Candida auris has rapidly emerged as a health care-associated and multidrug-resistant pathogen of global concern. In this work, we examined the relative expression of the four C. auris genes with the highest degree of homology to Candida albicans CDR1 and MDR1 among three triazole-resistant clinical isolates as compared to the triazole-susceptible genome reference clinical isolate.
KEYWORDS: CDR1, Candida, Candida auris, antifungal resistance, drug efflux, efflux pumps, triazole
ABSTRACT
Candida auris has rapidly emerged as a health care-associated and multidrug-resistant pathogen of global concern. In this work, we examined the relative expression of the four C. auris genes with the highest degree of homology to Candida albicans CDR1 and MDR1 among three triazole-resistant clinical isolates as compared to the triazole-susceptible genome reference clinical isolate. We subsequently utilized a novel Cas9-mediated system for genetic manipulations to delete C. auris CDR1 and MDR1 in both a triazole-resistant clinical isolate and a susceptible reference strain and observed that MICs for all clinically available triazoles decreased as much as 128-fold in the CDR1 deletion strains. The findings of this work reveal for the first time that C. auris CDR1 and MDR1 are more highly expressed among triazole-resistant clinical isolates of C. auris and that the overexpression of CDR1 is a significant contributor to clinical triazole resistance.
INTRODUCTION
Candida auris, originally identified in 2009, has rapidly emerged as a health care-associated and multidrug-resistant pathogen of global concern (1–4). Although clinical experience and epidemiological data remain insufficient to support clinical breakpoints or epidemiological cutoff values for C. auris, the Centers for Disease Control and Prevention (CDC) has put forth tentative breakpoints to assist the assessment of MICs for clinical isolates of C. auris. Applying these tentative breakpoints, clinical isolates of C. auris are often found to be resistant to multiple classes of antifungal agents, and isolates resistant to all clinically available antifungal agents have been identified. In the United States, where the triazole antifungal fluconazole constitutes >70% of all inpatient antifungal use, >90% of clinical isolates of C. auris are considered fluconazole resistant when applying the tentative breakpoint suggested by the CDC (≥32 mg/liter) (5–7). Most of these fluconazole-resistant clinical isolates of C. auris have been found to harbor mutations in the gene encoding the sterol demethylase enzyme inhibited by the triazoles, ERG11, and the most commonly identified mutations (encoding F126L, Y132F, and K143R) have been demonstrated to contribute to triazole resistance in other species of Candida. Furthermore, when C. auris ERG11 alleles harboring mutations encoding the Y132F or K143R amino acid substitution were episomally expressed in Saccharomyces cerevisiae, fluconazole MICs were observed to increase (8). However, fluconazole-susceptible isolates of C. auris (fluconazole MIC, 1 to 8 mg/liter) harboring the Y132F or K143R mutation have been reported, suggesting the presence of additional mechanisms of triazole resistance yet to be identified (1, 9). Additionally, recent studies examining the efflux pump activity of C. auris compared with Candida glabrata found that clinical isolates of C. auris exhibited significantly higher (up to 7-fold higher) efflux pump activity than C. glabrata (10). This is particularly intriguing because in C. glabrata, clinical triazole resistance is essentially entirely attributable to the activity of three ATP-binding cassette (ABC)-type efflux pumps (Cdr1, Pdh1, and Snq2) (11). The recently updated annotated C. auris genome contains homologs of ABC-type and major facilitator superfamily-type efflux pump genes from Candida albicans (CDR1 and MDR1, respectively); however, these C. auris efflux pump-encoding genes remain uncharacterized.
In this work, we examined the relative expression of the four C. auris genes with the highest degree of homology to C. albicans CDR1 and MDR1 among three triazole-resistant clinical isolates (AR0388, AR0389, and AR0390) as compared to the triazole-susceptible genome reference clinical isolate AR0387 (B8441). We subsequently utilized a novel Cas9-ribonucleoprotein (RNP)-mediated transformation system incorporating 50- to 70-base regions of microhomology introduced by PCR to individually delete CDR1 and MDR1 from triazole-susceptible isolate AR0387 and triazole-resistant clinical isolate AR0390 (which harbors an ERG11 mutation encoding the K143R amino acid substitution), observed to overexpress both of these genes. We then demonstrated that the MICs for all clinically available triazole agents were significantly decreased in the CDR1 deletion strains in the AR0390 fluconazole-resistant background (up to 128-fold reduction), whereas only itraconazole MICs were observed to decrease by 2-fold in the MDR1 deletion strains in the same background. Taken together, the findings of this work reveal for the first time that C. auris CDR1 and MDR1 are more highly expressed among triazole-resistant clinical isolates of C. auris and that the overexpression of CDR1 is a significant contributor to clinical triazole resistance.
RESULTS
C. auris CDR1 and MDR1 are more highly expressed among triazole-resistant clinical isolates.
In an effort to identify efflux pump-encoding genes that may participate in clinical triazole resistance among isolates of C. auris, the two C. auris genes in the recently updated B8441 C. auris genome (https://www.ncbi.nlm.nih.gov/assembly/GCA_002759435.2) with the highest degree of homology to C. albicans genes CDR1 and MDR1 were identified by BLAST. The C. auris genes with the highest degree of homology to C. albicans CDR1 were B9J08_000164 and B9J08_002451, here referred to as CDR1 and CDR2, respectively; and the C. auris genes with the highest degree of homology for C. albicans MDR1 were B9J08_003981 and B9J08_000368, here referred to as MDR1 and MDR2, respectively. To determine whether the constitutive expression of these four C. auris efflux pump-encoding genes may be greater among fluconazole-resistant isolates (when applying the tentative breakpoints set forth by the CDC) of C. auris, we utilized the CDC and FDA Antibiotic Resistance Isolate Bank (AR Bank) collection of C. auris isolates and reverse transcription-quantitative PCR (RT-qPCR). The three clinical isolates of C. auris from the South Asian clade with fluconazole MIC greatly exceeding the tentative breakpoint of ≥32 mg/liter (AR0388, AR0389, and AR0390; fluconazole MIC, ≥256 mg/liter) were then selected for evaluation of CDR1, CDR2, MDR1, and MDR2 expression relative to the fluconazole-susceptible C. auris isolate of the South Asian clade AR0387 (B8441). Isolate AR0382, which is also of the South Asian clade, was not included because it exhibits an intermediary fluconazole MIC of 16 mg/liter and a strong aggregative phenotype not observed among the other South Asian isolates in this collection (12). Additionally, because both fluconazole-susceptible and fluconazole-resistant isolates (applying the tentative FDA breakpoints) from the South American, African, and East Asian clades were not available from the AR Bank panel, isolates from these clades were not included.
RNA was isolated from each clinical isolate following growth in yeast extract-peptone-dextrose (YPD) medium at 30°C prepared as previously described, and the expression of CDR1, CDR2, MDR1, and MDR2 was evaluated compared with that observed in isolate AR0387, using the ΔΔCT (threshold cycle) method and the C. auris ACT1 housekeeping gene (B9J08_000486). All measurements were taken in biological and technical triplicates. The relative expression of CDR1 was notably higher in all fluconazole-resistant isolates (3.4- to 7.3-fold that of AR0387), whereas the relative expression of MDR1 was higher only in fluconazole-resistant isolates AR0388 and AR0390 (Fig. 1). However, expression of MDR1 in these two resistant isolates reached only 2.3- and 3.9-fold that of susceptible control isolate AR0387. Additionally, the relative expressions of both CDR2 and MDR2 were not elevated in any of the fluconazole-resistant isolates.
FIG 1.
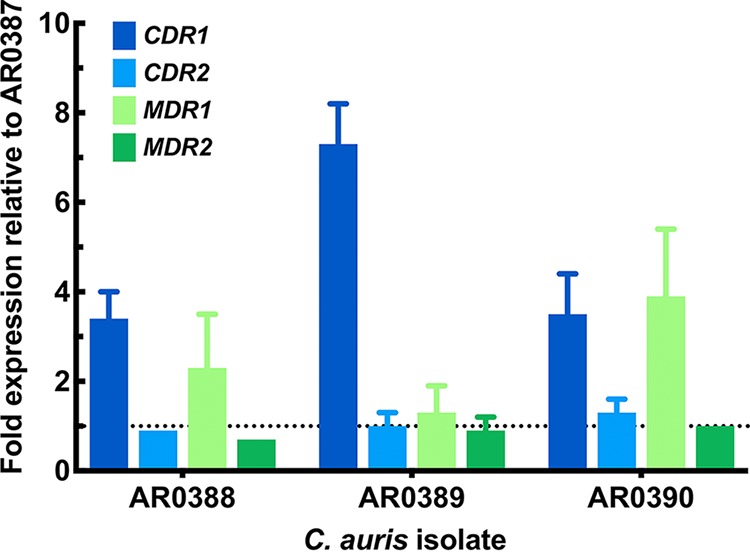
Relative expression of C. auris CDR1, CDR2, MDR1, and MDR2 in triazole-resistant isolates relative to AR0387. Experiments were performed in technical and biological triplicate; data shown as mean ± standard error of the mean.
Deletion of C. auris CDR1 restores fluconazole susceptibility in a highly resistant clinical isolate.
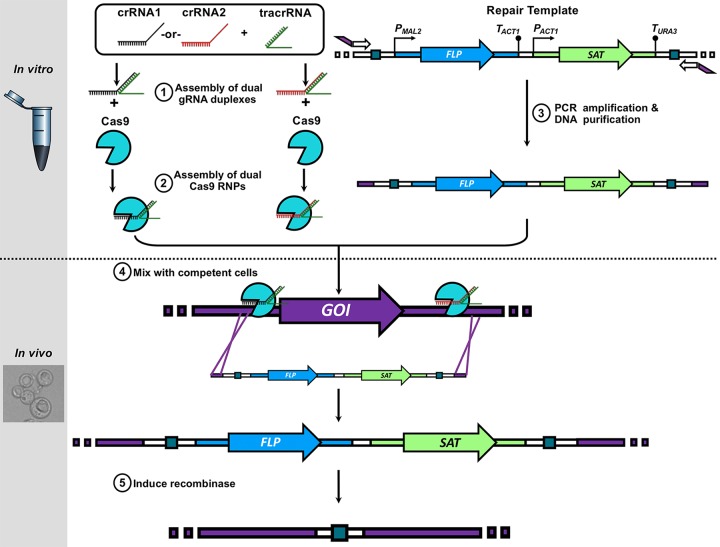
Because the constitutive expression of C. auris CDR1 and MDR1 was observed to be higher among multiple fluconazole-resistant clinical isolates of C. auris, we sought to delineate the direct impact of these two efflux pump-encoding genes on the MICs of all clinically available triazole antifungals. To accomplish this, we created CDR1 and MDR1 deletion strains in fluconazole-susceptible (AR0387) and fluconazole-resistant (AR0390) backgrounds using a novel Cas9-RNP-mediated transformation system we developed that utilizes dual Cas9 RNP to facilitate efficient target recombination with only 50 bases of PCR-introduced microhomology (Fig. 2). Notably, the fluconazole-resistant isolate AR0390 was selected because it was observed to more highly express CDR1 and MDR1 relative to AR0387. Two independent positive transformants for each gene of interest in each background were constructed using the SAT-FLP system as previously described (13). Susceptibility testing was then performed for fluconazole, voriconazole, isavuconazole, itraconazole, and posaconazole following modified Clinical and Laboratory Standards Institute methods for AR0387, AR0390, and the all-efflux pump deletion strains in each background. MIC determinations were performed in biological triplicate and were the lowest concentrations of drug that inhibit growth by 50% as determined by absorbance at optical density at 600 nm (OD600).
FIG 2.
Schematic of Cas9-RNP editing technique incorporating microhomology introduced by PCR for creation of efflux pump gene deletion strains.
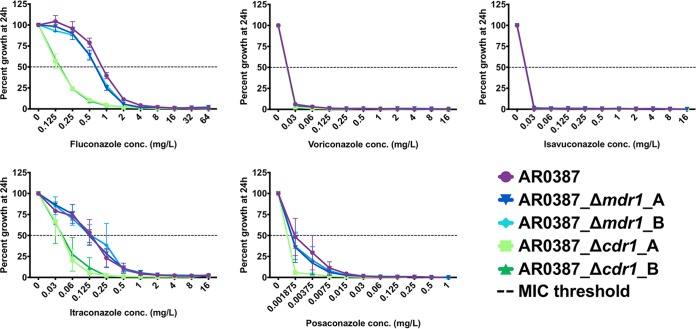
Fluconazole-susceptible clinical isolate AR0387 was observed to have a fluconazole MIC of 1 mg/liter, which is 32-fold below the tentative CDC breakpoint of ≥32 mg/liter (Fig. 3). The itraconazole MIC of this isolate was 0.25 mg/liter, and the MICs of all other triazoles were less than or equal to the lowest concentration tested (0.03 mg/liter for voriconazole and isavuconazole and 0.001875 mg/liter for posaconazole). In the CDR1 deletion strains in the AR0387 background, both fluconazole and itraconazole MICs decreased by 4-fold. In the MDR1 deletion strains constructed in this background, all triazole MICs were unchanged.
FIG 3.
Percentage growth of C. auris CDR1 and MDR1 deletion strains in the AR0387 background with escalating concentrations of triazoles measured at 24 h. All MIC determinations were performed in biological triplicate, and growth relative to the respective untreated controls was assessed by absorbance at OD600.
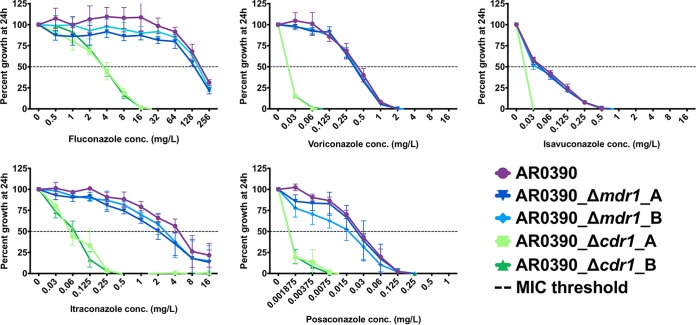
The triazole MICs for the fluconazole-resistant clinical isolate AR0390 were 256, 0.5, 0.06, 8, and 0.03mg/liter for fluconazole, voriconazole, isavuconazole, itraconazole, and posaconazole, respectively. In the CDR1 deletion strains in the AR0390 background, fluconazole and itraconazole MICs were dramatically decreased (64- to 128-fold) (Fig. 4). MICs for voriconazole, isavuconazole, and posaconazole were also decreased in the CDR1 deletion strains (≥2- to ≥16-fold); however, the full extent of these MIC reductions could not definitively be enumerated because the MICs in the CDR1 deletion strains were measured to be at or below the lowest concentration of these agents tested. In the MDR1 deletion strains in the AR0390 background, slight reductions in growth relative to the parental isolate were observed when testing fluconazole, itraconazole, and posaconazole susceptibilities. However, only in the case of itraconazole did this difference in relative growth result in a reduction in MIC (2-fold).
FIG 4.
Percentage growth of C. auris CDR1 and MDR1 deletion strains in the AR0390 background with escalating concentrations of triazole measured at 24 h. All MIC determinations were performed in biological triplicate, and growth relative to the respective untreated controls was assessed by absorbance at OD600.
DISCUSSION
C. auris is an emerging multidrug-resistant pathogen of great clinical concern. While a large proportion of clinical C. auris isolates are observed to have greatly elevated antifungal MICs relative to other more common species of Candida, the genetic and molecular basis of this clinical antifungal resistance remains unknown. In this work, we sought to characterize the contribution of efflux pump-encoding genes to triazole antifungal resistance in clinical isolates of C. auris. To accomplish this, we examined the relative expression of the four C. auris genes with the highest degree of homology to C. albicans CDR1 and MDR1 among three triazole-resistant clinical isolates (AR0388, AR0389, and AR0390) compared with the triazole-susceptible genome reference clinical isolate AR0387 (B8441). All of these clinical isolates are of the South Asian clade of C. auris, and all of these isolates are available as part of the CDC and FDA AR Isolate Bank collection.
Analysis of C. auris efflux pump gene expression, performed using RT-qPCR, revealed the C. auris gene with the highest degree of homology to C. albicans CDR1 to be more highly expressed among all three triazole-resistant isolates, at 3.4- to 7.3-fold higher than the expression measured in AR0387. This level of increased CDR1 expression is similar to that previously reported among fluconazole-resistant clinical isolates of C. albicans with elevated levels of CDR1 expression. Subsequently, deletion of CDR1 in fluconazole-susceptible AR0387 and fluconazole-resistant AR0390 clinical isolate backgrounds resulted in decreases in all triazole MICs not already equal to or below the lowest concentration of drug tested. Importantly, the magnitude of reduction in triazole MIC was significantly larger in the fluconazole-resistant AR0390 background than in the fluconazole-susceptible AR0387 background (64-fold versus 4-fold MIC reduction for both fluconazole and itraconazole). Additionally, deletion of CDR1 in the AR0390 background resulted in a fluconazole MIC that was 8-fold below the tentative breakpoint of ≥32 mg/liter (MIC, 4 mg/liter), and the MIC for all other triazole agents tested fell to concentrations less than or equal to the MIC obtained for the fluconazole-susceptible clinical isolate AR0387. Importantly, the restoration of triazole susceptibility occurred without correction of the K143R-encoding ERG11 mutation present in isolate AR0390. Thus, deletion of CDR1 alone in this fluconazole-resistant clinical isolate of C. auris was sufficient to abrogate triazole resistance.
Analysis of efflux pump gene expression also revealed the C. auris gene with the highest degree of homology to C. albicans MDR1 to be more highly expressed in triazole-resistant clinical isolates AR0388 and AR0390. However, only a 2.3- to 3.9-fold increase in expression was observed in these clinical C. auris isolates, which was considerably lower than the >100-fold increase in expression of C. albicans MDR1 that has been reported among isolates with gain-of-function mutations in the transcriptional regulator MRR1, where C. albicans MDR1 is known to be an effector of triazole resistance (14). Upon deletion of MDR1 in the AR0387 and AR0390 backgrounds, small differences in the percentage of growth in the presence of fluconazole were observed; however, this difference in growth was not large enough in either background to result in a change in MIC. Conversely, upon deletion of MDR1 in the fluconazole-resistant AR0390 background, itraconazole MICs were observed to decrease by 2-fold (4 versus 8 mg/liter in the parental AR0390 clinical isolate). Lastly, no differences in the expression of C. auris CDR2 or MDR2 were observed between the fluconazole-susceptible and fluconazole-resistant clinical isolates of C. auris.
Taken together, the findings of this study demonstrate for the first time that fluconazole-resistant clinical isolates of C. auris exhibit elevated levels of CDR1 and MDR1 expression and that CDR1 significantly contributes clinical resistance to the entire class of triazole antifungals. Further research is needed to identify the genetic determinants of increased CDR1 expression in clinical isolates of C. auris and to determine whether differences in CDR1 expression exist between fluconazole-susceptible and fluconazole-resistant isolates of C. auris from additional clades.
MATERIALS AND METHODS
Isolates, strains, and growth media used in this study.
The clinical C. auris isolates AR0387, AR0388, AR0389, and AR0390 were obtained from the CDC and FDA AR Isolate Bank collection of C. auris isolates. All clinical isolates and constructed strains were stored as frozen stocks in 50% glycerol at −80°C. All clinical isolates and strains were routinely grown in YPD (1% yeast extract, 2% peptone, and 2% dextrose) broth in a shaking incubator at 30°C unless otherwise indicated for specific experimental conditions.
MIC determination.
Antifungal susceptibilities for fluconazole, voriconazole, isavuconazole, itraconazole, and posaconazole were determined for all clinical isolates and strains by modified Clinical and Laboratory Standards Institute document M27 methodology utilizing broth microdilution RPMI (15). Each antifungal was obtained from the appropriate manufacturer, and stocks were prepared in dimethyl sulfoxide. After 24 h of incubation at 35°C, a BioTek Synergy 2 microplate reader (BioTek, Winooski, VT) was used to read absorbance at 600 nm. MIC was determined to be the lowest concentration at which 50% inhibition of growth was obtained. All susceptibility testing was performed in biological triplicate.
Reverse RT-qPCR.
C. auris overnight cultures were initiated by inoculating 2 ml YPD with cells transferred directly from frozen glycerol stock via sterile dowel. Cultures were incubated overnight at 30°C in an environmental shaking incubator. Fifty-milliliter conical tubes containing 5 ml YPD were each inoculated with 50 μl of each saturated overnight culture. These cultures were then incubated at 30°C for 3 h in an environmental shaking incubator, after which cells were collected by centrifugation, supernatants were poured off, and the cell pellets were stored at −80°C until RNA isolation was performed. As directed in the manufacturer’s instructions, the RevertAid RT kit (Thermo Scientific) was utilized to synthesize cDNA. PCR master mix and SYBR green were utilized to amplify C. auris ACT1 and CDR1, CDR2, MDR1, and MDR2 from cDNA by PCR per the manufacturer’s instructions. Table S1 in the supplemental material lists the gene-specific primers used for PCR. Conditions used for PCR were as follows: AmpliTaq Gold activation at 95°C for 10 min, 40 cycles of denaturation at 95°C for 15 s, and annealing/extension at 60°C. The dissociation curve and CT were determined using the 7500 detection real-time PCR system (Applied Biosystems). Changes in gene expression among isolates were then calculated using the 2-ΔΔCT method. All experiments were performed in triplicate from biological triplicates. As previously described, ΔCT values were used to calculate the standard error (16).
Cas9-RNP-mediated transformations.
Transformation of C. auris isolates was performed by electroporation as previously described, with modifications (17). For deletion of the genes of interest, repair templates containing the previously described SAT1-FLP cassette were prepared by PCR. Briefly, the entire SAT1-FLP cassette, including flanking FLP recognition target (FRT), sites was amplified from the plasmid pBSS2 using 5′ and 3′ primers, which introduced 50 to 70 bases of homology to the sequences immediately upstream and downstream of the genes of interest. Primers used are listed in Table S1. PCR products were purified using the Gene Clean II kit (MP Biomedicals). Approximately 1 μg of the purified repair template was then mixed with 4 μM of Cas9-RNP complexes targeting the sequences immediately upstream and immediately downstream of the open reading frame of the gene of interest and subsequently added to 40 μl of electrocompetent C. auris cells prepared as previously described (17). Electroporation was performed using the C. albicans protocol on a GenePulsar Xcell (Bio-Rad). Cells were then allowed to recover in YPD, incubating at 30°C in environmental shaking incubator for 4 to 6 h before being plated on YPD plates containing 200 μg/ml of nourseothricin for selection of transformants containing the SAT1-FLP cassette. After confirmation of transformants by PCR screening for integration of the cassette at the loci of the gene of interest and absence of the native open reading frame, positive transformants were grown overnight in YPM (1% yeast extract, 2% peptone, and 2% maltose) to induce the FLP recombinase and loss of the SAT1-FLP cassette. Strains identified as having lost nourseothricin resistance by replica plating on YPD with and without 200 μg/ml of nourseothricin were then confirmed by PCR screening for absence of the SAT1-FLP cassette, once again confirming the absence of the gene of interest.
Supplementary Material
ACKNOWLEDGMENTS
We thank the CDC for providing the C. auris isolates used in this study as part of the CDC and FDA Antibiotic Resistance Isolate Bank program.
This work was supported by NIH NIAID grant R01 A1058145 awarded to P.D.R.
We have no conflicting financial or personal interests to disclose.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.00057-19.
REFERENCES
- 1.Lockhart SR, Etienne KA, Vallabhaneni S, Farooqi J, Chowdhary A, Govender NP, Colombo AL, Calvo B, Cuomo CA, Desjardins CA, Berkow EL, Castanheira M, Magobo RE, Jabeen K, Asghar RJ, Meis JF, Jackson B, Chiller T, Litvintseva AP. 2017. Simultaneous emergence of multidrug-resistant Candida auris on 3 continents confirmed by whole-genome sequencing and epidemiological analyses. Clin Infect Dis 64:134–140. doi: 10.1093/cid/ciw691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Satoh K, Makimura K, Hasumi Y, Nishiyama Y, Uchida K, Yamaguchi H. 2009. Candida auris sp. nov., a novel ascomycetous yeast isolated from the external ear canal of an inpatient in a Japanese hospital. Microbiol Immunol 53:41–44. doi: 10.1111/j.1348-0421.2008.00083.x. [DOI] [PubMed] [Google Scholar]
- 3.Adams E, Quinn M, Tsay S, Poirot E, Chaturvedi S, Southwick K, Greenko J, Fernandez R, Kallen A, Vallabhaneni S, Haley V, Hutton B, Blog D, Lutterloh E, Zucker H, Candida Auris Investigation W. 2018. Candida auris in healthcare facilities, New York, USA, 2013-2017. Emerg Infect Dis 24:1816–1824. doi: 10.3201/eid2410.180649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eyre DW, Sheppard AE, Madder H, Moir I, Moroney R, Quan TP, Griffiths D, George S, Butcher L, Morgan M, Newnham R, Sunderland M, Clarke T, Foster D, Hoffman P, Borman AM, Johnson EM, Moore G, Brown CS, Walker AS, Peto TEA, Crook DW, Jeffery KJM. 2018. A Candida auris outbreak and its control in an intensive care setting. N Engl J Med 379:1322–1331. doi: 10.1056/NEJMoa1714373. [DOI] [PubMed] [Google Scholar]
- 5.Centers for Disease Control and Prevention. 2017. Candida auris clinical update - September 2017. https://www.cdc.gov/fungal/diseases/candidiasis/c-auris-alert-09-17.html. Accessed 16 February 2018.
- 6.Centers for Disease Control and Prevention. 2017. Recommendations for identification of Candida auris. https://www.cdc.gov/fungal/candida-auris/recommendations.html. Accessed 16 August 2018.
- 7.Vallabhaneni S, Baggs J, Tsay S, Srinivasan AR, Jernigan JA, Jackson BR. 2018. Trends in antifungal use in US hospitals, 2006–12. J Antimicrobial Chemother 73:2867–2875. doi: 10.1093/jac/dky270:dky270-dky270. [DOI] [PubMed] [Google Scholar]
- 8.Healey KR, Kordalewska M, Jimenez Ortigosa C, Singh A, Berrio I, Chowdhary A, Perlin DS. 2018. Limited ERG11 mutations identified in isolates of Candida auris directly contribute to reduced azole susceptibility. Antimicrob Agents Chemother 62:e01427-18. doi: 10.1128/AAC.01427-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chowdhary A, Prakash A, Sharma C, Kordalewska M, Kumar A, Sarma S, Tarai B, Singh A, Upadhyaya G, Upadhyay S, Yadav P, Singh PK, Khillan V, Sachdeva N, Perlin DS, Meis JF. 2018. A multicentre study of antifungal susceptibility patterns among 350 Candida auris isolates (2009-17) in India: role of the ERG11 and FKS1 genes in azole and echinocandin resistance. J Antimicrob Chemother 73:891–899. doi: 10.1093/jac/dkx480. [DOI] [PubMed] [Google Scholar]
- 10.Ben-Ami R, Berman J, Novikov A, Bash E, Shachor-Meyouhas Y, Zakin S, Maor Y, Tarabia J, Schechner V, Adler A, Finn T. 2017. Multidrug-resistant Candida haemulonii and C. auris, Tel Aviv, Israel. Emerg Infect Dis 23. doi: 10.3201/eid2302.161486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Whaley SG, Zhang Q, Caudle KE, Rogers PD. 2018. Relative contribution of the ABC transporters Cdr1, Pdh1, and Snq2 to azole resistance in Candida glabrata. Antimicrob Agents Chemother 62:e01070-18. doi: 10.1128/AAC.01070-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pathirana RU, Friedman J, Norris HL, Salvatori O, McCall AD, Kay J, Edgerton M. 2018. Fluconazole-resistant Candida auris is susceptible to salivary histatin 5 killing and to intrinsic host defenses. Antimicrob Agents Chemother 62:e01872-17. doi: 10.1128/AAC.01872-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reuss O, Vik A, Kolter R, Morschhauser J. 2004. The SAT1 flipper, an optimized tool for gene disruption in Candida albicans. Gene 341:119–127. doi: 10.1016/j.gene.2004.06.021. [DOI] [PubMed] [Google Scholar]
- 14.Morschhauser J, Barker KS, Liu TT, Bla BWJ, Homayouni R, Rogers PD. 2007. The transcription factor Mrr1p controls expression of the MDR1 efflux pump and mediates multidrug resistance in Candida albicans. PLoS Pathog 3:e164. doi: 10.1371/journal.ppat.0030164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clinical and Laboratory Standards Institute. 2017. Reference method for broth dilution antifungal susceptibility testing of yeast; approved standard—4th ed. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 16.Dunkel N, Liu TT, Barker KS, Homayouni R, Morschhauser J, Rogers PD. 2008. A gain-of-function mutation in the transcription factor Upc2p causes upregulation of ergosterol biosynthesis genes and increased fluconazole resistance in a clinical Candida albicans isolate. Eukaryot Cell 7:1180–1190. doi: 10.1128/EC.00103-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grahl N, Demers EG, Crocker AW, Hogan DA. 2017. Use of RNA-protein complexes for genome editing in non-albicans Candida species. mSphere 2:e00218-17. doi: 10.1128/mSphere.00218-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.