LETTER
Increased prevalence of infections caused by Gram-negative pathogens that are multidrug resistant has prompted the reconsideration of polymyxins as therapeutic options. Resistance to polymyxins is due to mutations in the lipid A synthesis (1) that can be caused by the acquisition of mcr (2). Genes mcr-1 through mcr-8 (3, 4) and multiple subtypes have been reported to encode proteins that share 30 to 70% amino acid identity.
We previously reported the prevalence of mcr-1 among Escherichia coli and Klebsiella pneumoniae isolates collected worldwide during 2014 and 2015 by the SENTRY Program (5). As an ongoing effort, colistin-resistant E. coli and K. pneumoniae clinical isolates collected in 2016 were screened for the presence of mcr, and a new mcr-1 variant was characterized.
Among 11,493 E. coli and K. pneumoniae isolates tested, 199 (1.7%) were resistant to colistin per EUCAST criteria (6) and considered non-wild type by the CLSI (7) epidemiological cutoff value. Isolates displaying colistin MIC values of ≥4 mg/liter (resistant/non-wild type) were screened for the mcr-1 and mcr-2 genes by PCR. All isolates carrying mcr were submitted to whole-genome sequencing.
A total of 12 isolates were mcr-1 positive, including 10 E. coli isolates (2 in the United States, 3 [clonal] in Venezuela, 3 in Peru, 1 in Colombia, and 1 in Poland) and 2 K. pneumoniae isolates (1 each in Spain and Italy) (Table 1) recovered from invasive infections (Table 1). Colistin MIC values ranged from 4 to >8 mg/liter. No isolate yielded positive results for mcr-2.
TABLE 1.
Characteristics of mcr-1-producing isolates
| Organism and country | MLSTa | Infection typeb | MIC (mg/liter) forc: |
Other resistance genes | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CST | CAZ | CRO | FEP | TZP | IPM | CIP | GEN | TOB | TET | TGC | SXT | ||||
| Escherichia coli | |||||||||||||||
| Colombia | 131 | BSI | 8 (R) | 0.12 (S) | ≤0.06 (S) | ≤0.12 (S) | 2 (S) | 0.25 (S) | ≤0.03 (S) | 1 (S) | 1 (S) | 2 (S) | 0.12 (S) | >4 (R) | ant(3′)-Ia, dfrA1 |
| Perud | 95 | BSI | 4 (R) | 0.12 (S) | ≤0.06 (S) | ≤0.12 (S) | 1 (S) | ≤0.12 (S) | >4 (R) | 1 (S) | 1 (S) | >16 (R) | 0.12 (S) | 1 (S) | ant(3′)-Ia, aph(6)-Ia, aph(6)-Id, blaTEM-1, dfrA1, fosA, qnrB19, sul2, tet(A) |
| Peru | 7954 | SSSI | 4 (R) | 0.12 (S) | ≤0.06 (S) | ≤0.12 (S) | 2 (S) | ≤0.12 (S) | >4 (R) | >8 (R) | 4 (S) | >16 (R) | 0.25 (S) | >4 (R) | aac(3)-IIa, aph(3′)-Ia, aph(6)-Ia, catA1, dfrA12, sul1, sul2, blaTEM-1, tet(A) |
| Peru | 1485 | SSSI | 4 (R) | >8 (R) | >8 (R) | >16 (R) | 2 (S) | ≤0.12 (S) | 1 (S) | 1 (S) | 1 (S) | >16 (R) | 0.12 (S) | >4 (R) | blaCTX-M-55, sul2, tet(A) |
| Poland | 410 | UTI | 4 (R) | 8 (R) | >8 (R) | 8 (R) | 1 (S) | ≤0.12 (S) | >4 (R) | 1 (S) | 0.5 (S) | >16 (R) | 0.5 (S) | >4 (R) | aadA2, aph(6)-Ia, blaCTX-M-15, blaTEM-1, dfrA12, sul1 |
| USA | 58 | BSI | 4 (R) | 0.25 (S) | >8 (R) | 2 (S) | 2 (S) | ≤0.12 (S) | 0.06 (S) | 1 (S) | 1 (S) | >16 (R) | 0.25 (S) | >4 (R) | aadA2, ant(3′)-Ia, blaCTX-M-14, blaTEM-1, cmlA1, dfrA12, floR, sul2, sul3, tet(A) |
| USA | 1148 | UTI | 4 (R) | >8 (R) | >8 (R) | 2 (S) | 2 (S) | ≤0.12 (S) | >4 (R) | 0.25 (S) | 0.5 (S) | >16 (R) | 0.12 (S) | >4 (R) | aadA2, aph(3′)-Ia, blaSHV-12, blaTEM-1, dfrA12, qnrB19, sul3 |
| Venezuelae | 744 | BSI | 4 (R) | 2 (S) | >8 (R) | 4 (R) | 1 (S) | ≤0.12 (S) | >4 (R) | 0.5 (S) | 1 (S) | >16 (R) | 0.25 (S) | >4 (R) | aadA5, ant(3′)-Ia, aph(3′)-Ia, aph(3′)-IIa, aph(6)-Ia, aph(6)-Id, blaCTX-M-65, blaTEM-1, lnu(G), catA1, cmlA1, floR, sul3, tetB, dfrA12, dfrA17 |
| Venezuelae | 744 | SSSI | 4 (R) | 1 (S) | >8 (R) | 2 (S) | 1 (S) | ≤0.12 (S) | >4 (R) | 0.25 (S) | 0.5 (S) | >16 (R) | 0.12 (S) | >4 (R) | aadA5, ant(3′)-Ia, aph(3′)-Ia, aph(3′)-IIa, aph(6)-Ia, aph(6)-Id, blaCTX-M-65, blaTEM-1, lnu(G), catA1, cmlA1, floR, sul3, tetB, dfrA12, dfrA17 |
| Venezuelae | 744 | PIHP | 8 (R) | 0.12 (S) | ≤0.06 (S) | ≤0.12 (S) | 2 (S) | ≤0.12 (S) | 4 (R) | 0.5 (S) | 0.5 (S) | >16 (R) | 0.25 (S) | >4 (R) | ant(3′)-Ia, aph(3′)-Ia, aph(3′)-IIa, aph(6)-Ia, aph(6)-Id, blaTEM-1, lnu(G), catA1, cmlA1, floR, sul3, tetB, dfrA12 |
| Klebsiella pneumoniae | |||||||||||||||
| Italy | 219 | UTI | >8 (R) | >8 (R) | >8 (R) | >16 (R) | 4 (S) | 0.25 (S) | 1 (S) | 0.5 (S) | 1 (S) | >16 (R) | 0.5 (S) | >4 (R) | aadA2, aph(3′)-Ia, aph(6)-Ia-like, aph(6)-Id, blaCTX-M-15, blaSHV-1, dfrA12, mph(A), oqxA10, oqxB5, qnrS1, sul1, sul2, tet(A) |
| Spain | 806 | BSI | >8 (R) | 0.12 (S) | ≤0.06 (S) | ≤0.12 (S) | 2 (S) | ≤0.12 (S) | ≤0.03 (S) | 0.25 (S) | 0.25 (S) | 4 (S) | 0.5 (S) | ≤0.5 (S) | aph(6)-Ia, aph(6)-Id, blaSHV-1, fosA |
MLST, multilocus sequence type.
BSI, bloodstream infection; SSSI, skin and skin structure infection; UTI, urinary tract infection; PIHP, pneumonia in hospital patient.
CST, colistin; CAZ, ceftazidime; CRO, ceftriaxone; FEP, cefepime; TZP, piperacillin-tazobactam; IPM, imipenem; CIP, ciprofloxacin; GEN, gentamicin; TOB, tobramycin; TET, tetracycline; TGC, tigecycline; SXT, trimethoprim-sulfamethoxazole; ND, not determined; R, resistant per CLSI/EUCAST criteria; S, susceptible per CLSI/EUCAST criteria.
E. coli harboring mcr-1.11.
E. coli isolates from Venezuela were clonal and displayed identical resistance gene profiles.
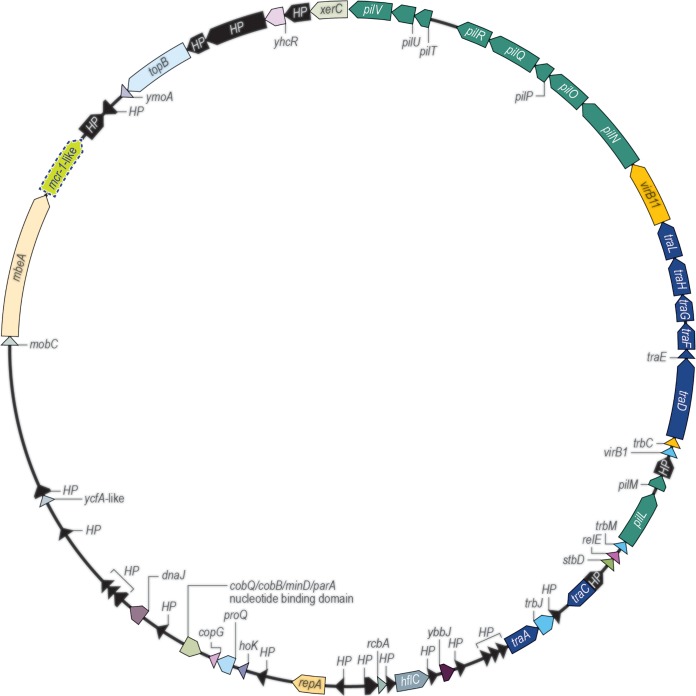
One E. coli isolate from Peru (sequence type 95 [ST95]) carried mcr-1 displaying an insertion of valine in amino acid position 6 and was designated mcr-1.11. This isolate showed susceptible phenotypes to β-lactams, aminoglycosides, tigecycline, and trimethoprim-sulfamethoxazole but resistance to tetracycline and quinolones (Table 1). The mcr-1.11 gene was located on a 63-kb IncI2 plasmid carrying no other resistance genes (GenBank accession number KY853650). Two genetically unrelated E. coli isolates (ST7954 and ST1485) from the same hospital that carried mcr-1 displayed the same plasmid structure (Table 1; Fig. 1).
FIG 1.
Schematic representation of IncI2 plasmids carrying mcr-1-like genes detected among E. coli isolates from a Peruvian medical center.
The mcr-1.11 gene cloned in an E. coli background exhibited colistin and polymyxin B MIC results (2 to 8 mg/liter) similar to those of mcr-1 (2 to 4 mg/liter). The mcr-1.11 gene likely emerged via spontaneous mutation within a plasmid structure that is endemic to the medical center in Peru, as seems common among mcr-like genes (1).
Unlike the vast majority of reports of mcr-like genes from animal-based sources, our results show a global prevalence of colistin resistance and mcr-1 among isolates collected from important human infections. Similar to the 2014 to 2015 survey, isolates carrying mcr-1 were identified in only 0.1% of the isolates tested; however, this study emphasizes its worldwide dissemination. Furthermore, our results and others (8, 9) suggest that these genes are prone to mutations as they spread, as was observed in the isolates from Peru.
A much higher prevalence of mcr-like genes from specific geographic locations (1, 10, 11) accompanied by the first report of mcr-1 in Pseudomonas aeruginosa (12) located on a chromosome demonstrate the ability of this gene to disseminate. Continued monitoring of mcr genes is warranted, but the development of global policies that might decrease the use of agents important to treat human infections or agents known to coselect for these resistance genes (13) seems prudent.
(These data were presented at the ASM Microbe 2018 meeting in poster format [Sunday-468].)
ACKNOWLEDGMENTS
This study was performed by JMI Laboratories. No outside funding was received for this study or preparation of the manuscript.
JMI Laboratories contracted to perform services in 2017 for Achaogen, Allecra Therapeutics, Allergan, Amplyx Pharmaceuticals, Antabio, API, Astellas Pharma, AstraZeneca, Athelas, Basilea Pharmaceutica, Bayer AG, Becton, Dickinson and Co., Boston Pharmaceuticals, CEM-102 Pharma, Cempra, Cidara Therapeutics, Inc., CorMedix, CSA BioTech, Cutanea Life Sciences, Inc., Entasis Therapeutics, Inc., Geom Therapeutics, Inc., GSK, Iterum Pharma, Medpace, Melinta Therapeutics, Inc., Merck & Co., Inc., MicuRx Pharmaceuticals, Inc., N8 Medical, Inc., Nabriva Therapeutics, Inc., NAEJA-RGM, Novartis, Paratek Pharmaceuticals, Inc., Pfizer, Polyphor, Ra Pharma, Rempex, Riptide Bioscience, Inc., Roche, Scynexis, Shionogi, Sinsa Labs, Inc., Skyline Antiinfectives, Sonoran Biosciences, Spero Therapeutics, SymbioticA, Synlogic, Synthes Biomaterials, TenNor Therapeutics, Tetraphase, The Medicines Company, Theravance Biopharma, VenatoRx Pharmaceuticals, Inc., Wockhardt, Yukon Pharma, Zai Laboratory, and Zavante Therapeutics, Inc.
We have no speakers’ bureau or stock options to declare.
REFERENCES
- 1.Poirel L, Jayol A, Nordmann P. 2017. Polymyxins: antibacterial activity, susceptibility testing, and resistance mechanisms encoded by plasmids or chromosomes. Clin Microbiol Rev 30:557–596. doi: 10.1128/CMR.00064-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu Y-Y, Wang Y, Walsh TR, Yi L-X, Zhang R, Spencer J, Doi Y, Tian G, Dong B, Huang X, Yu L-F, Gu D, Ren H, Chen X, Lv L, He D, Zhou H, Liang Z, Liu J-H, Shen J. 2016. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis 16:161–168. doi: 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- 3.Schwarz S, Johnson AP. 2016. Transferable resistance to colistin: a new but old threat. J Antimicrob Chemother 71:2066–2070. doi: 10.1093/jac/dkw274. [DOI] [PubMed] [Google Scholar]
- 4.Wang R, van Dorp L, Shaw LP, Bradley P, Wang Q, Wang X, Jin L, Zhang Q, Liu Y, Rieux A, Dorai-Schneiders T, Weinert LA, Iqbal Z, Didelot X, Wang H, Balloux F. 2018. The global distribution and spread of the mobilized colistin resistance gene mcr-1. Nat Commun 9:1179. doi: 10.1038/s41467-018-03205-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Castanheira M, Griffin MA, Deshpande LM, Mendes RE, Jones RN, Flamm RK. 2016. Detection of mcr-1 among Escherichia coli clinical isolates collected worldwide as part of the SENTRY Antimicrobial Surveillance Program in 2014 and 2015. Antimicrob Agents Chemother 60:5623–5624. doi: 10.1128/AAC.01267-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.EUCAST. 2018. Breakpoint tables for interpretation of MICs and zone diameters. http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_8.0_Breakpoint_Tables.pdf.
- 7.Clinical and Laboratory Standards Institute. 2018. Performance standards for antimicrobial susceptibility testing; 28th informational supplement. CLSI M100-S28. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 8.Zhao F, Feng Y, Lu X, McNally A, Zong Z. 2017. Remarkable diversity of Escherichia coli carrying mcr-1 from hospital sewage with the identification of two new mcr-1 variants. Front Microbiol 8:2094. doi: 10.3389/fmicb.2017.02094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ling Z, Yin W, Li H, Zhang Q, Wang X, Wang Z, Ke Y, Wang Y, Shen J. 2017. Chromosome-mediated mcr-3 variants in Aeromonas veronii from chicken meat. Antimicrob Agents Chemother 61:e01272-17. doi: 10.1128/AAC.01272-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu Y, Zhong LL, Srinivas S, Sun J, Huang M, Paterson DL, Lei S, Lin J, Li X, Tang Z, Feng S, Shen C, Tian GB, Feng Y. 2018. Spread of MCR-3 colistin resistance in China: an epidemiological, genomic and mechanistic study. EBioMedicine 34:139–157. doi: 10.1016/j.ebiom.2018.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eiamphungporn W, Yainoy S, Jumderm C, Tan-Arsuwongkul R, Tiengrim S, Thamlikitkul V. 2018. Prevalence of the colistin resistance gene mcr-1 in colistin-resistant Escherichia coli and Klebsiella pneumoniae isolated from humans in Thailand. J Glob Antimicrob Resist 15:32–35. doi: 10.1016/j.jgar.2018.06.007. [DOI] [PubMed] [Google Scholar]
- 12.Snesrud E, Maybank R, Kwak YI, Jones AR, Hinkle MK, McGann P. 2018. Chromosomally encoded mcr-5 in colistin-nonsusceptible Pseudomonas aeruginosa. Antimicrob Agents Chemother 62:e00679-18. doi: 10.1128/AAC.00679-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu F, Zeng X, Hinenoya A, Lin J. 2018. MCR-1 confers cross-resistance to bacitracin, a widely used in-feed antibiotic. mSphere 3:e00411-18. doi: 10.1128/mSphere.00411-18. [DOI] [PMC free article] [PubMed] [Google Scholar]