Polymyxins are used as a last-line therapy against multidrug-resistant (MDR) New Delhi metallo-β-lactamase (NDM)-producing Klebsiella pneumoniae. However, polymyxin resistance can emerge with monotherapy; therefore, novel strategies are urgently needed to minimize the resistance and maintain their clinical utility.
KEYWORDS: Klebsiella pneumoniae, drug repurposing, polymyxins, zidovudine
ABSTRACT
Polymyxins are used as a last-line therapy against multidrug-resistant (MDR) New Delhi metallo-β-lactamase (NDM)-producing Klebsiella pneumoniae. However, polymyxin resistance can emerge with monotherapy; therefore, novel strategies are urgently needed to minimize the resistance and maintain their clinical utility. This study aimed to investigate the pharmacodynamics of polymyxin B in combination with the antiretroviral drug zidovudine against K. pneumoniae. Three isolates were evaluated in static time-kill studies (0 to 64 mg/liter) over 48 h. An in vitro one-compartment pharmacokinetic/pharmacodynamic (PK/PD) model (IVM) was used to simulate humanized dosage regimens of polymyxin B (4 mg/liter as continuous infusion) and zidovudine (as bolus dose thrice daily to achieve maximum concentration of drug in broth [Cmax] of 6 mg/liter) against K. pneumoniae BM1 over 72 h. The antimicrobial synergy of the combination was further evaluated in a murine thigh infection model against K. pneumoniae 02. In the static time-kill studies, polymyxin B monotherapy produced rapid and extensive killing against all three isolates followed by extensive regrowth, whereas zidovudine produced modest killing followed by significant regrowth at 24 h. Polymyxin B in combination with zidovudine significantly enhanced the antimicrobial activity (≥4 log10 CFU/ml) and minimized bacterial regrowth. In the IVM, the combination was synergistic and the total bacterial loads were below the limit of detection for up to 72 h. In the murine thigh infection model, the bacterial burden at 24 h in the combination group was ≥3 log10 CFU/thigh lower than each monotherapy against K. pneumoniae 02. Overall, the polymyxin B-zidovudine combination demonstrates superior antimicrobial efficacy and minimized emergence of resistance to polymyxins.
INTRODUCTION
Antimicrobial resistance is a significant threat to human health globally (1). Multidrug-resistant (MDR) New Delhi metallo-β-lactamase (NDM)-producing Klebsiella pneumoniae has been highlighted by the World Health Organization (WHO) as a priority pathogen that poses critical need of new antibiotics (1, 2). Polymyxins (i.e., polymyxin B and colistin) are increasingly used as a last-line therapy for infections caused by NDM-producing MDR K. pneumoniae (3–5). After intravenous administration, polymyxin B and colistin display poor pharmacokinetics/pharmacodynamics in the lungs (6–8), potentially due to the binding to lung surfactant (9, 10). Furthermore, polymyxin monotherapy can lead to regrowth, which is particularly problematic in infections with high bacterial densities, such as pneumonia (3, 11, 12). Given the relatively high mutation frequency and dose-limiting nephrotoxicity of polymyxins (4), novel strategies are urgently needed to preserve their efficacy against life-threatening infections caused by NDM-producing MDR K. pneumoniae with minimal development of resistance (3, 13, 14).
The use of synergistic combinations of antibiotics with FDA-approved nonantibiotics has been proposed as a promising alternative to improve the clinical efficacy of polymyxins against these problematic MDR Gram-negative pathogens (13, 15–19). To date, a number of studies have shown that polymyxin B in combination with FDA-approved nonantibiotics drugs (e.g., ascorbic acid [20], benserazide [20], chloroxine [20], closantel [16], loperamide [21], tamoxifen [17], tegaserod [20], mitomycin C [20], mitotane [19], ivacaftor [15], and silver nanoparticles [18]) display synergistic killing activity against MDR Pseudomonas aeruginosa and Acinetobacter baumannii. However, only several studies investigated the efficacy of polymyxin combinations with nonantibiotic drugs against NDM-producing MDR K. pneumoniae (13, 17, 18). Zidovudine is a nucleoside reverse transcriptase inhibitor with activity against the human immunodeficiency virus (HIV) (22) and has also been shown to display antibacterial activity against K. pneumoniae (23–26). Zidovudine is purported to exert its antimicrobial activity via interfering with bacterial DNA replication (24, 27). Given that polymyxins permeabilize the outer membrane of Gram-negative pathogens (4), it is highly likely that polymyxin exposure enhances the antimicrobial activity of zidovudine by increasing the intracellular concentration, thereby allowing more zidovudine molecules to interact with their intracellular targets (13). The primary objective of this study was to investigate the pharmacodynamics of polymyxin B in combination with zidovudine against NDM-producing MDR K. pneumoniae using in vitro static time-kill, an in vitro one-compartment pharmacokinetic/pharmacodynamic (PK/PD) model (IVM), and a neutropenic mouse thigh infection model. Furthermore, a mechanism-based PK/PD model (MBM) was developed to characterize the time course and extent of synergic bacterial killing. This is the first preclinical PK/PD study to systematically examine the in vitro and in vivo PK/PD of the polymyxin-zidovudine combination to combat NDM-producing MDR K. pneumoniae.
RESULTS
MICs and in vitro static time-kill studies.
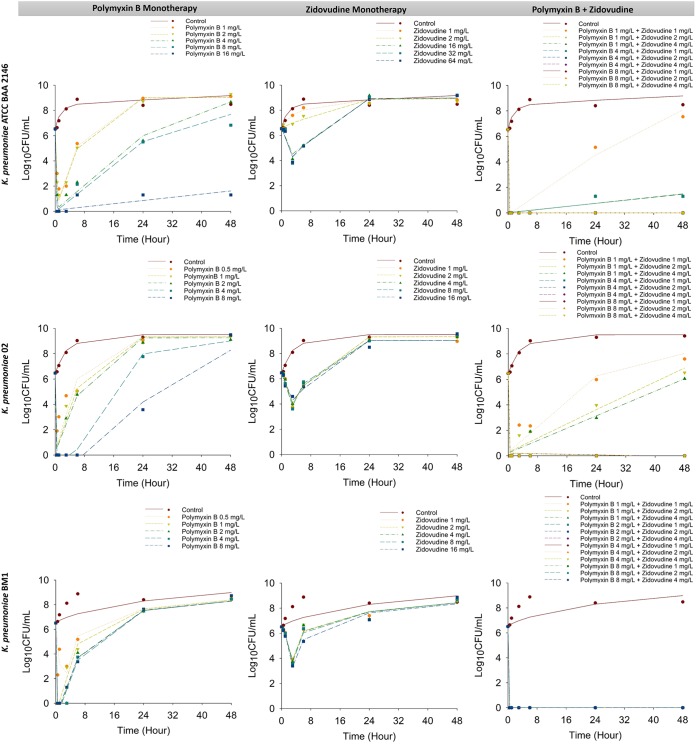
MICs of polymyxin B and zidovudine are summarized in Table 1. All three studied clinical isolates were susceptible to polymyxin B with an MIC of 0.5 mg/liter. Figure 1 shows the static time-kill kinetics of polymyxin B and zidovudine alone and in combination. Polymyxin B monotherapy produced rapid and extensive bacterial killing within 1 h with ≥3 log10 killing at 1 mg/liter and ≥6 log10 killing at 8 mg/liter against all isolates. Despite the initial killing, significant bacterial regrowth was observed as early as 24 h at all polymyxin B concentrations examined; within 24 h, greater than ∼4 log10 regrowth was observed for all polymyxin B concentrations against all strains except K. pneumoniae ATCC BAA 2146 treated with 16 mg/liter polymyxin B. At 24 h, for the most polymyxin B-treated group, the bacterial regrowth approached that observed in the control group (Fig. 1). On the other hand, zidovudine monotherapy produced excellent bacterial killing with a reduction of ∼2 to 3 log10 CFU/ml within 3 h posttreatment. However, substantial bacterial regrowth occurred across all strains and for all zidovudine concentrations. No significant differences in total bacterial counts were observed between treated and growth control groups at 24 and 48 h. The combination of polymyxin B (≥4 mg/liter) and zidovudine (≥1 mg/liter) significantly increased the extent of bacterial killing observed within the first hour by >5 log10 CFU/ml and remained synergistic up to 48 h for all examined strains. Notably, the combination was able to delay the bacterial regrowth significantly compared with that of either polymyxin B or zidovudine as a monotherapy. Synergistic bacterial killing was achieved with the lowest combination concentrations (i.e., 1 mg/liter polymyxin B and 1 mg/liter zidovudine) but was followed by significant regrowth for K. pneumoniae ATCC BAA 2146 and K. pneumoniae 02. Surprisingly, no regrowth was observed for K. pneumoniae BM1 even with the lowest combination concentrations. Overall, synergy was observed with all polymyxin B-zidovudine combinations against the three NDM-producing K. pneumoniae isolates over 48 h.
TABLE 1.
MICs of polymyxin B (PMB) and zidovudine (ZID) for NDM-producing K. pneumoniaea
| MDR strain | Polymyxin susceptibility | PMB MIC (mg/liter) | ZID MIC (mg/liter) |
|---|---|---|---|
| K. pneumoniae ATCC BAA 2146 | S | 0.5 | 64 |
| K. pneumoniae 02 | S | 0.5 | 2 |
| K. pneumoniae BM1 | S | 0.5 | 1 |
All isolates were polymyxin heteroresistant, which is defined as the existence within a polymyxin-susceptible isolate (MIC, ≤2 mg/liter), of subpopulations able to grow in the presence of 4 mg/liter polymyxin B (3, 16). All isolates were MDR, defined as nonsusceptible to ≥1 treating agent in ≥3 antimicrobial categories (53). There are no established CLSI or EUCAST breakpoints for polymyxin B and zidovudine against K. pneumoniae. EUCAST breakpoints for colistin were applied: Susceptibility and resistance to polymyxin B were defined as MICs of ≤2 mg/liter and >2 mg/liter, respectively (42).
FIG 1.
Static time-kill results for polymyxin B in combination with zidovudine against K. pneumoniae ATCC BAA 2146 (upper panel), K. pneumoniae 02 (middle panel), and K. pneumoniae BM1 (lower panel). Marks represent observed viable counts and lines represent individual fitted viable counts.
In vitro one-compartment PK/PD model.
Figure 2 shows the IVM time-kill kinetics of polymyxin B and zidovudine alone and in combination against K. pneumoniae BM1 over 72 h. Polymyxin B monotherapy (4 mg/liter as continuous infusion) resulted in rapid and extensive bacterial killing within 0.5 h posttreatment that was sustained until 28 h. Despite good killing, ≥4 log10 regrowth was observed at 48 h and 72 h. Zidovudine monotherapy (given as a bolus thrice daily to achieve maximum concentration of drug in broth [Cmax] of 6 mg/liter) produced rapid and extensive killing with a reduction of ∼4 log10 CFU/ml within 4 h followed by regrowth (≥4 log10) after 6 h. At 24 h, the antimicrobial activity of zidovudine monotherapy was completely diminished and the viable count was comparable to that of the growth control. The combination produced a rapid and extensive synergistic killing within 0.5 h and the total bacterial counts were below the limit of detection for the entire 72 h against K. pneumoniae BM1 (Fig. 2).
FIG 2.
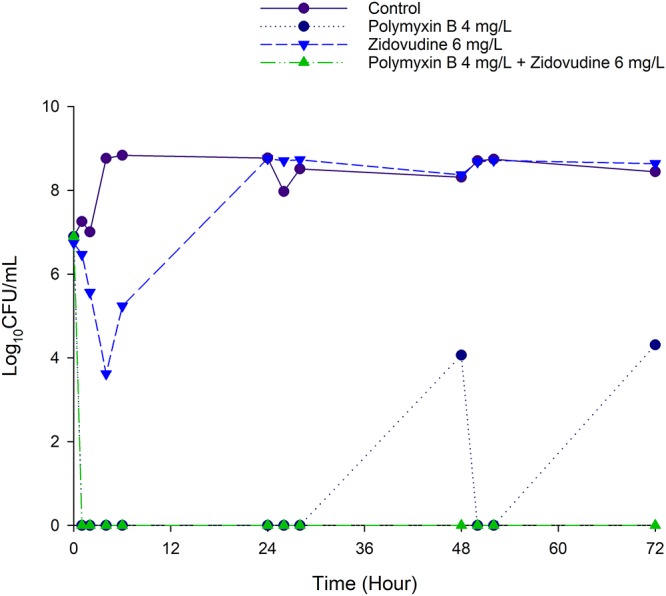
Killing kinetics of polymyxin B (4 mg/liter as continuous infusion) and zidovudine (bolus dose given 8 hourly to achieve Cmax of 6 mg/liter) alone and in combination against K. pneumoniae BM1 in the IVM with an inoculum of ∼107 CFU/ml.
Mechanism-based PK/PD modeling of the mono- and combination therapy.
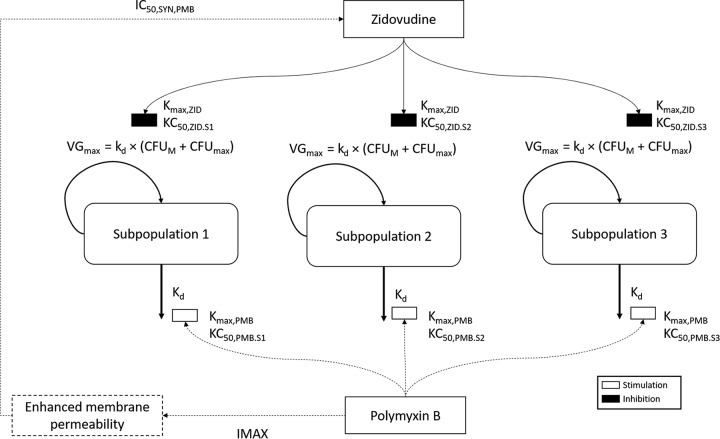
A mechanism-based PK/PD model (MBM) was developed to describe the time course of bacterial dynamics (killing and regrowth) for both mono- and combination therapy (Fig. 3). The MBM described the pharmacodynamics well (r2 ≥ 0.95) for the observed versus individually fitted log10 viable count for all three isolates (see Fig. S1 in the supplemental material). The MBM consisted of two to three subpopulations with different susceptibilities to polymyxin B and zidovudine and different initial inocula for the respective subpopulation. K. pneumoniae ATCC BAA 2146 and BM1 were described by two subpopulations (i.e., susceptible and resistant subpopulations for both antibiotics), whereas K. pneumoniae 02 was described by a three-subpopulation model (i.e., susceptible, intermediate, and resistant subpopulations for both antibiotics). For modeling purposes, the intermediate and resistant subpopulations were defined as subpopulations with a KC50 (the concentration of drug causing 50% of the maximum rate of killing [Kmax]) greater than the KC50 of the susceptible subpopulation (see Table S1 in the supplemental material). Subpopulation synergy was incorporated into the MBM by allowing the subpopulations to have different degrees of susceptibility to both zidovudine and polymyxin B. For K. pneumoniae ATCC BAA 2146 and BM1, subpopulations 2 and 3 were implemented, as they were susceptible to polymyxin B but resistant to zidovudine and vice versa. On the other hand, an intermediate subpopulation was needed for K. pneumoniae 02 to fully describe the data; hence, subpopulation 2 was implemented as intermediate to polymyxin B and resistant to zidovudine, while subpopulation 3 as resistant to polymyxin B and intermediate to zidovudine. The natural death rate constant (Kd) was assumed to be the same for all subpopulations within each isolate. In the current model, polymyxin B was assumed to increase the bacterial death rate constant using a maximum effect (Emax) model, and zidovudine was assumed to decrease the bacterial growth rate by acting on the maximal velocity of bacterial growth (VGmax) of each subpopulation. The maximal killing rate constants (Kmax) of polymyxin B and zidovudine were assumed to be same for all subpopulations. Mechanistic synergy due to polymyxin B enhancing the intracellular concentration of zidovudine was expressed as a decrease in KC50,ZID of the respective subpopulations with increasing polymyxin B concentrations. For all three isolates, the polymyxin B concentration required for half-maximal permeabilization of the outer membrane was estimated (IC50,SYN,PMB, 0.5 to 10.8 mg/liter) (Table S1). The population mean parameter estimates of the final model were relatively precise and unbiased (Table S1).
FIG 3.
Mechanism-based model describing the killing activity of polymyxin B and zidovudine alone and in combination against NDM-producing K. pneumoniae. The parameters are presented in Table S1 in the supplemental material. Polymyxin B was assumed to enhance the rate of bacterial death based on the Emax model, while zidovudine was assumed to reduce the rate of bacterial growth by decreasing the VGmax of each subpopulation. Mechanistic synergy due to polymyxin B enhancing the intracellular concentration of zidovudine was expressed as a decrease in KC50 of the respective subpopulations with increasing polymyxin B concentration.
Pharmacodynamics of polymyxin B and zidovudine mono- and combination therapy in a neutropenic mouse thigh infection model.
Figure 4 shows the antimicrobial efficacy of polymyxin B (10 mg/kg thrice daily) and zidovudine (200 mg/kg thrice daily) mono- and combination therapy against NDM-producing K. pneumoniae 02 in a neutropenic mouse thigh infection model. At 24 h, both polymyxin B and zidovudine monotherapies led to increased bacterial burden by ∼1 log10 CFU/thigh, compared with that of the growth control at 0 h. Polymyxin B (10 mg/kg thrice daily) in combination with zidovudine (200 mg/kg thrice daily) significantly increased the bacterial killing at 24 h by approximately ≥1 log10 CFU/thigh killing compared with the control at 0 h or ≥3 log10 CFU/thigh compared with each monotherapy at 24 h (Fig. 4).
FIG 4.
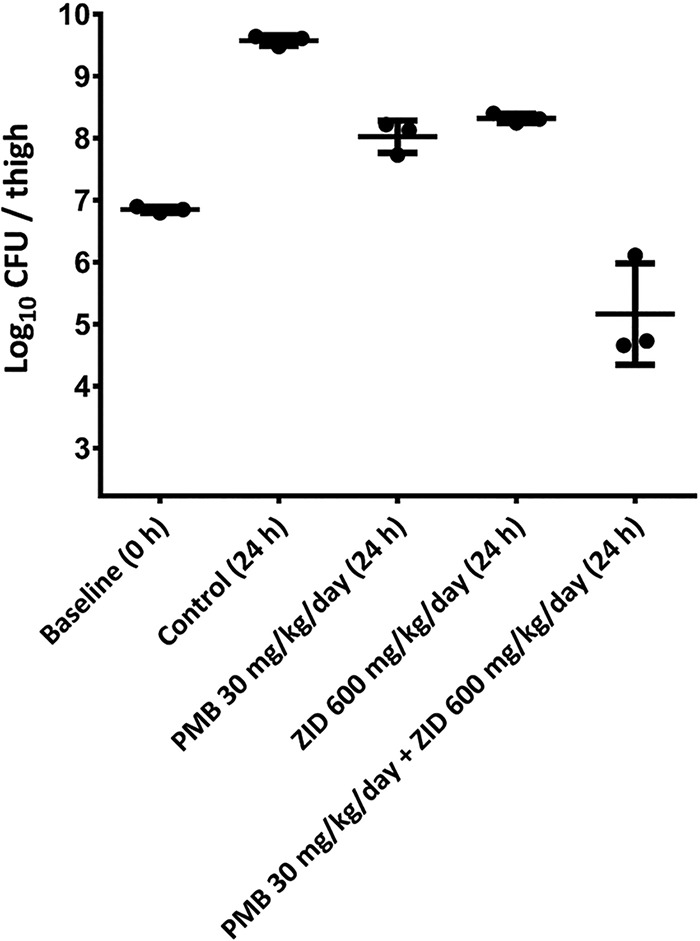
Efficacy of systemically administered polymyxin B (PMB) and zidovudine (ZID) mono- and combination therapy against NDM-producing K. pneumoniae 02 in a neutropenic murine thigh infection model. Data are mean ± standard deviation (n = 3). The y axis starts from the limit of detection (2.23 log10 CFU/thigh).
DISCUSSION
The present study is the first to systematically examine the PK/PD of polymyxin B in combination with the antiretroviral drug zidovudine against NDM-producing MDR K. pneumoniae using static-time-kill, IVM, and murine thigh infection models. The concentrations of polymyxin B employed in the IVM are clinically achievable in patients following the currently recommended dosage regimens (5, 28). Zidovudine is an FDA-approved medication used to treat HIV (23, 29–31). The primary antiretroviral mode of action of zidovudine involves the inhibition of HIV reverse transcriptase (22). Zidovudine is also purported to exert an antimicrobial activity via interfering with bacterial DNA replication (24, 27). Total plasma concentrations of 1 to 4 mg/liter zidovudine are achieved in patients after standard dosage regimens (29, 32, 33). Super-therapeutic concentrations were employed for both polymyxin B and zidovudine in the static time-kill experiment (Fig. 1) for PK/PD modeling purposes, as well as to evaluate the clinical potential of intensive dosing.
Polymyxin B monotherapy was effective against all three NDM-producing K. pneumoniae isolates in the static time-kill studies (Fig. 1). However, the antimicrobial activity was diminished beyond 3 h and significant regrowth was observed by 24 and 48 h. Zidovudine monotherapy only produced modest antibacterial activity against all three clinical isolates within 6 h; however, this was followed by significant regrowth at 24 and/or 48 h (Fig. 1). This regrowth phenomenon was consistent with previous observations in which Escherichia coli and Salmonella enterica serovar Typhimurium developed zidovudine resistance after short-term exposure (27). Excitingly, the combination of polymyxin B and zidovudine displayed substantially enhanced antibacterial activity against all three NDM isolates. Early bacterial killing to the undetectable level was observed with the combination (e.g., 4 mg/liter polymyxin B and 1 mg/liter zidovudine) against all three isolates (Fig. 1). As nephrotoxicity is a major dose-limiting adverse effect of intravenous polymyxin B in patients, dose escalation is not a viable option, and the synergy observed at the low clinically achievable concentration of polymyxin B (1 mg/liter) is ideal for optimizing the use of the combination in patients (5, 30, 34).
An MBM was developed for the three clinical isolates to evaluate and quantify the time course of bacterial killing by polymyxin B and zidovudine mono- and combination therapies (Fig. 3). The proposed MBM utilized a capacity-rated limited growth model, and polymyxin B was assumed to enhance the rate of natural death of bacteria, as previously reported (35). Zidovudine was assumed to slow the bacterial replication rate and was implemented in the model as a decrease in VGmax. The final proposed MBM provided a satisfactory fit (R2 ≥ 0.95) (Fig. S1) and well described the time course of bacterial growth and killing due to mono- and combination therapies. The MBM incorporated both the subpopulation and mechanistic synergy to describe the enhanced antimicrobial activity of the combination therapy. Exclusion of either synergy mechanism resulted in a model that could not be estimated (R2 < 0.5) (data not shown). Mechanistic synergy was incorporated in the MBM as an increase in the susceptibility of the respective subpopulation to zidovudine with increased polymyxin B concentrations (Fig. 3). Our proposed mechanistic synergy was supported by the mechanistic data from the polymyxin-mitotane combination against A. baumannii (36) and our fractional inhibitory concentration (FIC results) (see Figures S2 to S4 in the supplemental material). The increased permeability of the outer membrane was demonstrated by the decreased zidovudine MIC in the presence of increasing polymyxin B concentrations (Fig. S2 to S4). Subpopulation synergy was incorporated into the MBM by allowing the subpopulations to have different degrees of susceptibility to both zidovudine and polymyxin B. Further investigations are needed to directly quantify different bacterial subpopulations to describe their susceptibility to polymyxin B and zidovudine. Despite experimental and statistical evidence supporting the proposed mechanisms of synergy, systems biology studies are currently being conducted in our laboratory to elucidate the mechanisms of synergy and potential mechanisms of resistance. In-depth knowledge of the mechanistic killing and resistance mechanisms will allow us to refine our proposed model and subsequently facilitate the translation of this promising combination for future clinical applications.
Since the proposed MBM was developed based on static time-kill data, it is important to validate the model by assessing its ability to predict bacterial killing by the combination therapy in the IVM. In comparison with the static time-kill results, the IVM was able to closely mimic the PK of polymyxin B and zidovudine in humans. Through simulations, the model was capable of predicting the bacterial killing by the combination observed in the one-compartment IVM. In agreement with the observations in static time-kill data (Fig. 1), polymyxin B monotherapy (given as continuous infusion) was initially effective against K. pneumoniae BM1 but followed by regrowth, while zidovudine monotherapy (bolus dose every 8 h) produced modest killing and was associated with extensive regrowth. The polymyxin B-zidovudine combination was synergistic, and the total bacterial count remained below the limit of detection for 72 h (Fig. 2). Finally, our neutropenic mouse thigh infection results confirmed the in vivo antimicrobial synergy of polymyxin B in combination with zidovudine against K. pneumoniae (Fig. 4). In vivo studies are important for translational antibiotic dose optimization (37). The dosage regimen of polymyxin B was chosen based on its PK in critically ill patients and animal scaling (5), while the zidovudine dosage regimen was based on LD50 in rodents (38). In the murine thigh infection model, at 24 h, both polymyxin B and zidovudine monotherapy led to increased bacterial burden by ∼1 log10 CFU/thigh, compared with those at 0 h (Fig. 4). Excitingly, antimicrobial synergy was detected with the combination at 24 h, with nearly 1 to 2 log10 CFU/thigh reduction compared with the initial bacterial burden before the treatment (Fig. 4). Given the lack of a validated PK model describing the PK of zidovudine in mice (39), simulations to predict bacterial killing in mice using the MBM developed here were not performed.
Based on our previously published single-dose PK study (40), 30 mg/kg of body weight per day polymyxin B would achieve an area under the concentration-time curve for the free, unbound fraction of a drug (fAUC) of 6.5, which is close to the fAUC target of 8.2 ± 6.1 for 1 log10 CFU/thigh reduction in a murine thigh infection model for isolates with polymyxin B MIC of 0.5 mg/liter. (Fig. 4). The combination of polymyxin B with zidovudine significantly enhanced the antimicrobial efficacy; in the presence of zidovudine, even a polymyxin B fAUC of 6.5 was able to achieve ≥4 log10 CFU/thigh reduction at 24 h in a murine infection model against NDM-producing K. pneumoniae 02 (polymyxin B MIC of 0.5 mg/liter) (Fig. 4). The PK/PD index of zidovudine remains unknown; therefore, the antimicrobial efficacy of zidovudine cannot be interpreted based on the PK/PD index targets. Overall, these results highlight the clinical potential of the polymyxin B-zidovudine combination to combat problematic infections caused by NDM-producing MDR K. pneumoniae. With future clinical data on zidovudine, our MBM can be combined with human population PK models to perform Monte Carlo simulations for rational optimization of the dosage regimens for the combination therapy in patients.
To the best of our knowledge, our study provides the first preclinical PK/PD evidence for the potential of polymyxin-zidovudine combination against NDM-producing MDR K. pneumoniae. This synergistic combination significantly enhanced antimicrobial activity and reduced bacterial regrowth. Further investigation in humans is warranted for the translation into clinical settings.
MATERIALS AND METHODS
Chemicals and bacterial strains.
Polymyxin B (sulfate; batch number BCBD1065V; Sigma-Aldrich, Australia) solution was freshly prepared in sterile Milli-Q water before the experiments. Zidovudine (Sigma-Aldrich) was dissolved in dimethyl sulfoxide (DMSO; Sigma-Aldrich) and then diluted with sterile Milli-Q water to ensure a final DMSO concentration of ≤5% (vol/vol) (14). Three isolates of NDM-producing MDR K. pneumoniae were employed in this study. K. pneumoniae ATCC BAA 2146 is a polymyxin-heteroresistant strain from the American Type Culture Collection (Rockville, MD, United States) and was originally isolated from human urine. K. pneumoniae 02 and K. pneumoniae BM1 (formerly designated KP1 [41]) are polymyxin-heteroresistant clinical isolates.
MICs.
MICs of polymyxin B and zidovudine were determined for all isolates using broth microdilution in cation-adjusted Mueller-Hinton broth (CAMHB; Mg2+ at 12.2 mg/liter and Ca2+ at 23.0 mg/liter; Oxoid, Hampshire, England) (3). The susceptibility and resistance to polymyxin B were defined as MICs of ≤2 mg/liter and >2 mg/liter, respectively (42). There are no set clinical breakpoints for zidovudine against Enterobacteriaceae by either Clinical and Laboratory Standards Institute (CLSI) or European Committee on Antimicrobial Susceptibility Testing (EUCAST).
Static time-kill experiments.
Static time-kill experiments were conducted to evaluate the antibacterial activity of polymyxin B and zidovudine alone and in combination against NDM-producing K. pneumoniae (3, 14). All experiments were performed with an initial inoculum of ∼106 CFU/ml in 20 ml CAMHB in 50-ml pyrogen-free and sterile polypropylene tubes (Thermo Fisher, Melbourne, Australia). Polymyxin B and zidovudine monotherapy and its combination were evaluated over a range of concentrations (0 to 64 mg/liter), as illustrated in Fig. 1. Serial samples (50 μl) were collected at 0, 0.5, 1, 3, 6, 24, and 48 h for viable bacterial quantification on nutrient agar plates, and the limit of detection was 20 CFU/ml (equivalent to one colony per plate). All bacterial samples were washed with 0.9% saline to minimize any potential drug carryover. At 24 h, the bacterial suspension was centrifuged at 3,400 × g at 37°C for 10 min and resuspended in 20 ml CAMHB. A ProtoCOL automated colony counter (Symbiosis, Cambridge, United Kingdom) was used to quantify bacteria after a 24-h incubation at 37°C.
In vitro one-compartment PK/PD model experiment.
An IVM was employed to examine the antimicrobial efficacy of polymyxin B and zidovudine alone and in combination against K. pneumoniae BM1 over 72 h (14, 43). Four reservoirs were employed, namely, (i) a control reservoir without any antibiotic; (ii) polymyxin B monotherapy; (iii) zidovudine monotherapy; and (iv) polymyxin B-zidovudine combination therapy. Each reservoir contained 80 ml of CAMHB and was maintained at 37°C. PK of polymyxin B in critically ill patients was mimicked in the IVM (5, 28). Polymyxin B was added into the diluent reservoirs and delivered as a continuous infusion to achieve a central reservoir concentration of 4 mg/liter. Zidovudine was added into the central reservoir every 8 h via bolus administration using an automated syringe pump (New Era Pump Systems, NY, USA) to achieve a Cmax of 6 mg/liter. The same dosage regimens of each antibiotic were simulated for the combination therapy. Serial samples were collected from the reservoirs for viable counting at 0, 1, 2, 4, 6, 24, 26, 28, 48, 50, 52, and 72 h, and the limit of detection was 20 CFU/ml.
Mechanism-based PK/PD modeling of mono- and combination therapy.
An MBM was developed based on time-kill data of polymyxin B and zidovudine mono- and combination therapy to describe the rate and extent of bacterial killing observed in the static time-kill studies. Bacterial cells were partitioned into three preexisting subpopulations with different susceptibilities to polymyxin B and zidovudine. The number of subpopulations necessary to describe the data was based on model discrimination performed using the Akaike information criterion (AIC) or the log likelihood ratio test (reported as −1 × log likelihood in S-ADAPT), biological plausibility of the parameter estimates, visual inspection of the fitted function, and goodness of fit plots (35).
For each subpopulation, the rate of replication was modeled as capacity limited and dependent on the CFU at which the rate of replication is half-maximal (CFUm) and the maximal velocity of bacterial growth (VGmax) (44). The VGmax was parameterized as
| (1) |
Kd is the natural bacterial death rate constant.
The total bacterial population is described by:
| (2) |
Kd was characterized by as a first-order elimination rate constant. Polymyxin B and zidovudine killing activity was described by a sigmoidal Emax model as described in equations 3 and 4.
| (3) |
| (4) |
Kmax,PMB and Kmax,ZID are the maximum killing rate constants of polymyxin B and zidovudine, respectively. KC50,PMB,ii and KC50,ZID,ii are the concentrations of polymyxin B and zidovudine, respectively, resulting in 50% of Kmax for the iith subpopulation, and γ is the Hill coefficient. SYNEFF is the outer membrane remodeling effect as a result of polymyxin B, as described in equation 8.
The killing by polymyxin B (KPMB,ii) was assumed to enhance the natural bacterial death rate constant based on the bacterial subpopulation, while the killing by zidovudine (KZID,ii) was assumed to inhibit the rate of replication by inhibiting DNA replication. The differential equations for each bacterial subpopulation are outlined below.
| (5) |
| (6) |
| (7) |
Mechanism-based modeling of the synergy.
To implement mechanistic synergy, polymyxin B was assumed to permeabilize the outer membrane, thereby increasing the intracellular concentration of zidovudine (45, 46). This was implemented in the MBM by estimating a lower KC50 in the presence of polymyxin B as represented by the following:
| (8) |
IMAXii is the maximum fractional decrease of KC50,ZID by polymyxin B via outer membrane disruption, and CPMB is the polymyxin B concentration causing 50% of IMAXii for the iith subpopulation.
Initial conditions.
The initial inoculum of all subpopulations and mutation frequency (MUT) for the less susceptible subpopulation was estimated. The initial inoculum of subpopulation 2 was estimated as a fraction of the total initial inoculum (log10 CFU0). The initial condition for subpopulation 1 was implemented as (1 – MUT,S2 – MUT,S3) × CFU0, while the initial condition for subpopulation 2 was computed as MUT,S2 × CFU0.
Observation.
All viable counts were transformed to a log10 scale and simultaneously fitted using an additive error model in the log10 scale. The between-curve variability was fixed to a very small value (coefficient of variation, 10%) (47). All viable counts below the limit of detection were plotted as zero, as previously described (45, 46).
Estimation.
The Monte Carlo parametric expectation maximization algorithm (MC-PEM) (pmethod, 4) was used to comodel the time course of bacterial killing and regrowth observed in static time-kill studies in S-ADAPT (48) facilitated by S-ADAPT TRAN (49, 50). The final model was assessed by model discrimination (using AIC or the log likelihood ratio test), the goodness of fit, the visual inspection of diagnostic plots, and the biological plausibility and precision of the estimated parameters (45–47).
Mouse thigh infection model.
The antimicrobial synergy of the polymyxin B-zidovudine combination against NDM-producing K. pneumoniae 02 was evaluated in a neutropenic murine thigh infection model. All animal experiments were approved by the Monash University Animal Ethics Committee and conducted in accordance with the Australian Code of Practice for the Care and Use of Animals for Scientific Purposes. Female Swiss mice (8 to 10 weeks old) were employed in the neutropenic murine thigh infection model. Mice were injected intraperitoneally (i.p.) with cyclophosphamide on days −4 (150 mg/kg) and −1 (100 mg/kg) to induce neutropenia (6, 7, 35, 51, 52). On day 0, mice were injected 50 μl of an early logarithmic phase bacterial suspension (∼107 CFU/ml) into each thigh to achieve an inoculum of ∼106 CFU/ml. Neutropenic mice infected with NDM-producing K. pneumoniae 02 were treated with saline or antibiotics 2 h post-bacterial inoculation. There were a total of four treatment groups, namely, (i) 0.9% saline-treated group as the control, (ii) 10 mg/kg polymyxin B thrice daily (8 hourly; maximum daily dose, 30 mg/kg/day), (iii) 200 mg/kg zidovudine thrice daily (8 hourly; maximum daily dose, 600 mg/kg/day), and (iv) combination of 10 mg/kg polymyxin B thrice daily and 200 mg/kg zidovudine thrice daily. Polymyxin B was administered via subcutaneous injection, while zidovudine was administered via i.p. injection. The polymyxin B dose was selected to mimic the PK of polymyxin B in humans (5, 28), whereas zidovudine doses were based on the LD50 in rodents (38). At 0 and 24 h, the bacterial burden was determined. Mice were humanely killed, and thighs were aseptically removed and homogenized. The homogenate was filtered and subsequently serially diluted with 0.9% saline and spiral plated onto nutrient agar with subsequent incubation at 35°C for 24 h. Colonies were counted using a ProtoCOL colony counter, and CFU values were expressed as log10 CFU/thigh. The limit of detection was 164 CFU/thigh (equivalent to one colony per plate). Tukey’s multiple comparison test was used to compare the groups at 24 h and a P value of <0.05 was considered statistically significant.
Supplementary Material
ACKNOWLEDGMENTS
J.L., G.G.R., A.F., and T.V. are supported by a research grant from the National Institute of Allergy and Infectious Diseases of the National Institutes of Health (R01 AI111965). Y.-W.L. and M.-L.H. are recipients of the 2018 Faculty Bridging Fellowship, Monash University. J.L. is an Australian National Health Medical Research Council (NHMRC) Principal Research Fellow and T.V. is an Australian NHMRC Industry Career Development Level 2 Research Fellow.
We would like to dedicate this paper to Alan Forrest, who made significant contributions to antimicrobial PK/PD.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Allergy and Infectious Diseases or the National Institutes of Health.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.02176-18.
REFERENCES
- 1.World Health Organization. 2014. Antimicrobial resistance: 2014 global report on surveillance. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 2.Peirano G, Pillai DR, Pitondo-Silva A, Richardson D, Pitout JD. 2011. The characteristics of NDM-producing Klebsiella pneumoniae from Canada. Diagn Microbiol Infect Dis 71:106–109. doi: 10.1016/j.diagmicrobio.2011.06.013. [DOI] [PubMed] [Google Scholar]
- 3.Abdul Rahim N, Cheah SE, Johnson MD, Yu H, Sidjabat HE, Boyce J, Butler MS, Cooper MA, Fu J, Paterson DL, Nation RL, Bergen PJ, Velkov T, Li J. 2015. Synergistic killing of NDM-producing MDR Klebsiella pneumoniae by two “old” antibiotics-polymyxin B and chloramphenicol. J Antimicrob Chemother 70:2589–2597. doi: 10.1093/jac/dkv135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li J, Nation RL, Turnidge JD, Milne RW, Coulthard K, Rayner CR, Paterson DL. 2006. Colistin: the re-emerging antibiotic for multidrug-resistant Gram-negative bacterial infections. Lancet Infect Dis 6:589–601. doi: 10.1016/S1473-3099(06)70580-1. [DOI] [PubMed] [Google Scholar]
- 5.Cheah S-E, Li J, Bergen PJ, Nation RL. 2016. Polymyxin pharmacokinetics and pharmacodynamics, p 221–260. In Rotschafer JC, Andes DR, Rodvold KA (ed), Antibiotic pharmacodynamics. Springer, New York, New York, NY. [Google Scholar]
- 6.Lin Y-W, Zhou Q, Cheah S-E, Zhao J, Chen K, Wang J, Chan H-K, Li J. 2016. Pharmacokinetics/pharmacodynamics of pulmonary delivery of colistin against Pseudomonas aeruginosa in a mouse lung infection model. Antimicrob Agents Chemother 61:e02025-16. doi: 10.1128/aac.02025-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lin Y-W, Zhou Q, Onufrak NJ, Wirth V, Chen K, Wang J, Forrest A, Chan H-K, Li J. 2017. Aerosolized polymyxin B for treatment of respiratory tract infections: Determination of pharmacokinetic/pharmacodynamic indices for aerosolized polymyxin B against Pseudomonas aeruginosa in a mouse lung infection model. Antimicrob Agents Chemother 61:e00211-17. doi: 10.1128/aac.00211-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nation RL, Garonzik SM, Thamlikitkul V, Giamarellos-Bourboulis EJ, Forrest A, Paterson DL, Li J, Silveira FP. 2017. Dosing guidance for intravenous colistin in critically-ill patients. Clin Infect Dis 64:565–571. doi: 10.1093/cid/ciw839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang JX, Blaskovich MA, Pelingon R, Ramu S, Kavanagh A, Elliott AG, Butler MS, Montgomery AB, Cooper MA. 2015. Mucin binding reduces colistin antimicrobial activity. Antimicrob Agents Chemother 59:5925–5931. doi: 10.1128/AAC.00808-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schwameis R, Erdogan-Yildirim Z, Manafi M, Zeitlinger M, Strommer S, Sauermann R. 2013. Effect of pulmonary surfactant on antimicrobial activity in vitro. Antimicrob Agents Chemother 57:5151–5154. doi: 10.1128/AAC.00778-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arpin C, Noury P, Boraud D, Coulange L, Manetti A, André C, M'Zali F, Quentin C. 2012. NDM-1-producing Klebsiella pneumoniae resistant to colistin in a French community patient without history of foreign travel. Antimicrob Agents Chemother 56:3432–3434. doi: 10.1128/AAC.00230-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kumarasamy KK, Toleman MA, Walsh TR, Bagaria J, Butt F, Balakrishnan R, Chaudhary U, Doumith M, Giske CG, Irfan S, Krishnan P, Kumar AV, Maharjan S, Mushtaq S, Noorie T, Paterson DL, Pearson A, Perry C, Pike R, Rao B, Ray U, Sarma JB, Sharma M, Sheridan E, Thirunarayan MA, Turton J, Upadhyay S, Warner M, Welfare W, Livermore DM, Woodford N. 2010. Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study. Lancet Infect Dis 10:597–602. doi: 10.1016/S1473-3099(10)70143-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schneider EK, Reyes-Ortega F, Velkov T, Li J. 2017. Antibiotic–non-antibiotic combinations for combating extremely drug-resistant Gram-negative “superbugs.” Essays Biochem 61:115–125. doi: 10.1042/EBC20160058. [DOI] [PubMed] [Google Scholar]
- 14.Lin Y-W, Yu HH, Zhao J, Han M-L, Zhu Y, Akter J, Wickremasinghe H, Walpola H, Wirth V, Rao GG, Forrest A, Velkov T, Li J. 2018. Polymyxin B in combination with enrofloxacin exerts synergistic killing against extensive drug-resistant Pseudomonas aeruginosa. Antimicrob Agents Chemother 62:e00028-18. doi: 10.1128/aac.00028-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schneider EK, Azad MAK, Han M-L, (Tony) Zhou Q, Wang J, Huang JX, Cooper MA, Doi Y, Baker MA, Bergen PJ, Muller MT, Li J, Velkov T. 2016. An “unlikely” pair: the antimicrobial synergy of polymyxin B in combination with the cystic fibrosis transmembrane conductance regulator drugs KALYDECO and ORKAMBI. ACS Infect Dis 2:478–488. doi: 10.1021/acsinfecdis.6b00035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tran TB, Cheah S-E, Yu HH, Bergen PJ, Nation RL, Creek DJ, Purcell A, Forrest A, Doi Y, Song J, Velkov T, Li J. 2016. Anthelmintic closantel enhances bacterial killing of polymyxin B against multidrug-resistant Acinetobacter baumannii. J Antibiot (Tokyo) 69:415–421. doi: 10.1038/ja.2015.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hussein MH, Schneider EK, Elliott AG, Han M, Reyes-Ortega F, Morris F, Blaskovich MAT, Jasim R, Currie B, Mayo M, Baker M, Cooper MA, Li J, Velkov T. 2017. From breast cancer to antimicrobial: combating extremely resistant Gram-negative “superbugs” using novel combinations of polymyxin B with selective estrogen receptor modulators. Microb Drug Resist 23:640–650. doi: 10.1089/mdr.2016.0196. [DOI] [PubMed] [Google Scholar]
- 18.Jasim R, Schneider EK, Han M, Azad MA, Hussein M, Nowell C, Baker MA, Wang J, Li J, Velkov T. 2017. A fresh shine on cystic fibrosis inhalation therapy: antimicrobial synergy of polymyxin B in combination with silver nanoparticles. J Biomed Nanotechnol 13:447–457. doi: 10.1166/jbn.2017.2355. [DOI] [PubMed] [Google Scholar]
- 19.Tran TB, Wang J, Doi Y, Velkov T, Bergen PJ, Li J. 2018. Novel polymyxin combination with antineoplastic mitotane improved the bacterial killing against polymyxin-resistant multidrug-resistant gram-negative pathogens. Front Microbiol 9:721. doi: 10.3389/fmicb.2018.00721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ejim L, Farha MA, Falconer SB, Wildenhain J, Coombes BK, Tyers M, Brown ED, Wright GD. 2011. Combinations of antibiotics and nonantibiotic drugs enhance antimicrobial efficacy. Nat Chem Biol 7:348–350. doi: 10.1038/nchembio.559. [DOI] [PubMed] [Google Scholar]
- 21.Brown D. 2015. Antibiotic resistance breakers: can repurposed drugs fill the antibiotic discovery void? Nat Rev Drug Discov 14:821–832. doi: 10.1038/nrd4675. [DOI] [PubMed] [Google Scholar]
- 22.Furman PA, Barry DW. 1988. Spectrum of antiviral activity and mechanism of action of zidovudine. An overview. Am J Med 85:176–181. [PubMed] [Google Scholar]
- 23.Doleans-Jordheim A, Bergeron E, Bereyziat F, Ben-Larbi S, Dumitrescu O, Mazoyer MA, Morfin F, Dumontet C, Freney J, Jordheim LP. 2011. Zidovudine (AZT) has a bactericidal effect on Enterobacteria and induces genetic modifications in resistant strains. Eur J Clin Microbiol Infect Dis 30:1249–1256. doi: 10.1007/s10096-011-1220-3. [DOI] [PubMed] [Google Scholar]
- 24.Lewin CS, Allen RA, Amyes SGB. 1990. Mechanisms of zidovudine resistance in bacteria. J Med Microbiol 33:235–238. doi: 10.1099/00222615-33-4-235. [DOI] [PubMed] [Google Scholar]
- 25.Ng SMS, Sioson JSP, Yap JM, Ng FM, Ching HSV, Teo JWP, Jureen R, Hill J, Chia CSB. 2018. Repurposing zidovudine in combination with tigecycline for treating carbapenem-resistant Enterobacteriaceae infections. Eur J Clin Microbiol Infect Dis 37:141–148. doi: 10.1007/s10096-017-3114-5. [DOI] [PubMed] [Google Scholar]
- 26.Hu Y, Coates AHYJ. 2015. Combination comprising zidovudine and polymyxin. Patent WO2014147405A1.
- 27.Lewin CS, Allen R, Amyes SG. 1990. Zidovudine-resistance in Salmonella typhimurium and Escherichia coli. J Antimicrob Chemother 25:706–708. doi: 10.1093/jac/25.4.706. [DOI] [PubMed] [Google Scholar]
- 28.Zavascki AP, Goldani LZ, Cao G, Superti SV, Lutz L, Barth AL, Ramos F, Boniatti MM, Nation RL, Li J. 2008. Pharmacokinetics of intravenous polymyxin B in critically ill patients. Clin Infect Dis 47:1298–1304. doi: 10.1086/592577. [DOI] [PubMed] [Google Scholar]
- 29.Capparelli EV, Englund JA, Connor JD, Spector SA, McKinney RE, Palumbo P, Baker CJ. 2003. Population pharmacokinetics and pharmacodynamics of zidovudine in HIV‐infected infants and children. J Clin Pharmacol 43:133–140. doi: 10.1177/0091270002239821. [DOI] [PubMed] [Google Scholar]
- 30.Coates AHYJ. 2014. Zidovudine combination therapies for treating microbial infections. Patent WO2015114340A1
- 31.Luzier A, Morse GD. 1993. Intravascular distribution of zidovudine: role of plasma proteins and whole blood components. Antiviral Res 21:267–280. doi: 10.1016/0166-3542(93)90032-E. [DOI] [PubMed] [Google Scholar]
- 32.Gitterman SR, Drusano GL, Egorin MJ, Standiford HC. 1990. Population pharmacokinetics of zidovudine. Clin Pharmacol Ther 48:161–167. doi: 10.1038/clpt.1990.131. [DOI] [PubMed] [Google Scholar]
- 33.Zhou X-J, Sheiner LB, D’Aquila RT, Hughes MD, Hirsch MS, Fischl MA, Johnson VA, Myers M, Sommadossi J-P, Allergy TNIo, Investigators IDACTGP. 1999. Population pharmacokinetics of nevirapine, zidovudine, and didanosine in human immunodeficiency virus-infected patients. Antimicrob Agents Chemother 43:121–128. doi: 10.1128/AAC.43.1.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zavascki AP, Nation RL. 2017. Nephrotoxicity of polymyxins: is there any difference between colistimethate and polymyxin B? Antimicrob Agents Chemother 61:e02319-16. doi: 10.1128/AAC.02319-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin YW, Zhou QT, Han ML, Onufrak NJ, Chen K, Wang J, Forrest A, Chan HK, Li J. 2017. Mechanism-based pharmacokinetic/pharmacodynamic modeling of aerosolized colistin in a mouse lung infection model. Antimicrob Agents Chemother 62:e01965-17. doi: 10.1128/aac.01965-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tran TB, Bergen PJ, Creek DJ, Velkov T, Li J. 2018. Synergistic killing of polymyxin B in combination with the antineoplastic drug mitotane against polymyxin-susceptible and -resistant Acinetobacter baumannii: a metabolomic study. Front Pharmacol 9:359. doi: 10.3389/fphar.2018.00359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao M, Lepak AJ, Andes DR. 2016. Animal models in the pharmacokinetic/pharmacodynamic evaluation of antimicrobial agents. Bioorg Med Chem 24:6390–6400. doi: 10.1016/j.bmc.2016.11.008. [DOI] [PubMed] [Google Scholar]
- 38.Ayers KM, Tucker WE Jr, Hajian G, de Miranda P. 1996. Nonclinical toxicology studies with zidovudine: acute, subacute, and chronic toxicity in rodents, dogs, and monkeys. Toxicol Sci 32:129–139. doi: 10.1093/toxsci/32.2.129. [DOI] [PubMed] [Google Scholar]
- 39.Chow H-H. 1997. A physiologically based pharmacokinetic model of zidovudine (AZT) in the mouse: model development and scale-up to humans. J Pharm Sci 86:1223–1228. doi: 10.1021/js970243y. [DOI] [PubMed] [Google Scholar]
- 40.Landersdorfer CB, Wang J, Wirth V, Chen K, Kaye KS, Tsuji BT, Li J, Nation RL. 2018. Pharmacokinetics/pharmacodynamics of systemically administered polymyxin B against Klebsiella pneumoniae in mouse thigh and lung infection models. J Antimicrob Chemother 73:462–468. doi: 10.1093/jac/dkx409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sidjabat HE, Silveira FP, Potoski BA, Abu‐Elmagd KM, Adams‐Haduch JM, Paterson DL, Doi Y. 2009. Interspecies spread of Klebsiella pneumoniae carbapenemase gene in a single patient. Clin Infect Dis 49:1736–1738. doi: 10.1086/648077. [DOI] [PubMed] [Google Scholar]
- 42.EUCAST. 2014. Breakpoint tables for interpretation of MICs and zone diameters, version 4.0. European Committee on Antimicrobial Susceptibility Testing, Växjö, Sweden. http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/Breakpoint_table_v_4.0.pdf.
- 43.Deris ZZ, Yu HH, Davis K, Soon RL, Jacob J, Ku CK, Poudyal A, Bergen PJ, Tsuji BT, Bulitta JB, Forrest A, Paterson DL, Velkov T, Li J, Nation RL. 2012. The combination of colistin and doripenem is synergistic against Klebsiella pneumoniae at multiple inocula and suppresses colistin resistance in an in vitro pharmacokinetic/pharmacodynamic model. Antimicrob Agents Chemother 56:5103–5112. doi: 10.1128/AAC.01064-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Meagher AK, Forrest A, Dalhoff A, Stass H, Schentag JJ. 2004. Novel pharmacokinetic-pharmacodynamic model for prediction of outcomes with an extended-release formulation of ciprofloxacin. Antimicrob Agents Chemother 48:2061–2068. doi: 10.1128/AAC.48.6.2061-2068.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yadav R, Bulitta JB, Wang J, Nation RL, Landersdorfer CB. 2017. Evaluation of pharmacokinetic/pharmacodynamic model-based optimized combination regimens against multidrug-resistant Pseudomonas aeruginosa in a murine thigh infection model using humanized dosing schemes. Antimicrob Agents Chemother 61:e01268-17. doi: 10.1128/aac.01268-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yadav R, Landersdorfer CB, Nation RL, Boyce JD, Bulitta JB. 2015. Novel approach to optimize synergistic carbapenem-aminoglycoside combinations against carbapenem-resistant Acinetobacter baumannii. Antimicrob Agents Chemother 59:2286–2298. doi: 10.1128/AAC.04379-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Landersdorfer CB, Ly NS, Xu H, Tsuji BT, Bulitta JB. 2013. Quantifying subpopulation synergy for antibiotic combinations via mechanism-based modeling and a sequential dosing design. Antimicrob Agents Chemother 57:2343–2351. doi: 10.1128/AAC.00092-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bauer RJ, Guzy S, Ng C. 2007. A survey of population analysis methods and software for complex pharmacokinetic and pharmacodynamic models with examples. AAPS J 9:E60–E83. doi: 10.1208/aapsj0901007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bulitta JB, Landersdorfer CB. 2011. Performance and robustness of the Monte Carlo importance sampling algorithm using parallelized S-ADAPT for basic and complex mechanistic models. AAPS J 13:212–226. doi: 10.1208/s12248-011-9258-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bulitta JB, Ly NS, Yang JC, Forrest A, Jusko WJ, Tsuji BT. 2009. Development and qualification of a pharmacodynamic model for the pronounced inoculum effect of ceftazidime against Pseudomonas aeruginosa. Antimicrob Agents Chemother 53:46–56. doi: 10.1128/AAC.00489-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lin YW, Zhou QT, Han ML, Chen K, Onufrak NJ, Wang J, Turnidge JD, Howden BP, Forrest A, Chan HK, Li J. 2017. Elucidating the pharmacokinetics/pharmacodynamics of aerosolized colistin against multidrug-resistant Acinetobacter baumannii and Klebsiella pneumoniae in a mouse lung infection model. Antimicrob Agents Chemother 62:e01790-17. doi: 10.1128/aac.01790-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cheah S-E, Wang J, Turnidge JD, Li J, Nation RL. 2015. New pharmacokinetic/pharmacodynamic studies of systemically administered colistin against Pseudomonas aeruginosa and Acinetobacter baumannii in mouse thigh and lung infection models: smaller response in lung infection. J Antimicrob Agents 70:3291–3297. doi: 10.1093/jac/dkv267. [DOI] [PubMed] [Google Scholar]
- 53.Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, Harbarth S, Hindler JF, Kahlmeter G, Olsson-Liljequist B, Paterson DL, Rice LB, Stelling J, Struelens MJ, Vatopoulos A, Weber JT, Monnet DL. 2012. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect 18:268–281. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.