Abstract
Objective(s):
Resistance to carbapenems is the principal reason for the continuing utilization of colistin as a last resort choice for treating the infections resulted from multidrug carbapenem-resistant Pseudomonas aeruginosa (CRPA) isolates. The assessment of antimicrobial resistance pattern, the prevalence of carbapenem-resistance determinants, and molecular epidemiology of colistin-resistant isolates among CRPA strains were the aims of the present research.
Materials and Methods:
The current cross-sectional research was conducted on 269 CRPA isolates collected from various clinical samples from 2013 to 2016. After performing identification tests, disk diffusion as well as MIC methods were used for testing sensitivity to the antibiotics. Modified Hodge Test (MHT) was utilized to produce carbapenemase. PCR technique identified beta-lactamase classes A, B, and D genes.
Results:
In total, from 269 CRPA, five isolates (1.3%) were resistant to colistin. It was found that blaNDM-1, blaIMP-1, blaVIM-2, and blaOXA-10 genes were present in 40%, 40%, 20%, and 100% of colistin-resistant isolates, respectively. DLST type 25-11 is a significant cluster of colistin-resistant P. aeruginosa isolates.
Conclusion:
The appearance of colistin-resistant isolates in CRPA carrying blaNDM-1 with multiple carbapenem-resistant genes shows the great problem in the treatment of P. aeruginosa infections.
Key Words: bla NDM-1, Colistin, Double-locus sequence typing, Drug resistance, Pseudomonas aeruginosa
Introduction
Pseudomonas aeruginosa is a highly opportunistic pathogen. Carbapenems are the antibiotics which are utilized for treating multidrug-resistant P.aeruginosa (MDRP) isolates. Carbapenem antibiotics used to be effective agents against MDRP when first presented. However, the growing prevalence of carbapenem-resistant P. aeruginosa (CRPA) has turned into a severe health problem recently (1-3). These strains lead to high mortality rates in patients infected by P. aeruginosa and there are also few effective drugs against them. Colistin is a key antimicrobial agent used to treat P. aeruginosa infections (1, 2). Resistance to carbapenems can be related to the production of carbapenemase enzymes such as serine carbapenemases (containing KPC and GES enzymes), metallo-β-lactamases (MBLs) such as IMP, VIM, and NDM enzymes, and oxacillinases (such as OXA enzymes) (4). blaIMP and blaVIM are the most frequently acquired MBLs. However, the recently emerged NDM-type (New Delhi metallo-β-lactamases) is becoming the most important carbapenemase (4, 5). Most blaNDM-1 strains are resistant to a wide range of antibiotic groups, for example to aminoglycosides, sulfonamides, fluoroquinolones, and macrolides (4, 6). The clinically used polymyxin is effective against NDM-positive organisms (7). Polymyxins are antibiotics with a general structure containing a cyclic peptide and comprise five chemically different compounds (A–E). Polymyxins B and E (colistin) are used for curing Gram-negative bacterial pathogens (8). The great amount of activity that colistin has against many species of bacterial infections has been described in the recent literature. However, the use of colistin has been limited because it has serious neurotoxicity and nephrotoxicity (9). Colistin acts by connecting itself to lipopolysaccharide in the outer membrane of Gram-negative bacteria (10, 11). Although colistin resistance mechanisms have not been exactly understood, two principal functions of resistance to colistin in bacteria are adaptation and mutation (11). Various molecular typing methods have been utilized for analyzing the epidemiology of P. aeruginosa. Among these methods, pulsed-field gel electrophoresis (PFGE) and multilocus sequence typing (MLST) are time-consuming and expensive and require specific technical abilities (12, 13). The recently described double-locus sequence typing (DLST) pattern permits us to gain a standardized and clear definition of types according to the partial sequencing of two extremely inconstant loci for typing P. aeruginosa isolates (12, 13). DLST has a remarkable discriminatory power and reproducibility and can detect high-risk endemic clones (13). Although reports of colistin-resistant cases are rare, its incidence is considered a serious menace. Hence, the present research has been designed to study the antimicrobial resistance pattern, the frequency of carbapenem-resistant determinants, and molecular epidemiology of colistin-resistant isolates among the CRPA collected from hospitalized patients in Iran by employing genotypic, typing, and phenotypic techniques.
Materials and Methods
Bacterial isolation and identification
This cross-sectional work was conducted in three main hospitals in Ahvaz, Tehran, and Isfahan, Iran within a three-years period from October 2013 to July 2016.
This research was approved by Ethics Committee of Ahvaz Jundishapur University of Medical Sciences, Iran and conformed to the declaration of Helsinki. Overall 236 non-duplicate carbapenem-resistant P. aeruginosa isolates were collected from different medical specimens. By using standard microbiological techniques and also the genotypic method (the presence of the gyrB gene) (15), the bacteria were detected. The confirmed isolates were kept at the temperature of -80 °C in Trypticase Soy Broth comprising 15% glycerol.
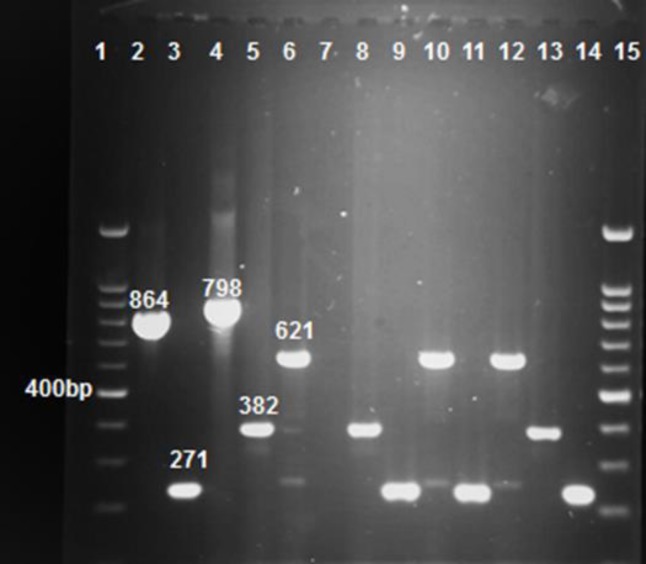
Figure 1.
Gel electrophoresis of multiplex PCR products following amplification with specific primers. Line 1 and 15 ladder, line 2, 3, 4, 5, and 6 positive control bla KPC, bla IMP, bla GES, bla VIM and bla NDM (864, 271, 798, 382, and 621 bp, respectively), line 7 deionized water as control negative, line 8-14 samples. All positive controls were provided by the Pasteur Institute, Iran
Testing antimicrobial sensitivity
The antibiotic sensitivity of all the isolates was tested by employing the Kirby-Bauer’s technique as suggested by the CLSI (16). The twelve standard antibiotic disks used include: IPM; imipenem (10 μg), MEM; meropenem (10 μg), ETP; ertapenem (10 μg), CTX; cefotaxime (30 μg), CT; ceftazidime (30 μg), FEP; cefepime (30 μg), GEN; gentamicin (10 μg), AN; amikacin (30 μg), TZP; piperacillin/tazobactam (100/10 μg), CIP; ciprofloxacin (5 μg), CST; colistin (10 μg), and ATM; aztreonam (30 μg) (Mast Group Ltd, UK). Isolates with resistance against a minimum of three groups of antibacterial agents were considered as MDR (17). The MHT was carried out to detect the carbapenemase-generating strains as recommended by CLSI (16). To detect ESBL-producing isolates, the combined disk technique by disks of ceftazidime (30 mg) with (10 mg) and without clavulanic acid (Mast Group Ltd, UK) was used. A growth in the area diameter of ≥5 mm around ceftazidime disc with and without clavulanic acid was assumed to be a positive result for ESBL production (18).
E-test
The E-test (imipenem 0.002-32μg/ml and colistin 0.064-1024 μg/ml) (Liofilchem, Roseto degli Abruzzi, Italy) was conducted based on the guidelines of the manufacturer. The tests were considered positive for imipenem and colistin when the ratio was ≥ 8 μg/ml (16). The E-test method was used to specify the minimum inhibitory concentrations (MICs) of imipenem and colistin (for colistin, MIC was detected only in resistant isolates by using Kirby-Bauer’s technique).
PCR amplification of carbapenem resistant genes
A DNA extraction set (Sinaclon, Iran) was employed for DNA extraction from the colistin-resistant isolates based on the guidelines of the manufacturer. PCR amplification for the detection of blaNDM, blaIMP, blaVIM, blaKPC, blaGES, blaSPM, and blaOXA-10 was done using specific primers as described previously (Table 1) (19). In this study, a pentaplex PCR assay was used in a thermal cycler (Eppendorf AG, Germany), with an initial denaturation of 4 min at the temperature of 94 °C followed by 30 cycles of a denaturation of 60 sec at the temperature of 94 °C, annealing 56 °C for blaOXA-10, 59 °C for blaSPM, and 55 °C pentaplex PCR and extension of 60 sec at 72 °C, with a single final extension of 7 min at 72 °C (19). Bioneer Company (Bioneer, Daejeon, South Korea) accomplished the sequencing of the amplicons. BLAST in NCBI was employed to analyze the nucleotide sequences.
Table 1.
Primers used in this study
| Primers | Sequence (5′–3′) | Product size (bp) | Reference |
|---|---|---|---|
| ms172 | GGATTCTCTCGCACGAGGT TACGTGACCTGACGTTGGTG |
400 | |
| ms217 | TTCTGGCTGTCGCGACTGAT GAACAGCGTCTTTTCCTCGC |
350 | |
| bla NDM-1 | GGTTTGGCGATCTGGTTTTC CGGAATGGCTCATCACGATC |
621 | |
| bla IMP | GGAATAGAGTGGCTTAATTC GGTTTAACAAAACAACCACC |
233 | |
| bla VIM | GTTTGGTCGCATATCGCAAC AATGCGCAGCACCAGGATAG |
382 | |
| bla SPM | AAAATCTGGGTACGCAAACG ACATTATCCGCTGGAACAGG |
271 | |
| bla KPC | CGTCTAGTTCTGCTGTCTTG CTTGTCATCCTTGTTAGGCG |
798 | |
| bla OXA-10 | ATTATCGGCCTAGAAACTGG CTTACTTCGCCAACTTCTCTG |
170 |
Double-locus sequence typing method
On the DLST website, the full procedure for DLST technique of P. aeruginosa is accessible. In brief, nucleotide extracts were utilized for PCR production of both ms172 and ms217 loci by employing particular primers (Table 1). The typical gel electrophoresis was performed, and gels marked with ethidium bromide (Sinaclon, Iran) were surveyed below UV light for the existence of one detectable perfect band for each PCR. The sizes of DNA sequences varied among strains. Bioneer Corporation (Bioneer, Daejeon, South Korea) purified and sequenced the PCR products. For allele assignment, the sequences were submitted to www.dlst.org. Two numbers were granted to each strain and represented its DLST type. In case no result for allele assignment was obtained, it was considered null (13, 20).
Statistical analysis
SPSSTM software, version 19 (IBM Corp, USA) was employed for the statistical analysis.
Results
In total, 236 isolates of carbapenem-resistant P. aeruginosa were collected from 369 patents. The outcomes of the antibiotic sensitivity pattern demonstrated that 90.7% and 90.7% of the isolates had resistance against imipenem and meropenem, respectively. Table 2 presents the complete outcomes of the antibiotic resistance pattern for all CRPA isolates. Most CRPA isolates were collected from urine samples [28 isolates (21%)], followed by trachea samples (19 isolates (16.3%)), and wound samples [18 isolates (15.5%)]. The number of ESBLs in CRPA carrying MBL was 13 (11.2%) of which one was a colistin-resistant isolate. Five CRPA isolates resistance against colistin were collected from urine, blood, and trachea specimens from ICU and gynecology wards. Nevertheless, two of these isolates were carrying blaNDM-1 gene and three of them had other MBL genes. All colistin-resistant isolates were carrying blaOXA-10 gene. However, blaKPC, bla GES, and blaSPM genes could not be identified in the colistin-resistant isolates. Interestingly, one of the colistin-resistant isolates had resistance against all kinds of antibiotics. Colistin-resistant isolates demonstrated an immense rate of resistance (100%) to ertapenem, ceftazidime, and ciprofloxacin antibiotics. 80% of colistin-resistant isolates were also MDR. By employing the E-test, it was found that Eieyhty percent of colistin-resistant isolates were resistant to imipenem (MIC ≥ 8 mg/ml). The sources of colistin-resistant isolates, MBLs genes, antibiogram patterns, specimens, and wards have been given in Table 3. In the current research, the DLST method was used in five colistin-resistant P. aeruginosa isolates obtained from various hospital wards during a period of 3 years. The majority cluster included three patients (60%) infected or colonized by the similar genotype DLST 25-11.
Table 2.
Antimicrobial sensitivity of CRPA isolates
| Antimicrobial agent | The number of CRPA isolates |
Number of sensitive persons (%) |
Number of intermediate) %( | Number of resistant persons (%) |
|---|---|---|---|---|
| imipenem | 236 | 12(5.1) | 10(4.2) | 214(90.7) |
| meropenem | 236 | 11(4.7) | 11(4.7) | 214(90.7) |
| ertapenem | 236 | 18(7.6) | 10 (4.2) | 208(88.2) |
| piperacillin-tazobactam | 236 | 47(19.9) | 56(23.7) | 133(56.4) |
| cefepime | 236 | 27(11.5) | 18(7.6) | 191(80.9) |
| amikacin | 236 | 67(28.4) | 15(6.4) | 154(65.2) |
| colistin | 236 | 231(97.9) | 0 | 5(2.1%) |
| ciprofloxacin | 236 | 25(10.6) | 11(4.7) | 200(84.7) |
| gentamicin | 236 | 35(14.8) | 0 | 201(85.2) |
| ceftazidime | 236 | 44(18.6) | 11(4.7) | 181(76.7) |
| cefotaxime | 236 | 3(1.3) | 22(9.3) | 211(89.4) |
| aztreonam | 236 | 47(19.9) | 86(36.4) | 103(43.7) |
Table 3.
The presence of non-sensitive isolates to colistin isolates carrying NDM-1 and other carbapenemase genes in CRPA isolates
| City/ Hospital | Gender | Sample/ward | carbapenemase genes |
aDLST type | bMHT | cMIC (mg/ml) CST | dMIC (mg/ml) IPM | eESBL | fMDR | Pattern antibiogram |
|||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NDM-1 | VIM | IMP | KPC | GES | SPM | OXA-10 | IMP | MEM | ETP | TZP | CEP | AN | CST | CIP | GEN | CAZ | CTX | AZT | |||||||||
| Isfahan/ General | M | Urine/ ICU | + | - | + | - | - | - | + | 25-11 | + | 8 | ≥32 | + | + | R | R | R | I | R | R | R | R | R | R | R | R |
| Ahvaz/ General | M | Urine/ Urology | - | - | + | - | - | - | + | 25-11 | + | 32 | ≥32 | - | + | R | R | R | R | I | R | R | R | R | R | R | R |
| Ahvaz/ General | M | Blood/ ICU | + | + | - | - | - | - | + | 25-11 | + | 16 | ≥32 | - | + | R | R | R | R | I | R | R | R | R | R | R | R |
| Tehran/ General | F | Trachea/Women | - | - | - | - | - | - | + | 5-91 | + | 8 | ≥32 | - | + | R | R | R | R | R | R | R | R | R | R | R | R |
| Ahvaz/ General | F | Trachea/Women | - | - | - | - | - | - | + | null | + | 32 | 4 | - | - | I | I | R | S | R | S | R | R | S | R | S | S |
Resistance rate of non-sensitive isoleates to colistin P.aeruginosa to tested antibiotics imipenem (IPM), meropenem (MEM), ertapenem (ETP), piperacillin-tazobactam (TZP), cefepime (CFP), amikacin (AN), colistin (CST), ciprofloxacin (CIP), gentamicin (GEN), ceftazidime (CAZ), cefixime (CTX), aztreonam (AZT).
aDouble-locus sequence typing, bmodified hodge test, , cminimum inhibitory concentration of colistin, dminimum inhibitory concentration of imipenem, eextended-spectrum β-lactamases,fmulti-drug resistant.
Discussion
In the present research, the presence of blaNDM-1 in colistin-resistant isolates in university teaching hospitals of Iran has been reported. To date, there has only been one report about the prevalence of blaNDM-1 in patients (21) and as far as we know, the present work represents the first report about the occurrence of colistin-resistant P. aeruginosa isolates co-existing with blaNDM-1 or other carbapenemase genes in Iran. Although reports are rare, its incidence is important because P. aeruginosa is an organism with a potent colonization ability in the hospital (22). As far as we know, this is the first study of colistin-resistant isolates with DLST type 25-11 in CRPA clinical isolates co-harboring blaNDM-1 identified in Iran and the second report on the detection of colistin-resistant isolates with blaNDM-1, the first one being reported by Mataseje et al. from North America (23). In other studies, conducted earlier in the northwest of Iran, Goli (11) and Saderi and Owlia (24) reported 4.8% and 9.1% resistance of isolates against colistin, respectively. These rates are slightly higher than the results obtained in our study.
However, reports from other countries explained that resistance to colistin varies from 0% (25) to 31.7% (26). This discrepancy can be due to the misuse of drugs, dissimilar policies of hospitals for controlling the infection, sanitation, and topographical distribution.
Another notable aspect in our results was the emergence and dissemination of colistin-resistant strains in three cities of Iran (Ahvaz, Tehran, and Isfahan) which showed the importance of prescribing antibiotics and optimizing effective infection control policies in our healthcare settings. Our research has also shown that besides colistin which acts as the antibiotic of choice for treating infections caused by CRPA isolates, amikacin is a very effective antibiotic as well. In line with our results, Liu et al. noted a very large amount of sensitivity to amikacin (91%) among clinical P. aeruginosa isolates (27). However, in contrast to our findings, Goli et al. revealed that piperacillin/tazobactam had the highest amount of activity against MDR strains of P. aeruginosa (11). Data from the previous studies showed that one of the colistin-resistant strains had resistance against all kinds of antibiotics, collected from trachea and carrying blaOXA-10. Furthermore, molecular typing of the isolates using DLST revealed a distinct pattern with 5-91 DLST type. A study from an Iranian burn hospital showed that the mortality rates of patients diseased by MBL-producing P. aeruginosa were higher than those infected by non-MBL-producing strains (28). In DLST method, the main cluster comprised 3 patients (60%) infected or colonized by the genotype DLST 25-11 which is important in MLST technique and Cholley et al. (13) considered it to be equal to ST-244. DLST is a novel and promising technique and can be utilized for future endemic superintendence of P. aeruginosa isolates. This typing pattern was founded on the fractional sequencing of two extremely inconstant loci and permitted us to gain a clear and constant classification of the varieties. DLST is an excellent technique for endemic studies of P. aeruginosa strains (12, 13, 20). The great stability, typability, and discriminatory power of DLST considerably decrease analysis prices and working time. All previous DLST published studies (12, 13, 20, 29, 30) only considered the genotyping of P. aeruginosa strains gathered from various samples. However, an absolutely experimental study with regard to colistin-resistant P. aeruginosa isolates or isolates presenting carbapenem-resistant genes is still unaccessible.
Conclusion
The high frequency of MDR and colistin-resistant P. aeruginosa carrying blaNDM-1 indicated the importance of routine surveillance over infection control since this antibiotic is the last line of curing infections resulted from MDR P. aeruginosa. Moreover, performing MIC sensitivity testing and combined drug therapy is recommended. Molecular typing of the isolates suggested that DLST type 25-11 was a dominant clone and DLST 5-91 was a high-risk clone with resistance to all used antibiotics. Therefore, further studies for accurate and specific use of this antibacterial agent which can help control the dissemination of colistin-resistant strains are needed.
Acknowledgment
The results described in this paper were part of a PhD thesis of mojtaba shahin, supported by a grant (No OG-94166) from Ahvaz Jundishapur University of Medical Sciences and Cellular and Molecular Research Center. This study was approved by the Medical Ethics Committee of Ahvaz Jundishapur University of Medical Sciences in Iran (permit number: IR.AJUMS.REC.1395.227).
Conflicts of Interest
The authors declare that there are no conflicts of interest.
References
- 1.Vaez H, Moghim S, Nasr Esfahani B, Ghasemian Safaei H. Clonal relatedness among imipenem-resistant Pseudomonas aeruginosa isolated from ICU-hospitalized patients. Crit Care Res Pract. 2015;2015:1–5. doi: 10.1155/2015/983207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mikucionyte G, Zamorano L, Vitkauskiene A, López-Causapé C, Juan C, Mulet X, et al. Nosocomial dissemination of VIM-2-producing ST235 Pseudomonas aeruginosa in Lithuania. Eur J Clin Microbiol Infect Dis. 2016;35:195–200. doi: 10.1007/s10096-015-2529-0. [DOI] [PubMed] [Google Scholar]
- 3.Leylabadlo HE, Asgharzadeh M, Aghazadeh M. Dissemination of carbapenemases producing Gram negative bacteria in the Middle East. Iran J Microbiol. 2015;7:226–246. [PMC free article] [PubMed] [Google Scholar]
- 4.Merie Queenan A, Bush K. carbapenemases: the versatile B-lactamases. Clin Microbiol Rev. 2007;20:440–458. doi: 10.1128/CMR.00001-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jovčić B, Lepšanović Z, Begović J, Filipić B, Kojić M. Two copies of blaNDM-1 gene are present in NDM-1 producing Pseudomonas aeruginosa isolates from Serbia. Antonie van Leeuwenhoek. 2014;105:613–618. doi: 10.1007/s10482-013-0094-z. [DOI] [PubMed] [Google Scholar]
- 6.Halaji M, Rezaei A, Zalipoor M, Faghri J. Investigation of class I, II, and III Integrons among Acinetobacter Baumannii isolates from hospitalized patients in Isfahan, Iran. Oman Med J. 2018;33:37–42. doi: 10.5001/omj.2018.07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wei W-J, Yang H-F, Ye Y, Li J-B. New Delhi metallo-β-lactamase-mediated carbapenem resistance: origin, diagnosis, treatment and public health concern. Chin Med J. 2015;128:1969–1976. doi: 10.4103/0366-6999.160566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee J-Y, Na IY, Park YK, Ko KS. Genomic variations between colistin-susceptible and-resistant Pseudomonas aeruginosa clinical isolates and their effects on colistin resistance. J Antimicrob Chemother. 2014;69:1248–1256. doi: 10.1093/jac/dkt531. [DOI] [PubMed] [Google Scholar]
- 9.Falagas ME, Fragoulis KN, Kasiakou SK, Sermaidis GJ, Michalopoulos A. Nephrotoxicity of intravenous colistin: a prospective evaluation. Int J Antimicrob Agents. 2005;26:504–507. doi: 10.1016/j.ijantimicag.2005.09.004. [DOI] [PubMed] [Google Scholar]
- 10.Dhariwal A, Tullu M. Colistin: Re-emergence of the ‘forgotten’ antimicrobial agent. J Postgrad Med. 2013;59:208–215. doi: 10.4103/0022-3859.118040. [DOI] [PubMed] [Google Scholar]
- 11.Goli HR, Nahaei MR, Rezaee MA, Hasani A, Kafil HS, Aghazadeh M. Emergence of colistin resistant Pseudomonas aeruginosa at Tabriz hospitals, Iran. Iran J Microbiol. 2016;8:62–69. [PMC free article] [PubMed] [Google Scholar]
- 12.Basset P, Blanc D. Fast and simple epidemiological typing of Pseudomonas aeruginosa using the double-locus sequence typing (DLST) method. Eur J Clin Microbiol Infect Dis. 2014;33:927–932. doi: 10.1007/s10096-013-2028-0. [DOI] [PubMed] [Google Scholar]
- 13.Cholley P, Stojanov M, Hocquet D, Thouverez M, Bertrand X, Blanc DS. Comparison of double-locus sequence typing (DLST) and multilocus sequence typing (MLST) for the investigation of Pseudomonas aeruginosa populations. Diagn Microbiol Infect Dis. 2015;82:274–277. doi: 10.1016/j.diagmicrobio.2015.03.027. [DOI] [PubMed] [Google Scholar]
- 14.Luce E. Koneman’s color atlas and textbook of diagnostic microbiology. Plast Reconstr Surg. 2010;125:414–415. [Google Scholar]
- 15.Lavenir R, Jocktane D, Laurent F, Nazaret S, Cournoyer B. Improved reliability of Pseudomonas aeruginosa PCR detection by the use of the species-specific ecfX gene target. J Microbiol Methods. 2007;70:20–29. doi: 10.1016/j.mimet.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 16.Patel J, Cockerill F, Alder J, Bradford P, Eliopoulos G, Hardy D, et al. Performance standards for antimicrobial susceptibility testing; twenty-fourth informational supplement. CLSI standards for antimicrobial susceptibility testing. 2014;34:1–226. [Google Scholar]
- 17.Amjad A, Mirza I, Abbasi S, Farwa U, Malik N, Zia F. Modified Hodge test: A simple and effective test for detection of carbapenemase production. Iran J Microbiol. 2011;3:189–193. [PMC free article] [PubMed] [Google Scholar]
- 18.Qu T-t, Zhang J-l, Wang J, Tao J, Yu Y-s, Chen Y-g, et al. Evaluation of phenotypic tests for detection of Metallo-β-lactamase-producing Pseudomonas aeruginosa strains in China. J Clin Microbiol. 2009;47:1136–1142. doi: 10.1128/JCM.01592-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Poirel L, Walsh TR, Cuvillier V, Nordmann P. Multiplex PCR for detection of acquired carbapenemase genes. Diagn Microbiol Infect Dis. 2011;70:119–123. doi: 10.1016/j.diagmicrobio.2010.12.002. [DOI] [PubMed] [Google Scholar]
- 20.Pappa O, Beloukas A, Vantarakis A, Mavridou A, Kefala A-M, Galanis A. Molecular characterization and phylogenetic analysis of Pseudomonas aeruginosa isolates recovered from Greek aquatic habitats implementing the Double-Locus Sequence Typing Scheme. Microb Ecol. 2017;74:78–88. doi: 10.1007/s00248-016-0920-8. [DOI] [PubMed] [Google Scholar]
- 21.Shokri D, Khorasgani MR, Fatemi SM, Soleimani-Delfan A. Resistotyping, phenotyping and genotyping of New Delhi metallo-β-lactamase (NDM) among Gram-negative bacilli from Iranian patients. J Med Microbiol. 2017;66:402–411. doi: 10.1099/jmm.0.000444. [DOI] [PubMed] [Google Scholar]
- 22.Shanthi M, Sekar U, Kamalanathan A, Sekar B. Detection of New Delhi metallo beta lactamase-1 (NDM-1) carbapenemase in Pseudomonas aeruginosa in a single centre in southern India. Indian J Med Res. 2014;140:546–550. [PMC free article] [PubMed] [Google Scholar]
- 23.Mataseje L, Peirano G, Church D, Conly J, Mulvey M, Pitout J. Colistin-nonsusceptible Pseudomonas aeruginosa sequence type 654 with blaNDM-1 arrives in North America. Antimicrob Agents Chemother. 2016;60:1794–800. doi: 10.1128/AAC.02591-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saderi H, Owlia P. Detection of multidrug resistant (MDR) and extremely drug resistant (XDR) P aeruginosa isolated from patients in Tehran, Iran. Iran J Pathol. 2015;10:265–271. [PMC free article] [PubMed] [Google Scholar]
- 25.Maroui I, Barguigua A, Aboulkacem A, Ouarrak K, Sbiti M, Louzi H, et al. First report of VIM-2 metallo-β-lactamases producing Pseudomonas aeruginosa isolates in Morocco. J Infect Cemother. 2016;22:127–132. doi: 10.1016/j.jiac.2015.11.008. [DOI] [PubMed] [Google Scholar]
- 26.Gill MM, Usman J, Kaleem F, Hassan A, Khalid A, Anjum R, et al. Frequency and antibiogram of multi-drug resistant Pseudomonas aeruginosa. J Coll Physicians Surg Pak. 2011;21:531–534. [PubMed] [Google Scholar]
- 27.Bahar MA, Jamali S, Samadikuchaksaraei A. Imipenem-resistant Pseudomonas aeruginosa strains carry metallo-β-lactamase gene blaVIM in a level I Iranian burn hospital. Burns. 2010;36:826–830. doi: 10.1016/j.burns.2009.10.011. [DOI] [PubMed] [Google Scholar]
- 28.Liu P-Y, Weng L-L, Tseng S-Y, Huang C-C, Cheng C-C, Mao Y-C, et al. Colistin resistance of Pseudomonas aeruginosa isolated from snakes in Taiwan. Can J Infect Dis Med Microbiol. 2017;2017:1–5. doi: 10.1155/2017/7058396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tissot F, Blanc D, Basset P, Zanetti G, Berger M, Que Y-A, et al. New genotyping method discovers sustained nosocomial Pseudomonas aeruginosa outbreak in an intensive care burn unit. J Hosp Infect. 2016;94:2–7. doi: 10.1016/j.jhin.2016.05.011. [DOI] [PubMed] [Google Scholar]
- 30.Blanc D, Magalhaes BG, Abdelbary M, Prod’hom G, Greub G, Wasserfallen J, et al. Hand soap contamination by Pseudomonas aeruginosa in a tertiary care hospital: no evidence of impact on patients. J Hosp Infect. 2016;93:63–67. doi: 10.1016/j.jhin.2016.02.010. [DOI] [PubMed] [Google Scholar]
- 31.Nordmann P, Naas T, Poirel L. Global spread of carbapenemase-producing Enterobacteriaceae. Emerg Infect Dis. 2011;17:1791–1798. doi: 10.3201/eid1710.110655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fallah F, Borhan RS, Hashemi A. Detection of bla (IMP) and bla (VIM) metallo-β-lactamases genes among Pseudomonas aeruginosa strains. International . Int J Burns Trauma. 2013;3:122–124. [PMC free article] [PubMed] [Google Scholar]
- 33.Golshani Z, Sharifzadeh A. Prevalence of blaOxa10 type beta-lactamase gene in carbapenemase producing Pseudomonas aeruginosa strains isolated from oatients in Isfahan. Jundishapur J Microbiol. 2013;6:1–4. [Google Scholar]