Extended-spectrum β-lactamase-producing Enterobacteriaceae (EPE) are a major cause of bloodstream infections, and the colonization rate of EPE in the gut microbiota of individuals lacking prior hospitalization or comorbidities is increasing. In this study, we performed an in-depth investigation of the temporal dynamics of EPE and their plasmids during one year by collecting fecal samples from three patients initially seeking medical care for urinary tract infections.
KEYWORDS: antibiotic resistance, colonization, ESBL, Enterobacteriaceae, plasmid
ABSTRACT
Extended-spectrum β-lactamase-producing Enterobacteriaceae (EPE) are a major cause of bloodstream infections, and the colonization rate of EPE in the gut microbiota of individuals lacking prior hospitalization or comorbidities is increasing. In this study, we performed an in-depth investigation of the temporal dynamics of EPE and their plasmids during one year by collecting fecal samples from three patients initially seeking medical care for urinary tract infections. In two of the patients, the same strain that caused the urinary tract infection (UTI) was found at all consecutive samplings from the gut microbiota, and no other EPEs were detected, while in the third patient the UTI strain was only found in the initial UTI sample. Instead, this patient presented a complex situation where a mixed microbiota of different EPE strain types, including three different E. coli ST131 variants, as well as different bacterial species, was identified over the course of the study. Different plasmid dynamics were displayed in each of the patients, including the spread of plasmids between different strain types over time and the transposition of blaCTX-M-15 from the chromosome to a plasmid, followed by subsequent loss through homologous recombination. Small cryptic plasmids were found in all isolates from all patients, and they appear to move frequently between different strains in the microbiota. In conclusion, we could demonstrate an extensive variation of EPE strain types, plasmid composition, rearrangements, and horizontal gene transfer of genetic material illustrating the high dynamics nature and interactive environment of the gut microbiota during post-UTI carriage.
INTRODUCTION
Extended spectrum β-lactamases (ESBLs) are enzymes that degrade most β-lactam antibiotics, such as penicillins and cephalosporins, the most commonly used group of antibiotics worldwide. ESBL-producing Enterobacteriaceae (EPE), especially Escherichia coli and Klebsiella pneumoniae, constitute a burden on health care systems conferring excess mortality and prolonged hospital stay (1–4). When comparing bloodstream infections (BSI) caused by ESBL-producing E. coli isolates with BSI caused by susceptible isolates, the impact of resistance is estimated to >2,700 excess deaths in Europe and €18.1 million in additional health care costs annually, represented by >120,000 excess days of hospital stay (2). In general, a doubling in medical cost is estimated for infections caused by ESBL-producing isolates compared to susceptible isolates (5).
In addition to β-lactam resistance, most ESBL-producing bacteria contain other resistance determinants and are therefore multidrug resistant, which will further reduce treatment options. The most common ESBL genes today belong to the blaCTX-M group, which were first discovered in the 1980s and since then have spread to all continents and have become endemic worldwide (6, 7). ESBL genes are often carried on large plasmids, typically between 50 and 200 kb in size, mostly belonging to the IncF, IncA/C, IncL/M, IncN, and IncI1 groups (8). In Enterobacteriaceae, ESBLs are mainly encoded and disseminated by plasmids belonging to the IncF group, with a relatively narrow host range (9). Beside conserved regions such as the replicon, the composition of plasmids can be highly dynamic. Resistance plasmids typically contain large numbers of mobile genetic elements (e.g., insertion sequences, transposons, and integrons) associated with the resistance genes. Hence, plasmid composition can vary extensively within an Inc group due to transposition events and rearrangements driven by transposase activity or homologous recombination between identical copies of the same element (10).
The gut microbiota, a reservoir for Enterobacteriaceae, is an ideal place for horizontal dissemination of conjugative plasmids and the average number of different plasmids in a single E. coli has been estimated to be two to three (11). Conjugative transfer of resistance plasmids during human carriage has been described (12–14), but compared to the epidemiology of bacterial strains, relatively little is known of the epidemiology and evolution of plasmids over time in vivo. In order to better understand how plasmids change and how they spread between different bacteria in the gut during colonization, we performed an in-depth analysis of ESBL plasmid dynamics using samples from three patients from a previous study. The previous study focused on the duration of ESBL carriage postinfection in 61 patients seeking medical advice for various infections caused by EPE (15). For this study, three of the patients from the previous work were selected that were EPE culture positive on all five sampling occasions during the year after a urinary tract infection (UTI). The isolates from the three patients showed different profiles regarding bacterial species, strain type, and number of ESBL-producing strains isolated per time point during the course of the study. Our aim was to study the plasmid composition of ESBL-producing bacteria in the gastrointestinal tracts of carriers, as well as to study the short-term evolution of plasmids during the course of one year. A combination of long-read and short-read massive parallel sequencing enabled highly detailed information about the plasmids and the bacteria they resided in. Studying the total plasmid composition rather than an individual plasmid provides valuable insight of how plasmids carrying resistance genes are disseminated and evolve in vivo over time.
RESULTS
Characteristics of the selected isolates.
A prospective study of ESBL carriage after UTI was previously performed by Titelman et al. (15). From the same study, we chose isolates from three patients for further investigation of the ESBL-plasmid dynamics based on their EPE content over the time of carriage. Patients 2 and 7 displayed carriage of one ESBL-producing strain of E. coli over the whole study period and represent a possibility to study plasmid dynamics within a specified bacterial clone. A more complex picture could be observed for patient 4 with a highly varying EPE content over the sampling period. From this patient, samples obtained at 1, 3, and 6 months post-UTI, EPE isolates with different morphologies, were retrieved from selective agar plates indicating either cocolonization by completely unrelated EPE or the transfer of plasmid-borne ESBL genes within the gut microflora. Therefore, isolates representing different morphologies were analyzed from these samples, including both ESBL-producing K. pneumoniae and ESBL-producing E. coli. In all other samples only one single-colony morphology could be detected, and hence only one isolate was taken for these time points. This approach will account for bacteria with different colony morphologies but may underestimate variation within morphologically indistinguishable strains at each time point.
Sequencing and phylogenetic relatedness of strains.
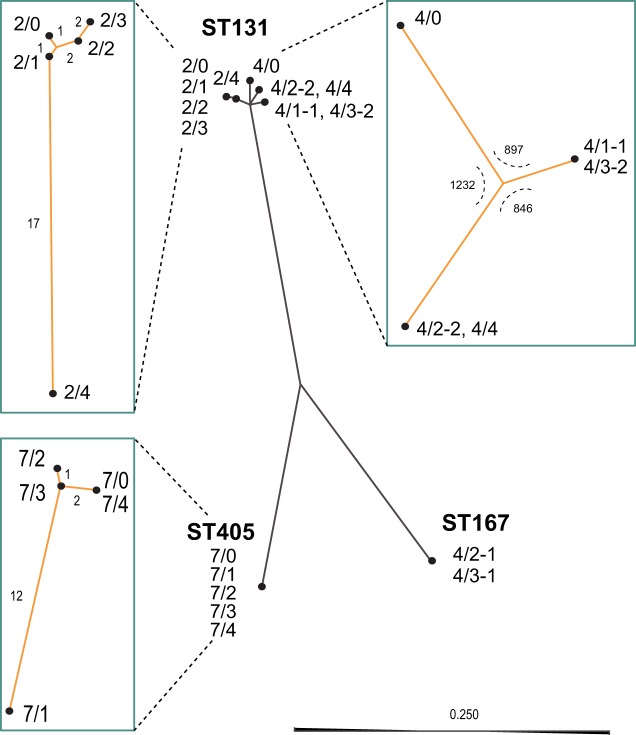
A combination of long-read Pacific Biosciences sequencing of index isolates representing each bacterial morphology complemented by short-read sequencing using Illumina MiSeq resulted in complete chromosome and plasmid sequences of all isolates. From the complete sequences we could identify known resistance genes and previously described chromosomal resistance mutations (see Table S2 in the supplemental material). From the chromosomal sequences we performed epidemiological strain typing by traditional multilocus sequence typing (MLST) for E. coli and K. pneumoniae (Table S2). We also made cross comparisons of the whole-genome sequences of all isolates from the individual patients to identify genetic variation between isolates (Table S3). To illustrate the genetic relatedness of all E. coli isolates, we performed a k-mer tree analysis, including the different MLSTs and separate single-nucleotide polymorphism (SNP) tree analyses for each MLST subset from the different patients (Fig. 1). This confirmed that patients 2 and 7 carried single strains over the whole sampling period with very little genetic variation over time (E. coli ST131 and E. coli ST405, respectively) and that patient 4 carried different strains of two different species, E. coli and K. pneumoniae (E. coli ST167, E. coli ST131, and K. pneumoniae ST147). Interestingly, there were three different ST131 morphotypes identified in patient 4 with ca. 1,000 SNPs between each and two different blaCTX-M phylogroups. Three of five samplings from patient 4 exhibited a mixed culture of various EPE strains, resulting in a total of nine genetically different EPE isolates from this patient (Fig. 1 and Table S2).
FIG 1.
Unrooted k-mer tree (solid dark gray lines) for all E. coli isolates and individual SNP-based trees (teal boxes) for each subgroup of isolates. The central tree is a neighbor-joining k-mer tree describing the genetic relatedness of the isolates based on the presence of 16-mer motifs throughout the complete chromosomes. The scale indicates Jensen-Shannon divergence length. Neighbor-joining SNP trees were made using complete genome alignments. The number of SNPs separating each node is indicated along the branches. For the tree of ST131 isolates from patient 4, the number of SNPs separating the isolates is indicated at the internal node separating the branches.
Resistance profiles and genetic resistance determinants of isolated bacteria.
MICs of 14 different antibiotics were determined for all isolates (Table 1). All isolates were resistant to cefotaxime (carrying either blaCTX-M-14, blaCTX-M-15, or blaCTX-M-27) in accordance with their ESBL phenotype. Interestingly, the first and last isolates from patient 2 (2/0 and 2/4) displayed significantly lower MICs for cefotaxime compared to the other three isolates of the same strain without detection of additional resistance genes. This trend was also observed for ertapenem and amdinocillin, indicating some genetic change regarding β-lactam resistance among the EPE isolates from this patient (see below). All isolates from the three patients were also resistant to sulfonamides, due to plasmid-encoded sul2 and/or sul1; trimethoprim because of different plasmid-encoded dfrA genes; and ciprofloxacin in the E. coli isolates due to plasmid encoded aac(6)-Ib-cr and/or mutations in gyrA and parC and in the K. pneumoniae isolates due to plasmid-encoded aac(6)-Ib-cr and qnrS1 and to chromosomal gyrA and parC mutations. High MICs toward erythromycin was displayed by 15/19 isolates through the acquired macrolide resistance determinant mphA. All isolates from patient 4 were also resistant to tetracycline [tetA(A) or tetA(B)] and some also to gentamicin [n = 5, aac(3′)IIa], nitrofurantoin (n = 4, mutations in nfsA and chromosomally encoded oqxAB), or displayed high MIC against kanamycin [n = 7, aac(6′)-Ib-cr, aph(3′)Ia], streptomycin (n = 4, strAB, aadA2, and aadA5), and chloramphenicol (n = 1, catA1). See Table S2 for more information about resistance genes and specific chromosomal mutations. The only tested antibiotics to which all isolates were sensitive were ertapenem, amdinocillin, and amikacin.
TABLE 1.
Resistance profiles of all isolates
| Patient and isolate | MIC (mg/liter) fora: |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CTX | ETP | MEC | TMP | SMZ | TET | ERY | CHL | CIP | STR | AMK | KAN | NIT | GEN | |
| Patient 2 | ||||||||||||||
| 2/0 | 32 | 0.008 | 0.25 | >32 | >1,024 | 1 | >256 | 4 | >32 | 8 | 2 | 2 | 8 | 0.5 |
| 2/1 | 128 | 0.03 | 0.5 | >32 | >1,024 | 1 | >256 | 4 | >32 | 8 | 2 | 2 | 8 | 0.5 |
| 2/2 | 128 | 0.03 | 0.5 | >32 | >1,024 | 1 | >256 | 4 | >32 | 4 | 2 | 2 | 8 | 0.5 |
| 2/3 | 128 | 0.03 | 0.5 | >32 | >1,024 | 1 | >256 | 4 | >32 | 4 | 2 | 2 | 8 | 0.5 |
| 2/4 | 32 | 0.008 | 0.25 | >32 | >1,024 | 1 | >256 | 4 | >32 | 4 | 2 | 2 | 8 | 0.5 |
| Patient 4 | ||||||||||||||
| 4/0 | 128 | 0.03 | 0.25 | >32 | >1,024 | 64 | >256 | 4 | >32 | 8 | 4 | 64 | 8 | 32 |
| 4/1-1 | 64 | 0.03 | 1 | >32 | >1,024 | 32 | >256 | 4 | >32 | 8 | 4 | >256 | 8 | 1 |
| 4/1-2 | 64 | 0.06 | 1 | >32 | >1,024 | >256 | 64 | >256 | >32 | 256 | 4 | 32 | 256 | 32 |
| 4/2-1 | 128 | 0.06 | 0.5 | >32 | >1,024 | >256 | 64 | 8 | >32 | 4 | 4 | 32 | 64 | 8 |
| 4/2-2 | 32 | 0.008 | 0.25 | >32 | >1,024 | 64 | >256 | 2 | >32 | 64 | 2 | 2 | 4 | 0.5 |
| 4/2-3 | 64 | 0.06 | 1 | >32 | >1,024 | >256 | 64 | 8 | >32 | 256 | 4 | 32 | 256 | 32 |
| 4/3-1 | 128 | 0.06 | 0.5 | >32 | >1,024 | >256 | 64 | 8 | >32 | 8 | 4 | 16 | 32 | 16 |
| 4/3-2 | 64 | 0.03 | 1 | >32 | >1,024 | 32 | >256 | 4 | >32 | 8 | 4 | >256 | 8 | 0.5 |
| 4/4 | 32 | 0.008 | 1 | >32 | >1,024 | 64 | >256 | 4 | >32 | 64 | 2 | 2 | 8 | 0.5 |
| Patient 7 | ||||||||||||||
| 7/0 | 32 | 0.03 | 0.25 | >32 | >1,024 | 2 | >256 | 8 | >32 | 8 | 2 | 2 | 4 | 0.5 |
| 7/1 | 32 | 0.06 | 0.25 | >32 | >1,024 | 2 | >256 | 32 | >32 | 8 | 4 | 2 | 2 | 0.5 |
| 7/2 | 32 | 0.06 | 0.25 | >32 | >1,024 | 4 | >256 | 8 | >32 | 4 | 2 | 2 | 4 | 0.25 |
| 7/3 | 32 | 0.06 | 0.25 | >32 | >1,024 | 2 | >256 | 8 | >32 | 8 | 2 | 2 | 4 | 0.25 |
| 7/4 | 32 | 0.06 | 0.25 | >32 | >1,024 | 2 | >256 | 8 | >32 | 8 | 2 | 2 | 4 | 0.5 |
No clinical breakpoints determined for Enterobacteriaceae (TET, ERY, STR, and KAN). Values indicated in boldface are greater than the clinical breakpoints for resistance according to EUCAST. CTX, cefotaxime; ETP, ertapenem; MEC, mecillinam; TMP, trimethoprim; SMZ, sulfamethoxazole; TET, tetracycline; ERY, erythromycin; CHL, chloramphenicol; CIP, ciprofloxacin; STR, streptomycin; AMK, amikacin; KAN, kanamycin; NIT, nitrofurantoin; GEN, gentamicin.
Cocolonization by multiple ST131 subtypes.
All five E. coli isolates from patient 2 and five of the nine isolates from patient 4 belonged to the B2 lineage of the internationally recognized expanded clonal complex E. coli ST131 (16). The five isolates from patient 2 had diverged slightly over the sampling time with a total of 25 unique SNPs between the isolates (Table S3), but the same plasmid was found in all isolates (see below). In contrast, cross comparison and phylogenetic analysis of the isolates from patient 4 showed that they were separated into three distinct subgroups where isolates 4/1-1 and 4/3-2 were identical, isolates 4/2-2 and 4/4 differed by one SNP, and the initial UTI isolate (4/0) clearly differed from the other two subgroups (Fig. 1). The genetic distance between the three subgroups were between 846 and 1,232 SNPs and, together with the difference in plasmid content (see below), show that patient 4 was colonized by distinctly different ST131 subtypes during the sampling period. Mapping of our ST131 isolates to an SNP tree using the ST131 data set from Matsumura et al. (17) clearly shows the different distributions of the isolates, with 2/0, 4/0, and 4/1-1 being part of the C2/H30Rx cluster, whereas 4/2-2 falls within the C1/H30R clade close to the C1-M27 subcluster in accordance with this isolate encoding blaCTX-M-27 (Fig. S1). Interestingly, only the ESBL plasmid of isolate 4/0 (p4/0.1) gave a best match to a plasmid of another ST131 isolate in GenBank, while all other ST131 plasmids found had best matches to plasmids from other E. coli sequence types (Table S4). This further corroborates the high plasmid variability found previously among the ST131 lineage.
Plasmidome analysis.
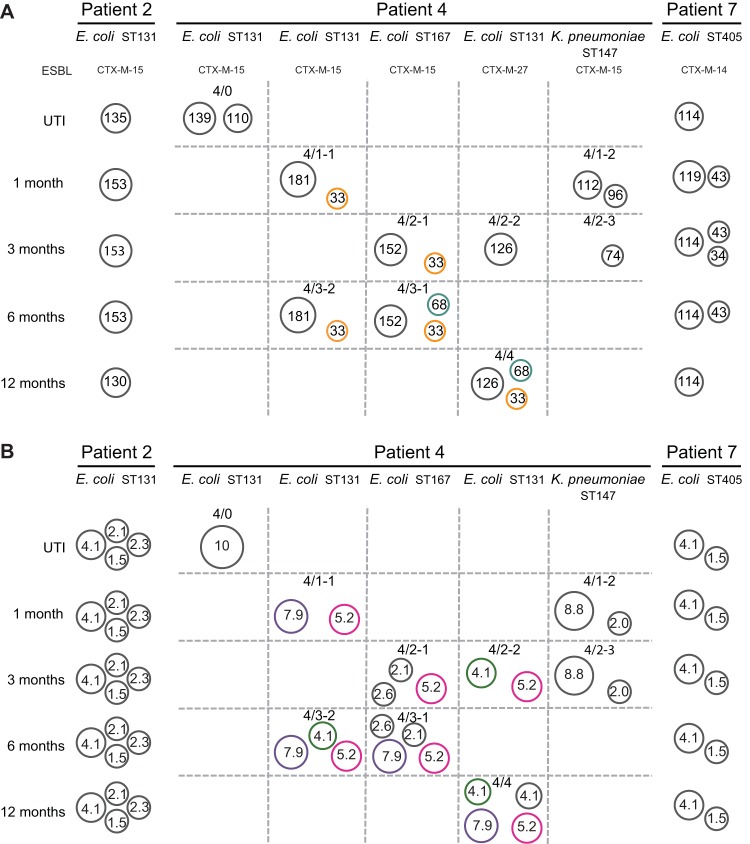
The plasmid contents of the isolates were completed by circularization of contigs from PacBio sequencing that did not assemble with the chromosomal sequences followed by BLASTn analysis of best matching references. Small plasmids (<10 kb) were also identified from de novo-assembled contigs of the MiSeq data not matching the consensus sequences generated by PacBio. This procedure identified 32 large (>10-kb) plasmids (Fig. 2 and Table S2). The number of large plasmids in each isolate ranged from one to three and total plasmid content was up to seven when including small cryptic plasmids (see below). All plasmids and putative plasmid transfer events are illustrated in Fig. 2.
FIG 2.
Summary of plasmidomes. (A) Large plasmids. A pBS512-33-like plasmid (orange) and a pVR50A-like plasmid (teal) were found in several different genetic backgrounds in patient 4. (B) Small cryptic plasmids. The 7.9-kb (purple), 5.2-kb (pink), and 4.1-kb (green) plasmids found in several different genetic backgrounds in patient 4 are indicated.
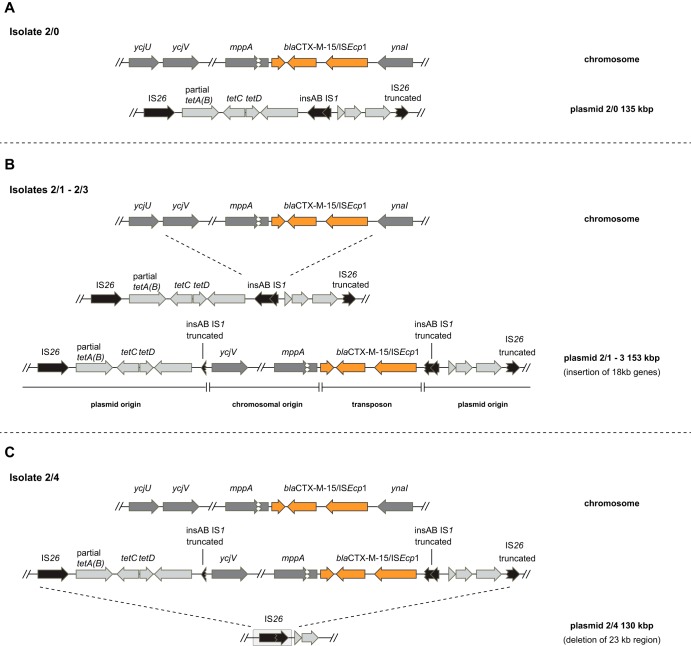
Although the UTI of patient 2 was successfully cleared, the E. coli ST131 strain that caused the infection could be isolated from the intestinal microflora in all subsequent samples. The UTI isolate carried one large multidrug resistance IncFIA/FIB/FII plasmid of 135 kb, p2/0.1, with the closest match to a plasmid from an E. coli ST648, EcIMT16316 (99% identity/80% query coverage, accession number CP023816) (Table S2). Interestingly, in isolate 2/0 the plasmid did not contain the blaCTX-M-15 gene, it was instead present on the chromosome inserted in the mppA gene (murein peptide permease A), together with its associated transposase ISEcp1b (Fig. 3A). However, the following isolates obtained at 1, 3, and 6 months post-UTI contained identical versions of the plasmid but with insertion of an 18-kb chromosomal region, including the blaCTX-M-15 ISEcp1b element (Fig. 3B). The insertion was most likely the result of a replicative transposition event initiated by the ISEcp1b transposase. The fifth sample, 2/4, contained the same plasmid but with a 22.6-kb deletion between an IS26 element upstream of the tetracycline resistance gene tetA(B) and a 96-bp IS26 remnant downstream of the blaCTX-M-15 gene (Fig. 3C). This deletion resulted in loss of the blaCTX-M-15 and tetA(B) genes from the plasmid. For all isolates, the chromosomal copy of the blaCTX-M-15 gene remained at the same site. This variation in ESBL gene composition also explains the variation in β-lactam susceptibility between the isolates from patient 2 with decreased susceptibility in the isolates with a second blaCTX-M-15 copy on the plasmid since increased expression can yield more beta-lactamase enzyme (see Discussion).
FIG 3.
Dynamics of blaCTX-M-15 movement between isolates in patient 2. The ISEcp1/blaCTX-M-15 transposition unit (orange) was inserted in the mppA gene in all five isolates from patient 2. (A and B) The initial UTI isolate 2/0 harbored a pEcIMT16316-like plasmid of 135 kbp (light gray) (A), while isolates 2/1, 2/2, and 2/3 all harbored a version of this plasmid with an insertion of 18 kbp of chromosomal genes (dark gray), including the ISEcp1/blaCTX-M-15 transposition unit (B). (C) This region of the plasmid and the flanking genes were deleted in the last isolate 2/4, through homologous recombination between identical flanking transposase genes (IS26 [black]).
All of the isolates from patient 7 contained a single ESBL-producing E. coli strain belonging to ST405. This strain carried an IncFIB/FII plasmid of 114 kb encoding dhfr17, sul-1, mphA, and aadA5 resistance genes. As in the EPE from patient 2, the ESBL gene (in this case blaCTX-M-14) was present on the chromosome, and no blaCTX-M-14 plasmid integration was seen in the isolates. However, in contrast to the other isolates, isolate 7/1 contained an erythromycin resistance ermABC operon integrated between directly repeated IS26 elements on the plasmid. If this difference between isolates represents an insertion or deletion through homologous recombination is not possible to determine from the sequence data. Isolate 7/1 also showed a substantial increase in chloramphenicol MIC (Table 1). Factors from the plasmids could be excluded since moving the plasmids to MG1655 gave no increase in MIC (data not shown). No obvious chromosomal mutations explaining this phenomenon could be identified, and further investigations are needed to elucidate the increase in MIC. Three of the isolates—7/1, 7/2, and 7/3—also harbored an additional plasmid of approximately 43 kb. This plasmid had a best match to pO157_Sal (77% identity/65% query coverage, accession number CP001927), which has been shown to affect the virulence, growth, and adaptation of an EHEC strain, E. coli O157:H7 (18). In addition, isolate 7/2 contained a third plasmid of 34 kb with high similarity to the Citrobacter freundii plasmid pN-Cit (99% identity/97% query coverage, accession number JQ996149). This plasmid was not found in any of the isolates from the other time points.
Extensive plasmid mobility facilitates spread of virulence and resistance genes.
The samples from patient 4 differed from the other two patients by containing ESBL-producing strains with different colony morphologies and a different genetic background. For these isolates, all ESBL genes were carried on plasmids. As seen from the chromosomal comparison, three different E. coli ST131 clones were present at different times in the gut microbiota of this patient. The sublineages also contained distinctly different large IncF multidrug resistance plasmids, further corroborating that they belong to different sublineages of ST131 (Fig. 2A and Table S2). The initial ST131 UTI isolate 4/0 contained a 139-kb blaCTX-M-15-encoding IncFIA/FII multidrug resistance plasmid most similar to the previously identified ST131 E. coli NCTC 13441 plasmid (100% identity/96% query coverage, accession number LT632321), and a second plasmid of 110 kb containing no resistance genes. This particular ST131 strain was not found in any of the other samples obtained from patient 4. Isolates 4/1-1 and 4/3-2 were genetically identical and shared an IncFIA/FIB/FII multidrug resistance plasmid of 181 kb encoding blaCTX-M-15, and a 33-kb IncX4 plasmid highly similar to pBS512_33 (99% identity/100% query coverage, accession number CP001059). Isolates 4/2-2 and 4/4 were of a third ST131 variant and differed by only 1 SNP on the chromosome and contained identical 126-kb IncFIA/FIB/FII plasmids encoding blaCTX-M-27. Isolates 4/2-1 and 4/3-1 were chromosomally identical and belonged to ST167 and contained identical versions of a 152-kb IncFIA/FIB/FII multidrug resistance plasmid encoding blaCTX-M-15. These isolates also contained identical copies of the 33-kb pBS512_33-like plasmid found in 4/1-1 and 4/3-2. The pBS512_33-like plasmid appears to have spread between different strains in patient 4 since identical plasmids were found in two different ST131 variants (4/1-1, 4/3-2, and 4/4) and also in the two ST167 (4/2-1 and 4/3-1), as illustrated in orange in Fig. 2A. In addition, 4/3-1 contained a 68-kb IncFII plasmid encoding the β-lactamase blaTEM-1, which also was found in 4/4 indicating that this plasmid had also spread between two different STs of E. coli. The two K. pneumoniae isolates differed by 2 SNPs on the chromosome and belonged to ST147. Isolate 4/1-2 had a 112-kb plasmid with no resistance genes and a 96-kb plasmid best matching pNDM-1fa (99% identity/53% query coverage, accession number CP014757). However, no blaNDM-1 gene was present on this plasmid, though there were a large number of other resistance genes, including blaCTX-M-15 (Table S2). Isolate 4/2-3 did not contain the 112-kb plasmid and had a smaller version of the pNDM-1fa-like plasmid of 74 kb that had lost the catB3 gene, most likely due to homologous recombination, but retained all other resistance genes. In addition to the deletion an inversion had also occurred between homologous regions.
High abundance and mobility of small cryptic plasmids.
Small cryptic plasmids (SCPs) of sizes up to 10 kb were found in all isolates (Table S5 and Fig. 2B). The presence of SCPs seems to be a common phenomenon in E. coli and K. pneumoniae isolates. It is not clear whether they have any phenotypic effect on the host bacterium since most SCPs only encode one or two genes, often involved in replication and plasmid mobility (19). In the five isolates from patient 2, four identical small cryptic plasmids were identified. Three of them were highly similar to SCPs identified in other Swedish E. coli plasmidomes previously described (20). Similarly, all five isolates from patient 7 contained the same two SCPs. Interestingly, a 1,552-bp plasmid with high sequence similarity to the previously sequenced pJJ1886 (100% identity/100% query coverage, accession number NC_022661) was present in all isolates from both patients 2 and 7. The pJJ1886 plasmid was originally found with four other plasmids in a highly virulent ST131 H30-Rx (21).
The SCPs of patient 4 were more diverse than for patients 2 and 7 in accordance with the diverse genetic backgrounds of the isolates (Table S5 and Fig. 2B). Several SCPs were found in isolates of different genetic backgrounds, indicating the extensive spread of these plasmids in the gut microbiome. In particular, two SCPs of 5.2 and 7.9 kb were found in multiple of the other E. coli isolates from patient 4. The 7.9-kb SCP was highly similar to pHUSEC41-3 (99% identity/100% query coverage, accession number NC_018997) previously isolated from a Shiga toxin-producing E. coli isolated in Germany (22). This plasmid displayed 15 open reading frame, four of which were related to ColE1 mobilization proteins (MobA to MobD), indicating a hitch-hiking capability with conjugative plasmids. A 4.1-kb plasmid identical to pMNCRE44_2 (100% identity/100% query coverage, accession number CP010878), previously found in a carbapenem-resistant E. coli strain (23), was found in three E. coli isolates. The K. pneumoniae isolates (4/1-2 and 4/2-3) showed two different SCPs of 8.8 and 2 kb that were not found in any of the E. coli isolates. In summary, the SCPs found in the isolates of patient 4 show extensive spread between E. coli of different strain types, whereas interspecies overlap between E. coli and K. pneumoniae was not seen.
Plasmid stability and fitness effects in vitro.
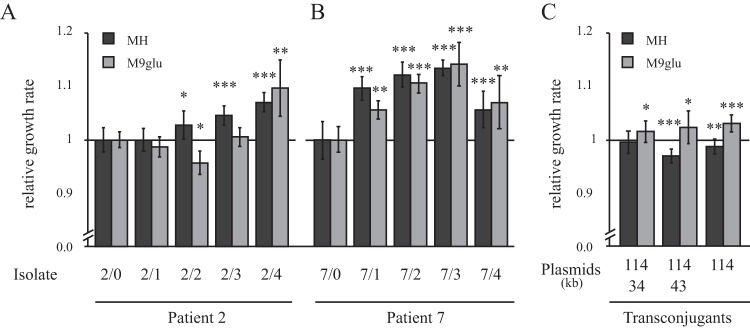
To determine whether the variation in plasmid content for the isolates from patient 7 were due to acquisition or loss of the plasmids, we performed in vitro stability experiments. This was done by serial passage of individual lineages of each isolate, followed by screening for plasmid presence over 500 generations of growth. By single cell passage on plates, the possible effect of growth differences between plasmid containing and plasmid free cells is reduced (12). Isolate 7/2 was chosen for studying the variable occurrence of the 34- and 43-kb plasmids in isolates from patient 7. The same stability assay was performed for isolate 2/1 to specifically study the stability of the incorporation of the blaCTX-M-15/ISEcp1b on the plasmid. No plasmid loss for isolates 2/1 and 7/2 was observed for any of the lineages, indicating that the plasmids are stable in their host cell at least under the laboratory conditions tested. Therefore, differences in occurrence between isolates of the same patient most likely represent new acquisition of plasmid rather than loss. Likewise, the presence of blaCTX-M-15 was persistent in isolate 2/1 indicating that the deletion found in isolate 2/4 is not a very frequent event. To assess whether acquisition of plasmids or plasmid presence of blaCTX-M-15 affected the fitness of the cell, relative growth rates were determined in two media, Mueller-Hinton medium and M9 glucose, as a proxy for fitness (Fig. 4A and B). By comparing all sequential isolates from patients 2 and 7, a general trend could be observed that the isolates increased their fitness over time. No correlation between plasmid presence of blaCTX-M-15 and fitness could be found, indicating that growth rate differences is likely due to chromosomal variations. In patient 7, the three isolates with extra plasmids grew significantly faster than the isolates without plasmids, which could indicate that the plasmids contribute a general fitness advantage to the cell (Fig. 4B). However, transfer of the plasmids to a common genetic background (MG1655) did not result in any major difference in fitness compared to the plasmid-free parental strain (Fig. 4C). The 114-kb plasmid, which was present in all isolates, gave a small increase in growth rate on its own in minimal medium but a small reduction in growth in rich medium. This indicates that chromosomal differences are responsible for the difference in fitness between the isolates. Transduction experiments were not successful due to phage resistance in the isolates, so alternative reconstruction experiments are required to conclusively determine any contribution of chromosomal differences.
FIG 4.
Relative growth rates of isolates from patients 2 (A) and 7 (B) in Mueller-Hinton medium (MH; dark gray columns) and M9 supplemented with 0.2% glucose (M9glu; light gray columns). Growth rates were normalized to the UTI isolate (set to 1) for each patient set of isolates. The standard deviations from five biological replicates are indicated. (C) Relative growth rate of transconjugants (MG1655) with plasmids from the isolates of patient 7. MG1655 without plasmid is set to 1 (not shown in the picture). The standard deviations from 16 biological replicates are indicated. Asterisks denote significant differences with respect to the UTI isolate (Student t test: *, P < 0.05; **, P < 0.01; ***, P < 0.001).
DISCUSSION
The prevalence of EPE carriage is increasing worldwide and consequently also in Sweden, where the incidence of infections caused by ESBL-producing E. coli and K. pneumoniae were 100 cases per 100,000 inhabitants in 2017 (24). In a recent study, asymptomatic EPE carriage was estimated to almost 5% of the Swedish population, where the main risk factor was travel to regions of high endemicity, such as Africa and Asia (25). Interestingly, E. coli ST131 were significantly more often found among patients contracting bloodstream infections than from asymptomatic carriers, and EPE colonizing asymptomatic carriers contained significantly fewer resistance genes, indicating a difference in risk of infections following colonization by different lineages. As in many parts of the world, the E. coli clone ST131 is the major cause of invasive EPE infections in Sweden (25). The reason for the success of the E. coli ST131 lineage is debated. Novais et al. (26) indicated that the spread and diversification of ST131 was driven by the acquisition of various IncF plasmids. On the other hand Zakour et al. (27) concluded that ST131 is first associated with acquired virulence, such as altered O antigen and attainment of prophages, and, second, not until recently, associated with antibiotic resistance, such as toward fluoroquinolones and β-lactams. The ST131 B2 clade C2 contain more antibiotic resistance genes, generally carried on plasmids, compared to other ST131 clades. These IncF plasmids are often associated with blaCTX-M genes, in particular the blaCTX-M-15 (28), which is consistent with patient 4 from our study, where three sublineages of ST131 were found of which two carried plasmid-borne blaCTX-M-15. In contrast, in the ST131 strain from patient 2, blaCTX-M-15 was present on the chromosome and in some isolates on the plasmid due to a transposition event from the chromosomal copy. Interestingly, insertion of blaCTX-M-15 in the mppA region has been observed before and the blaCTX-M gene is often flanked by IS26 (28), which in this study led to the deletion of the plasmid version of the blaCTX-M-15 gene in the last isolate from patient 2. Overall, the IncF plasmids examined in this study did not have a perfect match (between 53 and 96% sequence coverage) with other known plasmids illustrating the dynamic nature of influx and efflux of genes on these elements. As observed before, these changes are most pronounced when it comes to loss or gain of resistance genes in accordance with their frequent colocalization with transposable elements (10).
All the isolates analyzed here belonged to previously recognized sequence types associated with multidrug resistance and disease in humans. The E. coli ST405 lineage is, like ST131, described in the literature as a highly successful lineage associated with ESBL production and IncF plasmids (6, 29). Recently this multidrug-resistant ST group has been connected to spread of the plasmid-carried blaNDM genes (30, 31). E. coli ST167 has also been described as associated with ESBL production from Germany and Spain (32, 33). In China this strain type is not only reported carrying ESBL genes but also connected with carbapenemase production (34). The K. pneumoniae ST147 clone is an internationally dispersed K. pneumoniae clone associated with multidrug resistance and is known to harbor a plethora of resistance genes, including many different ESBL types (6, 35, 36).
The three patients were all diagnosed with UTI caused by EPE when included in the study. Although the number of patients is too small to draw any general conclusions regarding risk factors, some tentative indications can be pointed out. Patient 2 was previously healthy with small risk for acquisition of ESBL. The patient had been travelling to Gran Canaria, Spain, before contracting pyelitis, which was successfully treated with ciprofloxacin even though the isolated strain was shown to have a ciprofloxacin MIC of >32 mg/liter. Part of the stability of the ESBL strain and plasmidomes could be explained by lack of exposure to additional antibiotics and other risk factors such as hospitalization. We cannot conclude whether the ciprofloxacin treatment contributed to the persistence in colonization in the gut microbiota, but antibiotic treatment can selectively enrich resistant strains in vivo and prolong carriage (37, 38). The transposition of the ISEcp1/blaCTX-M-15 onto the plasmid shows a very dynamic nature of resistance genes that are associated with active transposases. The fact that the blaCTX-M-15 gene on the plasmid also contained insertion of the respective flanking chromosomal genes proved the direction of transfer. Increased gene copy number of blaCTX-M-15 has previously been shown to decrease susceptibility for the carbapenem ertapenem in combination with reduced outer membrane permeability (39). Although not reaching the clinical breakpoint, the ertapenem MIC increased with the additional copy of blaCTX-M-15 on the plasmid even without any additional mutations affecting porin expression among the isolates from patient 2. The 22.6-kb deletion seen in the plasmid of the fifth sample is not surprising given the lack of selective pressure and the possible cost of carrying more than one copy of this β-lactamase gene (39). Deletions originating between IS26 elements are very frequent and contribute to the dynamics of resistance cassettes in many plasmids (12).
Patient 7 had a suprapubic catheter and a history of previous UTIs with non-ESBL-producing bacteria. The patient was diagnosed with UTI caused by an EPE and treated with pivmecillinam. Prior to the UTI, the patient was admitted to a hospital in Sweden and treated with ciprofloxacin. During the course of the study, this patient had relapsing infections and received both pivmecillinam and nitrofurantoin treatment on several occasions. However, no additional resistance development was detected among the isolates studied here. Both pivmecillinam and nitrofurantoin have very small effects on the gut microbiota due to their pharmacokinetic properties (40, 41). The varying number of plasmids in the different isolates illustrate the dynamic population structure in the gut, where a varying presence of plasmids might reflect the normal flux of plasmids in bacteria. These plasmids seem to not affect the resistance against antibiotics, and it has yet to be shown whether they have any positive effects on the bacterial host.
Patient 4 contracted pyelitis with an EPE after travelling to Nepal. The infection was treated with ertapenem with a good clinical outcome. Prior to the infection, the patient had been hospitalized in Nepal, as well as in Sweden, and treated with tetracycline and ciprofloxacin. These factors could explain the diverse colonization with several different multidrug-resistant strains. Both tetracycline and ciprofloxacin are broad-spectrum antibiotics that affect the gut microbiota to a large extent enriching for bacteria that can survive these conditions. Genes conferring resistance to both tetracycline and ciprofloxacin were also found in the ESBL-producing bacteria from this patient showing high-level resistance. Traveling to a high-EPE-prevalence region such as Nepal and being in contact with health care are also factors that could explain the acquisition of several ESBL-producing strains and the presence of different ESBL genotypes (42). It cannot be excluded that some of the strains could have been acquired in Sweden, but since the estimated prevalence of EPE is much lower (5% in the Swedish population), that seems less likely (43).
These three different patient colonization scenarios illustrate the high complexity and interactive milieu that the gut microbiota represents. It is clear that there is not one answer for how resistance genes mobilize and persist. Multiple factors will affect the outcome and the persistence of resistance traits carried by the bacteria in the intestines. Antibiotic selection pressure, exposure to new infections/colonization, and the immune system, as well as the composition of bacterial species in the gut, will all contribute.
MATERIALS AND METHODS
Bacterial isolates and resistance testing.
EPE were isolated from three patients (designated patients 2, 4, and 7 in the previous study [15]) at five time points during the course of one year. The first clinical EPE sample from all patients was isolated from the urinary tract. At the specified time points 1, 3, 6, and 12 months after the initial clinical infection, self-collected fecal samples were obtained. EPE were identified with selective ChromID ESBL agar (bioMérieux, Craponne, France). Plates were streaked with transport swabs (Copan, Brescia, Italy) without prior broth enrichment. Colonies with distinct morphologies were selected and subjected to species identification and antimicrobial susceptibility testing, using VITEK2 (bioMérieux). One to three colonies representing each existing colony morphology were picked for the following analysis. In total, this generated 19 isolates of two different species (17 E. coli and 2 K. pneumoniae). ESBL production was confirmed with ESBL combination discs (Becton, Dickinson, NJ) or an ESBL Etest (bioMérieux) on Mueller-Hinton agar (Becton Dickinson) plates according to the manufacturers’ instructions. Susceptibility to 14 different antibiotics belonging to the major clinically important classes was determined by Etests (bioMérieux or Liofilchem, Roseto degli Abruzzi, Italy). Breakpoints for SIR-categorization were used according to EUCAST (http://www.eucast.org/clinical_breakpoints/).
The isolates were numbered according to the patient (patients 2, 4, and 7), sample number (0, 1, 2, 3, and 4), and morphology type if there were several in the same sample (i.e., 1 to 3). According to this labeling system, the UTI isolate named 2/0 is from patient 2, and there were no other morphotypes in this sample, while isolate 4/1-1 belongs to patient 4, is the first fecal sample obtained post-UTI, and represents the first of the morphologies in this sample, and isolate 4/1-2 is the second morphology in the same sample.
Whole-genome sequencing.
One isolate of each morphology (one each from patients 2 and 7 and five isolates from patient 4) were selected for whole-genome sequencing using the Pacific Biosciences (PacBio, Menlo Park, CA) technique. Whole-genome DNA was prepared from overnight cultures in Mueller-Hinton broth (MHB) using a Qiagen Genomic-Tip 100 according to the manufacturer’s instructions. PacBio sequencing was performed at the Science for Life Laboratory in Uppsala on an RSII system with one SMRT cell per genome. All 19 collected isolates were also whole-genome sequenced with an Illumina MiSeq instrument with two times 300-bp paired-end read lengths using the Nextera XT DNA library preparation kit (Illumina, San Diego, CA) according to the manufacturer´s instructions. The overall chromosomal coverage generated was >25-fold for all isolates.
Sequence assembly and bioinformatics analysis.
De novo assembly was performed for each data set with the CLC Genomics workbench v.9 (CLC Bio, Aarhus, Denmark), including the Microbial Genomics Pro Suite module. The contig sequences from the PacBio sequencing were circularized based on overlapping end-homology and mate-pair analysis and used as references for assembly of the Illumina reads from all other isolates belonging to the same strain. Complete chromosomal and plasmid sequences were obtained for all isolates. To verify that no sequences were missed between isolates, de novo assembly of nonmatched reads from combined reference assemblies for each data set was performed, and the remaining contigs were subjected to BLASTn searches and analyzed for gene content. All contigs and reference matches were tested against all isolates from the same patient. Reference assemblies were analyzed for single nucleotide polymorphisms, small deletion/insertion polymorphisms, and rearrangements between isolates from the same patient using the respective analysis tool in CLC Genomics Workbench. Small contigs from the Illumina plasmid sequencing with high coverage that represented full-length small cryptic plasmids were circularized by mate-pair analysis. Detection of resistance genes, plasmid replicons and virulence genes were done by submitting de novo-assembled contigs for each isolate to the online resources ResFinder (44), PlasmidFinder (45), and VirulenceFinder (46), respectively, at the Center for Genomic Epidemiology, DTU, Denmark. Analysis of chromosomal similarity between isolates from the same patient was done from the genome sequences by the MLST schemes for E. coli and K. pneumoniae according to Wirth et al. (47) and Diancourt et al. (48). All sequences have been deposited at GenBank under BioProject PRJNA413669 under accession numbers CP023820 to CP023858.
Plasmid stability.
In order to assess the stability of plasmids from two of the patients (2 and 7), a loss rate experiment was set up. Eighteen individual lineages from each isolate were restreaked on new MHA plates for 500 generations (with an approximation of 25 generations/passage), where the last single colony from each lineage was restreaked onto new plates daily. Every 100 generations the colony that was restreaked on new plates was also patched onto antibiotic plates to evaluate loss of the respective resistance plasmid in each lineage. At the end of the experiment for the lineages of isolate 2/1 cefotaxime MIC was determined with Etest in order to investigate loss of the blaCTX-M-15 from the plasmid. The presence of plasmids without resistance markers were verified by PCR using primers specific to the various plasmids (see Table S1) using a PCR program consisting of 3 min at 98°C for denaturation, followed by 25 cycles of denaturation 98°C (30 s), annealing 57°C (30 s), and elongation 72°C (30 s).
Growth rate measurements.
A BioscreenC MBR (Oy Growth Curve Ab, Ltd., Helsinki, Finland) was used to measure maximum growth rate at 37°C in MHB and in M9 minimal medium supplemented with 0.2% glucose (M9Glu; Sigma-Aldrich, Darmstadt, Germany), taking measurements of the optical density at 600 nm every 4 min. The values where exponential growth was observed were used (between 0.02 and 0.08).
Conjugation.
In order to transfer the plasmids from the clinical isolates (DA32103, DA32104, and DA32105) into E. coli MG1655 (DA5438) a two-step transfer was necessary with a ΔdapA auxotroph E. coli MG1655 (DA39037) as an intermediate recipient/donor. The mating of donor and recipient was conducted on Whatman Protran BA85 papers (GE Healthcare Life Sciences, Germany) on MHA plates containing 20 μg/ml of 2,6-diaminopimelic acid (DAP; Sigma-Aldrich, Darmstadt, Germany). Overnight cultures of donor and recipient were mixed in a 1:2 ratio. A dapA auxotrophy enables easy counterselection since without DAP DA39037 cannot grow. After overnight incubation, the bacteria were vortexed off the filter, and 100 μl of cell suspension was plated onto MHA (containing 20 μl/ml DAP, 15 μl of chloramphenicol, and 10 μl of trimethoprim in the case for selection of the intermediate transconjugant or only 10 μl of trimethoprim in the case for the selection of the final transconjugant) and incubated at 37°C overnight. Clones were restreaked and saved at –80°C. Clones were also whole-genome sequenced to verify that no mutations had occurred in the final recipient, as well as plasmids.
Phylogenetic analyses.
A k-mer tree was generated for the genome sequences for all E. coli isolates from the three patients, and SNP trees were made for each subset of the isolates with CLC Genomics workbench v.10 using the Microbial Genomics Module. The k-mer length used was 16 and a prefix of ATGAC was used for k-mer selection on both strands for each chromosome. Feature Frequency Profile estimate was done using Jensen-Shannon divergence (see, for example, reference 49). Tree construction was done using neighbor joining. SNP trees were prepared using complete chromosomal alignment of the isolates, including SNPs and single-nucleotide insertions/deletions using each initial UTI isolate as a reference for assembly. The prune distance for SNPs was set to 10, and the minimum z-score was set to 1.2. Phylogenetic relatedness to other ST131 isolates was determined using neighbor-joining tree construction with the data set (SRA accession DRA004266) from Matsumura et al. (17) with E. coli EC958 (NZ_HG941718) as the reference genome.
Supplementary Material
ACKNOWLEDGMENTS
We thank Petra J. Edquist and Chowdhury Mehedi Hasan for technical assistance and Karin Tegmark Wisell and Lionel Guy for constructive discussions and comments throughout the study.
This study was supported by grants from the Swedish Research Council Medicine and Health (K2013-99X-22208-01-5), the Carl Tryggers Foundation (CTS11:403), and the Magnus Bergvall Foundation to L.S.
The authors have declared that no competing interests exist.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.02201-18.
REFERENCES
- 1.Giske CG, Monnet DL, Cars O, Carmeli Y, on behalf of ReAct-Action on Antibiotic Resistance. 2008. Clinical and economic impact of common multidrug-resistant gram-negative bacilli. Antimicrob Agents Chemother 52:813–821. doi: 10.1128/AAC.01169-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de Kraker MEA, Wolkewitz M, Davey PG, Koller W, Berger J, Nagler J, Icket C, Kalenic S, Horvatic J, Seifert H, Kaasch A, Paniara O, Argyropoulou A, Bompola M, Smyth E, Skally M, Raglio A, Dumpis U, Kelmere AM, Borg M, Xuereb D, Ghita MC, Noble M, Kolman J, Grabljevec S, Turner D, Lansbury L, Grundmann H. 2011. Burden of antimicrobial resistance in European hospitals: excess mortality and length of hospital stay associated with bloodstream infections due to Escherichia coli resistant to third-generation cephalosporins. J Antimicrob Chemother 66:398–407. doi: 10.1093/jac/dkq412. [DOI] [PubMed] [Google Scholar]
- 3.de Kraker MEA. d, Davey PG, Grundmann H. 2011. Mortality and hospital stay associated with resistant Staphylococcus aureus and Escherichia coli bacteremia: estimating the burden of antibiotic resistance in Europe. PLoS Med 8:e1001104. doi: 10.1371/journal.pmed.1001104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schwaber MJ, Carmeli Y. 2007. Mortality and delay in effective therapy associated with extended-spectrum β-lactamase production in Enterobacteriaceae bacteremia: a systematic review and meta-analysis. J Antimicrob Chemother 60:913–920. doi: 10.1093/jac/dkm318. [DOI] [PubMed] [Google Scholar]
- 5.Nieminen O, Korppi M, Helminen M. 2017. Healthcare costs doubled when children had urinary tract infections caused by extended-spectrum β-lactamase-producing bacteria. Acta Paediatr 106:327–333. doi: 10.1111/apa.13656. [DOI] [PubMed] [Google Scholar]
- 6.D’Andrea MM, Arena F, Pallecchi L, Rossolini GM. 2013. CTX-M-type β-lactamases: a successful story of antibiotic resistance. Int J Med Microbiol 303:305–317. doi: 10.1016/j.ijmm.2013.02.008. [DOI] [PubMed] [Google Scholar]
- 7.Bevan ER, Jones AM, Hawkey PM. 2017. Global epidemiology of CTX-M β-lactamases: temporal and geographical shifts in genotype. J Antimicrob Chemother 72:2145–2155. doi: 10.1093/jac/dkx146. [DOI] [PubMed] [Google Scholar]
- 8.Carattoli A. 2011. Plasmids in Gram negatives: molecular typing of resistance plasmids. Int J Med Microbiol 301:654–658. doi: 10.1016/j.ijmm.2011.09.003. [DOI] [PubMed] [Google Scholar]
- 9.Carattoli A. 2009. Resistance plasmid families in Enterobacteriaceae. Antimicrob Agents Chemother 53:2227–2238. doi: 10.1128/AAC.01707-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.He S, Hickman AB, Varani AM, Siguier P, Chandler M, Dekker JP, Dyda F. 2015. Insertion sequence IS26 reorganizes plasmids in clinically isolated multidrug-resistant bacteria by replicative transposition. mBio 6:e00762–e00715. doi: 10.1128/mBio.00762-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sherley M, Gordon DM, Collignon PJ. 2003. Species differences in plasmid carriage in the Enterobacteriaceae. Plasmid 49:79–85. doi: 10.1016/S0147-619X(02)00107-5. [DOI] [PubMed] [Google Scholar]
- 12.Sandegren L, Linkevicius M, Lytsy B, Melhus Å, Andersson DI. 2012. Transfer of an Escherichia coli ST131 multiresistance cassette has created a Klebsiella pneumoniae-specific plasmid associated with a major nosocomial outbreak. J Antimicrob Chemother 67:74–83. doi: 10.1093/jac/dkr405. [DOI] [PubMed] [Google Scholar]
- 13.Trobos M, Lester CH, Olsen JE, Frimodt-Møller N, Hammerum AM. 2009. Natural transfer of sulphonamide and ampicillin resistance between Escherichia coli residing in the human intestine. J Antimicrob Chemother 63:80–86. doi: 10.1093/jac/dkn437. [DOI] [PubMed] [Google Scholar]
- 14.Göttig S, Gruber TM, Stecher B, Wichelhaus TA, Kempf VAJ. 2015. In vivo horizontal gene transfer of the carbapenemase OXA-48 during a nosocomial outbreak. Clin Infect Dis 60:1808–1815. doi: 10.1093/cid/civ191. [DOI] [PubMed] [Google Scholar]
- 15.Titelman E, Hasan CM, Iversen A, Nauclér P, Kais M, Kalin M, Giske CG. 2014. Faecal carriage of extended-spectrum β-lactamase-producing Enterobacteriaceae is common 12 months after infection and is related to strain factors. Clin Microbiol Infect 20:O508–O515. doi: 10.1111/1469-0691.12559. [DOI] [PubMed] [Google Scholar]
- 16.Forde BM, Ben Zakour NL, Stanton-Cook M, Phan M-D, Totsika M, Peters KM, Chan KG, Schembri MA, Upton M, Beatson SA. 2014. The complete genome sequence of Escherichia coli EC958: a high quality reference sequence for the globally disseminated multidrug resistant E. coli O25b:H4-ST131 clone. PLoS One 9:e104400. doi: 10.1371/journal.pone.0104400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matsumura Y, Pitout JDD, Gomi R, Matsuda T, Noguchi T, Yamamoto M, Peirano G, DeVinney R, Bradford PA, Motyl MR, Tanaka M, Nagao M, Takakura S, Ichiyama S. 2016. Global Escherichia coli sequence type 131 clade with blaCTX-M-27 gene. Emerg Infect Dis 22:1900–1907. doi: 10.3201/eid2211.160519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao H, Chen C, Xiong Y, Xu X, Lan R, Wang H, Yao X, Bai X, Liu X, Meng Q, Zhang X, Sun H, Zhao A, Bai X, Cheng Y, Chen Q, Ye C, Xu J. 2013. Global transcriptional and phenotypic analyses of Escherichia coli O157:H7 strain Xuzhou21 and its pO157_Sal cured mutant. PLoS One 8:e65466. doi: 10.1371/journal.pone.0065466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Burian J, Guller L, Mačor M, Kay WW. 1997. Small cryptic plasmids of multiplasmid, clinical Escherichia coli. Plasmid 37:2–14. doi: 10.1006/plas.1996.1273. [DOI] [PubMed] [Google Scholar]
- 20.Brolund A, Franzén O, Melefors Ö, Tegmark-Wisell K, Sandegren L. 2013. Plasmidome-analysis of ESBL-producing Escherichia coli using conventional typing and high-throughput sequencing. PLoS One 8:e65793. doi: 10.1371/journal.pone.0065793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Andersen PS, Stegger M, Aziz M, Contente-Cuomo T, Gibbons HS, Keim P, Sokurenko EV, Johnson JR, Price LB. 2013. Complete genome sequence of the epidemic and highly virulent CTX-M-15-producing H30-Rx subclone of Escherichia coli ST131. Genome Announc 1:e00988-13. doi: 10.1128/genomeA.00988-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Künne C, Billion A, Mshana SE, Schmiedel J, Domann E, Hossain H, Hain T, Imirzalioglu C, Chakraborty T. 2012. Complete sequences of plasmids from the hemolytic-uremic syndrome-associated Escherichia coli strain HUSEC41. J Bacteriol 194:532–533. doi: 10.1128/JB.06368-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson TJ, Hargreaves M, Shaw K, Snippes P, Lynfield R, Aziz M, Price LB. 2015. Complete genome sequence of a carbapenem-resistant extraintestinal pathogenic Escherichia coli strain belonging to the sequence type 131 H30R subclade. Genome Announc 3:e00272-15. doi: 10.1128/genomeA.00272-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Swedres-Svarm. 2017. Consumption of antibiotics and occurrence of resistance in Sweden. Solna/Uppsala ISSN1650-6332. SVA, Uppsala, Sweden. [Google Scholar]
- 25.Ny S, Löfmark S, Börjesson S, Englund S, Ringman M, Bergström J, Nauclér P, Giske CG, Byfors S. 2017. Community carriage of ESBL-producing Escherichia coli is associated with strains of low pathogenicity: a Swedish nationwide study. J Antimicrob Chemother 72:582–588. doi: 10.1093/jac/dkw419. [DOI] [PubMed] [Google Scholar]
- 26.Novais Â, Pires J, Ferreira H, Costa L, Montenegro C, Vuotto C, Donelli G, Coque TM, Peixe L. 2012. Characterization of globally spread Escherichia coli ST131 isolates (1991 to 2010). Antimicrob Agents Chemother 56:3973–3976. doi: 10.1128/AAC.00475-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zakour NLB, Alsheikh-Hussain AS, Ashcroft MM, Nhu NTK, Roberts LW, Stanton-Cook M, Schembri MA, Beatson SA. 2016. Sequential acquisition of virulence and fluoroquinolone resistance has shaped the evolution of Escherichia coli ST131. mBio 7:e00347-16. doi: 10.1128/mBio.00347-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stoesser N, Sheppard AE, Pankhurst L, De Maio N, Moore CE, Sebra R, Turner P, Anson LW, Kasarskis A, Batty EM, Kos V, Wilson DJ, Phetsouvanh R, Wyllie D, Sokurenko E, Manges AR, Johnson TJ, Price LB, Peto TEA, Johnson JR, Didelot X, Walker AS, Crook DW. 2016. Evolutionary history of the global emergence of the Escherichia coli epidemic clone ST131. mBio 7:e02162-15. doi: 10.1128/mBio.02162-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matsumura Y, Yamamoto M, Nagao M, Ito Y, Takakura S, Ichiyama S. 2013. Association of fluoroquinolone resistance, virulence genes, and IncF plasmids with extended-spectrum-β-lactamase-producing Escherichia coli sequence type 131 (ST131) and ST405 clonal groups. Antimicrob Agents Chemother 57:4736–4742. doi: 10.1128/AAC.00641-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.D’Andrea MM, Venturelli C, Giani T, Arena F, Conte V, Bresciani P, Rumpianesi F, Pantosti A, Narni F, Rossolini GM. 2011. Persistent carriage and infection by multidrug-resistant Escherichia coli ST405 producing NDM-1 carbapenemase: report on the first Italian cases. J Clin Microbiol 49:2755–2758. doi: 10.1128/JCM.00016-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang X, Feng Y, Zhou W, McNally A, Zong Z. 2018. Cryptic transmission of ST405 Escherichia coli carrying bla NDM-4 in hospital. Sci Rep 8:390. doi: 10.1038/s41598-017-18910-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schink A-K, Kadlec K, Kaspar H, Mankertz J, Schwarz S. 2013. Analysis of extended-spectrum-β-lactamase-producing Escherichia coli isolates collected in the GERM-Vet monitoring programme. J Antimicrob Chemother 68:1741–1749. doi: 10.1093/jac/dkt123. [DOI] [PubMed] [Google Scholar]
- 33.Oteo J, Diestra K, Juan C, Bautista V, Novais Â, Pérez-Vázquez M, Moyá B, Miró E, Coque TM, Oliver A, Cantón R, Navarro F, Campos J. 2009. Extended-spectrum β-lactamase-producing Escherichia coli in Spain belong to a large variety of multilocus sequence typing types, including ST10 complex/A, ST23 complex/A and ST131/B2. Int J Antimicrob Agents 34:173–176. doi: 10.1016/j.ijantimicag.2009.03.006. [DOI] [PubMed] [Google Scholar]
- 34.Huang Y, Yu X, Xie M, Wang X, Liao K, Xue W, Chan EW-C, Zhang R, Chen S. 2016. Widespread dissemination of carbapenem-resistant Escherichia coli sequence type 167 strains harboring blaNDM-5 in clinical settings in China. Antimicrob Agents Chemother 60:4364–4368. doi: 10.1128/AAC.00859-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kocsis B, Kocsis E, Fontana R, Cornaglia G, Mazzariol A. 2013. Identification of blaLAP-2 and qnrS1 genes in the internationally successful Klebsiella pneumoniae ST147 clone. J Med Microbiol 62:269–273. doi: 10.1099/jmm.0.050542-0. [DOI] [PubMed] [Google Scholar]
- 36.Papagiannitsis CC, Miriagou V, Giakkoupi P, Tzouvelekis LS, Vatopoulos AC. 2013. Characterization of pKP1780, a novel IncR plasmid from the emerging Klebsiella pneumoniae ST147, encoding the VIM-1 metallo-β-lactamase. J Antimicrob Chemother 68:2259–2262. doi: 10.1093/jac/dkt196. [DOI] [PubMed] [Google Scholar]
- 37.Dethlefsen L, Huse S, Sogin ML, Relman DA. 2008. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol 6:e280. doi: 10.1371/journal.pbio.0060280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Modi SR, Collins JJ, Relman DA. 2014. Antibiotics and the gut microbiota. J Clin Invest 124:4212. doi: 10.1172/JCI72333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Adler M, Anjum M, Berg OG, Andersson DI, Sandegren L. 2014. High fitness costs and instability of gene duplications reduce rates of evolution of new genes by duplication-divergence mechanisms. Mol Biol Evol 31:1526–1535. doi: 10.1093/molbev/msu111. [DOI] [PubMed] [Google Scholar]
- 40.Stewardson AJ, Gaïa N, François P, Malhotra-Kumar S, Delémont C, Martinez de Tejada B, Schrenzel J, Harbarth S, Lazarevic V. 2015. Collateral damage from oral ciprofloxacin versus nitrofurantoin in outpatients with urinary tract infections: a culture-free analysis of gut microbiota. Clin Microbiol Infect 21:344.e1–344.e11. doi: 10.1016/j.cmi.2014.11.016. [DOI] [PubMed] [Google Scholar]
- 41.Sullivan Å, Edlund C, Svenungsson B, Emtestam L, Nord CE. 2001. Effect of perorally administered pivmecillinam on the normal oropharyngeal, intestinal and skin microflora. J Chemother 13:299–308. doi: 10.1179/joc.2001.13.3.299. [DOI] [PubMed] [Google Scholar]
- 42.Baral P, Neupane S, Marasini BP, Ghimire KR, Lekhak B, Shrestha B. 2012. High prevalence of multidrug resistance in bacterial uropathogens from Kathmandu, Nepal. BMC Res Notes 5:38. doi: 10.1186/1756-0500-5-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.European Centre for Disease Prevention and Control. 2017. Antimicrobial resistance surveillance in Europe 2015: annual report of the European Antimicrobial Resistance Surveillance Network (EARS-Net). ECDC, Stockholm, Sweden. [Google Scholar]
- 44.Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, Aarestrup FM, Larsen MV. 2012. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Carattoli A, Zankari E, García-Fernández A, Larsen MV, Lund O, Villa L, Aarestrup FM, Hasman H. 2014. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother 58:3895–3903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Joensen KG, Scheutz F, Lund O, Hasman H, Kaas RS, Nielsen EM, Aarestrup FM. 2014. Real-time whole-genome sequencing for routine typing, surveillance, and outbreak detection of verotoxigenic Escherichia coli. J Clin Microbiol 52:1501–1510. doi: 10.1128/JCM.03617-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wirth T, Falush D, Lan R, Colles F, Mensa P, Wieler LH, Karch H, Reeves PR, Maiden MCJ, Ochman H, Achtman M. 2006. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol Microbiol 60:1136–1151. doi: 10.1111/j.1365-2958.2006.05172.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Diancourt L, Passet V, Verhoef J, Grimont PAD, Brisse S. 2005. Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J Clin Microbiol 43:4178–4182. doi: 10.1128/JCM.43.8.4178-4182.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sims GE, Jun S-R, Wu GA, Kim S-H. 2009. Alignment-free genome comparison with feature frequency profiles (FFP) and optimal resolutions. Proc Natl Acad Sci U S A 106:2677–2682. doi: 10.1073/pnas.0813249106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.