Abstract
The progression of spermatogenesis is precisely controlled by meiotic stage-specific genes, but the molecular mechanism for activation of such genes is still elusive. Here we found a novel testis-specific long noncoding RNA (lncRNA), Tesra, that was specifically expressed in the mouse testis at the Prss/Tessp gene cluster on chromosome 9. Tesra was transcribed downstream of Prss44/Tessp-4, starting within the gene, as a 4435-nucleotide transcript and developmentally activated at a stage similar to that for Prss/Tessp genes. By in situ hybridization, Tesra was found to be localized in and around germ cells and Leydig cells, being consistent with biochemical data showing its existence in cytoplasmic, nuclear, and extracellular fractions. Based on the finding of more signals in nuclei of pachytene spermatocytes, we explored the possibility that Tesra plays a role in transcriptional activation of Prss/Tessp genes. By a ChIRP assay, the Tesra transcript was found to bind to the Prss42/Tessp-2 promoter region in testicular germ cells, and transient overexpression of Tesra significantly activated endogenous Prss42/Tessp-2 expression and increased Prss42/Tessp-2 promoter activity in a reporter construct. These findings suggest that Tesra activates the Prss42/Tessp-2 gene by binding to the promoter. Finally, we investigated whether Tesra co-functioned with enhancers adjacent to another lncRNA, lncRNA-HSVIII. In the Tet-on system, Tesra transcription significantly increased activity of one enhancer, but Tesra and the enhancer were not interdependent. Collectively, our results proposed a potential function of an lncRNA, Tesra, in transcriptional activation and suggest a novel relationship between an lncRNA and an enhancer.
Keywords: long noncoding RNA, Prss/Tessp, spermatogenesis, enhancer, chromatin, transcriptional regulation
A novel long noncoding RNA, Tesra, that is specifically expressed in the mouse testis activates a spermatocyte-specific gene, Prss42/Tessp-2, in cooperation with an enhancer by binding to chromatin at the promoter and enhancing its activity.
Introduction
Spermatogenesis is the process to generate mature spermatozoa through three steps: mitosis, meiosis, and spermiogenesis. Mitotically proliferating diploid spermatogonia differentiate into primary spermatocytes, which divide twice with only one DNA replication to become haploid round spermatids through meiosis, and mature spermatozoa form by spermiogenesis [1,2]. This series of events is tightly controlled by various genes, and genes activated in primary spermatocytes during meiosis are critical for spermatogenesis [3–5]. Thus, elucidation of the mechanism of transcriptional activation at this stage is important for understanding spermatogenesis.
In the 1990s, several studies showed by generating transgenic mice that proximal promoters were sufficient for transcriptional activation of spermatocyte-specific genes [6–11], but studies conducted in the 21st century indicated or suggested the necessity of distal enhancers [12,13]. In agreement with the results of those studies, we identified a dual promoter−enhancer that functions in mouse spermatocytes to activate the spermatocyte-specific Tcam1 gene and drive male germ cell-specific lncRNA-Tcam1 and the ubiquitously expressed Smarcd2 gene [14]. In addition, recent genome-wide analyses have clearly shown that the testis expresses many more long noncoding RNAs (lncRNAs) than those expressed in other tissues [15,16], and it has been shown that the noncoding transcription occurs at every step of meiosis [17–24]. Because lncRNAs are key factors for gene activation in various cell types [25–29], they are likely to play some roles in spermatocyte-specific gene activation, but the function of testicular lncRNAs is not well understood.
Long noncoding RNAs are a class of noncoding RNAs longer than 200 nucleotides, and they have been found to be present in the nuclei, cytoplasm, and extracellular vesicles (EVs) in various tissues [30–35]. Generally, lncRNAs are not evolutionarily conserved, when compared with protein-coding genes [36–38], and show tissue-specific expression [15]. They are crucial regulators for transcription, RNA processing, translation, and construction of subnuclear structures in mammals [39–42]. In the case of transcriptional activation, lncRNAs often function in cooperation with enhancers, and enhancers are therefore located close to lncRNAs for which transcription is necessary for promoter–enhancer interactions [43,44]. Thus, in male germ cells, lncRNAs presumably contribute to transcriptional activation with enhancers.
We previously reported a testis-specific lncRNA, lncRNA-HSVIII, and two potential enhancers adjacent to it at the mouse Prss/Tessp gene cluster [45]. The Prss/Tessp cluster is located on mouse chromosome 9F2-F3 and consists of three paralogous genes, Prss42/Tessp-2, Prss43/Tessp-3, and Prss44/Tessp-4, encoding serine proteases [46]. They all are specifically activated in primary spermatocytes at the late pachytene stage, and Prss42/Tessp-2 and Prss43/Tessp-3 play important roles in the progression of meiosis and germ cell survival [46]. lncRNA-HSVIII is transcribed in a testis-specific manner downstream of the Prss42/Tessp-2 gene, and its localization changes from nuclei of primary spermatocytes to the cytosol of round spermatids during meiosis [45]. The two potential enhancers were found upstream and downstream of lncRNA-HSVIII, but, interestingly, they did not seem to be functionally related to lncRNA-HSVIII. In the process of our study, we noticed that other intergenic regions were transcribed in a testis-specific manner at the Prss/Tessp locus, and those lncRNAs might participate in gene activation during meiosis.
In this study, we identified another testis-specific lncRNA, Tesra (Tessp cluster lncRNA related to gene activation), at the Prss/Tessp locus. Tesra was localized in the nuclei, cytoplasm, and extracellular regions of germ cells, and nuclear Tesra bound to the Prss42/Tessp-2 promoter and increased its activity. In addition, an enhancer downstream of lncRNA-HSVIII co-functioned with Tesra to activate the Prss42/Tessp-2 gene promoter. These findings contribute to an understanding of transcriptional activation during spermatogenesis and suggest a novel relationship between an lncRNA and an enhancer.
Materials and methods
Animals
C57Bl/6Ncl mice (CLEA Japan Inc., Tokyo, Japan) were maintained on regular 14-h light/10-h dark cycles at 25°C and were given sufficient food and water. The experimental procedures used in this study were approved by the Institutional Animal Use and Care Committee at Hokkaido University.
Reverse transcription-polymerase chain reaction
Total RNA was extracted by ISOGEN, ISOGEN II, or ISOGEN-LS (Nippon Gene, Tokyo, Japan) according to the manufacturer's instructions. After treatment with TurboDNase (Life Technologies, Foster City, CA), the RNA was reverse-transcribed into complementary DNA (cDNA) using Superscript III (Life Technologies) based on the previously described method [47,48]. PCR was performed by using ExTaq (Takara, Kusatsu, Japan). Primer sequences are shown in Table 1.
Table 1.
Primers used in this study.
| Designation | Forward | Reverse |
|---|---|---|
| [RT-PCR/qRT-PCR] | ||
| Tesra PCR | 5′-CGTGCCTGTTTCTTTCCATT-3′ | 5′-CAGCCACCATGTTCTGTGAC-3′ |
| Tesra 2890bp | 5′-CGTGCCTGTTTCTTTCCATT-3′ | 5′-GTTGCATCTCCACCCATCA-3′ |
| Tesra full length | 5′-AGAGGTGTGAGGCTGGTGGG-3′ | 5′-TGAACATATTAAATGAGCTC-3′ |
| Gapdh | 5′-CATGACCACAGTCCATGCCATC-3′ | 5′-TAGCCCAAGATGCCCTTCAGTG-3′ |
| [5′RACE and 3′RACE] | ||
| GSP1 | 5′-CAGCCACCATGTTCTGTGAC-3′ | |
| GSP2 | 5′-GGCAACCTGGAAAGGAAAAT-3′ | |
| GSP3 | 5′-AATGGAAAGAAACAGGCACG-3′ | |
| GSP4 | 5′-CCTGAAGCTCACCCCTAGTA-3′ | |
| GSP5 | 5′-TTGATGGGTGGAGATGCAAC-3′ | |
| [RT-PCR/qRT-PCR with extracellular RNA] | ||
| Tesra exosome | 5′-ACCAGGGCCAGGAATTTATC-3′ | 5′-TGTTCCCAGTGTTGGCTGTA-3′ |
| lncRNA-HSVIII exosome | 5′-TTGTCATTGGGGCAGAAATA-3′ | 5′-CACCACAAAAACAAAGGGTG-3′ |
| [ChIRP-qPCR] | ||
| Rec8 promoter | 5′-CCTTGGCGAAGTCCTTGTTA-3′ | 5′-AATAGTCGCCAGCCAATCAC-3′ |
| B2m promoter | 5′-GGCCAGGGGTTTAACTTCTC-3′ | 5′-CCCTTGGGGTTTCTGCTTAT-3′ |
| Prss42/Tessp-2 promoter | 5′-GGTGCCTCTTTGGAGACCTA-3′ | 5′-GACCCCTTACTTCCTGTGGA-3′ |
| Prss44/Tessp-4 promoter | 5′-CTTCTGCTGTCTGGGAGTCA-3′ | 5′-AACGGGTGGTTTCATTTAGC-3′ |
| Prss43/Tessp-3 promoter | 5′-CTGCAGGTGACTCCACACTT-3′ | 5′-TTCCTCCTTCTGTCCCTCAG-3′ |
| intergenic Prss42/Tessp2-Prss44/Tessp-4 | 5′-CAGAGTCGCCTTCAAAATCC-3′ | 5′-TAGCCCAAATCAGCACCTTC-3′ |
| intergenic Prss44/Tessp4-Prss43/Tessp-3 | 5′-GAGGTACAGGCACAAGCACA-3′ | 5′-TAGCTGTGGCAGGCTTAGGT-3′ |
| Prss45/TESPL promoter | 5′-AGCACTAGGGGTGCTGTCAT-3′ | 5′-TGGACCCACTCTCGTCCTTA-3′ |
| Prss46 promoter | 5′-TCAGTGGAGACTGGCATGAA-3′ | 5′-CCTGTGACCTTGTCCCTTGT-3′ |
| Prss50/Tsp50 promoter | 5′-GCAGAGGAGGGTAGGGGTAT-3′ | 5′-TATGCCTCGCCTCAGCTAAT-3′ |
5′ and 3′ rapid amplification of cDNA ends
5′RACE (rapid amplification of cDNA ends) and 3′RACE were performed as previously described [48]. For 5′RACE, cDNA was generated by using total RNA from adult mouse testes and a gene-specific primer (GSP1) for reverse transcription. After the addition of oligodeoxycytidine by terminal deoxynucleotidyl transferase (Takara), the first PCR was performed with GSP2 and an abridged anchor primer. The second PCR was performed using GSP3 and an abridged universal amplification primer.
For 3′RACE, reverse transcription was performed using the oligo(dT)20 primer connected to an adaptor sequence. The first and second PCRs were performed using GSP4 and GSP5, respectively, with the adaptor primer.
All GSP sequences are shown in Table 1. Final PCR products were subcloned into a pBluescript II KS(+) vector (Stratagene, San Diego, CA) by the TA-cloning method, and their sequences were confirmed by DNA sequencing analysis.
Isolation of male germ cells, Sertoli cells, and Leydig cells
Isolation of male germ cells [46], Sertoli cells [49], and Leydig cells [14] was performed as previously described. Sertoli cells were harvested from 7- to 12-day-old testes, and germ and Leydig cells were harvested from adult testes.
Preparation of subcellular fractions
Cytoplasmic and nuclear fractions were obtained as previously described [45,48]. Briefly, the cells were lysed in NP-40 lysis buffer (10 mM Tris-HCl, 10 mM NaCl, 3 mM MgCl2, 0.5% NP-40, pH 7.5) on ice for 10 min, and after centrifugation at 1500 rpm for 5 min at 4°C, the supernatant was used as the cytoplasmic fraction. The precipitates were washed two or three times with NP-40 lysis buffer, and the resulting pellet was used as the nuclear fraction.
Quantitative RT-PCR
Complementary DNAs were prepared as described above. PCRs were performed by using Power SYBR Green PCR Master Mix for protein-coding genes (Life Technologies) and KOD SYBR qPCR Mix for lncRNAs (TOYOBO, Osaka, Japan), based on the previously described method [14,47]. The relative expression levels were normalized to endogenous Aip mRNA, except for quantification of extracellular lncRNAs. The levels of Tesra and lncRNA-HSVIII in the extracellular fraction and germ cells were examined with primer pairs, Tesra exosome and lncRNA-HSVIII exosome, and directly compared because the amplification efficiencies of primer pairs for these lncRNAs were similar. For quantitative detection of Tesra in cells and tissues, a primer pair, Tesra PCR, was used, and for extracellular lncRNAs, other primer sets were used (Table 1). The primers for Prss/Tessp cluster genes and Aip were described in our previous reports [46,47].
In situ hybridization
The RNA probes for detection of Tesra transcripts were prepared as follows. The full length of Tesra was obtained by RT-PCR with mouse testis cDNA and a primer pair of Tesra full length (Table 1) using KOD Fx Neo (TOYOBO) and was subcloned into pBluescript II KS(+) which was cut by EcoRV. After the DNA sequence had been confirmed, a part of Tesra (1545–2363) was isolated by restriction digestion with XbaI and PvuII, blunted by T4 DNA polymerase, and ligated into a blunted PstI site of a pGEM-3Zf(+) vector. The resulting plasmid was linearized with ApaI or NotI for making sense and antisense RNA probes, respectively. Digoxigenin (DIG)-labeled RNA probes were synthesized using a DIG RNA labeling kit (Roche Molecular Biochemicals, Mannheim, Germany) with SP6 RNA polymerase for a sense probe and T7 RNA polymerase for an antisense probe.
In situ hybridization with the Tyramide Signal Amplification (TSA) Plus system (PerkinElmer, Waltham, MA) was performed according to the procedure reported previously [45,50]. Briefly, adult mouse testes were fixed for 3 h at 4°C with 4% paraformaldehyde in phosphate buffered saline (PBS). After washing with PBS, the testes were dissected into two pieces with a razor blade under a dissecting microscope. The two pieces of testes were dehydrated, embedded in paraffin, and cut into 7-μm-thick sections. The sections were hybridized with 2 ng/μl of the DIG-labeled RNA probe overnight at 45°C. After washing, the sections were incubated with anti-DIG-horseradish peroxidase antibody (1:500 dilution; Roche) overnight at room temperature. The reaction with tyramide-Cy3 (1:50 dilution in 1X Plus Amplification Diluent [PerkinElmer], followed by 1:100 dilution with distilled water) was performed at room temperature for 20 min. To detect nuclei, the sections were incubated with 10 μg/ml Hoechst 33258 at room temperature for 10 min. The samples were mounted with a Fluoro-KEEPER Antifade Reagent (Nacalai Tesque, Kyoto, Japan) and observed under an LSM 5 LIVE confocal microscope (Carl Zeiss, Oberkochen, Germany).
Preparation of extracellular fractions and fractionation of extracellular vesicles
Adult testes were decapsulated, cut 20 times with scissors, and put in Dulbecco modified Eagle medium. The tube was inverted 10 times and kept at room temperature for 2–3 min, and the supernatant was collected into a new tube. The samples were spun at 1000 g for 15 min at 4°C and filtered through a 0.45-μm filter. Then each sample was centrifuged at 10,000 g for 30 min to remove large EVs, and the resulting supernatant was used as the extracellular fraction.
The fraction of EVs was collected from the extracellular fraction by using the EV-second® column (GL Sciences, Tokyo, Japan), which could divide the samples by size exclusion chromatography. After blocking with filtered fetal bovine serum, the column was washed with PBS six times, and the sample was loaded on it. Elution was done with PBS, and 10 continuous fractions (200 μl each) were collected.
Western blot analysis
A portion of the EV fraction (10 μl) was mixed with 4 × Laemmli sample buffer (#1610747, Bio-Rad Laboratories, Hercules, CA) containing 10% β-mercaptoethanol and boiled at 94°C for 5 min. The samples were resolved by SDS–polyacrylamide gel electrophoresis using Any kD Mini-PROTEAN® TGXTM Precast Gel (#4569036, Bio-Rad Laboratories) and then transferred onto polyvinylidene difluoride membranes (0.2-μm pore size; Bio-Rad Laboratories) via Trans-Blot® TurboTM Transfer System (#1704150, Bio-Rad Laboratories). The membranes were blocked with Blocking Reagent for WB (CSR-IS-CD-500W, Cosmo Bio Co., Ltd, Tokyo, Japan) at room temperature for 1 h and incubated with an anti-CD9 antibody (1:2000 dilution; ab92726, Abcam, Cambridge, MA) diluted in IMMUNO SHOT Platinum (CSR-IS-P-500, Cosmo Bio Co., Ltd) at 4°C overnight (Supplemental Table S1). The next day, the membranes were washed four times with 1 × Tris-buffered saline (20 mM Tris, 500 mM NaCl, pH 7.4) containing 0.1% Tween-20 (TBS-T) at room temperature and incubated with a secondary goat anti-rabbit immunoglobulin G-horseradish peroxidase conjugate (1:3000 dilution; #7074, Cell Signaling Technology, Danvers, MA) diluted in IMMUNO SHOT Platinum (Cosmo Bio Co., Ltd) at room temperature for 1 h. The membranes were washed again with TBS-T four times, and signals were detected using PierceTM ECL Plus Western Blotting Substrate (#32132, Thermo Fisher Scientific, Rockford, IL) and the Light Capture II system (AE-6981, ATTO Co., Tokyo, Japan).
Transmission electron microscopy
An EV fraction that was positive for CD9 was fixed by adding an equal volume of 4% paraformaldehyde (2% at a final concentration). Ten microliters of the fixed sample was attached on a mesh using an Exosome-TEM-easy kit (Bio 101, Carlsbad, CA) according to the manufacturer's protocol. The EVs were visualized by a transmission electron microscope (H-7650, Hitachi, Tokyo, Japan).
Chromatin isolation by RNA purification
Thirteen antisense oligo probes for Tesra were designed by using singlemoleculefish.com 18 (http://singlemoleculefish.com) as indicated in Table 2, and biotinylated at the 3′ end. Twenty million male germ cells were prepared from 21- to 22-day-old mice as described above. The cells in PBS were spun at 800–1000 rpm for 4 min, resuspended in 1% glutaraldehyde, and crosslinked for 10 min at room temperature on a rotator. The cross-linking reaction was quenched with a 1/10th volume of 1.25 M glycine at room temperature for 5 min. After washing the cells with PBS, a 10 × volume of the lysis buffer (final concentration of 50 mM Tris-Cl pH 7.0, 10 mM EDTA, 1% SDS) supplemented with phenylmethanesulfonyl fluoride, 1 × protease inhibitor (Wako Pure Chemicals, Osaka, Japan), and RNasin Plus (Promega, Madison, WI) was added to each tube. Then, the cell lysate was sonicated to shear the chromatin with 20% output for 10 s, 15 times, by using Ultrasonic Disruptor UD-201 (TOMY, Tokyo, Japan). The sample was spun at 15,000 rpm for 10 min at 4°C, and the supernatant was collected into another tube. One percent of this sample was taken as “input.” Then the sample was hybridized with the combined oligo probes at the final concentration of 100 pmol/ml in hybridization buffer (750 mM NaCl, 1% SDS, 50 mM Tris-Cl pH 7.0, 1 mM EDTA, 15% formamide) at 37°C for 4 h with shaking. Streptavidin beads (GE Healthcare, Uppsala, Sweden) (100 μl per 100 pmol probe) were washed with the lysis buffer three times and resuspended in the same buffer supplemented with phenylmethanesulfonyl fluoride, 1 × proteinase inhibitor, and RNasin Plus. After the hybridization reaction, the beads were added to the tube and the sample was incubated at 37°C for 30 min with shaking. The beads were washed with 1 ml wash buffer (2 × NaCl and sodium citrate and 0.5% SDS) five times and resuspended in 1 ml wash buffer. One hundred microliters was set aside for RNA isolation, and the rest of the suspension (900 μl) was used for DNA extraction. Each tube was centrifuged at 4000 rpm for 1 min at room temperature, and the beads were collected. For the RNA sample, ISOGEN II was added and total RNAs were purified for qRT-PCR. For the DNA sample, 150 μl DNA elution buffer (50 mM NaHCO3, 1% SDS) was added with 1.0 μl of 10 mg/ml RNase A (Nacalai Tesque) and 0.3 μl of 10 U/μl RNase H (Bio Academia, Osaka, Japan), and the sample was incubated at 37°C for 30 min with shaking. The tubes were centrifuged at 4000 rpm, and the supernatant was collected in a new tube. This process was carried out twice. Fifteen microliters of 10 mg/ml Proteinase K (Nacalai Tesque) was added to the combined supernatants, and the sample was incubated at 50°C for 45 min. After the incubation, DNA was extracted by phenol-chloroform-isoamylalcohol and collected by alcohol precipitation. The input sample was treated with Proteinase K and extracted in the same way. The collected DNAs were subjected to quantitative PCR (qPCR) using Power SYBR Green PCR Master Mix (Life Technologies) with primers listed in Table 1. The relative chromatin enrichment of each region was determined by calculating the ratio of the chromatin isolation by RNA purification (ChIRP) sample to the input sample, and the occupancy level was further normalized to the value at the Rec8 promoter, which was set to 1.0. As a control, we prepared an RNase(+) sample that was treated with RNase in the hybridization reaction to reduce Tesra transcripts.
Table 2.
Tiling oligo probes used for ChIRP assay.
| Designation | Reverse |
|---|---|
| [ChIRP tiling oligo] | |
| ChIRP-1 | 5′-CTAACATCTATCCTCTCCAA-3′ |
| ChIRP-2 | 5′-GCCTCTAAAGTCAGGAGAAC-3′ |
| ChIRP-3 | 5′-TCAAGGCAACCTGGAAAGGA-3′ |
| ChIRP-4 | 5′-TACCAGTGCTTGACTTAGTA-3′ |
| ChIRP-5 | 5′-GGCATTTCTTACTTCACACA-3′ |
| ChIRP-6 | 5′-CCAGTCTCTTAGGGATTTAT-3′ |
| ChIRP-7 | 5′-TACCTAACACTTCTTGCACT-3′ |
| ChIRP-8 | 5′-ACTGGAGCAGCAAGAATTGC-3′ |
| ChIRP-9 | 5′-GCTAGGCAAACTTCACCAAG-3′ |
| ChIRP-10 | 5′-TCCACCCATCAAGACATTAA-3′ |
| ChIRP-11 | 5′-AGAGAATTGGTGCCATGTTC-3′ |
| ChIRP-12 | 5′-CTACTGCTCCGATGAAATGT-3′ |
| ChIRP-13 | 5′-CAGTTTATTATCTGCTCACC-3′ |
Plasmid constructs
Tesra-OE: The full length of Tesra was subcloned into pBluescript II KS(+) as described above. The plasmid was digested with BamHI and EcoRI, and the Tesra fragment was inserted into a pcDNA3.1(+) vector which was cut by BamHI and EcoRI.
T-2pro-luc: This construct contained the luciferase gene driven by the Prss42/Tessp-2 promoter as previously described [45].
T-2pro-luc-up and T-2pro-luc-down: These constructs contained a 2.3-kb upstream or a 1.5-kb downstream sequence of lncRNA-HSVIII at the 3′ end of the luciferase gene in T-2pro-luc. They were both generated in our previous study [45].
Tesra-tet-on: The full length of Tesra was obtained and subcloned into pBluescript II KS(+) as described above but in the opposite direction. This plasmid was digested with SpeI and XhoI, and the Tesra fragment was inserted into a pPBhCMV*1-cHA-pA vector which was cut by SpeI and XhoI [51]. The pPBhCMV*1-cHA-pA plasmid and other vectors required to establish the Tet-on system were kind gifts from Dr Kazuhiro Murakami [51].
Cell culture and transfection
Hepa1–6 cells were cultured as previously described [14,47,52]. Transfection into Hepa1–6 cells was conducted according to the protocol of Genefectine (Genetrone Biotech, Seoul, South Korea) or GeneJuice transfection reagents (Merck, Darmstadt, Germany).
Overexpression of Tesra
For transient transfection, Tesra-OE was transfected into Hepa1–6 cells by using Genefectine. Selection was started 24 h after the transfection by treating the cells with 0.5–1.5 mg/ml G418. After selection for 3 days, the cells were dissolved in ISOGEN II, and total RNAs were purified for RT-PCR analyses. For a luciferase assay, T2pro-luc and pRL-CMV were co-transfected with Tesra-OE, and luciferase activity was measured 2 days later. In both experiments, pcDNA3.1(+) was used as a control.
For establishing the Tet-on system, we transfected Tesra-tet-on, pPBCAGrtTA-IN that bore a neomycin resistance gene, and pPyCAG-PBase into Hepa1–6 cells by GeneJuice. Twenty-four hours later, we selected successfully transfected cells by treating the cells with 2.0 mg/ml G418 for 24 days and established a stable cell line in which Tesra expression could be induced by the addition of Doxycycline (Dox). T2pro-luc, T-2pro-luc-up, or T-2pro-luc-down was transfected into these stable cells by GeneJuice, and 1.0 μg/ml Dox was added 1 day later. Twenty-four hours after the addition of Dox, the cells were used for a luciferase assay. The pRL-CMV vector (Promega) was co-transfected to normalize transfection efficiency.
Luciferase assay
The luciferase assay was conducted according to the protocol of Dual-Luciferase Reporter Assay System (Promega) as previously described [27,47].
Statistical analysis
Results were expressed as the average ± standard deviation (SD) of at least three independent experiments and were analyzed by the Student t test or one-way analysis of variance (ANOVA) followed by the Tukey post hoc test. A P value less than 0.05 was considered statistically significant.
Results
Identification and characterization of an lncRNA, Tesra, at the mouse Prss/Tessp locus
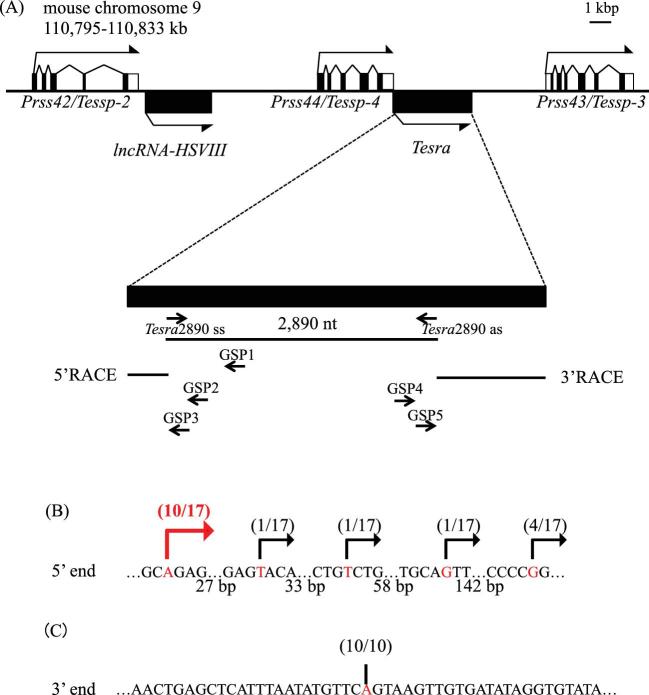
Based on transcriptome data (accession numbers: SRX135150, SRX135160, and SRX135162), we found lncRNA-HSVIII downstream of the Prss42/Tessp-2 gene in our previous study [45]. The data also indicated many reads mapped to an intergenic region between Prss44/Tessp-4 and Prss43/Tessp-3 genes in the testis, suggesting the presence of other testis-specific noncoding transcripts at this locus. Thus, we performed RT-PCR with total RNA from mouse testes, and a 2890-nucleotide sequence was successfully amplified (Figure 1A). We named this lncRNA Tesra and decided to determine its full length.
Figure 1.
Cloning of a novel lncRNA, Tesra. (A) A schematic view of the Prss/Tessp locus on mouse chromosome 9. Exons of three Prss/Tessp genes are depicted by the black and white boxes representing translated and untranslated regions, respectively. Two lncRNAs that do not contain any introns are also depicted by the black boxes. The bent arrows indicate the transcriptional direction. Below the gene structure, the genomic region transcribed into Tesra is enlarged. A 2890-nucleotide sequence was first detected as a transcribed sequence by RT-PCR. 5′RACE was performed with three primers (GSP1–GSP3), and 3′RACE was performed with GSP4 and GSP5 primers. The resulting RACE products are indicated by horizontal lines. (B) 5′-end variations of Tesra determined by 5′RACE. A sequence around the 5′ end of Tesra that overlaps with the 3′ untranslated region of the Prss44/Tessp-4 gene is shown. The number of nucleotides between each sequence array is presented below. A specific band was obtained by 5′RACE, and 17 subclones were sequenced. The position of the 5′ end of each subclone is indicated by the red letters and the bent arrows with the number of subclones. Five TSSs were detected, and the most upstream adenine was determined to be a main TSS (indicated by a red arrow). (C) 3′ end of Tesra determined by 3′RACE. A specific band was obtained by 3′RACE, and 10 subclones were checked by DNA sequencing. In all subclones, an adenine was identified as a main transcriptional termination site, as indicated by a red letter and a vertical line.
RACE analyses were performed to determine the 5′- and 3′-ends of Tesra. In 5′RACE experiments, one specific band was obtained after the second PCR, and five different transcriptional start sites (TSSs) were detected by sequencing of 17 subclones. Since 10 of the subclones showed an adenine as the TSS, we defined it as a major TSS of Tesra (Figure 1B, red bent arrow). By 3′RACE, one band was amplified in the second PCR, and a single transcriptional termination site was detected 4236 bp downstream of Prss44/Tessp-4 as a result of sequencing of 10 subclones (Figure 1C). Consequently, the full length of Tesra was 4435 nucleotides (DDBJ/EMBL/GenBank accession number: LC406701). It contained no intron and was presumed to have a poly(A)-tail because some subclones contained a chain of adenine nucleotides longer than oligo(dT). Interestingly, 199 bp of the full length of Tesra overlapped with the Prss44/Tessp-4 gene. Thus, a novel lncRNA, Tesra, was transcribed from the Prss/Tessp locus, and the full-length sequence was determined.
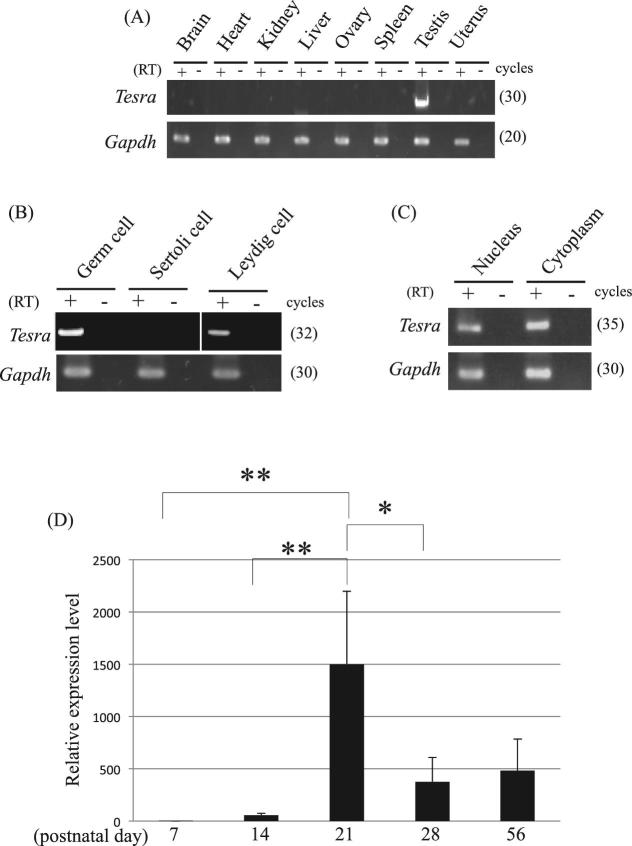
Expression and localization of Tesra
We examined the tissue specificity of Tesra by RT-PCR using eight mouse tissues. Tesra showed testis-specific expression as did the Prss/Tessp genes (Figure 2A) [46]. We next investigated the cell type specificity of Tesra in the testis, which contained three kinds of cells: germ cells, Sertoli cells, and Leydig cells. We prepared three fractions that were enriched in each type of cells. The Sertoli cell fraction was prepared by primary culture and basically contained no other types of cells. The germ and Leydig cell fractions were prepared from adult testes, and the major contaminants were Sertoli cells and germ cells, respectively. Tesra was detected in germ and Leydig cell fractions by RT-PCR but not in the Sertoli cell fraction (Figure 2B), suggesting that germ and Leydig cells express Tesra.
Figure 2.
Expression pattern of Tesra. (A) Tissue specificity of Tesra. Eight mouse tissues, as indicated, were collected from adult mice and used for RT-PCR. Reverse transcription was performed by using the oligo(dT) primer with (RT+) or without reverse transcriptase (RT–). Gapdh was detected as an internal control. The cycle number of each reaction is indicated in parenthesis. (B) Cell type specificity of Tesra in the testis. Mouse germ, Sertoli, and Leydig cell fractions were prepared and used for RT-PCR. Germ cells and Leydig cells were collected from adult mouse testes, while Sertoli cells were collected from 7- to 12-day-old testes and cultured for several days. The experiment was done and the data are presented as in (A). (C) Subcellular localization of Tesra in mouse male germ cells. Germ cells were collected and fractionated into nuclear and cytoplasmic fractions, and total RNAs were purified from both fractions. Gapdh was a positive control, in which both forward and reverse primers were designed within exon 6 of the gene. RT-PCR was done and the data are presented as in (A). The Tesra transcript was detected in both nuclear and cytoplasmic fractions. (D) Tesra expression during postnatal testis development. Mouse testes at postnatal days 7, 14, 21, 28, and 56 were collected, and total RNAs were purified and used for qRT-PCR analysis. The housekeeping Aip gene was examined as an internal control. The level of Tesra was normalized to Aip, and the value at day 7 was set to 1.0. The data are presented as means ± SD from three independent experiments, and the statistical significance was analyzed by one-way ANOVA followed by the Tukey post hoc test. *P < 0.05. **P < 0.01.
We then investigated the subcellular localization of Tesra. We prepared total RNAs from nuclear and cytoplasmic fractions from germ cells and performed RT-PCR. Tesra transcripts were present in both the nuclear and cytoplasmic fractions (Figure 2C). We also investigated the expression pattern in the mouse testis at postnatal days 7, 14, 21, 28, and 56. The Tesra signal was detected at a low level on postnatal day 7, and the expression level dramatically increased to a peak in the period from day 14 to day 21 (Figure 2D). The timing of an increase in Tesra transcription coincided with activation of the Prss/Tessp genes at the pachytene spermatocyte stage [46].
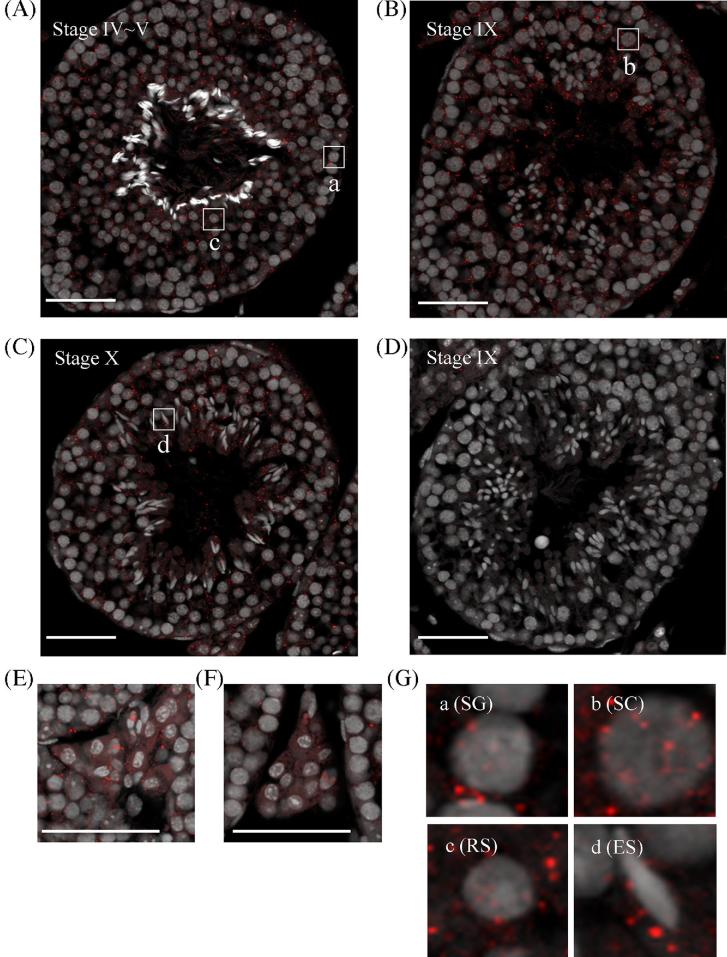
We further assessed Tesra expression during meiosis by in situ hybridization using the highly sensitive TSA method. Many more small dot-like signals were detected by the antisense probe than by the sense probe, and the signals were dispersed throughout seminiferous tubules (Figure 3A–D). At seminiferous epithelial stage IV–V, many signals were observed in the nuclei and cytoplasm of early to mid pachytene spermatocytes, and spermatogonia and round spermatids also contained dots in their nuclei and cytoplasm (Figure 3A and G). At stage IX, the number of signals was increased, and more dots were localized in the nuclei of late pachytene spermatocytes (Figure 3B and G). At stage X, the number of signals was slightly decreased, but red dots were still present in the nuclei and cytoplasm of pachytene spermatocytes (Figure 3C). Elongating spermatids also contained signals, but the distribution was biased toward the cytoplasm (Figure 3G). In interstitial regions, positive signals were present in Leydig cells, mostly in the cytoplasm (Figure 3E and F). These results indicated that Tesra was localized in both the nuclei and cytoplasm of spermatogonia, primary spermatocytes, and round spermatids and in the cytoplasm of elongating spermatids and Leydig cells. The number of signals was large at late epithelial seminiferous stages, especially in late pachytene spermatocytes compared to other cell types. Of note, Tesra signals were also observed in the lumen of seminiferous tubules, and similar results were obtained by using another antisense probe for Tesra (data not shown).
Figure 3.
Localization of Tesra in the mouse testis. In situ hybridization was performed in adult mouse testes by using the TSA system. Sections of mouse testes (7 μm) were hybridized with a DIG-labeled sense or antisense probe for Tesra. Positive signals were detected by reaction with tyramide-Cy3 (red), and nuclei were stained with Hoechst 33258 and shown as gray. Panels (A–D) show seminiferous tubules at indicated stages, and panels (E) and (F) show Leydig cells. Sections stained with the antisense probe are presented in (A–C) and (E), and those stained with the sense probe are shown in (D) and (F). Sections hybridized with the sense probe showed few signals. With the antisense probe, positive signals were observed in all seminiferous tubules, and localized in the nuclei and/or cytoplasm of spermatogonia, spermatocytes, spermatids, and Leydig cells. Enlarged pictures of each type of meiotic cells that are surrounded with frames, marked as a–d in (A–C), are indicated in (G). SG, spermatogonium; SC, spermatocyte; RS, round spermatid; ES, elongating spermatid. Scale bars, 50 μm.
To identify the scattered signals in the entire tubules and in the lumen by in situ hybridization, we tested the possibility that Tesra transcripts were present in extracellular regions. We collected the extracellular fraction of a testis, purified total RNA, and investigated Tesra expression by qRT-PCR. For comparison, we detected the expression of lncRNA-HSVIII which did not show scattered patterns by the method [45]. As a result, the Tesra signal was detected at a 4.6-fold higher level than lncRNA-HSVIII in the extracellular fraction whereas germ cells showed 1.8-fold difference (Figure 4A). We then attempted to isolate EV fractions containing exosomes since it has been shown that many extracellular lncRNAs are packaged within EVs to travel into neighboring or distant cells [53]. We applied the extracellular fraction to size exclusion chromatography and collected 10 fractions by elution. Western blot analysis indicated that a marker protein of exosomes, CD9, was mainly contained in fractions #6–#9 (Figure 4B), and the observation of a fraction by an electron microscope showed the presence of an EV (Figure 4C). We purified total RNAs from fractions #7 and #8, and the Tesra signal was clearly detected by RT-PCR in one of the fractions in duplicate experiments (Figure 4D). Therefore, some of the Tesra transcripts were likely to be excluded from the cells and associated with a particular size of EV, which probably resulted in detection of scattered signals by in situ hybridization.
Figure 4.
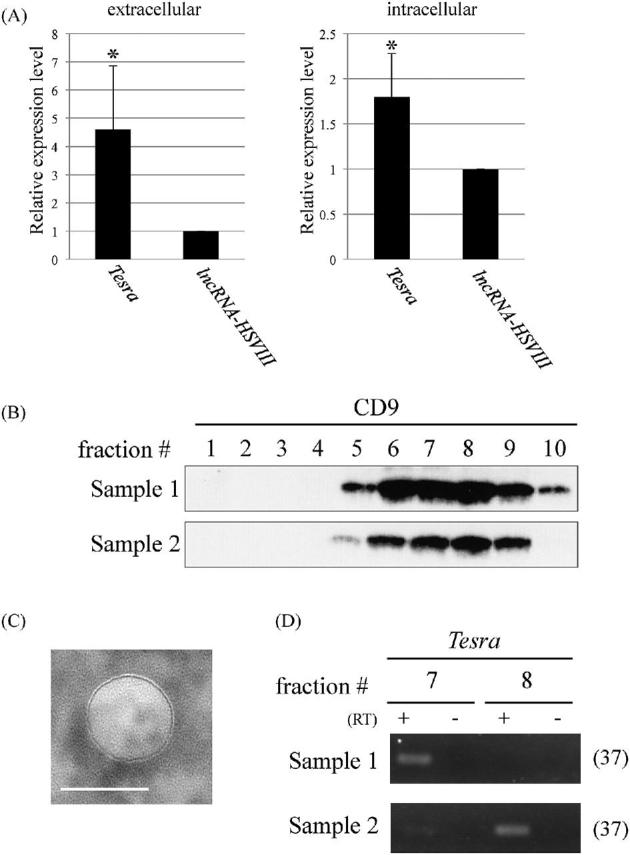
Extracellular localization of Tesra. (A) Expression of Tesra in the extracellular fraction (left) and germ cells (right) from adult mouse testes. The extracellular fraction was collected by immersing tissue pieces of adult testes in a cell culture medium, and total RNAs were purified. Germ cells were also investigated to measure the intracellular level. Complementary DNAs were synthesized with a random hexamer for the extracellular fraction and with the oligo(dT) primer for germ cells, and real-time PCR was performed to amplify Tesra and lncRNA-HSVIII. Since the amplification efficiencies were not so different, the expression levels were directly compared by setting the value of lncRNA-HSIII to 1.0. The data are presented as means ± SD from three independent experiments and were analyzed by the Student t test. *P < 0.05 relative to the control. (B) Western blot analysis of fractions collected by size exclusion chromatography. The extracellular fraction was applied to the column to separate EVs, and 10 fractions were eluted. A portion of each fraction was subjected to western blot analysis to detect the CD9 protein, a widely used exosome marker. We performed duplicate experiments, and both results are shown. Intense signals were detected in fractions #6-#9 of both samples. (C) Electron microscopic observation of an EV. A representative photograph of an EV in a fraction that was obtained by size exclusion chromatography and was positive for CD9. Similar vesicles were observed on the mesh. A scale bar, 100 nm. (D) RT-PCR with exosome fractions. Total RNAs were purified from fractions #7 and #8 of both samples in (B), and cDNAs were synthesized with a random hexamer. Tesra signals were detected by 37 cycles of PCR and agarose gel electrophoresis. In sample 1, the signal was observed in fraction #7, and in sample 2, both lanes showed the band, although the signal intensity was much higher in fraction #8.
Tesra binds to the Prss42/Tessp-2 promoter
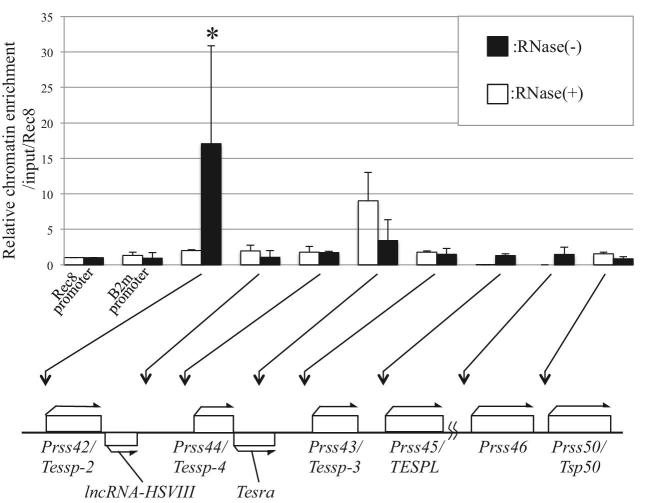
The fact that Tesra transcription was increased at a timing similar to that of Prss/Tessp gene activation (Figure 2D) and the fact that the transcripts were localized in nuclei of pachytene spermatocytes (Figure 3B and G) indicated that Tesra was possibly involved in activation of the Prss/Tessp genes during meiosis. To verify this possibility, we investigated whether the Tesra transcript interacted with a promoter region of any of the Prss/Tessp genes. For this purpose, we attempted to reveal the genome occupancy of Tesra at the Prss/Tessp locus (including three other Prss genes downstream of Prss43/Tessp-3) by performing a ChIRP assay [54,55]. We collected male germ cells from 21- to 22-day-old mice and hybridized the sonicated chromatin with biotinylated tiling oligos because the expression level of Tesra was the highest at these stages (Figure 2D). As a control, we prepared an RNase(+) sample that was treated with RNase in the hybridization reaction to reduce Tesra transcripts. We purified DNAs bound to the oligos and amplified genomic regions at the Prss/Tessp locus by qPCR. Rec8 and B2m promoters on different chromosomes from the Prss/Tessp cluster were examined as negative controls, and the occupancy level at the Rec8 promoter was set to 1.0. Co-isolation of Tesra transcripts with bound chromatin was confirmed by RNA isolation and qRT-PCR after ChIRP (data not shown).
Most regions showed no significant difference in genomic occupancy compared with the Rec8 promoter, but the Prss42/Tessp-2 promoter showed a significantly higher level of genome occupancy (Figure 5, black bars). Although the level was not significantly different from Rec8 promoter, the region transcribed into Tesra exhibited slightly higher occupancy. This was considered to be an artifact by direct interaction of tiling oligos with genome DNA, as evidenced by the control data (RNase+) showing a clear peak at this region (Figure 5, white bars). In contrast, no peak was observed at the Prss42/Tessp-2 promoter in the RNase(+) sample, indicating actual binding of Tesra transcripts to this region. Thus, the Tesra transcript interacts with the Prss42/Tessp-2 promoter at the Prss/Tessp locus in mouse germ cells, suggesting that Tesra plays a role in regulation of the expression of the Prss42/Tessp-2 gene.
Figure 5.
Tesra occupancy at the Prss/Tessp locus. ChIRP-qPCR for Tesra in mouse testicular germ cells was performed. Germ cells were collected from 21- to 22-day-old mouse testes, and nuclear extracts were prepared. Sonicated chromatin was hybridized with biotinylated tiling oligo probes, and the bound chromatin was collected by streptavidin beads. Purified genome DNAs were investigated by qPCR. Rec8 and B2m promoters that were located on different chromosomes from the Prss/Tessp cluster were amplified as negative controls. The value was normalized to that of the input sample, which was kept before hybridization, and further normalized to the level at the Rec8 promoter (=1.0). The relative chromatin enrichment at each position is shown by the black bars. The white bars show the data from the experiment with RNase in hybridization buffer as a negative control. Positions of amplicons are indicated by arrows below the graph. The data are presented as means ± SD from three independent experiments and were analyzed by one-way ANOVA followed by the Tukey post hoc test. *P < 0.05 compared to other regions.
Prss42/Tessp-2 gene activation by Tesra
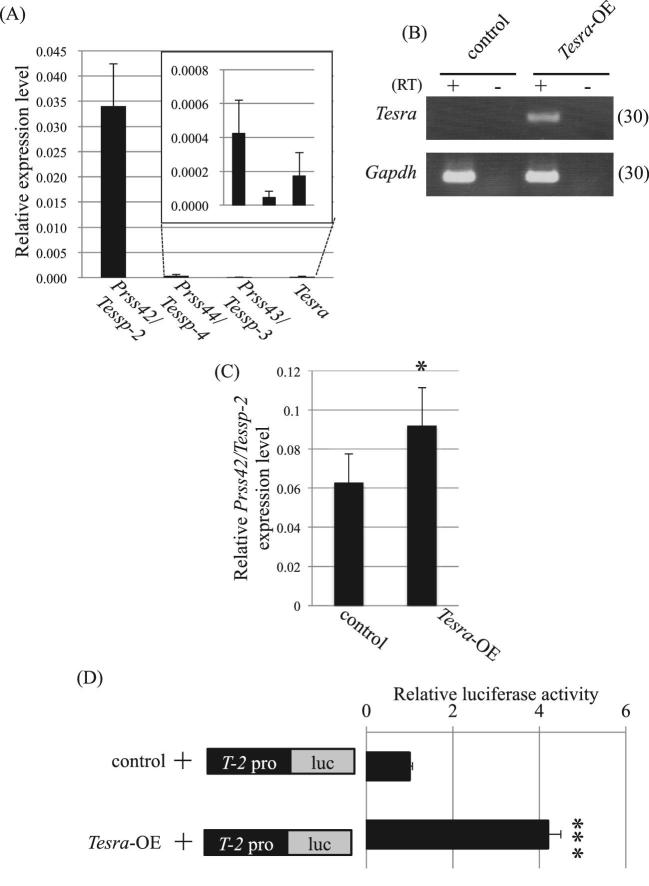
To assess the function of Tesra in Prss42/Tessp-2 regulation, we overexpressed Tesra in a mouse hepatic tumor cell line, Hepa1–6, that endogenously expressed Prss42/Tessp-2 mRNA (Figure 6A). We transfected Tesra-OE or a control vector, and after selection of successfully transfected cells with G418, endogenous Prss42/Tessp2 expression was evaluated by qRT-PCR. Tesra expression was dramatically induced in the cells with Tesra-OE compared with that in the control cells (Figure 6B), and the overexpression significantly increased endogenous Prss42/Tessp-2 expression (Figure 6C).
Figure 6.
Overexpression of Tesra increases Prss42/Tessp-2 expression and promoter activity. (A) Endogenous expression of Prss/Tessp cluster genes and Tesra in Hepa1–6 cells. qRT-PCR was performed with total RNAs from Hepa1–6 cells. Reverse transcription was done by using the oligo(dT) primer, and the Aip gene was examined as an internal control. Expression levels were normalized to Aip. (B) Successful overexpression of Tesra. Tesra-OE or the control vector was transiently transfected into Hepa1–6 cells, and after selection with G418, total RNA was purified from each sample. Complementary DNA was synthesized with the oligo(dT) primer, and PCR was conducted to detect Tesra and Gapdh expression. The cycle number is shown in parenthesis. A representative result from three experiments is shown. We could not see any difference among the three data sets. (C) Relative expression of Prss42/Tessp-2 mRNA by overexpression of Tesra. qRT-PCR was performed with the cDNAs prepared in (B). Data normalization was done by using Aip as an internal control. Transient overexpression of Tesra significantly increased Prss42/Tessp-2 expression in Hepa1–6 cells. The data are presented as means ± SD from three independent experiments and were analyzed by the Student t test. *P < 0.05 relative to the control. (D) Luciferase assay in Hepa1–6 cells transiently transfected with Tesra-OE. T-2pro-luc was transiently co-transfected with Tesra-OE or the control vector into Hepa1–6 cells, and luciferase activity was measured 2 days later. Relative luciferase activity is presented by setting the activity with the control to 1.0. Tesra overexpression significantly increased Prss42/Tessp-2 promoter activity. The data are presented as means ± SD from three independent experiments and were analyzed by the Student t test. ***P < 0.001 relative to the control.
We then carried out a transient reporter gene assay to determine whether Tesra increased Prss42/Tessp-2 promoter activity. We prepared a construct in which the luciferase gene was driven by a 1.6-kb Prss42/Tessp-2 promoter (T-2pro-luc), and co-transfected it with Tesra-OE into Hepa1–6 cells. As a result of measurement of luciferase activity 48 h later, Prss42/Tessp-2 promoter activity was significantly increased compared to that in the case of co-transfection with the control vector (Figure 6D). These data suggested that Tesra contributed to activation of the Prss42/Tessp-2 gene by increasing Prss42/Tessp-2 promoter activity.
Co-activation of Prss42/Tessp-2 promoter activity by an enhancer and Tesra
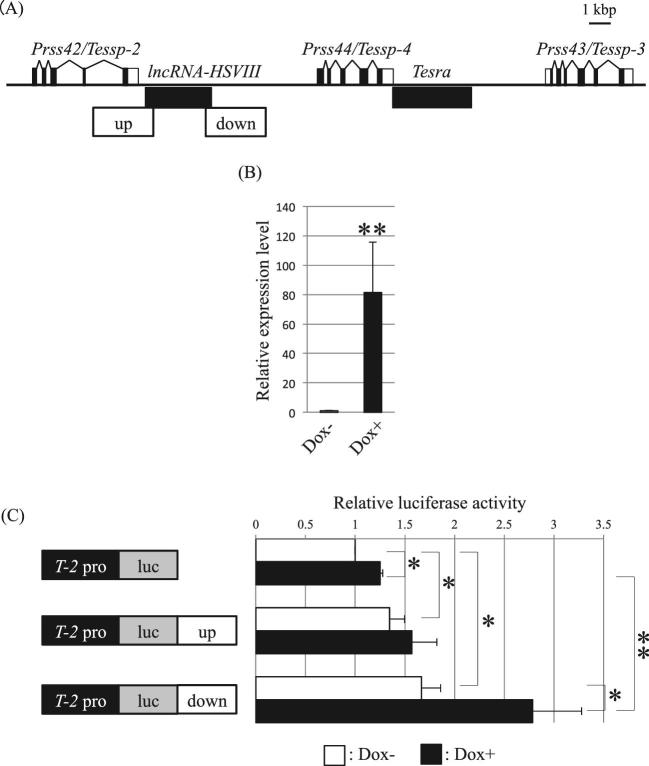
We previously reported that upstream and downstream sequences of lncRNA-HSVIII possessed enhancer activity for Prss42/Tessp-2 [45] (Figure 7A). We finally investigated whether Tesra was functionally related to these enhancers for Prss42/Tessp-2 activation. To answer this question, we established Tesra-inducible stable cells by the Tet-on system in Hepa1–6 cells. The stable cell line was established by co-transfection of Tesra-tet-on with other required vectors and selection with G418. Dox treatment and qRT-PCR verified that Tesra transcription was about 80-fold increased by culture for 24 h (Figure 7B).
Figure 7.
Tesra co-operatively increases Prss42/Tessp-2 promoter activity with the downstream enhancer. (A) A schematic view of the Prss/Tessp locus indicating positions of the enhancers. The Prss/Tessp locus is depicted as in Figure 1A. Two potential enhancers we identified before [45] are indicated with white boxes marked as “up” and “down.” (B) Successful induction of Tesra transcription in stable Hepa1–6 cells. We established stable Hepa1–6 cells that were responsive to Dox to induce Tesra transcription by the Tet-on system. Dox was added to the cells, and total RNAs were collected 24 h later. qRT-PCR was performed and the data are presented as in Figure 2D. Tesra transcription was significantly induced by the addition of Dox. The data are presented as means ± SD from three independent experiments and were analyzed by the Student t test. **P < 0.01 relative to Dox-. (C) Luciferase assay in stable Tesra-inducible Hepa1–6 cells. Each construct indicated on the left was transiently transfected into the cells. Dox was added 24 h after the transfection, and luciferase activity was measured after another 24 h (black bars). We also prepared cells that were transfected with luciferase constructs but not treated with Dox (white bars). The activity in the cells with T-2pro-luc without Dox was set to 1.0. The data are presented as means ± SD from three independent experiments, and the statistical significance was analyzed by one-way ANOVA followed by the Tukey post hoc test. *P < 0.05. **P < 0.01.
We transfected T-2pro-luc, T-2pro-luc-up, or T-2pro-luc-down constructs into the Tesra-inducible stable cell line and added Dox 24 h after the transfection. After another 24 h, we measured luciferase activity. Without Dox, we confirmed enhancer activity of both upstream and downstream sequences for the Prss42/Tessp-2 promoter (Figure 7C). The promoter activity was significantly increased by about 1.3-fold with T-2pro-luc-up and by about 1.7-fold with T-2pro-luc-down (Figure 7C, white bars). By the addition of Dox, we observed a further increase in Prss42/Tessp-2 promoter activity with T-2pro-luc-down; the activity was significantly increased by 2.2-fold (Figure 7C, black bars of T-2pro-luc vs T-2pro-luc-down). In contrast, promoter activity was not significantly changed in the cells with T-2pro-luc-up by Dox treatment (Figure 7C, black bars of T-2pro-luc vs T-2pro-luc-up). These results indicated that Tesra and the downstream enhancer could cooperatively but independently increase Prss42/Tessp-2 promoter activity.
Discussion
In this study, we identified a novel mouse testis-specific lncRNA, Tesra, that was expressed in both germ cells and Leydig cells but not in Sertoli cells. The subcellular localization was shown by in situ hybridization to be different depending on the cell type. Spermatogonia, primary spermatocytes, and round spermatids contained the Tesra transcript in both the nuclei and cytoplasm, while it was mostly present in the cytoplasm of elongating spermatids and Leydig cells. The number of Tesra signals was larger in pachytene spermatocytes than in other types of germ cells, being in agreement with the increase in Tesra expression in the testis between day 14 and day 21, when germ cells reach the stage of pachytene spermatocytes. Although some nuclear and cytoplasmic signals detected by in situ hybridization might be derived from extracellular transcripts, our data indicated the following expression pattern. Tesra transcription begins in spermatogonia at a low level and is dramatically enhanced when germ cells become mid or late pachytene spermatocytes. During this period, Tesra transcripts are localized in both the nuclei and cytoplasm, and such localization is maintained in round spermatids. In spermiogenesis, Tesra transcripts come to be localized in the cytoplasm of elongating spermatids. These findings suggest that Tesra has multiple functions during spermatogenesis.
Although we focused on nuclear Tesra in this study, in situ hybridization showed dot-like signals in cytoplasm, suggesting that Tesra transcripts are associated with some specific cytoplasmic structures. There are several granular structures composed of ribonucleoproteins in cytoplasm of mammalian cells. Stress granules appear in response to a variety of stress to repress translation [56], and similar RNA granules were observed in oocytes to control a timing of translation during oocyte maturation [57]. A processing body is another RNA granule which is related to translational repression and mRNA decay [58]. Because several genes were reported to be translationally arrested during male meiosis [59,60], cytoplasmic Tesra may also form a ribonucleoprotein complex and function in the regulation of translation.
It is interesting that Tesra transcipts were detected in extracellular fractions. We concluded extracellular localization based on (1) observation of scattered signals by in situ hybridization; (2) detection of Tesra in the extracellular fraction at a higher level than that of lncRNA-HSVIII, which never showed scattered signals, especially in the lumen [45]; and (3) detection of Tesra in some of the EV fractions. All of these findings support the existence of Tesra as extracellular RNA in the mouse testis, although Tesra may be present as fragmented RNAs rather than the full length as evidenced by more efficient detection of extracellular Tesra transcripts with the primer pair “Tesra exosome” than “Tesra PCR” (data not shown). Because only one of the two EV fractions showed the clear Tesra signal by RT-PCR, this transcript is probably contained in a specific population of EVs. In addition, the Tesra level relative to lncRNA-HSVIII in extracellular fractions was higher than that within germ cells, suggesting a specific mechanism of preferentially secreting Tesra out of the cell. It is known that the protein or miRNA profiling in EVs is different depending on the cell type or external stimulus [61,62] and the packaging of proteins or RNAs into EVs is a selective process [63,64]. In case of mRNAs, specific nucleotide sequences were reported to be involved in this selection [65], and some lncRNAs were suggested to be selectively sorted into EVs [66]. Therefore, Tesra transcripts may be packaged into EVs and secreted from germ cells by some specific mechanism.
Recent studies have shown that several lncRNAs are actually released into extracellular regions and enclosed within EVs such as exosomes [53,67]. So far, many extracellular lncRNAs have been reported to come from cancerous tissues, but nontumor tissues also secrete lncRNAs by EVs [68], suggesting the presence of more EV-associated lncRNAs in normal tissues. In the case of Tesra, its co-existence with an exosome marker strongly suggests that Tesra transcripts are associated with a particular size of EVs. However, the function of extracellular Tesra is unclear. Since some exosomal RNAs are related to cell-to-cell communication in distant or neighboring cells [53], it is possible that extracellular Tesra contributes to the progression of spermatogenesis through controlling germ cell-to-germ cell communication. More studies are necessary for elucidating the significance of extracellular Tesra.
To reveal the mechanism of transcriptional activation in primary spermatocytes, we used a Hepa1–6 cell line which was derived from a mouse hepatoma [69,70] instead of a spermatocyte-derived cell line, GC-2spd(ts) [71], because Hepa1–6 endogenously expressed Prss/Tessp cluster genes but GC-2spd(ts) did not (data not shown). However, while tumor cells were reported to partly share characteristics with testicular cells [72,73], they are different from spermatogenetic cells, and indeed, the expression levels of Prss/Tessp genes in Hepa1–6 cells were lower than those in the testis. This indicates the limitation of using Hepa1–6 cells for functional experiments, and our data suggest a potential function of Tesra. In vivo studies need to be done for concluding the function.
We focused on the function of Tesra in the nuclei of pachytene spermatocytes. In general, nuclear lncRNAs are often involved in transcriptional regulation of their neighboring genes [74–76], and the Prss/Tessp cluster genes are activated in pachytene spermatocytes. We therefore investigated the involvement of Tesra in their transcriptional activation. We obtained the following evidence that supported a positive role of Tesra in regulation of the Prss42/Tessp-2 gene. First, a ChIRP assay showed that Tesra significantly interacted with chromatin at the Prss42/Tessp-2 promoter in germ cells (Figure 5). This confirmed the in vivo interaction of the Tesra transcript with chromatin at the Prss42/Tessp-2 promoter. Second, Tesra significantly increased endogenous Prss42/Tessp-2 gene expression by transient overexpression in Hepa1–6 cells (Figure 6C). Third, a transient reporter gene assay showed that the overexpression also increased Prss42/Tessp-2 promoter activity in Hepa1–6 cells (Figure 6D). These findings suggest that Tesra contributes to transcriptional activation of the Prss42/Tessp-2 gene by enhancing promoter activity.
How does Tesra increase Prss42/Tessp-2 promoter activity? Many nuclear lncRNAs that participate in transcriptional activation recruit histone modifiers or transcription factors to chromatin regions of target genes [77]. For example, GClnc1 interacts with WDR5 and KAT2A, which form an epigenetic modification complex, to specify the histone modification pattern of target genes for transcriptional activation [78]. RMST interacts with hnRNPA2/B1 and SOX2 and plays a critical role in the binding of SOX2 to target gene promoters [79]. Similarly to these lncRNAs, Tesra may recruit transcription factors or histone modification enzymes to the Prss42/Tessp-2 promoter region. Alternatively, Tesra may function as a scaffold that interacts with multiple protein complexes, as reported for HOTTIP associated with the WDR5/MLL1 chromatin-modifying complex [80] and Rmrp associated with the DDX5–RORγt complex [81]. Further studies are required for understanding the detailed molecular mechanism concerning the function of Tesra.
Long noncoding RNAs often collaborate with enhancers to activate their target genes. For instance, one class of lncRNAs, enhancer-derived RNAs, is unidirectionally or bidirectionally transcribed at enhancer regions and mediates histone modification for active chromatin [82,83] and chromatin looping between the enhancer and promoter [84,85]. Some lncRNAs are not transcribed at enhancers but still affect the looping between distal enhancers and promoters [43]. In any case, these functional lncRNAs regulate the transcription of their target genes through their action on enhancers, and lncRNAs are therefore mostly considered to facilitate enhancer activity. In contrast, the transcriptional activation of Prss42/Tessp-2 by Tesra and the enhancer appears to be different from such examples. We previously reported enhancer activity of upstream and downstream sequences of lncRNA-HSVIII [45], but Tesra was transcribed at a distal region from these enhancers. In addition, a luciferase assay in a hepatoma cell showed that both Tesra and two enhancers could increase Prss42/Tessp-2 promoter activity independently (Figure 7). Each enhancer could individually activate the promoter, and Tesra also activated Prss42/Tessp-2 transcription in the absence of the enhancers. The downstream enhancer and Tesra just co-functioned for increasing Prss42/Tessp-2 promoter activity when they co-existed. These findings indicate that Tesra and the enhancer are not interdependent. To the best of our knowledge, there has been no report showing such a relationship between an lncRNA and an enhancer, and our data therefore suggest a novel mechanism of gene activation by an enhancer and an lncRNA.
Out of the two potential enhancers, Tesra, collaborated only with the downstream enhancer because the induction of Tesra transcription additively increased Prss42/Tessp-2 promoter activity with the downstream sequence but not with the upstream sequence (Figure 7). Consistent with those findings, it was actually the downstream sequence that was shown to interact with the Prss42/Tessp-2 promoter by a chromosome conformation capture assay [45]. Therefore, the two enhancers may function redundantly; the downstream enhancer may mainly function in the transcriptional activation of Prss42/Tessp-2, but when it is impaired, the gene may be activated by the upstream enhancer. Such a mechanism might have evolved due to the extreme importance of spermatogenesis for animals.
In combination with our previous study [45], we revealed that two factors potentially contribute to Prss42/Tessp-2 transcriptional activation. First, a downstream region of lncRNA-HSVIII, which interacted with the Prss42/Tessp-2 promoter by looping out the intervening region, could function as an enhancer. Second, lncRNA Tesra, which was transcribed downstream of Prss44/Tessp-4, bound to chromatin at the Prss42/Tessp-2 promoter to potentially increase its activity. Both factors activated Prss42/Tessp-2 transcription through increasing Prss42/Tessp-2 promoter activity. From these findings, we propose the following hypothetical model for Prss42/Tessp-2 gene activation during meiosis (Figure 8). When spermatogonia divide into primary spermatocytes, the chromatin at the Prss42/Tessp-2 promoter begins to interact with the lncRNA-HSVIII region at its 3′ end, and Prss42/Tessp-2 transcription is initiated. This interaction is maintained and even strengthened in late pachytene spermatocytes or secondary spermatocytes, and the downstream enhancer of lncRNA-HSVIII comes to physically contact the Prss42/Tessp-2 promoter to increase its transcriptional activity. Meanwhile, at a timing similar to that of chromatin interaction, Tesra transcripts are accumulated in the nuclei of pachytene spermatocytes and bind to the Prss42/Tessp-2 promoter directly or indirectly to positively affect its activity. These events may lead to a high level of transcriptional activation of the Prss42/Tessp-2 gene.
Figure 8.
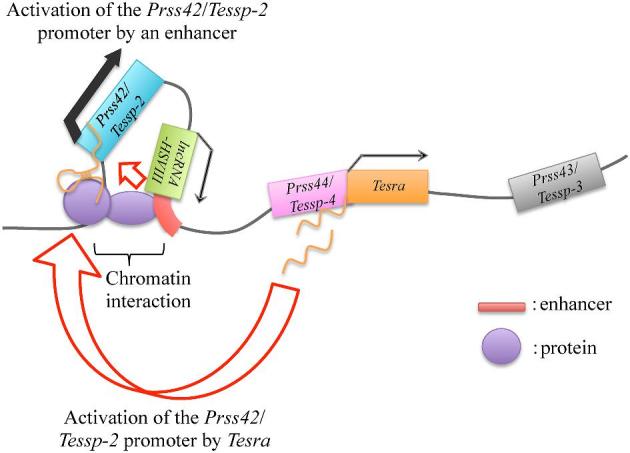
A hypothetical model for Prss42/Tessp-2 transcriptional activation. This figure indicates the hypothesized mechanism of Prss42/Tessp-2 gene activation based on our data. When spermatogonia divide into primary spermatocytes, the chromatin at the enhancer downstream of lncRNA-HSVIII begins to interact with that at the Prss42/Tessp-2 promoter, and thereby the enhancer is allowed to activate Prss42/Tessp-2 transcription. At a similar timing, Tesra transcripts are accumulated in the nuclei of primary spermatocytes and bind to chromatin at the Prss42/Tessp-2 promoter. The transcription level of Prss42/Tessp-2 is substantially enhanced by these two elements. There must be some proteins binding to Tesra transcripts and/or to the Prss42/Tessp-2 promoter.
Recent studies have revealed numerous lncRNAs in each stage of spermatogenesis, but only a few studies have shown the functions of testicular lncRNAs. Only a few mammalian lncRNAs have been found to be critical in male germ cell development, especially in the meiotic stage. For instance, testis-specific X-linked (Tsx), which is an X-linked lncRNA and is specifically expressed in pachytene spermatocytes, was shown to be crucial for the progression of meiosis and to possibly regulate Tsix during spermatogenesis [86]. R53 containing a SINE-B1 motif is expressed mainly in the nuclei of spermatocytes, and its reduction causes abnormal upregulation of the expression of Tnp1, Tnp2, and Prm1 genes, resulting in impairment of meiosis [87]. Tslrn1 is highly expressed in mouse primary spermatocytes, and mice with its knockout showed a significant reduction in spermatozoa [20]. Our group identified lncRNA-Tcam1 that is expressed at a high level in spermatocytes and suggested that it regulates immune-related genes for immune response during meiosis [88]. Downregulation of mrhl RNA is required for activation of Wnt signaling and the entry of spermatogonia into meiosis [89]. While these findings indicate the significance of lncRNAs in spermatogenesis, more lncRNAs have to be functionally analyzed when we consider the large number of lncRNAs identified in the testis. We successfully extended the knowledge of testicular lncRNAs by the present study.
In summary, we revealed the expression pattern and suggested the function of a novel lncRNA, Tesra. Tesra potentially contributes to transcriptional activation of the Prss42/Tessp-2 gene, and due to the significant role of this gene in the progression of meiosis from secondary spermatocytes to round spermatids [46], this lncRNA may play a role in spermatogenesis. Our results also suggested a novel relationship between an lncRNA and an enhancer, contribute to an understanding of how spermatogenesis is regulated, and provide an insight into the function of an lncRNA in reproduction.
Supplementary Data
Supplemental Table S1. Antibodies used for western blot analysis.
Supplementary Material
Acknowledgment
We thank Dr Kazuhiro Murakami for kindly giving us plasmids, pPBhCMV*1-cHA-pA, pPBCAGrtTA-IN, and pPyCAG-PBase. We also thank Ms Hiroko Yamamoto for helping us using an electron microscope. Natsumi Takei is a recipient of SUNBOR scholarship from SUNTORY Foundation for Life Sciences.
Notes
Conference presentation: Presented in part at the Consortium of Biological Sciences 2017, December 6–9, 2017, Kobe, Japan, 50th Annual Meeting of the Society for the Study of Reproduction, July 13–16, 2017, Marriott Wardman Park, Washington DC, Biochemistry and Molecular Biology 2015, December 1–4, 2015, Kobe, Japan, The 86th Annual Meeting of the Zoological Society of Japan, September 17–19, 2015, Niigata, Japan, and RNA Meeting 2015, July 15–17, 2015, Sapporo, Japan.
Edited by Dr. Sarah Kimmins
Footnotes
Grant support: This work was supported by Grants-in-aid for Scientific Research 15H04317 and 16K08939 and Grant-in-aid for JSPS Research Fellow 17J05929 from Japan Society for the Promotion of Science.
References
- 1. Hecht NB. Molecular mechanisms of male germ cell differentiation. Bioessays 1998; 20(7):555–561. [DOI] [PubMed] [Google Scholar]
- 2. Eddy EM. Male germ cell gene expression. Recent Prog Horm Res 2002; 57(1):103–128. [DOI] [PubMed] [Google Scholar]
- 3. O’Bryan MK, De Kretser D, Ivell R, Skakkebæk NE, Almstrup K, Leffers H. Mouse models for genes involved in impaired spermatogenesis. Int J Androl 2006; 29(1):76–89. [DOI] [PubMed] [Google Scholar]
- 4. Roy A, Matzuk MM. Deconstructing mammalian reproduction: using knockouts to define fertility pathways. Reproduction 2006; 131(2):207–219. [DOI] [PubMed] [Google Scholar]
- 5. Yan W. Male infertility caused by spermiogenic defects: lessons from gene knockouts. Mol Cell Endocrinol 2009; 306(1–2):24–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dix DJ, Rosario-Herrle M, Gotoh H, Mori C, Goulding EH, Barrett C V, Eddy EM. Developmentally regulated expression ofHsp70-2and aHsp70-2/lacZ transgene during spermatogenesis. Dev Biol 1996; 174(2):310–321. [DOI] [PubMed] [Google Scholar]
- 7. Salehi-Ashtiani K, Widrow RJ, Markert CL, Goldberg E. Testis-specific expression of a metallothionein I-driven transgene correlates with undermethylation of the locus in testicular DNA. Proc Natl Acad Sci USA 1993; 90(19):8886–8890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Nayernia K, Nieter S, Kremling H, Oberwinkler H, Engel W. Functional and molecular characterization of the transcriptional regulatory region of the proacrosin gene. J Biol Chem 1994; 269:32181–32186. [PubMed] [Google Scholar]
- 9. Li S, Zhou W, Doglio L, Goldberg E. Transgenic mice demonstrate a testis-specific promoter for lactate dehydrogenase, LDHC. J Biol Chem 1998; 273(47):31191–31194. [DOI] [PubMed] [Google Scholar]
- 10. Galcheva-Gargova Z, Tokeson JP, Karagyosov LK, Ebert KM, Kilpatrick DL. The rat proenkephalin germ line promoter contains multiple binding sites for spermatogenic cell nuclear proteins. Mol Endocrinol 1993; 7:979–991. [DOI] [PubMed] [Google Scholar]
- 11. Zhang LP, Stroud JC, Walter CA, Adrian GS, McCarrey JR. A gene-specific promoter in transgenic mice directs testis-specific demethylation prior to transcriptional activation in vivo. Biol Reprod 1998; 59(2):284–292. [DOI] [PubMed] [Google Scholar]
- 12. Lele KM, Wolgemuth DJ. Distinct regions of the mouse Cyclin A1 gene, Ccna1, confer male germ-cell specific expression and enhancer function. Biol Reprod 2004; 71(4):1340–1347. [DOI] [PubMed] [Google Scholar]
- 13. Hammoud SS, Low DHP, Yi C, Carrell DT, Guccione E, Cairns BR. Chromatin and transcription transitions of mammalian adult germline stem cells and spermatogenesis. Cell Stem Cell 2014; 15(2):239–253. [DOI] [PubMed] [Google Scholar]
- 14. Kurihara M, Shiraishi A, Satake H, Kimura AP. A conserved noncoding sequence can function as a spermatocyte-specific enhancer and a bidirectional promoter for a ubiquitously expressed gene and a testis-specific long noncoding RNA. J Mol Biol 2014; 426(17):3069–3093. [DOI] [PubMed] [Google Scholar]
- 15. Necsulea A, Soumillon M, Warnefors M, Liechti A, Daish T, Zeller U, Baker JC, Grützner F, Kaessmann H. The evolution of lncRNA repertoires and expression patterns in tetrapods. Nature 2014; 505(7485):635–640. [DOI] [PubMed] [Google Scholar]
- 16. Washietl S, Kellis M, Garber M. Evolutionary dynamics and tissue specificity of human long noncoding RNAs in six mammals. Genome Res 2014; 24(4):616–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bao J, Wu J, Schuster AS, Hennig GW, Yan W. Expression profiling reveals developmentally regulated lncRNA repertoire in the mouse male germline. Biol Reprod 2013; 89(5):107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sun J, Lin Y, Wu J. Long non-coding RNA expression profiling of mouse testis during postnatal development. PLoS ONE 2013; 8(10):e75750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Liang M, Li W, Tian H, Hu T, Wang L, Lin Y, Li Y, Huang H, Sun F. Sequential expression of long noncoding RNA as mRNA gene expression in specific stages of mouse spermatogenesis. Sci Rep 2015; 4(1):5966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wichman L, Somasundaram S, Breindel C, Valerio DM, McCarrey JR, Hodges CA, Khalil AM. Dynamic expression of long noncoding RNAs reveals their potential roles in spermatogenesis and fertility. Biol Reprod 2017; 97(2):313–323. [DOI] [PubMed] [Google Scholar]
- 21. Weng B, Ran ML, Chen B, He CQ, Dong LH, Peng FZ. Genome-wide analysis of long non-coding RNAs and their role in postnatal porcine testis development. Genomics 2017; 109(5–6):446–456. [DOI] [PubMed] [Google Scholar]
- 22. Zhang Y, Yang H, Han L, Li F, Zhang T, Pang J, Feng X, Ren C, Mao S, Wang F. Long noncoding RNA expression profile changes associated with dietary energy in the sheep testis during sexual maturation. Sci Rep 2017; 7(1):5180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ran M, Chen B, Li Z, Wu M, Liu X, He C, Zhang S, Li Z. Systematic identification of long noncoding RNAs in immature and mature porcine testes. Biol Reprod 2016; 94(4):77. [DOI] [PubMed] [Google Scholar]
- 24. Yang H, Wang F, Li F, Ren C, Pang J, Wan Y, Wang Z, Feng X, Zhang Y. Comprehensive analysis of long non-coding RNA and mRNA expression patterns in sheep testicular maturation. Biol Reprod 2018; 99(3):650–661. [DOI] [PubMed] [Google Scholar]
- 25. Yang L, Lin C, Jin C, Yang JC, Tanasa B, Li W, Merkurjev D, Ohgi KA, Meng D, Zhang J, Evans CP, Rosenfeld MG. lncRNA-dependent mechanisms of androgen-receptor-regulated gene activation programs. Nature 2013; 500(7464):598–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Amit-Avraham I, Pozner G, Eshar S, Fastman Y, Kolevzon N, Yavin E, Dzikowski R. Antisense long noncoding RNAs regulate var gene activation in the malaria parasite Plasmodium falciparum. Proc Natl Acad Sci USA 2015; 112(9):E982–E991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kimura AP, Yoneda R, Kurihara M, Mayama S, Matsubara S. A long noncoding RNA, lncRNA-Amhr2, plays a role in Amhr2 gene activation in mouse ovarian granulosa cells. Endocrinology 2017; 158(11):4105–4121. [DOI] [PubMed] [Google Scholar]
- 28. Jiang W, Liu Y, Liu R, Zhang K, Zhang Y. The lncRNA DEANR1 facilitates human endoderm differentiation by activating FOXA2 expression. Cell Rep 2015; 11(1):137–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Krawczyk M, Emerson BM. p50-associated COX-2 extragenic RNA (PACER) activates COX-2 gene expression by occluding repressive NF-?B complexes. eLife 2014; 3:e01776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bergmann JH, Li J, Eckersley-Maslin MA, Rigo F, Freier SM, Spector DL. Regulation of the ESC transcriptome by nuclear long noncoding RNAs. Genome Res 2015; 25(9):1336–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kadakkuzha BM, Liu X-A, McCrate J, Shankar G, Rizzo V, Afinogenova A, Young B, Fallahi M, Carvalloza AC, Raveendra B, Puthanveettil SV. Transcriptome analyses of adult mouse brain reveal enrichment of lncRNAs in specific brain regions and neuronal populations. Front Cell Neurosci 2015; 9:63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Carlevaro-Fita J, Rahim A, Guigó R, Vardy LA, Johnson R. Cytoplasmic long noncoding RNAs are frequently bound to and degraded at ribosomes in human cells. RNA 2016; 22(6):867–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ahadi A, Brennan S, Kennedy PJ, Hutvagner G, Tran N. Long non-coding RNAs harboring miRNA seed regions are enriched in prostate cancer exosomes. Sci Rep 2016; 6(1):24922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gui Y, Liu H, Zhang L, Lv W, Hu X. Altered microRNA profiles in cerebrospinal fluid exosome in Parkinson disease and Alzheimer disease. Oncotarget 2015; 6(35):37043–37053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lamichhane TN, Leung CA, Douti LY, Jay SM. Ethanol induces enhanced vascularization bioactivity of endothelial cell-derived extracellular vesicles via regulation of microRNAs and long non-coding RNAs. Sci Rep 2017; 7(1):13794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bergmann JH, Spector DL. Long non-coding RNAs: modulators of nuclear structure and function. Curr Opin Cell Biol 2014; 26:10–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem 2012; 81(1):145–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ulitsky I, Shkumatava A, Jan CH, Sive H, Bartel DP. Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell 2011; 147(7):1537–1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lai F, Orom UA, Cesaroni M, Beringer M, Taatjes DJ, Blobel GA, Shiekhattar R. Activating RNAs associate with mediator to enhance chromatin architecture and transcription. Nature 2013; 494(7438):497–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Atianand MK, Fitzgerald KA. Long non-coding rnas and control of gene expression in the immune system. Trends Mol Med 2014; 20(11):623–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yoon JH, Abdelmohsen K, Srikantan S, Yang X, Martindale JL, De S, Huarte M, Zhan M, Becker KG, Gorospe M. LincRNA-p21 suppresses target mRNA translation. Mol Cell 2012; 47(4):648–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Tripathi V, Ellis JD, Shen Z, Song DY, Pan Q, Watt AT, Freier SM, Bennett CF, Sharma A, Bubulya PA, Blencowe BJ, Prasanth SG et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell 2010; 39(6):925–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Xiang J-F, Yin Q-F, Chen T, Zhang Y, Zhang X-O, Wu Z, Zhang S, Wang H-B, Ge J, Lu X, Yang L, Chen L-L. Human colorectal cancer-specific CCAT1-L lncRNA regulates long-range chromatin interactions at the MYC locus. Cell Res 2014; 24(5):513–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Miao Y, Ajami NE, Huang T-S, Lin F-M, Lou C-H, Wang Y-T, Li S, Kang J, Munkacsi H, Maurya MR, Gupta S, Chien S et al. Enhancer-associated long non-coding RNA LEENE regulates endothelial nitric oxide synthase and endothelial function. Nat Commun 2018; 9(1):292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Yoneda R, Satoh Y, Yoshida I, Kawamura S, Kotani T, Kimura AP. A genomic region transcribed into a long noncoding RNA interacts with the Prss42/Tessp-2 promoter in spermatocytes during mouse spermatogenesis, and its flanking sequences can function as enhancers. Mol Reprod Dev 2016; 83(6):541–557. [DOI] [PubMed] [Google Scholar]
- 46. Yoneda R, Takahashi T, Matsui H, Takano N, Hasebe Y, Ogiwara K, Kimura AP. Three testis-specific paralogous serine proteases play different roles in murine spermatogenesis and are involved in germ cell survival during meiosis1. Biol Reprod 2013; 88(5):118. [DOI] [PubMed] [Google Scholar]
- 47. Matsubara S, Takahashi T, Kimura AP. Epigenetic patterns at the mouse prolyl oligopeptidase gene locus suggest the CpG island in the gene body to be a novel regulator for gene expression. Gene 2010; 465(1–2):17–29. [DOI] [PubMed] [Google Scholar]
- 48. Matsubara S, Kurihara M, Kimura AP. A long non-coding RNA transcribed from conserved non-coding sequences contributes to the mouse prolyl oligopeptidase gene activation. J Biochem 2014; 155 (4):243–256. [DOI] [PubMed] [Google Scholar]
- 49. Yoneda R, Kimura AP. A testis-specific serine protease, Prss41/Tessp-1, is necessary for the progression of meiosis during murine in vitro spermatogenesis. Biochem Biophys Res Commun 2013; 441(1):120–125. [DOI] [PubMed] [Google Scholar]
- 50. Takei N, Nakamura T, Kawamura S, Takada Y, Satoh Y, Kimura AP, Kotani T. High-sensitivity and high-resolution in situ hybridization of coding and long non-coding RNAs in vertebrate ovaries and testes. Biol Proced Online 2018; 20(1):6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Murakami K, Günesdogan U, Zylicz JJ, Tang WWC, Sengupta R, Kobayashi T, Kim S, Butler R, Dietmann S, Azim Surani M. NANOG alone induces germ cells in primed epiblast in vitro by activation of enhancers. Nature 2016; 529(7586):403–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kurihara M, Kimura AP. Characterization of the human TCAM1P pseudogene and its activation by a potential dual promoter-enhancer: comparison with a protein-coding mouse orthologue. FEBS Lett 2015; 589(4):540–547. [DOI] [PubMed] [Google Scholar]
- 53. Dragomir Mihnea, Chen Baoqing, Chalin GA. Exosomal lncRNAs as new players in cell-to-cell communication. Transl Cancer Res 2018; 7(S2):S243–S252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Chu C, Qu K, Zhong FL, Artandi SE, Chang HY. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol Cell 2011; 44(4):667–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Chu C, Quinn J, Chang HY. Chromatin isolation by RNA purification (ChIRP). J Vis Exp 2012; 61:e3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ross Buchan J. mRNP granules. RNA Biology 2014; 11(8):1019–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kotani T, Yasuda K, Ota R, Yamashita M. Cyclin b1 mRNA translation is temporally controlled through formation and disassembly of RNA granules. J Cell Biol 2013; 202(7):1041–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Luo Y, Na Z, Slavoff SA. P-Bodies: composition, properties, and functions. Biochemistry 2018; 57(17):2424–2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Morales CR, Kwon YK, Hecht NB. Cytoplasmic localization during storage and translation of the mRNAs of transition protein 1 and protamine 1, two translationally regulated transcripts of the mammalian testis. J Cell Sci 1991; 100(Pt 1):119–131. [DOI] [PubMed] [Google Scholar]
- 60. Baker CC, Gim BS, Fuller MT. Cell type-specific translational repression of Cyclin B during meiosis in males. Development 2015; 142:3394–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Zhang J, Li S, Li L, Li M, Guo C, Yao J, Mi S. Exosome and exosomal microRNA: trafficking, sorting, and function. Genomics Proteomics Bioinformatics 2015; 13:17–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Eldh M, Ekström K, Valadi H, Sjöstrand M, Olsson B, Jernås M, Lötvall J. Exosomes communicate protective messages during oxidative stress; possible role of exosomal shuttle RNA. PLoS ONE 2010; 5:e15353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Janas T, Janas MM, Sapoń K, Janas T. Mechanisms of RNA loading into exosomes. FEBS Lett 2015; 589:1391–1398. [DOI] [PubMed] [Google Scholar]
- 64. Gangoda L, Liem M, Ang CS, Keerthikumar S, Adda CG, Parker BS, Mathivanan S. Proteomic profiling of exosomes secreted by breast cancer cells with varying metastatic potential. Proteomics 2017; 17:1600370. [DOI] [PubMed] [Google Scholar]
- 65. Bolukbasi MF, Mizrak A, Ozdener GB, Madlener S, Ströbel T, Erkan EP, Fan JB, Breakefield XO, Saydam O. miR-1289 and “Zipcode”-like sequence enrich mRNAs in microvesicles. Mol Ther Nucleic Acids 2012; 1:e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Kim KM, Abdelmohsen K, Mustapic M, Kapogiannis D, Gorospe M. RNA in extracellular vesicles. WIREs RNA 2017; e1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Takahashi K, Yan IK, Wood J, Haga H, Patel T. Involvement of extracellular vesicle long noncoding RNA (linc-VLDLR) in tumor cell responses to chemotherapy. Mol Cancer Res 2014; 12:1377–1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Lu X, Bai D, Liu X, Zhou C, Yang G. Sedentary lifestyle related exosomal release of Hotair from gluteal-femoral fat promotes intestinal cell proliferation. Sci Rep 2017; 7:45648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Darlington GJ, Bernhard HP, Miller RA, Ruddle FH. Expression of liver phenotypes in cultured mouse hepatoma cells. J Natl Cancer Inst 1980; 64:809–819. [PubMed] [Google Scholar]
- 70. Darlington GJ. Liver cell lines. Methods Enzymol 1987; 151:19–38. [DOI] [PubMed] [Google Scholar]
- 71. Hofmann MC, Hess Ra, Goldberg E, Millán JL. Immortalized germ cells undergo meiosis in vitro. Proc Natl Acad Sci USA 1994; 91:5533–5537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Simpson AJG, Caballero OL, Jungbluth A, Chen Y-T, Old LJ. Cancer/testis antigens, gametogenesis and cancer. Nat Rev Cancer 2005; 5:615–625. [DOI] [PubMed] [Google Scholar]
- 73. Wang C, Gu Y, Zhang K, Xie K, Zhu M, Dai N, Jiang Y, Guo X, Liu M, Dai J, Wu L, Jin G et al. Systematic identification of genes with a cancer-testis expression pattern in 19 cancer types. Nat Commun 2016; 7:10499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Engreitz JM, Haines JE, Perez EM, Munson G, Chen J, Kane M, McDonel PE, Guttman M, Lander ES. Local regulation of gene expression by lncRNA promoters, transcription and splicing. Nature 2016; 539:452–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Dimitrova N, Zamudio JR, Jong RM, Soukup D, Resnick R, Sarma K, Ward AJ, Raj A, Lee JT, Sharp PA, Jacks T. LincRNA-p21 activates p21 in cis to promote polycomb target gene expression and to enforce the G1/S checkpoint. Mol Cell 2014; 54:777–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Wang L, Zhao Y, Bao X, Zhu X, Kwok YKY, Sun K, Chen X, Huang Y, Jauch R, Esteban MA, Sun H, Wang H. LncRNA Dum interacts with Dnmts to regulate Dppa2 expression during myogenic differentiation and muscle regeneration. Cell Res 2015; 25:335–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Sun Q, Hao Q, Prasanth K V. Nuclear long noncoding RNAs: key regulators of gene expression. Trends Genet 2018; 34:142–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Sun TT, He J, Liang Q, Ren LL, Yan TT, Yu TC, Tang JY, Bao YJ, Hu Y, Lin Y, Sun D, Chen YX et al. LncRNA GClnc1 promotes gastric carcinogenesis and may act as a modular scaffold of WDR5 and KAT2A complexes to specify the histone modification pattern. Cancer Discov 2016; 6:784–801. [DOI] [PubMed] [Google Scholar]
- 79. Ng SY, Bogu GK, Soh B, Stanton LW. The long noncoding RNA RMST interacts with SOX2 to regulate neurogenesis. Mol Cell 2013; 51:349–359. [DOI] [PubMed] [Google Scholar]
- 80. Cheng Y, Jutooru I, Chadalapaka G, Corton JC, Safe S. The long non-coding RNA HOTTIP enhances pancreatic cancer cell proliferation, survival and migration. Oncotarget 2015; 6:10840–10852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Huang W, Thomas B, Flynn RA, Gavzy SJ, Wu L, Kim S V., Hall JA, Miraldi ER, Ng CP, Rigo FW, Meadows S, Montoya NR et al. DDX5 and its associated lncRNA Rmrp modulate TH17 cell effector functions. Nature 2015; 528:517–522. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 82. Hah N, Murakami S, Nagari A, Danko CG, Lee Kraus W. Enhancer transcripts mark active estrogen receptor binding sites. Genome Res 2013; 23:1210–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Zhu Y, Sun L, Chen Z, Whitaker JW, Wang T, Wang W. Predicting enhancer transcription and activity from chromatin modifications. Nucleic Acids Res 2013; 41:10032–10043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Hsieh C-L, Fei T, Chen Y, Li T, Gao Y, Wang X, Sun T, Sweeney CJ, Lee G-SM, Chen S, Balk SP, Liu XS et al. Enhancer RNAs participate in androgen receptor-driven looping that selectively enhances gene activation. Proc Natl Acad Sci USA 2014; 111:7319–7324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Sanyal A, Lajoie BR, Jain G, Dekker J. The long-range interaction landscape of gene promoters. Nature 2012; 489:109–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Anguera MC, Ma W, Clift D, Namekawa S, Kelleher RJ III, Lee JT. Tsx produces a long noncoding RNA and has general functions in the germline, stem cells, and brain. PLoS Genet 2011; 7:e1002248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Nakajima R, Sato T, Ogawa T, Okano H, Noce T. A noncoding RNA containing a SINE-B1 motif associates with meiotic metaphase chromatin and has an indispensable function during spermatogenesis. PLoS ONE 2017; 12:e0179585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Kurihara M, Otsuka K, Matsubara S, Shiraishi A, Satake H, Kimura AP. A testis-specific long non-coding RNA, lncRNA-Tcam1, regulates immune-related genes in mouse male germ cells. Front Endocrinol 2017; 8:299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Akhade VS, Dighe SN, Kataruka S, Rao MRS. Mechanism of Wnt signaling induced down regulation of mrhl long non-coding RNA in mouse spermatogonial cells. Nucleic Acids Res 2016; 44:387–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.