Abstract
The human Fc-gamma receptors (FcγRs) link adaptive and innate immunity by binding immunoglobulin G (IgG). All human low-affinity FcγRs are encoded by the FCGR2/3 locus containing functional single nucleotide polymorphisms (SNPs) and gene copy number variants. This locus is notoriously difficult to genotype and high-throughput methods commonly used focus on only a few SNPs. We performed multiplex ligation-dependent probe amplification for all relevant genetic variations at the FCGR2/3 locus in >4,000 individuals to define linkage disequilibrium (LD) and allele frequencies in different populations. Strong LD and extensive ethnic variation in allele frequencies was found across the locus. LD was strongest for the FCGR2C-ORF haplotype (rs759550223+rs76277413), which leads to expression of FcγRIIc. In Europeans, the FCGR2C-ORF haplotype showed strong LD with, among others, rs201218628 (FCGR2A-Q27W, r2 = 0.63). LD between these two variants was weaker (r2 = 0.17) in Africans, whereas the FCGR2C-ORF haplotype was nearly absent in Asians (minor allele frequency <0.005%). The FCGR2C-ORF haplotype and rs1801274 (FCGR2A-H131R) were in weak LD (r2 = 0.08) in Europeans. We evaluated the importance of ethnic variation and LD in Kawasaki Disease (KD), an acute vasculitis in children with increased incidence in Asians. An association of rs1801274 with KD was previously shown in ethnically diverse genome-wide association studies. Now, we show in 1,028 European KD patients that the FCGR2C-ORF haplotype, although nearly absent in Asians, was more strongly associated with susceptibility to KD than rs1801274 in Europeans. Our data illustrate the importance of interpreting findings of association studies concerning the FCGR2/3 locus with knowledge of LD and ethnic variation.
Keywords: Fc-gamma receptor, FCGR polymorphism, linkage disequilibrium, Kawasaki disease (KD), immunogenetics
Introduction
The human cellular receptors for Immunoglobulin G (IgG), the Fc-gamma receptors (FcγR), have an important role in immunity by linking the adaptive and innate immune systems. Many genetic variations in the genes encoding FcγRs have been found to be associated with auto-immune (1–5), auto-inflammatory (6–8), and infectious diseases (9, 10), and with efficacy of immunotherapy in cancer patients (11–15). Several activating and one single inhibitory FcγR (FcγRIIb) exist, with differential expression on various leukocyte subsets (16, 17). Human FcγRs can be distinguished into one high-affinity receptor (FcγRI) and five low-affinity FcγRs (the different isoforms of FcγRII and FcγRIII) (16, 17). All five genes encoding the low-affinity FcγRs (FCGR2A, FCGR2B, FCGR2C, FCGR3A, and FCGR3B) are located in a complex gene cluster at chromosome 1q23.3. Many functionally relevant single nucleotide polymorphisms (SNPs) and copy number variants (CNVs) are described in the FCGR2/3 locus, leading to altered receptor functions ranging from different binding affinity to IgG to complete absence of expression of certain genes (17–19). The FCGR2/3 locus involves a segmental duplication, making it constitutively difficult to genotype because of the high degree of homology between the genes (18, 20). Due to the close proximity of all the five different FCGR2 and FCGR3 genes, the polymorphic variants in these genes are likely to be in strong Linkage Disequilibrium (LD). However, except for some incidental reports on LD between some of the SNPs (21–24), a comprehensive analysis of LD between the functional variants at this locus has not been previously performed.
One of the diseases in which only one genetic variant of the FCGR2/3 locus has been thoroughly studied is Kawasaki Disease (KD). KD is an acute systemic vasculitis that predominantly occurs in children <5 years (25). About 25% of untreated KD patients develop coronary artery aneurysms, which may lead to ischemic heart disease, myocardial infarction and sudden death at young age (26). Although the etiology of KD remains unknown, the general consensus is that KD reflects an abnormal inflammatory response to an unknown infectious trigger in genetically susceptible individuals. Standard treatment consists of a single infusion of high-dose intravenous immunoglobulins (IVIg) in combination with aspirin (27). Although the mechanism of action of IVIg in KD is unclear, early treatment shortens the duration of fever and reduces the incidence of coronary artery aneurysms to less than 5% (28). Since IVIg therapy is effective in the majority of patients, the receptors for IgG, the Fc-gamma Receptors (FcγRs), are of particular interest in KD research.
In our GWAS study on KD (6), we identified the FCGR2A-131H SNP (rs1801274) to be associated at genome-wide significance. This variant results in a substantial difference in the ability of FcγRIIa to bind the human IgG2 subclass (19). rs1801274 shows the strongest evidence of association with KD and this finding has been intensively studied and validated in a number of cohorts of varying ethnicity (6, 7, 29–34). Apart from the FCGR2A-H131R SNP (rs1801274), only a few other SNPs in this locus have been evaluated for KD susceptibility, without any significant association (29–31). Nevertheless, because of the sequence homology and the genetic complexity, a very large part of the FCGR2/3 locus was not covered in GWAS or other studies before. Hence, we postulated that other variants at the locus may also play a role in KD susceptibility, which could either be tagged by FCGR2A-131H (rs1801274), or act independently. To address this, we performed further fine-mapping of the FCGR2/3 gene cluster in a case-control as well as a family-based linkage study with a total of 1,028 patients with KD, and genotyped healthy control individuals of different ethnic groups to define LD and ethnic variation. We used a previously developed accurate multiplex ligation-dependent probe amplification (MLPA) assay covering all the functionally relevant SNPs and CNVs at the FCGR2/3 locus (5).
In the present study, including more than 4,000 individuals, we found marked ethnic differences in allele frequencies for most of the SNPs and CNVs. The most prominent difference was observed for the FCGR2C-ORF haplotype, which we have previously shown to result in expression of the activating FcγRIIc (35). In most individuals, FcγRIIc cannot be expressed as a result of a polymorphic stop codon in exon3 (rs759550223), but the expressed FCGR2C-ORF haplotype is associated with susceptibility to immune thrombocytopenic purpura (5). We now show that the FCGR2C-ORF haplotype is virtually absent in Asian and African populations. FCGR2C-ORF is in very strong LD with several other SNPs in the European population, but could be identified as a novel susceptibility haplotype for KD in this population, independent of the FCGR2A-H131R SNP. Our comprehensive analysis of the FCGR2/3 locus will greatly contribute to a better understanding of the relevance of the different FcγRs in inflammatory diseases.
Subjects and Methods
Study Populations
KD Cases
Unrelated KD cases were recruited from Australia, The Netherlands and the United States. All cases from Australia (109) and the United States (62) were also included in our previous GWAS (6), whereas the cases from the Netherlands (234) consisted of 166 cases from the GWAS and 68 new cases. There was no overlap with patients in the study previously reported by Biezeveld et al (30). The diagnosis of KD was based on the standard diagnostic clinical criteria from the American Heart Association.
Cohorts of Control Subjects
Europeans
Since no DNA of the control population in our previous GWAS was available, we genotyped a new group of unrelated controls of European descent, consisting of healthy individuals from Austria (478), Australia (156), The Netherlands (199), and the United Kingdom (86). All were of European descent by self-reported ethnicity (36, 37).
Chinese
The Chinese population consisted of 428 healthy individuals from Canada of Han-Chinese descent, all of which were grandparent-proven Han-Chinese.
African
The South African population consisted of 149 healthy blood donors of African descent by self-reported ethnicity as reported before (38). The Ethiopian population consisted of 142 healthy blood donors of African Ethiopian descent by self-reported ethnicity (38). The West African population consisted of 65 sickle-cell disease patients from the Netherlands, all of which were of West-African descent by self-reported ethnicity, including individuals from Ghana (52), Nigeria (4), Sierra Leone (4), Togo (3), and Cameroon (2). The Surinam population consisted of 78 sickle-cell disease patients of African Surinamese descent by self-reported ethnicity. The Antillean population consisted of 6 sickle-cell disease patients from the Netherlands who were from Curaçao and were of African Caribbean descent by self-reported ethnicity, and 68 healthy blood donors from Curaçao who were of African Caribbean descent by self-reported ethnicity as described previously (38).
Family-based association study
623 KD patients (none overlapping with the case control study) were included, consisting of KD patients from the United States (386, of which 348 complete trios and 38 incomplete trios, 153 European), Australia (104, all complete trios, 72 European) and the Netherlands (98, all complete trios, 82 European) and Italy (35, all complete trios, all Mediterranean). All KD patients in the family-based association study from the United States and Australia were included in our previous GWAS (6), the patients from the Netherlands and Italy were new.
In total, 4,091 individuals were genotyped. Table S1 provides an overview of all individuals. This study was carried out in accordance with the recommendations of the Kawasaki Study Protocol approved by the Medical Ethical Committee at the Academic Medical Centre in Amsterdam, the Netherlands, with written informed consent from all subjects. All subjects gave written informed consent in accordance with the Declaration of Helsinki. The protocol was approved by the Medical Ethical Committee at the Academic Medical Centre in Amsterdam, the Netherlands and by the medical ethical committees of the other participating centers.
Clinical Data
Clinical information was collected by review of the clinical KD registries. CAAs were defined based on the definition of the Japanese Ministry of Health or Z-scores >2.5 according to the Boston Z-score data. According to the definition of the Japanese Ministry of Health a coronary artery was considered abnormal if the diameter of the internal lumen was > in children younger than 5 years or > in a child aged 5 years or older, or if the internal diameter of a segment was at least 1.5 times larger than that of an adjacent segment. IVIg response was determined in the patients receiving treatment with IVIg within 11 days after the disease onset. Patients who received more than one dose of IVIg because of persistent or recrudescent fever more than 36 h after the initial IVIg dose were defined as IVIg non-responders.
Genotyping by MLPA and Construction of Haplotypes From MLPA Data
The MLPA assay was performed according to the manufacturer's protocol, essentially as described previously (5, 39) and is described in great detail in the Supplemental Methods.
Flow Cytometry, Gene Expression Microarray and RT-qPCR
Flow cytometry, gene expression microarray and RT-qPCR were performed as described in the Supplemental Methods.
Statistical Analysis
Genotype/Allele Frequencies and Linkage Disequilibrium
Differences in copy number and allele frequencies between (sub)populations and differences in allele frequencies between groups of individuals with normal, decreased and increased copy number were tested using Fisher's Exact test. Haplotype frequencies and linkage disequilibrium (expressed as r2 or D') between (multiallelic) markers were estimated in the populations and the parents from the KD trios using the gap package (40) (version 1.1-12).
Association With Susceptibility to Kawasaki Disease (KD)
In the case-control study, genotype frequencies were compared between KD cases and healthy controls using Fisher's exact test and odds ratios were estimated using (multiple) logistic regression. In the parent-affected offspring trios, the association between KD and the markers was examined using the (multimarker) FBAT (TDT) test statistic from the FBAT toolkit (41). Results from the case-control and KD trios were meta-analyzed using a fixed effect model and the generic inverse variance method following an approach described by Kazeem and Farrall (42) and using Review Manager software (Version 5, Cochrane Collaboration).
Comparison of Expression Levels
In case of multiple expression values per donor, the mean of these values was taken for the statistical analyses. Expressions between groups were compared using Mann-Whitney tests (two groups) or a Kruskal-Wallis test with post-hoc Mann-Whitney tests (>2 groups) using GraphPad Prism 6.02.
Apart from the TDT and meta-analyses and the expression analysis, all statistical analyses were carried out using R software (Version 3.0.3, R Core Team). A p-value below 0.05 was considered as statistically significant.
Results
Characterization of the FCGR2/3 Locus
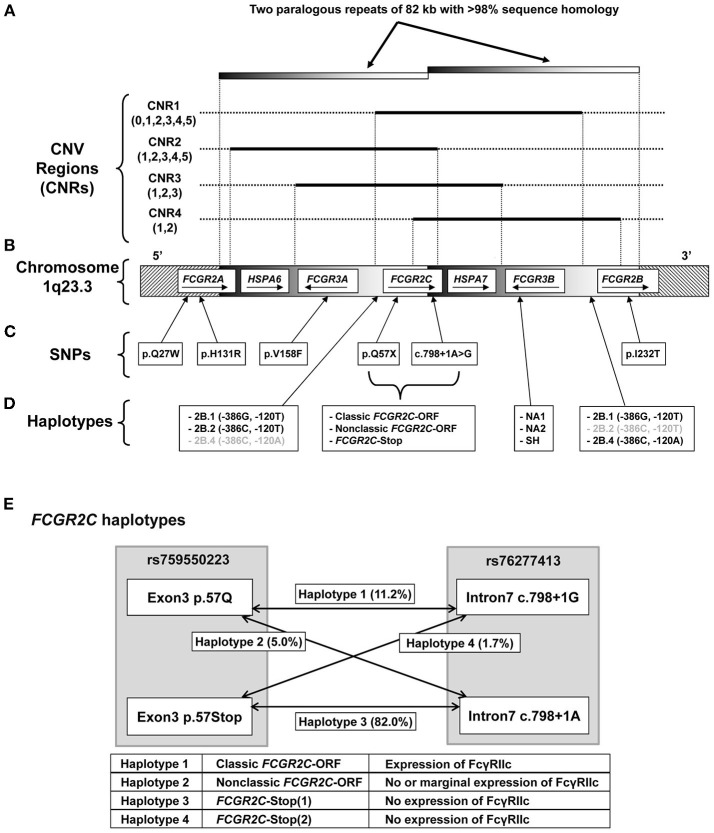
The FCGR2/3 locus is a complex region due to the presence of a large segmental duplication and copy number variants (CNV) (18, 43). MLPA was previously shown to accurately call copy number variation at the FCGR2/3 locus (5, 20). We used the MLPA to accurately identify all eight known functional SNPs and haplotypes, as well as the four CNV regions (CNRs), at the FCGR2/3 locus, which have previously been associated with various autoimmune and infectious diseases (Figure 1 and Table S2).
Figure 1.
Genomic organization of the FCGR2/3 locus. Overview of the FCGR2/3 locus at 1q23.3. (A) CNV at the locus occurs always in regions containing a series of genes, termed CNV regions (CNRs) (18, 21). Numbers between brackets indicate observed copy number for that CNR in the individuals tested in this study, black lines indicate the extent of the different CNRs. Gray shaded bars indicate the extent of two paralogous repeats of the locus. The novel rare CNR4 that we recently described (18) was found in 4 of the individuals included in this study, and was combined with the similar CNR1 in this study for reasons of simplicity. (B) Overview of the genes in the locus with their orientation. Depiction of genes is not to scale. (C) Functional SNPs at the locus, indicated by single letter amino acid code and amino acid position, except for the splice variant c.798+1A>G. All these SNPs were determined in this study. (D) Functional haplotypes at the locus. 2B.1, 2B.2, and 2B.4 are haplotypes of two SNPs (at nucleotide positions −386 and −120 relative to the start of translation) in the otherwise identical promoter regions of FCGR2B and FCGR2C (haplotypes in gray are very rarely found in that particular gene). FCGR3B NA1, NA2, and SH are haplotypes determined by six SNPs. These haplotypes determine the different allotypes of Human Neutrophil Antigen 1 (HNA1) involved in neutrophil alloimmunization, and respectively encode the HNA1a/HNA1b/HNA1c antigenic variants. (E) Schematic representation of the different haplotypes of FCGR2C haplotypes, which determine expression of FcγRIIc. FCGR2C-Stop (1) and FCGR2C-Stop (2) haplotypes are similar in function and expression and are taken together as FCGR2C-Stop throughout the manuscript. Percentages represent allele frequencies of the different haplotypes in European healthy controls.
Allele Frequencies of CNV and SNPs at the FCGR2/3 Locus Vary Among Different Ethnic Groups, Especially for the Classic and Nonclassic FCGR2C-ORF Haplotypes
The frequencies of many of the functional SNPs and CNVs have been reported to vary among different ethnic backgrounds (10, 21, 44–47), but information about the FCGR2C haplotypes is yet to be established. To explore differences in frequencies of SNPs and CNRs between several ethnic groups, we genotyped and compared large groups of healthy human subjects. Significant differences (P < 0.05) between ethnic groups were found for CNRs and for all SNPs except the FCGR3A-V158F SNP, which had no difference in frequency among all groups (Table 1). Analysis of subgroups within the European and African populations revealed subtle differences within the European population and marked differences within the African population (Table S3).
Table 1.
Frequencies of CNVs (CNRs, proportion of individuals with that number of copies is shown) and SNPs (allele frequencies are shown).
| Variant | European (n = 919) | Chinese (n = 428) | African (n = 508) | Fisher's exact | |
|---|---|---|---|---|---|
| CNR1 | |||||
| FCGR3B + FCGR2C | 0 copies | 0.00 | 0.00 | 0.00 | |
| 1 copy | 0.07 | 0.09 | 0.11 | ||
| 2 copies | 0.83 | 0.73 | 0.73 | ||
| 3 copies | 0.09 | 0.17 | 0.14 | ||
| 4 copies | 0.01 | 0.01 | 0.01 | <0.0001 | |
| CNR2 | |||||
| FCGR3A + FCGR2C | 1 copy | 0.01 | 0.01 | 0.01 | |
| 2 copies | 0.94 | 0.96 | 0.96 | ||
| 3 copies | 0.04 | 0.04 | 0.03 | ||
| 4 copies | 0.00 | 0.00 | 0.00 | 0.87 | |
| CNR3 | |||||
| FCGR3A + FCGR2C | 1 copy | 0.00* | 0.00 | 0.00* | |
| 2 copies | 1.00 | 0.98 | 1.00 | ||
| 3 copies | 0.00 | 0.02 | 0.00 | <0.001 | |
| FCGR2A | |||||
| 131 H | 0.54 | 0.67 | 0.44 | ||
| 131 R | 0.46 | 0.33 | 0.56 | <0.0001 | |
| 27 Q | 0.88 | 1.00 | 0.89 | ||
| 27 W | 0.12 | 0.00 | 0.11 | <0.0001 | |
| FCGR3A | |||||
| 158 F | 0.64 | 0.64 | 0.64 | ||
| 158 V | 0.36 | 0.36 | 0.36 | 0.94 | |
| FCGR2C | |||||
| Stop | 0.84 | 1.00 | 0.90 | ||
| Classic ORF | 0.11 | 0.00 | 0.02 | ||
| Nonclassic ORF | 0.05 | 0.00 | 0.08 | <0.0001 | |
| Promoter haplotype | 2B.1 | 0.89 | 1.00 | 0.95 | |
| 2B.2 | 0.11 | 0.00 | 0.05 | <0.0001 | |
| FCGR3B | |||||
| NA1 | 0.35 | 0.62 | 0.38 | ||
| NA2 | 0.62 | 0.38 | 0.46 | ||
| SH | 0.02 | 0.00 | 0.15 | <0.0001 | |
| FCGR2B | |||||
| 232I | 0.88 | 0.74 | 0.73 | ||
| 232T | 0.12 | 0.26 | 0.27 | <0.0001 | |
| Promoter haplotype | 2B.1 | 0.90 | 1.00 | 0.99 | |
| 2B.4 | 0.10 | 0.00 | 0.01 | <0.0001 | |
Fisher's exact test: Overall P for differences between populations for that variation is shown. P-values < 0.05 are shown in bold.
1 European and 1 West African individual showed a deletion of CNR3.
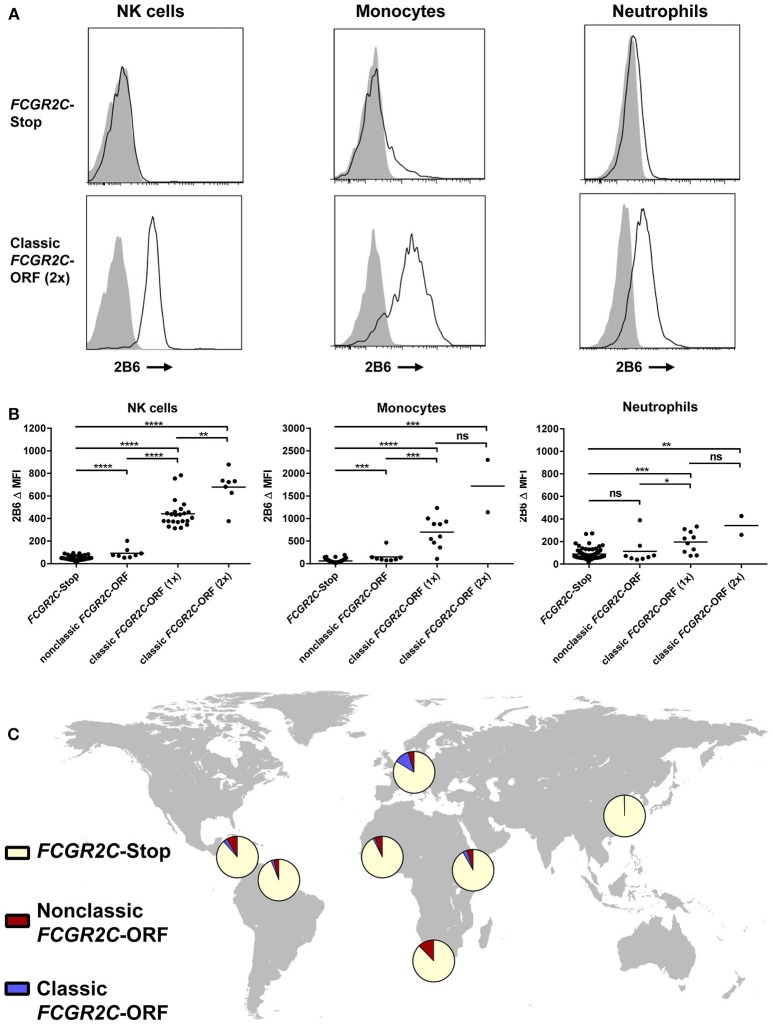
Among the groups included, the largest difference in allele frequency was revealed for the FCGR2C-haplotypes. FCGR2C consists of three haplotypes; the FCGR2C-Stop pseudogene that is not expressed as a result of the FCGR2C-Q57X SNP (rs759550223), its expressed counterpart, the so-called classic FCGR2C-ORF with an open reading frame at rs759550223, and the nonclassic FCGR2C-ORF, which has an open reading frame at rs759550223 but has an almost complete lack of expression as a result of a splice site mutation in intron7 (rs76277413) (35). Figure 1E gives a schematic overview of the haplotypes of FCGR2C. The classic FCGR2C-ORF haplotype results in the expression of FcγRIIc as an activating IgG receptor on myeloid cells and NK cells, as we have characterized previously (5, 48). We now formally demonstrate that the nonclassic FCGR2C-ORF haplotype can be determined by MLPA (see Supplemental Methods and Table S4 for a description), as expression of FcγRIIc is indeed low to absent in individuals genotyped as nonclassic FCGR2C-ORF by MLPA (Figure 2, gating strategy Figure S1). The slight difference in staining levels compared to individuals with the FCGR2C-stop variant shows that there is some residual expression of FcγRIIc protein, but this is less than 10% of the expression in classic FCGR2C-ORF individuals. These haplotypes were markedly different among different ethnic groups; the classic FCGR2C-ORF haplotype was virtually absent in Chinese (present in 2 out of 428 individuals, minor allele frequency <0.005%) and rare in the different African populations, whereas the nonclassic FCGR2C-ORF was more prevalent in African populations compared to Europeans (Table 1 and Figure 2C).
Figure 2.
Haplotypes of FCGR2C determine expression of FcγRIIc. (A) Representative histograms of staining with MoAb 2B6 on NK cells (left), monocytes (middle), and neutrophils (right) of an individual homozygous for FCGR2C-Stop variant (upper panel) and an individual homozygous for the FCGR2C-ORF variant (lower panel). 2B6 recognizes the extracellular domain of both FcγRIIb and FcγRIIc. In FCGR2C-Stop individuals, FcγRIIc cannot be expressed, therefore staining of 2B6 in those individuals must be FcγRIIb only. Black line: 2B6, gray shading: isotype control. (B) Summary of 2B6 staining, corrected for isotype control, on human NK cells (left), monocytes (middle) and neutrophils (right) of genotyped individuals. Y axis scale is different for monocytes than for the other cell types. Because FcγRIIb is also stained by 2B6, only cells that do not express FcγRIIb can be easily analyzed for FcγRIIc expression. Therefore, individuals with a deletion of CNR1 were excluded from the analysis of NK cells, and individuals with a 2B.4 promoter haplotype in FCGR2B were excluded from the analysis of monocytes and neutrophils, because these variants result in ectopic expression of FcγRIIb on NK cells (35), or myeloid cells (36), respectively. NK cell analysis: FCGR2C-Stop n = 93, nonclassic FCGR2C-ORF, including cases with 1 or 2 copies n = 8, FCGR2C-ORF(1x), with 1 copy of the classic FCGR2C-ORF haplotype, n = 23, FCGR2C-ORF(2x), with 2 copies n = 7. Monocyte and neutrophil analysis: FCGR2C-Stop n = 99, nonclassic FCGR2C-ORF, including cases with 1 or 2 copies n = 8, FCGR2C-ORF(1x), with 1 copy of the classic FCGR2C-ORF haplotype, n = 10, FCGR2C-ORF(2x), with 2 copies n = 2. Some individuals were analyzed more than once at different time points with similar results; means are shown for these. All individuals analyzed are of European descent except for five FCGR2C-Stop and two nonclassic FCGR2C-ORF individuals who were of African origin. (C) World map showing allele frequencies of FCGR2C haplotypes for different ethnic groups. MFI, median fluorescence intensity; ns, non-significant; *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001 as determined by Mann Whitney test.
Linkage Disequilibrium at the FCGR2/3 Locus Defined
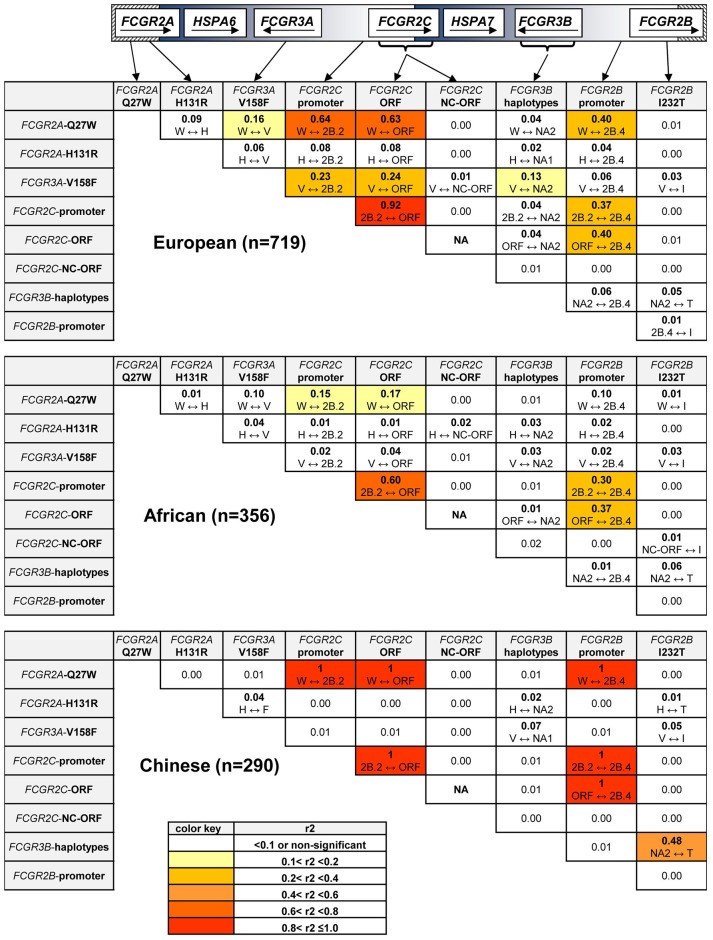
Because many functionally relevant SNPs in the FCGR2/3 locus are located in close proximity to each other, the SNPs in FCGR genes are likely to be in strong LD, which can greatly complicate the interpretation of genetic association studies. From the control samples of the different ethnic reference populations, we first calculated the background LD pattern based on the SNPs and haplotypes in the individuals that did not show CNV (r2 in Figure 3, D' in Figure S2).
Figure 3.
Linkage Disequilibrium at the FCGR2/3 locus. Linkage Disequilibrium for SNPs and haplotypes in individuals without CNV. r2 is shown for all combinations, which variant is linked to which variant is shown underneath. Values shown in bold are significantly different from 0 (p < 0.05). FCGR2C-ORF = classic FCGR2C-ORF haplotype vs. all other FCGR2C haplotypes. FCGR2C-NC-ORF = nonclassic FCGR2C-ORF haplotype vs. all other FCGR2C haplotypes. NA, not available, because the classic FCGR2C-ORF haplotype and nonclassic FCGR2C-ORF haplotype are mutually exclusive. Polymorphic amino acids are indicated by one-letter code.
In the European population, we found strong LD of the classic FCGR2C-ORF haplotype (rs759550223 and rs76277413) with several of the other SNPs in the region. First, the classic FCGR2C-ORF haplotype was in almost complete LD (r2 = 0.92) with the 2B.2 promoter in FCGR2C (rs149754834). Furthermore, there was strong LD between the classic FCGR2C-ORF variant and FCGR2A-27W (rs201218628, r2 = 0.63) and with the 2B.4 promoter haplotype in FCGR2B (rs143796418, r2 = 0.40). Weaker LD was observed for the classic FCGR2C-ORF haplotype with FCGR3A-158V (rs396991, r2 = 0.24) and FCGR2A-131H (rs1801274, r2 = 0.08).
In the Chinese population, LD for the classic FCGR2C-ORF haplotype appeared similar to the LD in Europeans, but this was based only on 2 individuals.
In the African population, LD was also found for the FCGR2C-ORF haplotype with several of the variants, but in general this LD was weaker than in Europeans (Figure 3, second panel).
The previously described LD between FCGR3A-158V (rs396991) and FCGR2A-131H (rs1801274) (21, 23) was confirmed in the European and African population, although relatively weak (r2 = 0.06). We show now that this LD was reversed in the Chinese population, i.e., FCGR3A-158F (rs396991) and FCGR2A-131H (rs1801274) were in weak LD (r2 = 0.04).
We then investigated LD between CNV and SNPs for all of the CNRs known at the locus. Because the standard measurements of LD (r2 and D') cannot be calculated in areas with CNV, we performed this analysis by calculating allele frequencies for groups of individuals with normal (2 copies), decreased (≤1 copies) or increased (≥3 copies) copy number of at least one CNR and analyzed significant differences by Fisher's exact test.
Results for CNR1 are shown in Table S5. For CNR1, strong LD was found between increased copy number and the nonclassic FCGR2C-ORF haplotype (rs759550223 and rs76277413), both in the European and African population. Increased copy number in CNR1 also revealed strong LD with the FCGR3B-SH (rs5030738) haplotype in the European, but not in the African population. Some other SNPs [FCGR2A-H131R (rs1801274); FCGR3A-V158F (rs396991); FCGR2B-I232T (rs1050501)] were also associated with changes in CNV in CNR1.
For the less prevalent CNR2, LD was found only for rs1050501 in the European population (All results for CNR2 are shown in Table S6).
For the rare CNR3, no statistically significant LD was found at all (data not shown).
Association of SNPs and CNV at the FCGR2/3 Locus With Susceptibility to KD
After defining the background allele frequencies and LD of the functional SNPs and CNV in the control groups, we then analyzed the full content of variants in the FCGR2/3 locus for susceptibility to KD, now also including the SNPs and CNV in the region that had not been covered in our previous GWAS study (6). We performed a case-control study in 405 KD cases and the cohort of 919 controls described above, all of European descent. For a family-based association study, 586 complete trios and 37 incomplete trios were genotyped. The characteristics of the KD patients are shown in Table S7.
Case-Control Study
Genotype and allele frequencies of CNVs and SNPs are shown in Table 2. Several significant differences between cases and controls were observed, the most significant being the classic FCGR2C-ORF (rs759550223 and rs76277413) (15.7% vs. 11.2%, P = 0.002). Other significantly associated SNPs were the 2B.2 promoter in FCGR2C (rs149754834) (15.3% vs. 10.8%, P = 0.009), the FCGR2A 27Q>W SNP (rs201218628) (15.3% vs. 11.9%, P = 0.014) and the 2B.4 promoter in FCGR2B (rs143796418) (12.7% vs. 10.0%, P = 0.047). These four significantly associated variants are in strong LD with each other (Figure 3). In a multiple logistic regression analysis that included all the variants, none were independently associated, but a backward regression analysis revealed the classic FCGR2C-ORF as the strongest predictor of KD susceptibility (data not shown).
Table 2.
Genotype and allele frequencies of functional genetic variants at the FCGR2/3 locus, comparing KD patients of European descent with healthy controls of European descent.
| Variant | Cases | Controls | Fisher | Single logistic regression (additive model) | Multiple logistic regression | ||
|---|---|---|---|---|---|---|---|
| (n = 405) | (n = 919) | All variants | 2 variants | ||||
| P-value | OR (95%LL-95%UL) | P-value | P-value | P-value | |||
| CNR1 | |||||||
| (FCGR2C + FCGR3B) | |||||||
| 0 copies | 1 | 1 | <2 vs. rest: | ||||
| 1 copy | 27 | 60 | 1.04 (0.66–1.66) | 0.853 | 0.719 | ||
| 2 copies | 348 | 768 | |||||
| 3 copies | 27 | 83 | >2 vs. rest: | ||||
| 4 copies | 2 | 7 | 0.533 | 0.71 (0.46–1.10) | 0.124 | 0.291 | |
| CNR2 | |||||||
| (FCGR2C + FCGR3A) | |||||||
| 1 copy | 3 | 11 | <2 vs. rest: | ||||
| 2 copies | 376 | 866 | 0.62 (0.17–2.22) | 0.459 | 0.491 | ||
| 3 copies | 25 | 41 | >2 vs. rest: | ||||
| 4 copies | 1 | 1 | 0.390 | 1.43 (0.87–2.37) | 0.162 | 0.256 | |
| CNR3 | |||||||
| (FCGR2C + FCGR3A) | |||||||
| 2 copies | 405 | 917 | >2 vs. rest: | ||||
| 3 copies | 0 | 2 | 1.000 | 0.00 (0.00-inf) | 0.973 | 0.973 | |
| FCGR2A Q27W | |||||||
| 289 | 713 | ||||||
| QW | 108 | 194 | |||||
| WW | 8 | 12 | 0.047 | ||||
| Allele frequency (W) | 15.3% | 11.9% | 1.35 (1.06–1.72) | 0.014 | 0.783 | ||
| FCGR2A H131R | |||||||
| HH | 122 | 269 | |||||
| HR | 211 | 463 | |||||
| RR | 72 | 187 | 0.559 | ||||
| Allele frequency (H) | 56.2% | 54.5% | 1.07 (0.91–1.27) | 0.408 | 0.857 | 0.927 | |
| FCGR3A V158F | |||||||
| 0 V (F, FF, FFF, FFFF) | 150 | 386 | |||||
| 1 V (V, VF, VFF) | 205 | 403 | |||||
| 2 V (VV, VVF, VVFF) | 47 | 128 | |||||
| 3 V (VVV) | 3 | 2 | 0.046 | ||||
| Allele frequency (V) | 37.0% | 35.5% | 1.08 (0.91–1.28) | 0.373 | 0.606 | ||
| FCGR2C promoter | |||||||
| 0 2B.2 | 286 | 717 | |||||
| 1 2B.2 | 110 | 185 | |||||
| 2 2B.2 | 9 | 16 | |||||
| 3 2B.2 | 0 | 1 | 0.017 | ||||
| Allele frequency (2B.2) | 15.3% | 11.5% | 1.37 (1.08–1.72) | 0.009 | NE | ||
| FCGR2C | |||||||
| ORF/Stop/NC-ORF | |||||||
| 0 ORF | 283 | 721 | |||||
| 1 ORF | 113 | 184 | |||||
| 2 ORF | 9 | 13 | |||||
| 3 ORF | 0 | 1 | 0.005 | ||||
| 0 NC-ORF | 389 | 853 | |||||
| 1 NC-ORF | 6 | 33 | |||||
| 2 NC-ORF | 10 | 33 | 0.059 | ||||
| Allele frequency (ORF) | 15.7% | 11.2% | 1.46 (1.16–1.85) | 0.002 | 0.093 | 0.002 | |
| Allele frequency (NC-ORF) | 3.1% | 5.2% | 0.72 (0.51–1.02) | 0.063 | 0.112 | ||
| Allele frequency (Stop) | 81.2% | 83.7% | 0.88 (0.74–1.04) | 0.136 | |||
| FCGR3B NA1/NA2/SH | |||||||
| 0 NA1 | 158 | 373 | |||||
| 1 NA1 | 201 | 430 | |||||
| 2 NA1 | 45 | 114 | |||||
| 3 NA1 | 1 | 2 | 0.754 | ||||
| 0 SH | 389 | 874 | |||||
| 1 SH | 16 | 45 | 0.481 | ||||
| Allele frequency (NA1) | 36.2% | 35.3% | 1.01 (0.85–1.20) | 0.933 | 0.537 | ||
| Allele frequency (NA2) | 63.8% | 64.7% | 0.94 (0.80–1.12) | ||||
| Allele frequency (SH) | 4.0% | 4.9% | 0.80 (0.45–1.43) | 0.450 | 0.247 | ||
| FCGR2B promoter | |||||||
| 0 2B.4 | 307 | 748 | |||||
| 1 2B.4 | 93 | 157 | |||||
| 2 2B.4 | 5 | 14 | 0.043 | ||||
| Allele frequency (2B.4) | 12.7% | 10.0% | 1.29 (1.00–1.67) | 0.047 | 0.834 | ||
| FCGR2B I232T | |||||||
| II | 322 | 697 | |||||
| IT | 76 | 201 | |||||
| TT | 7 | 21 | 0.359 | ||||
| Allele frequency (T) | 11.1% | 13.2% | 0.83 (0.64–1.06) | 0.141 | 0.189 | ||
For SNPs that are subject to CNV, several genotypes are pooled as indicated to combine all the different genotypes with the same copy number of 1 of the variants. For the tri-allelic haplotypes in FCGR2C and FCGR3B, this is done for two of the haplotypes separately. Fisher exact test was calculated on genotype frequencies as shown in the table. A single logistic regression analysis was performed for each (presumed) risk allele in an additive model. A multiple logistic regression analysis was performed on all variants (except the FCGR2C promoter haplotypes, which were left out of the multiple logistic regression analysis because of the near perfect LD with the classic FCGR2C-ORF haplotype) and on FCGR2A-H131R and classic FCGR2C-ORF alone. P-values < 0.05 are shown in bold.
We did not detect significant differences for any of the CNV regions, or for the other functional SNPs. Even though we detected a slight trend among the KD patients with higher frequency of the FCGR2A-131H (rs1801274) risk allele in the current study, this association found previously in GWAS and meta-analysis (6, 7, 33) was not replicated in this dataset of European patients and healthy controls. A multiple logistic regression analysis of only the FCGR2C-ORF and FCGR2A-131H revealed that the association of FCGR2C-ORF was independent of FCGR2A-131H (Table 2).
Family-Based Study on KD
In an attempt to confirm our findings, we performed a KD family-based association study in 623 family trios in which the child was diagnosed with KD. The transmission disequilibrium test (TDT) analysis revealed a significant association (P = 0.006) of FCGR2A-131H (rs1801274) (Table 3). For the FCGR2C-ORF haplotype (rs759550223 and rs76277413) and the other SNPs or CNRs tested, there was no evidence of association (except for the rare allele with two copies of FCCR3A on one chromosome, of which one was 158V and the other was 158F, which had only 18 informative families) (Table 3). Of note, the number of informative families for FCGR2C-ORF was also relatively small, as a result of the low prevalence of this variant (Table 1). Analysis of the families enabled us to construct complete haplotypes for all parental chromosomes, which confirmed the LD pattern observed in the cohort of healthy controls, both in parents without any CNV as in parents that did show CNV (Figure S3).
Table 3.
Transmission disequilibrium test for the different variants at the FCGR2/3 locus in a family-based association study.
| Allele/haplotype (on 1 chromosome) | Allele frequency | # families* | Z | P-value |
|---|---|---|---|---|
| CNR1 | ||||
| 0 (deletion) | 0.049 | 105 | 0.285 | 0.776 |
| 1 | 0.875 | 214 | −0.065 | 0.948 |
| 2 (duplication) | 0.074 | 133 | −0.338 | 0.735 |
| CNR2 | ||||
| 0 (deletion) | 0.006 | 13 | −0.277 | 0.782 |
| 1 | 0.976 | 61 | 0.378 | 0.705 |
| 2 (duplication) | 0.018 | 48 | −0.429 | 0.668 |
| FCGR2A Q27W | ||||
| Q | 0.891 | 210 | −0.328 | 0.743 |
| W | 0.109 | 210 | 0.328 | 0.743 |
| FCGR2A H131R | ||||
| H | 0.575 | 431 | 2.750 | 0.006 |
| R | 0.425 | 431 | −2.750 | 0.006 |
| FCGR3A V158F | ||||
| – | 0.005 | 11 | −0.302 | 0.763 |
| F | 0.642 | 395 | 0.483 | 0.629 |
| FF | 0.010 | 27 | 0.577 | 0.564 |
| VF | 0.006 | 18 | −2.828 | 0.005 |
| V | 0.331 | 397 | −0.088 | 0.930 |
| VV | 0.004 | 10 | 0 | 1.000 |
| Promoter FCGR2C | ||||
| - | 0.055 | 114 | 0.451 | 0.652 |
| 2B.1 | 0.748 | 349 | −0.242 | 0.809 |
| 2B.1-2B.1 | 0.089 | 164 | −0.818 | 0.414 |
| 2B.2 | 0.099 | 182 | 0.491 | 0.623 |
| FCGR2C ORF/Stop/NC-ORF** | ||||
| – | 0.055 | 115 | 0.268 | 0.788 |
| ORF | 0.100 | 184 | 1.120 | 0.263 |
| Stop | 0.743 | 354 | −0.241 | 0.810 |
| NC-ORF | 0.009 | 23 | −1.460 | 0.144 |
| Stop-stop | 0.075 | 143 | −0.477 | 0.633 |
| NC-ORF-NC-ORF | 0.007 | 19 | −0.229 | 0.819 |
| FCGR3B NA1/NA2/SH | ||||
| – | 0.053 | 108 | 0.186 | 0.853 |
| NA1 | 0.362 | 389 | 0.490 | 0.624 |
| NA1-NA2 | 0.051 | 96 | −0.198 | 0.843 |
| NA1-SH | 0.009 | 21 | 0.218 | 0.827 |
| NA2 | 0.508 | 396 | −0.439 | 0.660 |
| NA2-NA2 | 0.004 | 11 | −0.905 | 0.366 |
| SH | 0.004 | 12 | −1.155 | 0.248 |
| Promoter FCGR2B | ||||
| 2B.1 | 0.905 | 185 | −0.563 | 0.574 |
| 2B.4 | 0.090 | 172 | 0.946 | 0.344 |
| FCGR2B I232T | ||||
| I | 0.867 | 226 | 0.741 | 0.459 |
| T | 0.133 | 226 | −0.741 | 0.459 |
Number of informative families (i.e., at least one of the parents is heterozygous for the indicated allele or haplotype). Only alleles for which the number of informative families is >10 are shown.
ORF means classic FCGR2C-ORF haplotype, NC-ORF means nonclassic FCGR2C-ORF haplotype. Z; Z statistic, a positive Z indicates more transmission than expected, a negative Z indicates less transmission than expected, P indicates whether Z is significantly different from 0, P < 0.05 is considered significant.
Combined Analysis Reveals Both FCGR2A-131H and FCGR2C-ORF to be Significantly Associated With Susceptibility to KD
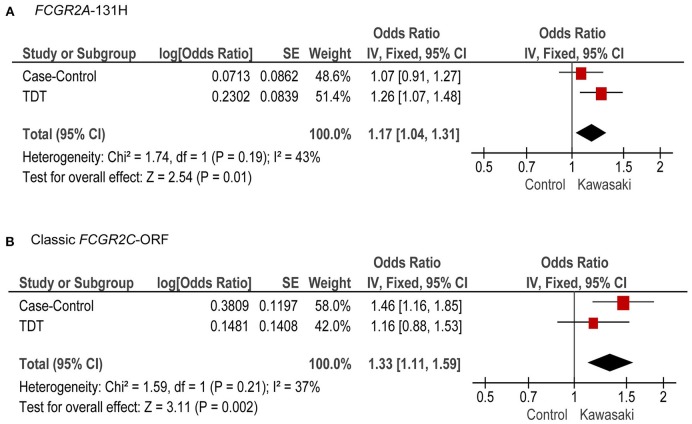
We performed a meta-analysis of the associations from both the case-control and familial TDT analyses, and we found the classic FCGR2C-ORF haplotype (rs759550223 and rs76277413, meta-P = 0.002) and the FCGR2A 131H (rs1801274, meta-P = 0.01) were both significantly associated with KD susceptibility (Figure 4).
Figure 4.
Meta-analysis of case-control and TDT for FCGR2A-131H and the classic FCGR2C-ORF haplotype. Combined OR (95% CI) and P-values from the case-control study and TDT analysis, for FCGR2A-131H (A) and classic FCGR2C-ORF (B).
mRNA for the FCGR2 Isoforms Is Upregulated in Acute KD Patients, in Contrast to the FCGR3 Isoforms
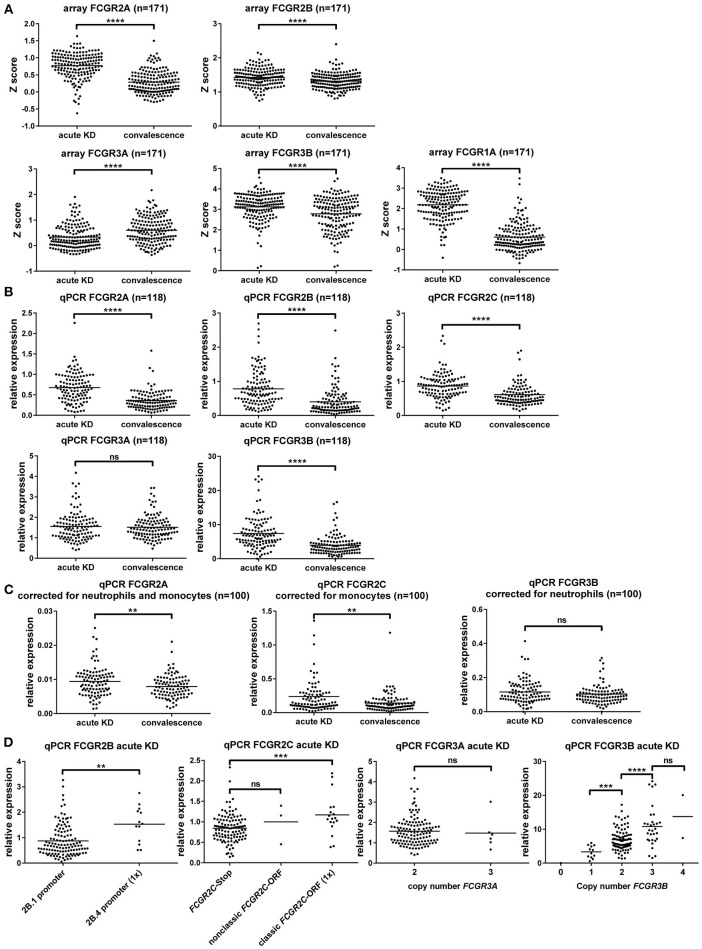
To determine whether alteration of expression levels of the low-affinity FcγRs plays a role in the pathophysiology of KD, we compared mRNA expression levels in KD patients in the acute and convalescent phase of the disease, using samples from a previous study (49). First, we compared Z scores for FCGR transcripts that were already present in the microarray for this study. In this analysis, we found FCGR2A, FCGR2B, FCGR3A, FCGR3B, and also FCGR1A, encoding the high-affinity FcγRI, to be all transcriptionally upregulated in acute KD (Figure 5A).
Figure 5.
Gene expression analysis of FcγRs shows upregulation of FcγRI and FcγRII, but not FcγRIII during acute KD. (A) Difference in expression intensity of various FCGR transcripts, as determined by RNA microarray, shown as Z scores (higher score indicating higher expression), in 171 subjects with KD in the acute and convalescent phase of the disease. (B–D) Relative expression of different FCGR transcripts detected by qPCR on whole blood, corrected for housekeeping genes GUS and GAPDH, as compared to one randomly chosen sample in the convalescence phase of KD. (B) Dot plots showing a comparison of the acute and convalescent phase of KD in 118 patients. (C) Dot plots showing a comparison of the acute and convalescent phase of KD for transcripts of FCGR2A, FCGR2C, and FCGR3B in 100 patients for which WBC differentials were known, after correction for the main cell type that expresses the transcript. (D) Comparison of genotypes for the expression of various transcripts in 135 patients with acute KD. FCGR2B: patients with only the 2B.1 promoter (n = 122) or with 1 copy of the 2B.4 promoter (n = 13) in FCGR2B. FCGR2C: patients with the FCGR2C-Stop haplotype (n = 114), patients with 1 copy of the classic FCGR2C-ORF haplotype (n = 18), patients with 1 or 2 copies of the nonclassic FCGR2C-ORF haplotype (n = 3). FCGR3A patients with 2 copies (n = 129), or 3 copies (n = 6) of the FCGR3A gene. FCGR3B: patients with 0 copies (n = 1), 1 copy (n = 12), 2 copies (n = 89), 3 copies (n = 31) or 4 copies (n = 2) of the FCGR3B gene. ns: non-significant; **p < 0.01; ***p < 0.001; ****p < 0.0001 as determined by paired t-test or Wilcoxon matched-pairs signed rank test (A–C) or students t-test or Mann Whitney test (D).
To confirm these findings and extend the analysis to FCGR2C, we then performed highly specific qPCRs for FCGRs on a selection of these patients from which RNA was still available. This confirmed that FCGR2A, FCGR2B and FCGR2C transcripts were all upregulated during acute KD (Figure 5B). FCGR3A was not differentially expressed between the acute and convalescent phase (Figure 5B) but FCGR3B seemed to be upregulated in the acute phase (Figure 5B). However, because acute KD could have resulted in a shift in leukocyte differentials and in our cohort a marked increase of neutrophil percentages was observed (data not shown), we applied a correction for percentages of different leukocyte subsets in the 100 patients for whom leukocyte differentials were available. In the case of FCGR3B, a correction for neutrophil percentages (Figure 5C) showed that the apparent upregulation was the result of the relative increase in neutrophils during acute KD and does not reflect a true increase in transcription. On the other hand, expression levels of FCGR2A and FCGR2C were increased in acute KD even after correction for shifts in white blood cell distribution (Figure 5C).
Comparison for several genetic differences known to influence expression levels showed marked differences (Figure 5D), confirming earlier reports and the validity of our analysis.
Discussion
In a comprehensive study using MLPA, we have analyzed the full collection of functionally defined SNPs and CNRs at the FCGR2/3 locus at an unprecedented level of detail. We report extensive LD in this notoriously difficult gene cluster, as well as large ethnic variation in different European, African and Asian subpopulations. Our findings are in line with previously published allele frequencies and CNV in different populations for this locus (21, 44, 50) and extend these findings with additional variants and populations. Applying this as the reference dataset, previously reported genetic association studies may need to be re-evaluated.
This is the first study to illustrate the relevance of a more detailed reference for a pediatric vasculitis. KD has a ten-fold increased prevalence in Japanese and other Asian populations compared to children of European descent. In multi-ethnic GWAS studies, the association of FCGR2A-131H(rs1801274) with KD susceptibility was detected across KD cohorts of different ethnic backgrounds, indicating that this common variant is an independent susceptibility marker in all groups, including the Asian and European populations (6, 7). We now show that within the European cohorts, the classic FCGR2C-ORF haplotype (rs759550223 and rs76277413) may be the most strongly associated FCGR gene variant with KD susceptibility. Evidence from low LD (r2 = 0.08) and conditional analyses identify the association of this classic FCGR2C-ORF haplotype to be independent of the previously identified FCGR2A-131H GWAS association. Interestingly, the classic FCGR2C-ORF, which is strongly associated with KD susceptibility in Europeans, was virtually non-existent in the Asian populations. This suggests that the increased prevalence of KD in Asian populations compared to European populations derives from factors other than the currently known genetic variation in FCGR genes.
The very strong LD of the classic FCGR2C-ORF haplotype with several other variants in the FCGR2/3 locus means that the interpretation of associations with this locus are more complex than previously appreciated. Classic FCGR2C-ORF is in strong LD with three other variants: the 2B.2 promoter in FCGR2C (rs149754834), FCGR2A-27W (rs201218628) and the 2B.4 haplotype in FCGR2B (rs143796418). Hence, all these variants could tag the classic FCGR2C-ORF and were also significantly associated with KD susceptibility in a single logistic regression analysis. However, when we analyzed all variants in a multiple logistic regression analysis, we found the classic FCGR2C-ORF to be the strongest predictor of KD susceptibility. The 2B.2 variant in FCGR2C was omitted from the multiple logistic regression analysis because of its near complete LD with classic FCGR2C-ORF. In fact, this variant can actually be only of biological relevance in the case of a classic FCGR2C-ORF haplotype, because with the other FCGR2C haplotypes, this 2B.2 promoter haplotype would reside in the promoter of an untranslated variant or FCGR2C (FCGR2C-Stop or nonclassic FCGR2C-ORF). It is unlikely that the tagging FCGR2A-Q27W SNP independently contributes to KD susceptibility, as it is a genetic variation for which a biological role has not been described (46). It lies outside the IgG-binding region of FcγRIIa and an analysis of expression levels revealed no influence on expression levels (Figure S4). However, genotyping the FCGR2A-Q27W SNP may be informative in genetic association studies, as it may be used as a tagging SNP for the classic FCGR2C-ORF as part of a susceptibility haplotype. The FCGR2A-Q27W SNP lies outside the copy number variable part of the FCGR2/3 locus and is straightforward to genotype.
We did not find a significant association of CNV of the locus for any of the different CNRs that have been described. This is in contrast with an earlier report that described an association of CNV in FCGR3B and in FCGR2C with susceptibility to KD (51). In our opinion, analysis of CNV of FCGR2C without information on the FCGR2C-ORF variant is futile, as CNV of FCGR2C per se does not correlate with expression levels, normally being a pseudogene (i.e., FCGR2C-Stop). On the other hand, CNV in the FCGR3B does have a potential biological role, as we confirmed with our qPCR analysis, which showed a direct effect of CNV of the FCGR3B gene on transcript levels of FCGR3B. Nevertheless, CNV of FCGR3B was not associated with KD susceptibility in our cohorts.
Transcript levels of FCGR2A have previously been shown to be increased in KD patients compared to febrile controls (52), and we now show that mRNA levels of all FCGR2 isoforms, as well as FCGR1A1 [encoding FcγRI (CD64)], are upregulated during the acute phase of KD, compared to paired convalescent samples of the same patients, which further underscores the importance of FcγRs in KD.
A striking finding of our study is the lack of a significant association of FCGR2A-131H in the case-control study, contrasting our previous GWAS findings (6). This discrepancy was not explained by a difference in allele frequency in the case group, but by a difference in allele frequency between the control groups tested. Both control groups were randomly selected individuals of European descent. A remarkable difference between the two control groups was that the control group of the GWAS consisted mainly of individuals from the United Kingdom, which in the present study have a significantly lower prevalence of the FCGR2A-131H than the other European groups (Table S3). Apparently, even within the European population, the selection of the control group may influence the results of association analyses. Although both control groups were randomly selected, we believe that the group used in the current study is more representative of the background population, since it consists of more controls from the countries of origin of the patients. Nevertheless, even with the new control group, in a combined meta-analysis with our TDT analysis, FCGR2A-131H was still significantly associated with KD susceptibility.
In addition to small differences within the European population, of more relevance were the significant differences in allele frequencies at the FCGR2/3 locus between the different ethnic groups. Our MLPA assay enabled us to look at the distribution of FCGR2C haplotypes in African, European and Chinese populations. We show that MLPA reliably distinguished the classic FCGR2C-ORF from the nonclassic FCGR2C-ORF haplotype that does not result in expression of FcγRIIc. Theoretically, only minimal errors in haplotype calling can occur for FCGR2C with the MLPA methods (calculated error rate of only 0.1%, Table S4), whereas Illumina whole-exome sequencing was unable to detect the rs759550223 SNP of the classic FCGR2C-ORF haplotype in all three individuals with this haplotype among ten individuals tested in total (error rate 30%) (18).
The classic FCGR2C-ORF haplotype is virtually absent from the Asian population, whereas in the African population, the non-expressed nonclassic ORF was much more prevalent than the classic FCGR2C-ORF. The absence of the classic FCGR2C-ORF in the Asian population is of particular interest because of the fact that there is a striking difference in the incidence of KD between children of Asian (69–308 per 100,000 children <5 years of age) (53) and of European descent (4–15 per 100,000 children <5 years of age) (54–56). Clearly, the FCGR2C-ORF is only a risk factor for KD susceptibility in European subjects, and cannot account for the increased incidence of KD in Asian children.
A potential limitation of our MLPA technology lies in the uncertainty of allocating the promoter haplotypes 2B.2 and 2B.4 to either FCGR2B or FCGR2C, but data previously generated by us and others (5, 36, 57, 58) show that our allocation approach is accurate in >95% of European individuals with at least one of the rare variants 2B.2 or 2B.4. The majority of individuals does not carry a rare variant and these individuals will be 100% accurately genotyped by MLPA.
Detailed knowledge of genetic linkage in IgG receptors has major implications for every other study on associations of FCGR2/3 polymorphisms with disease or therapeutic efficacy. For example, many studies investigating associations with therapeutic efficacy of therapeutic antibodies against cancer have found an association with the FCGR3A-158V variant (rs396991) (13–15, 59), which we now show to be in moderate LD with the classic FCGR2C-ORF (r2 = 0.24). Since the classic FCGR2C-ORF haplotype leads to expression of the activating FcγRIIc on NK cells, neutrophils, monocytes (Figure 2) and macrophages (17), it may contribute to killing of tumor cells by antibody-dependent cellular cytotoxicity by these cells, and could potentially be a stronger predictor of treatment success.
In conclusion, we have reported a novel association of the classic FCGR2C-ORF variant (rs759550223 and rs76277413) with susceptibility to KD in European patients, independent of the FCGR2A-131H (rs1801274), which is a separate susceptibility marker. Upregulation of the transcripts for both activating receptors encoded by these genes (respectively FcγRIIc and FcγRIIa) during acute KD further indicates their importance in KD pathophysiology. FcγRIIa and FcγRIIc are co-expressed by two circulating cell types, monocytes and neutrophils. Both cell types are actively recruited to arterial lesions in KD patients. Our data support a central role of the activating IgG receptors on these cell types in the pathophysiology of KD, whereas the SNPs in the inhibitory FcγRIIb were not associated. This suggests that inhibiting the function of activating FcγRs (which is a possible working mechanism of IVIg, the first-line treatment in KD) may be an important treatment goal in patients with this pediatric vasculitis during the acute phase of the disease.
Author Contributions
SN and CT performed experiments, analyzed data, wrote the manuscript and designed research. WB discussed data and designed research. MT performed statistical analysis. JG, LH, EP, AN, and JvdH performed experiments and analyzed data. RY, ML, VW, DB, A-LP, JE, RC, CS, JB, KF, and CvdS provided samples. TvdB, SD, and MH supervised research. MdB discussed data and designed research. TK supervised the study, wrote the manuscript and designed research. All authors contributed to the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Prof. Dirk Roos and Prof. Raoul Hennekam for critical reading of the manuscript, Dr. C. C. Khor, Dr. Joep Sins, Mrs. Aicha Ait Soussan, and Dr. Lonneke Haer-Wigman for help in sample collection, and the GERMS platform at Genome Institute Singapore for technical support. This work was supported by a grant from the Landsteiner Foundation for Bloodtransfusion Research (LSBR 0916) awarded to TK.
CT has received a grant from the Ter Meulen Fund, Royal Netherlands Academy of Arts and Sciences (TMF2012/227). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Contributor Information
The International Kawasaki Disease Genetics Consortium:
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2019.00185/full#supplementary-material
References
- 1.Lee YH, Ji JD, Song GG. Associations between FCGR3A polymorphisms and susceptibility to rheumatoid arthritis: a metaanalysis. J Rheumatol. (2008) 35:2129–35. 10.3899/jrheum.080186 [DOI] [PubMed] [Google Scholar]
- 2.Lee YH, Ji JD, Song GG. Fcgamma receptor IIB and IIIB polymorphisms and susceptibility to systemic lupus erythematosus and lupus nephritis: a meta-analysis. Lupus. (2009) 18:727–34. 10.1177/0961203309104020 [DOI] [PubMed] [Google Scholar]
- 3.Li LH, Yuan H, Pan HF, Li WX, Li XP, Ye DQ. Role of the Fcgamma receptor IIIA-V/F158 polymorphism in susceptibility to systemic lupus erythematosus and lupus nephritis: a meta-analysis. Scand J Rheumatol. (2010) 39:148–54. 10.3109/03009740903292304 [DOI] [PubMed] [Google Scholar]
- 4.Yuan H, Pan HF, Li LH, Feng JB, Li WX, Li XP, et al. Meta analysis on the association between FcgammaRIIa-R/H131 polymorphisms and systemic lupus erythematosus. Mol Biol Rep. (2009) 36:1053–8. 10.1007/s11033-008-9280-x [DOI] [PubMed] [Google Scholar]
- 5.Breunis WB, van Mirre E, Bruin M, Geissler J, de Boer M, Peters M, et al. Copy number variation of the activating FCGR2C gene predisposes to idiopathic thrombocytopenic purpura. Blood. (2008) 111:1029–38. 10.1182/blood-2007-03-079913 [DOI] [PubMed] [Google Scholar]
- 6.Khor CC, Davila S, Breunis WB, Lee YC, Shimizu C, Wright VJ, et al. Genome-wide association study identifies FCGR2A as a susceptibility locus for Kawasaki disease. Nat Genet. (2011) 43:1241–6. 10.1038/ng.981 [DOI] [PubMed] [Google Scholar]
- 7.Onouchi Y, Ozaki K, Burns JC, Shimizu C, Terai M, Hamada H, et al. A genome-wide association study identifies three new risk loci for Kawasaki disease. Nat Genet. (2012) 44:517–21. 10.1038/ng.2220 [DOI] [PubMed] [Google Scholar]
- 8.Asano K, Matsushita T, Umeno J, Hosono N, Takahashi A, Kawaguchi T, et al. A genome-wide association study identifies three new susceptibility loci for ulcerative colitis in the Japanese population. Nat Genet. (2009) 41:1325–9. 10.1038/ng.482 [DOI] [PubMed] [Google Scholar]
- 9.Chai L, Song YQ, Leung WK. Genetic polymorphism studies in periodontitis and Fcgamma receptors. J Periodontal Res. (2012) 47:273–85. 10.1111/j.1600-0765.2011.01437.x [DOI] [PubMed] [Google Scholar]
- 10.Adu B, Dodoo D, Adukpo S, Hedley PL, Arthur FK, Gerds TA, et al. Fc Gamma receptor IIIB (FcgammaRIIIB) polymorphisms are associated with clinical malaria in ghanaian children. PLoS ONE. (2012) 7:e46197. 10.1371/journal.pone.0046197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tamura K, Shimizu C, Hojo T, Akashi-Tanaka S, Kinoshita T, Yonemori K, et al. FcgammaR2A and 3A polymorphisms predict clinical outcome of trastuzumab in both neoadjuvant and metastatic settings in patients with HER2-positive breast cancer. Ann Oncol. (2011) 22:1302–7. 10.1093/annonc/mdq585 [DOI] [PubMed] [Google Scholar]
- 12.Hurvitz SA, Betting DJ, Stern HM, Quinaux E, Stinson J, Seshagiri S, et al. Analysis of Fcgamma receptor IIIa and IIa polymorphisms: lack of correlation with outcome in trastuzumab-treated breast cancer patients. Clin Cancer Res. (2012) 18:3478–86. 10.1158/1078-0432.CCR-11-2294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cartron G, Dacheux L, Salles G, Solal-Celigny P, Bardos P, Colombat P, et al. Therapeutic activity of humanized anti-CD20 monoclonal antibody and polymorphism in IgG Fc receptor FcgammaRIIIa gene. Blood. (2002) 99:754–8. 10.1182/blood.V99.3.754 [DOI] [PubMed] [Google Scholar]
- 14.Treon SP, Yang G, Hanzis C, Ioakimidis L, Verselis SJ, Fox EA, et al. Attainment of complete/very good partial response following rituximab-based therapy is an important determinant to progression-free survival, and is impacted by polymorphisms in FCGR3A in Waldenstrom macroglobulinaemia. Br J Haematol. (2011) 154:223–8. 10.1111/j.1365-2141.2011.08726.x [DOI] [PubMed] [Google Scholar]
- 15.Ahlgrimm M, Pfreundschuh M, Kreuz M, Regitz E, Preuss KD, Bittenbring J. The impact of Fc-gamma receptor polymorphisms in elderly patients with diffuse large B-cell lymphoma treated with CHOP with or without rituximab. Blood. (2011) 118:4657–62. 10.1182/blood-2011-04-346411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bruhns P. Properties of mouse and human IgG receptors and their contribution to disease models. Blood. (2012) 119:5640–9. 10.1182/blood-2012-01-380121 [DOI] [PubMed] [Google Scholar]
- 17.Nagelkerke SQ, Kuijpers TW. Immunomodulation by IVIg and the role of Fc-gamma receptors: classic mechanisms of action after all? Front Immunol. (2015) 5:674. 10.3389/fimmu.2014.00674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nagelkerke SQ, Tacke CE, Breunis WB, Geissler J, Sins JW, Appelhof B, et al. Nonallelic homologous recombination of the FCGR2/3 locus results in copy number variation and novel chimeric FCGR2 genes with aberrant functional expression. Genes Immun. (2015) 16:422–9. 10.1038/gene.2015.25 [DOI] [PubMed] [Google Scholar]
- 19.Bruhns P, Iannascoli B, England P, Mancardi DA, Fernandez N, Jorieux S, et al. Specificity and affinity of human Fcgamma receptors and their polymorphic variants for human IgG subclasses. Blood. (2009) 113:3716–25. 10.1182/blood-2008-09-179754 [DOI] [PubMed] [Google Scholar]
- 20.Hargreaves CE, Iriyama C, Rose-Zerilli MJ, Nagelkerke SQ, Hussain K, Ganderton R, et al. Evaluation of high-throughput genomic assays for the Fc gamma receptor locus. PLoS ONE. (2015) 10:e0142379 10.1371/journal.pone.0142379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Niederer HA, Willcocks LC, Rayner TF, Yang W, Lau YL, Williams TN, et al. Copy number, linkage disequilibrium and disease association in the FCGR locus. Hum Mol Genet. (2010) 19:3282–94. 10.1093/hmg/ddq216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hatjiharissi E, Hansen M, Santos DD, Xu L, Leleu X, Dimmock EW, et al. Genetic linkage of Fc gamma RIIa and Fc gamma RIIIa and implications for their use in predicting clinical responses to CD20-directed monoclonal antibody therapy. Clin Lymphoma Myeloma. (2007) 7:286–90. 10.3816/CLM.2007.n.004 [DOI] [PubMed] [Google Scholar]
- 23.Lejeune J, Thibault G, Ternant D, Cartron G, Watier H, Ohresser M. Evidence for linkage disequilibrium between Fcgamma RIIIa-V158F and Fcgamma RIIa-H131R polymorphisms in white patients, and for an Fcgamma RIIIa-restricted influence on the response to therapeutic antibodies. J Clin Oncol. (2008) 26:5489–91. 10.1200/JCO.2008.19.4118 [DOI] [PubMed] [Google Scholar]
- 24.Lejeune J, Piegu B, Gouilleux-Gruart V, Ohresser M, Watier H, Thibault G. FCGR2C genotyping by pyrosequencing reveals linkage disequilibrium with FCGR3A V158F and FCGR2A H131R polymorphisms in a Caucasian population. MAbs. (2012) 4:784–7. 10.4161/mabs.22287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kawasaki T. [Acute febrile mucocutaneous syndrome with lymphoid involvement with specific desquamation of the fingers and toes in children]. Arerugi. (1967) 16:178–222. [PubMed] [Google Scholar]
- 26.McCrindle BW, Rowley AH, Newburger JW, Burns JC, Bolger AF, Gewitz M, et al. Diagnosis, treatment, and long-term management of kawasaki disease: a scientific statement for health professionals from the american heart association. Circulation. (2017) 135:e927–e999. 10.1161/CIR.0000000000000484 [DOI] [PubMed] [Google Scholar]
- 27.Newburger JW, Takahashi M, Burns JC, Beiser AS, Chung KJ, Duffy CE, et al. The treatment of Kawasaki syndrome with intravenous gamma globulin. N Engl J Med. (1986) 315:341–7. 10.1056/NEJM198608073150601 [DOI] [PubMed] [Google Scholar]
- 28.Newburger JW, Takahashi M, Beiser AS, Burns JC, Bastian J, Chung KJ, et al. A single intravenous infusion of gamma globulin as compared with four infusions in the treatment of acute Kawasaki syndrome. N Engl J Med. (1991) 324:1633–9. 10.1056/NEJM199106063242305 [DOI] [PubMed] [Google Scholar]
- 29.Shrestha S, Wiener H, Shendre A, Kaslow RA, Wu J, Olson A, et al. Role of activating FcgammaR gene polymorphisms in Kawasaki disease susceptibility and intravenous immunoglobulin response. Circ Cardiovasc Genet. (2012) 5:309–16. 10.1161/CIRCGENETICS.111.962464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Biezeveld M, Geissler J, Merkus M, Kuipers IM, Ottenkamp J, Kuijpers T. The involvement of Fc gamma receptor gene polymorphisms in Kawasaki disease. Clin Exp Immunol. (2007) 147:106–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Taniuchi S, Masuda M, Teraguchi M, Ikemoto Y, Komiyama Y, Takahashi H, et al. Polymorphism of Fc gamma RIIa may affect the efficacy of gamma-globulin therapy in Kawasaki disease. J Clin Immunol. (2005) 25:309–13. 10.1007/s10875-005-4697-7 [DOI] [PubMed] [Google Scholar]
- 32.Chatzikyriakidou A, Aidinidou L, Giannopoulos A, Papadopoulou-Legbelou K, Kalinderi K, Fidani L. Absence of association of FCGR2A gene polymorphism rs1801274 with Kawasaki disease in Greek patients. Cardiol Young. (2014) 25:681–3. 10.1017/S1047951114000626 [DOI] [PubMed] [Google Scholar]
- 33.Duan J, Lou J, Zhang Q, Ke J, Qi Y, Shen N, et al. A genetic variant rs1801274 in FCGR2A as a potential risk marker for Kawasaki disease: a case-control study and meta-analysis. PLoS ONE. (2014) 9:e103329. 10.1371/journal.pone.0103329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lou J, Zhong R, Shen N, Lu XZ, Ke JT, Duan JY, et al. Systematic confirmation study of GWAS-identified genetic variants for Kawasaki disease in a Chinese population. Sci Rep. (2015) 5:8194. 10.1038/srep08194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van der Heijden J, Breunis WB, Geissler J, de Boer M, van den Berg TK, Kuijpers TW. Phenotypic variation in IgG receptors by nonclassical FCGR2C alleles. J Immunol. (2012) 188:1318–24. 10.4049/jimmunol.1003945 [DOI] [PubMed] [Google Scholar]
- 36.Tsang-A-Sjoe MWP, Nagelkerke SQ, Bultink IE, Geissler J, Tanck MW, Tacke CE, et al. Fc-gamma receptor polymorphisms differentially influence susceptibility to systemic lupus erythematosus and lupus nephritis. Rheumatology (Oxford). (2016) 55:939–48. 10.1093/rheumatology/kev433 [DOI] [PubMed] [Google Scholar]
- 37.Ellis JA, Ponsonby AL, Pezic A, Chavez RA, Allen RC, Akikusa JD, et al. CLARITY - ChiLdhood Arthritis Risk factor Identification sTudY. Pediatr Rheumatol Online J. (2012) 10:37. 10.1186/1546-0096-10-37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grootkerk-Tax MG, van Wintershoven JD, Ligthart PC, van Rhenen DJ, van der Schoot CE, Maaskant-van Wijk PA. RHD(T201R, F223V) cluster analysis in five different ethnic groups and serologic characterization of a new Ethiopian variant DARE, the DIII type 6, and the RHD(F223V). Transfusion. (2006) 46:606–15. 10.1111/j.1537-2995.2006.00759.x [DOI] [PubMed] [Google Scholar]
- 39.Breunis WB, van Mirre E, Geissler J, Laddach N, Wolbink G, van der Schoot E, et al. Copy number variation at the FCGR locus includes FCGR3A, FCGR2C and FCGR3B but not FCGR2A and FCGR2B. Hum Mutat. (2009) 30:E640–50. 10.1002/humu.20997 [DOI] [PubMed] [Google Scholar]
- 40.Zhao JH. gap: genetic analysis package. J Stat Softw. (2007) 23:1–18. 10.18637/jss.v023.i08 [DOI] [Google Scholar]
- 41.Rabinowitz D, Laird N. A unified approach to adjusting association tests for population admixture with arbitrary pedigree structure and arbitrary missing marker information. Hum Hered. (2000) 50:211–23 10.1159/000022918 [DOI] [PubMed] [Google Scholar]
- 42.Kazeem GR, Farrall M. Integrating case-control and TDT studies. Ann Hum Genet. (2005) 69(Pt 3):329–35. 10.1046/J.1469-1809.2005.00156.x [DOI] [PubMed] [Google Scholar]
- 43.Li X, Ptacek TS, Brown EE, Edberg JC. Fcgamma receptors: structure, function and role as genetic risk factors in SLE. Genes Immun. (2009) 10:380–9. 10.1038/gene.2009.35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Machado LR, Hardwick RJ, Bowdrey J, Bogle H, Knowles TJ, Sironi M, et al. Evolutionary history of copy-number-variable locus for the low-affinity Fcgamma receptor: mutation rate, autoimmune disease, and the legacy of helminth infection. Am J Hum Genet. (2012) 90:973–85. 10.1016/j.ajhg.2012.04.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Clatworthy MR, Willcocks L, Urban B, Langhorne J, Williams TN, Peshu N, et al. Systemic lupus erythematosus-associated defects in the inhibitory receptor FcgammaRIIb reduce susceptibility to malaria. Proc Natl Acad Sci USA. (2007) 104:7169–74. 10.1073/pnas.0608889104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van der Pol W, van de Winkel JG. IgG receptor polymorphisms: risk factors for disease. Immunogenetics. (1998) 48:222–32. 10.1007/s002510050426 [DOI] [PubMed] [Google Scholar]
- 47.Van Den Berg L, Myhr KM, Kluge B, Vedeler CA. Fcgamma receptor polymorphisms in populations in Ethiopia and Norway. Immunology. (2001) 104:87–91. 10.1046/j.1365-2567.2001.01284.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Meinderts SM, Sins JWR, Fijnvandraat K, Nagelkerke SQ, Geissler J, Tanck MW, et al. Non-classical FCGR2C haplotype is associated with protection from red blood cell allo-immunization in sickle cell disease. Blood. (2017) 130:2121–30. 10.1182/blood-2017-05-784876 [DOI] [PubMed] [Google Scholar]
- 49.Hoang LT, Shimizu C, Ling L, Naim AN, Khor CC, Tremoulet AH, et al. Global gene expression profiling identifies new therapeutic targets in acute Kawasaki disease. Genome Med. (2014) 6:541. 10.1186/s13073-014-0102-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lassauniere R, Tiemessen CT. Variability at the FCGR locus: characterization in Black South Africans and evidence for ethnic variation in and out of Africa. Genes Immun. (2016) 17:93–104. 10.1038/gene.2015.60 [DOI] [PubMed] [Google Scholar]
- 51.Makowsky R, Wiener HW, Ptacek TS, Silva M, Shendre A, Edberg JC, et al. FcgammaR gene copy number in Kawasaki disease and intravenous immunoglobulin treatment response. Pharmacogenet Genomics. (2013) 23:455–62. 10.1097/FPC.0b013e328363686e [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chang LS, Lo MH, Li SC, Yang MY, Hsieh KS, Kuo HC. The effect of FcgammaRIIA and FcgammaRIIB on coronary artery lesion formation and intravenous immunoglobulin treatment responses in children with Kawasaki disease. Oncotarget. (2017) 8:2044–52. 10.18632/oncotarget.13489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Makino N, Nakamura Y, Yashiro M, Sano T, Ae R, Kosami K, et al. Epidemiological observations of Kawasaki disease in Japan, 2013-2014. Pediatr Int. (2018) 60:581–7. 10.1111/ped.13544 [DOI] [PubMed] [Google Scholar]
- 54.Tacke CE, Breunis WB, Pereira RR, Breur JM, Kuipers IM, Kuijpers TW. Five years of Kawasaki disease in the Netherlands: a national surveillance study. Pediatr Infect Dis J. (2014) 33:793–7. 10.1097/INF.0000000000000271 [DOI] [PubMed] [Google Scholar]
- 55.Lin MT, Wu MH. The global epidemiology of Kawasaki disease: review and future perspectives. Glob Cardiol Sci Pract. (2017) 2017:e201720. 10.21542/gcsp.2017.20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Salo E, Griffiths EP, Farstad T, Schiller B, Nakamura Y, Yashiro M, et al. Incidence of Kawasaki disease in northern European countries. Pediatr Int. (2012) 54:770–2. 10.1111/j.1442-200X.2012.03692.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Su K, Wu J, Edberg JC, Li X, Ferguson P, Cooper GS, et al. A promoter haplotype of the immunoreceptor tyrosine-based inhibitory motif-bearing FcgammaRIIb alters receptor expression and associates with autoimmunity. I. Regulatory FCGR2B polymorphisms and their association with systemic lupus erythematosus. J Immunol. (2004) 172:7186–91. 10.4049/jimmunol.172.11.7186 [DOI] [PubMed] [Google Scholar]
- 58.Recke A, Vidarsson G, Ludwig RJ, Freitag M, Moller S, Vonthein R, et al. Allelic and copy-number variations of FcgammaRs affect granulocyte function and susceptibility for autoimmune blistering diseases. J Autoimmun. (2015) 61:36–44. 10.1016/j.jaut.2015.05.004 [DOI] [PubMed] [Google Scholar]
- 59.Wang W, Somers EB, Ross EN, Kline JB, O'Shannessy DJ, Schweizer C, et al. FCGR2A and FCGR3A genotypes correlate with farletuzumab response in patients with first-relapsed ovarian cancer exhibiting low CA125. Cytogenet Genome Res. (2017) 152:169–79. 10.1159/000481213 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.