A critical gap in tuberculosis (TB) treatment is detection of emergent drug resistance. We hypothesized that advanced phenotyping with whole-genome sequencing (WGS) will detect low-frequency Mycobacterium tuberculosis drug resistance.
KEYWORDS: South Africa, extensively drug-resistant tuberculosis, heteroresistance, phage, tuberculosis, whole-genome sequencing
ABSTRACT
A critical gap in tuberculosis (TB) treatment is detection of emergent drug resistance. We hypothesized that advanced phenotyping with whole-genome sequencing (WGS) will detect low-frequency Mycobacterium tuberculosis drug resistance. We assessed a reporter mycobacteriophage (Φ2GFP10) in vitro to detect drug-resistant subpopulations and predict M. tuberculosis bactericidal activity in this pilot study. Subsequently, we prospectively studied 20 TB patients with serial Φ2GFP10, Xpert MTB/RIF, and M. tuberculosis culture through end of treatment. WGS was performed, and single nucleotide polymorphisms (SNPs) were examined to detect mixed infection in selected M. tuberculosis isolates. Resistant M. tuberculosis isolates were detected at 1:100,000, and changes in cytometry-gated events were predictive of in vitro M. tuberculosis bactericidal activity using the Φ2GFP10 assay. Emergent drug resistance was detected in one patient by Φ2GFP10 at 3 weeks but not by conventional testing (M. tuberculosis culture and GeneXpert). WGS revealed a phylogeographically distinct extensively drug-resistant tuberculosis (XDR-TB) genome, identical to an XDR-TB isolate from the patient’s spouse. Variant lineage-specific SNPs were present early, suggesting mixed infection as the etiology of emergent resistance with temporal trends providing evidence for selection during treatment. Φ2GFP10 can detect low-frequency drug-resistant M. tuberculosis and with WGS characterize emergent M. tuberculosis resistance. In areas of high TB transmission and drug resistance, rapid screening for heteroresistance should be considered.
INTRODUCTION
Mycobacterium tuberculosis has overtaken HIV infection as a leading cause of death due to single infectious etiology worldwide (1–5). While overall global tuberculosis (TB) incidence has declined, our best estimates show drug-resistant TB rapidly increasing in both absolute numbers and as a proportion of all incident TB cases (2, 6, 7). Meanwhile TB incidence in HIV-endemic countries in southern Africa has reached levels not seen in the United States or Western Europe since the turn of the last century (2, 8–11).
Phenotypic diagnostics have potential to perform drug susceptibility testing (DST) for drugs with complex or uncharacterized genetic mechanisms of resistance, to quantify the level of resistance, to distinguish viable from nonviable M. tuberculosis, to measure treatment response, and to detect mixed infection (12, 13). Our group has extensive experience with the construction of genetically modified mycobacterium-specific viruses (mycobacteriophages) for the genetic manipulation of M. tuberculosis (14–16). We recently described the construction and diagnostic capabilities of a new, more powerful reporter phage, Φ2GFP10, which uses a more efficient promoter and more powerful fluorescence reporter to allow direct visualization of individual metabolically active bacilli using fluorescence-activated cell sorting (FACS), including in clinical sputum (17). The Φ2GFP10 phage allows for rapid M. tuberculosis detection and phenotypic DST in clinical sputum samples, including paucibacillary concentrations.
We performed preliminary in vitro experiments to determine the limits of detection of the Φ2GFP10 reporter phage for subpopulations of drug-resistant bacteria and to determine the correlation of inhibition of fluorescence expression after Φ2GFP10 infection in the presence of antibiotics with M. tuberculosis bactericidal activity. The correlate experiments in clinical samples were to look for heteroresistance in clinical TB samples and measure dynamic changes in mycobacterial viability in the sputum of TB patients on treatment. As an unanticipated occurrence, one TB patient developed amplification of drug resistance, which was detected by the reporter phage assay prior to detection by conventional means (M. tuberculosis culture and GeneXpert MTB/RIF). In the present study, we describe an approach utilizing phage infection, cell sorting, and whole-genome sequencing (WGS) to characterize emergent drug resistance in TB patients on treatment.
RESULTS
Detecting low-frequency drug-resistant M. tuberculosis subpopulations using a mycobacteriophage assay.
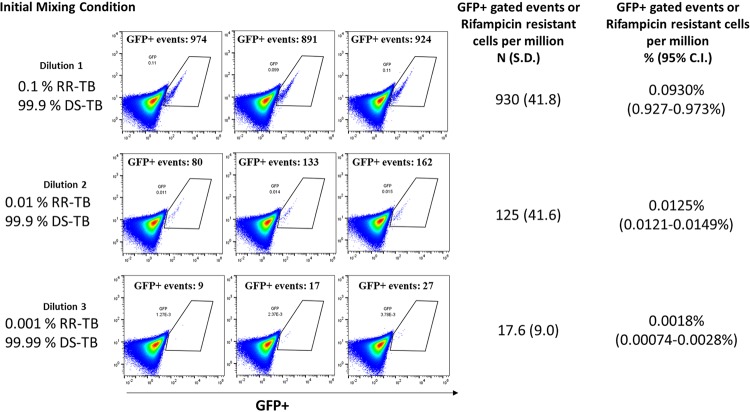
To determine the limits of detection for the Φ2GFP10 reporter phage, we designed a mixed-infection experiment using a drug-susceptible TB strain (mc26230) and a rifampin-resistant TB strain (mc27902). A liquid broth of M. tuberculosis in log-phase growth for both strains was prepared. The numbers of bacilli per ml were estimated using optical density measurements and a standard correlation scale. Initial mixing conditions of 1:1,000, 1:10,000, and 1:100,000 RR-TB to DS-TB wells were prepared. To determine the accuracy of the Φ2GFP10 reporter phage to recapitulate the initial mixing proportions, we prepared a rifampin and no-drug well (as described in Materials and Methods), as well as appropriate positive and negative controls. Φ2GFP10 phage infection, followed by flow cytometry and FACS gating, was performed as described in Materials and Methods. The number of FACS-gated events in the rifampin condition was used to approximate the number of rifampin-resistant bacilli, and the number of FACS-gated events in the no-drug condition approximated the number of total bacilli (drug susceptible and resistant). Using the rifampin condition as the numerator and the no-drug condition as the denominator, we determined the percentage of RR-TB in each experiment, which was extremely close to the initial condition down to the 1:100,000 dilution (R2 = 0.992) (Fig. 1).
FIG 1.
Recapitulation of initial mixing conditions of rifampin-resistant tuberculosis (RR-TB) with drug-susceptible tuberculosis (DS-TB) after phage infection and flow cytometry. Flow cytometry detects rare rifampin-resistant tuberculosis subpopulations. To estimate the limits of detection for the Ф2GFP10 reporter phage assay to detect resistant bacilli in mixed drug-susceptible/drug-resistant populations, we mixed attenuated drug-susceptible tuberculosis (mc26230) with attenuated rifampin-resistant tuberculosis (RR-TB) (mc27902), followed by serial dilutions, to create three mixed populations with 0.1, 0.01, and 0.001% RR-TB, respectively. We then incubated each initial mixing condition with Ф2GFP10 using previously described methods in the presence or absence of rifampin (17). FACS using experimentally constructed gates as previously described (17) were used to enumerate fluorescent events in the “Ф2GFP10 + M. tuberculosis” and “Ф2GFP10 + M. tuberculosis + rifampin conditions.” As previously shown (17), fluorescence in the presence of rifampin corresponds to rifampin resistance. We estimated the percentage of rifampin-resistant M. tuberculosis in each mixing condition by using the number of FACS-gated events in the “Ф2GFP10 + M. tuberculosis + rifampin” well as the numerator and the number of FACS-gated events in the “Ф2GFP10 + M. tuberculosis” well as the denominator. In this way, we were able to show that we were able to recapitulate the initial mixing conditions down to 1 drug-resistant M. tuberculosis organism per 100,000, with a high degree of correlation between phage-derived percentages and initial mixing conditions (R2 = 0.992).
Predicting M. tuberculosis bactericidal activity using a mycobacteriophage assay.
We hypothesized that fluorophage infection could be used as a rapid surrogate marker of potentially viable cells. To evaluate this hypothesis, we performed in vitro experiments to determine the potential of the mycobacteriophage assay to predict subsequent bactericidal activity. In liquid M. tuberculosis culture in the presence or absence of antimycobacterial agents, we compared dynamic changes in numbers of FACS-gated events after phage infection (representing metabolically active mycobacteria) with subsequent CFU in the presence of various antibiotics taken simultaneously from liquid culture at serial daily time points from time zero through day 7. Decreased enumerated FACS-gated events after phage infection was highly correlated mycobacterial killing, as measured by quantitative decreases in the M. tuberculosis CFU (R = 0.96, P < 0.001) (Fig. 2A).
FIG 2.
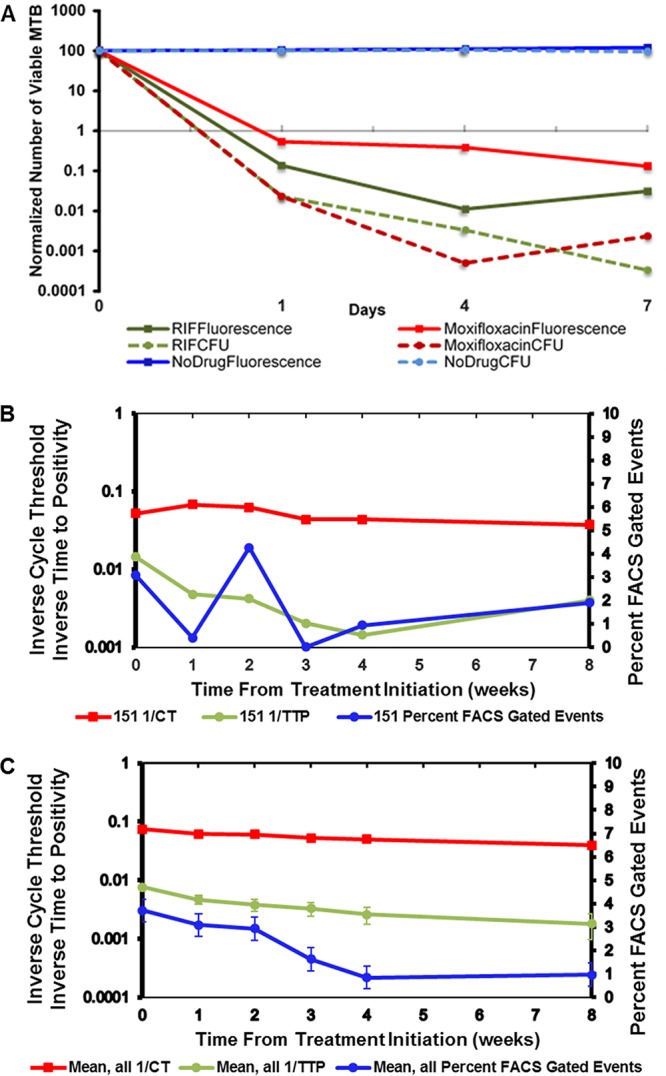
Fluoromycobacteriophage measures dynamic killing of M. tuberculosis in response to antibiotics in vitro and in ex vivo sputum samples from TB patients on treatment. (A) In vitro attenuated drug-susceptible M. tuberculosis (mc26230) was inoculated with rifampin (RIF), moxifloxacin, or no drug. Ф2GFP10 was added 16 h prior to each time point, and the number of fluorescent M. tuberculosis organisms was measured by flow cytometry (FACS) and by counting CFU in parallel. The data are normalized by dividing the M. tuberculosis counts by the baseline M. tuberculosis counts and plotted against a logarithmic scale. The data show that untreated M. tuberculosis remained viable in substantial numbers through day 7 by both FACS and CFU, while the levels of drug-susceptible M. tuberculosis treated with moxifloxacin or rifamycin decreased markedly. (B) Ex vivo sputum samples from AFB smear-positive TB patients (n = 20) from the initiation of treatment through 8 weeks of treatment. Sputa were sampled weekly through week 8. GeneXpert, culture DST, and phage assay (Ф2GFP10) were performed at baseline and at weeks 1, 2, 3, 4, and 8. Ф2GFP10 was added 16 h prior to each time point; the number of fluorescent M. tuberculosis was measured by flow cytometry (FACS), and the bacillary burden was estimated using the time to positivity in liquid M. tuberculosis growth media. The percentage FACS-gated events compared events in FACS-gated to all FACS events. The cycle threshold was derived from GeneXpert RT-PCR measurements. The time to positivity was the time to M. tuberculosis culture positivity in MGIT liquid culture media. The standard error of the mean is shown. Gated events and inverse times to positivity were highly correlated (R = 0.89, P = 0.033). (C) Detection of treatment failure by reporter phage. Emergent drug resistance was detected at week 4 in one patient (PID 151) by the reporter phage but not until week 8 by the GeneXpert MTB/RIF and the conventional proportion method DST. On conventional DST, the patient showed resistance to isoniazid, rifampin, ofloxacin, kanamycin, capreomycin, amikacin, and ethambutol at week 8 despite having drug-susceptible M. tuberculosis at treatment initiation. Baseline drug-susceptible TB with emergent XDR-TB in this patient was confirmed by independent DST done on separate clinical isolates collected by the provincial TB control system. The percentage of FACS-gated events, the time to positivity, and the cycle threshold were defined as described above.
Dynamic changes in FACS-gated M. tuberculosis in the sputa of TB patients on treatment.
To evaluate the ability of the reporter phage assay to detect dynamic changes on treatment, we recruited 20 participants with microbiologically confirmed drug-susceptible TB. The participants were followed with weekly study visits during the first month of standard TB chemotherapy, again at 8 weeks, and then clinically through the end of treatment, including serial symptom screening, adherence by self-report and pill counts, and HIV testing. In patients undergoing treatment, the numbers of fluorescent events after Φ2GFP10 infection and cell sorting decreased during treatment, correlating with inverse time to positivity and reflecting an M. tuberculosis treatment response.
All 20 participants were positive at baseline as determined by the GeneXpert MTB/RIF for the presence of M. tuberculosis, but all were negative for rifampin resistance. Drug susceptibility to rifampin and six other first- and second-line drugs was confirmed using standard protocols. Fifty-six percent of the patients whose HIV status was known were HIV infected, and 4 patients (20%) had previously received TB treatment.
We analyzed the kinetics FACS-gated events using the mycobacteriophage assay on serial samples from the 20 participants described above. When the number of FACS-gated events representing viable phage-infected bacteria expressing green fluorescent protein (GFP) was compared to the inverse of time to positivity in liquid TB growth media (a measure of mycobacterial burden), there was a high degree of correlation (R = 0.89, P = 0.033) (Fig. 2B), suggesting that the Φ2GFP10 assay may be a useful predictor of bacterial growth kinetics and, indirectly, sputum bacillary load.
Emergent drug resistance on treatment.
In one patient (PID 151) at week 4 of treatment the overall numbers Φ2GFP10 gated events substantially increased in the presence or absence of antimycobacterial agents (Fig. 2C). These findings suggested emergent drug resistance despite complete medication adherence by pill count and self-report. Subsequent drug-susceptibility testing on the isolate using Φ2GFP10 demonstrated an XDR-TB resistance pattern (resistance to isoniazid, rifamycin, kanamycin, and ofloxacin) (see Fig. S1 in the supplemental material). Conventional drug susceptibility testing and GeneXpert MTB/RIF revealed drug-resistant isolates only at week 8. While the overall cycle threshold (CT) values from the GeneXpert MTB/RIF were concordant with time to positivity and phage-gated events in predicting response to treatment (Fig. S2 and 3), analysis of the CT values from the GeneXpert MTB/RIF did not demonstrate any probe-specific reductions in CT values, confirming that the GeneXpert MTB/RIF was unable to detect this drug-resistant bacteria at earlier time points (Fig. S4).
The rapid amplification of resistance from drug susceptible to XDR in PID 151 in the absence of second-line drug exposure suggested mixed infection with a susceptible and resistant M. tuberculosis strain rather than acquired drug resistance. To exclude the possibility of laboratory cross contamination, we first obtained the drug susceptibility profile of the sample sent to the local diagnostic laboratory after 2 months of therapy. This independent sample was also reported as resistant to the six drugs tested (rifampin, isoniazid, ethambutol, streptomycin, ofloxacin, and kanamycin).
Genome sequencing to distinguish heteroresistance from evolved drug resistance.
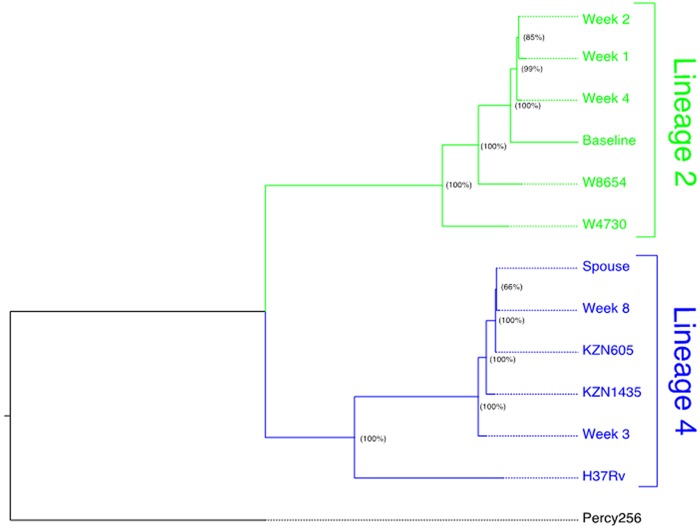
Using WGS data, we confirmed that PID 151 did not develop XDR-TB de novo during treatment but instead developed XDR-TB via primary or secondary polyclonal infection with a genetically distinct XDR-TB isolate. During follow-up, we discovered PID 151’s cohabiting spouse had been admitted to a TB referral hospital for treatment of XDR-TB. WGS-based phylogenetic reconstruction shows that isolates collected from PID 151 at early treatment time points (baseline; weeks 1, 2, and 4) clustered with drug-susceptible isolates from sub-Saharan Africa belonging to M. tuberculosis phylogeographic lineage 2 and isolates from later time points (weeks 3 and 8 and the spouse’s M. tuberculosis isolate) clustered with lineage 4 and with historical drug-resistant isolates from KwaZulu-Natal, South Africa. (Fig. 3).
FIG 3.
Genotyping demonstrates distinct M. tuberculosis lineages in a patient (PID 151) who acquired extensively drug-resistant tuberculosis (XDR-TB) on treatment (A) and analysis of heterozygous single- nucleotide polymorphisms reveals low frequency mixed infection from early in treatment (B). (A) Maximum-likelihood phylogenetic reconstruction. Whole-genome sequencing (WGS) data were obtained for baseline and week 1, 2, 3, 4, and 8 M. tuberculosis cultures from the patient who developed XDR-TB on treatment, as identified by the reporter phage assay (patient 151). Contact investigation determined that the patient’s spouse was previously diagnosed with XDR-TB, and we subsequently obtained WGS data for an M. tuberculosis isolate from the patient’s spouse. Phylogenetic reconstruction determined that initial isolates from patient 151 (baseline, week 1, and week 4) cluster distinctly with representative archival isolates from M. tuberculosis phylogeographic lineage 2 (W8654 and W4730) and isolates collected later during treatment (week 3 and week 8), as well as the isolate obtained from the patient’s spouse, cluster distinctly with archival MDR-TB and XDR-TB isolates collected in KwaZulu-Natal, South Africa (KZN1435 and KZN605). Node labels indicate bootstrap support (proportion of 100 bootstrap replicates) supporting each clade. (B) Whole-genome sequencing to detect mixed infection in early time points. To evaluate for potential evidence of polyclonal infection, we characterized sequence read heterozygosity at SNPs. We defined mixed variant calls as SNPs where reads matching the reference accounted for >25% of the total read depth. The numbers and proportions of all unfiltered SNPs meeting these criteria for each isolate are included column 2. We used representative archival strains to define nonoverlapping subsets of SNPs that are unique to either M. tuberculosis phylogeographic lineage 2 or lineage 4. Isolates phylogenetically grouped in lineage 2 in panel A share 362 unique SNPs; those grouped in lineage 4 share 425 unique SNPs. As a proxy estimate for the extent of admixture between the pretreatment L2 isolate and the L4 spousal isolate, we tabulated the number of L4-specific SNPs called for each isolate (column 3) and quantitated the average read depth at lineage 4-specific SNPs (column 4) for each isolate obtained during treatment. The normalized averaged read depth, i.e., the average read depth at lineage 4-specific SNPs divided by the average read depth for all isolates in a sample, is provided in column 6.
We then analyzed mixed variant calls as described in Materials and Methods. Sequences from weeks 1 to 8 exhibit a significant proportion of mixed variant calls, likely introduced by the presence of multiple M. tuberculosis subpopulations in the sequenced isolates, whereas no mixed variant calls were identified in the baseline sample. The overall numbers of mixed variant calls decreased as a proportion of all variants during the course of treatment (from 33% to 5% by Week 8). The number of lineage 4-specific SNPs that were called for each serial isolate increased progressively from four in the baseline isolate to 425 in the week 3 isolate, consistent with a dynamic polyclonal infection in which the lineage 4 isolate replaced the lineage 2 isolate under antibiotic pressure. Taken together, these lines of evidence suggest that PID 151 was infected with a drug-susceptible lineage 2 isolate and superinfected with an XDR-TB isolate, most likely from her spouse, which was subsequently selected for during antibiotic treatment for drug-susceptible TB.
When we looked for the presence of lineage-specific SNPs as variant reads, these were found at baseline (4) and increased from week 1; by week 3, lineage 4-specific SNPs had reached the maximum possible number (i.e., 425). Sequences from weeks 1 to 8 exhibit a significant proportion of mixed variant calls, likely introduced by the presence of multiple M. tuberculosis subpopulations in the sequenced isolates, whereas no mixed variant calls were identified in the baseline sample. Taken together, these lines of evidence suggest that PID 151 was infected with a drug-susceptible lineage 2 isolate and superinfected with an XDR-TB isolate, most likely from her spouse, which was subsequently selected for during antibiotic treatment for drug-susceptible TB.
DISCUSSION
It is increasingly recognized that mixed infection with M. tuberculosis is prevalent, particularly in settings with a high M. tuberculosis force of infection and HIV coinfection (18–20). Mixed infection with heteroresistant M. tuberculosis can rapidly lead to treatment failure and increased mortality due to positive selection for resistant M. tuberculosis subpopulations with infective antibiotics (21). We report here on the sequential emergence of TB drug resistance in a TB patient on treatment due to heteroresistance. Use of a novel reporter mycobacteriophage assay using a cell sorting strategy identified drug-resistant M. tuberculosis subpopulations of interest at substantially earlier time points compared to conventional methods. Our findings were confirmed and extended by WGS results of baseline and subsequent M. tuberculosis isolates, demonstrating an initial fully susceptible lineage 4 isolate that was replaced over time with an extensively drug-resistant lineage 2 isolate, confirming mixed infection as the likely etiology of the emergent drug resistance. As part of developing this clinical study, we performed proof-of-concept in vitro work demonstrating the correlation of phage-measured inhibition of fluorescence by antibiotics with subsequent M. tuberculosis killing, as measured by quantitative culture, and in vitro limits of mixed infection detection using the phage assay.
Although the reporter phage assay does not need to be linked to WGS, this may be an appealing approach. A phenotypic/genotypic approach could use high-throughput reporter phage screening to identify potentially M. tuberculosis heteroresistance to increase the efficiency of WGS by enriching for heteroresistance, potentially reducing false positives in WGS, and allow for recognition of heteroresistance due to antimycobacterial agents with mechanisms of drug resistance that are incompletely genetically characterized (e.g., bedaquiline and delaminid).
Fittingly, the original work on mixed TB infection in the human host was done using mycobacteriophage typing (22). An early report from 1976 examined 87 TB patients with M. tuberculosis isolated from two or more different organs and found evidence of different TB strains by differential phage infection in 3/87 (23). Modern study of mixed M. tuberculosis infection dates early studies using restriction fragment length polymorphisms (RFLP) with insertion sequence 6110 probes to fingerprint M. tuberculosis isolates (24, 25). RFLP was superseded by PCR-based methods and now whole-genome sequencing. With each progressively higher resolution technique, more frequent mixed infections have been revealed. Whole-genome sequencing and deep sequencing have revealed much more diverse heterogeneity than previously realized and herald a growing understanding that mixed infection and mixed infection with drug-resistant bacilli—heteroresistance—has both pathogen evolutionary and clinical implications.
The limitations of this study include epidemiologic, microbiologic, and sequencing limitations. From an epidemiologic standpoint, a larger, better-designed study is required to establish the role of combined phage phenotyping and whole-genome sequencing to clinically characterize heteroresistance. Microbiologic limitations include bias introduced by M. tuberculosis culturing, which may be responsible for some small inconsistencies seen in our results (e.g., week 3 isolate clustering with lineage 4). Sequencing directly from clinical sputum would obviate this source of bias but is currently technically challenging. Sequencing limitations include lack of high depth coverage at all SNP positions of interest; however, high-quality reads revealing emergent lineage-specific SNPs provides convincing evidence for early heteroresistance.
In high-burden TB settings, drug-resistant TB can and does emerge during treatment and may be due to the amplification of drug resistance, mixed infection (heteroresistance), or intercurrent infection. There are currently no widely accepted or feasible means to detect heteroresistance early as it emerges early in treatment. In the present study, we detected emergent heteroresistance using an advanced phenotypic reporter phage assay and whole-genome sequencing to detect heteroresistance and demonstrated presence of mixed infection in early in treatment. Larger studies are needed to define the role and utility of such an approach.
MATERIALS AND METHODS
Laboratory methods.
(i) Phage infection and flow cytometry gating. Methods describing the generation of the Φ2GFP10 phage have been previously published (26, 27). Briefly, the phage construct consists of an optimized mycobacteriophage backbone with a constitutive L5 promoter upstream from an mVenus reporter gene which produces GFP. After phage infection in a viable M. tuberculosis bacillus GFP expression is a biomarker for metabolic activity detectable by fluorescence microscopy, flow cytometry, or a fluorescent plate reader. In the presence of an effective antimycobacterial agent, translation is disrupted, GFP is not expressed, and fluorescence is not detected.
After Φ2GFP10 phage infection, the M. tuberculosis was characterized and enumerated using flow cytometry after infection (BD Accuri C6 flow cytometer [Becton Dickinson, Franklin Lakes, NJ]) using automated settings. Appropriate flow cytometry gates were experimentally derived. Viable M. tuberculosis were measured by enumerating fluorescent events by flow cytometry gating, and cutoffs for positive results were defined as previously described (26, 27).
(ii) Drug susceptibility testing with the phage assay. Methods describing drug susceptibility testing using the phage assay have been previously described (26, 27). Briefly, M. tuberculosis culture was set out in a 96-well plate with no-antibiotic controls and antibiotics of interest (isoniazid, rifampin, ofloxacin, and kanamycin). After overnight incubation, the Φ2GFP10 phage was added to all antibiotic-treated wells and one of the no-drug controls. A laboratory-grown M. tuberculosis culture was used as a positive control in each plate. Flow cytometry was performed after 24 h. Fluorescence in the “M. tuberculosis culture + phage” condition versus the negative control indicates the presence of metabolically active M. tuberculosis. Fluorescence in the “M. tuberculosis culture + phage + antibiotic” condition indicates M. tuberculosis that is resistant to the antibiotic of interest.
(iii) Measuring bactericidal activity of antimycobacterial drugs in vitro using Φ2GFP10 phage. To estimate the ability of Φ2GFP10 phage to predict bactericidal activity of antimycobacterial drugs, we compared the number of viable bacteria determined after Φ2GFP10 phage infection to the number of CFU in the presence of antibiotics. Attenuated drug-susceptible M. tuberculosis (mc26230) was inoculated with rifampin (RIF), moxifloxacin, or no drug. Φ2GFP10 was added 16 h prior to each time point, and the number of fluorescent M. tuberculosis organisms was measured by flow cytometry (FACS) and by counting the CFU in parallel. Data were normalized by baseline M. tuberculosis counts and plotted against a logarithmic scale.
Clinical methods.
As a pilot study to determine the phage assay’s ability to detect changes in M. tuberculosis populations during treatment, consecutive TB suspects were enrolled at an outpatient municipal chest clinic in Durban, South Africa. Adult pulmonary TB suspects were eligible to participate if they were not currently taking TB medications and had a documented positive sputum acid-fast bacillus (AFB) smear result in the clinic system. Demographic, symptom, and clinical data were recorded by study staff, including prior TB history. HIV counseling and testing was offered to all patients. Extensively drug-resistant tuberculosis (XDR-TB) was defined as resistance to isoniazid, rifampin, any of the fluoroquinolones, and any of the second-line injectable agents. Written informed consent was obtained from all participants.
The primary objective in the clinical pilot study was to measure dynamic changes in Φ2GFP10 reporter phage FACS-gated events during treatment in clinical sputum of TB patients. Early morning sputum samples, adherence, and clinical data were obtained at baseline and at weeks 1, 2, 3, 4, and 8. M. tuberculosis culture (BD MGIT), M. tuberculosis drug susceptibility testing using solid agar testing, Gene Xpert MTB RIF, and an Φ2GFP10 assay were performed at all time points. Patients were followed through end of TB treatment. Approval for the study was obtained from ethics review boards at the University of KwaZulu-Natal, Albert Einstein College of Medicine, Columbia University Medical Center, the KwaZulu-Natal Department of Health, and the eThekwini Municipality.
Whole-genome sequencing and data analysis.
Single M. tuberculosis colonies were selected from agar growth media and genomic M. tuberculosis DNA was isolated from clinical samples using standard methods. Library preparation and WGS were performed as previously described on the Illumina MiSeq (28). Raw sequencing reads were trimmed for low-quality bases, filtered for duplicate reads, and aligned to the H37Rv M. tuberculosis reference genome (NC_000962.3) using the Burrows-Wheeler Aligner. Variant calls were obtained using SAMtools. Variants in the highly variable PE/PPE gene family, variants in repeat regions or mobile elements, and variants within 15 bp of an insertion or deletion were excluded from analysis. The mean read depth at all SNPs ranged from 6.8 to 43.02. High-quality SNPs were defined as those with PHRED-scaled quality scores of >100. Mixed variant calls were defined as any SNP where reads matching the reference accounted for >25% of the total read depth. Lineage-specific SNPs were defined with respect to well-characterized clinical isolates from KwaZulu-Natal (KZN605 for lineage 4 and W8654 for lineage 2).
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by National Institutes of Health/National Institute of Allergy and Infectious Disease R01 (1R01AI114900 [M.H.L., A.P., and M.R.O.]), K23 (AI098479-01A1 [M.R.O.]), and T32 (T32AI007061 [T.B.]). This research was also supported by a Doris Duke Clinical Scientist Career Development Award and the Louis V. Gerstner Foundation (M.R.O.), by the Potts Memorial Foundation Research Fellowship/Howard Hughes Medical Institute (P.J.), by the Howard Hughes Medical Institute (A.P., M.H.L., and W.R.J.), and by the National Institute of Allergy and Infectious Diseases (T32AI007061 to T.S.B.).
There are no conflicts of interest to declare.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.01834-18.
REFERENCES
- 1.Horsburgh CR., Jr 2000. The global problem of multidrug-resistant tuberculosis: the genie is out of the bottle. JAMA 283:2575–2576. [DOI] [PubMed] [Google Scholar]
- 2.World Health Organization. 2017. World Health Organization: global tuberculosis report 2017. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 3.Friedrich MJ. 2017. Tuberculosis update 2017. JAMA 318:2287. doi: 10.1001/jama.2017.18477. [DOI] [PubMed] [Google Scholar]
- 4.Friedrich MJ. 2017. Drug-resistant tuberculosis predicted to increase in high-burden countries. JAMA 318:231. doi: 10.1001/jama.2017.9086. [DOI] [PubMed] [Google Scholar]
- 5.Zumla A, Raviglione M, Hafner R, von Reyn CF. 2013. Tuberculosis. N Engl J Med 368:745–755. doi: 10.1056/NEJMra1200894. [DOI] [PubMed] [Google Scholar]
- 6.Musa BM, Adamu AL, Galadanci NA, Zubayr B, Odoh CN, Aliyu MH. 2017. Trends in prevalence of multi drug resistant tuberculosis in sub-Saharan Africa: a systematic review and meta-analysis. PLoS One 12:e0185105. doi: 10.1371/journal.pone.0185105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zignol M, van Gemert W, Falzon D, Sismanidis C, Glaziou P, Floyd K, Raviglione M. 2012. Surveillance of anti-tuberculosis drug resistance in the world: an updated analysis, 2007–2010. Bull World Health Organ 90:111–119D. doi: 10.2471/BLT.11.092585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tafuma TA, Burnett RJ, Huis In’t Veld D. 2014. National guidelines not always followed when diagnosing smear-negative pulmonary tuberculosis in patients with HIV in Botswana. PLoS One 9:e88654. doi: 10.1371/journal.pone.0088654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van Schalkwyk C, Mndzebele S, Hlophe T, Garcia Calleja JM, Korenromp EL, Stoneburner R, Pervilhac C. 2013. Outcomes and impact of HIV prevention, ART and TB programs in Swaziland: early evidence from public health triangulation. PLoS One 8:e69437. doi: 10.1371/journal.pone.0069437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Loveday M, Padayatchi N, Voce A, Brust J, Wallengren K. 2013. The treatment journey of a patient with multidrug-resistant tuberculosis in South Africa: is it patient-centred? Int J Tuber Lung Dis 17:56–59. doi: 10.5588/ijtld.13.0101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shah NS, Auld SC, Brust JC, Mathema B, Ismail N, Moodley P, Mlisana K, Allana S, Campbell A, Mthiyane T, Morris N, Mpangase P, van der Meulen H, Omar SV, Brown TS, Narechania A, Shaskina E, Kapwata T, Kreiswirth B, Gandhi NR. 2017. Transmission of extensively drug-resistant tuberculosis in South Africa. N Engl J Med 376:243–253. doi: 10.1056/NEJMoa1604544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cohen T, van Helden PD, Wilson D, Colijn C, McLaughlin MM, Abubakar I, Warren RM. 2012. Mixed-strain Mycobacterium tuberculosis infections and the implications for tuberculosis treatment and control. Clin Microbiol Rev 25:708–719. doi: 10.1128/CMR.00021-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cohen T, Chindelevitch L, Misra R, Kempner ME, Galea J, Moodley P, Wilson D. 2016. Within-host heterogeneity of Mycobacterium tuberculosis infection is associated with poor early treatment response: a prospective cohort study. J Infect Dis 213:1796–1799. doi: 10.1093/infdis/jiw014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jacobs WR Jr, Tuckman M, Bloom BR. 1987. Introduction of foreign DNA into mycobacteria using a shuttle phasmid. Nature 327:532–535. doi: 10.1038/327532a0. [DOI] [PubMed] [Google Scholar]
- 15.Banerjee A, Dubnau E, Quemard A, Balasubramanian V, Um KS, Wilson T, Collins D, de Lisle G, Jacobs WR. Jr, 1994. inhA, a gene encoding a target for isoniazid and ethionamide in Mycobacterium tuberculosis. Science 263:227–230. doi: 10.1126/science.8284673. [DOI] [PubMed] [Google Scholar]
- 16.Jain P, Thaler DS, Maiga M, Timmins GS, Bishai WR, Hatfull GF, Larsen MH, Jacobs WR. 2011. Reporter phage and breath tests: emerging phenotypic assays for diagnosing active tuberculosis, antibiotic resistance, and treatment efficacy. J Infect Dis 204:S1142–S1150. doi: 10.1093/infdis/jir454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jain P, Hartman TE, Eisenberg N, O’Donnell MR, Kriakov J, Govender K, Makume M, Thaler DS, Hatfull GF, Sturm AW, Larsen MH, Moodley P, Jacobs WR Jr. 2012. Φ2GFP10, a high-intensity fluorophage, enables detection and rapid drug susceptibility testing of Mycobacterium tuberculosis directly from sputum samples. J Clin Microbiol 50:1362–1369. doi: 10.1128/JCM.06192-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tarashi S, Fateh A, Jamnani FR, Siadat SD, Vaziri F. 2017. Prevalence of Beijing and Haarlem genotypes among multidrug-resistant Mycobacterium tuberculosis in Iran: systematic review and meta-analysis. Tuberculosis (Edinb) 107:31–37. doi: 10.1016/j.tube.2017.03.005. [DOI] [PubMed] [Google Scholar]
- 19.Tarashi S, Fateh A, Mirsaeidi M, Siadat SD, Vaziri F. 2017. Mixed infections in tuberculosis: the missing part in a puzzle. Tuberculosis (Edinb) 107:168–174. doi: 10.1016/j.tube.2017.09.004. [DOI] [PubMed] [Google Scholar]
- 20.Shin SS, Modongo C, Ncube R, Sepako E, Klausner JD, Zetola NM. 2015. Advanced immune suppression is associated with increased prevalence of mixed-strain Mycobacterium tuberculosis infections among persons at high risk for drug-resistant tuberculosis in Botswana. J Infect Dis 211:347–351. doi: 10.1093/infdis/jiu421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zwerling A, Dowdy D, von Delft A, Taylor H, Merritt MW. 2017. Incorporating social justice and stigma in cost-effectiveness analysis: drug-resistant tuberculosis treatment. Int J Tuber Lung Dis 21:69–74. doi: 10.5588/ijtld.16.0839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stead WW, Bates JH. 1969. Primary tuberculosis from the far east: transmission by a veteran to two civilians. Ann Intern Med 70:707–711. [DOI] [PubMed] [Google Scholar]
- 23.Bates JH, Stead WW, Rado TA. 1976. Phage type of tubercle bacilli isolated from patients with two or more sites of organ involvement. Am Rev Respir Dis 114:353–358. doi: 10.1164/arrd.1976.114.2.353. [DOI] [PubMed] [Google Scholar]
- 24.van Embden JD, Cave MD, Crawford JT, Dale JW, Eisenach KD, Gicquel B, Hermans P, Martin C, McAdam R, Shinnick TM. 1993. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J Clin Microbiol 31:406–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van Rie A, Victor TC, Richardson M, Johnson R, van der Spuy GD, Murray EJ, Beyers N, Gey van Pittius NC, van Helden PD, Warren RM. 2005. Reinfection and mixed infection cause changing Mycobacterium tuberculosis drug-resistance patterns. Am J Respir Crit Care Med 172:636–642. doi: 10.1164/rccm.200503-449OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jain P, Weinrick BC, Kalivoda EJ, Yang H, Munsamy V, Vilcheze C, Weisbrod TR, Larsen MH, O’Donnell MR, Pym A, Jacobs WR. Jr, 2016. Dual-reporter mycobacteriophages (ϕ2DRMs) reveal preexisting Mycobacterium tuberculosis persistent cells in human sputum. mBio 7:e01023-16. doi: 10.1128/mBio.01023-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.O’Donnell MR, Pym A, Jain P, Munsamy V, Wolf A, Karim F, Jacobs WR Jr, Larsen MH. 2015. A novel reporter phage to detect tuberculosis and rifampin resistance in a high-HIV-burden population. J Clin Microbiol 53:2188–2194. doi: 10.1128/JCM.03530-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O'Neill MB, Mortimer TD, Pepperell CS. 2015. Diversity of Mycobacterium tuberculosis across evolutionary scales. PLoS Pathog 11:e1005257. doi: 10.1371/journal.ppat.1005257. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.