Pseudomonas aeruginosa colonizes the sputum of most adult cystic fibrosis patients, forming difficult-to-eradicate biofilms in which bacteria are protected in their self-produced extracellular polymeric substance (EPS) matrices. EPS provide biofilms with viscoelastic properties, causing time-dependent relaxation after stress-induced deformation, according to multiple characteristic time constants.
KEYWORDS: Pseudomonas aeruginosa, biofilm, recalcitrance, viscoelasticity
ABSTRACT
Pseudomonas aeruginosa colonizes the sputum of most adult cystic fibrosis patients, forming difficult-to-eradicate biofilms in which bacteria are protected in their self-produced extracellular polymeric substance (EPS) matrices. EPS provide biofilms with viscoelastic properties, causing time-dependent relaxation after stress-induced deformation, according to multiple characteristic time constants. These time constants reflect different biofilm (matrix) components. Since the viscoelasticity of biofilms has been related to antimicrobial penetration but not yet bacterial killing, this study aims to relate killing of P. aeruginosa, in its biofilm mode of growth, by three antimicrobials to biofilm viscoelasticity. P. aeruginosa biofilms were grown for 18 h in a constant-depth film fermenter, with mucin-containing artificial sputum medium (ASM+), artificial sputum medium without mucin (ASM−), or Luria-Bertani (LB) broth; this yielded 100-μm-thick biofilms that differed in their amounts of matrix environmental DNA (eDNA) and polysaccharides. Low-load compression testing, followed by three-element Maxwell analyses, showed that the fastest relaxation component, associated with unbound water, was most important in LB-medium-grown biofilms. Slower components due to water with dissolved polysaccharides, insoluble polysaccharides, and eDNA were most important in the relaxation of ASM+-grown biofilms. ASM−-grown biofilms showed intermediate stress relaxation. P. aeruginosa in LB-medium-grown biofilms was killed most by exposure to tobramycin, colistin, or an antimicrobial peptide, while ASM+ provided the most protective matrix, with less water and most insoluble polysaccharides and eDNA. In conclusion, stress relaxation of P. aeruginosa biofilms grown in different media revealed differences in matrix composition that, within the constraints of the antimicrobials and growth media applied, correlated with the matrix protection offered against different antimicrobials.
INTRODUCTION
Gram-negative Pseudomonas aeruginosa biofilms play an important role in chronic wound infections, otitis media, biomaterial-associated infections, and cystic fibrosis (CF) pneumonia (1). CF is characterized by the formation of thick mucus layers in the lungs, which makes them a suitable environment for P. aeruginosa to form biofilms (2). Approximately 80% of all adult CF patients are chronically infected with mucoid P. aeruginosa, which results in chronic illness and potentially death (3). Biofilm infections, including CF, are difficult to treat because the infecting bacteria surround themselves in a self-produced matrix of extracellular polymeric substances (EPS) (4). This can result in up to 1,000 times greater recalcitrance to antimicrobials than planktonic bacteria possess (5). Multiple mechanisms have been described for this recalcitrance of bacteria in a biofilm mode of growth, such as reduced metabolic activity, the presence of persister cells, and hampered penetration of antimicrobials into biofilms (3). The EPS matrix in P. aeruginosa biofilms mainly consists of water, proteins, lipids, environmental DNA (eDNA), and polysaccharides (6). The hallmark of CF infections caused by P. aeruginosa is the overproduction of polysaccharides, which negatively affects the survival of CF patients (7) because it facilitates strong bacterial binding, thus hampering clearance from the lungs as well as providing protection against the host immune system and antimicrobials.
EPS provide biofilms with viscoelastic properties. Elasticity relates to an immediate return of a material to its original shape after stress application, while viscoelasticity is the time-dependent, partial resumption of the original shape of a material after deformation. The time-dependent resumption of the biofilm shape after stress application can be subjected to a Maxwell analysis (8–10) to identify different stress relaxation processes occurring in a biofilm. Empirically, stress relaxation in biofilms has been divided into fast relaxation (stress relaxation time range, 0 to 5 s) due to fast flow of water with its low viscosity, a slow component (>100 s) related to repositioning of bacterial cells, and an intermediate component (5 to 100 s) caused by flow of more viscous EPS. Chlorohexidine penetration and bacterial killing in oral biofilms were related to biofilm viscoelasticity, decreasing with increasing prevalence of the fastest, water-attributable component and increasing with increasing prevalence of the slowest component associated with bacterial rearrangement (8). Accordingly, the viscoelasticity of a biofilm has been called a virulence factor (11). More detailed principal-component analysis attributed stress relaxation time ranges to three principal components, one involving water and soluble polysaccharides (0.01 to 3 s), one involving EPS components such as insoluble polysaccharides (3 and 70 s), and a principal component exclusively involving eDNA (10 to 25 s) (9). Collectively, these relatively fast components were related inversely to slow stress relaxation with time range constants of >70 s, due to bacterial cell rearrangement (9).
Although the viscoelastic properties of biofilms have been related to the combined effects of antimicrobial penetration and killing, which jointly define “recalcitrance,” no direct relationship between the viscoelasticity of a biofilm and antimicrobial killing has been established. Therefore, the aim of this study was to relate the viscoelasticity of P. aeruginosa biofilms to the killing of biofilm inhabitants by tobramycin, colistin, or an antimicrobial peptide at different concentrations. To this end, P. aeruginosa biofilms were grown in a constant-depth film fermenter (CDFF) in mucin-containing artificial sputum medium (ASM+), bearing similarity to the lung environment (12), artificial sputum medium without mucin (ASM−), or Luria-Bertani (LB) broth, a standard, high-nutrient, laboratory medium. The viscoelasticity of the biofilms would be determined from the stress relaxation of deformed biofilms and subsequent Maxwell analyses of the relaxation time constants.
RESULTS
Growth rates and antimicrobial susceptibility of planktonic P. aeruginosa in different media.
No differences in the growth rates of planktonic mucoid P. aeruginosa ATCC 39324, a clinical CF isolate, were observed when bacteria were grown in ASM+, ASM−, or LB broth (Fig. 1a). The minimal bactericidal concentrations (MBCs) against planktonic P. aeruginosa ATCC 39324 grown in different media are shown in Fig. 1b. Tobramycin and colistin yielded the same MBCs regardless of the growth medium used; for the antimicrobial peptide AA-230, however, the MBCs for P. aeruginosa grown in ASM+ and ASM− were 4 times higher than that for bacteria grown in LB broth.
FIG 1.
Planktonic growth curves and MBCs for P. aeruginosa ATCC 39324 in ASM+, ASM−, and LB medium. (a) Numbers of CFU per milliliter in planktonic cultures as a function of time in different growth media. Growth curves were performed in duplicate, with error bars denoting the difference between the two experiments. (b) MBCs for planktonic P. aeruginosa upon 24-h exposure to the different antimicrobials in PBS. MBC values were determined in triplicate with separately grown bacterial cultures, which yielded no differences in MBC values.
Characteristics and matrix composition of differently grown P. aeruginosa biofilms.
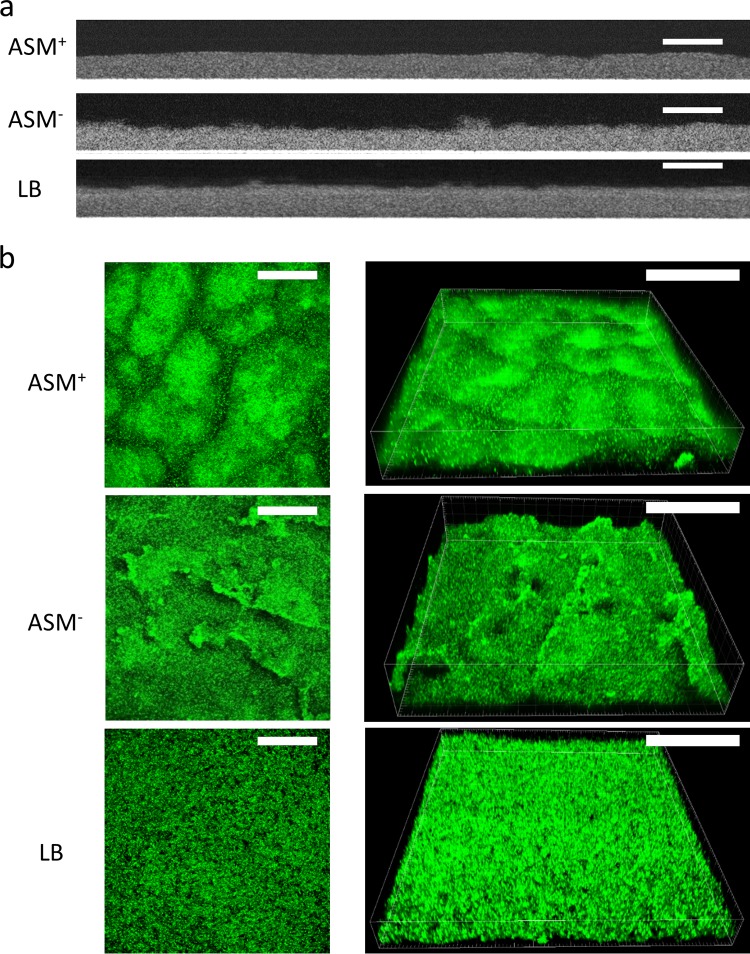
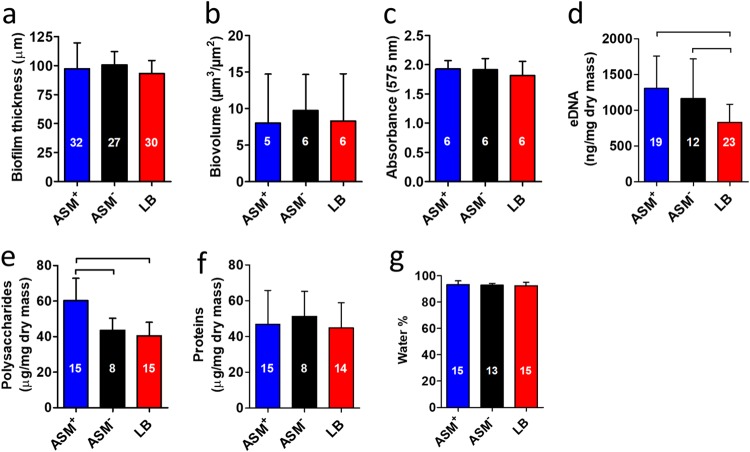
Biofilms of P. aeruginosa ATCC 39324 were grown in a CDFF with ASM+, ASM−, or LB broth for 18 h, employing wells with a depth of 100 μm (13). Biofilms were imaged using optical coherence tomography (OCT) and using confocal laser scanning microscopy (CLSM) after staining (Fig. 2a and b, respectively). Two-dimensional cross-sectional OCT images (Fig. 2a) confirmed that, on average, all biofilms grown were 100 μm thick (Fig. 3a), irrespective of the growth medium applied. Standard deviations over the thickness of biofilms grown in ASM+ with a surplus of mucin (22%; over three different CDFF runs, taking 10 biofilms from each run) were on average 2-fold larger than those of biofilms grown in the absence of a surplus of mucin (11% in both ASM− and LB medium). In CLSM images, biofilms grown in ASM+ showed a heterogeneous distribution of microcolonies surrounded by microchannels (Fig. 2b). Biofilms grown in LB medium had a highly homogeneous structure, without microcolonies and with less obvious microchannels. Biofilms grown in ASM− displayed an intermediate structure, compared to the biofilms grown in ASM+ and LB medium. COMSTAT analysis demonstrated no significant differences in the biovolumes of the biofilms (Fig. 3b). Metabolic activity of biofilms grown in ASM+, ASM−, and LB medium also showed no significant differences (Fig. 3c). Concentrations of eDNA (Fig. 3d) were similar in ASM+ and ASM− biofilms and higher than in LB medium biofilms. Polysaccharide concentrations (Fig. 3e) were similar in ASM− and LB medium biofilms but highest in ASM+ biofilms. No differences in protein concentrations (Fig. 3f) and water contents (Fig. 3g) were found. Significant differences in P. aeruginosa biofilm characteristics are summarized in Table 1.
FIG 2.
Microscopic images of P. aeruginosa ATCC 39324 biofilms grown in ASM+, ASM−, and LB medium. (a) Two-dimensional, cross-sectional, OCT images. Scale bars, 200 μm. (b) CLSM two-dimensional overlay (left) and three-dimensional (right) images of SYTO9-stained biofilms, yielding green-fluorescent bacteria. Scale bars, 100 μm.
FIG 3.
Characteristics and matrix composition of P. aeruginosa ATCC 39324 biofilms grown in ASM+, ASM−, and LB medium. (a) Thickness of the biofilms measured by OCT. (b) Biovolume of the biofilms obtained from COMSTAT analysis of CLSM images. (c) Metabolic activity of the biofilms measured with MTT. (d) eDNA presence in the biofilms, isolated with phenol-chloroform and measured with the nanodrop method. (e) Polysaccharide presence in the biofilms, measured using the anthrone-sulfuric acid colorimetric assay. (f) Total protein concentration in the biofilms. (g) Water content, obtained from a comparison of the weight of hydrated and dried biofilms and expressed as a percentage of the hydrated biofilm weight. Error bars denote standard deviations over n (numbers given in the columns) different biofilms, taken from different pans in three separate CDFF runs, except for data in panels b and c, which were taken from different pans in two separate CDFF runs. Significant differences (P < 0.05, ANOVA with Tukey’s post hoc analysis) between groups are indicated.
TABLE 1.
Summary of statistically significant differences in the characteristics of P. aeruginosa ATCC 39324 biofilms grown in ASM+, ASM−, or LB medium and their killing by antimicrobials (taking tobramycin, colistin, and the antimicrobial peptide AA-230 together)
| Biofilm characteristic | Differencea |
|---|---|
| Matrix eDNA | ASM+ = ASM− > LB |
| Matrix polysaccharides | ASM+ > ASM− = LB |
| Stress relaxation time | |
| <0.75 s | LB > ASM− > ASM+ |
| 0.75 s to <3 s | ASM+ > ASM− > LB |
| 3 s to <10 s | ASM+ > LB; ASM+ = ASM−; LB = ASM− |
| 10 s to <25 s | ASM+ > LB; ASM+ = ASM−; LB = ASM− |
| Antimicrobial killing | LB > ASM− ≥ ASM+ |
=, no significant difference; >, significant difference (P < 0.05).
Viscoelastic properties of differently grown P. aeruginosa biofilms.
Biofilms were compressed within 1 s to 80% of their initial thickness, equivalent to a strain (ε) of 0.2. Normalized stress on the biofilms required to maintain the same deformation decreased with time (Fig. 4a), showing relatively slow stress relaxation for ASM+-grown biofilms, while LB-medium-grown biofilms relaxed fastest. All biofilms showed nearly full stress relaxation toward 100 s. Stress relaxation as a function of time was fitted to a three-element Maxwell model. Inclusion of more Maxwell elements did not yield a better quality of the fit (Fig. 4b). LB-medium-grown biofilms showed significantly greater relative importance of the fastest time constant range (<0.75 s), compared with ASM+- and ASM−-grown biofilms, with ASM+-grown biofilms showing the least relative importance of the fastest relaxation time range (Fig. 4c). The relative importance of the other relaxation time constants ranging up to 25 s was highest for ASM+-grown biofilms and lowest for LB-medium-grown biofilms (Table 1).
FIG 4.
Stress relaxation analysis of P. aeruginosa ATCC 39324 biofilms grown in ASM+, ASM−, or LB medium. (a) Examples of the normalized stress in compressed biofilms (strain of 0.2) as a function of relaxation time. Stress at time zero amounted 2.2 kPa for all biofilms, regardless of the growth medium. (b) Quality of fitting of the stress relaxation data to a generalized Maxwell model as a function of the number of elements included in the model. The quality of the fit is indicated by chi-squared values. (c) Distribution of the relative importance of individual Maxwell elements (three-element model) for differently grown P. aeruginosa biofilms over different relaxation time constant ranges. Each data point represents a single measurement of 30 biofilms, taking 10 biofilms from different pans in three separate CDFF runs. Median values are indicated by horizontal lines. Significant differences (P < 0.05, ANOVA with Dunn’s post hoc analysis) between groups are indicated.
Antimicrobial killing in differently grown P. aeruginosa biofilms.
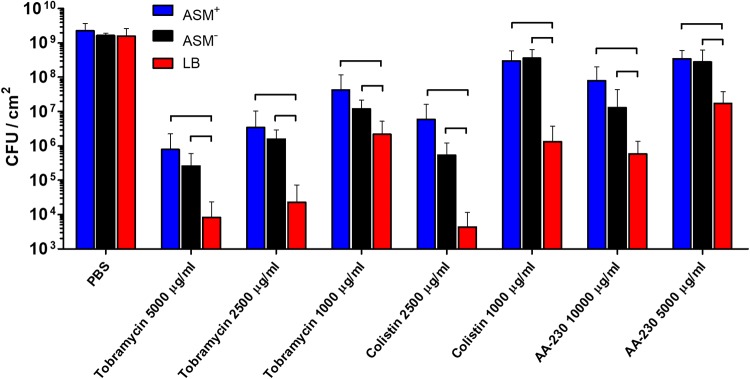
Biofilms were exposed for 24 h to phosphate-buffered saline (PBS) or PBS containing tobramycin, colistin, or the antimicrobial peptide AA-230, at concentrations well above their MBCs toward planktonic P. aeruginosa ATCC 39324 (Fig. 1b). The tobramycin concentrations applied were 1,000, 2,500, and 5,000 μg/ml, equivalent to 62, 156, and 313 times the MBC, respectively. Colistin was applied at concentrations of 1,000 and 2,500 μg/ml, equivalent to 8 and 20 times the MBC, respectively. For tobramycin and colistin, MBC-fold concentrations were independent of the growth medium (Fig. 1b); for the antimicrobial peptide AA-230, however, the concentrations applied (5,000 and 10,000 μg/ml) yielded different MBC-fold concentrations for bacteria grown in ASM+ and ASM− (39 and 78 times the MBC, respectively), compared with bacteria grown in LB medium (156 and 313 times the MBC, respectively). After antimicrobial exposure, biofilms were dispersed and the number of CFU in the biofilms was counted, taking PBS as a control. Control biofilms contained an average of 1.8 × 109 CFU/cm2, regardless of the growth medium used (Fig. 5). All antimicrobial exposures resulted in significant decreases in CFU per square centimeter, compared to biofilms after PBS exposure, with a clear dose-response relationship. In general, ASM+- and ASM−-grown biofilms showed significantly higher numbers of CFU, i.e., lower killing by antimicrobials, than did LB-medium-grown biofilms. No significant differences between ASM+- and ASM−-grown biofilms were observed.
FIG 5.
Numbers of CFU per square centimeter in P. aeruginosa ATCC 39324 biofilms grown in ASM+, ASM−, or LB medium after 24-h exposure to different concentrations of tobramycin, colistin, or the antimicrobial peptide AA-230 in PBS, with PBS as a control. Error bars denote standard deviations for at least nine different biofilms, taken from different pans in three separate CDFF runs. Significant differences (P < 0.05, ANOVA with Tukey’s post hoc analysis) between groups are indicated.
DISCUSSION
This study demonstrates that P. aeruginosa ATCC 39324 biofilms grown to a thickness of 100 μm in a CDFF possess different matrix compositions when grown in different growth media (Fig. 3) and allow different degrees of killing of the bacterial inhabitants by antimicrobials (Fig. 5). Maxwell analyses showed that the fastest relaxation component, associated with unbound water, was most important in LB-medium-grown biofilms (Fig. 4c), in the absence of obvious microchannels (Fig. 2b). Slower stress relaxation components due to water with dissolved polysaccharides, insoluble polysaccharides, and eDNA were most important in the relaxation of ASM+-grown biofilms. ASM−-grown biofilms showed intermediate stress relaxation. P. aeruginosa cells were killed most by exposure to tobramycin, colistin, or an antimicrobial peptide in LB-medium-grown biofilms, possibly due to the transport options provided by water. Biofilm growth in ASM+ provided the most protective matrix, with less unbound water and the most insoluble polysaccharides and eDNA, which maximally hampered penetration and killing. Interestingly, this statement coincides with the observation of microchannels in ASM+-grown biofilms (Fig. 2b). Since microchannels by definition have a transport function (14), this suggests that the matrix composition may be more important than the possession of clear channel-like structures for the transport of antimicrobials in a biofilm. In conclusion, stress relaxation analysis (Fig. 4) of P. aeruginosa biofilms grown in different media revealed differences in matrix composition (Fig. 3) that, within the constraints of the antimicrobials and growth media used, correlated with the matrix protection offered against different antimicrobials (Fig. 5). Without the use of a CDFF, it would have been impossible to carry out this study, because the use of different growth media would have yielded biofilms with different thicknesses (15–18).
Two artificial sputum media, mimicking the environment of the lungs of CF patients, and a nutrient-rich, laboratory medium (LB medium) were used. Biofilms grown in the artificial sputum media possessed more matrix eDNA and polysaccharides than did biofilms grown in LB medium. The possession of more eDNA and polysaccharides yielded different stress relaxation behavior of the biofilms, with greater importance of time relaxation ranges of 3 to 10 s and 10 to 25 s, respectively. This is fully in line with previous analyses of stress relaxation time ranges of biofilms from a wide variety of different strains and species, with known matrix amounts of eDNA and polysaccharides (9). Both eDNA and polysaccharides act as glue in biofilms, and growth in artificial sputum medium accordingly gave more compact biofilms with condensed microcolonies, in comparison with biofilms grown in LB medium (Fig. 2b), in line with literature findings (15, 19). The arrangement of bacteria in more compact, condensed microcolonies limits their possibility to rearrange during stress relaxation, which explains their slower relaxation (Fig. 4a). Although growth media that mimic the environment of the lungs were selected, it is virtually impossible to select a substratum material on which to grow the biofilms that also mimics the in vivo situation. However, considering the use of CDFF-grown biofilms compressed by the scraper action and the limited calling distance of quorum-sensing molecules in a biofilm, it may be expected that, over the 100-μm thickness of the Pseudomonas biofilms studied, the influences of the substratum might have averaged out (20).
LB-medium-grown biofilms demonstrated a stronger influence of water (relaxation times of <0.75 s) than did biofilms grown in artificial sputum media (Table 1), although dry weight measurements of the percentage of water in the biofilms were too insensitive to reflect differences with biofilms grown in other media (Fig. 3a). It should be noted that relaxation time constants up to 3 s were previously taken together (9), while we here separate this relaxation time constant range into two relaxation time constant ranges, attributing the time constant range between 0.75 and 3 s to more viscous water with dissolved polysaccharides. Through this distinction, the role of undissolved polysaccharides as a glue becomes reflected in stress relaxation analyses of biofilms.
Tobramycin, colistin, and AA-230 are all hydrophilic antimicrobials and have similar positive charges at physiological pH (21–23). The molecular weight of tobramycin is 468 g/mol, that of colistin is 1,156 g/mol, and that of AA-230 is 2,578 g/mol; this makes it interesting to compare bacterial killing by these antimicrobials at equivalent molar concentrations. At equivalent molar concentrations between 1.9 and 2.1 μM, corresponding to tobramycin, colistin, and AA-230 concentrations of 1,000, 2,500, and 5,000 μg/ml, respectively, colistin showed higher levels of killing than did tobramycin and AA-230, which was unexpected (24) because colistin, with its higher molecular weight, would diffuse more slowly into a biofilm at a similar molar concentration. Moreover, expressing antimicrobial concentrations in MBC-fold equivalents, a molar concentration of 2.1 μM colistin corresponds to an MBC-fold concentration of only 20× MBC, far lower than the MBC-fold concentrations of tobramycin and AA-230 at similar molar concentrations. These differences in killing efficacy are likely due to the different modes of killing; tobramycin works on the inhibition of protein synthesis and colistin on the destruction of the bacterial cell membrane (22, 25); the antimicrobial peptide AA-230, with the highest molecular weight, has a similar mode of action and accordingly showed less killing than did colistin, due to hampered penetration. Penetration of all three cationic antimicrobials would likely be hampered by the high concentrations of eDNA and polysaccharides in the biofilms grown in artificial sputum media, due to electrostatic double-layer attraction with negatively charged bacterial cell surface components (23, 26–29). Moreover, eDNA in the biofilms induces production of spermidine and aminoarabinose on the outer membrane, thereby reducing the permeability to aminoglycosides such as tobramycin (27) and leading to decreased killing efficacy in biofilms grown in the artificial sputum media. In addition, the presence of mucin can give rise to a higher tolerance for tobramycin (19). However, this effect is probably of minor importance, as differences in the killing of P. aeruginosa biofilms grown in artificial sputum medium with or without mucin were small (Fig. 5).
In conclusion, (i) viscoelastic properties of P. aeruginosa biofilms grown in a CDFF and in different media differ due to possession of different amounts of water, (in)soluble polysaccharides, and eDNA and, (ii) within the constraints of the antimicrobials and growth media used, these properties were related to antimicrobial bacterial killing in the biofilm. More unbound water and less EPS, i.e., polysaccharides and eDNA, as in LB-medium-grown biofilms, facilitated greater killing than a less aqueous matrix with more EPS, as in ASM+-grown biofilms.
MATERIALS AND METHODS
Bacterial cultures and media.
P. aeruginosa ATCC 39324, a clinical CF isolate (mucoid phenotype), was cultured on a blood agar plate and a single colony was used to inoculate 10 ml of tryptone soya broth (TSB) (Oxoid, Basingstoke, England) for aerobic incubation at 37°C. After 24 h, this preculture was added to 200 ml of TSB and incubated aerobically at 37°C for 16 h, with rotary shaking at 150 rpm, after which bacteria were harvested by centrifugation (5,000 × g for 5 min at 10°C). Bacterial pellets were washed two times with 10 ml sterile PBS (10 mM potassium phosphate, 150 mM NaCl [pH 7.0]), bacteria were resuspended in 10 ml sterile PBS, and bacterial concentrations were determined using a Bürker-Türk counting chamber.
Planktonic growth rates.
In order to investigate whether P. aeruginosa growth rates were similar in the different media, bacteria were suspended to 104 CFU/ml in 40 ml of ASM+ (12) (4 g/liter DNA, 5 g/liter mucin, 5 ml/liter egg yolk emulsion, 4.75 g/liter Casamino Acids, 0.25 g/liter l-tryptophan, 5 g/liter NaCl, 2.2 g/liter KCl [pH 7.0]), ASM−, or LB medium and incubated at 37°C, with rotary shaking at 150 rpm. At time zero and after 3, 6, and 24 h, 200-μl aliquots were taken and serially diluted 10-fold in sterile PBS. Two 10-μl droplets of each dilution were spotted on a tryptone soya agar plate and incubated for 24 h at 37°C, after which the CFU were counted and expressed as CFU per milliliter.
Minimal bactericidal concentrations.
Two-fold serial dilutions of the antimicrobials from 512 to 1 μg/ml were made in a 96-well plate in ASM+, ASM−, and LB medium, each with a total volume of 100 μl. For control, wells filled with growth medium in the absence of antimicrobials were used. Bacteria were diluted to a concentration of 2 × 106 bacteria/ml in either ASM+, ASM−, or LB medium, and 100 μl of bacterial suspension was added to each well to yield a total volume in the well of 200 μl. The plates were incubated statically at 37°C; after 24 h, 30-μl aliquots were spotted on tryptone soya agar plates, after which the plates were incubated at 37°C for 24 h. The lowest antimicrobial concentration not showing visible bacterial colonies was taken as the MBC.
Biofilm growth.
Biofilms were grown in a sterile CDFF (13, 30) at 37°C on stainless steel disks. The sterile disks were placed in each of the five wells of a pan, and 15 pans were placed in the turntable of the CDFF. The thickness of the biofilms in the pans was controlled by setting the well depth to leave 100 μm above the disks for the biofilm to grow. An amount of 200 ml of bacterial suspension in TSB containing 5 × 107 bacteria/ml was introduced into the CDFF over 1 h, while the turntable was rotating at 3 rpm. Rotation was stopped for 30 min to allow bacterial adhesion before the growth medium (ASM+, ASM−, or LB medium) was introduced and rotation was continued. The biofilms were grown for 18 h, and the medium flow was continuous at a flow rate of 16 ml/h. After 18 h, disks with adhering biofilms were aseptically taken out of the pans for further experiments.
Miscellaneous properties of differently grown biofilms.
To determine the average thickness of biofilms, OCT (Ganymede-II; Thorlabs, Lubeck, Germany) was used. Biofilms were submerged in PBS, and a series of two-dimensional, cross-sectional images (1,500 by 372 pixels) of the biofilms were generated. The average thickness of the biofilm was derived from the two-dimensional cross-sectional images using Otsu thresholding (31).
Biofilms were stained using SYTO9 live stain (bacLight; Invitrogen, Breda, The Netherlands) for 30 min in the dark to reveal the biofilm structure. After staining, biofilms were immersed in PBS and images were obtained with a confocal laser scanning microscope (Leica TCS-SP2; Leica Microsystems GmbH, Heidelberg, Germany) with a 40× water objective lens. Images were analyzed using Fiji (32), Imaris (Bitplane, Belfast, UK), and COMSTAT 2.1 (33, 34).
In order to investigate whether P. aeruginosa biofilms grown in the different media had similar metabolic activities, biofilms were exposed at 37°C for 2.5 h to 200 μl 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) (0.75 mg/ml) dissolved in sterile PBS. In metabolically active bacteria, MTT is intracellularly reduced to formazan. Following the incubation, biofilms were washed with PBS, 1 ml of isopropanol was added to dissolve the formazan crystals inside the bacterial cells, and the optical density of the solution was measured at 575 nm using a FLUOstar Optima plate reader (BMG Labtech GmbH, Offenburg, Germany).
Matrix composition.
For eDNA determination, 1 ml eDNA extraction buffer (10 mM EDTA, 0.9% NaCl) was added to individual biofilms taken over three CDFF runs (five biofilms per CDFF run), vortex-mixed, and resuspended until the biofilm was fully detached from the substratum disk. Dispersed biofilms were centrifuged (5,000 × g for 5 min at 10°C) to remove intact bacterial cells along with intracellular DNA. eDNA isolation was performed using the phenol-chloroform method (9). RNase was added to the eDNA after isolation and the mixture was incubated for 30 min at 37°C, after which the concentration of eDNA was measured using the ratio of absorbances at 260 nm and 280 nm, with the nanodrop method.
For both polysaccharide and protein determinations, five biofilms from different pans within one CDFF run were pooled, resuspended in 500 μl sterile PBS, and vortex-mixed for 1 min to detach the biofilms from the substratum disks; this was performed in triplicate, with separate CDFF runs. Resuspended biofilms were centrifuged (5,000 × g for 5 min at 10°C) to remove bacterial cells, after which 400 μl of supernatant was collected. The supernatant was immediately placed on ice and used for both polysaccharide and protein determinations.
The protein concentration was measured with the Pierce bicinchoninic acid (BCA) protein assay kit (Thermo Scientific, Waltham, MA, USA), using the microplate procedure. Briefly, 25 μl of sample from the supernatant or bovine serum albumin (25 to 2,000 μg/ml bovine albumin was used for the calibration curve) was added to 200 μl of working reagent and mixed for 30 s. Plates were incubated at 37°C for 30 min, after which the plates were cooled to room temperature and absorbance at 560 nm was measured using a FLUOstar Optima plate reader.
Polysaccharide determination was performed using the colorimetric assay for glucose-based carbohydrates (35). For the glucose-based carbohydrate method, 40 μl of supernatant or a glucose solution (1 to 4,096 μg/ml was used for the calibration curve) was added to a 96-well plate and placed in the refrigerator for 15 min. One hundred microliters of anthrone solution (2 mg/ml in H2SO4) was added to the wells and mixed, and the plates were incubated at 92°C for 3 min, after which the plates were placed in a water bath at room temperature for 15 min. Absorbance at 590 nm was measured and compared against the glucose calibration curve with a FLUOstar Optima plate reader.
To determine the water percentage in the biofilms, the wet weight of the disk and biofilm was measured using an analytical balance (model XP105DR; Mettler Toledo, Columbus, OH, USA). Prior to weighing, the bottom and sides of the disks were carefully dried with a tissue, after which their weights were measured. The disks with biofilms were then dried at 60°C in a vacuum oven for at least 24 h and weighed again. Subsequently, the dried biofilm was removed from the disk and the disk alone was weighed. With these data, the dry weight and water percentage of the biofilms were calculated.
Viscoelastic properties of P. aeruginosa biofilms.
The viscoelastic properties of the biofilms were determined by using a low-load compression tester (LLCT) (36). To this end, a small part of a substratum disk was cleaned to enable determination of the substratum surface using a LLCT plunger (2.5-mm diameter). After the position of the substratum surface was determined, the position of the biofilm surface was determined by compressing the biofilm with a small “touch level” of 0.01 g. Next, the biofilm was compressed to 80% of its original thickness (strain of 0.2) within 1 s and held constant for 100 s while stress relaxation was measured. Stress relaxation [σ(t)] was measured as a function of time t for 100 s and was normalized with respect to strain, according to
| (1) |
Normalized stress relaxation as a function of time E(t) was fitted to a three-element generalized Maxwell model by using the Solver tool in Microsoft Excel 2010, according to
| (2) |
where τi (i.e., ηi/Ei) is the relaxation time constant, Ei is the spring constant, and ηi is the viscosity term for each Maxwell element i. In order to indicate the relative importance of each relaxation process, RIi was calculated for each element as
| (3) |
and elements were placed in six relaxation time ranges.
Antimicrobial exposure of P. aeruginosa biofilms and bacterial killing.
Biofilms were exposed to different concentrations of tobramycin (1,000, 2,500, and 5,000 μg/ml) or colistin sodium methanesulfonate (1,000 and 2,500 μg/ml), both purchased from Sigma-Aldrich Chemie GmbH (Steinheim, Germany), and the antimicrobial peptide AA-230 (5,000 and 10,000 μg/ml), supplied by Adenium Biotech (Copenhagen, Denmark) and synthesized by PolyPeptide (Malmö, Sweden). Tobramycin and colistin are both used against P. aeruginosa infections in CF patients. All antimicrobials were dissolved in sterile PBS. A 20-μl drop of sterile PBS or antimicrobial solution was pipetted onto the biofilm. The plate was sealed with Parafilm to prevent evaporation and was incubated for 24 h at 37°C. Concentrations well above the MBC of each antimicrobial for planktonically grown P. aeruginosa were chosen. After antimicrobial exposure, biofilms were washed with sterile PBS, and 1 ml of sterile PBS was added and vortex-mixed for 1 min, to disrupt the biofilm and to detach the bacteria from the substratum disk. Finally, dispersions were sonicated in a sonication bath for 5 min to disrupt bacterial aggregates, which did not cause bacterial death (data not shown). Samples were serially diluted 10-fold, and two 10-μl drops of each dilution were spotted on tryptone soya agar and incubated at 37°C. After 24 h, colonies were counted and the number of CFU per square centimeter of substratum surface was calculated.
Statistical analysis.
Statistical analysis was performed with GraphPad Prism v5.00 for Windows (GraphPad Software, La Jolla, CA, USA). Differences in biofilm thickness, viscoelastic properties, biofilm recalcitrance to antimicrobials, and EPS components were evaluated after normality testing. Analysis of variance (ANOVA) was performed to test significance between groups, with either Dunn’s post hoc test or Tukey’s post hoc test, depending on normal or nonnormal distribution. Data with P values of <0.05 were accepted as significant. All data reported represent means with standard deviations, unless stated otherwise.
ACKNOWLEDGMENTS
The research leading to these results received funding from the European Union Seventh Framework Program (FP7/2007-2013) under grant agreement 604182 and was carried out within the project FORMAMP. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
R.T.R. and W.W. collected and analyzed the data presented. R.T.R., H.C.V.D.M., H.J.B., and P.K.S. prepared the outline of the manuscript and wrote the text. The text was critically reviewed by W.W. and E.D.D.J.
H.J.B. is also a director of a consulting company, SASA BV (Thesinge, The Netherlands). The authors declare no potential conflicts of interest with respect to authorship and/or publication of this article.
Opinions and assertions contained herein are those of the authors and are not to be construed as necessarily representing the views of their respective employers.
REFERENCES
- 1.Rybtke M, Hultqvist LD, Givskov M, Tolker-Nielsen T. 2015. Pseudomonas aeruginosa biofilm infections: community structure, antimicrobial tolerance and immune response. J Mol Biol 427:3628–3645. doi: 10.1016/j.jmb.2015.08.016. [DOI] [PubMed] [Google Scholar]
- 2.Damron FH, Goldberg JB. 2012. Proteolytic regulation of alginate overproduction in Pseudomonas aeruginosa. Mol Microbiol 84:595–607. doi: 10.1111/j.1365-2958.2012.08049.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ciofu O, Tolker-Nielsen T, Jensen PØ, Wang H, Høiby N. 2015. Antimicrobial resistance, respiratory tract infections and role of biofilms in lung infections in cystic fibrosis patients. Adv Drug Deliv Rev 85:7–23. doi: 10.1016/j.addr.2014.11.017. [DOI] [PubMed] [Google Scholar]
- 4.Bjarnsholt T, Alhede M, Alhede M, Eickhardt-Sørensen SR, Moser C, Kühl M, Jensen PØ, Høiby N. 2013. The in vivo biofilm. Trends Microbiol 21:466–474. doi: 10.1016/j.tim.2013.06.002. [DOI] [PubMed] [Google Scholar]
- 5.Ceri H, Olson ME, Stremick C, Read RR, Morck D, Buret A. 1999. The Calgary biofilm device: new technology for rapid determination of antibiotic susceptibilities of bacterial biofilms. J Clin Microbiol 37:1771–1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Flemming H-C, Wingender J. 2010. The biofilm matrix. Nat Rev Microbiol 8:623–633. doi: 10.1038/nrmicro2415. [DOI] [PubMed] [Google Scholar]
- 7.May TB, Shinabarger D, Maharaj R, Kato J, Chu L, Devault JD, Roychoudhury S, Zielinski NA, Berry A, Rothmel RK, Misra TK, Chakrabarty AM. 1991. Alginate synthesis by Pseudomonas aeruginosa: a key pathogenic factor in chronic pulmonary infections of cystic fibrosis patients. Clin Microbiol Rev 4:191–206. doi: 10.1128/CMR.4.2.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.He Y, Peterson BW, Jongsma MA, Ren Y, Sharma PK, Busscher HJ, van der Mei HC. 2013. Stress relaxation analysis facilitates a quantitative approach towards antimicrobial penetration into biofilms. PLoS One 8:e63750. doi: 10.1371/journal.pone.0063750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peterson BW, van der Mei HC, Sjollema J, Busscher HJ, Sharma PK. 2013. A distinguishable role of eDNA in the viscoelastic relaxation of biofilms. mBio 4:e00497-13. doi: 10.1128/mBio.00497-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peterson BW, Busscher HJ, Sharma PK, van der Mei HC. 2014. Visualization of microbiological processes underlying stress relaxation in Pseudomonas aeruginosa biofilms. Microsc Microanal 20:912–915. doi: 10.1017/S1431927614000361. [DOI] [PubMed] [Google Scholar]
- 11.Peterson BW, He Y, Ren Y, Zerdoum A, Libera MR, Sharma PK, van Winkelhoff A-J, Neut D, Stoodley P, van der Mei HC, Busscher HJ. 2015. Viscoelasticity of biofilms and their recalcitrance to mechanical and chemical challenges. FEMS Microbiol Rev 39:234–245. doi: 10.1093/femsre/fuu008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sriramulu DD, Lu H, Lam JS. 2005. Microcolony formation: a novel biofilm model of Pseudomonas aeruginosa for the cystic fibrosis lung. J Med Microbiol 54:667–676. doi: 10.1099/jmm.0.45969-0. [DOI] [PubMed] [Google Scholar]
- 13.Rozenbaum RT, Woudstra W, de Jong ED, van der Mei HC, Busscher HJ, Sharma PK. 2017. A constant depth film fermenter to grow microbial biofilms. Nat Protoc Exch doi: 10.1038/protex.2017.024. [DOI] [Google Scholar]
- 14.Donlan RM. 2002. Biofilms: microbial life on surfaces. Emerg Infect Dis 8:881–890. doi: 10.3201/eid0809.020063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fung C, Naughton S, Turnbull L, Tingpej P, Rose B, Arthur J, Hu H, Harmer C, Harbour C, Hassett DJ, Whitchurch CB, Manos J. 2010. Gene expression of Pseudomonas aeruginosa in a mucin-containing synthetic growth medium mimicking cystic fibrosis lung sputum. J Med Microbiol 59:1089–1100. doi: 10.1099/jmm.0.019984-0. [DOI] [PubMed] [Google Scholar]
- 16.Hancock V, Witsø IL, Klemm P. 2011. Biofilm formation as a function of adhesin, growth medium, substratum and strain type. Int J Med Microbiol 301:570–576. doi: 10.1016/j.ijmm.2011.04.018. [DOI] [PubMed] [Google Scholar]
- 17.Choi NY, Kim BR, Bae YM, Lee SY. 2013. Biofilm formation, attachment, and cell hydrophobicity of foodborne pathogens under varied environmental conditions. J Korean Soc Appl Biol Chem 56:207–220. doi: 10.1007/s13765-012-3253-4. [DOI] [Google Scholar]
- 18.Klausen M, Heydorn A, Ragas P, Lambertsen L, Aaes-Jørgensen A, Molin S, Tolker-Nielsen T. 2003. Biofilm formation by Pseudomonas aeruginosa wild type, flagella and type IV pili mutants. Mol Microbiol 48:1511–1524. doi: 10.1046/j.1365-2958.2003.03525.x. [DOI] [PubMed] [Google Scholar]
- 19.Landry RM, An D, Hupp JT, Singh PK, Parsek MR. 2006. Mucin-Pseudomonas aeruginosa interactions promote biofilm formation and antibiotic resistance. Mol Microbiol 59:142–151. doi: 10.1111/j.1365-2958.2005.04941.x. [DOI] [PubMed] [Google Scholar]
- 20.Ren Y, Wang C, Chen Z, Allan E, van der Mei HC, Busscher HJ. 2018. Emergent heterogeneous micro-environments in biofilms: substratum surface heterogeneity and bacterial adhesion force-sensing one. FEMS Microbiol Rev 42:259–272. doi: 10.1093/femsre/fuy001. [DOI] [PubMed] [Google Scholar]
- 21.Pfeifer C, Fassauer G, Gerecke H, Jira T, Remane Y, Frontini R, Byrne J, Reinhardt R. 2015. Purity determination of amphotericin B, colistin sulfate and tobramycin sulfate in a hydrophilic suspension by HPLC. J Chromatogr B Analyt Technol Biomed Life Sci 990:7–14. doi: 10.1016/j.jchromb.2015.02.043. [DOI] [PubMed] [Google Scholar]
- 22.Bergen PJ, Li J, Rayner CR, Nation RL. 2006. Colistin methanesulfonate is an inactive prodrug of colistin against Pseudomonas aeruginosa. Antimicrob Agents Chemother 50:1953–1958. doi: 10.1128/AAC.00035-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Drew KRP, Sanders LK, Culumber ZW, Zribi O, Wong GCL. 2009. Cationic amphiphiles increase activity of aminoglycoside antibiotic tobramycin in the presence of airway polyelectrolytes. J Am Chem Soc 131:486–493. doi: 10.1021/ja803925n. [DOI] [PubMed] [Google Scholar]
- 24.Forier K, Messiaen AS, Raemdonck K, Nelis H, De Smedt S, Demeester J, Coenye T, Braeckmans K. 2014. Probing the size limit for nanomedicine penetration into Burkholderia multivorans and Pseudomonas aeruginosa biofilms. J Control Release 195:21–28. doi: 10.1016/j.jconrel.2014.07.061. [DOI] [PubMed] [Google Scholar]
- 25.Kotra LP, Haddad J, Mobashery S. 2000. Aminoglycosides: perspectives on mechanisms of action and resistance and strategies to counter resistance. Antimicrob Agents Chemother 44:3249–3256. doi: 10.1128/AAC.44.12.3249-3256.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chiang WC, Nilsson M, Jensen PØ, Høiby N, Nielsen TE, Givskov M, Tolker-Nielsen T. 2013. Extracellular DNA shields against aminoglycosides in Pseudomonas aeruginosa biofilms. Antimicrob Agents Chemother 57:2352–2361. doi: 10.1128/AAC.00001-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wilton M, Charron-Mazenod L, Moore R, Lewenza S. 2016. Extracellular DNA acidifies biofilms and induces aminoglycoside resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother 60:544–553. doi: 10.1128/AAC.01650-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nichols WW, Dorrington SM, Slack MPE, Walmsley HL. 1988. Inhibition of tobramycin diffusion by binding to alginate. Antimicrob Agents Chemother 32:518–523. doi: 10.1128/AAC.32.4.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hentzer M, Teitzel GM, Balzer GJ, Molin S, Givskov M, Matthew R, Heydorn A, Parsek MR. 2001. Alginate overproduction affects Pseudomonas aeruginosa biofilm structure and function. J Bacteriol 183:5395–5401. doi: 10.1128/JB.183.18.5395-5401.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hope CK, Wilson M. 2006. Biofilm structure and cell vitality in a laboratory model of subgingival plaque. J Microbiol Methods 66:390–398. doi: 10.1016/j.mimet.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 31.Hou J, Veeregowda DH, van de Belt-Gritter B, Busscher HJ, van der Mei HC. 2018. Extracellular polymeric matrix production and relaxation under fluid shear and mechanical pressure in Staphylococcus aureus biofilms. Appl Environ Microbiol 84:e01516-17. doi: 10.1128/AEM.01516-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez J-Y, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A. 2012. Fiji: an open-source platform for biological-image analysis. Nat Methods 9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heydorn A, Nielsen AT, Hentzer M, Sternberg C, Givskov M, Ersbøll BK, Molin S. 2000. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology 146:2395–2407. doi: 10.1099/00221287-146-10-2395. [DOI] [PubMed] [Google Scholar]
- 34.Vorregaard M. 2008. Comstat2: a modern 3D image analysis environment for biofilms. Master’s thesis. Technical University of Denmark, Kongens Lyngby, Denmark. [Google Scholar]
- 35.Laurentin A, Edwards CA. 2003. A microtiter modification of the anthrone-sulfuric acid colorimetric assay for glucose-based carbohydrates. Anal Biochem 315:143–145. doi: 10.1016/S0003-2697(02)00704-2. [DOI] [PubMed] [Google Scholar]
- 36.Paramonova E, Kalmykowa OJ, van der Mei HC, Busscher HJ, Sharma PK. 2009. Impact of hydrodynamics on oral biofilm strength. J Dent Res 88:922–926. doi: 10.1177/0022034509344569. [DOI] [PubMed] [Google Scholar]