Abstract
Objective(s):
Acinetobacter pittii has become an emerging opportunistic noscomial pathogen worldwide with multi-drug resistance. In the present study, an A. pittii strain was isolated from bronchoalveolar lavage fluid sample harboring both OXA-58 and NDM-1carbapenemase producing genes. The mechanisms of carbapenem resistance of the A. pittii strain was investigated.
Materials and Methods:
Carbapenemase producing genes were examined by PCR and DNA sequencing. S1-PFGE was used to localize carbapenemase encoding genes. Filter mating and electrotransformation were used to investigate the transferability of such carbapenemase encoding genes between different strains. Genetic surroundings of blaOXA-58 and blaNDM-1 genes were detected as well.
Results:
The A. pittii strain, carrying both OXA-58 and NDM-1 carbapenemase encoding genes, was resistant to all β-lactam antibiotics, while suscepitible to ciprofloxacin, levofloxacin, tobramycin, cotrimoxazole and tigecycline. Southern blot hybridization for the blaOXA-58 and blaNDM-1 gene indicated that the two genes locate in the same plasmid with molecular weight of 310.1-336.5kb. BlaOXA-58 was located in an ISAba3-blaOXA-58-ISAba3-like structure, and the blaNDM-1 gene cluster was embedded in an ISAba125-aphA6- blaNDM-1-bleMBL-ΔtrpF-dsbC-cutA structure sequentially.
Conclusion:
In the present study, it is first reported an A. pittii clincal strain in China, co-harboring OXA-58 and NDM-1 carbapenemase producing genes residing on a same plasmid. In hospital and community settings, it is of great significance and urgence to increase the surveillance of these kinds of organisms.
Key Words: Acinetobacter pittii, Carbapenemase, Co-harboring, NDM-1, OXA-58
Introduction
Acinetobacter spp. are frequent pathogens responsible for nosocomial infections, including Acinetobacter baumannii as the predominant species followed by Acinetobacter pittii and Acinetobacter nosocomialis (1). Since the phenomenon of carbapenem resistance increasingly emergences, there is a big challenge of multidrug resistant Acinetobacter in treating hospital infections (2, 3).
To our knowledge, the increasing expression of OXA type carbapenemase genes is the most frequent mechanism of carbapenem resistance in Acinetobacter. It mainly includes the intrinsic blaOXA-51-like gene as well as the horizontally acquired genes such as blaOXA-23-like, blaOXA-24-like and blaOXA-58-like genes (4, 5). The blaOXA-23-like genes are much more prevalent than the blaOXA-24-like and blaOXA-58-like genes. All of them are capable of yielding carbapenem resistance in a high level and result in serious local outbreaks (6-8). Moreover, the expression of metallo-β-lactam (MBL) carbapenemase also plays an important role in carbapenem resistance of Acinetobacter, including IMP, VIM, SIM, and NDM detected previously in Acinetobacter (9). Although these MBLs genes were less detected than OXAs, their carbapenemase activities were typically much higher (10, 11). Especially, since 2008, there have been an increasing number of reports about the dissemination of NDM-1-producing Acinetobacter spp in many countries and it resulted in a major threat for clinical treatments in view of its highly frequent co-occurrence with other resistance genes (12-14). Recently, the global spread of blaNDM-1-harboring A. pittii strains is fierce. With these powerful genes, as the reservoirs for dissemination, they are able to transfer highly across various bacterial species (15-17).
In this study, we investigated the antibiotic susceptibility, genetic environment, and transferability of a single clinical A. pittii isolate co-harboring blaNDM-1 and blaOXA-58 genes on a same plasmid, in order to improve awareness of the urgency of carbapenemase-producing A. pittii isolates in China.
Materials and Methods
Bacterial isolates
The A. pittii was isolated from a bronchoalveolar lavage fluid sample of a 56-year-old man suffering from chronic obstructive pulmonary disease (COPD). It was initially identified as Acinetobacter calcoaceticus baumannii by Vitek 2 system (bioMérieux, Marcy l’Etoile, France). To confirm the identity of this strain, a fragment of the 16S rRNA gene was amplified using primers 27-forward (5’- AGA GTT TGA TCC TGG CTC AG -3’) and 1492-reverse (5’- GGT TAC CTT GTT ACG ACT T -3’) by PCR[10] and the resultant PCR product sequenced as A. pittii with 100% identity, designated as AB34.
Antimicrobial susceptibility testing
The minimum inhibitory concentration (MIC) of various antibiotics was detected on the Vitek 2 system (bioMérieux, Marcy l’Etoile, France).
According to the CLSI clinical breakpoints (2017; CLSI Document M100-S27), antimicrobial susceptibility was interpreted. The following antibiotics were investigated in the present study: amikacin, ciprofloxacin, ampicillin/sulbactam, ceftazidime, imipenem, cotrimoxazol, tobramycin, piperacillin/tazobactam, cefepime, gentamicin, levofloxacin, meropenem, rifampin and tigecycline. Quality control for the MIC analysis was carried out with Pseudomonas aeruginosa ATCC 27853 and Escherichia coli ATCC 25922.
Screening of carbapenemases encoding genes
Amplification of carbapenemases encoding genes was perfomed, including OXA-51, OXA-23, OXA-58, OXA-143, OXA-24, NDM-1, GIM-1, SPM-1, IMP-1, SIM-1, VIM-1. These primers are shown in Table 1. All amplicons were sequenced using the ABI PRISM Big Dye Terminator Cycle Sequencing Ready Reaction kit (Applied Biosystems -2- Inc, USA).
Table 1.
Primer sequences of the carbapenemases encoding genes
| Target genes | Name of primers | Sequence | Amplicon size | Reference |
|---|---|---|---|---|
| OXA-51-live | OXA51_Mup | 5'-TAATGCTTTGATCGGCCTTG-3' | 353 | (18) |
| OXA51_Mdw | 5'-TGGATTGCACTTCATCTTGG-3' | |||
| OXA-23-live | OXA23_Mup | 5'-GATCGGATTGGAGAACCAGA-3' | 501 | (18) |
| OXA23_Mdw | 5'-ATTTCTGACCGCATTTCCAT-3' | |||
| OXA-24-live | OXA24_Mup | 5'-GGTTAGTTGGCCCCCTTAAA-3' | 246 | (18) |
| OXA24_Mdw | 5'-AGTTGAGCGAAAAGGGGATT-3 | |||
| OXA-58-live | OXA58_Mup | 5'-AAGTATTGGGGCTTGTGCTG-3' | 599 | (19) |
| OXA58_Mdw | 5'-CCCCTCTGCGCTCTACATAC-3' | |||
| OXA-143-live | OXA143_Mup | 5'-TGGCACTTTCAGCAGTTCCT-3' | 149 | (20) |
| OXA143_Mdw | 5'-TAATCTTGAGGGGGCCAACC-3' | |||
| SIM | SIM-F | 5'-TACAAGGGATTCGGCATCG-3' | 570 | (18) |
| SIM-R | 5'-TAATGGCCTGTTCCCATGTG-3' | |||
| SPM | SPM-F | 5'-AAAATCTGGGTACGCAAACG-3' | 271 | (18) |
| SPM-R | 5'-ACATTATCCGCTGGAACAGG-3' | |||
| GIM | GIM-F | 5'-TCGACACACCTTGGTCTGAA-3' | 477 | (18) |
| GIM-R | 5'-AACTTCCAACTTTGCCATGC-3' | |||
| IMP | IMP-F | 5'-GAAGGCGTTTATGTTCATAC-3' | 587 | (18) |
| IMP-R | 5'-GTACGTTTCAAGAGTGATGC-3' | |||
| VIM | VIM-F | 5'-GTTTGGTCGCATATCGCAAC-3' | 389 | (18) |
| VIM-R | 5'-AATGCGCAGCACCAGGATAG-3' | |||
| NDM-1 | NDM-F | 5'-GCAGCTTGTCGGCCATGCGGGC-3' | 782 | (18) |
| NDM-R | 5'-GGTCGCGAAGCTGAGCACCGCAT-3' |
S1-PFGE and southern blot
The total bacterial DNA was first prepared in agarose plugs, digested with S1 nuclease (Takara, Japan) and further separated by PFGE, as reported previously (21). The DNA fragments were transferred horizontally to a nylon membrane (Millipore, USA), hybridized with digoxigen in-labeled blaOXA-58 and blaNDM-1 probe and detected using a nitroblue tetrazolium/5-bromo-4-chloro-3’-indolyl-phosphate color detection kit (Roche Applied Sciences, USA).
Filter mating experiment
Filter mating experiment was performed with the rifampin-resistant EC600 and azide-resistant E. coli J53 as the recipient strains. The transconjugants were selected on Mueller-Hinton agar plates containing [ampicillin (50 mg/l) and rifampicin (1024 mg/l)] or [ampicillin (50 mg/l) and NaN3 (200 mg/l)], respectively, and incubated for 16–18 hr at 37 °C. The successful transconjugants were selected on Mueller-Hinton agar incorporating the same concentration of antibiotics mentioned above. The transformants would be confirmed the resistant genes by PCR.
Plasmid construction and electrotransformation
The plasmid DNA of isolate AB34 was extracted, digested by restriction enzyme EcoRI or SacI and then cloned into the cloning vector pPet28a. The conjugant was electrotransformed to E. coli DH5α competent cells and selected on Mueller-Hinton agar plates containing ampicillin (50 mg/L) in order to obtain the E. coli clone expressing the corresponding carbapenemase enzyme.
Genetic surroundings detection
A total genomic sample of A. pittii strain AB34 was extracted and purified using the Wizard Genomic DNA purification kit (Promega Corporation, Madison, WI) according to the manufacturer’s instructions. DNA concentration was estimated using a Qubit dsDNA HS Assay Kit and a Qubit 3.0 Fluorometer (Invitrogen, Thermo Fisher Scientific, USA). Extracted DNA was then sequenced with a standard 2×125 paired-end runs protocol on an Illumina HiSeq 2000 (Illumina, San Diego, CA, USA). The quality of the high-throughput sequence data was assessed by FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Raw sequence reads were then de novo assembled using
Plasmid SPAdes 3.9.0 (http://bioinf.spbau.ru/spades)(22), in order to identify plasmid contigs, and the quality was assessed by QUAST (http://quast.bioinf. spbau.ru).
Nucleotide sequence accession numbers
The blaOXA-58 or blaNDM-1 nucleotide sequences are available in GenBank under the accession number KF208466 or KF208467, respectively.
Results
Susceptibility results
The isolate AB34 was resistant to all β-lactams including ceftazidime, cefepime and carbapenems as well as ampicillin/sulbactam inhibitor combinations, but remained susceptible to cotrimoxazole, tobramycin, ciprofloxacin, levofloxacin and tigecycline (Table 2).
Table 2.
MIC for the Acinetobacter pittii AB34
| Antibiotics | MIC(μg/mL) | Interpretation |
|---|---|---|
| amikacin | 24 | I |
| ciprofloxacin | 0.19 | S |
| imipenem | ≥32 | R |
| cotrimoxazole | <=20 | S |
| tobramycin | <=1 | S |
| ampicillin/sulbactam | ≥32 | R |
| gentamicin | ≥256 | R |
| levofloxacin | 2 | S |
| ceftazidime | ≥64 | R |
| cefepime | ≥256 | R |
| meropenem | ≥32 | R |
| tigecycline | 2 | S |
Detection of carbapenemases encoding genes
Only OXA-58(599bp) and NDM-1(720bp) were detected in this strain. The amplified products were confirmed by sequencing. BLAST version 2.2.24 (http://blast.ddbj.nig.ac.jp/) was used to process the sequencing data and identify genes.
Location of bla OXA-58 and bla NDM-1 genes
After PFGE (Figure 1) and southern blot hybridization (Figure 2), it was found that both blaOXA-58 and blaNDM-1 genes resided on a same 310.1-336.5kb plasmid. Horizontal transfer of the two carbapenem resistance determinants from AB34 to EC600 or E. coli J53 was not detected in filter mating.
Figure 1.

S1-PFGE. Lane M, DNA Marker; lane 1, strain AB34
Figure 2.

Southern blot hybridization of OXA-58 and NDM-1 genes. Lane 1, OXA-58 hybridization; lane 2, NDM-1hybridization; lane M, DNA Marker
Genetic surroundings
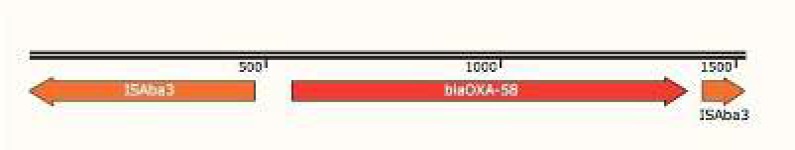
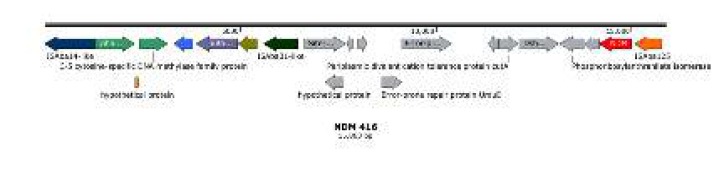
A BLAST search against all completely sequenced blaOXA-58 and blaNDM-1-genes-co-harboring plasmids in GenBank (http://www.ncbi.nlm.nih.gov/GenBank/) showed that the blaOXA-58 gene in AB34 was located down-stream of the shortened ISAba3 gene, and up-stream of a complete copy of ISAba3 gene (Figure 3). The blaNDM-1 gene cluster of AB34 was arranged sequentially as ISAba125, aphA6, blaNDM-1, bleMBL, ΔtrpF, dsbD, cutA1, GroES, UmuD, hypothetical protein, site-specific DNA methylase-like, ISAba31-like, dihydrofolate reductase putative membrane protein, C-5 cytosine-specific DNA methylase family protein, ISAba14-like from right to left (Figure 4).
Figure 3.
Analysis of blaOXA-58-carrying composite transposon in Acinetobacter Pittii AB34 genome. Genes and transcription orientations are indicated by arrows
Figure 4.
Analysis of blaNDM-1-carrying composite transposon in Acinetobacter Pittii AB34 genome. Genes and transcription orientations are indicated by arrows
Discussion
Since the last decade, carbapenemase-producing Acinetobacter spp have disseminated rapidly throughout the world, posing an urgent threat to public health (1, 23). A. pittii, formerly named Acinetobacter genomic species 3, is increasingly recognized as a clinically important pathogen within the Acinetobacter calcoaceticus– A. baumannii complex, which addresses a particular concern due to its competency to acquire multidrug resistance against a wide range of antimicrobial agents (24).
The first known OXA-58-producing Acinetobacter strain was isolated in France in 2003 (9). It shares less than 50% of amino-acid homology with oxacillinase. OXA-58 is a widely spread carbapenem-hydrolyzing class D β-lactamases (CHDLs) that has been reported in Acinetobacter spp. from Europe (25), Argentina (26), Australia (27), the United States (28) and many Asian countries (29). Though OXA-58 shows only low carbapenem-hydrolyzing activity in vitro, the insertion sequence upstream of blaOXA-58 enhances its transcription greatly and mediates resistance to carbapenems (30-32). It is speculated that the insertion of other IS element into ISAba3-like could generate a hybrid promoter to enhance the transcription of blaOXA-58 and mediate greater carbapenem resistance than the intact ISAba3-like element as previously reported (32, 33). However, in China, the most common carbapenamase-producing type of A. baumannii is OXA-23,while OXA - 58 is rarely reported (34).
The NDM-1 gene encodes an enzyme that hydrolyses and inactivates all β-lactam antibiotics including carbapenems, except for aztreonam, and thus induces resistance to carbapenems (35). A. baumannii carrying NDM-1 have been reported from clinical and environmental isolates in several countries (36-39). Not only Acinetobacter spp. act as reservoirs for blaNDM genes in non-human settings, as recently shown in several Chinese studies with identification of NDM-1-producers among A. calcoaceticus and Acinetobacter junii from environmental samples from livestock farms (40), Acinetobacter johnsonii from hospital sewage (40) and Acinetobacter lwoffii from chickens(40), but also act as a source of blaNDM genes then horizontally transferred to enterobacterial species as evidenced (41).
It is noteworthy that coexistence of blaNDM and blaOXA has been described in Acinetobacter, e.g. blaOXA-23 and blaNDM-1 in A. baumannii from India (42) and the Czech Republic (43), and blaNDM-1, blaOXA-23 and blaIMP in A. baumannii from China (44). However, it remains unclear whether and how these co-existing carbapenemase genes are expressed to contribute to drug resistance.
A. pittii 44551 was recovered from a patient with gout combined with tuberculosis and was found to harbor the carbapenemase genes blaNDM-1 and blaOXA-58 on two different plasmids pNDM-44551 and pOXA58-44551, respectively, from China in 2015 (1). Emergence of ST119 A. pittii AP 882 co-harbouring NDM-1 and OXA-58 in Malaysia was reported as well, of which genes encoding NDM-1 and OXA-58 resided on an ca.140 kb mega plasmid and a 35 kb plasmid, respectively (45).
Similarly, in our present study, AB34 was isolated and detected co-harbouring OXA-58 and NDM-1carbapenemase producing resided on the same 310.1-336.5kb plasmid. However, horizontal transfer of carbapenem resistance determinants from AB34 to EC600 or E. coli J53 (AzR) was not detected in filter mating experiment. The up-stream and down-stream of OXA-58 gene in AB34 are ISAba3, which shows 99% similarity to A. pittii pOXA-58-44551 (1). It is reported that the structure of OXA-58 of A. pittii 44551 is 372F-ISAba3-like-blaOXA-58-ISAba3, where the blaOXA-58 contributed little to β-lactams resistance due to a lack of the blaOXA-58-driven promoter (1). An intact ISAba3-like element upstream of blaOXA-58 has been linked to a lower level of resistance to imipenem compared with blaOXA-58 with hybrid promoters such as IS6 family-ISAba3-like-blaOXA-58.
The upstream of NDM-1 of AB34 is ISAba125, while the down-streams are arranged sequentially as aphA6, blaNDM-1, bleMBL, ΔtrpF, dsbD, cutA1, GroES, UmuD, hypothetical protein, site-specific DNA methylase-like, ISAba31-like, dihydrofolate reductase putative membrane protein, C-5 cytosine-specific DNA methylase family protein, ISAba14-like, which was with 99% sequence identity against that of Acinetobacter lwoffii pNDM-BJ01 from Beijing, China (46). It is proved that the genetic surroundings of blaNDM-1 is an important vector to mediate to integration and transfer. It should be noted that the ISAba125 element upstream of blaNDM-1 is usually intact in Acinetobacter but often truncated in Enterobacteriaceae, suggesting the probable spread of the blaNDM-1 genetic platforms from Acinetobacter to Enterobacteriaceae (47-50).
Conclusion
This study has improved awareness of the urgence of carbapenemase-producing A. pittii isolates in China. Further investigations on the comparative genomic analysis of a large-scale sampling of A. pittii strains from a wide spatial and temporal range in the context of genomic epidemiological characteristics are currently on the way. These data highlight the molecular mechanisms contributing to the rapid development of antimicrobial resistance and will facilitate to expand our understanding of the global public health concern caused by Acinetobacter spp.
Acknowledgements
We are grateful to Professor Qing Yang, from Department of Laboratory Medicine, the First Affiliated Hospital of Zhejiang University, Hangzhou, China, for guiding us to carry out related experiments.
References
- 1.Zhou S, Chen X, Meng X, Zhang G, Wang J, Zhou D, et al. “Roar” of blaNDM-1 and “silence” of blaOXA-58 co-exist in Acinetobacter pittii. Sci Rep. 2015;5:8976. doi: 10.1038/srep08976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Poirel L, Bonnin RA, Nordmann P. Genetic basis of antibiotic resistance in pathogenic Acinetobacter species. IUBMB LIFE. 2011;63:1061–1067. doi: 10.1002/iub.532. [DOI] [PubMed] [Google Scholar]
- 3.Poirel L, Nordmann P. Carbapenem resistance in Acinetobacter baumannii: mechanisms and epidemiology. Clin Microbiol Infect. 2006;12:826–836. doi: 10.1111/j.1469-0691.2006.01456.x. [DOI] [PubMed] [Google Scholar]
- 4.Karah N, Sundsfjord A, Towner K, Samuelsen O. Insights into the global molecular epidemiology of carbapenem non-susceptible clones of Acinetobacter baumannii. Drug Resist Updat. 2012;15:237–247. doi: 10.1016/j.drup.2012.06.001. [DOI] [PubMed] [Google Scholar]
- 5.Dijkshoorn L, Nemec A, Seifert H. An increasing threat in hospitals: multidrug-resistant Acinetobacter baumannii. NAT REV MICROBIOL. 2007;5:939–951. doi: 10.1038/nrmicro1789. [DOI] [PubMed] [Google Scholar]
- 6.Mendes RE, Bell JM, Turnidge JD, Castanheira M, Jones RN. Emergence and widespread dissemination of OXA-23, -24/40 and -58 carbapenemases among Acinetobacterspp in Asia-Pacific nations: report from the SENTRY Surveillance Program. J Antimicrob Chemother. 2009;63:55–59. doi: 10.1093/jac/dkn434. [DOI] [PubMed] [Google Scholar]
- 7.Moro M, Nizzero P, Biancardi A, Baldan R, Scarpellini P, Curti C, et al. An outbreak caused by multidrug-resistant OXA-58-positive Acinetobacter baumannii in an intensive care unit in Italy. J Hosp Infect. 2008;68:97–99. doi: 10.1016/j.jhin.2007.10.007. [DOI] [PubMed] [Google Scholar]
- 8.Castanheira M, Wanger A, Kruzel M, Deshpande LM, Jones RN. Emergence and clonal dissemination of OXA-24- and OXA-58-producing Acinetobacter baumannii strains in Houston, Texas: report from the SENTRY Antimicrobial Surveillance Program. J Clin Microbiol. 2008;46:3179–3180. doi: 10.1128/JCM.00988-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maragakis LL, Perl TM. Acinetobacter baumannii: epidemiology, antimicrobial resistance, and treatment options. Clin Infect Dis. 2008;46:1254–1263. doi: 10.1086/529198. [DOI] [PubMed] [Google Scholar]
- 10.Cornaglia G, Giamarellou H, Rossolini GM. Metallo-beta-lactamases: a last frontier for beta-lactams? Lancet Infect Dis. 2011;11:381–393. doi: 10.1016/S1473-3099(11)70056-1. [DOI] [PubMed] [Google Scholar]
- 11.Gupta V. Metallo beta lactamases in Pseudomonas aeruginosa and Acinetobacter species. Expert Opin Investig Drugs. 2008;17:131–143. doi: 10.1517/13543784.17.2.131. [DOI] [PubMed] [Google Scholar]
- 12.Krizova L, Bonnin RA, Nordmann P, Nemec A, Poirel L. Characterization of a multidrug-resistant Acinetobacter baumannii strain carrying the blaNDM-1 and blaOXA-23 carbapenemase genes from the Czech Republic. J Antimicrob Chemother. 2012;67:1550–1552. doi: 10.1093/jac/dks064. [DOI] [PubMed] [Google Scholar]
- 13.Nemec A, Krizova L. Carbapenem-resistant Acinetobacter baumannii carrying the NDM-1 gene, Czech Republic, 2011. Euro Surveill. 2012:17. [PubMed] [Google Scholar]
- 14.Zhang C, Qiu S, Wang Y, Qi L, Hao R, Liu X, et al. Higher isolation of NDM-1 producing Acinetobacter baumannii from the sewage of the hospitals in Beijing. PLOS ONE. 2014;8:e64857. doi: 10.1371/journal.pone.0064857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang TW, Lauderdale TL, Liao TL, Hsu MC, Chang FY, Chang SC, et al. Effective transfer of a 47 kb NDM-1-positive plasmid among Acinetobacter species. J Antimicrob Chemother. 2015;70:2734–2738. doi: 10.1093/jac/dkv191. [DOI] [PubMed] [Google Scholar]
- 16.Kamolvit W, Derrington P, Paterson DL, Sidjabat HE. A case of IMP-4-, OXA-421-, OXA-96-, and CARB-2-producing Acinetobacter pittii sequence type 119 in Australia. J CLIN MICROBIOL. 2015;53:727–730. doi: 10.1128/JCM.02726-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pagano M, Poirel L, Martins AF, Rozales FP, Zavascki AP, Barth AL, et al. Emergence of NDM-1-producing Acinetobacter pittii in Brazil. Int J Antimicrob Agents. 2015;45:444–445. doi: 10.1016/j.ijantimicag.2014.12.011. [DOI] [PubMed] [Google Scholar]
- 18.Sun FJ, Shi HQ, Zhang XB, Fang YD, Chen YC, Chen JH, et al. Detection of carbapenemase-encoding genes among clinical isolates of Pseudomonas aeruginosa in a Chinese burn unit. J Burn Care Res. 2013;34:453–458. doi: 10.1097/BCR.0b013e3182700afd. [DOI] [PubMed] [Google Scholar]
- 19.Rodriguez-Martinez JM, Poirel L, Nordmann P. Molecular epidemiology and mechanisms of carbapenem resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2009;53:4783–4788. doi: 10.1128/AAC.00574-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Higgins PG, Poirel L, Lehmann M, Nordmann P, Seifert H. OXA-143, a novel carbapenem-hydrolyzing class D beta-lactamase in Acinetobacter baumannii. Antimicrob Agents Chemother. 2009;53:5035–5038. doi: 10.1128/AAC.00856-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou S, Chen X, Meng X, Zhang G, Wang J, Zhou D, et al. “Roar” of blaNDM-1 and “silence” of blaOXA-58 co-exist in Acinetobacter pittii. Sci Rep. 2015;5:8976. doi: 10.1038/srep08976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J COMPUT BIOL. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin MF, Lan CY. Antimicrobial resistance in Acinetobacter baumannii: From bench to bedside. World J Clin Cases. 2014;2:787–814. doi: 10.12998/wjcc.v2.i12.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang J, Chen Y, Jia X, Luo Y, Song Q, Zhao W, et al. Dissemination and characterization of NDM-1-producing Acinetobacter pittii in an intensive care unit in China. Clin Microbiol Infect. 2012;18:E506–E513. doi: 10.1111/1469-0691.12035. [DOI] [PubMed] [Google Scholar]
- 25.Poirel L, Marque S, Heritier C, Segonds C, Chabanon G, Nordmann P. OXA-58, a novel class D {beta}-lactamase involved in resistance to carbapenems in Acinetobacter baumannii. Antimicrob Agents Chemother. 2005;49:202–208. doi: 10.1128/AAC.49.1.202-208.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Coelho J, Woodford N, Afzal-Shah M, Livermore D. Occurrence of OXA-58-like carbapenemases in Acinetobacter spp collected over 10 years in three continents. Antimicrob Agents Chemother. 2006;50:756–758. doi: 10.1128/AAC.50.2.756-758.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peleg AY, Franklin C, Walters LJ, Bell JM, Spelman DW. OXA-58 and IMP-4 carbapenem-hydrolyzing beta-lactamases in an Acinetobacter junii blood culture isolate from Australia. Antimicrob Agents Chemother. 2006;50:399–400. doi: 10.1128/AAC.50.1.399-400.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Castanheira M, Wanger A, Kruzel M, Deshpande LM, Jones RN. Emergence and clonal dissemination of OXA-24- and OXA-58-producing Acinetobacter baumannii strains in Houston, Texas: report from the SENTRY Antimicrobial surveillance program. J Clin Microbiol. 2008;46:3179–3180. doi: 10.1128/JCM.00988-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mendes RE, Bell JM, Turnidge JD, Castanheira M, Jones RN. Emergence and widespread dissemination of OXA-23, -24/40 and -58 carbapenemases among Acinetobacter spp in Asia-Pacific nations: report from the SENTRY Surveillance Program. J Antimicrob Chemother. 2009;63:55–59. doi: 10.1093/jac/dkn434. [DOI] [PubMed] [Google Scholar]
- 30.Poirel L, Nordmann P. Genetic structures at the origin of acquisition and expression of the carbapenem-hydrolyzing oxacillinase gene blaOXA-58 in Acinetobacter baumannii. Antimicrob Agents Chemother. 2006;50:1442–1448. doi: 10.1128/AAC.50.4.1442-1448.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen TL, Wu RC, Shaio MF, Fung CP, Cho WL. Acquisition of a plasmid-borne blaOXA-58 gene with an upstream IS1008 insertion conferring a high level of carbapenem resistance to Acinetobacter baumannii. Antimicrob Agents Chemother. 2008;52:2573–2580. doi: 10.1128/AAC.00393-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen TL, Chang WC, Kuo SC, Lee YT, Chen CP, Siu LK, et al. Contribution of a plasmid-borne blaOXA-58 gene with its hybrid promoter provided by IS1006 and an ISAba3-like element to beta-lactam resistance in acinetobacter genomic species 13TU. Antimicrob Agents Chemother. 2010;54:3107–3112. doi: 10.1128/AAC.00128-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boo TW, Crowley B. Detection of blaOXA-58 and blaOXA-23-like genes in carbapenem-susceptible Acinetobacter clinical isolates: should we be concerned? J Med Microbiol. 2009;58:839–841. doi: 10.1099/jmm.0.008904-0. [DOI] [PubMed] [Google Scholar]
- 34.Liu LL, Ji SJ, Ruan Z, Fu Y, Fu YQ, Wang YF, et al. Dissemination of blaOXA-23 in Acinetobacter spp in China: main roles of conjugative plasmid pAZJ221 and transposon Tn2009. Antimicrob Agents Chemother. 2015;59:1998–2005. doi: 10.1128/AAC.04574-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yong D, Toleman MA, Giske CG, Cho HS, Sundman K, Lee K, et al. Characterization of a new metallo-beta-lactamase gene, bla(NDM-1), and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob Agents Chemother. 2009;53:5046–5054. doi: 10.1128/AAC.00774-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tran DN, Tran HH, Matsui M, Suzuki M, Suzuki S, Shibayama K, et al. Emergence of New Delhi metallo-beta-lactamase 1 and other carbapenemase-producing Acinetobacter calcoaceticus-baumannii complex among patients in hospitals in Ha Noi, Viet Nam. Eur J Clin Microbiol Infect Dis. 2017;36:219–225. doi: 10.1007/s10096-016-2784-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bharadwaj R, Joshi S, Dohe V, Gaikwad V, Kulkarni G, Shouche Y. Prevalence of New Delhi metallo-beta-lactamase (NDM-1)-positive bacteria in a tertiary care centre in Pune, India. Int J Antimicrob Agents. 2012;39:265–266. doi: 10.1016/j.ijantimicag.2011.09.027. [DOI] [PubMed] [Google Scholar]
- 38.Jones LS, Toleman MA, Weeks JL, Howe RA, Walsh TR, Kumarasamy KK. Plasmid carriage of bla NDM-1 in clinical Acinetobacter baumannii isolates from India. Antimicrob Agents Chemother. 2014;58:4211–4213. doi: 10.1128/AAC.02500-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen Y, Cui Y, Pu F, Jiang G, Zhao X, Yuan Y, et al. Draft genome sequence of an Acinetobacter genomic species 3 strain harboring a bla(NDM-1) gene. J Bacteriol. 2012;194:204–205. doi: 10.1128/JB.06202-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bonnin RA, Poirel L, Nordmann P. New Delhi metallo-beta-lactamase-producing Acinetobacter baumannii: a novel paradigm for spreading antibiotic resistance genes. Future Microbiol. 2014;9:33–41. doi: 10.2217/fmb.13.69. [DOI] [PubMed] [Google Scholar]
- 41.Poirel L, Bonnin RA, Nordmann P. Analysis of the resistome of a multidrug-resistant NDM-1-producing Escherichia coli strain by high-throughput genome sequencing. Antimicrob Agents Chemother. 2011;55:4224–4229. doi: 10.1128/AAC.00165-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Karthikeyan K, Thirunarayan MA, Krishnan P. Coexistence of blaOXA-23 with blaNDM-1 and armA in clinical isolates of Acinetobacter baumannii from India. J Antimicrob Chemother. 2010;65:2253–2254. doi: 10.1093/jac/dkq273. [DOI] [PubMed] [Google Scholar]
- 43.Krizova L, Bonnin RA, Nordmann P, Nemec A, Poirel L. Characterization of a multidrug-resistant Acinetobacter baumannii strain carrying the blaNDM-1 and blaOXA-23 carbapenemase genes from the Czech Republic. J Antimicrob Chemother. 2012;67:1550–1552. doi: 10.1093/jac/dks064. [DOI] [PubMed] [Google Scholar]
- 44.Chen Z, Qlu S, Wang Y, Wang Y, Liu S, Wang Z, et al. Coexistence of blaNDM-1 with the prevalent blaOXA23 and blaIMP in pan-drug resistant Acinetobacter baumannii isolates in China. CLIN INFECT DIS. 2011;52:692–693. doi: 10.1093/cid/ciq231. [DOI] [PubMed] [Google Scholar]
- 45.Ang GY, Yu CY, Cheong YM, Yin WF, Chan KG. Emergence of ST119 Acinetobacter pittii co-harbouring NDM-1 and OXA-58 in Malaysia. Int J Antimicrob Agents. 2016;47:168–169. doi: 10.1016/j.ijantimicag.2015.11.008. [DOI] [PubMed] [Google Scholar]
- 46.Hu H, Hu Y, Pan Y, Liang H, Wang H, Wang X, et al. Novel plasmid and its variant harboring both a bla(NDM-1) gene and type IV secretion system in clinical isolates of Acinetobacter lwoffii. Antimicrob Agents Chemother. 2012;56:1698–1702. doi: 10.1128/AAC.06199-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Poirel L, Dortet L, Bernabeu S, Nordmann P. Genetic features of blaNDM-1-positive Enterobacteriaceae. Antimicrob Agents Chemother. 2011;55:5403–5407. doi: 10.1128/AAC.00585-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wailan AM, Paterson DL. The spread and acquisition of NDM-1: a multifactorial problem. Expert Rev Anti Infect Ther. 2014;12:91–115. doi: 10.1586/14787210.2014.856756. [DOI] [PubMed] [Google Scholar]
- 49.Bogaerts P, Huang TD, Rezende DCR, Bouchahrouf W, Glupczynski Y. Could Acinetobacter pittii act as an NDM-1 reservoir for Enterobacteriaceae? J Antimicrob Chemother. 2013;68:2414–2415. doi: 10.1093/jac/dkt201. [DOI] [PubMed] [Google Scholar]
- 50.Poirel L, Naas T, Nordmann P. Diversity, epidemiology, and genetics of class D beta-lactamases. Antimicrob Agents Chemother. 2010;54:24–38. doi: 10.1128/AAC.01512-08. [DOI] [PMC free article] [PubMed] [Google Scholar]