Abstract
Arterial diseases including abdominal aortic aneurysm and atherosclerosis are biomechanical diseases characterized by significant changes in the structure and strength of the vessel wall. It is now established that local variations in fibrillar collagen and elastin matrix turnover is critical to arterial stiffening and progression of the disease. The collagen content in the aortic wall has nominally been quantified by biochemical assays and immunohistochemical analysis as the total amount because of the difficulty in separating the media and adventitia. In this work, we have developed an algorithm for automatic quantification of layer-specific collagen content from bright-field and polarized microscopic images of histological sections of mouse aorta stained with Picrosirius red (PSR) stain. The images were processed sequentially including separation of layers, erosion, segregation of regions, binarization, and quantification of pixel intensities to obtain collagen content in the media and adventitia separately. We observed that the automated algorithm rapidly and accurately quantified collagen content from a wide range of image quality compared with manual measurements particularly when the medial and adventitial layers overlap. Together, our algorithm will be of significant impact in the rapid, reliable, and accurate analyses of collagen distribution in histological sections of connective tissues.
Keywords: AAA, histology, stiffening
Introduction
Aortic stiffness is an independent predictor of cardiovascular and cerebrovascular mortality and morbidity even after accounting for the classical risk factors.1 Aortic stiffness increases not only in normal aging process but also in aortic diseases including aneurysm, dissection, and atherosclerosis. As aortic stiffness is one of the earliest detectable manifestations of the adverse functional changes in the blood vessel, evaluating the underlying structural changes is critical for detection and intervention of cardiovascular diseases.
The aortic wall is comprised of three layers of distinct composition and properties: intima, media, and adventitia. In healthy arteries, the intima is composed of a single layer of endothelial cells, a thin basal membrane, and a subendothelial layer composed of collagen fibrils; the media is composed of smooth muscle cells, a network of elastin, and collagen fibrils, which are separated by a few layers of circumferentially lamellar elastin sheets; and the adventitia is composed of thick bundles of collagen fibrils arranged in helical structures and fibroblasts.2 In healthy human aorta, the intimal, medial, and adventitial thickness is ~50 to 150 µm, 300 to 500 µm, and 500 to 1000 µm, respectively, and total thickness of 1 to 2 mm.3,4 Aortic stiffness is mainly determined by the major extracellular matrix (ECM) components of the arterial wall, that is, collagen and elastin, and less so by cellular components.5 The breakdown of elastin and fibrillar collagen is compensated by the deposition of uncoiled neo-collagen synthesized by the vascular smooth muscle cells (VSMC), which leads to the transfer of load to less compliant collagen. Thus, changes in collagen content can be important for the understanding of the etiology of increase in aortic stiffness.
Traditionally, the total collagen content of the aorta is estimated by either biochemical assay for hydroxyproline levels or by histological or immunohistochemical analysis of tissue sections.6 Although these assays provide the total collagen content, changes in individual layers of the aorta is often of immense interest. In abdominal aortic aneurysms (AAA), spatial variations in the ECM distribution is correlated with segmental stiffening which in turn determines disease progression and severity.7 The Picrosirius red (PSR) staining provides a convenient, inexpensive, fade-resistant, and reproducible staining of collagen in tissue sections, and is preferred over traditional trichrome stains. The PSR staining method relies on the elongated, anionic structure of the Sirius red dye molecule–binding parallel to cationic collagen fibers, thus enhancing the natural birefringence of collagen bundles.8 The PSR staining technique has been widely used for qualitative or quantitative description of changes in collagen content, not only of the aorta but also of numerous other connective tissues including skin and myocardial scars wherein relative distribution between different regions of the tissue is of interest.9,10 In this work, we describe an automated methodology to quantify collagen content separately in the medial and adventitial layers of mouse aortic sections from bright-field and polarized microscopy images. Combining information from these two microscopy techniques allows for rapid and reliable quantification of localized collagen content in histological sections.
Methods
Sample Processing
Murine aorta were explanted from C57BL/6J mice and incubated in 4% paraformaldehyde (PFA) at 4C for 2 hr. Thereafter, aorta were transferred to a new 4% PFA in PBS solution supplemented with 20% sucrose and incubated overnight at 4C. The next day, the aorta were cut in 1 cm pieces and rinsed quickly in cold PBS. Next, samples were embedded in Tissue Plus OCT compound (Fisher Scientific) and immediately frozen down. Samples were stored at −80C until further processing. Similar procedures were followed for processing, sectioning, and imaging of murine femoral and carotid arteries, and images were analyzed, similar to aortic sections, as described below.
Sample Sectioning
Materials were allowed to equilibrate to the chamber temperature of −20C for 10 min. After attaching samples to the circular cryostat block, the tissue blocks were sectioned at 7 µm using a cryostat (CM3050 S, Leica Biosystems Inc., Wetzlar, Germany), and mounted on SuperFrost Ultra Plus (Fisher Scientific) glass slides. Sections were incubated in ice-cold acetone for 4 min and stored at −80C until further processing.
Picrosirius Red Staining
Sections were thawed and allowed to equilibrate to room temperature for 10 min. Then, sections were fixed in ice-cold acetone for 4 min again and allowed to dry. PSR staining was performed using Picrosirius Red Stain Kit (catalog number: ab150681, Abcam) according to the manufacturer instructions. In short, PSR solution was applied to completely cover the tissue section and incubated for 60 min. Sections were rinsed twice in acetic acid and once in absolute ethanol (Fisher Scientific). Sections were dehydrated with two washes in absolute ethanol and mounted using synthetic resin (catalog number: 1900231, Fisher Scientific). Samples were stored at room temperature until imaging.
Image Acquisition
PSR-stained sections were visualized utilizing a Leica microscope (Leica DM4000B, Leica Biosystems Inc., USA) and a polarized filter (Leica ICT/Pol, Leica Biosystems Inc.). Images of different aortic segments were obtained using DISKUS (Version 5.0.6.277, Hilgers technisches Buero, Koenigswinter, Germany) and saved as a pairwise results objects (bright-field/polarized light) for further processing.
Image Selection
Complementary images obtained by bright-field microscopy and polarized light microscopy of the same histological section are necessary for quantification of collagen content. The section must have a contiguous adventitia, which means that the red ring in the adventitia must be continuous with no breaks. The bright-field and polarized light images must also be aligned to properly use the bright-field image to distinguish between the adventitia and media. The lighting throughout the images must be relatively even to accurately discriminate between the background and the histological section. There must also be minimized debris on the histological slide to ensure the collagen is quantified accurately. Finally, the adventitia and media should not overlap as it may sometimes happen due improper processing.
Automated Analysis of Collagen Content Using MATLAB
Downloading and Running the Program
The program for automated collagen quantification can be found at https://git.io/vhsGV. The repository needs to be downloaded and extracted into the active MATLAB (MathWorks, Natick, MA) directory. The program was run within MATLAB by using the command “Automatic_Collagen_Quant_AAA_PSR.” The graphical user interface (GUI) was then available for use to quantify the collagen using bright-field and polarized images.
The program needs complementary pairs of bright-field and polarized images to quantify the collagen of the tissue section. The program supports various image types such as .JPG, .IMG, and .TIFF, but it is highly recommended to use uncompressed .TIFF files to maintain the integrity of the image. The bright-field image is loaded by clicking the “Load Bright Field” button in the GUI and selecting the corresponding image. A preview of the image and its file name will appear in the GUI for verification. The polarized image is loaded by clicking the “Load Polarized” button in the GUI and selecting the corresponding image. A preview of the image and its file name will be displayed next to the preview of the bright-field image. At this point, the program can quantify collagen content by pressing the “Process” button. However, some extra steps may be necessary to properly process the images depending on the conditions for image acquisition or the output format. (1) If the bright-field and polarized images are not properly aligned, the “Offset” button must be toggled on and the amount of offset in pixels should be entered for the X-Offset and Y-Offset. (2) Depending on the resolution of the image, the size of the structural elements used to process the bright-field image may also need to be changed. This process is elaborated on further in the “Results” section. (3) The data can be expressed as the surface area of the image if the “Scale Bar” button is toggled on, and the length of the scale bar and its length are entered as input into the GUI.
Uploading the Image
The bright-field and polarized images were uploaded into MATLAB. The images should ideally be uncompressed .TIFF images, which are preferable to best preserve the quality of the images, although the program does allow for the uploading of other image format types.
Preprocessing of Images
The uploaded images are adjusted to an array of m × n × 3 to account for images that may be in RGBa space, which outputs images in an m × n × 4 array. The green channel of the bright-field image was isolated by selecting the second of the three layers of the image array which resulted in a grayscale image. The polarized image was converted into grayscale and the contrast of the image was improved. The conversion into grayscale was achieved using the MATLAB function, rgb2gray, which converts the RGB values to grayscale by performing a weighted sum of the red (R), green (G), and blue (B) components using the following formula: 0.2989 × (R) + 0.5870 × (G) + 0.1140 (B). Improving the contrast was accomplished using the adapthisteq MATLAB function, which uses contrast-limited adaptive histogram equalization (CLAHE) algorithm. The algorithm operates on small regions of the image, referred to as tiles, rather than the image as a whole. The contrast of each tile is enhanced using a contrast transform function, and the neighboring tiles are combined using bilinear interpolation to eliminate the artificially created boundaries between the tiles.
Processing the Bright-field Image
The grayscale image obtained from the green channel of the image was then adaptively binarized using the imbinarize function with a sensitivity of 0.3. The locally adaptive threshold in MATLAB utilizes Bradley’s Method.11 This sensitivity was chosen to sufficiently thin the portions associated with the media, that would later be removed through erosion and dilation techniques, and to preserve the integrity of the adventitia. Two structural elements in the form of a disk were created with diameters of four and two pixels. The two-pixel structural element was used for erosion and the four-pixel structural element was used for dilation. These two structural elements are set to be two and four pixels by default, but the sizes can be changed within the GUI to account for different resolutions. The binarized image was eroded, dilated, and eroded to remove the small fibers associated with the media, while preserving the integrity of the image. The bwareaopen function was used to remove small objects that were not associated with the tissue and to remove any possible residual debris. This was done by determining the connected components throughout the image and their respective areas, and the connected components that were smaller than desired size were removed. By default, the program will remove any isolated components that are comprised of less than 600 pixels, but the threshold for the small objects can be changed within the GUI. The adventitia is now isolated with an empty center which will be used as the region of interest (ROI). The ROI was calculated by finding the center of the image and using the grayconnected function to select all the pixels within the borders of the adventitia that are now contiguous after removing the excess debris in the previous step. A mask was created by duplicating the ROI and scaling it to 80% of its original size. This mask was used to delete any extraneous debris in ROI.
Processing the Polarized Image
The polarized image, now in grayscale, was binarized using Otsu’s method by using imbinarize with global thresholding.12 The ROI was then used to isolate the collagen associated with the media and the extra debris in the middle that may appear during binarization was removed using the mask.
Quantifying Collagen
The total collagen was quantified by counting the number of white pixels in the binarized polarized image. The white pixels that were within the ROI were counted as collagen within the media. These values were then used to determine the percent of collagen locally. If the scale bar measurements are available, the size of the scale bar and the actual length of the scale bar may be used in their respective fields in the GUI, allowing the program to compute the surface area covered by collagen.
Manual Analysis of Collagen Content Using Adobe Photoshop
Two operators were trained to trace the outlines between adventitia and media in the image slices. The operators were given the same set of 15 images and manually drew a line to represent the border between the adventitia and media in the polarized light images. The outlines were done in Adobe Photoshop by creating a layer over the image and drawing an outline around the inside of the heavily red portions of the image, representing the border between adventitia and media. The operators were also timed to determine the amount of time that was required to trace the outlines. These outlines were then used to produce ROIs that were imported into MATLAB for collagen quantification using the same methods as previously described. The localized percentage of collagen within the automatically generated ROI and manually created ROI were then compared along a scatter plot to determine if the two measurements were comparable.
Statistics
We obtained 20 image sets from various parts of the mouse aorta, and they were analyzed for total collagen content by either automated program or manual methods. The data sets were plotted against each other, and linear regression (GraphPad Prism) was used to compare the quality of the fit.
Results and Discussion
We created a GUI to quantify collagen content and distribution in histological sections obtained from murine aorta. The overall process flow that is built into the automated program and the GUI are shown in Fig. 1A and B, respectively. Each PSR-stained image set consists of two images, each obtained from bright-field and polarized light microscopy, and the program superimposes these two images to demarcate the media and adventitia, identify the inner and outer boundaries of the two regions, and from the residual intensities, calculate the collagen content in these regions.
Figure 1.
(A) A simplified flowchart outlining the steps taken within the program to quantify the collagen. (B) The graphical user interface of the program used for quantifying collagen content in media and adventitia.
The bright-field images of PSR-stained sections show a distinct rich red outer ring, and an orange-yellow inner ring (Fig. 2A). The outer red stain is from the adventitia, which has the highest amount of collagen, thus stains red. The orange-yellow in the bright-field image is from the media. However, there is still some collagen within this layer of the vessel wall, but it is not possible to see using only by bright-field microscopy. Polarized light microscopy, allows for the observation of the collagen that is present within the media (Fig. 2B). Collagen stained with PSR is birefringent under polarized light and there are changes in the birefringence colors that range from green, yellow, or red. Although the origins of the color were initially thought to be representative of collagen subtypes (I vs. III based on yellow-red vs. green), and the fiber thickness, it is now believed that the birefringence colors largely depend on the orientation of the collagen with respect to orientation of the polarized light.13–15 Thus, polarized light microscopy in combination with PSR is most useful when used for quantification of the overall collagen content as the colors are easily differentiated from the black background.
Figure 2.
The original bright-field (A) and polarized light microscopy images (D); Grayscale representation of the green layer isolated from RGB composition and segmented from bright-field image (B), and from polarized image (E). (C) The segmented images from (B) were subjected to a cycle of erosion, dilation, and erosion to remove the pixel associated with the media while maintaining image integrity. Isolated islands were also removed to remove extraneous debris. (F) A visual representation of the polarized image separated by adventitia (green) and the media (white). The scale bar is 200 µm.
Processing of Ideal Cases
Image sets with perfect alignment of bright-field and polarized microscopy images, distinct adventitia and media, and proper lighting may be processed readily using the algorithm described in Fig. 1, and is shown stepwise in Fig. 2. First, the green channel from both bright-field and polarized microscopy images was isolated and converted into grayscale (Fig. 2C and D). The green channel was chosen because it allowed for the greatest contrast between adventitia and media when compared with the red and blue channels (Fig. S1). Once in grayscale, adaptive thresholding was performed on the bright-field image, as seen in Fig. 2C, and the thin fibers and debris were filtered out to establish the ROI, shown in Fig. 2D. Afterward, the polarized image was processed using a global threshold method (Fig. 2E), and the collagen was quantified using the ROI to establish the amounts of collagen in the media and adventitia (Fig. 2F). Finally, the results are displayed as the cross-sectional area containing collagen or as pixels positive for collagen content. We also followed similar image processing steps for bright-field and polarized images of PSR-stained sections of femoral and carotid arteries of mice. As shown in Figs. S2 and S3, respectively, our program successfully demarcates the media and adventitia without any noticeable overlap.
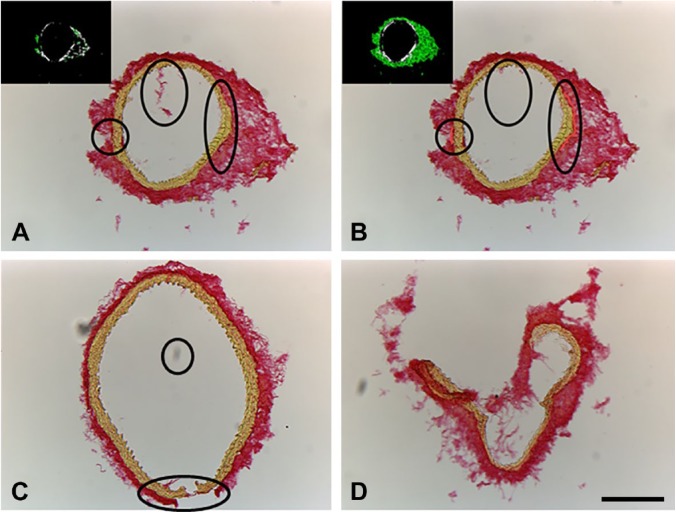
Processing of Non-ideal Cases
We observed that inherently most of the images were less than ideal, and required some extra processing steps (Fig. 3). These include non-contiguous adventitia, excessive debris in the sample, or misalignment between bright-field and polarized images. If these images were processed as is, neither total collagen content nor regional distribution could be obtained (Fig. 3A, inset). Fortunately, these common issues can be easily fixed by a few corrective actions, either in Adobe Photoshop before uploading the images into the GUI or in the GUI itself. (1) The adventitia can be bridged to make it contiguous using Adobe Photoshop. We used the eyedropper tool to sample the color of the adventitia, and used the pencil tool set at 8 pixels to bridge any gaps within the adventitia. (2) In case of excessive debris in the sample, we used the clone stamp tool to mask the debris and preserve the integrity of the image to not have an effect on future segmentation. (3) We observed a consistent 12 pixel vertical offset between the bright-field and polarized images, probably due to physical reasons while switching the filters in the microscope. To nullify this offset, the value of the offset can be obtained by manual alignment of one set of images. This value may be entered in the GUI before processing by clicking the check box next to “Offset” in the GUI and feeding in the x and y offsets in their respective fields. Once corrected, the collagen content can be quantified using the automated program (Fig. 3B and inset, Fig. 3C). Despite these corrections, some images may not be salvageable because of poor quality such as excessive overlap of adventitia and media or complete loss of tissue integrity (Fig. 3D). However, if the media and adventitia are intact, the presence of extraneous tissue will not affect the quantification, and hence preprocessing is not necessary (Fig. S3).
Figure 3.
Sample images that require preprocessing. (A) A non-ideal image with small separations within the adventitia and excess debris and the program is not able to establish a proper ROI. (B) The non-ideal case with the excess debris removed and the adventitia bridged to produce a proper ROI for collagen quantification. (C) Another non-ideal image with corrections. (D) A non-salvageable image. Abbreviation: ROI, region of interest. The scale bar is 200 µm.
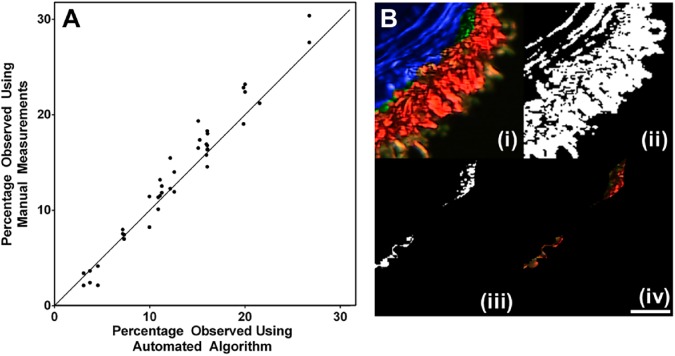
After preprocessing as necessary, we compared the workflow and the results obtained from automated measurements with manual measurements using Adobe Photoshop (Fig. 4A). To obtain the pixel distribution in adventitia and media separately by manual measurement, a user took an average of 4 min per image to define the ROI without quantifying the collagen. The manually defined ROI was then imported into MATLAB to locally quantify the collagen. On the contrary, the program can process the images in a matter of seconds and offers consistent measurements between samples. After preprocessing, the non-ideal cases produced results identical to that of ideal cases. The results obtained from the automated algorithm were virtually identical to those obtained by manual measurements at lower collagen levels, although at higher collagen levels, the manual measurements consistently overpredicts the collagen distribution. The reason is that the automated algorithm can work inherently much easier as the analysis is done on a pixel-by-pixel basis when compared with a human which is inherently much coarser, and this difference is exemplified in Fig. 4B. As the ROI is being defined by the stark contrast between the coloration in the layers, it is highly dependent on the ability to distinguish between the layers. When doing manual analysis, any number of factors can lead to inaccuracies such as the steadiness or fatigue of the hand, the accuracy and resolution of the mouse/trackpad itself, and the amount of time necessary to achieve the same kind of outline that is supplied by the program. In contrast, as the automated algorithm operates on a rigid set of rules, it is an ideal method for consistent demarcation between layers. For instance, in Fig. 4B, the number of pixels for the segment shown was determined to be 15,406 pixels out of the 219,375 pixels of the entire sample. The errors made using the manual method lead to an over-measurement of 565 pixels, shown in green.
Figure 4.
(A) Scatter plot demonstrating the similarity between manually defining the ROI and automatically defining the ROI with the program (n=20). The methods produced virtually identical results at lower collagen levels although the manual method overpredicted at higher collagen levels (R2 = 0.94). (B) Accuracy of the automatic program versus manual tracing. (i) A subsection of a polarized image with the ROI generated by the automated program (blue) and by manual tracing (green); (ii) Binarized image; (iii) The extra pixels that were counted after global thresholding in manual measurement but not automated algorithm; (iv) The extra pixels were pixels that should be attributed to adventitia by showing the pixels were associated with red color. Abbreviation: ROI, region of interest. The scale bar is 100 µm.
In summary, we have developed an automated algorithm for efficient, reliable, and rapid estimation of collagen content in the various regions of the aortic sections. This algorithm will enable comprehensive analyses of localized matrix remodeling in aortic sections that are routinely processed for insights into the mechanisms and functional consequences. Furthermore, our algorithm may be easily adapted for the histopathological analysis of non-homogeneously distributed fibrosis typical of myocardial infarction and other connective tissue disorders.
Supplemental Material
Supplemental material, DS_10.1369_0022155418814231 for An Automated Algorithm to Quantify Collagen Distribution in Aortic Wall by Dustin M. Nguyen, Markus U. Wagenhäuser, Dennis Mehrkens, Matti Adam, Philip S. Tsao and Anand K. Ramasubramanian in Journal of Histochemistry & Cytochemistry
Footnotes
Competing Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Author Contributions: DMN developed the algorithm and wrote the code, MUW, DM, and MA obtained and stained aortic sections, DMN, MUW, PST, and AKR designed the study, analyzed the data, and wrote the manuscript. All authors have read and approved the final manuscript.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by research grants from the National Institute of Health (R01 HL122939 and R56 HL135654) and the Department of Veterans Affairs (1I01BX002641-01A) to PST.
ORCID iD: Dustin M. Nguyen  https://orcid.org/0000-0001-6715-4627
https://orcid.org/0000-0001-6715-4627
Contributor Information
Dustin M. Nguyen, Department of Chemical and Materials Engineering, San José State University, San José, CA
Markus U. Wagenhäuser, Division of Cardiovascular Medicine, Stanford University School of Medicine, Stanford, CA VA Palo Alto Health Care System, Palo Alto, CA.
Dennis Mehrkens, Department of Cardiovascular Medicine, University Heart Center and Cologne Cardiovascular Research Center, University of Cologne, Cologne, Germany.
Matti Adam, Department of Cardiovascular Medicine, University Heart Center and Cologne Cardiovascular Research Center, University of Cologne, Cologne, Germany.
Philip S. Tsao, Division of Cardiovascular Medicine, Stanford University School of Medicine, Stanford, CA VA Palo Alto Health Care System, Palo Alto, CA.
Anand K. Ramasubramanian, Department of Chemical and Materials Engineering, San José State University, San José, CA.
Literature Cited
- 1. Cavalcante JL, Lima JAC, Redheuil A, Al-Mallah MH. Aortic stiffness: current understanding and future directions. J Am Coll Cardiol. 2011;57(14):1511–22. doi: 10.1016/j.jacc.2010.12.017. [DOI] [PubMed] [Google Scholar]
- 2. Tsamis A, Krawiec JT, Vorp DA. Elastin and collagen fibre microstructure of the human aorta in ageing and disease: a review. J R Soc Interface. 2013;10(83):20121004. doi: 10.1098/rsif.2012.1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Restrepo C, Strong JP, Guzmán MA, Tejada C. Geographic comparisons of diffuse intimal thickening of the aorta. Atherosclerosis. 1979;32:177–93. [DOI] [PubMed] [Google Scholar]
- 4. Rosero EB, Peshock RM, Khera A, Clagett P, Lo H, Timaran CH. Sex, race, and age distributions of mean aortic wall thickness in a multiethnic population-based sample. J Vasc Surg. 2011;53:950–7. [DOI] [PubMed] [Google Scholar]
- 5. Wagenseil JE, Mecham RP. Vascular extracellular matrix and arterial mechanics. Physiol Rev. 2009;89(3):957–89. doi: 10.1152/physrev.00041.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Coats WD, Whittaker P, Cheung DT, Currier JW, Han B, Faxon DP. Collagen content is significantly lower in restenotic versus nonrestenotic vessels after balloon angioplasty in the atherosclerotic rabbit model. Circulation. 1997;95:1293–300. [DOI] [PubMed] [Google Scholar]
- 7. Raaz U, Zöllner AM, Schellinger IN, Toh R, Nakagami F, Brandt M, Emrich FC, Kayama Y, Eken S, Adam M, Maegdefessel L, Hertel T, Deng A, Jagger A, Buerke M, Dalman RL, Spin JM, Kuhl E, Tsao PS. Segmental aortic stiffening contributes to experimental abdominal aortic aneurysm development. Circulation. 2015;131:1783–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wegner KA, Keikhosravi A, Eliceiri KW, Vezina CM. Fluorescence of picrosirius red multiplexed with immunohistochemistry for the quantitative assessment of collagen in tissue sections. J Histochem Cytochem. 2017;65(8):479–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Vogel B, Siebert H, Hofmann U. Determination of collagen content within picrosirius red stained paraffin-embedded tissue sections using fluorescence microscopy. MethodsX. 2015;2:124–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Street JM, Souza ACP, Alvarez-Prats A, Horino T, Hu X, Yuen PST, Star RA. Automated quantification of renal fibrosis with Sirius Red and polarization contrast microscopy. Physiol Rep. 2014;2(7):e12088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bradley D, Roth G. Adaptive thresholding using the integral image. J Graph Tools. 2007;12:13–21. [Google Scholar]
- 12. Otsu N. A Threshold selection method from gray-level histograms. IEEE Trans Syst Man Cybern. 1979;9(1):62–6. http://ieeexplore.ieee.org/document/4310076/. [Google Scholar]
- 13. Rich L, Whittaker P. Collagen and picrosirius red staining : a polarized light assessment of fibrillar hue and spatial distribution. Braz J Morphol Sci. 2005;22(2):97–104. http://jms.org.br/PDF/v22n2a06.pdf. [Google Scholar]
- 14. Coleman R. Picrosirius red staining revisited. Acta Histochem. 2011;113(3):231–3. [DOI] [PubMed] [Google Scholar]
- 15. Lattouf R, Younes R, Lutomski D, Naaman N, Godeau G, Senni K, Changotade S. Picrosirius red staining: a useful tool to appraise collagen networks in normal and pathological tissues. J Histochem Cytochem. 2014;62(10):751–8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, DS_10.1369_0022155418814231 for An Automated Algorithm to Quantify Collagen Distribution in Aortic Wall by Dustin M. Nguyen, Markus U. Wagenhäuser, Dennis Mehrkens, Matti Adam, Philip S. Tsao and Anand K. Ramasubramanian in Journal of Histochemistry & Cytochemistry