRP54 and Nfe2l1 was localized to the nuclei but unlike Nfe2l1, SRP54 was mainly expressed in the cytoplasm, especially of SN4741 (Figure S1). In human postmortem midbrain tissue, both SRP54 and Nfe2l1 were found expressed in the nuclei of dopamine neurons (Figure 4D,E). More of SRP54 was expressed in the cytoplasm and more of Nfe2l1, in the nuclei. These human data were replicated in another healthy subject (Figure S2). Consistently with the nuclear expression, ChIP-PCR analysis of naïve human SK-N-AS cells showed that both SRP54 and Nfe2l1 bound to E-8.7 in the SLC6A3 promoter (Figure 4F).
Discussion
This study identified a main enhancer, 5KSE, in the human SLC6A3 and cloned two TFs that functionally recognized the enhancer. SKSE carries two sets of tandem repeats, which could play more critical role in health and disease than simpler polymorphisms[25]. Another finding is the discovery of transcriptional antagonism between 5KSE and downstream VNTRs. The distribution of the main alleles at the three VNTRs considered here and their level of linkage disequilibrium have been described before[26].
Increased SLC6A3 activity can be a risk for psychiatric disorders
Previous studies have shown that environmental risks such as nicotine[27,28], various stressors[29,30], and high-fat diet[31] that all upregulate the DAT gene activity[24]. Particularly, cigarette smoking is considered as a gateway drug for substance use disorders[32]. Therefore, delineation of the molecular mechanisms for SLC6A3 upregulation is an urgent matter in order to understand genetic contribution of SLC6A3 to related neuropsychiatric disorders. Cloning of trans-activators, such as SRP54 and Nfe2l1, may provide an opportunity to elucidate signaling pathways to endogenous and external risks. In this regard, investigation of 5KSE may enhance our understanding of the role of SLC6A3 in brain function or pathophysiology.
Regulation of SLC6A3 does not follow a simple mode
For the last decades, the widely used genetic markers 3’VNTR and Int8VNTR both downregulated heterogeneous promoters in vitro. It remains unknown whether the silencing activity interacts with activating events. Findings from this study suggest for the first time that different cis-acting elements may interact with each other selectively in controlling the gene activity (Figure 5). An example of the selectivity is that E-8.7, not E-9.7, may attenuate the silencing activity from the downstream VNTRs, excluding a simple additive mode. However, E8.7 and E-9.7 together appear to follow an additive mode in enhancing the promoter activity (Figure 2C vs Figure 3B). Although E-9.7 and E-8.7 share 80.7% identity, it is unknown why E9.7 fails to attenuate the downstream VNTRs. An assumption is that different TFs bind differentially to these two sub-elements. Previously we have reported that 5’VNTR in 5KSE is correlated with the mRNA levels in healthy midbrain tissues[26] but the functionality of 5’VNTR is not revealed by this study. We failed to observe that 5’VNTR can regulate the promoter on its own, suggesting that additional mechanisms remain to be elucidated. Interestingly, the transcriptional antagonism between 5KSE and downstream VNTRs is different from a traditional competition model where different TFs compete for binding to the same cis-acting site[33–38]. That is, in SLC6A3, different TFs on different sites might compete for the same core promoter complex.
Figure 5: Transcriptional cis-antagonism in SLC6A3 (dashed arrow, 5’ to 3’).
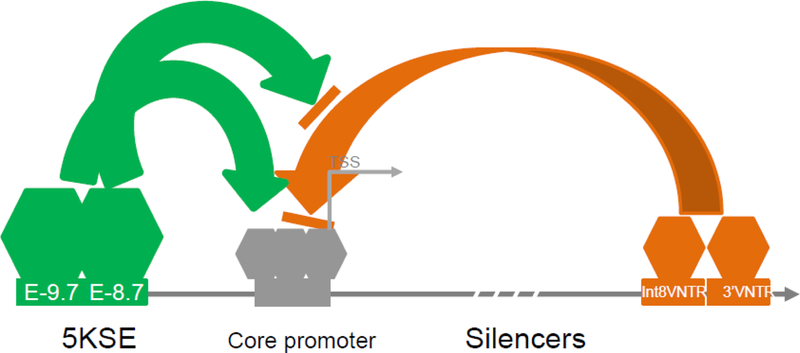
Blue, potential disease-specific pathway targeting; green, enhancing; brown, silencing; gray, core promoter; rectangles, cis-acting elements; hexagons, proteins.
Intragenic antagonism implies haplotype-dependent gene activity
In an additive regulatory mode, different cis-acting elements may function independently from each other so that the overall gene activity in a cell at a time is the additive result of these independent regulations. In case of polymorphic cis-acting elements such as VNTRs, each allele can be correlated with a gene expression level and serve as an effective genetic marker in a case-control association study. However, in the presence of intragenic antagonism, an activator-bound allele may not necessarily correlate with a higher gene activity due to attenuation by another site so that this functional marker may not serve as an effective genetic marker for “main effect”. In other words, haplotypes of the interacting sites would serve as effective markers in association studies[39–41]. Therefore, elucidating transcriptional antagonism is critical for an understanding of genetic etiology. In this case, E-8.7 carries one common polymorphism, rs1478435, and 12 other low frequency polymorphisms according to the 1000 Genomes Project[4]. Identifying two TFs for E-8.7, we still don’t know whether they target polymorphic sequences. Future study is warranted to clarify the TF binding sequences in E-8.7 and whether these TFs recognize any TFs on the downstream VNTRs.
E-9.7 is the most conserved sub-element here
Among the three sub-elements, E-9.7 is the most conserved in two ways. It finds significant homologies (88–89%) in sixteen other chromosomes (except 2, 5, 14, 16, 18, 20, 21 and Y) in the human genome. Among different species, it also shows the highest homologies. It is unclear why it has less function than E-8.7. As well, the function of E-10.3 remains cryptic. It is possible that these conserved sequences contribute to developmental regulation of SLC6A3 activity as the DAT gene is not active till E14 in rats[42]. This hypothesis is based on recent literature that conserved enhancers are developmentally important[43,44]. Consistently, Nef2l1 is already implicated in dopamine neuron development[45]. It remains unknown specifically how SLC6A3 is developmentally activated; investigation of these enhancing elements would help dissect the developmental mechanism.
SRP54 and Nfe2l1 are novel trans-activators for SLC6A3
SRP54, the Signal Recognition Particle 54, has a known function in which it helps to insert hydrophobic signaling peptide of a membrane protein into a membrane[46]. It has never been studied for dopamine neurons or as a TF; Nfe2l1 is a known TF albeit it has not been studied in dopamine neurons. It was reported of an Nfe2l1 binding consensus sequence[47] but this sequence is not matched with E-8.7, possibly because of cell type-specific binding [48,49]. Similar to the human postmortem observations, we have also confirmed that both proteins are expressed in the nuclei of dopamine neurons of mouse VTA and SNc (data not shown here). The dopaminergic significances of these proteins are unknown and warrant future investigation.
Limitations
SLC6A3 has a human-specific array of cis-acting elements, which limits valuable utilization of experimental systems for in vivo validations of intragenic transcriptional antagonism. To better understand the intragenic antagonism, we would need to identify all related TFs including their binding sequences in SLC6A3 but currently we have an incomplete understanding.
Conclusions
5KSE is a large effect enhancer and exerts its function via previously unknown trans-activators including SRP54 and Nfe2l1. SLC6A3 harbors an intragenic transcriptional antagonism between upstream 5KSE and downstream VNTRs, emphasizing a haplotypic activity of SLC6A3.
Supplementary Material
ACKNOWLEDGMENTS
This work is supported by NIH grants DA021409 and DA031573 to ZL; YZ, JZ, XC and NX were supported by Chinese Government Visiting Scholarships.
Footnotes
Conflict of Interest: None
References
- 1.Hughes JR, Roberts N, McGowan S, Hay D, Giannoulatou E, Lynch M, De Gobbi M, Taylor S, Gibbons R, Higgs DR (2014) Analysis of hundreds of cis-regulatory landscapes at high resolution in a single, high-throughput experiment. Nature genetics 46 (2):205–212. doi: 10.1038/ng.2871 [DOI] [PubMed] [Google Scholar]
- 2.Maston GA, Evans SK, Green MR (2006) Transcriptional regulatory elements in the human genome. Annual review of genomics and human genetics 7:29–59. doi: 10.1146/annurev.genom.7.080505.115623 [DOI] [PubMed] [Google Scholar]
- 3.Noonan JP, McCallion AS (2010) Genomics of long-range regulatory elements. Annual review of genomics and human genetics 11:1–23. doi: 10.1146/annurev-genom-082509-141651 [DOI] [PubMed] [Google Scholar]
- 4.Sudmant PH, Rausch T, Gardner EJ, Handsaker RE, Abyzov A, Huddleston J, Zhang Y, Ye K, Jun G, Fritz MH, Konkel MK, Malhotra A, Stutz AM, Shi X, Casale FP, Chen J, Hormozdiari F, Dayama G, Chen K, Malig M, Chaisson MJP, Walter K, Meiers S, Kashin S, Garrison E, Auton A, Lam HYK, Mu XJ, Alkan C, Antaki D, Bae T, Cerveira E, Chines P, Chong Z, Clarke L, Dal E, Ding L, Emery S, Fan X, Gujral M, Kahveci F, Kidd JM, Kong Y, Lameijer EW, McCarthy S, Flicek P, Gibbs RA, Marth G, Mason CE, Menelaou A, Muzny DM, Nelson BJ, Noor A, Parrish NF, Pendleton M, Quitadamo A, Raeder B, Schadt EE, Romanovitch M, Schlattl A, Sebra R, Shabalin AA, Untergasser A, Walker JA, Wang M, Yu F, Zhang C, Zhang J, Zheng-Bradley X, Zhou W, Zichner T, Sebat J, Batzer MA, McCarroll SA, Mills RE, Gerstein MB, Bashir A, Stegle O, Devine SE, Lee C, Eichler EE, Korbel JO (2015) An integrated map of structural variation in 2,504 human genomes. Nature 526 (7571):75–81. doi: 10.1038/nature15394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, Marchini JL, McCarthy S, McVean GA, Abecasis GR (2015) A global reference for human genetic variation. Nature 526 (7571):68–74. doi: 10.1038/nature15393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jaber M, Jones S, Giros B, Caron MG (1997) The dopamine transporter: a crucial component regulating dopamine transmission. Movement disorders : official journal of the Movement Disorder Society 12 (5):629–633. doi: 10.1002/mds.870120502 [DOI] [PubMed] [Google Scholar]
- 7.Kristensen AS, Andersen J, Jorgensen TN, Sorensen L, Eriksen J, Loland CJ, Stromgaard K, Gether U (2011) SLC6 neurotransmitter transporters: structure, function, and regulation. Pharmacological reviews 63 (3):585–640. doi: 10.1124/pr.108.000869 [DOI] [PubMed] [Google Scholar]
- 8.Amara SG, Kuhar MJ (1993) Neurotransmitter transporters: recent progress. Annual review of neuroscience 16:73–93. doi: 10.1146/annurev.ne.16.030193.000445 [DOI] [PubMed] [Google Scholar]
- 9.Lin Z, Canales JJ, Bjorgvinsson T, Thomsen M, Qu H, Liu QR, Torres GE, Caine SB (2011) Monoamine transporters: vulnerable and vital doorkeepers. Progress in molecular biology and translational science 98:1–46. doi: 10.1016/b978-0-12-385506-0.00001-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vandenbergh DJ, Persico AM, Hawkins AL, Griffin CA, Li X, Jabs EW, Uhl GR (1992) Human dopamine transporter gene (DAT1) maps to chromosome 5p15.3 and displays a VNTR. Genomics 14 (4):1104–1106 [DOI] [PubMed] [Google Scholar]
- 11.Guindalini C, Howard M, Haddley K, Laranjeira R, Collier D, Ammar N, Craig I, O’Gara C, Bubb VJ, Greenwood T, Kelsoe J, Asherson P, Murray RM, Castelo A, Quinn JP, Vallada H, Breen G (2006) A dopamine transporter gene functional variant associated with cocaine abuse in a Brazilian sample. Proceedings of the National Academy of Sciences of the United States of America 103 (12):4552–4557. doi: 10.1073/pnas.0504789103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Franke B, Vasquez AA, Johansson S, Hoogman M, Romanos J, Boreatti-Hummer A, Heine M, Jacob CP, Lesch KP, Casas M, Ribases M, Bosch R, Sanchez-Mora C, Gomez-Barros N, Fernandez-Castillo N, Bayes M, Halmoy A, Halleland H, Landaas ET, Fasmer OB, Knappskog PM, Heister AJ, Kiemeney LA, Kooij JJ, Boonstra AM, Kan CC, Asherson P, Faraone SV, Buitelaar JK, Haavik J, Cormand B, Ramos-Quiroga JA, Reif A (2010) Multicenter analysis of the SLC6A3/DAT1 VNTR haplotype in persistent ADHD suggests differential involvement of the gene in childhood and persistent ADHD. Neuropsychopharmacology : official publication of the American College of Neuropsychopharmacology 35 (3):656–664. doi: 10.1038/npp.2009.170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao Y, Xiong N, Liu Y, Zhou Y, Li N, Qing H, Lin Z (2013) Human dopamine transporter gene: differential regulation of 18-kb haplotypes. Pharmacogenomics 14 (12):1481–1494. doi: 10.2217/pgs.13.141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fuke S, Sasagawa N, Ishiura S (2005) Identification and characterization of the Hesr1/Hey1 as a candidate trans-acting factor on gene expression through the 3’ non-coding polymorphic region of the human dopamine transporter (DAT1) gene. Journal of biochemistry 137 (2):205– 216. doi: 10.1093/jb/mvi020 [DOI] [PubMed] [Google Scholar]
- 15.Sacchetti P, Brownschidle LA, Granneman JG, Bannon MJ (1999) Characterization of the 5’-flanking region of the human dopamine transporter gene. Brain research Molecular brain research 74 (1–2):167–174 [DOI] [PubMed] [Google Scholar]
- 16.Zhao Y, Zhou Y, Xiong N, Lin Z (2012) Identification of an intronic cis-acting element in the human dopamine transporter gene. Molecular biology reports 39 (5):5393–5399. doi: 10.1007/s11033-011-1339-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kouzmenko AP, Pereira AM, Singh BS (1997) Intronic sequences are involved in neural targeting of human dopamine transporter gene expression. Biochemical and biophysical research communications 240 (3):807–811. doi: 10.1006/bbrc.1997.7754 [DOI] [PubMed] [Google Scholar]
- 18.Liu K, Yu J, Zhao J, Zhou Y, Xiong N, Xu J, Wang T, Bell RL, Qing H, Lin Z (2017) (AZI2)3’UTR Is a New SLC6A3 Downregulator Associated with an Epistatic Protection Against Substance Use Disorders. Molecular neurobiology doi: 10.1007/s12035-017-0781-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sacchetti P, Mitchell TR, Granneman JG, Bannon MJ (2001) Nurr1 enhances transcription of the human dopamine transporter gene through a novel mechanism. Journal of neurochemistry 76 (5):1565–1572 [DOI] [PubMed] [Google Scholar]
- 20.Wang J, Bannon MJ (2005) Sp1 and Sp3 activate transcription of the human dopamine transporter gene. Journal of neurochemistry 93 (2):474–482. doi: 10.1111/j.1471-4159.2005.03051.x [DOI] [PubMed] [Google Scholar]
- 21.Martinat C, Bacci JJ, Leete T, Kim J, Vanti WB, Newman AH, Cha JH, Gether U, Wang H, Abeliovich A (2006) Cooperative transcription activation by Nurr1 and Pitx3 induces embryonic stem cell maturation to the midbrain dopamine neuron phenotype. Proceedings of the National Academy of Sciences of the United States of America 103 (8):2874–2879. doi: 10.1073/pnas.0511153103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stotz A, Linder P (1990) The ADE2 gene from Saccharomyces cerevisiae: sequence and new vectors. Gene 95 (1):91–98 [DOI] [PubMed] [Google Scholar]
- 23.Fromont-Racine M, Rain JC, Legrain P (1997) Toward a functional analysis of the yeast genome through exhaustive two-hybrid screens. Nature genetics 16 (3):277–282. doi: 10.1038/ng0797-277 [DOI] [PubMed] [Google Scholar]
- 24.Kennedy JL, Xiong N, Yu J, Zai CC, Pouget JG, Li J, Liu K, Qing H, Wang T, Martin E, Levy DL, Lin Z (2016) Increased Nigral SLC6A3 Activity in Schizophrenia Patients: Findings From the Toronto-McLean Cohorts. Schizophrenia bulletin 42 (3):772–781. doi: 10.1093/schbul/sbv191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hannan AJ (2018) Tandem repeats mediating genetic plasticity in health and disease. Nature reviews Genetics doi: 10.1038/nrg.2017.115 [DOI] [PubMed] [Google Scholar]
- 26.Zhou Y, Michelhaugh SK, Schmidt CJ, Liu JS, Bannon MJ, Lin Z (2014) Ventral midbrain correlation between genetic variation and expression of the dopamine transporter gene in cocaine-abusing versus non-abusing subjects. Addiction biology 19 (1):122–131. doi: 10.1111/j.1369-1600.2011.00391.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li S, Kim KY, Kim JH, Kim JH, Park MS, Bahk JY, Kim MO (2004) Chronic nicotine and smoking treatment increases dopamine transporter mRNA expression in the rat midbrain. Neuroscience letters 363 (1):29–32. doi: 10.1016/j.neulet.2004.03.053 [DOI] [PubMed] [Google Scholar]
- 28.Hadjiconstantinou M, Duchemin AM, Zhang H, Neff NH (2011) Enhanced dopamine transporter function in striatum during nicotine withdrawal. Synapse (New York, NY) 65 (2):91–98. doi: 10.1002/syn.20820 [DOI] [PubMed] [Google Scholar]
- 29.Filipenko ML, Alekseyenko OV, Beilina AG, Kamynina TP, Kudryavtseva NN (2001) Increase of tyrosine hydroxylase and dopamine transporter mRNA levels in ventral tegmental area of male mice under influence of repeated aggression experience. Brain research Molecular brain research 96 (1–2):77–81 [DOI] [PubMed] [Google Scholar]
- 30.Mantsch JR, Baker DA, Funk D, Le AD, Shaham Y (2016) Stress-Induced Reinstatement of Drug Seeking: 20 Years of Progress. Neuropsychopharmacology : official publication of the American College of Neuropsychopharmacology 41 (1):335–356. doi: 10.1038/npp.2015.142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ong ZY, Muhlhausler BS (2011) Maternal “junk-food” feeding of rat dams alters food choices and development of the mesolimbic reward pathway in the offspring. FASEB journal : official publication of the Federation of American Societies for Experimental Biology 25 (7):2167–2179. doi: 10.1096/fj.10-178392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kandel ER, Kandel DB (2014) Shattuck Lecture. A molecular basis for nicotine as a gateway drug. The New England journal of medicine 371 (10):932–943. doi: 10.1056/NEJMsa1405092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Amendt BA, Sutherland LB, Russo AF (1999) Transcriptional antagonism between Hmx1 and Nkx2.5 for a shared DNA-binding site. The Journal of biological chemistry 274 (17):11635–11642 [DOI] [PubMed] [Google Scholar]
- 34.Bougarne N, Paumelle R, Caron S, Hennuyer N, Mansouri R, Gervois P, Staels B, Haegeman G, De Bosscher K (2009) PPARalpha blocks glucocorticoid receptor alpha-mediated transactivation but cooperates with the activated glucocorticoid receptor alpha for transrepression on NF-kappaB. Proceedings of the National Academy of Sciences of the United States of America 106 (18):7397–7402. doi: 10.1073/pnas.0806742106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hoppe KL, Francone OL (1998) Binding and functional effects of transcription factors Sp1 and Sp3 on the proximal human lecithin:cholesterol acyltransferase promoter. Journal of lipid research 39 (5):969–977 [PubMed] [Google Scholar]
- 36.Ilsley MD, Gillinder KR, Magor GW, Huang S, Bailey TL, Crossley M, Perkins AC (2017) Kruppel-like factors compete for promoters and enhancers to fine-tune transcription. Nucleic acids research 45 (11):6572–6588. doi: 10.1093/nar/gkx441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Joseph SR, Palfy M, Hilbert L, Kumar M, Karschau J, Zaburdaev V, Shevchenko A, Vastenhouw NL (2017) Competition between histone and transcription factor binding regulates the onset of transcription in zebrafish embryos. eLife 6. doi: 10.7554/eLife.23326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pierce SL, England SK (2010) SK3 channel expression during pregnancy is regulated through estrogen and Sp factor-mediated transcriptional control of the KCNN3 gene. American journal of physiology Endocrinology and metabolism 299 (4):E640–646. doi: 10.1152/ajpendo.00063.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Culverhouse R, Suarez BK, Lin J, Reich T (2002) A perspective on epistasis: limits of models displaying no main effect. American journal of human genetics 70 (2):461–471. doi: 10.1086/338759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hemani G, Knott S, Haley C (2013) An evolutionary perspective on epistasis and the missing heritability. PLoS genetics 9 (2):e1003295. doi: 10.1371/journal.pgen.1003295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zuk O, Hechter E, Sunyaev SR, Lander ES (2012) The mystery of missing heritability: Genetic interactions create phantom heritability. Proceedings of the National Academy of Sciences of the United States of America 109 (4):1193–1198. doi: 10.1073/pnas.1119675109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fujita M, Shimada S, Nishimura T, Uhl GR, Tohyama M (1993) Ontogeny of dopamine transporter mRNA expression in the rat brain. Brain research Molecular brain research 19 (3):222–226 [DOI] [PubMed] [Google Scholar]
- 43.Nord AS, Blow MJ, Attanasio C, Akiyama JA, Holt A, Hosseini R, Phouanenavong S, Plajzer-Frick I, Shoukry M, Afzal V, Rubenstein JL, Rubin EM, Pennacchio LA, Visel A (2013) Rapid and pervasive changes in genome-wide enhancer usage during mammalian development. Cell 155 (7):1521–1531. doi: 10.1016/j.cell.2013.11.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Osterwalder M, Barozzi I, Tissieres V, Fukuda-Yuzawa Y, Mannion BJ, Afzal SY, Lee EA, Zhu Y, Plajzer-Frick I, Pickle CS, Kato M, Garvin TH, Pham QT, Harrington AN, Akiyama JA, Afzal V, Lopez-Rios J, Dickel DE, Visel A, Pennacchio LA (2018) Enhancer redundancy provides phenotypic robustness in mammalian development. Nature 554 (7691):239–243. doi: 10.1038/nature25461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Villaescusa JC, Li B, Toledo EM, Rivetti di Val Cervo P, Yang S, Stott SR, Kaiser K, Islam S, Gyllborg D, Laguna-Goya R, Landreh M, Lonnerberg P, Falk A, Bergman T, Barker RA, Linnarsson S, Selleri L, Arenas E (2016) A PBX1 transcriptional network controls dopaminergic neuron development and is impaired in Parkinson’s disease. The EMBO journal 35 (18):1963–1978. doi: 10.15252/embj.201593725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Janda CY, Li J, Oubridge C, Hernandez H, Robinson CV, Nagai K (2010) Recognition of a signal peptide by the signal recognition particle. Nature 465 (7297):507–510. doi: 10.1038/nature08870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Morrish F, Giedt C, Hockenbery D (2003) c-MYC apoptotic function is mediated by NRF-1 target genes. Genes & development 17 (2):240–255. doi: 10.1101/gad.1032503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Morikawa M, Koinuma D, Tsutsumi S, Vasilaki E, Kanki Y, Heldin CH, Aburatani H, Miyazono K (2011) ChIP-seq reveals cell type-specific binding patterns of BMP-specific Smads and a novel binding motif. Nucleic acids research 39 (20):8712–8727. doi: 10.1093/nar/gkr572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gertz J, Reddy TE, Varley KE, Garabedian MJ, Myers RM (2012) Genistein and bisphenol A exposure cause estrogen receptor 1 to bind thousands of sites in a cell type-specific manner. Genome research 22 (11):2153–2162. doi: 10.1101/gr.135681.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


