Abstract
The fertility of sex-reversed XY female mice is severely impaired by a massive loss of oocytes and failure of meiotic progression. This phenomenon remains an outstanding mystery. We sought to determine the molecular etiology of XY oocyte dysfunction by generating sex-reversed females that bear genetic ablation of Sry, a vital sex determination gene, on an inbred C57BL/6 background. These mutant mice, termed XYsry− mutants, showed severe attrition of germ cells during fetal development, resulting in the depletion of ovarian germ cells prior to sexual maturation. Comprehensive transcriptome analyses of primordial germ cells (PGCs) and postnatal oocytes demonstrated that XYsry− females had deviated significantly from normal developmental processes during the stages of mitotic proliferation. The impaired proliferation of XYsry− PGCs was associated with aberrant β-catenin signaling and the excessive expression of transposable elements. Upon entry to the meiotic stage, XYsry− oocytes demonstrated extensive defects, including the impairment of crossover formation, the failure of primordial follicle maintenance, and no capacity for embryo development. Together, these results suggest potential molecular causes for germ cell disruption in sex-reversed female mice, thereby providing insights into disorders of sex differentiation in humans, such as “Swyer syndrome,” in which patients with an XY karyotype present as typical females and are infertile.
Keywords: sex-reversed mice, infertility of XY female, XY PGCs/oocytes, transcriptome analysis, germ cell depletion
We report the molecular etiology of XY oocyte dysfunction in sex-reversed female mice harboring Sry deletion. The results show potential molecular causes for germ cell disruption providing insights into disorders of sex differentiation in humans.
Introduction
Widely conserved in mammals, the gene Sex determining region Y (Sry) encodes a key protein for the sex determination of gonads, which have the potential to develop in one of two mutually exclusive, sex-specific directions. SRY shunts these “bipotential” gonads into male sex-inducing testis formation [1, 2]. In mice, Sry expression occurs in gonadal somatic cells of the genital ridges at embryonic day (E) 11.0–12.0 [1, 3], and triggers the activation of a male sex differentiation program by inducing expression of the genes SRY-related high-mobility group box 9 (Sox9) and fibroblast growth factor 9 (Fgf9) [1, 2].
Notably, Sry mutations are known to result in male-to-female sex-reversed individuals. Mouse models of these sex-reversed XY female mice were first produced from chimeric mice via embryonic stem (ES) cells that carried a mutation at the gene testis-determining region of the Y chromosome (Tdy), induced using a retroviral vector [4–6]. More recently, a TALEN-mediated gene editing procedure has been used to target specific Y chromosome genes, resulting in the production of anatomical XY females via deletion of the HMG domain of Sry [7, 8]. These animals were found to be either infertile or subfertile [8–12]. XY female mice have also been produced by the deletion of autosomal Sry-related genes. For example, the deletion of Sox9 in the gonadal somatic cells of male embryos resulted in their development into XY females [9]. Additionally, the alteration of histone epigenetic signatures through deletion of the JmjC-containing H3K9 demethylase Jmjd1a, which positively regulates Sry expression, also resulted in XY female mice [10]. Furthermore, it is known that male-to-female sex-reversed mice are produced when the Mus musculus domesticus or poschiavinus-type Y chromosome is transferred to the C57BL/6 (B6) background, as in the B6-YAKR, B6-YPOS, or B6-YTIR models [11–15]. Specifically, these XY female mice exhibit an anatomically female phenotype with ovary formation; however, they are infertile or subfertile, exhibiting extensive loss and dysfunction of germ cells in the ovary [4, 7, 8, 16–20]. Overall, it is evident that XY females form well-constructed fetal gonads consisting of germ cell cysts and surrounding somatic cells, including pre-granulosa cells and stromal cells, but fertilized XY oocytes are not capable of early embryonic development [8, 18, 21, 22].
In spite of a growing number of studies of male-to-female sex-reversed mice, a consensus has failed to converge on the phenotypic features of XY females due to the variety of genetic backgrounds analyzed in these studies. Furthermore, little is known about the molecular etiology of oocyte dysfunction and impaired fertility in XY females. Thus, to overcome the confounding influence of variable genetic backgrounds, and to identify molecular mechanisms that underlie the fertility problems of male-to-female sex-reversed mice, we produced Sry-mutated XYsry− females using the CRISPR/Cas9 system on an inbred C57BL/6 background. To define germ cell dysfunction in XYsry− females, we performed comprehensive transcriptomic analyses of germ cells and gonadal somatic cells. The present study shows potential molecular causes that underlie germ cell disruption in sex-reversed mice, demonstrating that primordial germ cells (PGCs) of sex-reversed mice substantially deviate from normal developmental processes in stages of mitotic proliferation prior to entry into the stage of meiotic differentiation.
Materials and methods
Animals and ethic statements
This study was carried out in strict accordance with the Tokyo University of Agriculture Guide for Care and Use of Laboratory
Animals. The protocol was approved by the Committee on the Ethics of Animal Experiments of the Tokyo University of Agriculture (Permit Number: 260064SE). At the time of sample collection, all animals were sacrificed by cervical dislocation, and all efforts were made to minimize suffering.
Generation of Sry mutant mice (XYsry− females) by CRISPR/Cas9 system
To generate the Sry targeting vector, we used a pX330-U6-Chimeric BB-CBh-hSpCas9 (pX330) plasmid, kindly provided by Dr Masahito Ikawa (Research Institute for Microbial Diseases, Osaka University). An sgRNA sequence was designated within the SRY HMG-box domain (5′–TGGTGTGGTCCCGTGGTGAG–3′), inserted annealed double-strand DNA with four overhangs into the BbsI site of the pX330 plasmid. C57BL/6 female mice were superovulated and mated with C57BL/6 male mice; then, zygotes were collected from ampulla of the oviducts. The microinjection of the Sry-targeting pX330 plasmid (2 ng/μl) into male pronuclei of PN3-5 stage zygotes was carried out under standard procedures. The injected zygotes were cultured in potassium simplex optimized medium (KSOM) at 37°C and embryos that developed into the two-cell stage were transferred into the oviducts of pseudopregnant ICR female mice (Supplementary Figure S1).
Germ cells and gonadal somatic cells collection
To collect E13.5 PGCs, postneonatal day 1 (P1) oocytes, and gonadal somatic cells, gonads from 13.5 dpc and 1 dpp post-embryo transfer were digested in 1 mg/ml collagenase solution (Wako, Oosaka, Japan) at 37°C for 40 min, followed by treatment with 0.25% trypsin-EDTA solution (0.53 mM; Sigma, MO, USA) at 37°C for 15 min. After adding foetal bovine serum, a single-cell suspension was obtained by gentle pipetting. Cells were then incubated in a 1:50 dilution of PE-conjugated anti-SSEA1 (for labeling of E13.5 PGCs; 560142, BD Pharmingen, NJ, USA) or PE (Phycoerythrin)-conjugated anti-CD117 (for labeling of P1 oocytes, 105807, BioLegend, CA, USA) antibody. PE-positive cells (PGCs and oocytes) and PE-negative cells (gonadal somatic cells) were isolated and collected using a FACSAria II cell sorter (BD Bioscience, NJ, USA, Supplementary Figure S1). Ovulated MII oocytes were extracted from the oviducts of XX and XYsry− female mice according to the methods described in our previous report [23]. Considering the variability in the genotype of the XYsry− females, when collecting PGCs, neonatal oocytes, gonadal somatic cells, and MII oocytes, we supplied pooled gonadal cell suspensions and MII oocytes from, respectively, four and five XYsry− female individuals at each time point. MII oocytes were also collected from superovulated XX and XYsry− females at 4 weeks of age for single-oocyte RNA-seq.
RNA isolation, RNA-seq library preparation, and sequencing
Total RNA of PGCs, oocytes, and gonadal somatic cells was isolated using an RNeasy Micro Kit (QIAGEN, Hilden, FRG) with DNase treatment. Complementary DNA synthesis and pre-amplification were performed with total RNA (10 ng) using, respectively, a SMARTer Ultra Low Input RNA Kit and an Advantage 2 PCR Kit (Clontech, CA, USA), both according to the manufacturers’ instructions. Pre-amplified cDNA was fragmented into 200-bp fragments using an S2 sonicator (Covaris, MA, USA) and then used to construct sequencing libraries using a NEBNext Ultra DNA Library Prep Kit according to the manufacturer's protocol (New England BioLabs, MA, USA). Two biological replicates were used for each sample.
To perform single-cell RNA-seq analyses of 4-week-old XX and XYsry− MII oocytes, single and bulk (n = 20–22) MII oocytes from oviducts of respective genotypes were picked by glass pipettes and were immediately lysed in Clontech Lysis Buffer. The whole lysate was supplied for cDNA synthesis and RNA-seq library preparation with, respectively, a SMARTer Ultra Low Input RNA Kit (Clontech, CA USA) and Nextera XT library prep kit (Illumina, CA, USA), each according to the manufacturer's instructions. Indexed libraries were pooled (10 nM each) and sequenced using an Illumina HiSeq 2500 System under single-end, 100-bp conditions.
Statistical analyses
All bulk RNA-seq data were collected from two replicates. To detect differentially expressed genes between XX and XYsry− female samples, we employed two criteria; fold change (FC) > 2 and moderated t-test with a Benjamini-Hochberg false-discovery rate (FDR) of P < 0.05. For comparison of gene expression levels in multiple groups, one-way ANOVA and Tukey–Kramer tests were used. Statistically significantly differences in populations between two samples were detected by chi-squared and Fisher exact tests. A Student t-test was used to compare mean values of cell number and oocyte diameters between two samples. A Fisher exact test was also used for detection of significantly enriched Gene Ontology (GO) term and KEGG pathway. Exact numbers of animals used and data analyses are described in the figure legends. All statistics were performed by using StrandNGS software (Agilent, CA, USA), R packages or the DAVID web tool. Details of other standard methods and reagents are described in the “Supplementary Experimental Procedures.”
Accession number
The RNA-seq data from this study have been deposited in the DDBJ under the accession numbers DRA005345 and DRA005822.
Results
Phenotype of Sry-targeted mice
To identify molecular mechanisms that underlie the fertility problems of sex-reversed XY female mice on an inbred C57BL/6 background, we deleted a portion of the Sry sequence adjacent to the transcription start site via the CRISPR/Cas9 system (Supplementary Figure S2A). Sequence analyses detected either small deletions, ranging from 1 to 14 bps (n = 63, 95%), or large insertions of ≥ 500 bps (n = 3, 5%) in the Sry gene (Supplementary Figure S2B and C). Of these, 7 bp deletions (n = 23, 41%) were the most frequent. In total, we generated and examined 65 sex-reversed XYsry− female mice for our analyses. Detailed information regarding deleted bases and position is shown in Supplementary Figure S2C. As designed the deletion started from 3 bases after a specific protospacer adjacent motif (PAM) sequences. Importantly, by converting DNA nucleotide sequences to amino acid sequences, we confirmed that all mutant lines contain frameshift mutations within the HMG-box coding region of Sry and resulting in loss of function. This result revealed that our CRISPR strategy is extremely efficient for producing functionally Sry-deficient mice (37.8%, 65 of 172 XY pups, Supplementary Figure S2D) in comparison with gene targeting by homologous recombination using ES cells.
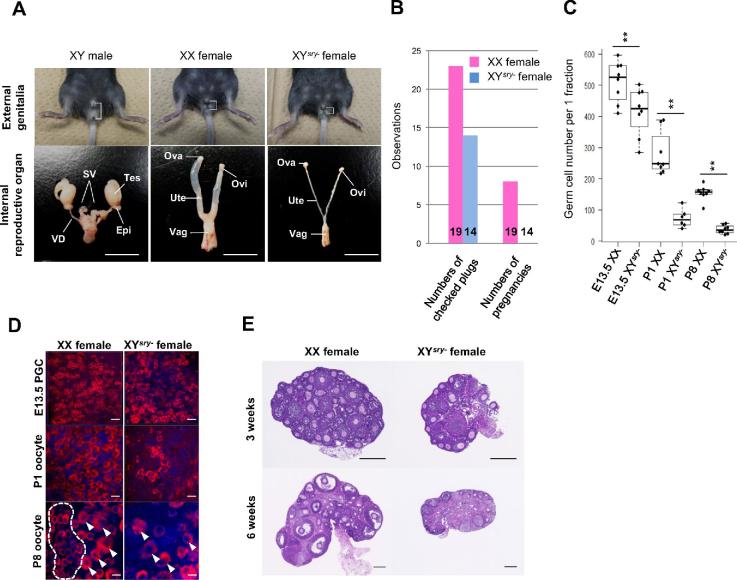
In all cases, these mice presented as anatomically normal females, although we detected aplasia in their reproductive organs (Figure 1A). Moreover, we confirmed that our XYsry− females showed estrus cycle through vaginal smear checks (data not shown). Therefore, it was suggested that XYsry− females are hormonally receptive to males. Mating tests of 14 of the XYsry− females (8–10 weeks of age) with normal XY males revealed that the sex-reversed mice were uniformly infertile (Figure 1B). To determine the cause of infertility, we examined the numbers of PGCs and oocytes in XX and XYsry− females. Strikingly, we noted a significant decrease in PGCs by E13.5 in the XYsry− females (Figure 1C); subsequent oocyte numbers at P1 were decreased to one-fifth that of controls, and P8 oocyte numbers were decreased to one-sixth that of controls. Immunofluorescence staining of DDX4, a germ cell marker, indicated that fewer germ cells were apparent in XYsry− female gonads. In particular, most primordial follicle oocytes were lost by P8 in XYsry− females (Figure 1D). The decrease in PGCs in XYsry− female ovaries progressed quickly after mid gestation, finally resulting in ovarian atrophy accompanied by oocyte depletion by 6 weeks of age (Figure 1E). These results revealed that the infertility of sex-reversed XYsry− female mice was due to a complete depletion of germ cells.
Figure 1.
XYsry− female mice evince infertility and oocyte depletion. (A) Representative images of external genitalia (top) and internal reproductive organs (bottom) of an XY male (left), an XX female (mid), and an XYsry− female (right) at 8 weeks of age, derived from the same litter. XYsry− female mice were characterized by a decreased ano-genital distance (white bracket) and a feminized internal reproductive structure. Tes, testis; Epi, epididymis; SV, seminal vesicle; VD, vas deferens; Ova, ovary; Ovi, oviduct; Ute, uterus; Vag; vagina. Scale bar, 1 cm. (B) Fertility tests of XX and XYsry− females. The left bar graph shows the total number of observed plugs; the right bar graph illustrates the total number of pregnancies. The number of tested XX and XYsry− females is shown at the bottom of respective bars. (C) Boxplot of the total oocyte number per one fraction (x: 212.2 μm × y: 212.2 μm × z: 25.0 μm) in XX and XYsry− female gonads (Student t-test, **P < 0.01). (D) Representative images of immunostained germ cells at different developmental stages (E13.5, P1, and P8) in XX and XYsry− female gonads. PGCs and oocytes were stained with an antibody against a germ-cell-specific marker (DDX4, shown in red) and counterstained with DAPI (blue). White dashed circles and arrowheads indicate primordial follicle pools (oocyte reserves) and primary follicles (which begin growth and development) at P8. Scale bar, 20 μm; magnification, ×40. (E) Representative hematoxylin and eosin-stained sections of XX and XYsry− female ovaries at 3 and 6 weeks of age. Scale bar, 200 μm.
Transcriptome analysis of XYsry− germ cells
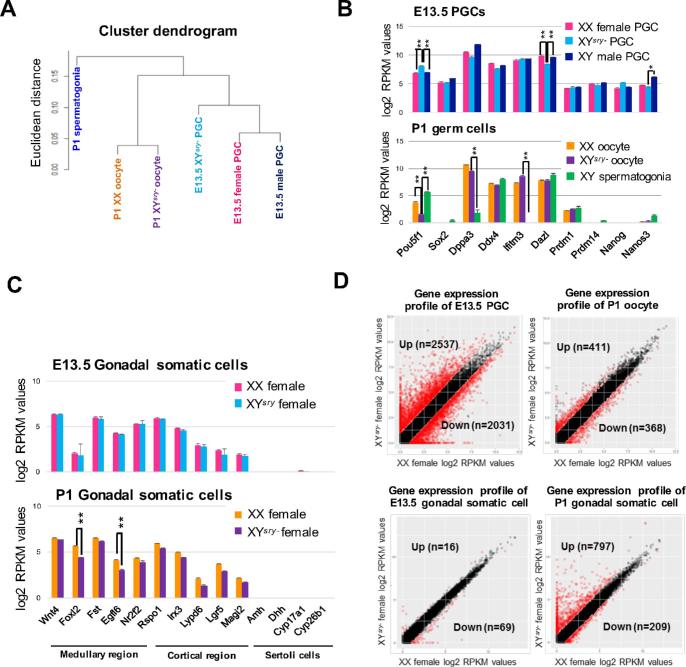
To understand the mechanisms that underlie the depletion of germ cells in XYsry− females, we conducted transcriptome analyses of germ cells and gonadal somatic cells. RNA-seq datasets provided accurate gene expression profiles; summaries of the gene expression profiles are shown in Supplementary Tables S2 and S3. Hierarchical cluster analyses revealed that gene expression profiles segregated into two groups representing E13.5 PGCs and P1 oocytes, and that both profiles of the XYsry− female germ cells could be clearly distinguished from those of XX females and XY males (Figure 2A, Supplementary Figure S3). Previously, we demonstrated pronounced sex-specific gene expression profiles in E13.5 female and male PGCs, and we determined groups of female PGC-specific expressed genes (FSGs) and male PGC-specific expressed genes (MSGs) [24]. Using these lists of FSGs and MSGs, the present transcriptome data revealed that many FSGs were significantly upregulated in XYsry− PGCs at E13.5 (n = 245/651), while many MSGs were significantly downregulated (n = 157/428: data not shown).
Figure 2.
Extensive transcriptional changes in XYsry− germ cells. (A) Unsupervised hierarchical cluster of all transcript profiles (n = 36 172) from six RNA-seq datasets. (B) Ten germline-specific gene expression patterns (mean ± SEM) from each germ cell RNA-seq dataset at E13.5 (top) and P1 (bottom) with two replicates per sample. Asterisks indicate statistically significant differences between XX WT and XYsry− females (moderated t-test, **P < 0.01). (C) Gonadal somatic cell-marker gene expression patterns (five medullary cell markers, five cortical cell markers, and four Sertoli cell markers; mean ± SEM) from each gonadal somatic cell RNA-seq dataset at E13.5 (top) and P1 (bottom) with two replicates per sample. Asterisks indicate statistically significant differences between WT and XYsry− females (moderated t-test, **P < 0.01). (D) The upper two panels illustrate scatter plots of log2 RPKM values of WT (X-axis) and XYsry− germ cell RNA-seq samples (Y-axis) obtained at E13.5 and P1. The lower two panels illustrate scatter plots of log2 RPKM values of WT (X-axis) and XYsry– gonadal somatic cells RNA-seq samples (Y-axis) obtained at E13.5 and P1. Red circles indicate genes found to be differentially expressed (≥twofold difference in the expression level; moderated t-test, significance level of ≤0.05).
Transcriptome profiles were very similar between XX and XYsry- females, while the several marker genes for PGCs (Figure 2B) and gonadal somatic cells (Figure 2C) in XYsry− females were differentially expressed. The expression of POU domain, class 5, transcription factor 1 (Pou5f1), which encodes a transcription factor necessary for germ cell differentiation and survival [25], was significantly upregulated in E13.5 PGCs and downregulated in P1 oocytes of XYsry− females. As well, deleted in azoospermia-like (Dazl), which encodes an RNA-binding protein critical for the mitosis-to-meiosis transition [26], was significantly decreased in XYsry− PGCs (Figure 2B). In P1 XYsry- female gonadal somatic cells, expression of forkhead box L2 (Foxl2), a gene that encodes a forkhead transcription factor required for female sex differentiation and ovarian function [27], and expression of EGF-like-domain, multiple 6 (Egfl6), which encodes an epidermal growth factor implicated in ovarian differentiation [28], were both significantly decreased (Figure 2C). As expected, the expression of representative Sertoli cell-marker genes was no longer detectable in XYsry− females (Figure 2C). These findings demonstrate that the dysregulation of several genes required for germ cells and gonadal somatic cells was associated with germ cell defects in XYsry− females.
Next, we focused on differentially expressed genes in E13.5 XYsry− PGCs, which were screened based on two parameters: (1) a FC of > 2, and (2) statistical significance determined using a moderated t-test with a Benjamini–Hochberg FDR of <0.05 (Figure 2D). In E13.5 XYsry−PGCs, 2537 (21% of all transcripts) were upregulated and 2031 (17% of all transcripts) genes were downregulated (Figure 2D, Supplementary Table S4). In P1 oocytes, the number of upregulated genes decreased to 411 (3.2%), and the number of downregulated genes decreased to 368 (2.9%; Figure 2D, Supplementary Table S5). Of the genes that underwent significant changes in expression, most were classified as protein-coding (84%–98% of all transcripts, Supplementary Figure S4A). Interestingly, we observed an enriched proportion of noncoding RNAs (11.3%) among the downregulated genes of E13.5 XYsry− PGCs (Supplementary Figure S4A). We list the 20 genes with the greatest statistical significance in the PGCs and oocytes of XYsry- females in Supplementary Figure S4B and C. Relatively low numbers of differentially expressed genes (16 upregulated and 69 downregulated) were detected in gonadal somatic cells of E13.5 XYsry− females (Figure 2D, Supplementary Table S6). However, we observed a marked increase in the numbers of differentially expressed genes in the P1 stage, particularly for upregulated genes (797 upregulated and 209 downregulated, Figure 2D; Supplementary Table S7). These results demonstrate that sex-reversed germ cells and gonadal environments conceal transcriptional anomalies prior to obvious phenotypic defects.
These findings raised the possibility that significant changes to the epigenome lead to global gene expression changes. To test this possibility, we examined the expression of DNA methylation- and histone modification-related genes in E13.5 XYsry− PGCs. No significant changes were found amid DNA methylation- and demethylation-related genes, save for Dnmt3b and Tet3, whose expression were very low even in WT female PGCs. However, expression of genes related to histone modifications implicated in both active transcription and repression underwent significant changes in many cases (Supplementary Figure S5). While Suv39h1, an H3K9 methyltransferase, was downregulated in the XYsry− females, Kdm4a, an H3K9 demethylase, was upregulated in XYsry− females. Such changes in expression have the potential to decrease H3K9 methylation in the germ cells of XYsry− females. In addition, Kdm6a, an H3K27 demethylase, was downregulated, while Ring1a, an H2AK119 ubiquitin ligase, was upregulated.
Y-linked genes
Contrary to XY sex-reversed female mice, XO female mice are fertile [29, 30], although this fertility does not apply to humans. Therefore, it can be assumed that the immediate cause of XYsry− infertility is deleterious effects from the Y chromosome which result in transcriptional anomalies for autosomal genes. To investigate this hypothesis, we examined the expression levels of Y-linked genes in XYsry− germ cells at the E13.5 and P1 stages via RNA-seq analyses, comparing Y-linked gene expression patterns based on the different subcellular conditions between oocytes and spermatogonial cells. Single-copy genes located in the Y chromosome MSYp region (schematic shown in Supplementary Figure S6A) were expressed at similar levels to those of XY male germ cells, although the expression levels of Zfy1, Elf2s3y, and Uty were significantly decreased in E13.5 XYsry− PGCs (Supplementary Figure S6B). The expression of multicopy genes such as Rbmy1a1 was also significantly decreased in XYsry− germ cells, especially in P1 oocytes. These results raise the possibility that the expression of particular Y-linked genes in XYsry− females is deleterious in XYsry− female germ cells.
Causes of primordial germ cell and oocyte depletion
The number of PGCs at E13.5, when proliferation has ceased [25], was already significantly lower in XYsry− females (Figure 1C), suggesting that XYsry– PGCs evinced decreased proliferation compared with that of XX females and, thus, could not reach the maximum number. Furthermore, the number of PGCs in XYsry− females decreased to one-fifth of XX females at the P1 stage. Gene ontology and functional pathway analysis using differentially expressed gene sets provided valuable information toward understanding the mechanisms that underlie germ cell depletion in XYsry− females (Supplementary Figure S7, Supplementary Tables S8 and S9). Up- and downregulated genes were enriched for the following categories of biological processes (Supplementary Figure S7A and B): For upregulated genes, the categories were “Tissue morphogenesis” and “Enzyme linked receptor protein”; for downregulated genes, the categories were “DNA metabolism,” “Cell cycle process,” and “Chromosome organization.” GO term interaction analysis formed three large networks with regard to “Regulation of gene expression,” “Tissue morphogenesis,” and “Cell cycle and cell division” (Supplementary Figure S7C).
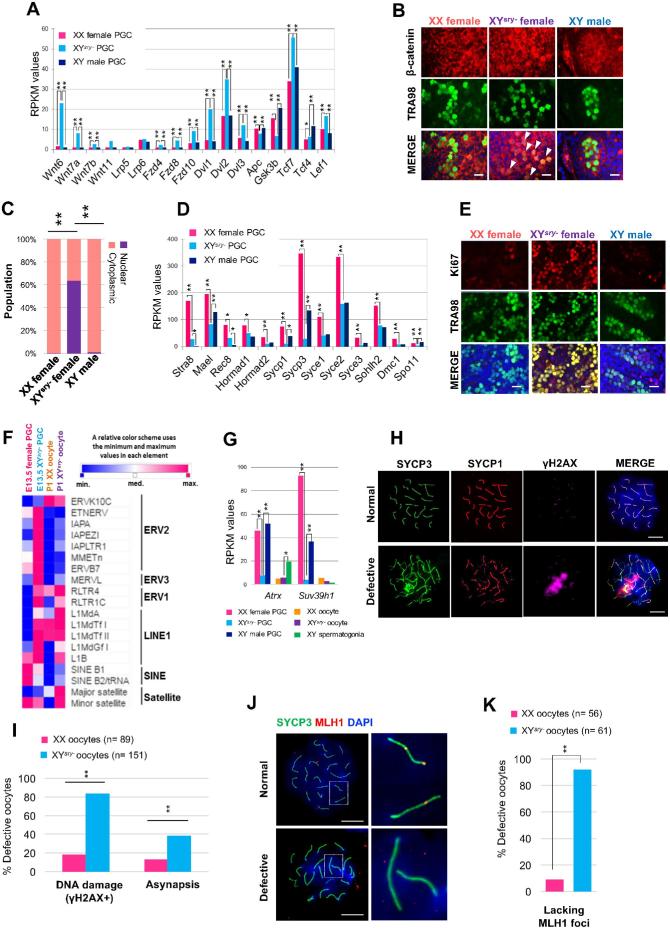
In particular, many “Wnt/β-catenin signal”-related genes were upregulated (Figure 3A, Supplementary Table S4) in PGCs of XYsry− females at E13.5. Although expression of β-catenin (Ctnnb1) was not changed (Supplementary Table S2), immunofluorescence staining revealed that the β-catenin protein was specifically located in the nuclei of PGCs in XYsry− females but not in XX females (Figure 3B and C). This observation suggests that β-catenin has an adverse effect on cell proliferation. Conversely, our observation of downregulation of many genes related to meiosis suggests that the transition of PGCs to meiosis at E13.5 was delayed in XYsry− females (Figure 3D, Supplementary Table S4). In support of this, immunofluorescence of Ki67, which is expressed by proliferating cells in all phases of the active cell cycle (G1, S, G2, and M), revealed that many germ cells in XYsry− females are still actively proliferating at E14.5 (Figure 3E). At this stage, XX female germ cells enter meiosis while XY male germ cells become quiescent; thus, the expression of a proliferation marker in E14.5 XYsry– germ cells might cause the delayed entry into meiosis.
Figure 3.
Meiotic defects of XYsry− germ cells that accompany the impairment of cell cycle progression and the activation of retrotransposable elements. (A) Expression patterns of components of the Wnt signaling pathway from each E13.5 PGC RNA-seq dataset with two replicates per sample. Asterisks indicate statistically significant differences between RPKM values in E13.5 female, XYsry−, and male PGCs (Tukey–Kramer test, *P < 0.05, **P < 0.01). (B) Representative images of immunostained E13.5 gonads. PGCs were stained with antibodies against a germ-cell-specific marker (TRA98, green) and β-catenin (red), and counterstained with DAPI (blue). White arrowheads show XYsry− PGCs that evince stabilized localization of β-catenin in nuclei. Scale bar, 20 μm; magnification, ×40. (C) The pattern of subcellular localization of β-catenin in E13.5 PGCs from XX female and XYsry−, and an XY male. Localization patterns were assigned as predominantly nuclear or cytoplasmic. (D) Expression patterns of meiosis-related genes from each E13.5 PGC RNA-seq dataset with two replicates per sample. Asterisks indicate statistically significant differences between RPKM values in E13.5 female, XYsry−, and male PGCs (Tukey–Kramer test, *P < 0.05, **P < 0.01). (E) Representative images of immunostained E14.5 gonads. PGCs were stained with anti-TRA98 and anti-Ki67 (cell proliferation marker, red) antibodies, and counterstained with DAPI. Scale bar, 20 μm; magnification, ×40. (F) Expression of representative retrotransposable elements in E13.5 and P1 germ cells. Data are represented by a heatmap of the relative expression levels of each retrotransposable element. (G) Expression of Suv39h1 and Atrx from each germ cell RNA-seq dataset with two replicates per sample. Asterisks indicate statistically significant differences between RPKM values in XX female, XYsry−, and male germ cells (Tukey–Kramer test, *P < 0.05, **P < 0.01). (H) Immunofluorescence analysis of E17.5 pachytene oocytes from XX and XYsry− females (n = 3 for each) stained with anti-γH2AX (magenta), anti-SYCP3 (green), and anti-SYCP1 (red) antibodies, and counterstained with DAPI. Representative images, termed “Normal” and “Defective,” were derived from, respectively, XX and XYsry− oocytes. Scale bar, 10 μm; magnification, ×60. (I) The percentage of defective oocytes with DNA damage (γH2AX+) and extensive chromosomal asynapsis. Asterisks indicate statistically significant differences between XX and XYsry− oocytes (Fisher exact test, **P < 0.01). (J) Immunofluorescence analysis of E17.5 pachytene stage oocytes from XX and XYsry− females (n = 3 for each) for MLH1. Oocytes were stained with anti-SYCP3 (green) and anti-MLH1 (red) antibodies, and counterstained with DAPI. Representative images, termed “Normal” and “Defective,” were derived from, respectively, XX and XYsry− oocytes. Magnified images of dashed rectangles are shown to the right. Scale bar, 10 μm; magnification, × 60. (K) Percentage of defective oocytes lacking MLH1 foci. Asterisks indicate statistically significant differences between XX and XYsry− oocytes (Fisher exact test, **P < 0.01).
Another interesting feature of the gene expression change in PGCs of XYsry− females at E13.5 is the elevation of apoptosis-related genes. For example, the Tnf family, including tumor necrosis factor receptor superfamily member 12A (Tnfrs12a), evinced elevated expression (Supplementary Figure S8A, Supplementary Table S4). Additionally, mRNA expression of the cell death makers Tnfrsf21, Traf3, Traf4, Traf6, Ripk2, and Casp8 were increased in XYsry− PGCs as well. However, the apoptotic cell populations were not significantly changed in E13.5 XYsry− PGCs compared with WT controls (Supplementary Figure S8B and C).
Interestingly, expression levels of many endogenous retrovirus (ERVs) and LINE-1 retrotransposons were notably elevated in E13.5 XYsry− PGCs (Figure 3F). ERV1 and satellite DNA were upregulated in P1 oocytes. Furthermore, α-thalassemia mental retardation X-linked (Atrx), which is involved in retrotransposable element silencing, and suppressor of variegation 3–9 homolog 1 (Suv39h1), which is an H3K9 methyltransferase involved in chromatin remodeling of constitutive heterochromatin structures, were both markedly repressed in E13.5 XYsry− PGCs (Figure 3G, Supplementary Table S4).
Next, we asked whether the aberrant expression of retrotransposable elements is associated with meiotic defects in the fetal germ cells of XYsry– females. To this end, we performed immunofluorescence analyses of chromosome spreads from E17.5 oocytes. At this stage, a majority of cells are in the midst of the pachytene stage, when synapsis of homologous chromosomes has completed as determined by immunofluorescence against synaptonemal complex protein 3 (SYCP3), a structural component of meiotic chromosome axes. Immunofluorescence against histone H2AX phosphorylated at Serine 139 (γH2AX), a marker of DNA damage signaling, demonstrated that 83.3% of XYsry− oocytes evinced γH2AX foci compared to 18.2% in the XX oocytes (Figure 3H and I), suggesting that an ectopic accumulation of DNA damage signaling takes place in XYsry− oocytes. Based on immunofluorescence against SYCP3 and SYCP1, a central structural component of meiotic chromosome axes, chromosomal asynapsis was detected in 38.1% of XYsry− oocytes, compared to 11.9% in the XX oocytes (Figure 3H and I). These results suggest that asynapsis was increased in XYsry− oocytes in response to ectopic DNA damage signaling. Furthermore, immunofluorescence against MLH1, a marker of sites of recombination crossover formation, revealed that 91.8% of XYsry− oocytes were lacking MLH1 foci compared to 8.9% in the XX oocytes (Figure 3J and K), indicating an extensive impairment of crossover formation in XYsry− oocytes.
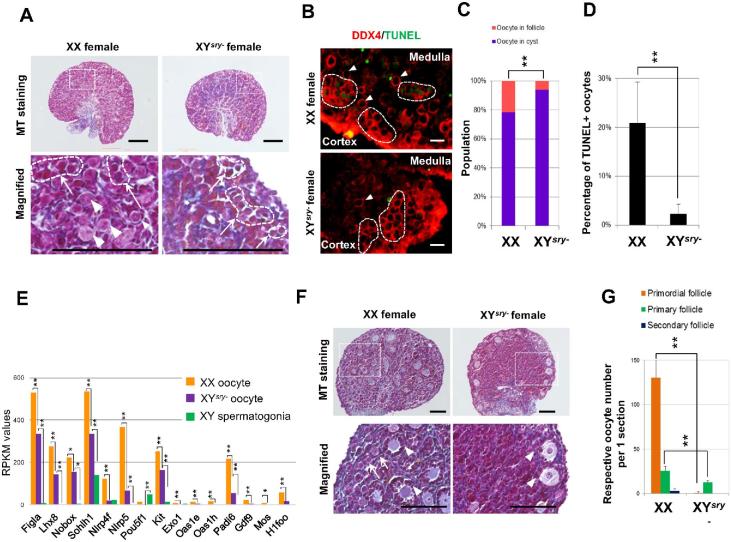
In accord with widespread meiotic defects, histopathological analysis (Figure 4A) and immunofluorescence staining against DDX4 (Figure 4B, Supplementary Figure S9) demonstrated that only a few oocytes were observed in the cortical region of P1 XYsry− ovaries, and barely any oocytes were detected in the medullary region (Supplementary Figure S9). Masson trichrome (MT) staining showed that connective tissues and collagen fibers were increased in the medullary region of XYsry− ovary (Figure 4A). Interestingly, XYsry− oocytes in the cyst were apparently smaller than those of XX females, and the proportion of the oocytes in the cyst was significantly high (Figure 4C), suggesting that oocyte cyst breakdown was largely impaired. Furthermore, a high proportion of apoptotic oocytes was detected in the cortical region of control XX ovaries, although only a few were detected in XYsry− ovaries at P1 (Figure 4B and D, Supplementary Figure S9). These observations were supported by transcriptome data. Many “Cell adhesion” -related genes and folliculogenesis-related genes were dysregulated in P1 XYsry− oocytes (Figure 4E, Supplementary Table S8). GO analysis revealed that upregulated genes in the P1 XYsry− oocytes were enriched with particular annotations, including “Cell adhesion” and “Negative regulation of apoptosis” (Supplementary Table S8). Furthermore, histopathological analysis of the P8 ovary demonstrated that only a few growing primary follicles exist in the cortical region, and the primordial oocyte reserve mostly disappeared from the entirety of P8 XYsry− ovaries (Figure 4F and G). These findings suggest that oocyte cyst breakdown and subsequent maintenance of oocyte pools were largely impaired in neonatal XYsry− ovaries.
Figure 4.
Slowing of oocyte-cyst breakdown and elimination of primordial follicles in neonatal XYsry- ovaries. (A) Representative Masson trichrome (MT) staining of ovarian sections of XX and XYsry− females at P1. Bottom panels are magnified images of dashed rectangles depicted in the upper panels. White arrows/dashed circles indicate oocyte cysts; arrowheads indicate primordial follicles. Scale bar, 100 μm. (B) TUNEL assay. Representative images of immunostained ovaries from XX and XYsry− females at P1. Ovaries were stained with an antibody against a germ-cell-specific marker (DDX4, red) and TUNEL (an apoptotic cell marker, green). White arrows/dashed circles indicate oocyte cysts; arrowheads indicate primordial follicles. Scale bar, 20 μm; magnification, ×40. (C) Population percentages of oocytes in either cysts or oocytes in follicles from XX and XYsry− ovaries at P1. Asterisks indicate statistically significant differences between the groups (chi-squared test:, **P < 0.01, n = 3). (D) Percentages of TUNEL-positive oocytes from P1 XX and P1 XYsry- ovarian sections. Percentages of apoptotic oocytes (TUNEL-positive) are shown as mean ± standard error of the mean. Asterisks indicate a statistically significant difference between the groups (Student t-test, **P < 0.01, n = 3). (E) Expression patterns of folliculogenesis-related genes from each P1 germ cell RNA-seq dataset with two replicates per sample. Asterisks indicate statistically significant differences in XX oocytes, XYsry− oocytes, and XY spermatogonia (Tukey–Kramer test, *P < 0.05, **P < 0.01). (F) Representative MT sections of ovaries of P8 XX and XYsry− females. Bottom panels represent magnified images of dashed rectangles in the upper panels. White arrows indicate primordial follicles; white arrowheads indicate primary follicles. Scale bar, 100 μm. (G) Numbers of respective oocytes per one ovarian section at P8. Asterisks indicate a statistically significant difference between XX and XYsry− oocytes (Student t-test, **P < 0.01, n = 3).
Mature oocyte dysfunction
XYsry− females were infertile but possessed antral follicle oocytes at 3 weeks of age (Figure 1E). To determine the capabilities for fertilization and embryo development of XYsry– antral follicle oocytes after ovulation, we first examined whether the XYsry− females respond to exogenous gonadotropins administered at 4 weeks. The ovulated XYsry− oocyte diameters (including the zona pellucida) were similar to those of WT oocytes (Supplementary Figure S10A), although the number of ovulated oocytes was less than half of the WT (XX n = 3: 51.5 ± 2.5 (mean ± SEM) vs XYsry− n = 3: 20.5 ± 9.1 (mean ± SEM), Student t-test P < 0.01). MII oocytes evinced aberrant spindle formation: diffused poles, skewed spindles, disrupted spindles, and misaligned and decondensed chromosomes (Supplementary Figure S10B and C). After in vitro fertilization (n = 3), 34.7% (16/46) of the MII oocytes showed the capability for fertilization and formed male and female pronuclei. Ten developed to the 4-cell stage, and one grew to a blastocyst (Supplementary Figure S10D). Immunostaining against OCT4 showed that OCT4-positive cells were observed even in the outer cell layer of the XYsry− female-derived blastocyst (Supplementary Figure S10E). This result suggests that differentiation into inner cell mass cells and trophectoderm cells failed in the XYsry− female-derived embryo.
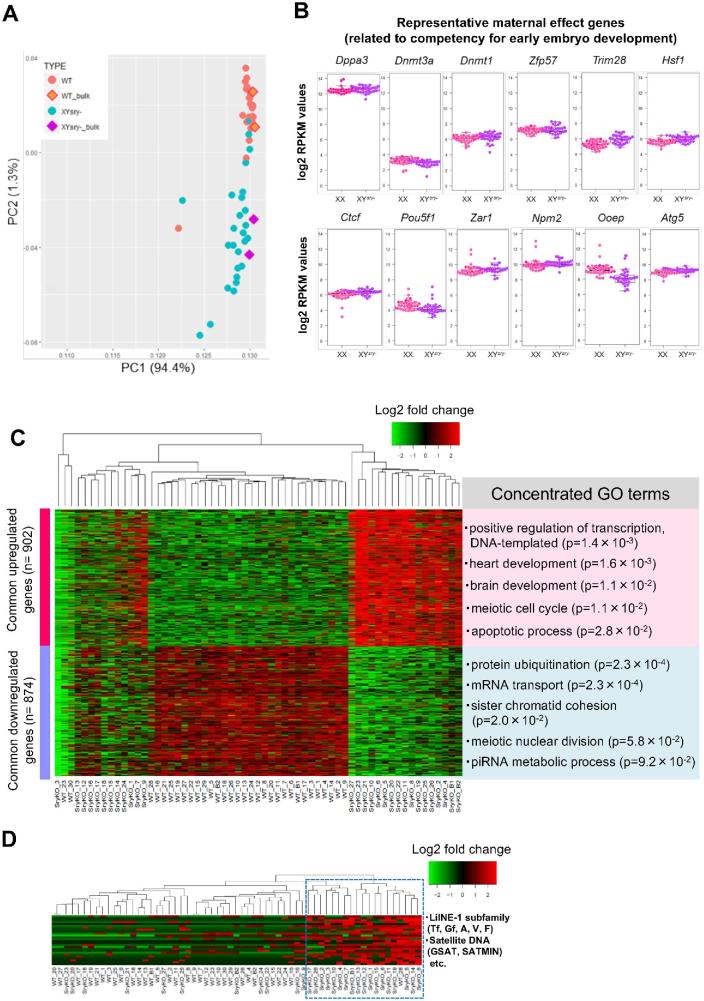
To unravel molecular mechanisms underlying the mature oocyte dysfunction in XYsry− females, we conducted single-oocyte transcriptome analyses using 27 XYsry− and 30 XX oocytes. Principal component analysis revealed almost identical gene expression profiles in XX single-oocyte libraries (Figure 5A, Supplementary Table S10). However, each XYsry− oocyte profile largely varied for the second principal component (PC2) (Figure 5A). The maternal effect genes, which are important for early development, were expressed at similar levels to those of XX oocytes (Figure 5B). K-means clustering analyses revealed that 15 XYsry− oocytes were classified into a cluster distinct from the WT cluster consisting of 29 oocytes (Figure 5C). The other mutant oocytes formed a subcluster that differed from the WT cluster. GO analysis using differentially expressed gene sets from XYsry− oocytes provided terms related to oocyte dysfunctions (Figure 5C); for example, upregulated genes included the categories “meiotic cell cycle” and “apoptotic process”; on the other hand, downregulated genes included the categories “protein ubiquitination,” “sister chromatid cohesion,” and “meiotic nuclear division.” Furthermore, retrotransposable elements such as LINE-1, ERV, and satellite DNA were highly expressed in 18 XYsry− oocytes, and only one WT oocyte was classified in this cluster (Figure 5D, Supplementary Table S12). Thus, single-oocyte RNA-seq data demonstrated that most XYsry− MII oocytes exhibited extensive transcriptional anomalies, which likely impair the progression of the second meiotic division and early development.
Figure 5.
Gene expression profiling of MII oocytes from XYsry− females via single-cell transcriptome analysis. (A) Bidimensional principal component analysis of gene expression profiles of XX (n = 30) and XYsry− (n = 27) single oocytes. The bulk oocyte (from 20 oocytes, n = 2) data are shown in the square. The first principal component (PC1) captures 94.4% of the gene expression variability, and the second principal component (PC2) captures 1.3% of the gene expression variability. (B) Boxplots of log2 RPKM values for 12 representative maternal effect genes in XX (pink) and XYsry− oocytes (purple). With respect to the expression of each gene, no significant difference was observed between the two groups. (C) Heatmap of the relative expression of the 902 common upregulated and 874 common downregulated genes expressed in each XYsry− MII oocyte as identified by k-means clustering. Representative GO terms and the enrichment P values are shown at right (Fisher exact test). WT, XX female; SryKO, XYsry− female. (D) Heatmap shows relative expression of 20 LINE-1 subfamilies, 3 ERV2 elements, and 3 forms of satellite DNA in XYsry− MII oocytes as identified using k-means clustering. The blue dashed rectangle indicates a cluster comprised of highly expressed retorotransposable elements. Notably, XYsry− MII oocytes were highly assigned to this cluster (18/19 (94.7%), Fisher exact test; P < 0.01). WT, XX female; SryKO, XYsry− female.
Discussion
Paradoxically, germ cells from any type of sex-reversed mice—i.e. both XX males and XY females—exhibit morphologically normal sexual features yet, in many cases, germ cell development fails and fertility is severely or entirely impaired [4, 7, 16–19, 31–33]. Here, by combining a detailed phenotypic analysis using contemporary molecular cytogenetic and histological methods with transcriptomic profiling, we provide new insights to understand the basis of germ cell failure.
Impaired primordial germ cell proliferation and the mitosis-to-meiosis transition in XYsry− germ cells
The decrease in XYsry− PGCs follows the transition from the mitotic stage to meiosis—it is conceivable that the Wnt/β-catenin signaling pathway is involved in the decline of PGC proliferation in XYsry− females; this is because a large number of genes that comprise the pathway evince significant increases in expression (Figure 3A). Although the expression of β-catenin itself was not increased, nuclear localization of β-catenin was observed in the XYsry− PGCs. It has been reported that the stable localization of β-catenin in the nuclei of mitotic-stage PGCs blocks the transition from the G1 to S stages, thereby leading to inhibitory effects on germ cell proliferation in mice [34]. Furthermore, the nuclear localization of β-catenin is inhibited by phosphorylation of a portion of the protein encoded by exon 3, and this phosphorylation is catalyzed by glycogen synthase kinase 3β (GSK-3b) [35]. Therefore, the finding that Gsk3b expression was decreased in XYsry− PGCs suggests that β-catenin localization into the nuclei of these cells arises from a lack of inhibition, leading to likely and extensive impacts on gene regulation. In tumor cells, overexpression of β-catenin induces a delay in the cell cycle as well as apoptosis, independent of the actions of Tcf and Lef1 [36, 37]. Therefore, we infer that the Wnt/β-catenin signaling pathway comprises a critical factor in the decreased proliferation of PGCs in XYsry− females.
In mice, PGCs reach their maximum numbers (∼12 000) and enter the prophase stage of meiosis I at E13.5–14.5; afterwards, they continuously decrease in number, resulting in a population of less than one-third of the maximum number of cells around the time of birth [25, 38, 39]. In the present study, we show that most germ cells in XYsry− ovaries were positive for Ki67, suggesting that these ovaries undergo mitotic proliferation. Previous studies demonstrated that expression of pluripotent-related genes, such as Pou5f1 and Nanog, prevented meiotic gene expression and entrance into the meiotic phase [28]; furthermore, Dazl-deficient female PGCs maintained Ki67 signals into even E15.5 [29]. Our transcriptome data demonstrated that expression levels of Pou5f1 were significantly upregulated, while expression levels of Dazl were significantly downregulated in XYsry– PGCs (Figure 2B). These results suggest that up- and downregulation of, respectively, Pou5f1 and Dazl in XYsry− PGCs led to impairment of the mitosis-to-meiosis transition and prolonged the mitotic phase into and beyond E13.5–14.5.
Furthermore, excessive activation of retrotransposable elements preceded meiotic defects in XYsry− pachytene oocytes. We have demonstrated that a marked increase in the expression of LINE-1 and ERV retrotransposons (>10-fold) in XYsry− females is associated with germ cell depletion. Forced enhancement of LINE-1 expression is involved in fetal oocyte attrition [40]. Consistent with previous findings, excessive activation of transposable elements, including LINE-1, in E13.5 XYsry– germ cells precedes such meiotic defects as the ectopic accumulation of DNA damage signaling, increased frequencies of asynapsis, and extensive impairment of crossover formation (Figure 3F, H–K). Moreover, expression levels of Atrx and Suv39h1, which are involved in the suppression of retrotransposable elements via trimethylation of H3K9 and the promotion of heterochromatin formation, were significantly decreased in XYsry– germ cells. These data raise the possibility that the failure to silence retrotransposable elements, as a consequence of downregulation of Atrx and Suv39h1, is followed by meiotic defects.
Deleterious effects of the Y chromosome for XYsry− oogenesis and fertility
The fertility of XO female mice strongly indicates that genes expressed from the Y chromosome are involved in oocyte depletion and impaired fertility in XYsry− females [32]. However, other than the function of Sry, little is known about the function of these genes. To date, mice that bear many types of Y chromosome mutations have been reported [18, 33, 41]. Using a genetic model of XO mice carrying genes located on the short arm of the Y chromosome, the functions of the Y-associated genes have been investigated [18]. As a Y-linked transcriptional regulator, Zfy2 is essential for promoting Meiosis II in male germ cells [33, 42], and ectopic Zfy2 expression in XO oocytes led to frequent Meiosis II defects and preimplantation embryo arrest [22]. Infertility of XO mice expressing Zfy2 indicates that Zfy2 can serve as a malignant factor if it is expressed in oocytes [18].
In our single-oocyte RNA-seq analyses, Zfy2 was expressed in 66.7% (18/27) of XYsry− MII oocytes (Supplementary Table S11). This finding supports a scenario whereby ectopic expression of Zfy2 in XYsry– MII oocytes contributed to oocyte dysfunction and infertility. In spermatogenesis, germ cells that fail to synapse their homologous chromosomes, or that fail to undergo meiotic sex chromosome inactivation, are eliminated by apoptosis during the mid-pachytene stage [33, 43]. Zfy2 was proposed to be a surveillance gene, which acts to remove cells with unpaired chromosomes during meiosis [33]. Therefore, it is intriguing to speculate that ectopic expression of Zfy2 in XYsry− females is involved in the depletion of pachytene oocytes rife with asynapsis.
Moreover, the other five MSYp genes (Zfy1, Eif2s3y, Ube1y1, Uty, Ddx3y) are also expressed in XYsry− mature MII oocytes. As a result of GO analysis using the DAVID web tools [44], it was revealed that Ddx3y and Eif2s3y have functions associated with, respectively, “Chromosome segregation” and “Translation.” Therefore, it was suggested that expression of Y-linked genes in XYsry− oocytes led to frequent meiosis II defects and preimplantation developmental arrest [22]. Transcriptome analyses at E13.5 and P1 revealed that the Zfy1, Ube1y1, Smcy, Eif2s3y, Uty, and Ddx3y genes expressed in XY male PGCs were also expressed in XYsry− PGCs (Supplementary Figure S6B). These findings suggest that Y-linked genes might have detrimental effects on fetal and perinatal oogenesis.
Another possible explanation for impaired oogenesis in XYsry− females is the presence of asynapsed XY chromosomes. In XO female mice, it was shown that oocyte numbers are reduced to nearly half that of XX females in neonatal and developmental periods [45, 46]. Recently, Cloutier et al. showed that oocyte elimination in XO mice is linked to the presence of γH2AX on asynapsed chromosomes [30]. Consistent with this notion, we found that 16.7% of XYsry− oocytes did not evince γH2AX signals (Figure 3I). Previous studies demonstrated that the X chromosome of XO pachytene oocytes frequently engages in nonhomologous self-synapsis, thereby evading the meiotic silencing of unsynapsed chromatin [33]. Thus, it is possible that γH2AX-negative XYsry− oocytes are capable of meiotic progression by nonhomologous self-synapsis of, respectively, the X and Y chromosomes.
Failure of oocyte cyst breakdown and follicular maintenance in XYsry− females
Oocyte loss in postnatal XYsry− females was drastic and distinct; thus, oocyte cyst breakdown may be closely related. It is known that Foxl2-positive medullary granulosa cells are important for precise oocyte cyst breakdown and primordial follicle formation [47, 48]. Knockout of Foxl2 in granulosa cells impairs cyst breakdown by inhibiting granulosa differentiation and the proper establishment of basal lamina around the forming follicle [27]. In this study, expression of Foxl2 and apoptotic oocytes were significantly decreased in XYsry− female gonadal somatic cells compared with WT (Figures 2C and 4D). This might be related to the excessive proliferation of connective tissues and collagen fibres that we observed in the medullary region of P1 XYsry− ovaries (Figure 4A). Thus, regression of granulosa cells and a decrease in Foxl2 expression might be involved in the impairment of oocyte cyst breakdown and primordial follicle formation in neonatal XYsry− ovaries. However, the major elimination of XYsry− female germ cells occurred in the fetal stage via delayed cell cycling, meiotic defects, and excess transposable element activation. On the other hand, decreased apoptotic oocyte in the P1 stage merely indicated the failure of oocyte cyst breakdown. Therefore, we believe that decreased oocyte numbers and decreased apoptotic oocytes at P1 are independent phenomena. Furthermore, it was reported that Foxl2 deficiency was also associated with premature ovarian failure [27, 49]. These results suggest that follicles in the cortical region undergo aberrant activation via decreased expression of Foxl2 in XYsry− ovaries, leading to the precocious exhaustion of oocytes prior to the pubertal stage.
Conclusion
The present study suggests potential molecular causes that underlie germ cell loss in XYsry− sex-reversed female mice. These results should contribute to our understanding of disorders of sex development in humans, such as Swyer syndrome, in which patients with an XY karyotype present as anatomically female and exhibit infertility consequent to mutations in Sry and Map3k1 [50].
Supplementary data
Supplementary Figure S1. Schematic representation of germ cell collection by FACS for RNA-seq. Illustrations of experimental procedures for the generation of XYsry− female mice and collection of germ cells by FACS for RNA-seq. Approximately 2 to 3 pl of 2 ng/μl Sry-targeting pX330 plasmid solution was injected into the male pronuclei of zygotes, which were then cultured in KSOM. Cells that reached the 2-cell embryo stage were transferred into the oviducts of recipient mice. XYsry− female gonads at the E13.5 and P1 stages were identified by PCR amplification of Xist and Zfy loci and suspended by collagenase and trypsin-EDTA treatments. PGCs and oocytes were stained with phycoerythrin (PE)-conjugated anti-SSEA1 and anti-CD117 (KIT) antibodies. Following immunostaining, PE-positive cells were collected via FACS sorting. Confocal microscopy images of collected PGCs and oocytes.
Supplementary Figure S2. Generation of Sry-mutated mice via the CRISPR/Cas9 system and types of mutations. (A) Schematic diagram of the mouse Sry locus. Start codon, HMG box domain coding region, gRNA sequence, and PAM sequence were highlighted in red, blue, orange, and green, respectively. TSS: Transcription start site. (B) Precision of targeted mutation within Sry locus was confirmed by Sanger sequencing. The number of XYsry− female samples is shown with respect to each mutation type. (C) CAS9-mediated NHEJ mutation types and frequency at HMG box domain coding region of Sry. Indel mutation sites were highlighted in red underline. (D) Summary of XYsry− female mice production efficiency.
Supplementary Figure S3. Hierarchical clustering analysis of transcriptome datasets from E13.5 and P1 germ cells. Unsupervised hierarchical clustering of all transcripts (n = 36 172) from 12 RNA-seq datasets. The heat map shows relative expression levels.
Supplementary Figure S4. Gene expression profiles of differentially expressed genes in XYsry− germ cells. (A) Pie charts showing numbers of each reference gene type as defined in the UCSC Genome Browser for differentially expressed genes. Expression values of the top 20 differentially expressed genes in E13.5 (B) and P1 (C) XYsry− germ cells: upregulated genes (left panel) and downregulated genes (right panel).
Supplementary Figure S5. Unsupervised hierarchical clustering of gene expression for 37 representative epigenetic regulator genes from E13.5 PGC RNA-seq datasets. The heatmap shows relative gene expression levels.
Supplementary Figure S6. Expression of Y-linked genes in E13.5 PGCs and P1 oocytes of XYsry− females. (A) Schematic diagram of the mouse Y chromosome showing the male-specific region of the Y short arm (MSYp; represented at scale in the magnified view), the male-specific region of the Y long arm (MSYq), and the pseudoautosomal region (PAR). The MSYp region has seven single copy genes, two duplicated genes, and one multicopy gene. The MSYq region carries several multicopy genes. The PAR is the chromosomal region where sex (X-Y) chromosome pairing occurs. (B) Expression patterns of Y-linked genes from E13.5 and P1 germ cell RNA-seq datasets.
Supplementary Figure S7. GO and network analyses using differentially expressed genes in E13.5 XYsry− PGCs. (A, B) Pie charts illustrate the relative distribution of GO terms that describe the biological processes of upregulated genes (A) and downregulated genes (B). (C) Cytoscape visualization of REViGO clustering results of significantly enriched GO categories of differentially expressed genes. Strongly enriched GO terms are shown as nodes, and pairwise similarities are shown as edges. (D) Expression of differentially expressed genes related to the Wnt receptor signaling pathway (GO: 0016055), apoptosis (GO: 0006915), and cell cycle process (GO: 0022402). The genes are ranked in the order of decreasing fold-change (FC) values.
Supplementary Figure S8. Detection of apoptotic cells in E13.5 gonads. (A) Expression of apoptosis-related genes from each E13.5 PGC RNA-seq dataset. Asterisks indicate statistically significant differences between XX female, XYsry− and XY male PGCs (Tukey–Kramer test, *P < 0.05, **P < 0.01). (B) Left: representative images of immunostained E13.5 gonads. PGCs were stained with anti-TRA98 and anti-cCASP3 antibodies, and counterstained with DAPI. Arrowheads show apoptotic germ cells detected by the anti-cCASP3 antibody. Scale bar, 20 μm; magnification, ×40. Right: the percentage of apoptotic cells is shown in the right panel and expressed as the mean ± standard error of the mean. (C) Left: representative images of immunostained E13.5 gonads. PGCs were stained with anti-TRA98 and anti-cPARP antibodies, and counterstained with DAPI. Arrowheads show apoptotic germ cells detected by the anti-cPARP antibody. Scale bar, 20 μm; magnification, ×40. Right: the percentage of apoptotic PGCs are shown. Right: the percentage of cPARP-positive cells (number of cPARP-positive cells/number of TRA98-positive cells) is expressed as the mean ± standard error of the mean.
Supplementary Figure S9. Slowing of oocyte-cyst breakdown and elimination of primordial follicles in the neonatal XYsry− ovaries. Representative images of immunostained P1 ovaries in XX and XYsry− females. Ovaries were stained with ant-DDX4 antibody and TUNEL, and counterstained with DAPI (blue). White dashed lines indicate cortex–medulla boundaries. Arrowheads indicate TUNEL-positive cells. Scale bar, 50 μm. ×20 magnification.
Supplementary Figure S10. Defects of spindle formation and chromosomal alignment in MII oocytes derived from XYsry− females, and the developmental capability of XYsry− MII oocytes. (A) Dot plots of MII oocyte diameters from XX and XYsry– females. No significant difference was observed between XX and XYsry− MII oocytes (Student t-test, P = 0.0895). (B) Representative images of immunostained MII oocytes in XX and XYsry− females. MII oocytes were stained with anti-α-tubulin and counterstained with DAPI. Normal spindles (Type I) were observed in only the XX group. The α-tubulin images show a barrel-shaped microtubule spindle in a position parallel to the oolemma and evince two clearly defined spindle poles. The condensed chromosomes were aligned on the equatorial plate. Aberrant spindles (Type II–Type IV) were observed solely in the XYsry- group (Type II: bulkier spindle with diffused spindle poles, Type III: skewed spindle, Type IV: disrupted spindle and misaligned chromsomes; misaligned and de-condensed chromosomes). Scale bar, 20 μm. (C) Left: percentage of MII oocytes containing each type of meiotic spindle from XX and XYsry– MII oocytes. The definitions of Type I, II, III, and IV spindles were described in (B). Right: percentage contribution of aligned and misaligned chromosome from XX and XYsry– MII oocytes. Aligned and misaligned chromosomes were defined in (B). (D) Fertilization and developmental rates of the described oocytes derived from XX and XYsry− females. Asterisks indicate statistically significant differences between the groups (Chi-squared test, **P < 0.01). (E) Representative images of immunostained E4.5 blastocysts derived from XX and XYsry− females. Blastocysts were stained with anti-OCT4 and counterstained with DAPI.
Supplementary Table S1. Alignment and quantification statistics for each RNA-seq library sample.
Supplementary Table S2. Gene expression profiling of germ cells from WT and XYsry− female mice by RNA-seq analysis.
Supplementary Table S3. Gene expression profiling of gonadal somatic cells from WT and XYsry− female mice by RNA-seq analysis.
Supplementary Table S4. List of differentially expressed genes in E13.5 XYsry− PGCs.
Supplementary Table S5. List of differentially expressed genes in P1 XYsry− oocytes.
Supplementary Table S6. List of differentially expressed genes in E13.5 XYsry− female gonadal somatic cells.
Supplementary Table S7. List of differentially expressed genes in P1 XYsry− female gonadal somatic cells.
Supplementary Table S8. Results of GO and KEGG pathway analyses using differentially expressed genes from XYsry− germ cells.
Supplementary Table S9. Results of GO and KEGG pathway analyses using differentially expressed genes from XYsry− female gonadal somatic cells.
Supplementary Table S10. Alignment and quantification statistics for single-cell RNA-seq analyses.
Supplementary Table S11. Gene expression profiling of MII oocytes obtained from XYsry− females via single-cell transcriptional analyses.
Supplementary Table S12. List of differentially expressed genes common to each XYsry− MII oocyte identified by k-means clustering.
Acknowledgments
We thank Drs. Hidehiko Ogawa, Yayoi Obata, and Hisato Kobayashi for helpful comments, and Dr Asuka Kamio, Takumi Yoshioka for assistance with NGS data collection and data analyses. Also, we are grateful for the gift of pX330 plasmid from Dr Masahito Ikawa.
Notes
Edited by Dr. Monika A. Ward, PhD, University of Hawaii John A. Burns School of Medicine
Footnotes
Grant Support: This project was supported by Grants-in-Aid for Scientific Research from the Japanese Science and Technology Agency and by CREST from the Japanese Agency for Medical Research and Development to Tomohiro Kono.
Author contributions
TK and AS conceived and designed the experiments. TW provided instructions regarding the construction of the CRISPR/Cas9 targeting plasmid. AS, CN, and YS performed microinjection experiments; and AS, CN, and KGA carried out phenotypic analyses of generated XYsry− females. AS and YK performed RNA-seq and data analyses. TK, AS, SHN, and KGA wrote the paper. All authors discussed the results and commented on the manuscript.
Competing financial interests
The authors declare no competing financial interests.
References
- 1. Kashimada K, Koopman P. Sry: the master switch in mammalian sex determination. Development 2010; 137(23):3921–3930. [DOI] [PubMed] [Google Scholar]
- 2. Tanaka SS, Nishinakamura R. Regulation of male sex determination: genital ridge formation and Sry activation in mice. Cell Mol Life Sci 2014; 71(24):4781–4802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Miyamoto Y, Taniguchi H, Hamel F, Silversides DW, Viger RS. A GATA4/WT1 cooperation regulates transcription of genes required for mammalian sex determination and differentiation. BMC Mol Biol 2008; 9(1):44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lovell-Badge R, Robertson E. XY female mice resulting from a heritable mutation in the primary testis-determining gene, Tdy. Development 1990; 109(3):635–646. [DOI] [PubMed] [Google Scholar]
- 5. Gubbay J, Collignon J, Koopman P, Capel B, Economou A, Münsterberg A, Vivian N, Goodfellow P, Lovell-Badge R. A gene mapping to the sex-determining region of the mouse Y chromosome is a member of a novel family of embryonically expressed genes. Nature 1990; 346(6281):245–250. [DOI] [PubMed] [Google Scholar]
- 6. Gubbay J, Vivian N, Economou A, Jackson D, Goodfellow P, Lovell-Badge R. Inverted repeat structure of the Sry locus in mice. Proc Natl Acad Sci USA 1992; 89(17):7953–7957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kato T, Miyata K, Sonobe M, Yamashita S, Tamano M, Miura K, Kanai Y, Miyamoto S, Sakuma T, Yamamoto T, Inui M, Kikusui T et al. Production of Sry knockout mouse using TALEN via oocyte injection. Sci Rep 2013; 3(1):3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wang H, Hu Y-C, Markoulaki S, Welstead GG, Cheng AW, Shivalila CS, Pyntikova T, Dadon DB, Voytas DF, Bogdanove AJ, Page DC, Jaenisch R. TALEN-mediated editing of the mouse Y chromosome. Nat Biotechnol 2013; 31(6):530–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lavery R, Lardenois A, Ranc-Jianmotamedi F, Pauper E, Gregoire EP, Vigier C, Moreilhon C, Primig M, Chaboissier MC. XY Sox9 embryonic loss-of-function mouse mutants show complete sex reversal and produce partially fertile XY oocytes. Dev Biol 2011; 354(1):111–122. [DOI] [PubMed] [Google Scholar]
- 10. Kuroki S, Matoba S, Akiyoshi M, Matsumura Y, Miyachi H, Mise N, Abe K, Ogura A, Wilhelm D, Koopman P, Nozaki M, Kanai Y et al. Epigenetic regulation of mouse sex determination by the histone demethylase Jmjd1a. Science 2013; 341(6150):1106–1109. [DOI] [PubMed] [Google Scholar]
- 11. Correa SM, Washburn LL, Kahlon RS, Musson MC, Bouma GJ, Eicher EM, Albrecht KH. Sex reversal in C57BL/6J XY mice caused by increased expression of ovarian genes and insufficient activation of the testis determining pathway. PLoS Genet 2012; 8(4):e1002569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Washburn LL, Albrecht KH, Eicher EM. C57BL/6J-T-associated sex reversal in mice is caused by reduced expression of a Mus domesticus Sry allele. Genetics 2001; 158(4):1675–1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Coward P, Nagai K, Chen D, Thomas HD, Nagamine CM, Lau YF. Polymorphism of a CAG trinucleotide repeat within Sry correlates with B6.YDom sex reversal. Nat Genet 1994; 6(3):245–250. [DOI] [PubMed] [Google Scholar]
- 14. Eicher EM, Washburn LL, Whitney JB 3rd, Morrow KE. Mus poschiavinus Y chromosome in the C57BL/6J murine genome causes sex reversal. Science 1982; 217(4559):535–537. [DOI] [PubMed] [Google Scholar]
- 15. Lee CH, Taketo T. Normal onset, but prolonged expression, of Sry gene in the B6.YDOM sex-reversed mouse gonad. Developmental Biology 1994; 165(2):442–452. [DOI] [PubMed] [Google Scholar]
- 16. Taketo T. The role of sex chromosomes in mammalian germ cell differentiation: can the germ cells carrying X and Y chromosomes differentiate into fertile oocytes? Asian J Androl 2015; 17(3):360–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mahadevaiah SK, Lovell-Badge R, Burgoyne PS, Tdy-negative XY. Tdy-negative XY, XXY and XYY female mice: breeding data and synaptonemal complex analysis. Reproduction 1993; 97(1):151–160. [DOI] [PubMed] [Google Scholar]
- 18. Vernet N, Szot M, Mahadevaiah SK, Ellis PJ, Decarpentrie F, Ojarikre OA, Á R, Taketo T, Burgoyne PS. The expression of Y-linked Zfy2 in XY mouse oocytes leads to frequent meiosis 2 defects, a high incidence of subsequent early cleavage stage arrest and infertility. Development 2014; 141(4):855–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Vanderhyden BC, Macdonald EA, Merchant-Larios H, Fernandez A, Amleh A, Nasseri R, Taketo T. Interactions between the oocyte and cumulus cells in the ovary of the B6.YTIR sex-reversed female mouse. Biol Reprod 1997; 57(3):641–646. [DOI] [PubMed] [Google Scholar]
- 20. Park EH, Taketo T. Onset and progress of meiotic prophase in the oocytes in the B6.YTIR sex-reversed mouse ovary. Biol Reprod 2003; 69(6):1879–1889. [DOI] [PubMed] [Google Scholar]
- 21. Wong J, Luckers L, Okawara Y, Pelletier R, Taketo T. Follicular development and atresia in the B6.YTIR sex-reversed mouse ovary. Biol Reprod 2000; 63(3):756–762. [DOI] [PubMed] [Google Scholar]
- 22. Villemure M, Chen HY, Kurokawa M, Fissore RA, Taketo T. The presence of X- and Y-chromosomes in oocytes leads to impairment in the progression of the second meiotic division. Dev Biol 2007; 301(1):1–13. [DOI] [PubMed] [Google Scholar]
- 23. Obata Y, Kono T. Maternal primary imprinting is established at a specific time for each gene throughout oocyte growth. J Biol Chem 2002; 277(7):5285–5289. [DOI] [PubMed] [Google Scholar]
- 24. Sakashita A, Kawabata Y, Jincho Y, Tajima S, Kumamoto S, Kobayashi H, Matsui Y, Kono T. Sex specification and heterogeneity of primordial germ cells in mice. PLoS ONE 2015; 10(12):e0144836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yokobayashi S, Liang C-Y, Kohler H, Nestorov P, Liu Z, Vidal M, van Lohuizen M, Roloff TC, Peters AHFM. PRC1 coordinates timing of sexual differentiation of female primordial germ cells. Nature 2013; 495(7440):236–240. [DOI] [PubMed] [Google Scholar]
- 26. Gill ME, Hu YC, Lin Y, Page DC. Licensing of gametogenesis, dependent on RNA binding protein DAZL, as a gateway to sexual differentiation of fetal germ cells. Proc Natl Acad Sci USA 2011; 108(18):7443–7448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Uda M, Ottolenghi C, Crisponi L, Garcia JE, Deiana M, Kimber W, Forabosco A, Cao A, Schlessinger D, Pilia G. Foxl2 disruption causes mouse ovarian failure by pervasive blockage of follicle development. Hum Mol Genet 2004; 13(11):1171–1181. [DOI] [PubMed] [Google Scholar]
- 28. Chen H, Palmer JS, Thiagarajan RD, Dinger ME, Lesieur E, Chiu H, Schulz A, Spiller C, Grimmond SM, Little MH, Koopman P, Wilhelm D. Identification of novel markers of mouse fetal ovary development. PLoS ONE 2012; 7(7):e41683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Probst FJ, Cooper ML, Cheung SW, Justice MJ. Genotype, phenotype, and karyotype correlation in the XO mouse model of Turner Syndrome. J Hered 2008; 99(5):512–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cloutier JM, Mahadevaiah SK, ElInati E, Nussenzweig A, Tóth A, Turner JM. Histone H2AFX links meiotic chromosome asynapsis to prophase I oocyte loss in mammals. PLoS Genet 2015; 11(10):e1005462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ma K, Mallidis C, Bhasin S. The role of Y chromosome deletions in male infertility. Eur J Endocrinol 2000; 142(5):418–430. [DOI] [PubMed] [Google Scholar]
- 32. Vernet N, Mahadevaiah SK, Ellis PJ, de Rooij DG, Burgoyne PS. Spermatid development in XO male mice with varying Y chromosome short-arm gene content: evidence for a Y gene controlling the initiation of sperm morphogenesis. Reproduction 2012; 144(4):433–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Vernet N, Mahadevaiah SK, Ojarikre OA, Longepied G, Prosser HM, Bradley A, Mitchell MJ, Burgoyne PS. The Y-encoded gene zfy2 acts to remove cells with unpaired chromosomes at the first meiotic metaphase in male mice. Curr Biol 2011; 21(9):787–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kimura T, Nakamura T, Murayama K, Umehara H, Yamano N, Watanabe S, Taketo MM, Nakano T. The stabilization of beta-catenin leads to impaired primordial germ cell development via aberrant cell cycle progression. Dev Biol 2006; 300(2):545–553. [DOI] [PubMed] [Google Scholar]
- 35. Wu D, Pan W. GSK3: a multifaceted kinase in Wnt signaling. Trends Biochem Sci 2010; 35(3):161–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kim K, Pang KM, Evans M, Hay ED. Overexpression of beta-catenin induces apoptosis independent of its transactivation function with LEF-1 or the involvement of major G1 cell cycle regulators. Mol Biol Cell 2000; 11(10):3509–3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Olmeda D, Castel S, Vilaró S, Cano A. Beta-catenin regulation during the cell cycle: implications in G2/M and apoptosis. Mol Biol Cell 2003; 14(7):2844–2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pepling ME, Spradling AC. Mouse ovarian germ cell cysts undergo programmed breakdown to form primordial follicles. Dev Biol 2001; 234(2):339–351. [DOI] [PubMed] [Google Scholar]
- 39. Lei L, Spradling AC. Mouse oocytes differentiate through organelle enrichment from sister cyst germ cells. Science 2016; 352(6281):95–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Malki S, van der Heijden GW, O’Donnell KA, Martin SL, Bortvin A. A role for retrotransposon LINE-1 in fetal oocyte attrition in mice. Dev Cell 2014; 29(5):521–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Royo H, Polikiewicz G, Mahadevaiah SK, Prosser H, Mitchell M, Bradley A, de Rooij DG, Burgoyne PS, Turner JM. Evidence that meiotic sex chromosome inactivation is essential for male fertility. Curr Biol 2010; 20(23):2117–2123. [DOI] [PubMed] [Google Scholar]
- 42. Vernet N, Mahadevaiah SK, Yamauchi Y, Decarpentrie F, Mitchell MJ, Ward MA, Burgoyne PS. Mouse Y-linked Zfy1 and Zfy2 are expressed during the male-specific interphase between meiosis I and meiosis II and promote the 2nd meiotic division. PLoS Genet 2014; 10(6):e1004444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Burgoyne PS, Mahadevaiah SK, Turner JM. The consequences of asynapsis for mammalian meiosis. Nat Rev Genet 2009; 10(3):207–216. [DOI] [PubMed] [Google Scholar]
- 44. Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009; 4(1):44–57. [DOI] [PubMed] [Google Scholar]
- 45. Burgoyne PS, Baker TG. Perinatal oocyte loss in XO mice and its implications for the aetiology of gonadal dysgenesis in XO women. Reproduction 1985; 75(2):633–645. [DOI] [PubMed] [Google Scholar]
- 46. Burgoyne PS, Baker TG. Oocyte depletion in XO mice and their XX sibs from 12 to 200 days post partum. Reproduction 1981; 61(1):207–212. [DOI] [PubMed] [Google Scholar]
- 47. Tingen C, Kim A, Woodruff TK. The primordial pool of follicles and nest breakdown in mammalian ovaries. Mol Hum Reprod 2009; 15(12):795–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wang C, Zhou B, Xia G. Mechanisms controlling germline cyst breakdown and primordial follicle formation. Cell Mol Life Sci 2017; 74(14):2547–2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Crisponi L, Deiana M, Loi A, Chiappe F, Uda M, Amati P, Bisceglia L, Zelante L, Nagaraja R, Porcu S, Ristaldi MS, Marzella R et al. The putative forkhead transcription factor FOXL2 is mutated in blepharophimosis/ptosis/epicanthus inversus syndrome. Nat Genet 2001; 27(2):159–166. [DOI] [PubMed] [Google Scholar]
- 50. Pearlman A, Loke J, Le Caignec C, White S, Chin L, Friedman A, Warr N, Willan J, Brauer D, Farmer C, Brooks E, Oddoux C et al. Mutations in MAP3K1 cause 46,XY disorders of sex development and implicate a common signal transduction pathway in human testis determination. Am J Hum Genet 2010; 87(6):898–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.