Abstract
Aims
Nationwide large‐scale genetic and outcome studies in cohorts with hypertrophic cardiomyopathy (HCM) have not been previously published.
Methods and results
We sequenced 59 cardiomyopathy‐associated genes in 382 unrelated Finnish patients with HCM and found 24 pathogenic or likely pathogenic mutations in six genes in 38.2% of patients. Most mutations were located in sarcomere genes (MYBPC3, MYH7, TPM1, and MYL2). Previously reported mutations by our study group (MYBPC3‐Gln1061Ter, MYH7‐Arg1053Gln, and TPM1‐Asp175Asn) and a fourth major mutation MYH7‐Val606Met accounted for 28.0% of cases. Mutations in GLA and PRKAG2 were found in three patients. Furthermore, we found 49 variants of unknown significance in 31 genes in 20.4% of cases. During a 6.7 ± 4.2 year follow‐up, annual all‐cause mortality in 482 index patients and their relatives with HCM was higher than that in the matched Finnish population (1.70 vs. 0.87%; P < 0.001). Sudden cardiac deaths were rare (n = 8). Systolic heart failure (hazard ratio 17.256, 95% confidence interval 3.266–91.170, P = 0.001) and maximal left ventricular wall thickness (hazard ratio 1.223, 95% confidence interval 1.098–1.363, P < 0.001) were independent predictors of HCM‐related mortality and life‐threatening cardiac events. The patients with a pathogenic or likely pathogenic mutation underwent an implantable cardioverter defibrillator implantation more often than patients without a pathogenic or likely pathogenic mutation (12.9 vs. 3.5%, P < 0.001), but there was no difference in all‐cause or HCM‐related mortality between the two groups. Mortality due to HCM during 10 year follow‐up among the 5.2 million population of Finland was studied from death certificates of the National Registry, showing 269 HCM‐related deaths, of which 32% were sudden.
Conclusions
We identified pathogenic and likely pathogenic mutations in 38% of Finnish patients with HCM. Four major sarcomere mutations accounted for 28% of HCM cases, whereas HCM‐related mutations in non‐sarcomeric genes were rare. Mortality in patients with HCM exceeded that of the general population. Finally, among 5.2 million Finns, there were at least 27 HCM‐related deaths annually.
Keywords: Hypertrophic cardiomyopathy, Genetics, Targeted sequencing, Mutation, Mortality, Outcome
Introduction
Hypertrophic cardiomyopathy (HCM), the most common monogenic heart disease with dominant mode of inheritance, is caused primarily by mutations in genes encoding sarcomere proteins. The two major genes responsible for HCM are cardiac myosin‐binding protein C (MYBPC3) and beta‐myosin heavy chain (MYH7) accounting for over a half of the HCM cases.1 The utilization of large‐scale sequencing methods has increased the spectrum of the putative HCM‐related genes into non‐sarcomeric genes, but two recent studies have suggested that mutations in non‐sarcomeric genes are rare cause of HCM.2, 3
The genetic background of HCM may be variable across different populations. To our knowledge, only one study investigating a large panel of cardiomyopathy‐related genes in a national HCM cohort has been published.4 The Finnish population has unique genetic background, originated from geographical, linguistic, and cultural isolation of the population.5 Consequently, in Finland, the distribution of rare genetic variants is different from that in other European populations, and a founder effect is frequently observed in the genetics of monogenic diseases.5 We have previously identified three Finnish founder mutations in sarcomere genes (MYBPC3‐Gln1061Ter, MYH7‐Arg1053Gln, and TPM1‐Asp175Asn), accounting for 24% of all cases with HCM in Finland.6, 7 However, there are no previous studies investigating large panels of cardiomyopathy‐related genes using next‐generation sequencing in Finnish patients with HCM.
In early reports derived from referral centres in Europe and the USA, HCM‐related mortality was reported to be as high as 6% per year, but in a more recent report, the prognosis of HCM has been shown to be comparable with the general population.8 Risk stratification and the use of implantable cardiac defibrillator (ICD) in high‐risk patients has improved the prognosis of HCM.8 In a cross‐sectional setting, HCM in Finns is characterized by a relatively benign phenotype,9 but longitudinal observational studies on the natural history and the outcome of HCM have not been previously investigated in Finnish patients with HCM.
In Finland, forensic pathologists of the National Mortality Registry review all deaths and the cause of death. Autopsy is performed in a high percentage of deaths and in all cases of sudden and unclear deaths. To our knowledge, there are no previous studies on nationwide HCM‐related deaths.
The aim of our study was, therefore, firstly, to investigate the genetic basis of HCM in the nationwide cohort of Finnish patients by screening 59 cardiomyopathy‐associated genes with targeted sequencing. Secondly, we investigated HCM‐related mortality and adverse cardiovascular events in a prospective outcome study including Finnish index patients and relatives with HCM. Thirdly, the number of HCM‐related deaths among the 5.2 million population of Finland was examined from records and death certificates of the National Registry during the 10 year follow‐up.
Patients and methods
The diagnosis of hypertrophic cardiomyopathy
The diagnosis of HCM was based on left ventricular hypertrophy (LVH) ≥15 mm in probands and ≥13 mm in relatives on two‐dimensional echocardiography or cardiac magnetic resonance imaging in the absence of other obvious causes for LVH.
Study cohorts and ethical approval
Our study includes three cohorts: (i) FinHCM Genetic Study (n = 382), which includes consecutive unrelated Finnish patients with HCM, as previously described,6, 10 (ii) FinHCM Outcome Study, which consists of 482 Finnish patients with HCM (339 probands and 143 relatives), and (iii) HCM Mortality Study (n = 573) based on the National Mortality Registry including all inhabitants of Finland, who deceased between 2001 and 2010, and whose cause of death was HCM.
The study protocols were in agreement with the Declaration of Helsinki and were approved by the Ethics Committee of the Kuopio University Hospital. All patients gave written informed consent. The permission to use mortality data was accepted by the National Registry of Finland (Statistics Finland).
Genetic analyses of the FinHCM Genetic Study
FinHCM Genetic Study includes consecutive unrelated Finnish patients with HCM (n = 382), recruited from 12 Finnish hospitals (all five university hospitals and seven central hospitals), as previously described.6, 10 Participants of the study underwent a screening of 59 cardiomyopathy‐related genes using the Illumina MiSeq‐sequencer (Supporting Information, Table S1 ). DNA was extracted from peripheral blood leucocytes for genetic analyses. The sequencing panel was designed to include all genes suggested to be associated with non‐syndromic cardiomyopathies at the time of the study onset. Sequencing methods applied in this study have been previously reported,11 and the details are presented in the Supporting Information. All pathogenic (P)/likely pathogenic (LP) variants were confirmed by Sanger sequencing. If an HCM‐related genetic variant was found in the index patient, it was screened in the relatives of the patients, when possible.
Classification of the filtered variants
Variants that underwent the filtering process as described in the Supporting Information were assigned as P, LP, variant of uncertain significance (VUS), likely benign, benign, or a variant with conflicting interpretations of pathogenicity (CON) according to the ClinVar archive (www.ncbi.nlm.nih.gov/clinvar; accessed 21 November 2017). For variants assigned as P, LP, CON favouring pathogenicity (P and/or LP + VUS), or VUS, the allele frequency in Finns and non‐Finnish Europeans was obtained from the Genome Aggregation Database (gnomAD browser beta; http://gnomad.broadinstitute.org; accessed 21 November 2017).
FinHCM Outcome Study
The FinHCM Outcome Study includes 482 Finnish patients (mean age 50.8 ± 15.0 years) with HCM (339 probands, of whom 333 were included also in the genetic study, and 143 relatives). At baseline, the participants of the FinHCM Outcome Study underwent a clinical and echocardiographic evaluation as previously described.6, 10 A 24 h Holter registration and bicycle ergometer test was performed in majority of the patients. In many cases, cardiac magnetic resonance imaging was performed to verify the diagnosis of HCM.
The baseline of the Finnish Outcome Study was defined by the time of initial recruitment (between October 1994 and April 2010). The follow‐up period was until 1 January 2011 in all patients or until the time of death if it occurred before the end of follow‐up. The mean follow‐up time was 6.7 ± 4.2 years (range 0.1–16.2 years). The follow‐up data were collected from hospital medical records by the physician recruiting the patient at baseline. Death records and certificates were obtained from the National Mortality Registry (Statistics Finland) and were available in all but one patient. Causes of death were reclassified as (i) HCM‐related sudden death, (ii) HCM‐related heart failure, (iii) HCM‐related atrial fibrillation and embolic stroke, and (iv) non‐HCM‐related death according to data from death certificates and medical records by two experienced cardiologists (P. J. and J. K.). If the death could not be classified as HCM‐related because of another possible cause (e.g. evidence of acute coronary syndrome related to sudden death) or the cause of death was multifactorial, the death was classified as HCM as a possible contributing factor. Autopsy was performed in 26 (47%) of the deaths.
HCM Mortality Study
HCM Mortality Study (n = 573) included all inhabitants of Finland who deceased between 2001 and 2010 and whose principal cause of death was HCM (ICD10 Diagnosis Number I42.1 or I42.2), or HCM was a contributing factor to death, according to death records from the National Mortality Registry. Autopsy was performed in 62% of deaths. In the present study, two experienced cardiologists (P. J. and J. K.) reviewed the death certificates and reclassified the cause of death as in the FinHCM Outcome Study (see aforementioned).
Statistical analyses
Statistical analyses were performed with the IBM SPSS Statistics 22.0 software (IBM, New York, NY, USA) and R statistical software, version 3.1.1. Data are presented as mean ± standard deviation for continuous variables and n (%) for categorical variables. Demographic and clinical parameters listed in Supporting Information, Table S5 were tested with the Cox regression analysis to investigate the factors predicting HCM‐related mortality or life‐threatening cardiac events (adequate ICD shock therapy, successful cardiopulmonary resuscitation, or cardiac transplantation). The variables that were statistically significant (P < 0.05) in the univariate analysis were further analyzed with a multivariate Cox proportional hazard model. Comparisons between two groups (P/LP vs. no P/LP mutation) were made by using the χ 2 or Mann–Whitney U test, when appropriate.
We applied Kaplan–Meier method to investigate survival. The observed survival of the FinHCM outcome cohort was compared with the expected survival of the Finnish general population (n = 5.2 million) between 1995 and 2010 matched for age and sex. Observed and expected surviving fractions were compared by using one‐sample log‐rank test.
Results
FinHCM Genetic Study
Clinical characteristics
Patients included in the FinHCM Genetic Study (235 male and 147 female; mean age 53 ± 14 years) had mostly moderate LVH with normal left ventricular (LV) dimensions and ejection fraction (Supporting Information, Table S2 ). LV outflow tract obstruction (pressure gradient >30 mmHg) was present in 23 patients (6%). The clinical phenotype of HCM in Finland, particularly in those with founder mutations MYBPC3‐Gln1061Ter12, 13, 14 and TPM1‐Asp175Asn,15, 16, 17, 18, 19 has previously been extensively reported.6, 9, 20
Genetic analyses
After filtering, a total of 230 different rare variants in 50 genes were found in 349 of 382 patients (Figure 1 and Supporting Information, Tables S1 and S3 ). Altogether, 24 P/LP variants were found in four sarcomere (MYBPC3, MYH7, MYL2, and TPM1) and two non‐sarcomeric genes (GLA and PRKAG2) in 146 patients (38.2%; Figure 1 and Table 1). A P/LP mutation was most often found in MYBPC3 (70/382, 18.3%) or MYH7 (48/382, 12.6%). The most frequent variants were MYBPC3‐Gln1061Ter (11.3%),21 MYH7‐Arg1053Gln (7.9%),22 and TPM1‐Asp175Asn (6.3%),10 which have previously been reported by our group, and MYH7‐Val606Met (2.6%). Altogether, these four sarcomere mutations accounted for 28% of the HCM cases and 73% of cases with a P/LP mutation. Mutations (P/LP) in non‐sarcomeric genes GLA and PRKAG2 were found in two and one subjects, respectively (Table 1). In the carriers of the two GLA mutations, a familial Fabry disease was found. None of the study participants had more than one P/LP variant. The allele counts of the P/LP mutations were zero or very low in both Finns and non‐Finnish Europeans according to the Genome Aggregation Database. A P/LP variant was found in 119 of 143 (83.2%) of clinically affected relatives that were included in the FinHCM Outcome Study.
Figure 1.
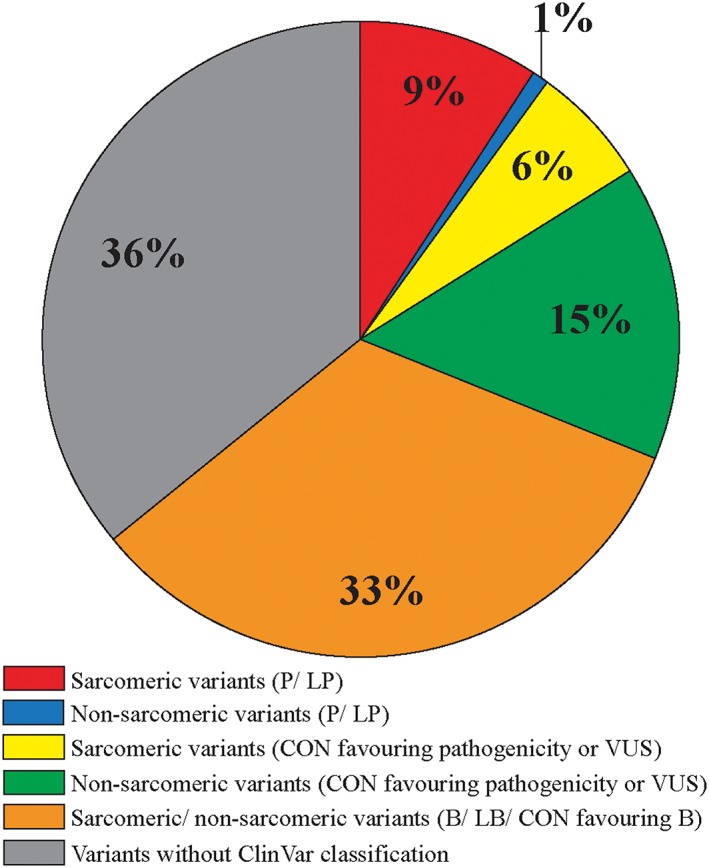
Percentage distribution of genetic variants in 382 Finnish index patients with hypertrophic cardiomyopathy. B, benign; CON, variant with conflicting interpretations of pathogenicity; LB, likely benign; LP, likely pathogenic; P, pathogenic; VUS, variant of uncertain significance.
Table 1.
P/LP mutations associated with cardiomyopathy found in the FinHCM Genetic Study (n = 382)
| Gene | Nucleotide change | Amino acid change | No. of patients with variant, n (%) | Class (ClinVar) | Allele count in Finns (gnomAD) | Allele count in European (non‐Finnish) (gnomAD) |
|---|---|---|---|---|---|---|
| GLA | c.658C>T | p.Arg220Ter | 1 | P | NA | NA |
| c.1228A>G | p.Thr410Ala | 1 | P | NA | NA | |
| MYBPC3 | c.655‐2A>Ca | 3 | NAb | NA | NA | |
| c.927‐2A>G | 1 | P | 0/3494 | 1/14 966 | ||
| c.1227‐13G>Aa | 3 | LP | NA | NA | ||
| c.1358dupC | p.Val454fs | 1 | P/LP | NA | NA | |
| c.1505_1509delGGTTC | p.Arg502fs | 3 | LP | NA | NA | |
| c.1575T>G | p.Tyr525Ter | 2 | P/LP | NA | NA | |
| c.2373dupG | p.Trp792Valfs | 4 | P | 0/17 280 | 3/67 494 | |
| c.2556_2557delCGinsTCTa , c | p.Gly853Leufs | 4 | P | NA | NA | |
| c.3181C>Ta | p.Gln1061Ter | 43 (11.3) | P | 3/16 578 | 0/126 472 | |
| c.3190+5G>A | 5 | P/LP | 0/12 874 | 2/111 318 | ||
| c.3296_3297delGG; c.3295dupG | p.Gly1099fs/p.Tyr1100fs | 1 | P/LP | 3/19 554 | 0/94 384 | |
| MYH7 | c.1178C>T | p.Ala393Val | 1 | LP | NA | NA |
| c.1816G>A | p.Val606Met | 10 (2.6) | P/LP | 0/3492 | 1/14 998 | |
| c.1987C>T | p.Arg663Cys | 1 | P/LP | NA | NA | |
| c.2155C>Ta | p.Arg719Trp | 1 | P | 0/3492 | 1/14 998 | |
| c.2207T>C | p.Ile736Thr | 1 | P | NA | NA | |
| c.2539_2541delAAG | p.Lys847Del | 2 | LP | NA | NA | |
| c.2770G>A | p.Glu924Lys | 2 | P/LP | NA | NA | |
| c.3158G>Aa | p.Arg1053Gln | 30 (7.9) | LP | 17/25 794 | 0/126 704 | |
| MYL2 | c.173G>A | p.Arg58Gln | 1 | P/LP | 2/22 298 | 0/111 682 |
| PRKAG2 | c.905G>A | p.Arg302Gln | 1 | P | NA | NA |
| TPM1 | c.523G>Aa | p.Asp175Asn | 24 (6.3) | P | 4/25 748 | 1/126 700 |
| Total | 146 (38.2) |
Altogether, 49 VUS or CON variants favouring pathogenicity were detected in 31 different genes in 78 patients (20.4%; Figure 1 and Supporting Information, Tables S3 and S4 ). Of these patients, 58 (15.2%) did not have a P/LP mutation. Variants were detected in both sarcomeric (predominantly MYBPC3 and MYH7; 14 variants in 26 patients) and non‐sarcomeric genes (35 variants in 26 genes in 52 patients). Most of the variants (44/49) were missense variants in single patients. All VUS had zero to low allele counts in the Genome Aggregation Database. For 82 rare variants, the ClinVar classification was not available.
FinHCM Outcome Study
Baseline clinical characteristics
Baseline characteristics of most of the patients of the FinHCM Outcome Study have been reported previously.6, 7, 9, 10, 21 Baseline clinical characteristics of all study patients are shown in Supporting Information, Table S5 . The mean maximal thickness of LV on echocardiography was 20 ± 5 mm, and LV dimensions and ejection fraction were within the normal limits in most patients (data not shown). Of the patients, 4 and 5% had a history of pacemaker or ICD implantation, respectively. Only one patient had a cardiac resynchronization therapy device implanted before baseline. None of the patients had undergone heart transplantation before baseline.
Deaths and cardiac events during the 6.7 year follow‐up
The incidence of cardiovascular events in the FinHCM Outcome Study are shown in Supporting Information, Table S6 . The overall survival was 89% (all‐cause mortality 1.70% per year), and 55 patients (11%) died during the follow‐up (Figures 2 and 3 ). All‐cause mortality of the HCM outcome cohort was higher than that of age‐matched and sex‐matched general Finnish population of 5.2 million between 1995 and 2010 (Figure 3 ).
Figure 2.
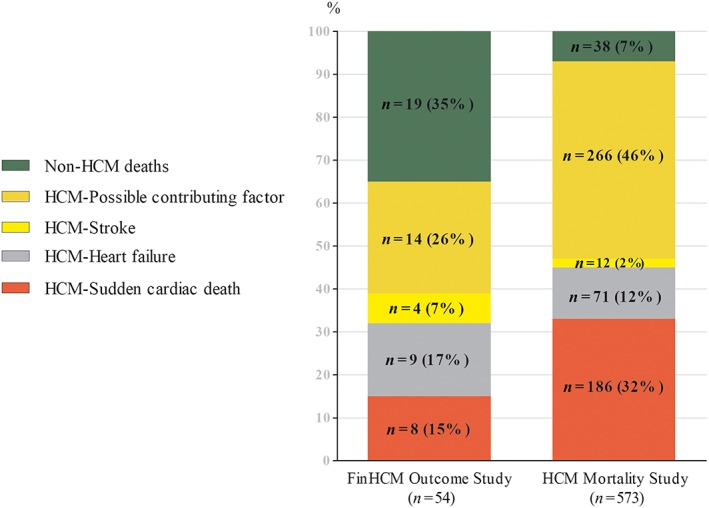
Distribution of the causes of death in the FinHCM Outcome Study (54 deaths) and in the HCM Mortality Study (573 deaths). HCM, hypertrophic cardiomyopathy.
Figure 3.
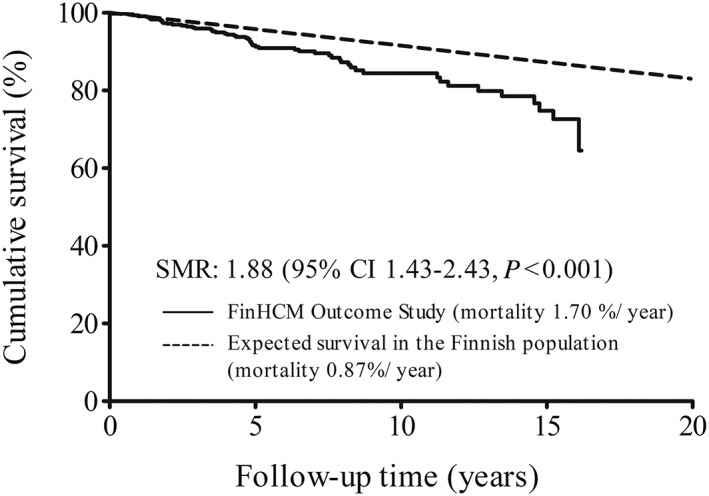
Kaplan–Meier survival in the FinHCM Outcome Study (n = 482) compared with expected all‐cause mortality in the general Finnish population matched for age and sex. CI, confidence interval; SMR, standardized mortality ratio.
Of all deaths of the FinHCM Outcome Study, 38% were HCM‐related (eight sudden cardiac deaths (SCDs), nine heart failures, and four strokes; Figure 2 and Supporting Information, Table S7 ). The overall HCM‐related mortality was 4.4% (0.65% per year). Of those who suffered HCM‐related death, about half had a P/LP mutation. In 14 cases (25%), HCM was a contributing factor to death. The cause of death was not related to HCM in 35% of the patients.
A life‐threatening cardiac event occurred in nine patients during the follow‐up (an adequate ICD shock therapy in four patients, a successful cardiopulmonary resuscitation in one patient, and cardiac transplantation in four patients; three of these patients died later during the follow‐up). Altogether, 27 subjects suffered an HCM‐related death or life‐threatening HCM event, giving the overall event rate 5.6% (0.85% per year) (Supporting Information, Table S6 ). An ICD was implanted in 9% of study subjects. The indication for ICD was secondary prevention in 27% and primary prevention in 63% of implantations. Bradycardia pacemaker or cardiac resynchronization therapy was implanted in only few subjects. LV outflow tract obstruction was treated with either surgical myectomy or alcohol septal ablation in 4% of patients, respectively. Paroxysmal or chronic atrial fibrillation (AF) was present in 20% of patients. New‐onset heart failure was diagnosed in 11% of subjects. Coronary heart disease and cerebrovascular events were uncommon.
Predictors of hypertrophic cardiomyopathy‐related death and life‐threatening hypertrophic cardiomyopathy events
In univariate Cox analysis, advanced functional class (3–4), history of AF, history of ventricular fibrillation/cardiac arrest, pacemaker implantation, diastolic or systolic heart failure at baseline, maximal LV wall thickness, low LV ejection fraction, and the presence of AF at Holter registration were predictors of combined HCM‐related mortality and life‐threatening HCM events (Supporting Information, Table S8 ). In multivariate Cox proportional hazard analysis, only a history of systolic heart failure and maximal LV wall thickness remained statistically significant.
Differences between patients with or without a pathogenic/likely pathogenic mutation
Of the 482 patients with HCM, 255 (136 probands and 119 relatives; 52.9%) had a P/LP mutation shown in Table 1. The patients with a P/LP mutation were more often women (60.7 vs. 47.3%, P = 0.004), were younger (47.8 ± 14.6 vs. 54.1 ± 14.8 years, P < 0.001), had greater maximal LV wall thickness (20.2 ± 5.3 vs. 18.9 ± 4.0 mm, P = 0.011), and had smaller LV end‐diastolic (44.4 ± 6.7 vs. 47.4 ± 6.9 mm, P < 0.001) and end‐systolic (27.8 ± 6.5 vs. 29.8 ± 7.6 mm, P = 0.016) dimensions on echocardiography than those without a P/LP mutation.
During the follow‐up, the patients with a P/LP mutation underwent an ICD implantation more often than patients without a P/LP mutation (12.9 vs. 3.5%, P < 0.001). All‐cause/HCM‐related mortality or combined HCM mortality and life‐threatening events did not differ between the two groups (data not shown).
HCM Mortality Study
Between 2001 and 2010, 573 individuals (56% male, age at death 68.1 ± 16.6 years; 6% <40 years of age) in Finland suffered HCM‐related death. HCM was assigned as the principal cause of death by the National Mortality Registry in 393 of these patients (69%). Two cardiologists of the present study reclassified HCM as the principal cause of death in 269 of 573 cases (47% of all deaths, age at death 63.3 ± 16.5 years; Figure 2 ), including 186 sudden cardiac (32%), 71 heart failure (12%), and 12 stroke deaths (2%), respectively. In 266 subjects, HCM (46%) was reclassified as a contributing factor to death.
Discussion
Principal findings
Targeted sequencing of 59 genes associated with cardiomyopathies revealed P/LP mutations in 38% of HCM cases in the nationwide FinHCM Genetic Study and VUS or CON variants favouring pathogenicity in 20%. The four most frequent sarcomeric mutations accounted for 28% of the HCM cases, indicating that these few major mutations explain a substantial number of HCM cases in Finland. In contrast, P/LP variants in non‐sarcomeric genes were rare and found only in GLA and PRKAG2 genes in three patients. In the FinHCM Outcome Study, annual all‐cause mortality exceeded that of the age‐matched and gender‐matched general Finnish population. History of systolic heart failure and maximal LV wall thickness were independent predictors of HCM‐related mortality and life‐threatening cardiac events. The patients with a P/LP mutation underwent an ICD implantation more often than patients without a P/LP mutation, but all‐cause/HCM‐related mortality or combined HCM mortality and life‐threatening events did not differ between the two groups. Finally, among 5.2 million Finns, there were at least 27 HCM‐related deaths annually.
FinHCM Genetic Study
A P/LP mutation was identified in 38% of the cases, mainly in sarcomere genes. P/LP variants in non‐sarcomeric genes were rare. There are only few studies on targeted sequencing of comprehensive cardiomyopathy gene panels in large patient populations with HCM2, 4, 23, 24, 25 and only one in a national HCM cohort.4 Back in 2003, a French group reported a disease‐causing mutation in 63% (124/197) of unrelated HCM patients who underwent a screening of only nine core sarcomeric protein genes.26 In their study, the pathogenicity of variants was determined by traditional methods (e.g. co‐segregation with HCM and absence in controls). In a more recent report, in the screening of 41 genes, a potential HCM‐ related variant was found in 64% of 223 patients, but pathogenicity of variants was not validated against any currently available genetic databases.4 In another recent study, in 31 genes tested, P/LP variants were found in 39.9% of cases almost exclusively in sarcomeric genes.2 Expanded gene panels do not appear to substantially increase sensitivity of HCM testing beyond gene panels including eight core sarcomere genes, GLA, PRKAG2, and LAMP2.23 Generally, because of the variability in patient cohorts, genetic methodology, and interpretation of results, the comparison of the results of published studies with targeted sequencing is challenging.
The present study highlights the fundamental role of four sarcomeric mutations in the pathogenesis of HCM in the Finnish population. We have previously reported three major Finnish HCM‐causing mutations, MYBPC3‐Gln1061Ter, MYH7‐Arg1053Gln, and TPM1‐Asp175Asn, which are rare in other populations.6, 7 Now, we report another major mutation, MYH7‐Val606Met, in 2.6% of patients. These four mutations explained almost one‐third of the HCM cases and more than two‐thirds of the cases with a P or LP mutation. Likewise, in a nationwide HCM study in Iceland, a single MYBPC3 mutation c.927‐2A>G accounted for 58% of all HCM cases.27 The occurrence of distinct major mutations for HCM in genetically isolated populations emphasizes the need for national genetic studies in HCM.27, 28
In our study, 15% of the patients had a VUS or CON variant favouring pathogenicity without another likely causal variant for HCM. Moreover, for a considerable number of rare variants, ClinVar classification was not available. Some of these variants might be actual disease‐causing mutations, and further studies are needed to determine their causality. Finally, a considerable number of patients did not have a P/LP, VUS, or CON variant in any of the genes tested. Further studies are needed to investigate if these patients have mutations in unknown genes for HCM, in regulatory non‐coding regions of already identified causal genes, or LVH‐inducing variants in multiple genes.
FinHCM Outcome Study
In the FinHCM Outcome Study, the annual all‐cause mortality (1.70%) was higher than that expected (0.87%) in the matched general Finnish population, and annual HCM‐related mortality was 0.65%. There are only a few previous mortality studies including mainly US patient populations with HCM.29, 30 All‐cause mortality in the present study is in agreement with a study of US HCM patients29 but contrasts a retrospective study of 706 HCM patients, where the overall survival was not different from that of matched White US population.30 The annual HCM‐related mortality in the present study is comparable with HCM‐related mortality reported in other adult HCM cohorts.29, 30 A rather favourable outcome in our study is likely to be explained by the inclusion of relatives, who usually have milder disease than probands. Furthermore, if the 14 deaths classified as contributors to death were classified as HCM deaths, HCM‐related mortality would have been higher.
Sudden cardiac death and heart failure were the major mechanisms of HCM‐related death in the present study, followed by embolic stroke. In a previous study, the mechanisms of HCM deaths were most often SCD and heart failure.29
A history of systolic heart failure and maximal LV wall thickness, both well‐known risk factors for HCM‐related death,8 were independent predictors of combined HCM‐related death and life‐threatening events. Many of the other risk factors for SCD did not predict HCM‐related mortality and life‐threatening HCM events in the present study, possibly because of small number of deaths.
The presence of a P/LP mutation was associated with female gender, younger age, thicker LV wall, and smaller LV dimensions. The phenotypic expression of the two major mutations MYBPC3‐Gln1061Ter and TPM1‐D175N has been investigated in detail in our previous study.6 Although there was no difference in HCM‐related mortality or combined HCM‐related mortality and life‐threatening events, patients with a P/LP mutation were more likely to undergo an ICD implantation. In a recent study, it was reported that young age at diagnosis and the presence of a sarcomere mutation are associated with an adverse clinical outcome.31
HCM Mortality Study
The HCM Mortality Study shows that in Finland, with a population of 5.2 million, at least 27 deaths annually are attributed to HCM. To our knowledge, there are no previous nationwide studies on HCM‐related deaths. About 70% of the reclassified HCM‐related deaths in Finland were sudden. Undoubtedly, post‐mortem diagnosis of HCM at autopsy may be challenging. However, our findings suggest that a considerable number of subjects suffer an HCM‐related death in Finland and that the proportion of SCDs compared with other causes of death is higher at the population level than that in patient populations participating in clinical studies. As the HCM Mortality Study cohort was based on patients with an HCM‐related death according to death certificates, the actual HCM‐related mortality at the population level might be even higher. For example, the low frequency of embolic stroke in the HCM Mortality Study cohort might be explained by that not all fatal embolic strokes suffered by HCM patients were classified as HCM‐related in death certificates.
Clinical implications
Screening of a panel of 59 cardiomyopathy genes identifies a causative mutation in about 40% of Finnish patients with HCM. Screening of the four major mutations identifies genetic cause for HCM in about 30% and should be a primary genetic testing in Finland, even if large‐scale genetic panels are used.
All‐cause mortality exceeded the expected all‐cause mortality in the general Finnish population, but the rate of HCM‐related deaths and life‐threatening HCM events was relatively low. However, patients with marked cardiac hypertrophy or heart failure should be monitored carefully to reduce HCM‐related mortality. Finally, a considerable number of HCM‐related deaths, particularly SCDs, in the Finnish population suggest that there are undiagnosed and untreated patients with HCM who would benefit of more effective diagnostic and treatment options.
Strengths and limitations of the study
First, a comprehensive panel of 59 genes associated with cardiomyopathy was used in the screening of the large nationwide FinHCM genetic cohort, but not all putative genes associated with HCM were investigated. Thus, some disease‐causing mutations may have been missed. However, as the frequency of P/LP mutations in other than the eight core sarcomeric genes is very low in the present study and few recently published reports,2, 3 major mutations were probably not missed.
Second, pathogenicity of the genetic variants was determined according to the present ClinVar archive classification. The ClinVar classification, however, was not available for all rare variants, and some of the variants classified as variants with unknown significance may be disease causing.
Third, for variants assigned as P, LP, or VUS, the allele frequencies in Finns and non‐Finnish Europeans were obtained from the Genome Aggregation Database. Only few previous studies have included population‐based allele frequencies to investigate rare protein‐altering variants for HCM.2, 4
Fourth, for the present study, medical records and mortality data from the National Mortality Registry were available for almost all subjects and deaths, and autopsy was performed in a considerable number of deaths, increasing reliability of our findings.
Conclusions
By using targeted genetic testing of a large cardiomyopathy panel, a causative mutation was found in approximately 40% of Finnish patients with HCM. In Finnish patients, the genetic profile of HCM is unique, as four major mutations in the sarcomeric genes account for one‐third of all cases and three‐quarters of cases with P or LP mutation. HCM‐related mortality in patients with diagnosed HCM was low, but all‐cause mortality was higher than that expected in the general population. Finally, according to the National Registry, at least five HCM‐related deaths per million inhabitants occur annually in Finland.
Conflict of interest
None declared.
Funding
This work was supported by the Academy of Finland, the Finnish Foundation for Cardiovascular Research, and the Kuopio University Hospital (EVO grants to J.K.).
Supporting information
Data S1. Supporting information.
Table S1. List of the 59 genes included in the FinHCM Genetic Study screening.
Table S2. Clinical characteristics of the FinHCM Genetic Study (n=382).
Table S3. A list of all 382 patients with HCM included in the FinHCM Genetic Study, and the rare variants (GMAF > 1 %) detected in each patient.
Table S4. Variants with conflicting evidence favouring pathogenicity, or with uncertain significance found in the FinHCM Genetic Study (n=382).
Table S5. Baseline characteristics of the patients of the FinHCM Outcome Study (n=482).
Table S6. New events in the FinHCM Outcome Study (n=482).
Table S7. HCM‐related deaths and their circumstances in the FinHCM Outcome Study.
Table S8. Univariate and multivariate predictors (Cox regression) of combined HCM‐related mortality and life‐threatening HCM events in the FinHCM Outcome Study.
Acknowledgements
We thank Raija Miettinen, MSc, Satu Nenonen, RN, Eila Ruotsalainen, RN, Sini Weckström, RN, and Minna Rautiainen, RN, for the assistance in data collection and biostatistician Tuomas Selander for the assistance in statistical analyses.
Jääskeläinen P., Vangipurapu J., Raivo J., Kuulasmaa T., Heliö T., Aalto‐Setälä K., Kaartinen M., Ilveskoski E., Vanninen S., Hämäläinen L., Melin J., Kokkonen J., Nieminen M. S., The FinHCM Study Group , Laakso M., and Kuusisto J. (2019) Genetic basis and outcome in a nationwide study of Finnish patients with hypertrophic cardiomyopathy, ESC Heart Failure, 6: 436–445. 10.1002/ehf2.12420.
References
- 1. Burke MA, Cook SA, Seidman JG, Seidman CE. Clinical and mechanistic insights into the genetics of cardiomyopathy. J Am Coll Cardiol 2016; 68: 2871–2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Walsh R, Buchan R, Wilk A, John S, Felkin LE, Thomson KL, Chiaw TH, Loong CC, Pua CJ, Raphael C, Prasad S, Barton PJ, Funke B, Watkins H, Ware JS, Cook SA. Defining the genetic architecture of hypertrophic cardiomyopathy: re‐evaluating the role of non‐sarcomeric genes. Eur Heart J 2017; 38: 3461–3468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mademont‐Soler I, Mates J, Yotti R, Espinosa MA, Pérez‐Serra A, Fernandez‐Avila AI, Coll M, Méndez I, Iglesias A, Del Olmo B, Riuró H, Cuenca S, Allegue C, Campuzano O, Picó F, Ferrer‐Costa C, Álvarez P, Castillo S, Garcia‐Pavia P, Gonzalez‐Lopez E, Padron‐Barthe L, Díaz de Bustamante A, Darnaude MT, González‐Hevia JI, Brugada J, Fernandez‐Aviles F, Brugada R. Additional value of screening for minor genes and copy number variants in hypertrophic cardiomyopathy. PLoS One 2017; 12: e0181465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lopes LR, Zekavati A, Syrris P, Hubank M, Giambartolomei C, Dalageorgou C, Jenkins S, McKenna W, Uk10k Consortium , Plagnol V, Elliott PM. Genetic complexity in hypertrophic cardiomyopathy revealed by high‐throughput sequencing. J Med Genet 2013; 50: 228–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lim ET, Würtz P, Havulinna AS, Palta P, Tukiainen T, Rehnström K, Esko T, Mägi R, Inouye M, Lappalainen T, Chan Y, Salem RM, Lek M, Flannick J, Sim X, Manning A, Ladenvall C, Bumpstead S, Hämäläinen E, Aalto K, Maksimow M, Salmi M, Blankenberg S, Ardissino D, Shah S, Horne B, McPherson R, Hovingh GK, Reilly MP, Watkins H, Goel A, Farrall M, Girelli D, Reiner AP, Stitziel NO, Kathiresan S, Gabriel S, Barrett JC, Lehtimäki T, Laakso M, Groop L, Kaprio J, Perola M, McCarthy MI, Boehnke M, Altshuler DM, Lindgren CM, Hirschhorn JN, Metspalu A, Freimer NB, Zeller T, Jalkanen S, Koskinen S, Raitakari O, Durbin R, MacArthur DG, Salomaa V, Ripatti S, Daly MJ, Palotie A, Sequencing Initiative Suomi (SISu) Project . Distribution and medical impact of loss‐of‐function variants in the Finnish founder population. PLoS Genet 2014; 10: e1004494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jääskeläinen P, Heliö T, Aalto‐Setälä K, Kaartinen M, Ilveskoski E, Hämäläinen L, Melin J, Nieminen MS, Laakso M, Kuusisto J, for the FinHCM Study Group . Two founder mutations in the alpha‐tropomyosin and the cardiac myosin‐binding protein C genes are common causes of hypertrophic cardiomyopathy in the Finnish population. Ann Med 2013;45:85–90. [DOI] [PubMed] [Google Scholar]
- 7. Jääskeläinen P, Heliö T, Aalto‐Setälä K, Kaartinen M, Ilveskoski E, Hämäläinen L, Melin J, Kärkkäinen S, Peuhkurinen K, Nieminen MS, Laakso M, FinHCM Study Group , Kuusisto J. A new common mutation in the cardiac beta‐myosin heavy chain gene in Finnish patients with hypertrophic cardiomyopathy. Ann Med 2014; 46: 424–429. [DOI] [PubMed] [Google Scholar]
- 8. Maron BJ, Rowin EJ, Casey SA, Maron MS. How hypertrophic cardiomyopathy became a contemporary treatable genetic disease with low mortality: shaped by 50 years of clinical research and practice. JAMA Cardiol 2016; 1: 98–105. [DOI] [PubMed] [Google Scholar]
- 9. Jääskeläinen P, Miettinen R, Kärkkäinen P, Toivonen L, Laakso M, Kuusisto J. Genetics of hypertrophic cardiomyopathy in eastern Finland: few founder mutations with benign or intermediary phenotypes. Ann Med 2004; 36: 23–32. [DOI] [PubMed] [Google Scholar]
- 10. Jääskeläinen P, Soranta M, Miettinen R, Saarinen L, Pihlajamäki J, Silvennoinen K, Tikanoja T, Laakso M, Kuusisto J. The cardiac beta‐myosin heavy chain gene is not the predominant gene for hypertrophic cardiomyopathy in the Finnish population. J Am Coll Cardiol 1998; 32: 1709–1716. [DOI] [PubMed] [Google Scholar]
- 11. Löf C, Patyra K, Kuulasmaa T, Vangipurapu J, Undeutsch H, Jaeschke H, Pajunen T, Kero A, Krude H, Biebermann H, Kleinau G, Kühnen P, Rantakari K, Miettinen P, Kirjavainen T, Pursiheimo JP, Mustila T, Jääskeläinen J, Ojaniemi M, Toppari J, Ignatius J, Laakso M, Kero J. Detection of novel gene variants associated with congenital hypothyroidism in a Finnish patient cohort. Thyroid 2016; 26: 1215–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jørgenrud B, Jalanko M, Heliö T, Jääskeläinen P, Laine M, Hilvo M, Nieminen MS, Laakso M, Hyötyläinen T, Orešič M, Kuusisto J. The metabolome in Finnish carriers of the MYBPC3‐Q1061X mutation for hypertrophic cardiomyopathy. PLoS One 2015; 10: e0134184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tarkiainen M, Sipola P, Jalanko M, Heliö T, Laine M, Järvinen V, Häyrinen K, Lauerma K, Kuusisto J. Cardiovascular magnetic resonance of mitral valve length in hypertrophic cardiomyopathy. J Cardiovasc Magn Reson 2016; 18: 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jalanko M, Tarkiainen M, Sipola P, Jääskeläinen P, Lauerma K, Laine M, Nieminen MS, Laakso M, Heliö T, Kuusisto J. Left ventricular mechanical dispersion is associated with nonsustained ventricular tachycardia in hypertrophic cardiomyopathy. Ann Med 2016; 48: 417–427. [DOI] [PubMed] [Google Scholar]
- 15. Sipola P, Lauerma K, Husso‐Saastamoinen M, Kuikka JT, Vanninen E, Laitinen T, Manninen H, Niemi P, Peuhkurinen K, Jääskeläinen P, Laakso M, Kuusisto J, Aronen HJ. First‐pass MR imaging in the assessment of perfusion impairment in patients with hypertrophic cardiomyopathy and the Asp175Asn mutation of the alpha‐tropomyosin gene. Radiology 2003; 226: 129–137. [DOI] [PubMed] [Google Scholar]
- 16. Hedman A, Hartikainen J, Vanninen E, Laitinen T, Jääskeläinen P, Laakso M, Peuhkurinen K, Kuusisto J. Inducibility of life‐threatening ventricular arrhythmias is related to maximum left ventricular thickness and clinical markers of sudden cardiac death in patients with hypertrophic cardiomyopathy attributable to the Asp175Asn mutation in the alpha‐tropomyosin gene. J Mol Cell Cardiol 2004; 36: 91–99. [DOI] [PubMed] [Google Scholar]
- 17. Sipola P, Lauerma K, Jääskeläinen P, Laakso M, Peuhkurinen K, Manninen H, Aronen HJ, Kuusisto J. Cine MR imaging of myocardial contractile impairment in patients with hypertrophic cardiomyopathy attributable to Asp175Asn mutation in the alpha‐tropomyosin gene. Radiology 2005; 236: 815–824. [DOI] [PubMed] [Google Scholar]
- 18. Sipola P, Peuhkurinen K, Lauerma K, Husso M, Jääskeläinen P, Laakso M, Aronen HJ, Risteli J, Kuusisto J. Myocardial late gadolinium enhancement is associated with raised serum amino‐terminal propeptide of type III collagen concentrations in patients with hypertrophic cardiomyopathy attributable to the Asp175Asn mutation in the alpha tropomyosin gene: magnetic resonance imaging study. Heart 2006; 92: 1321–1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kuusisto J, Kärjä V, Sipola P, Kholová I, Peuhkurinen K, Jääskeläinen P, Naukkarinen A, Ylä‐Herttuala S, Punnonen K, Laakso M. Low‐grade inflammation and the phenotypic expression of myocardial fibrosis in hypertrophic cardiomyopathy. Heart 2012; 98: 1007–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kuusisto J, Sipola P, Jääskeläinen P, Naukkarinen A. Current perspectives in hypertrophic cardiomyopathy with the focus on patients in the Finnish population: a review. Ann Med 2016; 48: 496–508. [DOI] [PubMed] [Google Scholar]
- 21. Jääskeläinen P, Kuusisto J, Miettinen R, Kärkkäinen P, Kärkkäinen S, Heikkinen S, Peltola P, Pihlajamäki J, Vauhkonen I, Laakso M. Mutations in the cardiac myosin‐binding protein C gene are the predominant cause of familial hypertrophic cardiomyopathy in eastern Finland. J Mol Med 2002; 80: 412–422. [DOI] [PubMed] [Google Scholar]
- 22. Kärkkäinen S, Heliö T, Jääskeläinen P, Miettinen R, Tuomainen P, Ylitalo K, Kaartinen M, Reissell E, Toivonen L, Nieminen MS, Kuusisto J, Laakso M, Peuhkurinen K. Two novel mutations in the beta‐myosin heavy chain gene associated with dilated cardiomyopathy. Eur J Heart Fail 2004; 6: 861–868. [DOI] [PubMed] [Google Scholar]
- 23. Alfares AA, Kelly MA, McDermott G, Funke BH, Lebo MS, Baxter SB, Shen J, McLaughlin HM, Clark EH, Babb LJ, Cox SW, DePalma SR, Ho CY, Seidman JG, Seidman CE, Rehm HL. Results of clinical genetic testing of 2,912 probands with hypertrophic cardiomyopathy: expanded panels offer limited additional sensitivity. Genet Med 2015; 17: 880–888. [DOI] [PubMed] [Google Scholar]
- 24. Rubattu S, Bozzao C, Pennacchini E, Pagannone E, Musumeci BM, Piane M, Germani A, Savio C, Francia P, Volpe M, Autore C, Chessa L. A next‐generation sequencing approach to identify gene mutations in early‐ and late‐onset hypertrophic cardiomyopathy patients of an Italian cohort. Int J Mol Sci 2016; 17: E1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Walsh R, Thomson KL, Ware JS, Funke BH, Woodley J, McGuire KJ, Mazzarotto F, Blair E, Seller A, Taylor JC, Minikel EV, Exome Aggregation Consortium , MacArthur DG, Farrall M, Cook SA, Watkins H. Reassessment of Mendelian gene pathogenicity using 7,855 cardiomyopathy cases and 60,706 reference samples. Genet Med 2017; 19: 192–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Richard P, Charron P, Carrier L, Ledeuil C, Cheav T, Pichereau C, Benaiche A, Isnard R, Dubourg O, Burban M, Gueffet JP, Millaire A, Desnos M, Schwartz K, Hainque B, Komajda M, EUROGENE Heart Failure Project . Hypertrophic cardiomyopathy: distribution of disease genes, spectrum of mutations, and implications for a molecular diagnosis strategy. Circulation 2003; 107: 2227–2232. [DOI] [PubMed] [Google Scholar]
- 27. Adalsteinsdottir B, Teekakirikul P, Maron BJ, Burke MA, Gudbjartsson DF, Holm H, Stefansson K, DePalma SR, Mazaika E, McDonough B, Danielsen R, Seidman JG, Seidman CE, Gunnarsson GT. Nationwide study on hypertrophic cardiomyopathy in Iceland: evidence of a MYBPC3 founder mutation. Circulation 2014; 130: 1158–1167. [DOI] [PubMed] [Google Scholar]
- 28. Alders M, Jongbloed R, Deelen W, van den Wijngaard A, Doevendans P, Ten Cate F, Regitz‐Zagrosek V, Vosberg HP, van Langen I, Wilde A, Dooijes D, Mannens M. The 2373insG mutation in the MYBPC3 gene is a founder mutation, which accounts for nearly one‐fourth of the HCM cases in the Netherlands. Eur Heart J 2003; 24: 1848–1853. [DOI] [PubMed] [Google Scholar]
- 29. Maron BJ, Rowin EJ, Casey SA, Link MS, Lesser JR, Chan RH, Garberich RF, Udelson JE, Maron MS. Hypertrophic cardiomyopathy in adulthood associated with low cardiovascular mortality with contemporary management strategies. J Am Coll Cardiol 2015; 65: 1915–1928. [DOI] [PubMed] [Google Scholar]
- 30. Hebl VB, Miranda WR, Ong KC, Hodge DO, Bos JM, Gentile F, Klarich KW, Nishimura RA, Ackerman MJ, Gersh BJ, Ommen SR, Geske JB. The natural history of nonobstructive hypertrophic cardiomyopathy. Mayo Clin Proc 2016; 91: 279–287. [DOI] [PubMed] [Google Scholar]
- 31. Ho CY, Day SM, Ashley EA, Michels M, Pereira AC, Jacoby D, Cirino AL, Fox JC, Lakdawala NK, Ware JS, Caleshu CA, Helms AS, Colan SD, Girolami F, Cecchi F, Seidman CE, Sajeev G, Signorovitch J, Green EM, Olivotto I. Genotype and lifetime burden of disease in hypertrophic cardiomyopathy: insights from the Sarcomeric Human Cardiomyopathy Registry (SHaRe). Circulation 2018; 138: 1387–1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1. Supporting information.
Table S1. List of the 59 genes included in the FinHCM Genetic Study screening.
Table S2. Clinical characteristics of the FinHCM Genetic Study (n=382).
Table S3. A list of all 382 patients with HCM included in the FinHCM Genetic Study, and the rare variants (GMAF > 1 %) detected in each patient.
Table S4. Variants with conflicting evidence favouring pathogenicity, or with uncertain significance found in the FinHCM Genetic Study (n=382).
Table S5. Baseline characteristics of the patients of the FinHCM Outcome Study (n=482).
Table S6. New events in the FinHCM Outcome Study (n=482).
Table S7. HCM‐related deaths and their circumstances in the FinHCM Outcome Study.
Table S8. Univariate and multivariate predictors (Cox regression) of combined HCM‐related mortality and life‐threatening HCM events in the FinHCM Outcome Study.


