Abstract
To improve cancer patient outcome significantly, we must understand the mechanisms regulating stem-like cancer cells, which have been implicated as a cause of metastasis and treatment resistance. The transcription factor C/EBPδ can exhibit pro- and anti-tumorigenic activities, but the mechanisms underlying the complexity of its functions are poorly understood. Here, we identify a role for breast cancer cell intrinsic C/EBPδ in promoting phenotypes that have been associated with cancer stem cells (CSC). While C/EBPδ expression is not abundant in most metastatic breast cancers, our data support a pro-tumorigenic role of C/EBPδ when expressed in subsets of tumor cells and/or through transient activation by the tumor microenvironment or loss of substrate adhesion. Using genetic mouse models and human breast cancer cell lines, we show that deletion or depletion of C/EBPδ reduced expression of stem cell factors and stemnness markers, sphere formation and self-renewal, along with growth of tumors and established experimental metastases in vivo. C/EBPδ is also known as a mediator of the innate immune response, which is enhanced by hypoxia and interleukin-6 (IL-6) signaling, two conditions that also play important roles in cancer progression. Our mechanistic data reveal C/EBPδ as a link that engages two positive feed-back loops, in part by directly targeting the IL-6 receptor (IL6RA) gene, and, thus, amplifying IL-6 and HIF-1 signaling. This study provides a molecular mechanism for the synergism of tumor micro-environmental conditions in cancer progression with potential implications for the targeting of cancer stem cells.
Keywords: hypoxia, IL-6, breast cancer, cancer stem cells, C/EBP
Introduction
It is well established that most solid tumors display significant clonal heterogeneity, which contributes to malignant progression37. Cells with the ability to initiate new tumors in pre-clinical models are termed tumor-initiating cells (TICs) and are considered a type of cancer stem cell (CSCs). Despite controversies concerning the term “cancer stem cell”, there is surmounting evidence that cancer cells with tumor-initiating and/or stem cell-like properties are highly malignant and support metastatic spread39, 44, 6. Thus, more insights are needed to identify critical signaling molecule(s) that contribute to this highly malignant cell phenotype.
CSCs express at least a subset of proteins that also promote stemness in normal stem cells, including Notch and the so-called OSKM reprogramming factors OCT4 (POU5F1), SOX2, KLF4 and MYC12, 14. Cells with CSC characteristics are enriched within populations with specific cell surface markers such as CD44+/CD24- or also ALDH activity, which also contribute directly to the CSC properties40. Hypoxia and inflammation can stimulate the generation of CSCs, in part through induction of NOTCH15, 19. Hypoxia activates the hypoxia-inducible factor 1 alpha (HIF-1α), which can contribute to the expression of stemness factors26. The inflammatory cytokine interleukin-6 (IL-6) activates the signal transducer and activator of transcription 3 (STAT3), which leads to epithelial-mesenchymal transition (EMT) and the generation of CSCs7, 25, 38, 43. While the HIF-1 and STAT3 pathways can intersect, to our knowledge, the potential synergistic activation of both pathways has not been explored mechanistically.
Hypoxia and IL-6 also induce the transcription factor CCAAT/enhancer binding protein delta (C/EBPd, CEBPD)4. In turn, C/EBPd promotes HIF-1α expression2, 45 and directly activates IL-6 expression21, functions that are part of its role as a mediator of the innate immune response by myeloid cells4. Studies in human cancers showed that C/EBPd is overexpressed and associated with poor outcome in urothelial carcinoma41, and proposed C/EBPδ as a critical factor in high-grade glioblastoma8. C/EBPd supports mammary tumor metastasis in the MMTV-Neu transgenic mouse model2, a phenotype that can be explained at least in part through functions in the tumor host that promote tumor lymphangiogenesis, angiogenesis and myeloid-suppressor cells29, 30. On the other hand, C/EBPd is also expressed in normal breast epithelium and lowgrade, hormone-receptor positive breast cancer, and has been likened to a tumor suppressor – primarily due to functions in growth arrest, the DNA damage response, and cell death4. These observations suggest that the cellular context and/or microenvironment may modulate the specific effects of C/EBPd activity. However, a direct, tumor cell intrinsic role of C/EBPd in human cancer cells has not been demonstrated to date in pre-clinical models. Here we report a tumor promoting role of C/EBPd by supporting signaling pathways and expression of genes that are associated with cancer stem cell-like phenotypes in breast cancer cells. Our study also shows how conditions in the tumor microenvironment mechanistically synergize to promote the malignancy of cancer cells and provides evidence that targeting of C/EBPd may attenuate growth of tumors and metastases.
Results
C/EBPδ promotes cancer stem cell-associated features in mouse and human breast cancer cells.
C/EBPδ null mutant mice exhibit a reduced incidence of lung metastasis from MMTV-Neu mouse mammary tumors2. To gain further insight into this phenotype we assessed the circulating tumor cells (CTCs) by two independent methods and found that tumor-bearing Cebpd KO mice harbored fewer CTCs compared to controls (Fig. 1a, Fig.S1a). The generation of CTCs has been linked to cancer cell stemness 31. Quantification of CD61+:CD49f+ cells, which are enriched for TICs22, revealed that Cebpd KO tumors also contained fewer such cells compared to controls (Fig. 1b). Furthermore, sphere formation efficiency (SFE), which often correlates with tumor initiating capability44, was reduced among Cebpd null tumor cells. Assessment of self-renewal showed that the SFE of Cebpd WT cells increased with passages, while that of Cebpd KO cells decreased (Fig. 1c). These data show that C/EBPδ promotes the generation or maintenance of cells with stem cell-like characteristics in this mouse mammary tumor model.
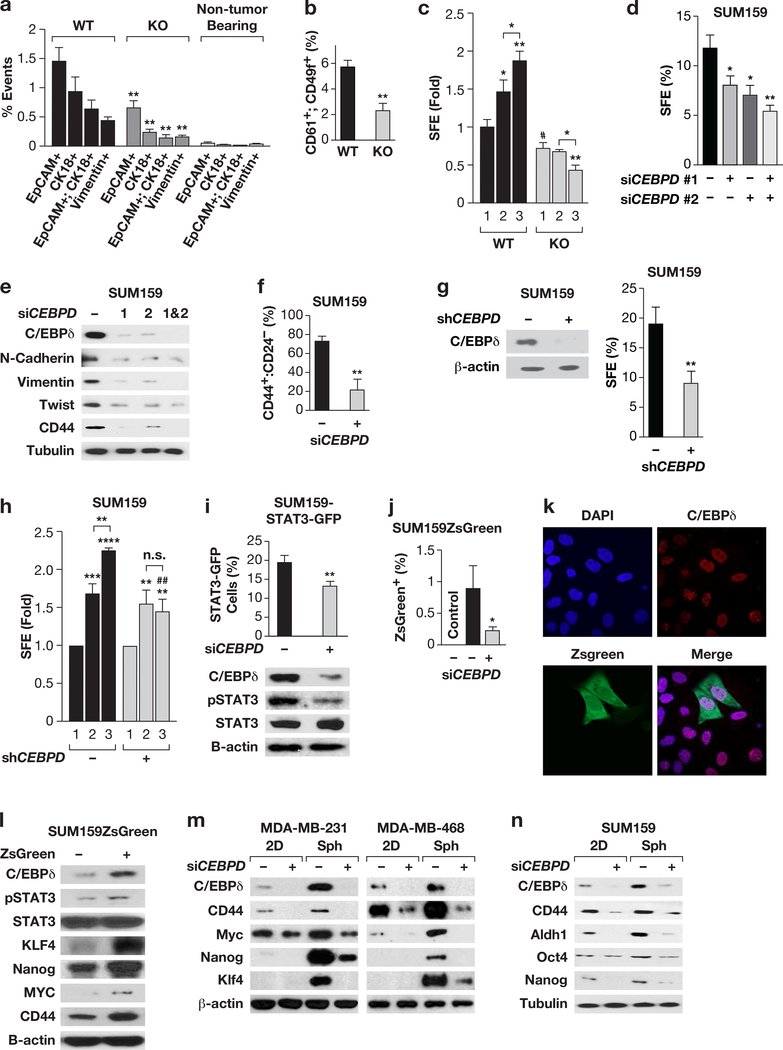
Figure 1. C/EBPδ promotes CSC-like phenotypes in MMTV-Neu mouse mammary tumor cells and human breast cancer cell lines.
(a) Flow-cytometric quantification of epithelial (EpCam+, CK18+)-, and mesenchymal(Vimentin+)-like CTCs42 from peripheral blood of tumorbearing wild-type (WT) and Cebpd-deficient (KO) MMTV-Neu transgenic mice. Data represent the mean ± S.E.M; n=5, **P<0.01, two-tailed unequal variance t-test. Non-tumor bearing Cebpd ko/+ mice were used as negative controls (n=3). (b) Flow-cytometric quantification of CD61+:CD49f+ cells in tumors from mice as in panel (a). Data represent the mean ± S.E.M; n=8, **P<0.01, two-tailed unequal variance t-test. (c) SFE of cells dissociated from tumors and cultured in suspension for 5 days (1st generation) and further cultured for 2nd and 3rd generation spheres (mean ± S.E.M; n=3, *P<0.05, **P<0.01, #P<0.05, two-tailed unequal variance t-test). (d) SFE of SUM159 cells after transfection with Control or two independent CEBPD siRNA oligos (1, 2) alone or in combination (mean ± S.E.M; n=3, *P<0.05, **P<0.01, two-tailed unequal variance t-test). (e) Western analysis of the indicated proteins in SUM159 cells 2d after transfection as in panel (d). (f) Flow-cytometric quantification of CD44+:CD24- cells in SUM159 cell cultures 3d after transfection of siControl and siCEBPD siRNA oligos (mean ± S.E.M; n=3, **P<0.01, two-tailed unequal variance t-test). (g) Representative Western analysis of C/EBPδ expression in SUM159 cells with stable depletion of C/EBPδ (+, shCEBPD) or control cells (-, shControl) as well as SFE (% of cells seeded) after 4 days of culture in suspension. Similar results were obtained with transient silencing of CEBPD (Fig. S1e). (h) Fold change in SFE by 2nd and 3rd generation spheres of SUM159 cells with stable depletion of CEBPD or shControl cells. Data represent the mean ± S.E.M; n=3, **P<0.01, ***P<0.001, ****P<0.0001; ##P<0.01, effect of shCEBPD; n.s., not significant, two-tailed unequal variance t-test. (i) Quantification of STAT3-GFP+ populations from SUM159 cells 72 h after transfection with Control or CEBPD siRNA and the western analysis as indicated (mean ± S.E.M; n=3, **P<0.01, two-tailed unequal variance t-test). (j) Flow-cytometric quantification of green fluorescent cells in SUM159ZsGreen cells transfected with Control or CEBPD siRNA for 72 h. No green cells were detected in control SUM159 cells without ZsGreen (Control). Data represent the mean ± S.E.M; n=3, *P<0.05, two-tailed unequal variance t-test. (k) Representative immunocytochemistry of C/EBPδ expression in SIM159ZsGreen cells. Nuclei were stained with DAPI. (l) Western analysis of the indicated proteins in SUM159ZsGreen sorted by GFP expression. (m, n) Western analysis of the indicated cell lines transfected with Control (−) or CEBPD siRNA (+) for 24 h followed by separation into culture on plastic (2D) or as spheres (Sph.) for 4 days.
Across human breast epithelial cell lines, C/EBPδ is highly expressed in untransformed MCF-10A cells compared to several breast cancer lines35. However, we found that C/EBPδ was also highly expressed in vitro and in vivo (Fig. S1b-c) in SUM159 triple-negative breast cancer (TNBC) cells, which are known to express many stem cell markers32. C/EBPδ knockdown with two independent siRNAs in SUM159 cells significantly reduced their SFE (Fig. 1d) and expression of the CD44 receptor as well as the mesenchymal markers N-cadherin, Vimentin and Twist (Fig. 1e). CEBPD-silencing also reduced the number of CD44+:CD24- cells (Fig. 1f, Fig. S1d), that are enriched for CSCs44, as well as the SFE and self-renewal of sphere-forming cells (Fig. 1g-h). While 60–70% of SUM159 cells were CD44+:CD24-, less than 20% formed spheres, which is in agreement with a fraction of cells exhibiting expression of a STAT3-reporter that are highly enriched for TICs43. In our hands, 19% of SUM159 STAT3-GFP cells expressed the STAT3 reporter, and CEBPD silencing reduced the number of STAT3-activated cells (Fig. 1i, Fig. S1f). As an alternative model system, we analyzed SUM159 cells with the cODC-ZsGreen reporter (SUM159ZsG), which is constitutively degraded by the proteasome but stabilized in cells with low proteasome activity and high tumor initiating and metastatic activity20. CEBPDsilencing also reduced the number of ZsGreen+ cells in this line (Fig. 1j). Expression analysis showed that C/EBPδ was present in most cells including green cells (Fig. 1k), and quantitatively enriched in ZsGreen+ cells along with stemness factors (Fig. 1l).
The TNBC cell lines MDA-MB-231 and MDA-MB-468 express low levels of C/EBPδ (Fig. S1b). However, culture of cells as spheres, which increases the fraction of TICs1, induced expression of C/EBPδ along with CD44, Myc, Nanog and KLF4 in a C/EBPδ-dependent manner, as also seen in SUM159 (Fig. 1m-n). Thus, despite differences in basal expression levels, C/EBPδ contributes to expression of stemness factors in all three cell lines. Taken together, these results show that C/EBPδ is expressed in cells with stemness reporter activity, albeit not exclusively so, and enhances cancer stem cell-like gene expression and sphere formation in TNBC lines.
C/EBPδ promotes tumor growth, lung colonization, and stemness gene expression in vivo
To assess the role of C/EBPδ in SUM159 cells in vivo, we generated cells with doxycycline (Dox)-dependent expression of CEBPD-shRNA and confirmed that Dox reduced the SFE only in cells with CEBPD-shRNA, while cell proliferation was not affected (Fig. S2a-c). The initial tumor onset and growth was similar between the two cell populations (Fig. S2d). However, subsequent Dox treatment inhibited the growth specifically of tumors expressing CEBPD-shRNA (Fig. 2a-b), and reduced expression of CD44, Myc, and KLF4 along with C/EBPδ (Fig. 2c). In contrast, Dox did not affect C/EBPδ expression in tumors from shControl cells (Fig. S2e). Furthermore, SFE was decreased in cells from Dox-treated shCEBPD-SUM159 tumors (Fig. 2d). These data show that C/EBPδ promotes the growth of SUM159 tumors and expression of stemness-associated genes in vivo.
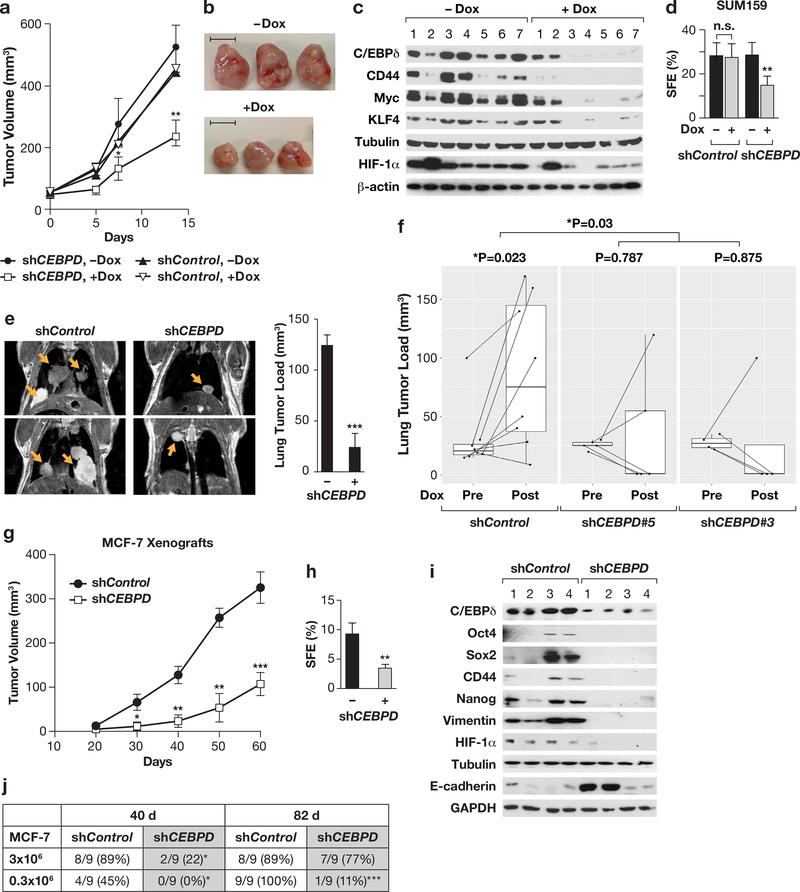
Figure 2. C/EBPδ promotes primary tumor growth and experimental metastasis in vivo.
(a) Tumor volumes of SUM159 cells with Dox-inducible shRNAs at the indicated times of treatment, started at 50mm3 (n=13, *P<0.05, **P<0.01, by exact logistic regression in the SAS logistic procedure). (b) Representative tumors from panel (a) at endpoint (day 14). Scale bar = 1 cm. (c) Western analysis of the indicated proteins in seven independent tumors per group as in panel (b). (d) SFE of cells dissociated from tumors as in panel (b), cultured for 5 days (n=5–7, **P<0.01, two-tailed unequal variance t-test). (e) Representative magnetic resonance images of mouse thoraxes 8 weeks after tail vein injection and quantification of the tumor load in lungs by ImageJ from those mice that showed lesions, i.e. 3/4 for shControl and 4/9 for shCEBPD. Data represent the mean ± S.E.M; n=3 shControl, n=4 shCEBPD, ***P<0.001, two-tailed unequal variance t-test. (f) Quantification of lung tumor load by MRI imaging before (Pre) and after four weeks (Post) of doxycycline treatment using two SUM159 cell lines with independent Doxinducible CEBPD-shRNAs or shControl (shControl, n=8; shCEBPD#5, n=5; shCEBPD#3, n=4). The indicated P values were generated with the Paired Wilcoxon rank test. Dox significantly reduced the lung tumor load in the combined shCEBPD mice compared to controls (P=0.03 by Wilcoxon rank test of log-fold changes in tumor load). (g) Tumor volume measurements of MCF-7 stable cells (shControl and shCEBPD) injected orthotopically into athymic nude mice. Data are from three independent sets of cells as described in Fig. S2i-j (Batch I, n=14; Batch II, n=4; Batch III; n=5; mean ±SEM; *P<0.05, ** P<0.01, *** P<0.001, by exact logistic regression in the SAS logistic procedure). (h) SFE of single cell suspensions from similar-sized MCF-7 xenograft tumors, cultured for 5 days (n=5, **P<0.01, two-tailed unequal variance ttest). (i) Western analysis of the indicated proteins in four independent tumors as in panel (g). (j) Tumor initiation by MCF-7 cells (3×106 or 0.3×106) with stable depletion of C/EBPδ (shCEBPD) or control cells (shControl) injected orthotopically. Data are from two experiments and indicate the presence of tumors from palpable stage at the indicated times after injection (n=9, *P<0.05, ***P<0.001, by exact logistic regression in the SAS logistic procedure).
To address the role of C/EBPδ in the establishment of experimental lung metastases, we injected SUM159 cells by the tail vein. Mice injected with C/EBPδ-depleted cells survived significantly longer than the control cohort (Fig. S2f). Microscopic quantification showed a significant reduction in lung colonies by CEBPD-silenced cells (Fig. S2g). Magnetic resonance imaging of mice that did not display overt symptoms 64 days after tumor cell injection revealed several large nodules in the lungs of 3/4 (75%) control mice compared to single and/or smaller nodules in 4/9 (44%) mice that had received C/EBPδ-depleted cells (Fig. 2e). Using the Dox-inducible system (Fig. S2h), we found by MRI that silencing of CEBPD in established lung colonies caused regression of the lung lesions in most mice, while the lung tumor burden continued to increase in most Dox-treated shControl mice (Fig. 2f). In agreement with the decreased incidence of lung metastasis in Cebpd-deficient MMTV-Neu mice2, these data indicate that C/EBPδ contributes to the survival of CTCs and/or the establishment and maintenance of breast tumor cell growth in lungs.
Previously, we reported that abundant C/EBPδ expression is associated with estrogen receptor (ER)+ BC and low tumor grade, and that C/EBPδ attenuates proliferation and motility of ER+ MCF-7 cells in culture27. When assessed in vivo, however, CEBPD-silenced MCF-7 tumors grew significantly more slowly than controls and mice therefore survived longer (Fig. 2g and S2k-l), harbored fewer sphere-forming cells (Fig. 2h) and expressed lower levels of CD44, Vimentin, and stemness-related transcription factors, while in some cases expression of Ecadherin was elevated (Fig. 2i). C/EBPδ expression was elevated in MCF-7 tumors compared to cells grown on plastic (Fig. S2m), possibly due to the more hypoxic environment. C/EBPδdepletion also significantly reduced the tumor take rate when 10-times fewer cells were injected (Fig. 2j), suggesting a reduction of TICs in C/EBPδ-depleted MCF-7 cells.
Analysis of ER+ cells reveals a role for C/EBPδ in hypoxia- and IL-6-induced stem cell-like features
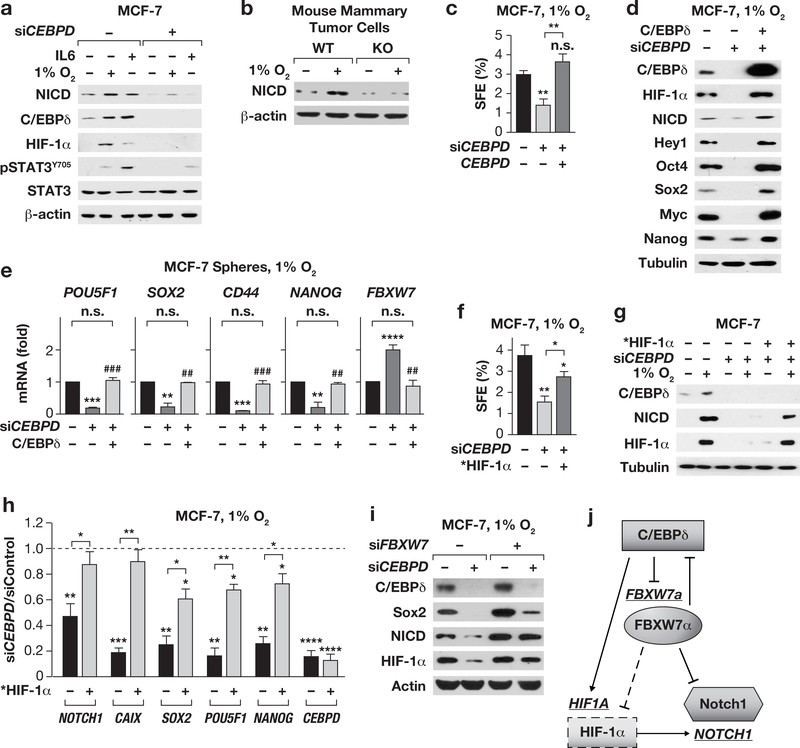
Consistent with the known role of C/EBPδ in HIF-1a expression2, 45, we also noticed reduced HIF-1a protein in CEBPD-depleted tumors (Fig. 2c and 2i). This observation is indicative of the more hypoxic environment in vivo compared to cell culture, which prompted us to address the potential role of C/EBPδ in mediating hypoxia-mediated stemness in ER+ cells. Indeed, CEBPD-depletion had no effect on the basal levels of E-cadherin or N-cadherin in MCF-7 and T47D cells but compromised the hypoxia-mediated downregulation of E-cadherin as well as the upregulation of mesenchymal N-cadherin and vimentin (Fig. 3a). The SFE and number of CD44+:CD24- cells is much lower in MCF-7 compared to SUM159 but can be increased by hypoxia or treatment with IL-617, 34. Silencing of C/EBPδ caused a modest reduction in the basal level SFE and CD44+:CD24- cells but was more critical in the context of hypoxia and IL-6 (Fig. 3b-c).
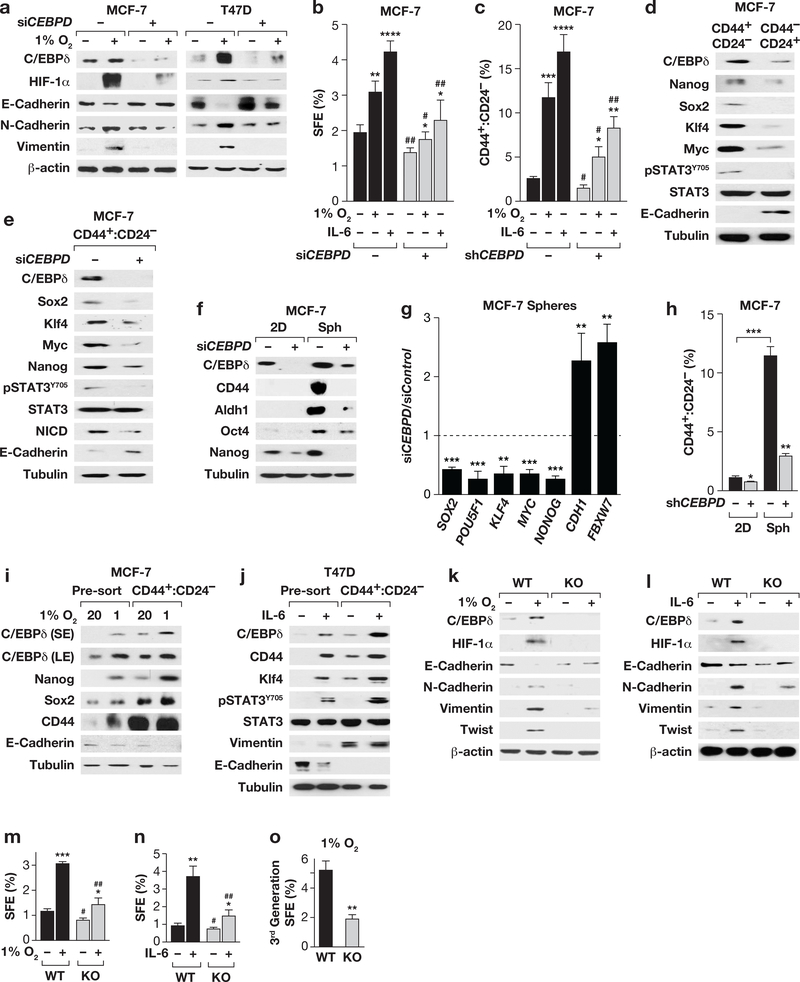
Figure 3. C/EBPδ promotes hypoxia and IL-6-induced CSC-related features in breast cancer cells.
(a) Western analysis of MCF-7 and T47D cells transfected with Control (−) or CEBPD siRNA followed by culture ± 1% O2 for 3 days. (b) SFE of MCF-7 cells transfected and cultured for 4 days in suspension ± IL-6 (100 ng/ml) or 1% O2. Data represent the mean ± S.E.M; n=6, untreated, n=3, treated. *P<0.05, **P<0.01, ****P<0.0001; #P<0.05, ##P<0.01, effect of siCEBPD, two-tailed unequal variance t-test. (c) Flow-cytometric quantification of CD44+:CD24- cells in MCF-7 cells with stable depletion of C/EBPδ (shCEBPD, +) or shControl cells (−) cultured ± 1% O2 or IL-6 (50 ng/ml) for 10 days. Data represent the mean ± S.E.M; n=3, *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001; #p<0.05, ##p<0.01, effect of shCEBPD, two tailed unequal variance t-test. Representative flow cytometry scatter-plots of CD44/CD24 expressing MCF-7 cells are shown in Fig. S2n. (d) Western analysis of the indicated proteins in MCF-7 cell subpopulations as indicated. (e) Western analysis of the CD44+:CD24- population sorted from MCF-7 cells 48 h after transfection with Control or CEBPD siRNA. (f) Western analysis of MCF-7 cells transfected with Control (−) or CEBPD siRNA (+) for 24 h followed by culture on plastic (2D) or as spheres (Sph) for 4 days. (g) qPCR analysis of the indicated genes in MCF-7 cells transfected with siRNAs against Control or CEBPD followed by culture in suspension for 4 days. Data represent the mean ± S.E.M; n=3, ** P<0.01, ***P<0.001, two-tailed unequal variance t-test. Data are shown as relative to the levels in siControl cells. (h) Flow-cytometric quantification of CD44+:CD24- cells in MCF-7 cells treated as described in panel (f). Data represent the mean ± S.E.M; n=3, *P<0.05, **P<0.01, ***P<0.001, two-tailed unequal variance t-test. (i) Western analysis of the indicated proteins in MCF-7 cells cultured ±1% O2 for 10 days followed by FACS isolation of CD44+:CD24- cells, compared to unsorted (pre-sort). (j) Western analysis of the indicated proteins in T47D cells ± IL-6 (50 ng/ml, 10 days), prepared as in panel (i) (S/LE, short/long exposure). (k&l) Western analysis of the indicated proteins in primary tumor cells after 3 days of culture at 1% O2 or with IL-6 (100 ng/ml) as indicated. (m&n) SFE of cells dissociated from tumors and cultured in suspension at 1% O2 or with IL-6 (100 ng/ml) for 5 days as indicated. Data represent the mean ± S.E.M; n=3, *P<0.05, **P<0.01, ***P<0.001; #p<0.05, ##p<0.01, genotype effect within treatment, two-tailed unequal variance t-test. (o) SFE of primary tumor cells from 3rd generation spheres cultured in suspension at 1% O2. Data represent the mean ± S.E.M; n=3, **P<0.01, two-tailed unequal variance t-test.
Expression analysis showed that C/EBPδ was enriched in CD44+:CD24- cells compared to CD44-:CD24+, which are considered non-CSCs25, 46, along with Nanog, Sox2, Klf4, Myc and also pSTAT3, while E-cadherin was enriched in non-CSCs (Fig. 3d). Similar results were obtained at the level of mRNA, which also showed an inverse correlation of C/EBPδ with Ecadherin (CDH1) and FBXW7 (Table S2). C/EBPδ knockdown reduced the expression of all of these stemness-associated factors including Notch-intracellular domain 1 (NICD) within the CD44+:CD24- MCF-7 cell fraction, while E-cadherin expression was increased (Fig. 3e). Similar results were obtained in MCF-7 cell spheres at the level of protein (Fig. 3f) and mRNA, which also showed increased expression of CDH1 and FBXW7 in C/EBPδ-depleted MCF-7 spheres (Fig. 3g). This result is consistent with the significant reduced number of CD44+:CD24- cells when CEBPD-silenced MCF-7 cells were cultured as spheres (Fig. 3h). Hypoxia or IL-6 increased C/EBPδ expression along with several stemness factors not only in the bulk population but also in the CD44+:CD24- cells of MCF-7 and T47D cells (Fig. 3i-j). Collectively, these data show that C/EBPδ expression is not unique to a specific subpopulation of cells, but its expression level correlates with and directly or indirectly enhances expression of genes that have been associated with cancer stem cell activity.
Our finding that C/EBPδ promotes the cadherin switch, stemness-related gene expression, and sphere formation in response to hypoxia and IL-6 was also confirmed in the luminal MMTV-Neu mouse model (Fig. 3k-l). Hypoxia and IL-6 induced a switch from Ecadherin to N-cadherin and expression of Vimentin and Twist in control cells. These responses were significantly blunted in Cebpd KO cells, including HIF-1α induction by IL-6. Cebpd null cells exhibited a modest reduction in baseline SFE but a greater reduction under hypoxia or IL-6 treatment (Fig. 3m-n). Furthermore, the self-renewal of sphere forming cells (Fig. 1c) could not be rescued by hypoxia (Fig. 3o). Lastly, these findings were not unique to luminal cells, as hypoxia or IL-6 also increased the fraction of SUM159 ZsGreen+ CSCs and HIF-1α expression in a C/EBPδ-dependent manner (Fig. S3a-b). Taken together, these results show that C/EBPδ promotes EMT, sphere formation, abundance of CD44+/CD24-cells, and expression of stemness factors in response to hypoxia and IL-6. Furthermore, the data suggest a role for C/EBPδ in the cross-talk between hypoxia and IL-6.
C/EBPδ acts upstream of IL-6 signaling through activation of the IL6RA gene
We had noticed that C/EBPδ promoted STAT3 phosphorylation in MCF-7 CD44+/CD24-cells in the absence of exogenous IL-6 treatment (Fig. 3d-e). Consistent with findings in macrophages21, we observed that C/EBPδ supported IL6 gene expression in SUM159 and MCF7 cells as well as mouse mammary tumor cells at both the basal level and in response to hypoxia (Fig. S4a-c). Therefore, we hypothesized that C/EBPδ stimulates STAT3 phosphorylation through activation of IL-6 expression. However, ectopic IL-6 could only partially rescue STAT3 phosphorylation (and HIF-1α accumulation) in CEBPD-silenced MCF-7 cells under hypoxia (Fig. 4a). This defect in the IL-6 response was also seen at the level of JAK2 phosphorylation in MCF-7 and T47D cells (Fig. 4b). In SUM159 cells, the high basal level phosphorylation of JAK2 and STAT3 was also reduced upon CEBPD-silencing (Fig. 4c). These results led us to assess the expression of the IL-6 receptor. C/EBPδ depletion reduced the mRNA levels of the common IL-6 cytokine family receptor subunit gp130 (IL6ST) in SUM159 cells but not in MCF7 cells, while expression of the IL-6 specific gp80 subunit (IL-6Rα, IL6RA) was reduced in both cell lines (Fig. 4d). Breast CSCs are known to express the IL-6 receptor 25 and we also found higher levels of IL6RA mRNA in CD44+:CD24- MCF-7 cells compared to CD44-/CD24+ cells (Table S2). IL-6Rα protein expression was not detectable by Western analysis. However, the secreted form sIL-6Rα28, was reduced in media from CEBPD-silenced MCF-7 cells and SUM159 cells with either transient or Dox-induced knockdown of C/EBPδ (Fig. 4e-g). On the other hand, knockdown of FBXW7 in MCF-7 cells increased sIL-6Rα production (Fig. 4e), which is consistent with its role in directly targeting C/EBPδ for degradation3. The IL6RA promoter harbors two potential C/EBP binding sites, and ChIP analysis in MCF-7 cells demonstrated that C/EBPδ could bind this region (Fig. 4h). In MCF-7 and T47D cells, ectopic sIL-6Rα supplementation completely rescued IL-6-induced pSTAT3 accumulation in C/EBPδdepleted cells (Fig. 4i). However, sphere formation in response to IL-6 was only partially rescued (Fig. 4j), indicating that C/EBPδ promoted sphere formation not only by supporting sIL6Rα production. Taken together, these data demonstrate that C/EBPδ contributes to IL-6mediated STAT3 phosphorylation by activating IL-6Rα expression and thereby enhancing IL6/STAT3-mediated signaling and phenotypes in breast cancer cells.
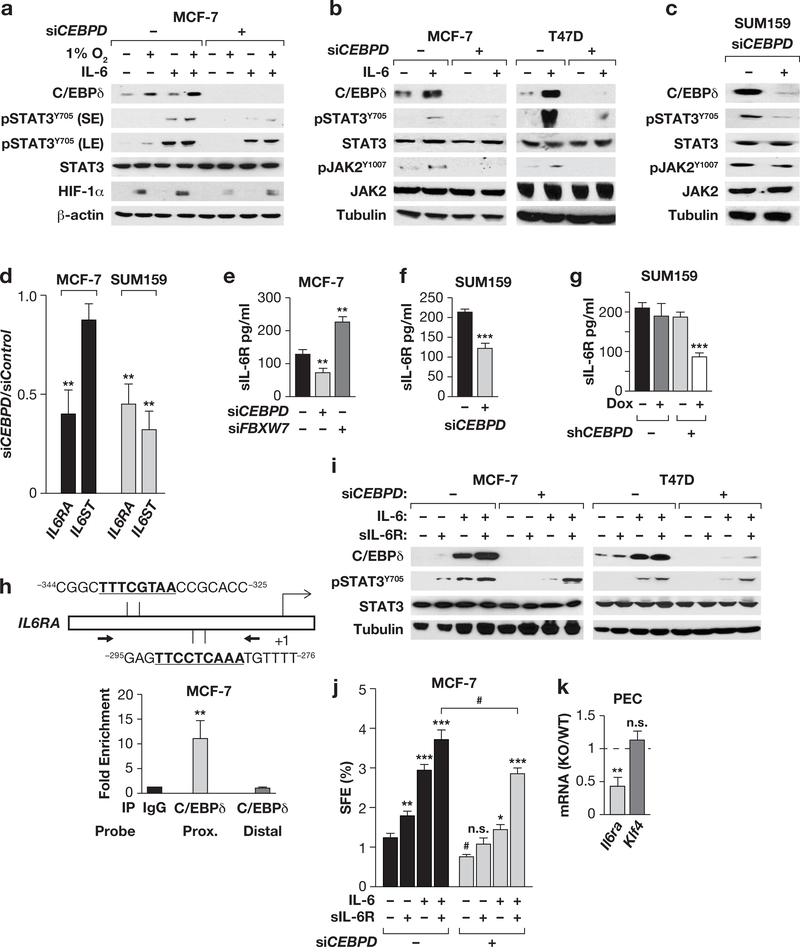
Figure 4. C/EBPδ promotes IL-6 signaling through activation of the IL-6 receptor gene IL6RA.
(a) Western analysis of the indicated proteins in MCF-7 cells ± CEBPD siRNA and cultured ±1% O2 or IL-6 (200 ng/ml) for 8 h (S/LE, short/long exposure). (b) Western analysis of the indicated proteins in MCF-7 and T47D cells ± CEBPD siRNA and cultured ±IL-6 (100 ng/ml, 24 h). (c) Western analysis of the indicated proteins in SUM159 cells 48 h after nucleofection with Control or CEBPD siRNA. (d) qPCR analysis of IL6RA and IL6ST mRNA in MCF-7 and SUM159 cells 48 h after nucleofection with Control or CEBPD siRNA. Data represent the mean ± S.E.M; n=3, **P<0.01, by two-tailed unequal variance t-test. (e&f) Quantification of IL-6Ra protein in the conditioned medium from cells 48 h after transfection with the indicated siRNA. Data represent the mean ± S.E.M; n=3, ** P<0.01, ***P<0.001, two-tailed unequal variance t-test. See Fig. S4d for silencing controls. (g) Quantification of sIL-6Rα in the conditioned media from SUM159 cells with Dox-inducible Control and CEBPD shRNA cells treated ± Dox for 24 h (n=3; ***P<0.001). (h) Schematic showing two C/EBP binding motifs in the IL6R promoter and relative location of qPCR primers for ChIP analysis (bottom panel) with MCF-7 chromatin and a C/EBPd-specific antibody relative to IgG control. Distal: negative control from the CD44 gene. Data represent the mean ± S.E.M; n=3, **P<0.01, twotailed unequal variance t-test. (i) Western analysis of the indicated proteins in MCF-7 and T47D cells ±CEBPD siRNA after 16 h of treatment with IL-6 and/or sIL-6R (200 ng/ml each). (j) SFE of MCF-7 cells under conditions as shown in panel (i). Data represent the mean ± S.E.M; n=3, *P<0.05, **P<0.01, ***P<0.001; #p<0.05, effect of siCEBPD; n.s., not significant, two-tailed unequal variance t-test. (k) qPCR analysis of Il6ra and Klf4 mRNA levels in freshly isolated peritoneal exudate cells (PEC) from Cebpd WT and KO mice; Klf4 is shown as a negative control as it was not altered (n=4; **P<0.01, two-tailed unequal variance t-test.).
C/EBPδ and IL-6 signaling are also critical for myeloid cell activation. Therefore, we investigated their relationship in mouse peritoneal cells comprising >70% myeloid cells 3, and found that Il6ra expression was reduced in Cebpd knockout cells (Fig. 4k). These data indicate that the role of C/EBPδ in the expression of the ILRA gene is not limited to breast cancer cells.
C/EBPδ enhances NOTCH1/NICD expression through regulation of HIF-1α and FBXW7
One of the final executors of many stemness-inducing signaling pathways is the intracellular domain of Notch, NICD12. In MCF-7 cells, both IL-6 and hypoxia induced NOTCH1 mRNA (Fig. S5a) and NICD protein (Fig. 5a) in a C/EBPδ-dependent manner. Also in mammary tumor cells C/EBPδ was necessary for NICD induction by hypoxia (Fig. 5b). Given the known role of HIF-1 in NOTCH1 expression49, we asked whether ectopic HIF-1α would rescue stemnessrelated phenotypes in C/EBPδ-depleted cells under hypoxia. First, we verified that ectopic C/EBPδ expression along with CEBPD siRNA not only rescued sphere formation (Fig. 5c), but also restored induction of NICD, its target Hey1, and several other stemness factors and prevented the induction of FBXW7 (Fig. 5d-e).
Figure 5. C/EBPδ augments NOTCH1/NICD expression through HIF-1α and FBXW7α.
(a) Western analysis of the indicated proteins in MCF-7 cells ± CEBPD siRNA cultured ±1% O2 or IL-6 (100 ng/ml) for 24 h. (b) Western analysis of primary MMTV-Neu tumor cells from Cebpd WT and KO mice cultured ± 1% O2 for 3 days. (c) SFE of MCF-7 cells transfected with siRNA and a C/EBPδ expression construct or vector, cultured in suspension at 1% O2 for 4 days. Data represent the mean ± S.E.M; n=3, **P<0.01, n.s., not significant, two-tailed unequal variance ttest. (d) Western analysis and (e) qPCR analysis of cells as in panel (c) (n=3, **P<0.01, ***P<0.001, ****P<0.001, compared to siControl; ##P<0.01, ###P<0.001, effect of C/EBPδ overexpression; n.s., not significant). (f) SFE as in panel (c) of MCF-7 cells transfected with siRNA along with mutant *HIF-1α expression plasmids as indicated (n=3, *P<0.05, **P<0.01, two-tailed unequal variance t-test). (g) Western analysis and (h) qPCR analysis of MCF-7 spheres as in panel (f).Data represent the mean ± S.E.M; n=3, *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001, two-tailed unequal variance t-test. (i) Western analysis of MCF-7 cells transfected with the indicated siRNA followed by culture in suspension at 1%O2 for 4 days. See Fig.S5b for silencing controls. (j) Model of the regulatory circuit of the indicated genes (underlined) and resultant proteins (shapes). The model is supported by the data in this manuscript plus published reports (see in-text citations).
Next, we tested the effect of ectopic HIF-1α by transfecting a P402A/P564A-mutated HIF-1α (*HIF-1α) protein, which is more stable than wild-type HIF-1α18. In contrast to the complete rescue by ectopic C/EBPδ, expression of *HIF-1α only partially rescued the SFE in C/EBPδ-depleted cells under hypoxia (Fig. 5f), although HIF-1α protein levels were reconstituted in hypoxic C/EBPδ-depleted cells (Fig. 5g) as well as expression of the targets NOTCH1 and CAIX (Fig. 5h). However, expression of NICD protein (Fig. 5g) and mRNAs for SOX2, POU5F1, and NANOG were only partially rescued (Fig. 5h). These results showed that C/EBPδ induced NOTCH1 mRNA expression through HIF1α, but additional mechanisms act at the protein level. C/EBPδ enhances HIF-1α expression by inhibiting expression of FBXW72, which also targets NICD for degradation10. Accordingly, co-silencing of FBXW7 rescued HIF-1α and NICD protein expression in C/EBPδ-depleted cells (Fig. 5i). Together, these data support a model by which C/EBPδ promotes NOTCH1 mRNA expression through HIF-1α, while NICD protein degradation is attenuated through inhibition of FBXW7 expression (Fig. 5j).
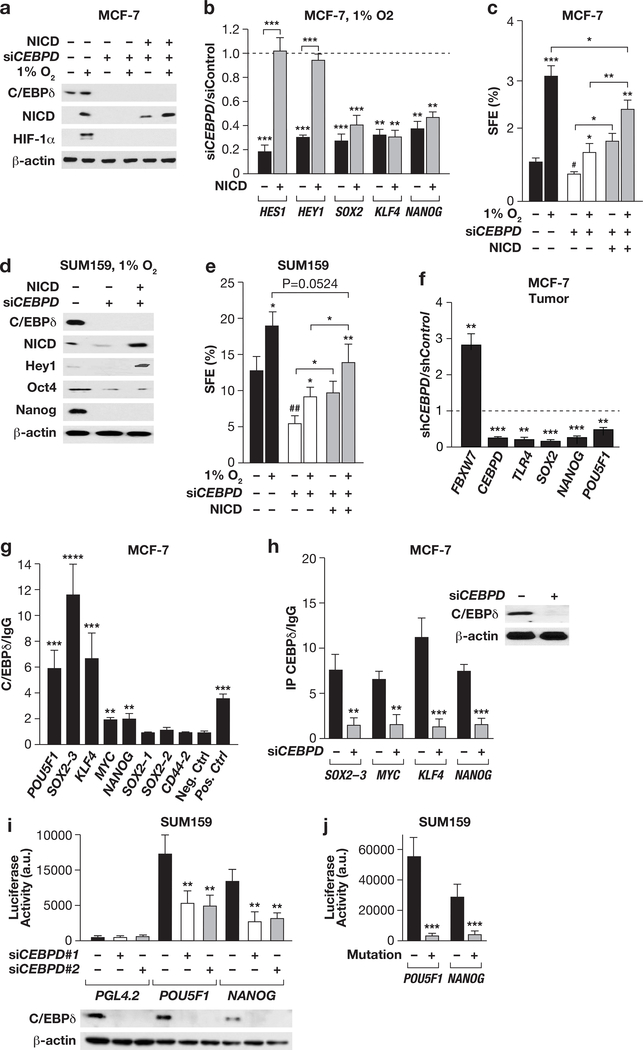
C/EBPδ directly targets the promoters of stemness and reprogramming factors
We next assessed whether ectopic NICD would rescue stemness features of C/EBPδ-depleted cells. A NICD expression construct restored NICD protein levels in CEBPD-silenced cells, but – as expected - did not recover HIF-1α expression (Fig. 6a). Ectopic NICD completely rescued mRNA expression of its targets HEY1 and HEY2 in CEBPD-silenced cells, but not of SOX2, KLF4, and NANOG (Fig. 6b). Accordingly, SFE under hypoxia was only partially rescued by ectopic NICD (Fig. 6c). Similar results were obtained in SUM159 cells (Fig. 6d-e). While the lack of complete rescue could be in part due to still impaired HIF-1α expression, these data also prompted us to investigate if C/EBPδ regulated some of the stemness genes directly. The promoters of SOX2, KLF4, POU5F1, MYC, and NANOG all harbor C/EBP binding motifs (Table S3). In agreement with C/EBPδ-dependent mRNA expression of these genes in MCF-7 spheres (Fig. 3g) and xenograft tumors (Fig. 6f), ChIP analysis confirmed that most of the predicted sites were bound by C/EBPδ in MCF-7 (Fig. 6g-h). Promoter reporter assays further support a role for C/EBPδ in the regulation of NANOG and POU5F1 as CEBPD-silencing (Fig. 6i) or mutation of the C/EBP binding sites (Fig. 6j) significantly reduced promoter reporter gene expression in SUM159 cells. Collectively, these results indicate that C/EBPδ can directly regulate the promoters of several stemness-promoting factors.
Figure 6. C/EBPδ directly targets the promoters of several stemness factors.
(a) Western analysis of the indicated proteins in MCF-7 cells transfected with siRNA and NICD expression plasmids or controls as indicated and cultured in suspension ± 1%O2 for 4 days. (b) qPCR analysis of the indicated mRNAs in MCF-7 spheres as in panel (a). Data represent the mean ± S.E.M; n=3, **P<0.01, ***P<0.001, two-tailed unequal variance t-test. (c) SFE of MCF-7 as in panel (a). Data represent the mean ± S.E.M; n=3, *P<0.05, **P<0.01, ***P<0.001; #P<0.05, siControl vs siCEBPD, two-tailed unequal variance t-test. (d) Western analysis of the indicated proteins in SUM159 cells transfected and cultured as in panel (a). (e) SFE of SUM159 cells as in panel (c). Data represent the mean ± S.E.M; n=3, *P<0.05, **P<0.01; ##p<0.01, siControl vs siCEBPD, two-tailed unequal variance t-test. (f) qPCR analysis of the indicated genes from MCF-7 control and shCEBPD xenograft tumors (n=6, **P<0.01, ***P<0.001). (g) qPCR analysis of DNA fragments in ChIP assays with MCF-7 chromatin and C/EBPd-specific antibodies relative to IgG as control. Neg. Ctrl.: from KLF4 locus; Pos. Ctrl.: TLR4. Data represent the mean ± S.E.M; n=3, **P<0.01, ***P<0.001, ****P<0.0001, two-tailed unequal variance t-test. (h) qPCR analysis of ChIP assays with IgG or C/EBPδ–specific antibodies and chromatin from MCF-7 cells 72 h after transfection with Control or CEBPD siRNA (n=3, **P<0.01, ***P<0.001). The inset shows Western analysis of C/EBPδ and β-actin in representative cell lysates. (i) Luciferase reporter activity in SUM159 cells transfected with the indicated promoter reporter constructs or PGL4.2 vector control along with scrambled control siRNA (−) or one of two independent CEBPD siRNAs. Data represent the mean ± S.E.M; n=3, **P<0.01. The Western shows CEBPD silencing in representative extracts. (j) Luciferase reporter activities expressed from POU5F1 and NANOG promoters ± mutations of the C/EBP binding motif (as shown in Table S3) 72 h after transfection into SUM159 cells (n=3, ***P<0.001).
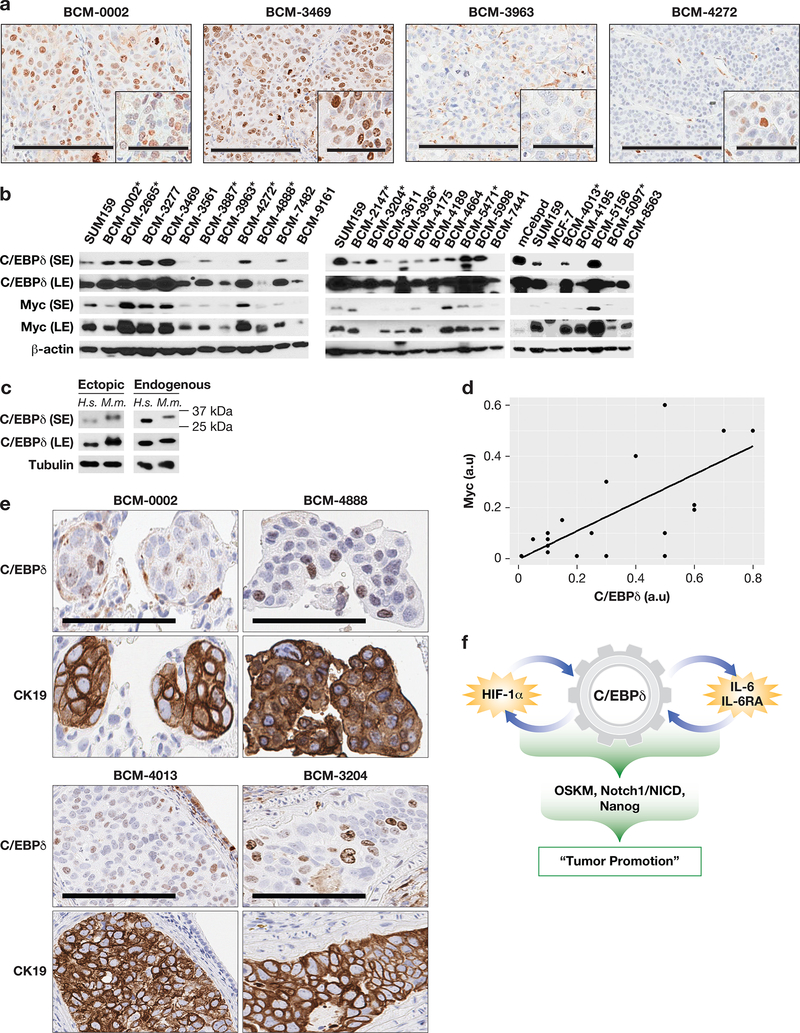
C/EBPδ is expressed in metastatic patient-derived xenografts (PDXs) and lung metastases
To revisit C/EBPδ expression in human cancer and determine if C/EBPδ was detectable in metastatic breast cancer tissues, we turned to PDX models because this resource allows the confirmation of immunostaining results by Western analysis. We assessed 26 PDX models representing mostly TNBCs, 11 of which are metastatic in mice at a rate of 7–66% 48 (and Table S1). As expected27, the majority of these tissues did not exhibit abundant C/EBPδ staining. Fig. 7a illustrates C/EBPδ expression in two of the metastatic models (BCM-0002 and −4272), lack of nuclear detection in one specimen (BCM-3963), and abundant expression in BCM-3469. Western data (Fig. 7b-c) agreed with these results and also showed that human C/EBPδ was expressed in most models. In agreement with a previous report16, tissue level expression of stemeness-factors did not correlate with each other, nor with C/EBPδ (data not shown). However, a highly significant positive relationship was observed between the expression of C/EBPδ and Myc, which has been linked to CSC phenotypes and tumor progression13, 47 (Fig. 7d). Of the 11 PDXs that are metastatic, four expressed C/EBPδ levels similar to SUM159 (BCM-3887, −3204, −5471, and −4013), and three expressed higher amounts (BCM-0002, −2665, and −4272). Notably, BCM-0002 was established from a brain metastasis. Collectively, these data show that C/EBPδ can be expressed at low levels and/or in a subset(s) of cells within metastatic PDX models.
Figure 7. C/EBPδ is expressed in metastatic patient-derived xenografts (PDXs) and lung metastases.
(a) C/EBPδ immunostaining in four PDX models. Scale bars = 200, respectively 60 μm. (b) Western analysis of the indicated proteins in PDX tissues as indicated compared to SUM159 cells in culture (*metastatic in mice; S/LE, short/long exposure). (c) Western analysis of C/EBPδ showing the migration differences between human (H.s.) and mouse (M.m.) C/EBPδ. Ectopic proteins are from HEK293 cells transfected with the respective expression constructs. Endogenous proteins are in extracts from SUM159 human breast cancer cells and the mouse monocytic cell line RAW264.7. Tubulin served as loading control. (d) Correlation plot of ImageJ quantifications of the LE signals for C/EBPδ and Myc from panel (b), normalized to β-actin (a.u., arbitrary units; P=1.032×10−6, Linear Regression). (e) Immunostaining of C/EBPδ and CK19 on parallel section of lung metastases from mice bearing endpoint tumors of the indicated PDX models (scale bar = 100 μm). (f) Model illustrating how the integration of the two positive feed-back loops of HIF-1 and IL-6 signaling together with C/EBPd’s direct targeting of stemness genes can coordinate and amplify the induction of stemness-like features and promote tumor growth (OSKM: Oct4, Sox2, Klf4, Myc).
Next, we assessed C/EBPδ expression in lung lobes of 2–3 mice each from 11 models. CK19 and/or pan-cytokeratin staining demonstrated the presence of varying numbers of metastases in 1–3 mice from eight models. Most of these represented micrometastases and the majority did not exhibit C/EBPδ staining (data not shown). However, C/EBPδ positive cells were detectable in some metastases from seven models, as shown by the examples in Fig. 7e. Thus, while C/EBPδ expression was not frequent in metastases, its expression appears compatible with metastatic tumor cell growth.
Discussion
Although C/EBPδ had often been associated with growth arrest and differentiation in cell culture assays 4, our results demonstrate that human breast cancer cell intrinsic C/EBPδ promotes the growth of primary tumors and experimental metastases in preclinical assays. Tumors that express C/EBPδ have greater numbers of cells with CSC cell surface marker expression and sphere formation capability. Furthermore, self-renewal of sphere formation is supported by C/EBPδ, along with expression of several stemness-promoting factors. Specifically, we find that hypoxia and IL-6 signaling mechanistically synergize by recruiting C/EBPδ as a link between two positive feedback loops (Fig. 7f), which results in the concerted activation of HIF-1, STAT3 and C/EBPδ. This pathway includes inhibition of the FBXW7a tumor suppressor program and activation of the Notch pathway (see Figure 5j), which all together are likely to drive the generation and maintenance of CSCs. Furthermore, C/EBPδ activates expression of the chemokine receptor CXCR43, which has also important roles in CSCs9. FBXW7 regulates normal tissue stem cell balance and inhibits cancer cell stemness 10, 33. We suggest that targeting of C/EBPδ for degradation3 may be part of FBXW7’s function to promote CSCs 33.
In the context of tumor inflammation, the IL-6/STAT3 pathway can convert nonCSCs into CSCs and stimulate the expansion of CSC populations 7, 25, 38, 43. Accordingly, the IL-6Rα receptor is overexpressed in breast CSCs and correlates with poor prognosis 25. Despite this important role of IL-6, nothing is known about the transcriptional regulation of its receptor IL6RA in epithelial cells. We found that C/EBPδ activates IL6RA, adding a new element to the positive feedback loop between IL-6 and C/EBPδ. Thus, C/EBPδ may establish both an autocrine and paracrine pathway to support the development of CSCs. Furthermore, we suggest that some reported functions of C/EBPδ downstream of IL-6 (as a STAT3-induced gene) in other cell types may need to be re-evaluated in light of its role in IL-6Rα expression.
In the context of hypoxia, HIF-1 induces the expression of pro-inflammatory cytokines in the tumor microenvironment 24. However, their crosstalk in establishing an IL-6 autocrine loop had not been fully appreciated. We found that C/EBPδ mediated at least in part hypoxia-induced STAT3 phosphorylation as well as IL-6-induced HIF-1α expression. Conceivably, C/EBPδ may fulfill this role also in the context of the innate immune response. Given that Notch and HIF-1 cooperate to maintain an undifferentiated state 15, our observations reveal how C/EBPδ may serve as a focal point for the generation and amplification of CSCs.
The stem cell phenotype requires the cooperation of several factors as well as the tight regulation of their dosage, because high expression of a given stemness factor can also drive lineage specific differentiation 36. Such consideration may reconcile our current data with C/EBPδ’s expression in a subset of normal breast epithelial cells and lower grade, hormonereceptor positive cancers that correlates with longer patient survival 27, and which may be related to different functions within more differentiated cells. In this regard, the pattern of C/EBPδ expression in breast cancer is similar to that of ERa, the target of first line therapy in ER+ cancers. In ER+ cell lines, C/EBPδ ‘s role in supporting stemness features was most pronounced under hypoxia, IL-6 treatment, and loss of substrate adhesion. Thus, conditions in the tumor microenvironment, as well as the cellular context, have profound effects on C/EBPδ functions, which may also explain its role in supporting MCF-7 cell tumor growth in vivo (this study), but not their proliferation in culture 27. Traditionally, lower grade, ER+ cancers could not be established as PDX models 11. Future mechanistic studies will have to address how C/EBPδ functions are modulated in this context.
In the absence of hypoxia or ectopic IL-6, C/EBPδ supported expression of mesenchymal markers and stemness only in the TNBC cell lines. Analysis of PDX tissues demonstrated that C/EBPδ can be expressed in TNBC specimen including metastatic models, notably also one that had been established from a brain metastasis. The pattern of expression suggests that C/EBPδ may be expressed in specific subclones within these heterogeneous tissues or reflect transient expression. Different subsets of CSCs can co-exist and interconvert and may only need to exist transiently in order to contribute to metastasis 23, 44. Thus, even low levels of C/EBPδ protein may still be sufficient to promote stemness features, especially during transient and acute activation by hypoxia/inflammation and possibly during transition as a circulating tumor cell.
In conclusion, this study shows that by linking and amplifying hypoxia and IL-6 signaling, C/EBPδ orchestrates the synergistic actions of microenvironmental conditions on tumor cell intrinsic signaling pathways that promote cancer cell dedifferentiation, mesenchymal transition, and stem cell-like properties. Our gene silencing/deletion approaches provide proof of principle that C/EBPδ inhibition can reduce tumor growth.
Materials and Methods
See Supplementary Information for details on standard reagents and methodologies.
Sphere cultures.
2,500–10,000 cells were plated on ultra-low attachment plates with MammoCult medium (STEMCELL Technologies, cat.#05621&05622) for 4–5 Days. SFE is expressed as % of cells seeded, evaluating 150–500 μm-sized spheres by Gelcount (Oxford Optronix, UK). For self-renewal assays, the spheres were dissociated into single-cell suspension and re-plated.
Silencing of CEBPD gene expression.
In addition to genetic gene deletion, collectively six different regions of the CEBPD gene were targeted by RNA-interference; with siRNAs, constitutive shRNAs and Dox-inducible shRNAs targeting unique regions (Fig. S6); and complemented by several reconstitution assays. Unless indicated otherwise, cells were passaged 24 h after transfection with si/shRNA, and the effect of si/shRNAs on protein/RNA expression was assessed ≥2 days after nucleofection. Every experiment included non-specific si/shRNA as control.
Patient-derived xenografts.
PDX models were established and maintained as previously reported 48. Western blot analysis was from transplant generations 2–13, and within 0–3 generations of the samples used to obtain immunohistochemistry data. Analysis of lung metastases was from mice with primary tumors of ~1 cm3 in volume. Animal care for mice bearing PDX tumors was in accordance with the NIH Guide for the Care and Use of Experimental Animals.
Statistical analysis.
Unless stated otherwise, quantitative data were analysed by the two-tailed unequal variance t-test and are shown as the mean±S.E.M. The number of samples (n) refers to biological replicates. See Supplementary Information for further details.
Supplementary Material
Acknowledgements
We are thankful for superb support through services provided by Leidos Biomedical Research, Inc., FNLCR (Laboratory Animal Sciences Program, Protein Expression Laboratory, Illustrations and Graphical Support); and the NCI/CCR cores (Small Animal Imaging, Flow Cytometry, Optical Microscopy and Analysis Laboratory); and Data Management Services, Inc for assistance with Statistics. We thank Linda Miller, Suzanne Specht, Rena Mao, and the student interns for their valuable contributions; Heidi Dowst for providing clinical information related to the PDX models; and all the investigators who kindly shared their valuable reagents (see Methods) or provided critical comments on the manuscript.
Competing Interest Statement
This research was supported by the Intramural Research Program of the NIH, National Cancer Institute, in part with Federal Funds from the Frederick Cancer Institute (NIH) under contract no. HHSN261200800001E. L.E.D. and M.T.L. were supported by BCRF Founders Fund Grant CPRIT DP150069 and V-Foundation grant T2014–010. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Footnotes
Conflict of interest. M.T.L. is a limited partner in StemMed Ltd, and a Manager in StemMed Holdings, its general partner. He also holds an equity stake in Tvardi Therapeutics Inc. L.E.D. is a compensated employee of StemMed Ltd.
Supplementary Information is available at the Oncogene’s website.
REFERENCES
- 1.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A 2003; 100: 3983–3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balamurugan K, Wang JM, Tsai HH, Sharan S, Anver M, Leighty R et al. The tumour suppressor C/EBPdelta inhibits FBXW7 expression and promotes mammary tumour metastasis. Embo J 2010; 29: 4106–4117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balamurugan K, Sharan S, Klarmann KD, Zhang Y, Coppola V, Summers GH et al. FBXW7alpha attenuates inflammatory signalling by downregulating C/EBPdelta and its target gene Tlr4. Nature communications 2013; 4: 1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Balamurugan K, Sterneck E. The Many Faces of C/EBPdelta and their Implications in Inflammation and Cancer. Int J Biol Sci 2013; 9: 917–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Balamurugan K HIF-1 at the crossroads of hypoxia, inflammation, and cancer. Int J Cancer 2016; 138: 1058–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brower V Cancer Stem Cell Hypothesis Evolves With Emerging Research. J Natl Cancer Inst 2016; 108. [DOI] [PubMed] [Google Scholar]
- 7.Chang Q, Daly L, Bromberg J. The IL-6 feed-forward loop: a driver of tumorigenesis. Semin Immunol 2014; 26: 48–53. [DOI] [PubMed] [Google Scholar]
- 8.Chen JC, Alvarez MJ, Talos F, Dhruv H, Rieckhof GE, Iyer A et al. Identification of causal genetic drivers of human disease through systems-level analysis of regulatory networks. Cell 2014; 159: 402–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cojoc M, Peitzsch C, Trautmann F, Polishchuk L, Telegeev GD, Dubrovska A. Emerging targets in cancer management: role of the CXCL12/CXCR4 axis. Onco Targets Ther 2013; 6: 1347–1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davis RJ, Welcker M, Clurman BE. Tumor suppression by the Fbw7 ubiquitin ligase: mechanisms and opportunities. Cancer Cell 2014; 26: 455–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dobrolecki LE, Airhart SD, Alferez DG, Aparicio S, Behbod F, Bentires-Alj M et al. Patient-derived xenograft (PDX) models in basic and translational breast cancer research. Cancer Metastasis Rev 2016; 35: 547–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Espinoza I, Pochampally R, Xing F, Watabe K, Miele L. Notch signaling: targeting cancer stem cells and epithelial-to-mesenchymal transition. Onco Targets Ther 2013; 6: 1249–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fallah Y, Brundage J, Allegakoen P, Shajahan-Haq AN. MYC-Driven Pathways in Breast Cancer Subtypes. Biomolecules 2017; 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Friedmann-Morvinski D, Verma IM. Dedifferentiation and reprogramming: origins of cancer stem cells. EMBO reports 2014; 15: 244–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gustafsson MV, Zheng X, Pereira T, Gradin K, Jin S, Lundkvist J et al. Hypoxia requires notch signaling to maintain the undifferentiated cell state. Dev Cell 2005; 9: 617–628. [DOI] [PubMed] [Google Scholar]
- 16.Gwak JM, Kim M, Kim HJ, Jang MH, Park SY. Expression of embryonal stem cell transcription factors in breast cancer: Oct4 as an indicator for poor clinical outcome and tamoxifen resistance. Oncotarget 2017; 8: 36305–36318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harrison H, Rogerson L, Gregson HJ, Brennan KR, Clarke RB, Landberg G. Contrasting hypoxic effects on breast cancer stem cell hierarchy is dependent on ER-alpha status. Cancer Res 2013; 73: 1420–1433. [DOI] [PubMed] [Google Scholar]
- 18.Kageyama Y, Koshiji M, To KK, Tian YM, Ratcliffe PJ, Huang LE. Leu-574 of human HIF-1alpha is a molecular determinant of prolyl hydroxylation. FASEB J 2004; 18: 10281030. [DOI] [PubMed] [Google Scholar]
- 19.Korkaya H, Liu S, Wicha MS. Regulation of cancer stem cells by cytokine networks: attacking cancer’s inflammatory roots. Clin Cancer Res 2011; 17: 6125–6129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lagadec C, Vlashi E, Frohnen P, Alhiyari Y, Chan M, Pajonk F. The RNA-binding protein Musashi-1 regulates proteasome subunit expression in breast cancer- and gliomainitiating cells. Stem Cells 2014; 32: 135–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Litvak V, Ramsey SA, Rust AG, Zak DE, Kennedy KA, Lampano AE et al. Function of C/EBPdelta in a regulatory circuit that discriminates between transient and persistent TLR4-induced signals. Nat Immunol 2009; 10: 437–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lo PK, Kanojia D, Liu X, Singh UP, Berger FG, Wang Q et al. CD49f and CD61 identify Her2/neu-induced mammary tumor-initiating cells that are potentially derived from luminal progenitors and maintained by the integrin-TGFbeta signaling. Oncogene 2012; 31: 2614–2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luo M, Brooks M, Wicha MS. Epithelial-mesenchymal plasticity of breast cancer stem cells: implications for metastasis and therapeutic resistance. Curr Pharm Des 2015; 21: 1301–1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mamlouk S, Wielockx B. Hypoxia-inducible factors as key regulators of tumor inflammation. Int J Cancer 2013; 132: 2721–2729. [DOI] [PubMed] [Google Scholar]
- 25.Marotta LL, Almendro V, Marusyk A, Shipitsin M, Schemme J, Walker SR et al. The JAK2/STAT3 signaling pathway is required for growth of CD44(+)CD24(−) stem celllike breast cancer cells in human tumors. J Clin Invest 2011; 121: 2723–2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mathieu J, Zhang Z, Zhou W, Wang AJ, Heddleston JM, Pinna CM et al. HIF induces human embryonic stem cell markers in cancer cells. Cancer Res 2011; 71: 4640–4652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mendoza-Villanueva D, Balamurugan K, Ali HR, Kim SR, Sharan S, Johnson RC et al. The C/EBPdelta protein is stabilized by estrogen receptor alpha activity, inhibits SNAI2 expression and associates with good prognosis in breast cancer. Oncogene 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mihara M, Hashizume M, Yoshida H, Suzuki M, Shiina M. IL-6/IL-6 receptor system and its role in physiological and pathological conditions. Clin Sci (Lond) 2012; 122: 143159. [DOI] [PubMed] [Google Scholar]
- 29.Min Y, Ghose S, Boelte K, Li J, Yang L, Lin PC. C/EBP-delta regulates VEGF-C autocrine signaling in lymphangiogenesis and metastasis of lung cancer through HIF1alpha. Oncogene 2011; 30: 4901–4909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Min Y, Li J, Qu P, Lin PC. C/EBP-delta positively regulates MDSC expansion and endothelial VEGFR2 expression in tumor development. Oncotarget 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mitra A, Mishra L, Li S. EMT, CTCs and CSCs in tumor relapse and drug-resistance. Oncotarget 2015; 6: 10697–10711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Prat A, Parker JS, Karginova O, Fan C, Livasy C, Herschkowitz JI et al. Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Breast Cancer Res 2010; 12: R68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rustighi A, Zannini A, Tiberi L, Sommaggio R, Piazza S, Sorrentino G et al. Prolylisomerase Pin1 controls normal and cancer stem cells of the breast. EMBO molecular medicine 2014; 6: 99–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sansone P, Storci G, Tavolari S, Guarnieri T, Giovannini C, Taffurelli M et al. IL-6 triggers malignant features in mammospheres from human ductal breast carcinoma and normal mammary gland. J Clin Invest 2007; 117: 3988–4002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sarkar TR, Sharan S, Wang J, Pawar SA, Cantwell CA, Johnson PF et al. Identification of a Src tyrosine kinase/SIAH2 E3 ubiquitin ligase pathway that regulates C/EBPdelta expression and contributes to transformation of breast tumor cells. Mol Cell Biol 2012; 32: 320–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Seymour T, Twigger AJ, Kakulas F. Pluripotency Genes and Their Functions in the Normal and Aberrant Breast and Brain. Int J Mol Sci 2015; 16: 27288–27301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tabassum DP, Polyak K. Tumorigenesis: it takes a village. Nat Rev Cancer 2015; 15: 473–483. [DOI] [PubMed] [Google Scholar]
- 38.Taniguchi K, Karin M. IL-6 and related cytokines as the critical lynchpins between inflammation and cancer. Semin Immunol 2014; 26: 54–74. [DOI] [PubMed] [Google Scholar]
- 39.Valent P, Bonnet D, De Maria R, Lapidot T, Copland M, Melo JV et al. Cancer stem cell definitions and terminology: the devil is in the details. Nat Rev Cancer 2012; 12: 767–775. [DOI] [PubMed] [Google Scholar]
- 40.Visvader JE, Lindeman GJ. Cancer stem cells: current status and evolving complexities. Cell Stem Cell 2012; 10: 717–728. [DOI] [PubMed] [Google Scholar]
- 41.Wang YH, Wu WJ, Wang WJ, Huang HY, Li WM, Yeh BW et al. CEBPD amplification and overexpression in urothelial carcinoma: a driver of tumor metastasis indicating adverse prognosis. Oncotarget 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Watanabe M, Uehara Y, Yamashita N, Fujimura Y, Nishio K, Sawada T et al. Multicolor detection of rare tumor cells in blood using a novel flow cytometry-based system. Cytometry A 2014; 85: 206–213. [DOI] [PubMed] [Google Scholar]
- 43.Wei W, Tweardy DJ, Zhang M, Zhang X, Landua J, Petrovic I et al. STAT3 signaling is activated preferentially in tumor-initiating cells in claudin-low models of human breast cancer. Stem Cells 2014; 32: 2571–2582. [DOI] [PubMed] [Google Scholar]
- 44.Wei W, Lewis MT. Identifying and targeting tumor-initiating cells in the treatment of breast cancer. Endocr Relat Cancer 2015; 22: R135–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yamaguchi J, Tanaka T, Eto N, Nangaku M. Inflammation and hypoxia linked to renal injury by CCAAT/enhancer-binding protein delta. Kidney Int 2015; 88: 262–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yan W, Chen Y, Yao Y, Zhang H, Wang T. Increased invasion and tumorigenicity capacity of CD44+/CD24-breast cancer MCF7 cells in vitro and in nude mice. Cancer cell international 2013; 13: 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang A, Qin S, Schulte BA, Ethier SP, Tew KD, Wang GY. MYC Inhibition Depletes Cancer Stem-like Cells in Triple-Negative Breast Cancer. Cancer Res 2017; 77: 66416650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang X, Claerhout S, Prat A, Dobrolecki LE, Petrovic I, Lai Q et al. A renewable tissue resource of phenotypically stable, biologically and ethnically diverse, patient-derived human breast cancer xenograft models. Cancer Res 2013; 73: 4885–4897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zou J, Li P, Lu F, Liu N, Dai J, Ye J et al. Notch1 is required for hypoxia-induced proliferation, invasion and chemoresistance of T-cell acute lymphoblastic leukemia cells. J Hematol Oncol 2013; 6: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.