Abstract
Gastric cancer (GC) is the fourth most common malignant neoplasm and the second leading cause of cancer death. Identification of key molecular signaling pathways involved in gastric carcinogenesis and progression facilitates early GC diagnosis and the development of targeted therapies for advanced GC patients. Emerging evidence has revealed a close correlation between forkhead box (FOX) proteins and cancer development. However, the prognostic significance of forkhead box S1 (FOXS1) in patients with GC and the function of FOXS1 in GC progression remain undefined. In this study, we found that upregulation of FOXS1 was frequently detected in GC tissues and strongly correlated with an aggressive phenotype and poor prognosis. Functional assays confirmed that FOXS1 knockdown suppressed cell proliferation and colony numbers, with induction of cell arrest in the G0/G1 phase of the cell cycle, whereas forced expression of FOXS1 had the opposite effect. Additionally, forced expression of FOXS1 accelerated tumor growth in vivo and increased cell migration and invasion through promoting epithelial–mesenchymal transition (EMT) both in vitro and in vivo. Mechanistically, the core promoter region of FOXS1 was identified at nucleotides −660~ +1, and NFKB1 indirectly bind the motif on FOXS1 promoters and inhibit FOXS1 expression. Gene set enrichment analysis revealed that the FOXS1 gene was most abundantly enriched in the hedgehog signaling pathway and that GLI1 expression was significantly correlated with FOXS1 expression in GC. GLI1 directly bound to the promoter motif of FOXS1 and significantly decreased FOXS1 expression. Finally, we found that miR-125a-5p repressed FOXS1 expression at the translational level by binding to the 3′ untranslated region (UTR) of FOXS1. Together, these results suggest that FOXS1 can promote GC development and could be exploited as a diagnostic and prognostic biomarker for GC.
Introduction
Gastric cancer (GC) is the fourth most common malignant neoplasm and the second leading cause of cancer death. With approximately one million diagnosed cases and over 700,000 deaths recorded annually, GC is the third most common cause of cancer deaths worldwide1, and is frequently undiagnosed until a relatively advanced stage. The current optimal approach for GC therapy is surgical resection with curative intent and adjuvant chemotherapy or radiotherapy. However, the recurrence rate of GC remains high2 and clinical responses to other inhibitors for GC at the genetic level have been disappointing3. Because of the high molecular heterogeneity in GC, additional diagnostic and prognostic biomarkers and therapeutic targets are needed for improving clinical outcomes.
Fox proteins are a family of evolutionarily conserved transcriptional regulators defined by a common DNA-binding domain (DBD) termed the forkhead box or winged helix domain, and involved in a wide spectrum of biological processes, such as metabolism, development, differentiation, proliferation, apoptosis, migration, invasion and longevity4,5. Furthermore, FOX genes are differentially expressed in a large number of cancers; their role can be either oncogenic or tumor suppressive, depending on the family member and cell type6. FOXO3a modulates WNT/β-catenin signaling and suppresses EMT in prostate cancer cells7, while FOXM1 promotes pancreatic cancer EMT and metastasis via upregulating the expression of the urokinase plasminogen activator system8. The FOXM1 transcription factor network is activated in over 84% of cases of high-grade serous ovarian cancers via involvement in the homologous recombination DNA damage and repair pathway9, and FoxC2 has been linked to tumorigenesis and the progression of colorectal cancers through the Akt/GSK-3β/Snail pathway10, while FOXF2 suppresses gastric cancer through a novel FOXF2-IRF2BPL-β-catenin signaling axis11. Currently, FOXS1 is known to be expressed only in the sensory nervous system, and the role and involved mechanism of FOXS1 in gastric cancers have been rarely reported.
The Hedgehog (Hh) signaling pathway is crucial for growth and is often associated with human cancers12. Glioma-associated oncogene 1 (Gli1), a key transcription factor and a terminal effector of the HH cascade, has been shown to affect EMT in GC cancer cell lines and to influence lymphatic metastasis13. Although GLI1 elicits signal amplification by regulating target genes, the number of confirmed targets of the GLI1 transcription factor, for example, Patched (PTCH1 and PTCH2) and Human Hedge-hog-Interacting Protein (HHIP), is surprisingly small14. Yumei Diao et al. recently reported that FOXS1 was one of the top targets of the GLI1 gene and was shown to act in a negative feedback loop limiting the cellular effects of GLI1 in medulloblastoma and rhabdomyosarcoma cells15. However, the interplay of GLI1 and FOXS1 in GC cells has not been reported.
MicroRNAs (miRNAs) are a class of small 22-nucleotide-long endogenous noncoding RNAs that regulate the expression of genes at the posttranscriptional level by interacting with their 3′UTRs in a sequence-specific manner16, resulting in mRNA degradation or translational inhibition17. miRNAs are abnormally expressed in various cancers, and deregulated miRNA expression is strongly associated with tumor initiation, promotion and progression18. Therefore, it is urgently necessary to acquire knowledge regarding whether regulate the FOXS1 gene in GC and which miRNAs are involved.
Here, we determined the overexpression of FOXS1 in GC. FOXS1 identified as a target gene of GLI1 was most abundantly enriched in the hedgehog signal pathway. In addition, miR-125a-5p could bind the 3′UTR of FOXS1 and regulate the expression of FOXS1 via translational repression in gastric cancer cells, which may lead to further advancements in the knowledge of gastric cancer tumorigenesis.
Results
FOXS1 is highly expressed in gastric cancer
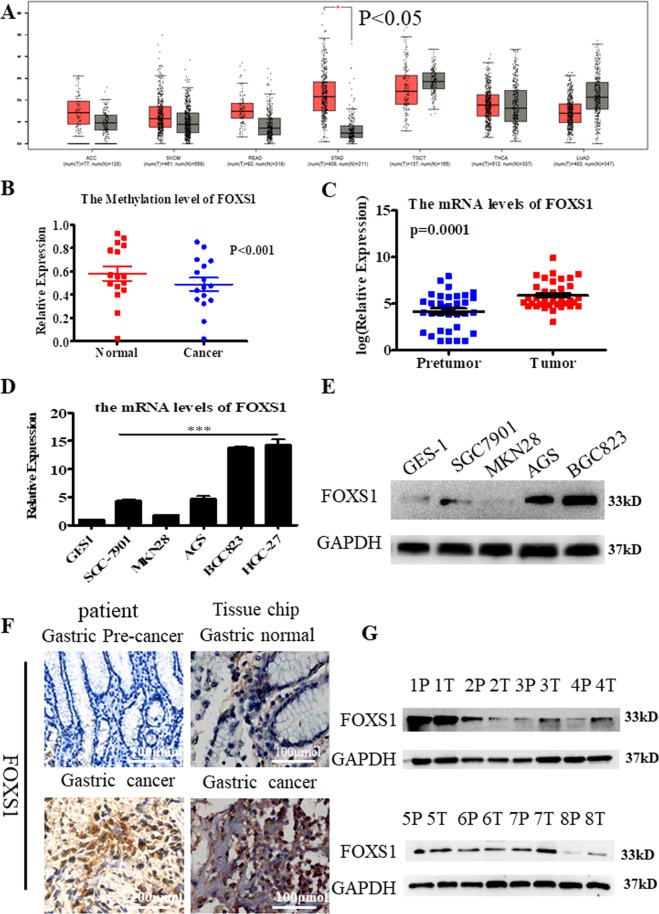
To examine the expression of FOXS1 in gastric cancer, we found that FOXS1 was significantly highly expressed in gastric cancer, but not in six other kinds of cancers by using GEPIA online software (Fig. 1A). By analyzing the cancer genome atlas (TCGA) data, we found that FOXS1 expression was significantly higher in gastric cancer samples than in noncancerous samples (normal = 38, cancer = 371, P < 0.001), consistent with the finding that FOXS1 expression was higher in in gastric cancer tissue than in normal gastric tissue in GSE19826 (N = 15, C = 12, P < 0.001), GSE13911 (N = 31, C = 38, P < 0.001) and GSE51575 (N = 26, C = 26, P < 0.001) datasets (Table 1). Using genome-wide methylation studies we identified lower FOXS1 methylation in gastric cancer, additional evidence indicating that FOXS1 is more highly expressed in gastric cancer tissues than in normal gastric tissues (P < 0.001, Fig. 1B). To validate the TCGA analysis results, we measured the expression of FOXS1 in 35 paired gastric cancer samples and paired precancer samples. The RT-PCR results showed that FOXS1 expression is significantly higher in gastric cancer tissues than in precancer tissues (P = 0.001, Fig. 1C). In addition, FOXS1 expression was significantly higher in gastric cancer cells than in GES-1 cells (normal gastric epithelial cell) at both the transcriptional (P < 0.001, Fig. 1D) and translational (Fig. 1E) levels. The immunohistochemistry (IHC) results also showed that FOXS1 expression was significantly higher in gastric cancer tissues than in the corresponding precancer tissues (Fig. 1F), consistent with the western blot (WB) analysis results shown in Fig. 1G. To further verify this result, IHC was performed on the tissue microarray chips containing 15 samples of early-stage, 55 samples of advanced-stage GC and 10 normal gastric tissue samples. The results showed that the average FOXS1 expression level was higher in 7 of the 15 samples of early-stage GC, and 45 of the 55 samples of advanced-stage GC than in the 10 normal tissues relative to the average expression of 10 normal tissues (Supplementary Figs S1–S3). These results suggest that FOXS1 expression can be a unique indicator of advanced gastric cancer.
Figure 1.
FOXS1 is high expression in gastric cancer. (A) FOXS1 expression levels in Adrenocortical carcinoma (ACC), skin cutaneous melanoma (SKCM), Stomach adenocarcinoma (STAD), Rectum adenocarcinoma (READ), Thyroid carcinoma (THCA), Testicular Germ Cell Tumors (TGCT), Thyroid carcinoma (THCA) and Lung adenocarcinoma (LUAD) using GEPIA online software. (B) The methylation levels of FOXS1 gene analyzing by TCGA sequencing data. (C) The mRNA levels of FOXS1 were analyzed by RT-PCR in 35 pairs of gastric cancer and adjacent nontumor tissues (n = 35). (D) The mRNA levels of FOXS1 were analyzed by RT-PCR in six kinds of gastric cancer cells (SGC7901, MKN28, AGS, BGC823 and HGC-27) and one gastric normal epithelial cell GES-1. (E) The protein levels of FOXS1 were analyzed by WB in four kinds of gastric cancer cells (SGC7901, MKN28, AGS and BGC823) and one gastric normal epithelial cell GES-1. (F) Left, immunohistochemical staining of FOXS1 in one paired samples of gastric cancer versus adjacent normal tissues; Right, immunohistochemical staining of FOXS1 in one sample of gastric cancer and non-adjacent gastric cancer normal tissues. (G) The protein levels of FOXS1 in 8 paired samples of gastric cancer versus adjacent normal tissues.
Table 1.
The FOXS1 gene expression in GEO data and TCGA data.
| FOXS1 | GSE19826 | GSE13911 | GSE51575 | TCGA | ||||
|---|---|---|---|---|---|---|---|---|
| N15 | C12 | N31 | C38 | N26 | C26 | N38 | C371 | |
| logFC | 1.74 | 0.645252 | 1.85 | 1.341 | ||||
| P value | 3.64E-04*** | 3.77E-02*** | 8.52e-08*** | <0.001*** | ||||
***P < 0.001 were considered statistically significant.
High FOXS1 expression indicates poor prognosis in gastric cancer patients
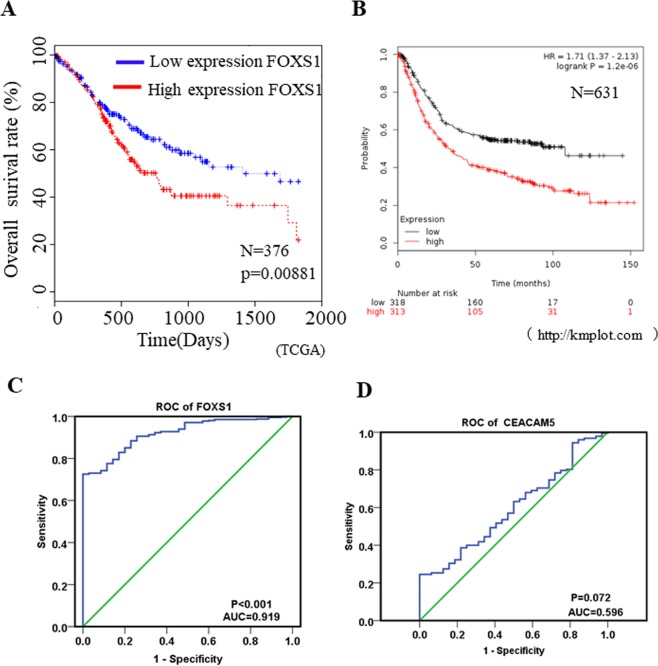
To explore the clinicpathological role of FOXS1 in gastric cancer, we classified 376 patients into two groups on the basis of the mean FOXS1 expression level by analyzing TCGA data. Patients with higher FOXS1 expression had significantly poorer prognoses than those with lower FOXS1 expression (P = 0.0081, Fig. 2A). The prognostic significance of FOXS1 mRNA assessed by the KM plotter application (http://kmplot.com/analysis/)19 also showed that upregulated FOXS1 expression was significantly correlated with poor survival and exhibited clear prognostic trend in 631 gastric cancer patients (P < 0.001, HR = 1.9; Fig. 2B). In addition, chi-square test showed that upregulated FOXS1 expression was correlated with patient age (P = 0.016), and T stage (P = 0.001). However, no significant differences were observed regarding patient sex, N stage, M stage, FIGO stage and the presence of residual tumor (Table 2). To assess whether FOXS1 could be used to predict gastric cancer development, we applied univariate and multivariate analyses. The univariate analysis results indicated that T stage (P = 0.023), N stage (P = 0.038), M stage (P = 0.003), age (P = 0.017), FIGO stage(I + II/III + IV) (P = 0.002) and FOXS1 expression (P = 0.009) influenced the survival rate of gastric cancer patients (Table 3), while the results of multivariate analysis applying a Cox proportional hazards model indicated that only age (P = 0.001), M stage (P = 0.022) and FOXS1 expression (P = 0.009) were independent impact factors for gastric cancer patients (Table 3). Furthermore, to evaluate the usefulness of FOXS1 in discriminating gastric cancer patients from healthy persons, we performed receiver-operating characteristic (ROC) analysis. The ROC analysis results revealed that FOXS1 could be a valuable biomarker for distinguishing gastric cancer patients from healthy persons, with an area under the curve (AUC) of 0.919 (Fig. 2C). Notably, the diagnostic value. of CEACAM5, a diagnostic marker for gastric cancer at the protein level that has frequently been used in the clinic20, for distinguishing gastric cancer patients from healthy persons was much lower (AUC = 0.596, P = 0.072) than that of FOXS1 (Fig. 2D). These results imply that FOXS1 exhibits powerful prognostic and diagnostic value for gastric cancer patients and can be a potential therapeutic target in gastric cancer.
Figure 2.
High expression of FOXS1 indicates poor prognosis for gastric cancer patients (A) Kaplan-Meier curves of patients with gastric cancer with low versus high expression of FOXS1 using TCGA sequencing data (N = 371; P = 0.00881). (B) Kaplan-Meier curves of patients with gastric cancer with low versus high expression of FOXS1 using online software Kaplan-Meier Plotter (http://kmplot.com) (N = 631; P < 0.001). (C) ROC curve of FOXS1was determined by SPSS and the AUC was 0.919, P < 0.001. (D) ROC curve of CEACAM5 was determined by SPSS and the AUC was 0.596, P = 0.072.
Table 2.
The correlation between FOXS1 gene expression level and patients’ clinical parameter.
| Number | FOXS1 expression Low / High | P-value | |||
|---|---|---|---|---|---|
| gender | female | 148 | 78 | 70 | 0.392 |
| male | 267 | 129 | 138 | ||
| age | < = 65 | 182 | 78 | 104 | 0.016** |
| > = 65 | 244 | 123 | 101 | ||
| Stage(T) | 1/2 | 110 | 69 | 41 | <0.001*** |
| 3/4 | 296 | 131 | 165 | ||
| Stage(N) | 0 | 124 | 66 | 58 | 0.270 |
| 1/2/3 | 273 | 129 | 144 | ||
| Stage(M) | 0 | 367 | 183 | 184 | 0.842 |
| 1 | 27 | 14 | 13 | ||
| Ajcc | 1/2 | 181 | 92 | 89 | 0.559 |
| 3/4 | 211 | 101 | 110 | ||
| residual tumor | Free | 330 | 161 | 169 | 0.077 |
| With | 34 | 22 | 12 | ||
Low/high by the sample mean. Pearson’s Chi-square tests.
**P < 0.01 and ***P < 0.001 were considered statistically significant.
Table 3.
Univariate and multivariate analyses of the contribution of FOXS1 on the survival of GC patients.
| Clinical parameters | Univariate analysis | Multivariate analysis | ||
|---|---|---|---|---|
| HRa (95%CI) | P value | HRa (95%CI) | P value | |
| FOXS1 | 1.001 (1.000–1.003) | 0.009** | 1.002 (1.000–1.003) | 0.009** |
| Age (years) | 1.022 (1.004–1.041) | 0.017** | 1.033 (1.014–1.052) | 0.001** |
| T stage (T1 + T2/T3 + T4) | 1.306 (1.038–1.643) | 0.023* | 1.203 (0.931–1.555) | 0.158 |
| N stage (No/Yes) | 1.259 (1.013–1.565) | 0.038* | 1.014 (0.732–1.404) | 0.934 |
| M stage (No/Yes) | 1.501 (1.153–1.955) | 0.003** | 1.433 (1.095–1.876) | 0.009** |
| Stage(I + II/III + IV) | 1.356 (1.119–1.642) | 0.002** | 1.254 (0.927–1.696) | 0.143 |
| Grade(I + II/III + IV) | 1.194 (0.986–1.446) | 0.069 | — | — |
| Tumor size(< = 10 mm/>10 mm) | 3.182 (1.614–2.671) | <0.001*** | — | — |
| Gender | 1.467 (0.978–2.199) | 0.064 | — | — |
aHR, hazad ratio. *P < 0.05, **P < 0.01, ***P < 0.001 were considered statistically significant.
FOXS1 regulates gastric cancer cell proliferation and colony formation
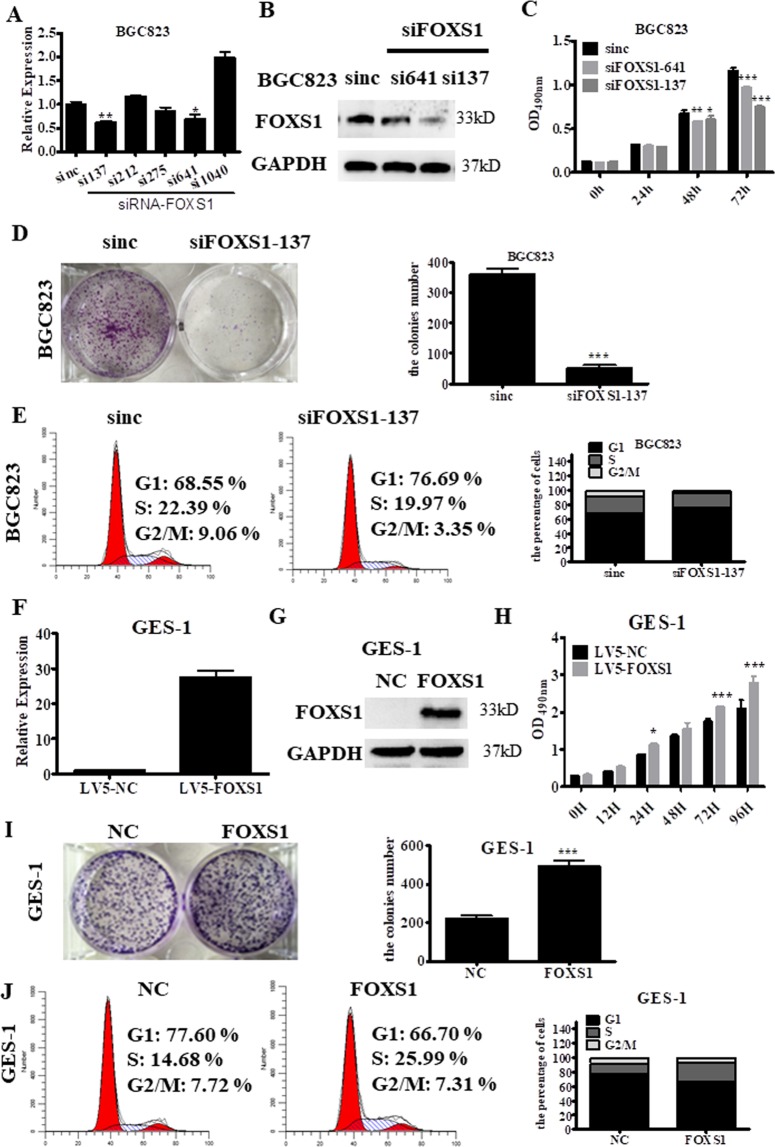
To clarify the functions of FOXS1, we designed five independent short interfering RNAs (siRNAs) targeting FOXS1 and transfected these siRNAs into BGC823 cells, which exhibit a relatively high level of FOXS1expression. The results showed that targeting FOXS1 with siRNA-137 or siRNA-641 effectively knocked down FOXS1 expression in BGC823 cells at both mRNA level (P < 0.01, P < 0.05, respectively; Fig. 3A) and the protein level (Fig. 3B). Cell proliferation assays revealed that silencing FOXS1 by transfection with siRNA-137 or siRNA-641 targeting FOXS1 significantly inhibited cell proliferation relative to that of the control cells (Fig. 3C). In addition, FOXS1 silencing significantly reduced the colony numbers in BGC823 cells (P < 0.001, Fig. 3D). Moreover, the flow cytometry (FCM) results showed that blocking FOXS1 expression in BGC823 cells led to an accumulation of cells in G1 phase and a decrease in the number of cells in the S and G2/M phases (Fig. 3E). In contrast, stable overexpression of FOXS1 in GES-1 cells and SGC7901 cells, which exhibit a relatively low level of FOXS1expression, significantly promoted cell proliferation (Fig. 3F–H and Supplementary Fig. S4A–C) and increased the colony numbers (Fig. 3I and Supplementary Fig. S4D). The flow cytometry (FCM) results further revealed a reduction in the G1-phase population and an increase in the S-phase population of GES-1 cells overexpressing FOXS1 (Fig. 3J). These data demonstrate that FOXS1 affects the G1-S transition in cell-cycle progression and increases the proliferative and colony-forming abilities of GC cells.
Figure 3.
FOXS1 regulates gastric cancer cell proliferation and the ability of colony formation. (A) The knockdown effect of siRNA targeting at five sites (137, 212, 275, 641 and 1040) of FOXS1 mRNA at mRNA levels were detected by RT-PCR. (B) The knockdown effect of siRNA targeting at two sites (137 and 641) of FOXS1 mRNA at protein levels were detected by WB. (C) The effect of FOXS1 knockdown by transfection with siRNA-137 or siRNA-641 targeting FOXS1 on cell viability was determined by Cell Proliferation Assay kits. (D) The effect of FOXS1 knockdown by transfection with siRNA-137 targeting FOXS1 on cell colony formation was analyzed by Crystal violet staining, and quantitative analysis of cell colony numbers was showed in the right panel. (E) The effect of FOXS1 knockdown on cell cycle distribution. (F,G) Overexpression effect of FOXS1 at mRNA levels was confirmed by RT-PCR and at protein levels was confirmed by WB. (H) Effect of FOXS1 overexpression on cell viability were detected by Cell Proliferation Assay kits. (I) The effect of FOXS1 overexpression on cell colony formation was analyzed by Crystal violet staining, and quantitative analysis of cell colony numbers was showed in the right panel. (J) The effect of FOXS1 overexpression on cell cycle distribution.
FOXS1 promotes gastric cancer cell migration, invasion and EMT
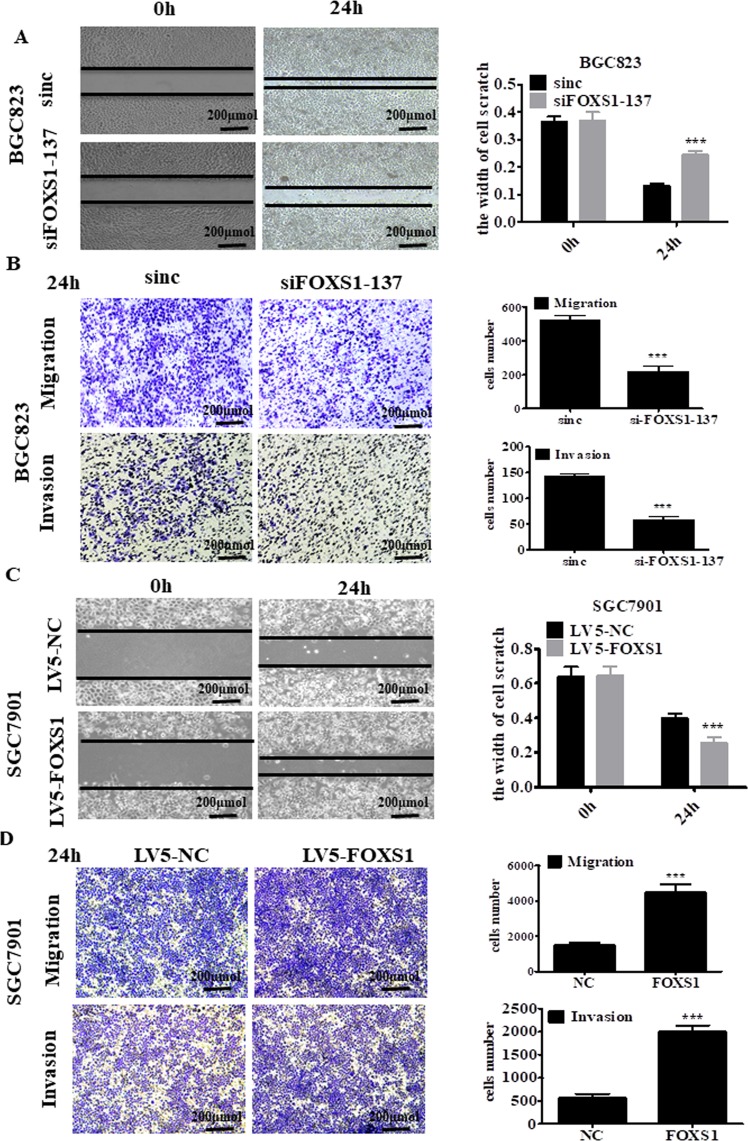
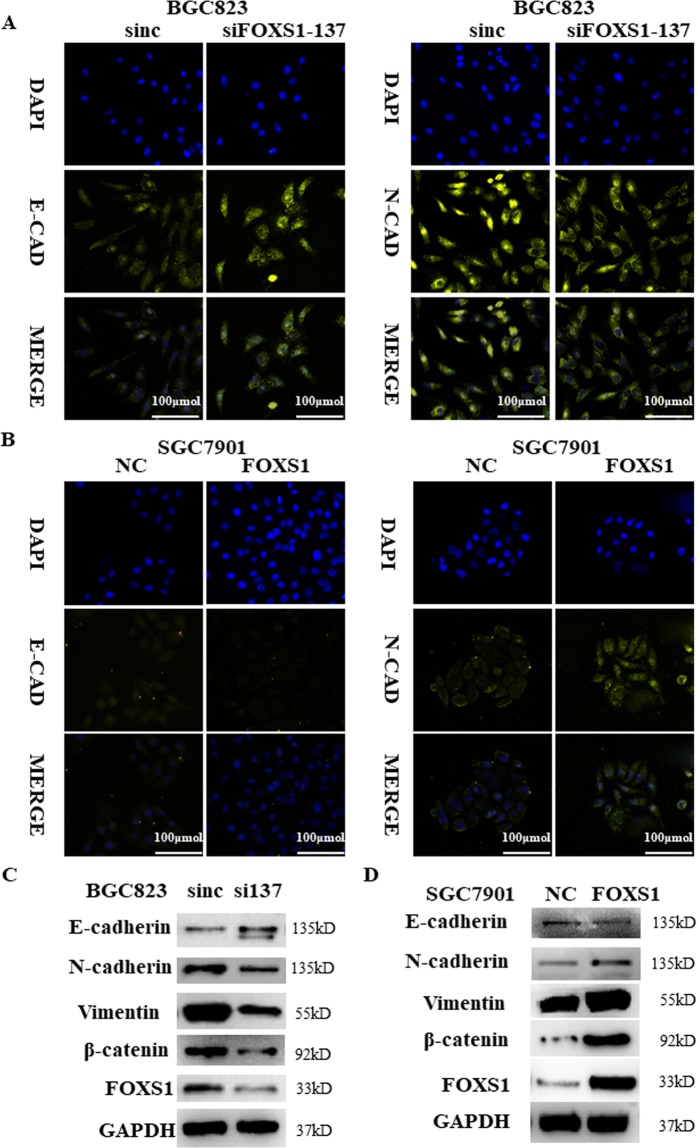
We further investigated the effect of FOXS1 on the migration and invasive abilities of gastric cancer cells using in vitro Transwell assays with or without a Matrigel matrix layer on the inserts. Knockdown of FOXS1 in BGC823 cells significantly suppressed the cell wound healing, migration and invasive abilities (Fig. 4A,B), while FOXS1 overexpression in SGC7901 cells significantly increased cell wound healing, migration and invasive abilities (Fig. 4C,D). To further prove the effect of FOXS1 on invasion and migration in gastric cancer, we performed WB analysis and immunofluorescence to measure the expression levels of EMT markers. The immunofluorescence results showed that FOXS1 knockdown increased the level of the epithelial marker E-cadherin (Fig. 5A left), but decreased the levels of the mesenchymal marker N-cadherin (Fig. 5A right). As predicted, FOXS1 overexpression produced the opposite results (Fig. 5B). Consistent with the above results, the WB analysis results showed that FOXS1 knockdown significantly inhibited the expression of N-cadherin, Vimentin and β-catenin, but increased E-cadherin expression. However, FOXS1 overexpression produced the inverse results (Fig. 5C,D). To further determine whether FOXS1 promotes EMT via the Wnt/β-catenin pathway, we next detected the expression of Wnt/β-catenin pathway related proteins (such as Cyclin-D1, and c-Myc)21. The RT-PCR results showed that FOXS1 overexpression significantly enhanced the gene expression of Cyclin-D1and c-Myc (Supplementary Fig. S5).
Figure 4.
FOXS1 promotes gastric cancer cell migration and invasion in vitro. (A,C) Effect of FOXS1 knockdown or FOXS1 overexpression on the wound-healing process in indicated cells and the scratch width were quantitative analyzed in the right panel. (B,D) Effect of FOXS1 knockdown or FOXS1 overexpression on metastatic ability of gastric cancer cells using in vitro migration and invasion transwell assays. Statistical analysis has shown in the right panel.
Figure 5.
FOXS1 promotes gastric cancer cell EMT in vitro. (A,B) The influence of FOXS1 knockdown or overexpression on EMT markers (E-cadherin and N-cadherin) were detected by immunofluorescence. (C,D) The influence of FOXS1 knockdown or overexpression on the proteins (E-cadherin, N-cadherin, Vimentin, β-catenin and GAPDH) were detected by WB.
FOXS1 promotes tumor growth and EMT in vivo
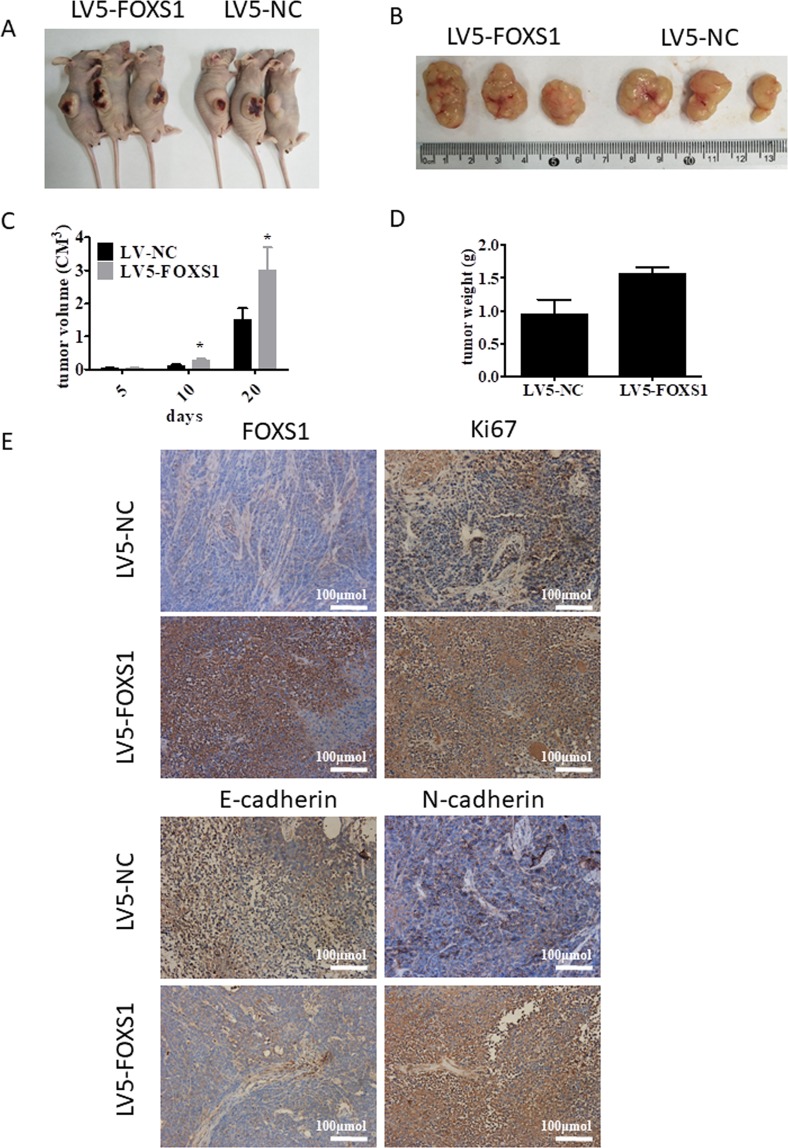
To further confirm whether FOXS1 could affect tumorigenesis and EMT in gastric cancer in vivo, SGC7901 cells infected with LV5-FOXS1 or control lentivirus were injected into the flank of nude mice. As shown in Fig. 6A, the indicated gastric cancer cells successfully formed gastric cancer tumors and the tumors shown in Fig. 6B were collected and measured. The results showed that FOXS1 overexpression increased tumor growth in terms of both size and weight in nude mice (Fig. 6C,D). The IHC results showed that FOXS1 was highly expressed in the LV5-FOXS1-infected SGC7901 cells, indicating that the LV5-FOXS1 lentivirus was successfully transduced into SGC7901 cells (Fig. 6E). In addition, positive Ki67 and N-cadherin staining was much stronger in FOXS1 overexpressing SGC7901 cells than in control cells, while E-cadherin expression was lower in FOXS1 overexpressing SGC7901 cells than in control cells (Fig. 6E). These in vivo data further demonstrated that FOXS1 can promote gastric cancer tumorigenesis and EMT events.
Figure 6.
FOXS1 promotes gastric cancer cell growth and altered the expression of EMT markers in vivo. (A) The nude mouse models with FOXS1 xenografts was successful developed by respectively injected SGC7901 cells infected with LV5-FOXS1 or LV-NC into each flank of nude mice. (B) A representative picture of the morphology of tumor xenografts after excision at 20 days of treatment. (C) FOXS1 overexpression accelerated tumor growth in nude mice. (D) The tumor weight in LV-FOXS1 group and LV-NC control group were quantitatively analyzed. (E) The FOXS1 expression in the FOXS1-infected tumors was confirmed by IHC staining. Cell proliferation in tumors isolated from FOXS1-overexpressing or control nude mice xenografts were determined by Ki-67 staining. EMT markers E-cadherin was decreased in FOXS1-infected tumors while N-cadherin expression was enhanced in FOXS1-infected tumors.
Analysis and identification of the FOXS1 promoter region and transcription factors
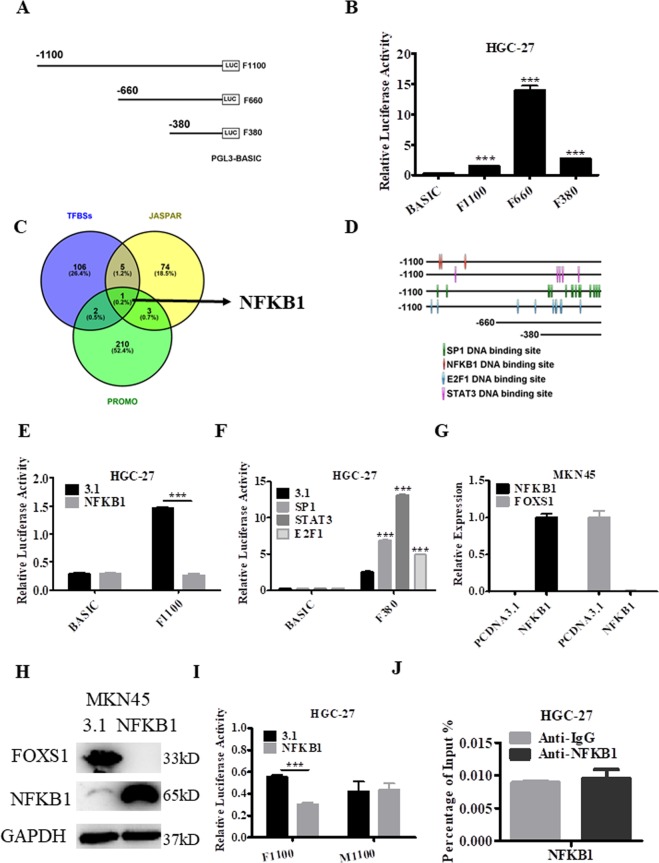
In the UCSC database, the promoter region of FOXS1was predicted to be located at chr20: 31,845,488-31,846,588, and three luciferase reporter vectors containing the indicated genomic fragments of the FOXS1gene were constructed (Fig. 7A). The results of the dual-luciferase assay showed that the luciferase activity was significantly increased in HGC27 cells transfected with F1100, F660 and F380 compared with that in HGC-27 cells transfected with the pGL3-Basic vector. In addition, the genomic region from nucleotides −660 to +1 in the FOXS1 gene had stronger promoter activity than other regions in this gene (Fig. 7B). To further investigate the potential regulators involved in FOXS1 expression, potential transcription factor binding sites in the FOXS1 promoter were identified using three online software packages: JASPAR (http://jaspar.genereg.net/), TFBSs (http://biogridlasagna.engr.uconn.edu/lasagna_search/) and PROMO (https://wwwbimas.cit.nih.gov/molbio/proscan/). As shown in Fig. 7C, NFKB1 binding sites were found in the promoter region of FOXS1 in all three independent databases. In addition, binding sites for the other three transcription factors SP1, STAT3 and E2F1 were analyzed by JASPAR along with the NFKB1 transcription factor binding sites. The results showed that the 3 potential NFKB1-binding sites were mostly located in the region between nucleotides −1100 ~ −660 in FOXS1 genes, while the 11 potential SP1-binding sites, the 4 potential STAT3-binding sites and the 7 potential E2F1-binding sites were mostly located in the region between nucleotides −380 ~ +1 in the FOXS1 genes (Fig. 7D). NFKB1 binding significantly inhibited luciferase activity (Fig. 7E), while SP1, STAT3 and E2F1 binding markedly enhanced luciferase activity driven by the promoter regions of the FOXS1 gene (Fig. 7F). These results led us to hypothesize that NFKB1 may function as a transcriptional repressor but SP1, STAT3 and E2F1 act as transcriptional activators coregulating FOXS1 expression. To further investigate whether exogenous NFKB1 negatively regulates FOXS1, an NFKB1 overexpression plasmid was transfected into MKN45 cells. The results showed that exogenous NFKB1 overexpression significantly inhibited FOXS1 expression at both the mRNA (Fig. 7G) and protein (Fig. 7H) levels, suggesting that exogenous NFKB1 may be an effective treatment strategy targeting FOXS1 in GC patients. Moreover, the NFKB1 binding motifs “GTGGATTTCC” in the promoter-reporter constructs were mutated to “GTGGATGTCC” in the promoter-reporter constructs of FOXS1 and the results showed that the effect of NFKB1 overexpression on luciferase activity driven by the F1100 region after mutation of NFKB1 binding motif sequence was abrogated (Fig. 7I). A further ChIP assay was performed and the results showed that NFKB1 can’t directly bound to the FOXS1 promoter region (Fig. 7J), indicating that NFKB1 deregulate the expression of FOXS1 in other special ways.
Figure 7.
The analysis and identification of FOXS1 core promoter. (A) Schematic diagram of FOXS1 upstream 1100 bp promoter reporter constructs. (B) Dual luciferase reporter assays were used to determine the core promoter activity region of FOXS1. (C,D) Prediction of transcription factors binding sites in the FOXS1promoter region using TFBSs, JASPAR and PROMO. (E,F) The influence of transcription factors on FOXS1 promoters were determined by dual luciferase reporter assays. (G) The expression of NFKB1 and FOXS1 after NFKB1 overexpression was determined by RT-PCR in MKN45 cells. (H) The expression of NFKB1 and FOXS1 after NFKB1 overexpression was determined by western blot in MKN45 cells. GAPDH was internal control. (I) The effect of transcription factors NFKB1 on FOXS1 promoters with NFKB-binding mutated sites were determined by dual luciferase reporter assays. (I) ChIP assays were performed to detect NFKB1 directly bound in the FOXS1 promoter region in HGC-27 cells.
The FOXS1 gene was most abundantly enriched in the Hh pathway and was identified as a GLI1 target gene in GC
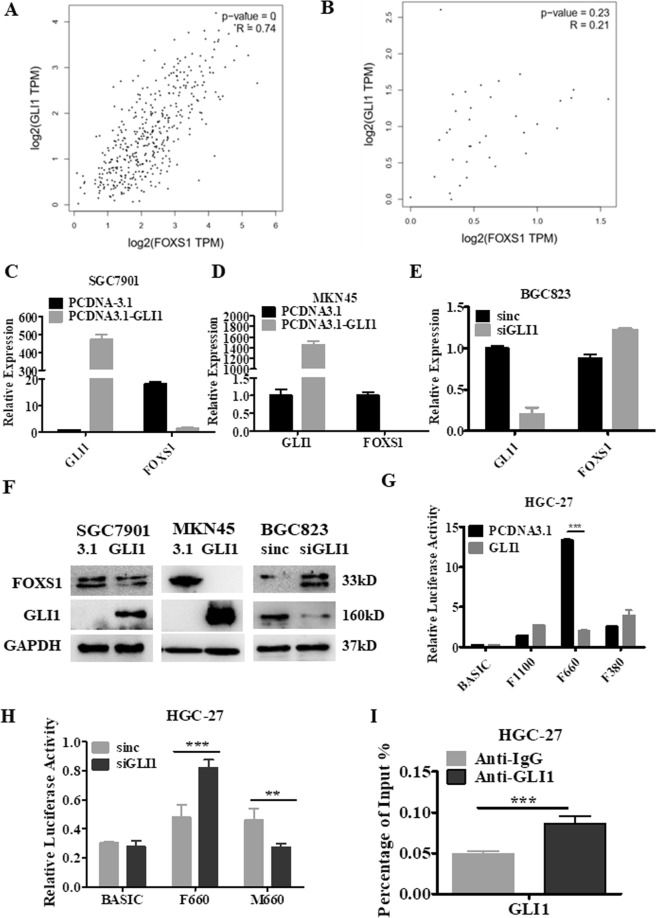
The RNA-seq data for patients in the TCGA database were used to divide patients into the FOXS1 high expression group and the FOXS1 low expression group according to the mean level of FOXS1 gene expression. Enrichment analysis was performed on the two groups of genes to screen statistically significant signaling pathways by GSEA (software http://software.broadinstitute.org/gsea/index.jsp). We found that the FOXS1 gene was significantly enriched in the KEGG_HEDGEHOG_SIGNALING_PATHWAY, KEGG_ECM_RECEPTOR_INTERACTION, KEGG_FOCAL_ADHESION, KEGG_VASCULAR_ SMOOTH MUSCLE_CONTRACTION and 11 other important signaling pathways (Table 4). Notably, FOXS1 was most abundantly enriched in the Hh signaling pathway (NES, 0.61; FDR, q = 0; FWER, p = 0.003). In GC, tumorigenicity and EMT are regulated through the activation of GLI1, a key member of the Hh signaling pathway13. Importantly, correlation analysis revealed that the expression of FOXS1 was strongly correlated (R = 0.74, P < 0.001) with the expression of GLI1 in the STAD tumor dataset (Fig. 8A), whereas FOXS1 expression was not correlated with GLI1 expression in the TCGA STAD normal dataset (Fig. 8B), indicating an interplay between GLI1 and FOXS1 expression in STAD tumors. Next, we determined the expression of GLI1 in three kinds of gastric cancer cells and GES-1 cells. The results showed that GLI1 had a high expression level in BGC823 cells, a moderate expression level in MKN45 and SGC7901 cells and a low expression level in GES-1 cells (Supplementary Fig. S6), an expression trend consistent with that of FOXS1. To further investigate whether FOXS1 is a target gene of GLI1 in STAD tumors, GLI1 exogenous overexpression and gene silencing experiments were performed. GLI1 overexpression significantly inhibited FOXS1 expression in both SGC7901 cells and MKN45 cells (Fig. 8C,8D and 8F left and middle), while GLI1 gene silencing upregulated FOXS1 expression in BGC823 cells (Fig. 8E and 8F right). Therefore, we hypothesized that the transcription factor GLI1 may be a transcriptional repressor of FOXS1. To investigate the potential GLI1/FOXS1 regulatory mechanism, dual luciferase assays were performed. Although GLI1 exerted a differential effect on luciferase activity driven by the FOXS1 promoter regions in HGC-27 and SGC7901 cells, GLI1 overexpression significantly decreased luciferase activity driven by the FOXS1 promoter at the core promoter region of F660 in both HGC-27 cells (Fig. 8G) and SGC7901 cells (Supplementary Fig. S7), while GLI1 silencing notably increased luciferase activity driven by the F660 region in HGC-27 cells (Fig. 8H). Moreover, we mutated the GLI1 binding motif “CACCACCCAG” to “CACAACCCAG” and found that the effect of GLI1 silencing on luciferase activity driven by the F660 region was reversed (Fig. 8H). A further ChIP experiment demonstrated that GLI1 directly bound to the FOXS1 promoter region (Fig. 8I). These findings indicate that GLI1 acts as a unique transcriptional repressor of FOXS1 by binding FOXS1 promoter regions in GC cells.
Table 4.
Statistically significant signaling pathway for FOXS1 gene enrichment.
| GS Details | NES | FDR q-val | FWER p-val |
|---|---|---|---|
| KEGG_HEDGEHOG_SIGNALING_PATHWAY | 0.61 | 0 | 0.003** |
| KEGG_ECM_RECEPTOR_INTERACTION | 0.67 | 0 | 0.005** |
| KEGG_FOCAL_ADHESION | 0.56 | 0 | 0.006** |
| KEGG_VASCULAR_SMOOTH_MUSCLE_CONTRACTION | 0.55 | 0 | 0.008** |
| KEGG_BASAL_CELL_CARCINOMA | 0.58 | 0 | 0.007** |
| KEGG_NEUROACTIVE_LIGAND_RECEPTOR_INTERACTION | 0.52 | 0 | 0.006** |
| KEGG_GAP_JUNCTION | 0.5 | 0 | 0.009** |
| KEGG_CELL_ADHESION_MOLECULES_CAMS | 0.56 | 0.004 | 0.015* |
| KEGG_DILATED_CARDIOMYOPATHY | 0.55 | 0.004 | 0.021* |
| KEGG_CYTOKINE_CYTOKINE_RECEPTOR_INTERACTION | 0.5 | 0.004 | 0.035* |
| KEGG_MELANOGENESIS | 0.46 | 0 | 0.034* |
| KEGG_CALCIUM_SIGNALING_PATHWAY | 0.47 | 0.004 | 0.042* |
| KEGG_HYPERTROPHIC_CARDIOMYOPATHY_HCM | 0.52 | 0.006 | 0.044* |
| KEGG_REGULATION_OF_ACTIN_CYTOSKELETON | 0.42 | 0.002 | 0.047* |
*P < 0.05, **P < 0.01, ***P < 0.001 were considered statistically significant.
Figure 8.
FOXS1 expression positively correlates with GLI1 expression and FOXS1 is a target gene of GLI1. The pearson correlation between FOXS1 and GLI1 expression was detected by analyzing TCGA data in STAD tumor samples (A) and in STAD normal samples (B). FOXS1 and GLI1 expression was detected by RT-PCR in SGC7901 (C) and MKN45 cells (D) transfected with pCDNA3-vector or pCDNA3-GLI1. (E) FOXS1 and GLI1 expression was detected by RT-PCR in BGC823 cells transfected with siRNA control or siRNA targeting GLI1. (F) The protein levels of FOXS1 and GLI1 was detected by western blot in SGC7901 and MKN45 cells transfected with pCDNA3-vector or pCDNA3-GLI1 or in BGC823 cells transfected with siRNA control or siRNA targeting GLI1. (G) Dual luciferase activity analysis of the four promoter activity regions of FOXS1 after transfection with pCDNA3-vector or pCDNA3-GLI1. (H) Dual luciferase activity analysis of the F660 and M660 promoter of FOXS1 after GLI1 knockdown. M660 refers to core promoter reporting vectors with GLI1 binding motif mutation sites. (I) ChIP assays were performed to detect GLI1 directly bound in the FOXS1 promoter region in HGC-27 cells.
miR-125a-5p downregulated FOXS1 expression through translational repression
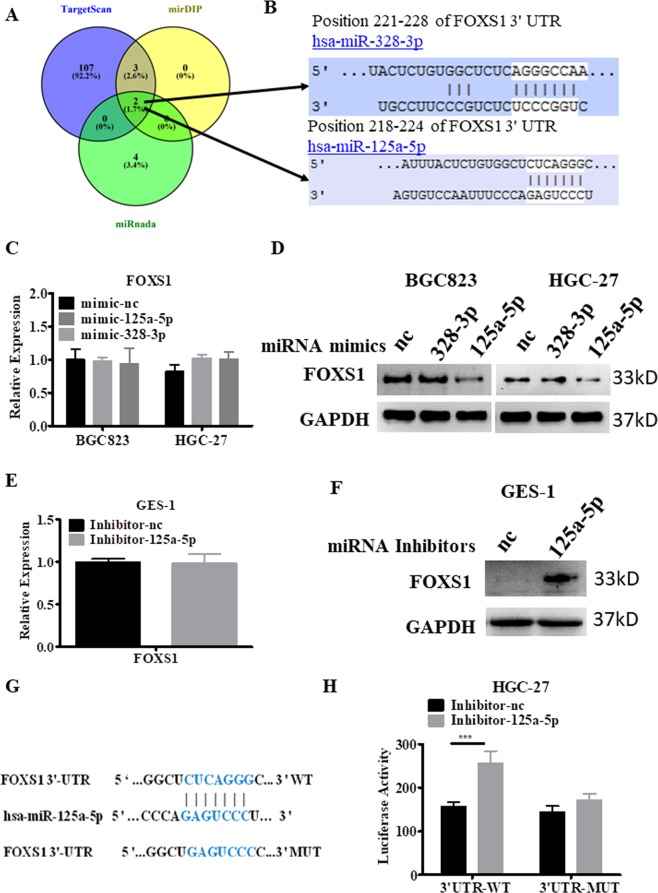
To investigate the miRNA-related mechanism of FOXS1 upregulation in gastric cancer, we used 3 independent databases miRanda, mirDIP, and TargetScan to computationally predict miRNAs that may be involved (Fig. 9A). Two miRNAs (miR-328-3p and miR-125a-5p) were identified in all three independent datasets (Fig. 9B). Then, two miRNA mimics of these miRNAs were transfected into BGC823 and HGC27 cells, which exhibit high expression of FOXS1. The RT-PCR results showed that although miR-328-3p and miR-125a-5p were successfully overexpressed (Supplementary Fig. S8A,B), no difference in the mRNA levels of FOXS1 were detected after transfection of the miR-328-3p or miR-125a-5p mimics (Fig. 9C). However, the protein level of FOXS1 was appreciably decreased in miR-125a-5p-transfected cells compared with that in negative control-transfected cells, while no difference was found in miR-328-3p-transfected cells (Fig. 9D). To confirm the above conclusion, miR-125a-5p inhibitors were synthesized and transfected into GES-1 cells, which exhibit high levels of miR-125a-5p expression22 and low levels of FOXS1 expression. The RT-PCR results showed that the expression of miR-125a-5p was inhibited after transfection with miR-125a-5p inhibitors, with no change in the mRNA levels of FOXS1 (Fig. 9E and Supplementary Fig. S8C). As shown in Fig. 9F, the FOXS1 protein levels were markedly upregulated after transfection with miR-125a-5p inhibitors in the normal gastric cell line GES1. To investigate whether FOXS1 was a direct target of miR-125a-5p, wild-type or mutant binding site fragments in the FOXS1 3′ UTR (Fig. 9G) were directly fused downstream of the firefly luciferase gene in the GP-miRGLO vector. Cotransfection of miR-125a-5p inhibitors with the GP-miRGLO-FOXS1-3′UTR wild-type luciferase reporter plasmid caused a marked increase in luciferase activity, whereas luciferase activity was not significantly affected by cotransfection of miR-125a-5p inhibitors with the GP-miRGLO-FOXS1-3′UTR mutant luciferase reporter plasmid (Fig. 9H). Collectively, these results suggest that miR-125a-5p directly targets FOXS1 in gastric cancer cells, deregulating FOXS1 expression through translational repression.
Figure 9.
MiR-125a-5p inhibits FOXS1 expression by translational repression. (A) Three independent miRNA target databases were used to predict the potential miRNAs. (B) Schematic representation of the potential miRNAs binding sites in 3′UTR of FOXS1. (C) RT-PCR results of the expression of FOXS1 in the miRNA mimics of miR-328-3p or miR-125a-5p transfection group in BGC823 and HGC-27cells. (D) Levels of FOXS1 protein were analyzed by WB after transfection with mimics of miR-328-3p or miR-125a-5pfor 72 hours. (E) RT-PCR results of the expression of FOXS1 in GES-1 cells transfected with miRNA inhibitors of miR-125a-5p or nc. (F) Levels of FOXS1 protein were analyzed by WB after transfection with inhibitors of miR-125a-5p for 72 hours. (G) Predicted binding sites and mutant binding site between miR-125a-5p and 3′UTR OF FOXS1. (H) HGC-27 cells were firstly transfected with miR-125a-5p inhibitors or inhibitors nc and next day transfected with a luciferase reporter-containing FOXS1 wild-type 3′UTR (3′UTR-WT) or FOXS1 mutant 3′UTR (3′UTR-MUT). Luciferase activity of the indicated cells were determined by Luciferase assay kits.
Discussion
Recent evidence suggests that FOX gene family members are important in a wide spectrum of biological processes4,23,24. However, the role of FOXS1, a new member of the FOX gene family25, in cancer has not been reported to date. In this study, we concluded that FOXS1 was highly expressed in gastric cancer. Tissue microarray revealed that FOXS1could be a unique indicator of advanced gastric cance. In addition, high FOXS1 expression indicated poor survival and differentiated gastric cancer from normal tissues with high sensitivity and specificity. FOXS1 expression was positively related to tumor size and advanced age and could be an independent prognostic factor. In addition, FOXS1 promoted cell proliferation, migration and invasion via the Wnt/β-catenin pathway, which may be coregulated by the transcriptional activators STAT3, SP1, and E2F1 and the transcriptional repressor NFKB1. Exogenous NFKB1 overexpression inhibited FOXS1 expression, suggesting a new treatment target for GC patients. In addition, FOXS1, identified as a target gene of GLI1, was most abundantly enriched in the Hh signal pathway. Finally, miR-125a-5p bound the 3′UTR of FOXS1 and regulated the expression of FOXS1 via translational repression in gastric cancer cells, which may lead to further advancements in the knowledge of gastric cancer tumorigenesis.
Given that Fox proteins control these essential developmental and homeostatic processes, a loss or gain of Fox function can alter cell fate and lead to tumorigenesis4,26–28. In our study, we found that FOXS1 was highly expressed in gastric cancer. In addition, FOXS1 overexpression in GES-1 and SGC7901 cells promoted cell proliferation and cell colony formation ability, while FOXS1 silencing in BGC823 produced the inverse results. These results are consistent with the report that FOXS1 and six other genes were positive impact factors serving as a prognostic signature to predict the survival of gastric cancer patients and to monitor the long-term treatment of gastric cancer29. Nevertheless, FOXS1 has the opposite characteristic in liver cancer; it is expressed at lower levels in most HCC tissues than in normal liver tissues. These decreased expression levels are correlated with tumor size, AJCC stage, and tumor differentiation, acting as an important factor for predicting the prognosis of HCC patients30. From the above results, we concluded that members of the FOX gene family, including FOXS1, can be oncogenes or tumor suppressor genes depending on the cancer type.
EMT also plays an important role in the invasion and metastasis of cancers31. As a result of EMT, tumor cells exhibit the characteristics of downregulated expression of epithelial-associated markers such as E-cadherin and upregulated expression of mesenchymal markers such as N-cadherin and Vimentin13. In our work, we found that FOXS1 expression was inversely correlated with the expression of epithelial markers (E-cadherin) and positively correlated with the expression of mesenchymal markers (vimentin and N-cadherin), indicating that FOXS1 could induce EMT in gastric cancer cells. A large body of evidence suggests that the canonical Wnt/β-catenin pathway plays an important role in inducing epithelial cancer cells to undergo EMT32,33. In our work, we found that FOXS1 silencing inhibited β-catenin expression while FOXS1 overexpression augmented β-catenin expression. In addition, wnt/β-catenin can regulate important tumor-related genes including c-Myc and Cyclin D121. We investigated the effect of FOXS1 on the Wnt/β-catenin signaling pathway, and found that FOXS1 overexpression significantly enhanced the mRNA levels of c-Myc and Cyclin D1. These results preliminarily indicate that FOXS1 promotes EMT via the Wnt/β-catenin signaling pathway.
Despite the extensive efforts to understand the contribution of FOXS1 to gastric cancer, the mediators responsible for FOXS1 upregulation in gastric cancer remain unidentified. In our work, the results of luciferase reporter assays showed that the core promoter regions of FOXS1 are located at nucleotides −660~ +1 in gastric cancer HGC-27 cells, but at −380 ~ +1 in SGC7901 cells (Supplementary Fig. S9A). In addition, FOXS1 has 11 potential SP1-binding sites, 4 potential STAT3-binding sites, and 7 potential E2F1-binding sites, which are mostly located in the −380 ~ +1 region of the FOXS1 gene, and 3 NFKB1-binding sites in the −1100 ~ −660 region. As predicted, the luciferase reporter assay showed that STAT3, E2F1, and SP1 enhanced luciferase activity driven by F380 in both HGC-27 cells and SGC7901 cells. Unexpectedly, NFKB1 inhibited luciferase activity driven by F1100 in HGC-27 cells but slightly increased luciferase activity driven by F1100 in SGC7901 cells (Supplementary Fig. S9B), suggesting that NFKB1 regulates FOXS1 expression in a manner dependent on the type of gastric cancer cells.
Glioma-associated oncogene 1 (GLI1) is one of the three GLI transcription factors within the Hh pathway34. Yan and coworkers found that Gli1 overexpression is a frequent event in gastric cancer tissues35,36,and is correlated with characteristics of a more aggressive phenotype, including poorly differentiated histology, advanced TNM stage, serosal involvement and lymph node metastasis35. In this study, we found that FOXS1 expression was significantly correlated with GLI1 expression in the STAD tumor dataset but was not correlated with GLI1 expression in the STAD normal dataset, suggesting that GLI1 and FOXS1 may exhibit interplay in gastric cancer. Moreover, Diao et al. recently published that both GLI1 and FOXS1 are highly expressed and positively correlated with GLI1 in medulloblastoma samples15, further suggesting that the existence of FOXS1/GLI1 interplay in human cancers may not be dependent on the type of human tumor. Interestingly, FOXS1 expression can reduce GLI1 transcriptional activity, and GLI1/FOXS1 exhibit direct protein–protein interaction in HEK293A cells15,37. In our study, we found that GLI1 inhibited FOXS1 expression by binding the promoter regions of FOXS1, suggesting that GLI1 and FOXS1, two oncogenes, may combine and suppress each other’s expression. This phenomenon suggests that there may be negative feedback between FOXS1 and GLI1 in human cancers that needs to be further studied.
Mature miRNAs are a large group of conserved, single-stranded, noncoding RNAs of approximately 22 nucleotides (nt) in length that act as posttranscriptional regulators of gene expression by binding to their target coding mRNA through imperfect complementarity at multiple sites in the 3′-UTR of a gene, resulting in cleavage, translational repression, or chromatin modification of the coding mRNA and thus interfering with gene expression38–44. It is worth noting that low expression of miR-125a-5p was previously reported to be an independent prognostic factor in gastric cancer and inhibits the proliferation, invasion and metastasis of human gastric cancer cells42. In addition, Cao Y. et al. reported that miR-125a-5p expression was lower in human gastric cancer cell lines (SGC7901, HGC27 and BGC823) than in the normal gastric mucosa cell line GES1, with a similar pattern in 82 pairs of cancer tissues and matched normal tissues, as evidenced by RT-PCR analysis22,42, while FOXS1 was highly expressed in the above gastric cancer cell lines and gastric cancer tissues, revealing that the endogenous FOXS1 and miR-125a-5p levels were inversely correlated. miR-125a-5p can bind the 3′-UTR of FOXS1 and repress FOXS1 expression at the transcriptional level, consistent with the finding that miR-125a-5p mimics were able to inhibit FoxN1 3′UTR luciferase activity and suppress FoxN1 expression39. These results reveal that FOXS1 is also a target gene of miR-125a-5p and that its high expression might be due to the loss of miR-125a-5p, providing novel insight into miRNA regulation of human gene expression.
In summary, we first systematically identified FOXS1 as a novel oncogene and pinpointed the related regulatory network involved in FOXS1 in gastric cancer cells, providing new insight into understanding the mechanism underlying gastric cancer development. The findings of this study may potentially lead to the development of meaningful prognostic biomarkers for gastric cancer, and specific intervention strategies targeting FOXS1 might provide new therapeutic targets for gastric cancer patients.
Materials and Methods
Ethical approval and informed consent
All experimental procedures were approved by the first affiliated hospital of Chongqing Medical University and Chongqing Medical University. Written informed consent was obtained for all patient samples. The gastric tumor tissues and adjacent tissues were collected from 35 patients with gastric cancer at first affiliated hospital of Chongqing Medical University (during 2016 and 2017). The tissue microarray chips of gastric cancer (ST8014) were bought from Alenabio Biological Technology Co., Ltd. The study protocol was approved by the ethics committees at Chongqing medical university. Gastric cancer was diagnosed based on H&E staining and immune phenotype.
Cell line and culture
The human gastric cancer cell lines (BGC823, MKN28, AGS, HGC-27 and SGC7901) were purchased from the Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China). GES-1 was purchased from Obio Technology (Shanghai) Corp., Ltd. All the cells except AGS cultured in F12K medium were cultured in RPMI 1640 medium supplemented with 10% of fetal bovine serum, 100 U/ml of penicillin and 100 mg/ml of streptomycin. The cells were grown at 37 °C in a humidified incubator with 5% CO2. I confirm that the methods were performed in accordance with the indicating guidelines and regulations.
Immunohistochemistry (IHC)
The human gastric cancer, adjacent tissue and tissues from heterologous tumor in nude mice were collected, fixed in 4% paraformaldehyde, embedded in paraffin, and sectioned (3μm). These paraffin sections were dewaxed, rehydrated and incubated in 3% H2O2 for 10 min to block endogenous peroxidase. Then the sections were autoclaved in 10 mM sodium citrate buffer (pH 6.0) for 15 min. Normal goat serum (10%) was used to block non-specific staining and then the tissue sections were exposed to indicated antibodies. For tissue microarray chips of gastric cancer versus gastric normal tissue, the target molecules were performed by using FOXS1 antibody. All of the staining were observed by at least 2 independent investigators blinded to the histopathologic features and patient data of the samples. I confirm that the methods were performed in accordance with the indicating guidelines and regulations.
RNA extraction, reverse transcription, and Real-time PCR (RT-PCR)
Total RNA was extracted from tissues or cells using the Eastep ® Super Total RNA Extraction Kit (Promega Biotechnology, China), and the reverse transcription was performed using the PrimeScript™ RT Master Mix according to the manufacturer’s instructions (Takara Biotechnology, China). RT-PCR was performed by using the SYBR Green PCR Kit (Bio-Rad, China) in triplicate in three independent experiments by the 2−ΔΔCt method. The relative expression of miR-125a-5p or miR-328-3p were normalized to U6 and detected by Hairpin-itTM miRNAs RT-PCR Quantitation Kit (Applied by Genepharma, Shang hai). RT-PCR primers sequences were showed as the Supplementary Table S1. I confirm that the methods were performed in accordance with the indicating guidelines and regulations.
Western Blot (WB)
Western blot analysis was performed as previously described45. The FOXS1 primary antibody were purchased from SIGMA (HPA042475). E-cadherin (3195), N-cadherin (13116), β-catenin (8480), Vimentin (5741) and anti-rabbit IgG, HRP-linked antibody (7074) were purchased from CST. I confirm that the methods were performed in accordance with the indicating guidelines and regulations.
Immunofluorescence (IF)
Cells were plated on 8-lm-thick chip, fixed in 4% ice-cold paraformaldehyde, and permeabilized using 0.5% Triton X-100/PBS for 20 min at room temperature. The cells were blocked with 10% albumin from bovine serum for 1 h and then incubated with anti-bodies against E-cadherin and N-cadherin (1:100, CST) overnight at 4 °C. After three washes, cells were incubated with Alexa Fluor 488-conjugated goat anti-rabbit secondary antibody (1: 1000; Invitrogen). The nuclei were counterstained with DAPI (1:100, Invitrogen), and cells were visualized with a laser scanning confocal microscope (Leica Microsystems, Wetzlar, Germany). I confirm that the methods were performed in accordance with the indicating guidelines and regulations.
Cell proliferation assay
Cells were plated (3000 per well) in a 96-well plate. The absorbs value was measured at 490 nm by using CellTiter 96®AQueous One Solution Cell Proliferation Assay kits (Promega, G3582) at indicating times. I confirm that the methods were performed in accordance with the indicating guidelines and regulations.
Wound healing assay
The cells were seeded in 6-well plates and cultured to 100% confluence. Monolayer cells were washed with PBS, scraped with a plastic 200-ul pipette tip and then incubated with fresh 1640 medium without serum. The wounded areas were photographed by phase contrast microscopy at 24 h. The assay was done in triplicate at least three times. I confirm that the methods were performed in accordance with the indicating guidelines and regulations.
SiRNA and transfection
The sequences of the siRNAs used to suppress FOXS1 expression and microRNA sequences were showed in Supplementary Table S2. Lipofectamine® RNAiMAX Reagent (Invitrogen) was used for transfection of miRNA or siRNA according to the manufacturer’s instructions. Lent virus5-GFP-FOXS1 was used to infect GES-1 or SGC7901 cells. After infection 24 h, cells were cultured in complete media containing puromycin (6 μg/ml) to generate a stable overexpressed cell line. I confirm that the methods were performed in accordance with the indicating guidelines and regulations.
Dual- luciferase reporter assay
Transfection and luciferase reporter assay were performed as previously described46. The full-length and the different fragment sequences were synthesized and then cloned into the pGL3-basic vector. Nucleotide sequences of all the cloned DNA fragments were confirmed by direct DNA sequencing. I confirm that the methods were performed in accordance with the indicating guidelines and regulations.
In vivo tumor angiogenesis assays
The animal study protocol was approved by the Animal Experimentation Ethics Committee of Chongqing Medical University. Six specific pathogen-free (SPF) BALB/c nude mice (4–6 week old) were obtained from the Institute of Medical Laboratory Animals, Chinese Academy of Medical Sciences. The mice were kept at 55 ± 5% humidity and 22–25 °C in Laboratory Animal Center of Chongqing medical university, fed with sterile water and food, and adaptively fed for 1 week before any experiment. SGC7901 cells (2 × 106) infected with LV5-NC or LV5-FOXS1 virus were injected in the femoral area of the mice (n = 3/group). The tumor was measured with calipers and the volume was calculated using the formula: (π/6) × 3, where x = the largest diameter. Three weeks after tumor inoculation, the mice were sacrificed and the tumors were extracted to determine tumor weight. Data are presented as the mean ± SD. At the end, mice were sacrificed, the tumors were collected, fixed in 4% formaldehyde, sectioned for IHC staining, and observed under a microscope (Olympus, Tokyo, Japan). I confirm that the methods were performed in accordance with the indicating guidelines and regulations.
Statistical analysis
All in vitro experiments were repeated three times or more, and data are presented as mean + SD. The student t test assumed two-tailed distributions to calculate statistical significance between groups. Survival curves were generated using the Kaplan–Meier method and compared using the log-rank tests. For analysis of correlation between FOXS1 levels and clinical features, Pearson’s chi-square tests were used. The independent prognostic factors were identified by the Cox proportional hazards regression model. ROC curve was generated with SPSS software. Differences were analyzed by GraphPad Prism 5. P-value < 0.05 was marked as statistically significant. P-value < 0.01 was indicated as highly statistically significant. P-value < 0.001 was indicated as extremely statistically significant difference. I confirm that the methods were performed in accordance with the indicating guidelines and regulations.
Supplementary information
Acknowledgements
We thank all individuals who take part in this research. This study was reviewed and approved by the Ethics Committee of The First Affiliated Hospital of Chongqing Medical University and Chongqing Medical University. All animal studies were approved by the Animal Experimentation Ethics Committee of Chongqing Medical University, Chongqing, China. All authors have read, approved the final manuscript and consent to publish. The authors declare that all data used in this study are available in the article and additional files. This work was supported by the Fund of Key Research and Development of Social and People’s Livelihood (cstc2018jscxmszdX0031 to Fangzhou Song) and Research Grant from Chongqing Medical University (4101050007).
Author Contributions
Conceived and designed the study: Sen Wang. Development of methodology: Wanfeng Zhang, Xue Leng, Jing Song. Acquisition of data: Sen Wang, Wanfeng Zhang, and Xue Leng. Analysis and interpretation of data: Wang Sen and Fang-zhou Song. Writing of the manuscript: Sen Wang, Kexin Wang. Administrative, technical, or material support: Geli Liu, Yan Ma, Yujing Wang, Xianqin Zhang, Yitao Wang, Haiyu Li, Kun Liu, Wei Zhang, Longke Ran, Guijun Qin, Fangzhou Song. All authors read and approved the final manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Longke Ran, Wanfeng Zhang, Xue Leng and Kexin Wang contributed equally.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-41717-w.
References
- 1.Kankeu Fonkoua, L. & Yee, N. S. Molecular Characterization of Gastric Carcinoma: Therapeutic Implications for Biomarkers and Targets. Biomedicines6, 10.3390/biomedicines6010032 (2018). [DOI] [PMC free article] [PubMed]
- 2.Ding K, et al. GSE1 predicts poor survival outcome in gastric cancer patients by SLC7A5 enhancement of tumor growth and metastasis. The Journal of biological chemistry. 2018;293:3949–3964. doi: 10.1074/jbc.RA117.001103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bijlsma MF, Sadanandam A, Tan P, Vermeulen L. Molecular subtypes in cancers of the gastrointestinal tract. Nature reviews. Gastroenterology & hepatology. 2017;14:333–342. doi: 10.1038/nrgastro.2017.33. [DOI] [PubMed] [Google Scholar]
- 4.Myatt SS, Lam EW. The emerging roles of forkhead box (Fox) proteins in cancer. Nature reviews. Cancer. 2007;7:847–859. doi: 10.1038/nrc2223. [DOI] [PubMed] [Google Scholar]
- 5.Wotton KR, Shimeld SM. Comparative genomics of vertebrate Fox cluster loci. BMC genomics. 2006;7:271. doi: 10.1186/1471-2164-7-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heglind M, et al. Lack of the central nervous system- and neural crest-expressed forkhead gene Foxs1 affects motor function and body weight. Molecular and cellular biology. 2005;25:5616–5625. doi: 10.1128/MCB.25.13.5616-5625.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu H, et al. FOXO3a modulates WNT/beta-catenin signaling and suppresses epithelial-to-mesenchymal transition in prostate cancer cells. Cellular signalling. 2015;27:510–518. doi: 10.1016/j.cellsig.2015.01.001. [DOI] [PubMed] [Google Scholar]
- 8.Huang C, et al. FOXM1c promotes pancreatic cancer epithelial-to-mesenchymal transition and metastasis via upregulation of expression of the urokinase plasminogen activator system. Clinical cancer research: an official journal of the American Association for Cancer Research. 2014;20:1477–1488. doi: 10.1158/1078-0432.ccr-13-2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fang, P. et al. Olaparib-induced Adaptive Response Is Disrupted by FOXM1 Targeting that Enhances Sensitivity to PARP Inhibition. Molecular cancer research: MCR, 10.1158/1541-7786.MCR-17-0607 (2018). [DOI] [PMC free article] [PubMed]
- 10.Li J, et al. FOXO4 and FOXD3 are predictive of prognosis in gastric carcinoma patients. Oncotarget. 2016;7:25585–25592. doi: 10.18632/oncotarget.8339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Higashimori, A. et al. Forkhead box F2 suppresses gastric cancer through a novel FOXF2-IRF2BPL-β-catenin signaling axis. Cancer Research, 10.1158/0008-5472.can-17-2403 (2018). [DOI] [PubMed]
- 12.Zeng C, et al. SPOP suppresses tumorigenesis by regulating Hedgehog/Gli2 signaling pathway in gastric cancer. J Exp Clin Cancer Res. 2014;33:75. doi: 10.1186/s13046-014-0075-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chong Y, et al. Galectin-1 from cancer-associated fibroblasts induces epithelial-mesenchymal transition through beta1 integrin-mediated upregulation of Gli1 in gastric cancer. J Exp Clin Cancer Res. 2016;35:175. doi: 10.1186/s13046-016-0449-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Katoh Y, Katoh M. Hedgehog Target Genes: Mechanisms of Carcinogenesis Induced by Aberrant Hedgehog Signaling Activation. Current Molecular Medicine. 2009;9:873–886. doi: 10.2174/156652409789105570. [DOI] [PubMed] [Google Scholar]
- 15.Diao Y, et al. Identification of novel GLI1 target genes and regulatory circuits in human cancer cells. Molecular oncology. 2018;12:1718–1734. doi: 10.1002/1878-0261.12366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lv Z, et al. Disruption of the c-Myc/miR-200b-3p/PRDX2 regulatory loop enhances tumor metastasis and chemotherapeutic resistance in colorectal cancer. Journal of translational medicine. 2017;15:257. doi: 10.1186/s12967-017-1357-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li Z, et al. A plasma mir-125a-5p as a novel biomarker for Kawasaki disease and induces apoptosis in HUVECs. PloS one. 2017;12:e0175407. doi: 10.1371/journal.pone.0175407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang T, et al. miR-508-3p concordantly silences NFKB1 and RELA to inactivate canonical NF-kappaB signaling in gastric carcinogenesis. Molecular cancer. 2016;15:9. doi: 10.1186/s12943-016-0493-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gyorffy B, Surowiak P, Budczies J, Lanczky A. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PloS one. 2013;8:e82241. doi: 10.1371/journal.pone.0082241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matsumura N, et al. Identification of novel molecular markers for detection of gastric cancer cells in the peripheral blood circulation using genome-wide microarray analysis. Experimental and therapeutic medicine. 2011;2:705–713. doi: 10.3892/etm.2011.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tian, L., Zhao, Z., Xie, L. & Zhu, J. MiR-361-5p inhibits the mobility of gastric cancer cells through suppressing epithelial-mesenchymal transition via the Wnt/beta-catenin pathway. Gene, 10.1016/j.gene.2018.06.095 (2018). [DOI] [PubMed]
- 22.Cao Y, et al. MicroRNA-125a-5p inhibits invasion and metastasis of gastric cancer cells by targeting BRMS1 expression. Oncology letters. 2018;15:5119–5130. doi: 10.3892/ol.2018.7983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hannenhalli S, Kaestner KH. The evolution of Fox genes and their role in development and disease. Nature reviews. Genetics. 2009;10:233–240. doi: 10.1038/nrg2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koon HB, Ippolito GC, Banham AH, Tucker PW. FOXP1: a potential therapeutic target in cancer. Expert opinion on therapeutic targets. 2007;11:955–965. doi: 10.1517/14728222.11.7.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jackson BC, Carpenter C, Nebert DW, Vasiliou V. Update of human and mouse forkhead box (FOX) gene families. HUMAN GENOMICS. VOL. 2010;4:345–352. doi: 10.1186/1479-7364-4-5-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Friedman JR, Kaestner KH. The Foxa family of transcription factors in development and metabolism. Cellular and molecular life sciences: CMLS. 2006;63:2317–2328. doi: 10.1007/s00018-006-6095-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carbajo-Pescador S, Mauriz JL, Garcia-Palomo A, Gonzalez-Gallego J. FoxO proteins: regulation and molecular targets in liver cancer. Current medicinal chemistry. 2014;21:1231–1246. doi: 10.2174/0929867321666131228205703. [DOI] [PubMed] [Google Scholar]
- 28.Agnihotri NS, Astekar M. The role of novel prognostic markers PROX1 and FOXC2 in carcinogenesis of oral squamous cell carcinoma. Journal of experimental therapeutics & oncology. 2018;12:171–184. [PubMed] [Google Scholar]
- 29.Wang, Z., Chen, G., Wang, Q., Lu, W. & Xu, M. Identification and validation of a prognostic 9-genes expression signature for gastric cancer. Oncotarget, 10.18632/oncotarget.17764 (2017). [DOI] [PMC free article] [PubMed]
- 30.Qiyu LDL, et al. is a prognostic factor for hepatocellular carcinoma. Int J Clin Exp Pathol. 2016;9:12761–12766. [Google Scholar]
- 31.Wang ZS, et al. Significance and prognostic value of Gli-1 and Snail/E-cadherin expression in progressive gastric cancer. Tumour biology: the journal of the International Society for Oncodevelopmental Biology and Medicine. 2014;35:1357–1363. doi: 10.1007/s13277-013-1185-1. [DOI] [PubMed] [Google Scholar]
- 32.Brabletz T, et al. Invasion and metastasis in colorectal cancer: epithelial-mesenchymal transition, mesenchymal-epithelial transition, stem cells and beta-catenin. Cells, tissues, organs. 2005;179:56–65. doi: 10.1159/000084509. [DOI] [PubMed] [Google Scholar]
- 33.Gao Y, et al. ALEX1 Regulates Proliferation and Apoptosis in Breast Cancer Cells. Asian Pacific journal of cancer prevention: APJCP. 2015;16:3293–3299. doi: 10.7314/APJCP.2015.16.8.3293. [DOI] [PubMed] [Google Scholar]
- 34.Briscoe J, Therond PP. The mechanisms of Hedgehog signalling and its roles in development and disease. Nature reviews. Molecular cell biology. 2013;14:416–429. doi: 10.1038/nrm3598. [DOI] [PubMed] [Google Scholar]
- 35.Abdel-Rahman O. Hedgehog pathway aberrations and gastric cancer; evaluation of prognostic impact and exploration of therapeutic potentials. Tumour biology: the journal of the International Society for Oncodevelopmental Biology and Medicine. 2015;36:1367–1374. doi: 10.1007/s13277-015-3216-6. [DOI] [PubMed] [Google Scholar]
- 36.Yan R, et al. Suppression of growth and migration by blocking the Hedgehog signaling pathway in gastric cancer cells. Cellular oncology (Dordrecht) 2013;36:421–435. doi: 10.1007/s13402-013-0149-1. [DOI] [PubMed] [Google Scholar]
- 37.Shimokawa T, et al. RNA editing of the GLI1 transcription factor modulates the output of Hedgehog signaling. RNA biology. 2013;10:321–333. doi: 10.4161/rna.23343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Malumbres M. miRNAs and cancer: an epigenetics view. Molecular aspects of medicine. 2013;34:863–874. doi: 10.1016/j.mam.2012.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu M, Sizova O, Wang L, Su DM. A Fine-Tune Role of Mir-125a-5p on Foxn1 During Age-Associated Changes in the Thymus. Aging and disease. 2017;8:277–286. doi: 10.14336/ad.2016.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wada R, Akiyama Y, Hashimoto Y, Fukamachi H, Yuasa Y. miR-212 is downregulated and suppresses methyl-CpG-binding protein MeCP2 in human gastric cancer. International journal of cancer. 2010;127:1106–1114. doi: 10.1002/ijc.25126. [DOI] [PubMed] [Google Scholar]
- 41.Guo X, Wu Y, Hartley RS. MicroRNA-125a represses cell growth by targeting HuR in breast cancer. RNA biology. 2009;6:575–583. doi: 10.4161/rna.6.5.10079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nishida N, et al. MicroRNA-125a-5p is an independent prognostic factor in gastric cancer and inhibits the proliferation of human gastric cancer cells in combination with trastuzumab. Clinical cancer research: an official journal of the American Association for Cancer Research. 2011;17:2725–2733. doi: 10.1158/1078-0432.CCR-10-2132. [DOI] [PubMed] [Google Scholar]
- 43.Wang G, Mao W, Zheng S, Ye J. Epidermal growth factor receptor-regulated miR-125a-5p–a metastatic inhibitor of lung cancer. The FEBS journal. 2009;276:5571–5578. doi: 10.1111/j.1742-4658.2009.07238.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ferretti E, et al. MicroRNA profiling in human medulloblastoma. International journal of cancer. 2009;124:568–577. doi: 10.1002/ijc.23948. [DOI] [PubMed] [Google Scholar]
- 45.Wang S, et al. Overaccumulation of p53-mediated autophagy protects against betulinic acid-induced apoptotic cell death in colorectal cancer cells. Cell death & disease. 2017;8:e3087. doi: 10.1038/cddis.2017.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang S, et al. Identification and analysis of the promoter region of the human HAS3 gene. Biochemical and biophysical research communications. 2015;460:1008–1014. doi: 10.1016/j.bbrc.2015.03.142. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.