The nimbleness of Neisseria gonorrhoeae to evade the effect of antibiotics has perpetuated the fight against antibiotic-resistant gonorrhea for more than 80 years. The ability to develop resistance to antibiotics is attributable to its indiscriminate nature in accepting and integrating exogenous DNA into its genome.
KEYWORDS: 23S rRNA mutation, azithromycin resistance, MTR mutations, Neisseria gonorrhoeae
ABSTRACT
The nimbleness of Neisseria gonorrhoeae to evade the effect of antibiotics has perpetuated the fight against antibiotic-resistant gonorrhea for more than 80 years. The ability to develop resistance to antibiotics is attributable to its indiscriminate nature in accepting and integrating exogenous DNA into its genome. Here, we provide data demonstrating a novel combination of the 23S rRNA A2059G mutation with a mosaic-multiple transferable resistance (mosaic-mtr) locus haplotype in 14 N. gonorrhoeae isolates with high-level azithromycin MICs (≥256 μg/ml), a combination that may confer more fitness than in previously identified isolates with high-level azithromycin resistance. To our knowledge, this is the first description of N. gonorrhoeae strains harboring this novel combination of resistance determinants. These strains were isolated at two independent jurisdictions participating in the Gonococcal Isolate Surveillance Project (GISP) and in the Strengthening the U.S. Response to Resistant Gonorrhea (SURRG) project. The data suggest that the genome of N. gonorrhoeae continues to shuffle its genetic material. These findings further illuminate the genomic plasticity of N. gonorrhoeae, which allows this pathogen to develop mutations to escape the inhibitory effects of antibiotics.
INTRODUCTION
Antimicrobial agents play a critical role in the fight against gonorrhea, a sexually transmitted disease caused by Neisseria gonorrhoeae. The use of antibiotics for gonorrhea management began in the mid-1930s (1–3). This date also marked the dawn of a perpetual battle against antibiotic-resistant N. gonorrhoeae (AR-Ng). Over the last 80+ years, N. gonorrhoeae has progressively developed resistance to all first-line antibiotics following their introduction (1). The therapeutic longevity of antibiotics is inversely dependent to the malleability of the genetics of the pathogen (3). Neisseria gonorrhoeae demonstrates a high degree of genetic plasticity and uses this plasticity as a mechanism to develop antimicrobial resistance.
In an effort to restrain the emergence and spread of AR-Ng, the United States and other countries adopted a combination therapy strategy consisting of ceftriaxone as a primary therapy and azithromycin as a secondary agent (4). In spite of this effort, some N. gonorrhoeae isolates continue to defy therapeutic intervention and evade the effect of antibiotics (5). Gonococcal isolates displaying reduced susceptibility to ceftriaxone and/or azithromycin have emerged around the globe (3, 6–10). In the United States, the percentage of isolates with reduced cephalosporin susceptibility declined after 2012; however, the number and percentage of gonococcal isolates with reduced susceptibility to azithromycin have steadily increased since then (11). The increase in the number of N. gonorrhoeae isolates displaying elevated MICs to azithromycin is typically associated with the accumulation of genetic mutations in the multiple transferable resistance (mtr) locus or in the 23S rRNA gene (10, 12–14).
The substitution of a nongonococcal mtr locus (mosaic-mtr) of a closely related Neisseria species into a N. gonorrhoeae strain can increase its azithromycin MIC to up to 2 μg/ml (13). The decrease in azithromycin sensitivity is more pronounced in N. gonorrhoeae isolates harboring 23S rRNA gene mutations. An A-to-G substitution at nucleotide position 2059 (Escherichia coli designation) and a C-to-T substitution at position 2611 (E. coli designation) in the N. gonorrhoeae 23S rRNA gene confer reduced susceptibility to azithromycin, with in vitro MICs up to >256 μg/ml and 16 μg/ml, respectively (15).
Mosaic-mtr substitutions and 23S rRNA mutation-mediated azithromycin resistance mechanisms rarely occur simultaneously in the same N. gonorrhoeae isolates. So far, only five N. gonorrhoeae isolates in the United States have been reported to contain the mosaic-mtr and C2611T haplotype (13). Also, to our knowledge, the combination of a mosaic-mtr and A2059G haplotype has not been reported for N. gonorrhoeae strains in the United States or elsewhere in the world to date. In this paper, we report the emergence of a novel haplotype carrying a mosaic-mtr and A2059G 23S rRNA in 14 N. gonorrhoeae isolates recovered recently by two independent jurisdictions participating in the Gonococcal Isolate Surveillance Project (GISP) and the Strengthening the U.S. Response to Resistant Gonorrhea (SURRG).
RESULTS
All 14 N. gonorrhoeae isolates shared a similar antibiogram and a high-level resistance to azithromycin.
The N. gonorrhoeae isolates were collected from patients who presented at health clinics participating in the national gonococcal surveillance program established by the Centers for Disease Control and Prevention (CDC). Clinics participating in the gonococcal surveillance program routinely culture N. gonorrhoeae for antimicrobial susceptibility testing (AST). Between August 2017 and August 2018, 14 isolates from urethral (n = 10), pharyngeal (n = 2), and rectal (n = 2) samples were determined to display high-level resistance to azithromycin through routine AST. These isolates were cultured from 12 patients at two clinics in two different cities. Two urethral and pharyngeal isolate pairs were recovered from two patients. Initial MICs obtained using Etest by the local public health labs (SURRG) on all 14 isolates were ≥256 μg/ml for azithromycin and 0.016 μg/ml for both cefixime and ceftriaxone (Table 1). The MICs were later confirmed at the AR Lab Network using agar dilution and at the CDC using both AST methods. The CDC and AR Lab Network labs also assessed the susceptibilities of these isolates to ciprofloxacin, gentamicin, penicillin, and tetracycline using agar dilution (Table 1). The MICs for these antibiotics were within one doubling dilution from each other by either AST method.
TABLE 1.
Modal MICs were determined using both Etest and agar dilution MICsa
| Isolate IDb | WGS ID | Isolate source | Antibiogram modal MIC (μg/ml) by drugc |
23S rRNA mutation |
mtr locus |
Ribosomal gene present |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AZI | CRO | CFX | CIP | PEN | GEN | TET | A2059G | C2611T | Mosaic-mtr | mtr120 | ΔAmtrR-p | G45D | H105Y | rplD | rplV | |||
| 3001042919 | LRRBGS_0305 | Rectum | >256 | 0.016 | 0.016 | 0.015 | 1 | 4 | 2 | 4/4 | No | Yes | C | No | No | No | No | No |
| 3001159873 | LRRBGS_0645 | Urethra | >256 | 0.016 | 0.016 | 0.016 | 0.5 | 8 | 2 | 4/4 | No | Yes | C | No | No | No | No | No |
| 3001159874 | LRRBGS_0646 | Pharynx | >256 | 0.016 | 0.016 | 0.016 | 0.5 | 8 | 2 | 4/4 | No | Yes | C | No | No | No | No | No |
| 3001038282 | GCWGS_2319 | Pharynx | >256 | 0.016 | 0.016 | 0.016 | 0.5 | 8 | 2 | 4/4 | No | Yes | C | No | No | No | No | No |
| 3001038283 | GCWGS_2318 | Urethra | >256 | 0.016 | 0.016 | 0.016 | 0.5 | 8 | 2 | 4/4 | No | Yes | C | No | No | No | No | No |
| 3001043590 | GCWGS_1523 | Urethra | >256 | 0.016 | 0.016 | 0.016 | 0.25 | 8 | 4 | 4/4 | No | Yes | C | No | No | No | No | No |
| 3001160582 | GCWGS_1822 | Urethra | >256 | 0.016 | 0.016 | 0.016 | 0.5 | 4 | 2 | 4/4 | No | Yes | C | No | No | No | No | No |
| 3001404582 | GCWGS_1833 | Urethra | >256 | 0.016 | 0.016 | 0.016 | 0.5 | 4 | 2 | 4/4 | No | Yes | C | No | No | No | No | No |
| 3001160639 | GCWGS_2471 | Urethra | >256 | 0.016 | 0.016 | 0.016 | 0.5 | 4 | 4 | 4/4 | No | Yes | C | No | No | No | No | No |
| 3001044133 | GCWGS_2472 | Rectum | >256 | 0.016 | 0.016 | 0.032 | 1 | 8 | 2 | 4/4 | No | Yes | C | No | No | No | No | No |
| 3001160716 | GCWGS_2475 | Urethra | >256 | 0.016 | 0.016 | 0.016 | 0.5 | 4 | 2 | 4/4 | No | Yes | C | No | No | No | No | No |
| 3001521670 | GCWGS_2582 | Urethra | >256 | 0.016 | 0.016 | 0.016 | 0.5 | 4 | 2 | 4/4 | No | Yes | C | No | No | No | No | No |
| 3001160833 | GCWGS_2706 | Urethra | >256 | 0.016 | 0.016 | 0.016 | 0.5 | 4 | 2 | 4/4 | No | Yes | C | No | No | No | No | No |
| 3001521660 | GCWGS_3184 | Urethra | >256 | 0.016 | 0.016 | 0.016 | 0.5 | 8 | 4 | 4/4 | No | Yes | C | No | No | No | No | No |
Azithromycin Etest MICs are reported; the maximum concentration for an azithromycin Etest strip is 256 μg/ml. Mutational analyses were achieved through Sanger sequencing and WGS.
ID, identifier.
AZI, azithromycin; CRO, ceftriaxone; CFX, cefixime; CIP, ciprofloxacin; PEN, penicillin; GEN, gentamicin; TET, tetracycline.
All 14 N. gonorrhoeae isolates harbored a novel combination of azithromycin resistance markers.
The DNA sequences of the 23S rRNA genes and the mtr locus were analyzed to determine whether these resistance mechanisms are present in these isolates and may explain the observed high level of azithromycin resistance. Across these loci, all 14 isolates had identical sequences. Analysis of the 23S rRNA gene revealed a single-nucleotide aberration, an A-to-G transversion mutation across all four alleles, at position 2059. No mutations were found at the 2611 position of the 23S rRNA gene (Table 1).
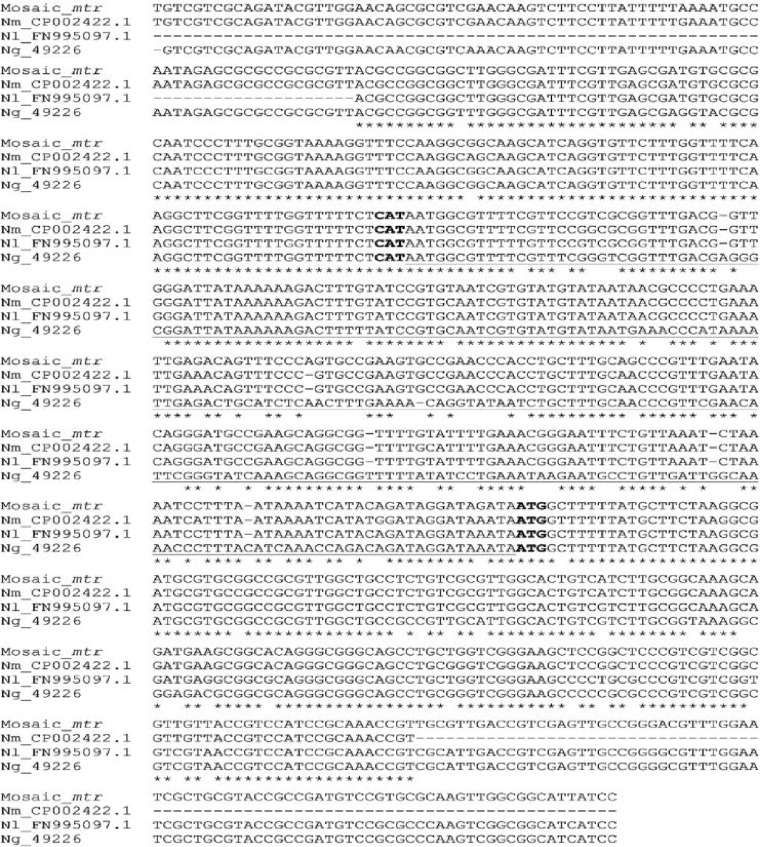
DNA analysis of the mtr locus revealed evidence of exogenous DNA integration at this site (Table 1). In all 14 isolates, the partially analyzed mtrC and mtrR genes and their associated promoter regions were replaced with nongonococcal counterparts (mosaic-mtr). The DNA sequence of this fragment (706 bp) showed high identity to N. meningitidis and Neisseria lactamica, at 97% and 96%, respectively (Fig. 1).
FIG 1.
Multiple-sequence alignment using CLUSTALW demonstrated that the gonococcal mosaic-mtr shared 97% and 96% sequence identity to N. meningitidis (Nm_CP002422.1) and N. lactamica (Nl_FN995097.1) mtr, respectively. In contrast, the mosaic-mtr shared 86% sequence identity to the mtr of the gonococcal strain ATCC 49226 (Ng_49226). The mtr promoter is underlined. The partial mtrR and mtrC genes flanking the mtr promoter are at the 3′ and 5′ ends, respectively. The start codons for the mtrR and mtrC genes are in bold.
Whole-genome sequencing (WGS) analysis confirmed the results of 23S rRNA (A2059G) mutation and a mosaic-mtr locus obtained by PCR and Sanger sequencing, and it revealed that the mosaicism seen in the mtr locus extended through a large part of the entire mtrCDE operon (data not shown). WGS analysis, however, did not reveal a −35 delA in the mtr promoter (ΔAmtrR-p) nor any additional nonsynonymous amino acid mutations in the mtrR gene (G45D and H105Y) or in the rplV and rplD ribosomal genes, mutations previously reported to have an influence on azithromycin MICs (Table 1) (10).
DISCUSSION
The sustained increase in the number of N. gonorrhoeae isolates with elevated MICs for azithromycin around the world in recent years threatens the effectiveness of azithromycin as part of the current treatment regimen (3, 6). In recent years, many countries have observed an increase in the prevalence of reduced susceptibility to azithromycin in N. gonorrhoeae. The percentage of isolates displaying an elevated MIC to azithromycin ranged from 4.7% to 32% in 2014 to 2017 (3, 6–10). Most of the isolates with elevated azithromycin MICs can be associated with mutations in either the 23S rRNA gene or the mtr locus (15). Moreover, an A-to-G transversion mutation at nucleotide position 2059 in the 23S rRNA can lead to high-level azithromycin resistance (i.e., MIC, ≥256 μg/ml).
In our study, we investigated 14 N. gonorrhoeae isolates displaying an MIC of ≥256 μg/ml for azithromycin. These 14 isolates were susceptible to ceftriaxone, cefixime, and ciprofloxacin but demonstrated intermediate resistance to penicillin and resistance to tetracycline. DNA sequence analysis revealed that all 14 isolates harbored the mosaic-mtr and the A2059G mutation in all four copies of their 23S rRNA gene. While investigators from many countries have reported on the A2059G mutation or the mosaic-mtr, none described these two haplotypes together (5, 10, 12–14, 16–25). The 14 isolates in this study are the first known isolates to have possessed both the A2059G mutation and the mosaic-mtr.
The dual azithromycin-attenuating mutations found in these 14 isolates were likely acquired through spontaneous and intragenus recombination. Genetic modification to gain antimicrobial resistance is a common phenomenon in N. gonorrhoeae (13, 15, 24). The A2059G point mutation in the 23S rRNA was likely acquired through spontaneous mutation. Chisholm et al. demonstrated that a single-allelic mutation at this residue could further transfer to the wild-type 23S rRNA alleles via endogenous homologous recombination under selection pressure (15). Conversely, the “mosaic” allele was likely acquired from other neisserial species through intragenus recombination (13, 24, 25). DNA sequences of mtr found in these 14 isolates shared high genetic identity to those of N. meningitidis and N. lactamica (Fig. 1). Furthermore, Wadsworth et al. demonstrated that the mtr from these two neisserial species, when transformed into N. gonorrhoeae, could recapitulate the azithromycin susceptibility pattern elicited by mosaic-mtr (13).
Although the A2059A mutation confers resistance to azithromycin, it has been posited that this same mutation exerts a fitness cost on N. gonorrhoeae isolates (14). This may explain why isolates with this genotype failed to transmit and persist in the population. In contrast, it has been speculated that mutations in the mtr locus may result in overexpression of the mtrCDE efflux pump and may increase fitness in N. gonorrhoeae (26, 27). Interestingly, the mosaic-mtr has been shown to increase MtrCDE protein expression in N. gonorrhoeae (13, 28). Thus, the A2059G-mosaic-mtr genotype might be more fit than the A2059G-alone genotype and perhaps is better able to sustain transmission of a gonococcal strain that confers high-level azithromycin resistance. Future exploration is needed to investigate this hypothesis.
The treatment and control strategies for N. gonorrhoeae infections have constantly evolved because of the organism’s ability to overcome the growth-inhibitory effect of first-line antibiotics. This bacterium develops resistance to antimicrobials at a high rate (1). As a result, the only recommended treatment against uncomplicated gonorrhea is dual therapy with an extended-spectrum cephalosporin (ceftriaxone) plus azithromycin. The ability to evade the effects of antibiotics displayed by N. gonorrhoeae is an attribute of the promiscuity in DNA uptake and the pliable nature of this bacterium’s genome to integrate exogenous DNA and/or its ability to develop mutations de novo. With the emergence of isolates possessing the dual azithromycin-attenuating genotype, A2059G-mosaic-mtr, N. gonorrhoeae isolates might have a mechanism to both harbor resistance and sustain transmission. More research on the fitness cost of these strains could further elucidate these hypotheses.
MATERIALS AND METHODS
Neisseria gonorrhoeae culture.
Gonococcal isolates were obtained through the national surveillance programs GISP (http://www.cdc.gov/std/gisp/) and SURRG. All isolates were propagated on GC medium base agar supplemented with 1% hemoglobin and/or 1% IsoVitaleX (BD, Franklin Lakes, NJ, USA), and were incubated at 36 ± 1°C and 5% CO2. Species identification of the gonococcal isolates was confirmed using matrix-assisted laser desorption ionization–time of flight mass spectrometry (Bruker Daltonics, Boston, MA, USA).
Antimicrobial susceptibility testing.
The MICs of all 14 N. gonorrhoeae isolates were assessed using the agar dilution and the Etest antimicrobial susceptibility testing (AST) methods, as described previously (29). MICs for cefixime, ceftriaxone, azithromycin, ciprofloxacin, gentamicin, penicillin, and tetracycline were determined for all isolates. Etest strips were purchased from bioMérieux (Durham, NC, USA), and antibiotic powders were purchased from Sigma-Aldrich (St. Louis, MO, USA). The susceptibility interpretations were based on CLSI interpretation guidelines (30) when available.
Molecular analysis of the 23S rRNA genes and the mtr locus.
(i) Sanger DNA sequencing. PCR and Sanger sequencing were employed to analyze the DNA sequence of the 23S rRNA genes as well as the promoter and coding regions of the mtrR and mtrC genes. Total genomic DNA was purified using the QIAamp DNA minikit, as described by the manufacturer (Qiagen, Valencia, CA, USA). Amplification and sequencing of the four 23S rRNA alleles were achieved using the primers previously described by Ng et al. (31). In brief, an allele-specific primer was paired with the gonrRNA-F primer to amplify each allele. Then, each allele-amplicon was sequenced bidirectionally using the gonrRNA-F and gonrRNA-R2 primers. The amplification of the partial mtrR and mtrC genes and their promoters was carried out using the forward primer NGmtrC-F (5′-TGT CGA TCT GAT ACA GCG GC-3′) and the previously described reverse primer MTR2 (32). The mtrR amplicon was pair-end sequenced with the NGmtrC-F and NGmtrC-R (5′-CCA AGA ACC TCC TTC GGC AT-3′) and the MTR1339 (33) and MTR2 primer pairs. The conditions for PCR and DNA sequencing were previously described (34).
(ii) Whole-genome sequencing. Whole-genome sequencing of N. gonorrhoeae isolates was performed either at the Tennessee Department of Health Laboratory or at the CDC. DNA extraction, library preparation, and Illumina-based sequencing were performed according to a standardized protocol (https://www.cdc.gov/pulsenet/pdf/PNL32-MiSeq-Nextera-XT.pdf).
Bioinformatics analysis. DNA sequences acquired through Sanger sequencing were visualized and processed using the Geneious program (Biomatters Ltd., San Francisco, CA, USA). The DNA sequences of the 23S rRNA gene and the mtr region from the N. gonorrhoeae strain ATCC 49226 were used as references. The mtr sequence identity was determined using BLASTn (https://blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastn&PAGE_TYPE=BlastSearch&LINK_LOC=blasthome). The mtr sequences of Neisseria meningitidis (GenBank accession no. CP002422.1), Neisseria lactamica (GenBank accession no. FN995097.1), N. gonorrhoeae (ATCC 49226), and the gonococcal mosaic-mtr were also aligned using CLUSTALW (https://www.genome.jp/tools-bin/clustalw).
WGS data were analyzed using a custom script written in Python version 3.6. Raw reads in FASTQ format were used as input for analysis on a per-isolate basis. The raw reads were first analyzed by FastQC version 0.11.5 (35) and then filtered and trimmed using Trim Galore version 0.3.7 (36) with the command line options –q 30 –length 50 –retain_unpaired and posttrimming FastQC. The output from Trim Galore was used as the input both de novo assembly using SPAdes version 3.7.0 with default settings and for reference mapping using breseq version 0.30 (37) with N. gonorrhoeae FA19 (GenBank accession no. CP012026) as the reference genome. In order to extract variants in the Neisseria gonorrhoeae 23S rRNA and count the number of copies in the genome, breseq was run a second time using the sequence for a single allele of the 23S rRNA gene from strain MS11 as the mapping reference (GenBank accession no. X67293) with the polymorphism-prediction option turned on. The de novo genome assembly and the output from breseq were used for the next steps in the analysis pipeline, which uses the PANDAS version 0.22 and Biopython (38) packages to extract variants and format the data.
ACKNOWLEDGMENTS
The members of the SURRG Working Group are Robert D. Kirkcaldy (CDC), Henrietta Hardin (Tennessee Department of Health), George Merkison (Mario County Public Health Department), Evelyn Nash (CDC), Kevin Pettus (CDC), Jeanine A. McLean (CDC), Robert P. Kohn (San Francisco Department of Public Health), and Kim M. Gernert (CDC).
We thank the Antimicrobial Exchange Branch (ARX), the GC AR Lab Network, the GISP, and the SURRG grantees for their contributions to N. gonorrhoeae antimicrobial resistance surveillance.
This study was funded by the CDC, partially with funds from the Combating Antibiotic-Resistant Bacteria (CARB) initiative.
The findings and conclusions of this article are those of the authors and do not necessarily represent the views of the Centers for Disease Control and Prevention.
REFERENCES
- 1.Unemo M. 2015. Current and future antimicrobial treatment of gonorrhoea–the rapidly evolving Neisseria gonorrhoeae continues to challenge. BMC Infect Dis 21:364. doi: 10.1186/s12879-015-1029-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hook EW III, Kirkcaldy RD. 2018. A brief history of evolving diagnostics and therapy for gonorrhea: lessons learned. Clin Infect Dis 67:1294–1299. doi: 10.1093/cid/ciy271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Whittles LK, White PJ, Paul J, Didelot X. 2018. Epidemiological trends of antibiotic resistant gonorrhoea in the United Kingdom. Antibiotics (Basel) 7:60. doi: 10.3390/antibiotics7030060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Workowski KA, Bolan GA. 2015. Sexually transmitted diseases treatment guidelines, 2015. MMWR Recomm Rep 2015 64:1–137. [PMC free article] [PubMed] [Google Scholar]
- 5.Eyre DW, Sanderson ND, Lord E, Regisford-Reimmer N, Chau K, Barker L, Morgan M, Newnham R, Golparian D, Unemo M, Crook DW, Peto TE, Hughes G, Cole MJ, Fifer H, Edwards A, Andersson MI. 2018. Gonorrhoea treatment failure caused by a Neisseria gonorrhoeae strain with combined ceftriaxone and high-level azithromycin resistance, England, February 2018. Euro Surveill 23:pii=1800323. doi: 10.2807/1560-7917.ES.2018.23.27.1800323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barbee LA, Soge OO, Katz DA, Dombrowski JC, Holmes KK, Golden MR. 2018. Increases in Neisseria gonorrhoeae with reduced susceptibility to azithromycin among men who have sex with men in Seattle, King County, Washington, 2012–2016. Clin Infect Dis 66:712–718. doi: 10.1093/cid/cix898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bazan JA, Williams Roberts M, Soge OO, Torrone EA, Dennison A, Ervin M, Hun S, Fields KS, Turner AN. 2018. Rapid increase in gonorrhea cases with reduced susceptibility to azithromycin in Columbus, Ohio. Sex Transm Dis 45:e5–e6. doi: 10.1097/OLQ.0000000000000765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buder S, Dudareva S, Jansen K, Loenenbach A, Nikisins S, Sailer A, Guhl E, Kohl PK, Bremer V. 2018. GORENET study group antimicrobial resistance of Neisseria gonorrhoeae in Germany: low levels of cephalosporin resistance, but high azithromycin resistance. BMC Infect Dis 18:44. doi: 10.1186/s12879-018-2944-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ryan L, Golparian D, Fennelly N, Rose L, Walsh P, Lawlor B, Mac Aogáin M, Unemo M, Crowley B. 2018. Antimicrobial resistance and molecular epidemiology using whole-genome sequencing of Neisseria gonorrhoeae in Ireland, 2014–2016: focus on extended-spectrum cephalosporins and azithromycin. Eur J Clin Microbiol Infect Dis 37:1661–1672. doi: 10.1007/s10096-018-3296-5. [DOI] [PubMed] [Google Scholar]
- 10.Wan C, Li Y, Le WJ, Liu YR, Li S, Wang BX, Rice PA, Su XH. 2018. Increasing resistance to azithromycin in Neisseria gonorrhoeae in eastern Chinese cities: resistance mechanisms and genetic diversity among isolates from Nanjing. Antimicrob Agents Chemother 62:e02499-17. doi: 10.1128/AAC.02499-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Centers for Disease Control and Prevention. 2016. Gonococcal Isolate Surveillance Project (GISP) profiles, Figure 3. Centers for Disease Control and Prevention, Atlanta, GA: https://www.cdc.gov/std/stats16/gisp2016/default.htm. [Google Scholar]
- 12.Wind CM, Bruisten SM, Schim van der Loeff MF, Dierdorp M, de Vries HJC, van Dam AP. 2017. A case-control study of molecular epidemiology in relation to azithromycin resistance in Neisseria gonorrhoeae isolates collected in Amsterdam, the Netherlands, between 2008 and 2015. Antimicrob Agents Chemother 61:e02374-16. doi: 10.1128/AAC.02374-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wadsworth CB, Arnold BJ, Sater MRA, Grad YH. 2018. Azithromycin resistance through interspecific acquisition of an epistasis-dependent efflux pump component and transcriptional regulator in Neisseria gonorrhoeae. mBio 9:e01419-18. doi: 10.1128/mBio.01419-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fifer H, Cole M, Hughes G, Padfield S, Smolarchuk C, Woodford N, Wensley A, Mustafa N, Schaefer U, Myers R, Templeton K, Shepherd J, Underwood A. 2018. Sustained transmission of high-level azithromycin-resistant Neisseria gonorrhoeae in England: an observational study. Lancet Infect Dis 18:573–581. doi: 10.1016/S1473-3099(18)30122-1. [DOI] [PubMed] [Google Scholar]
- 15.Chisholm SA, Dave J, Ison CA. 2010. High-level azithromycin resistance occurs in Neisseria gonorrhoeae as a result of a single point mutation in the 23S rRNA genes. Antimicrob Agents Chemother 54:3812–3816. doi: 10.1128/AAC.00309-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Galarza PG, Alcalá B, Salcedo C, Canigia LF, Buscemi L, Pagano I, Oviedo C, Vázquez JA. 2009. Emergence of high level azithromycin-resistant Neisseria gonorrhoeae strain isolated in Argentina. Sex Transm Dis 36:787–788. doi: 10.1097/OLQ.0b013e3181b61bb1. [DOI] [PubMed] [Google Scholar]
- 17.Katz AR, Komeya AY, Soge OO, Kiaha MI, Lee MV, Wasserman G, Maningas EV, Whelen AC, Kirkcaldy RD, Shapiro SJ, Bolan GA, Holmes KK. 2012. Neisseria gonorrhoeae with high-level Resistance to azithromycin; case report of the first isolate identified in the United States. Clin Infect Dis 54:841–843. doi: 10.1093/cid/cir929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Unemo M, Golparian D, Hellmark B. 2014. First three Neisseria gonorrhoeae isolates with high-level resistance to azithromycin in Sweden: a threat to currently available dual-antimicrobial regimens for treatment of gonorrhea? Antimicrob Agents Chemother 58:624–625. doi: 10.1128/AAC.02093-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Allen VG, Seah C, Martin I, Melano RG. 2014. Azithromycin resistance is coevolving with reduced susceptibility to cephalosporins in Neisseria gonorrhoeae in Ontario, Canada. Antimicrob Agents Chemother 58:2528–2534. doi: 10.1128/AAC.02608-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stevens K, Zaia A, Tawil S, Bates J, Hicks V, Whiley D, Limnios A, Lahra MM, Howden BP. 2015. Neisseria gonorrhoeae isolates with high-level resistance to azithromycin in Australia. J Antimicrob Chemother 70:1267–1268. doi: 10.1093/jac/dku490. [DOI] [PubMed] [Google Scholar]
- 21.Demczuk W, Martin I, Peterson S, Bharat A, Van Domselaar G, Graham M, Lefebvre B, Allen V, Hoang L, Tyrrell G, Horsman G, Wylie J, Haldane D, Archibald C, Wong T, Unemo M, Mulvey MR. 2016. Genomic epidemiology and molecular resistance mechanisms of azithromycin-resistant Neisseria gonorrhoeae in Canada from 1997 to 2014. J Clin Microbiol 54:1304–1313. doi: 10.1128/JCM.03195-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jacobsson S, Golparian D, Cole M, Spiteri G, Martin I, Bergheim T, Borrego MJ, Crowley B, Crucitti T, Van Dam AP, Hoffmann S, Jeverica S, Kohl P, Mlynarczyk-Bonikowska B, Pakarna G, Stary A, Stefanelli P, Pavlik P, Tzelepi E, Abad R, Harris SR, Unemo M. 2016. WGS analysis and molecular resistance mechanisms of azithromycin-resistant (MIC >2 mg/L) Neisseria gonorrhoeae isolates in Europe from 2009 to 2014. J Antimicrob Chemother 71:3109–3116. doi: 10.1093/jac/dkw279. [DOI] [PubMed] [Google Scholar]
- 23.Smolarchuk C, Wensley A, Padfield S, Fifer H, Lee A, Hughes G. 2018. Persistence of an outbreak of gonorrhoea with high-level resistance to azithromycin in England, November 2014–May 2018. Euro Surveill 23:pii=1800287. doi: 10.2807/1560-7917.ES.2018.23.23.1800287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Trembizki E, Doyle C, Jennison A, Smith H, Bates J, Lahra M, Whiley D, GRAND Study Investigators. 2014. A Neisseria gonorrhoeae strain with a meningococcal mtrR sequence. J Med Microbiol 63:1113–1115. doi: 10.1099/jmm.0.074286-0. [DOI] [PubMed] [Google Scholar]
- 25.Whiley DM, Kundu RL, Jennison AV, Buckley C, Limnios A, Hogan T, Enriquez R, El Nasser J, George CR, Lahra MM. 2018. Azithromycin-resistant Neisseria gonorrhoeae spreading amongst men who have sex with men (MSM) and heterosexuals in New South Wales, Australia, 2017. J Antimicrob Chemother 17:148–145. doi: 10.1093/jac/dky017. [DOI] [PubMed] [Google Scholar]
- 26.Ohneck EA, Zalucki YM, Johnson PJ, Dhulipala V, Golparian D, Unemo M, Jerse AE, Shafer WM. 2011. A novel mechanism of high-level, broad-spectrum antibiotic resistance caused by a single base pair change in Neisseria gonorrhoeae. mBio 2:e00187-11. doi: 10.1128/mBio.00187-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ohneck EA, Goytia M, Rouquette-Loughlin CE, Joseph SJ, Read TD, Jerse AE, Shafer WM. 2015. Overproduction of the MtrCDE efflux pump in Neisseria gonorrhoeae produces unexpected changes in cellular transcription patterns. Antimicrob Agents Chemother 59:724–726. doi: 10.1128/AAC.04148-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rouquette-Loughlin CE, Reimche JL, Balthazar JT, Dhulipala V, Gernert KM, Kersh EN, Pham CD, Pettus K, Abrams AJ, Trees DL, St. Cyr S, Shafer WM. 2018. Mechanistic basis for decreased antimicrobial susceptibility in a clinical isolate of Neisseria gonorrhoeae possessing a mosaic-like mtr efflux pump locus. mBio 9:e02281-18. doi: 10.1128/mBio.02281-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu H, Taylor TH Jr, Pettus K, Trees D. 2014. Assessment of Etest as an alternative to agar dilution for antimicrobial susceptibility testing of Neisseria gonorrhoeae. J Clin Microbiol 52:1435–1440. doi: 10.1128/JCM.02131-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Clinical and Laboratory Standards Institute. 2017. Performance standards for antimicrobial susceptibility testing, 27th ed. CLSI Supplement M100. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 31.Ng LK, Martin I, Liu G, Bryden L. 2002. Mutation in 23S rRNA associated with macrolide resistance in Neisseria gonorrhoeae. Antimicrob Agents Chemother 46:3020–3025. doi: 10.1128/AAC.46.9.3020-3025.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mavroidi A, Tzouvelekis LS, Kyriakis KP, Avgerinou H, Daniilidou M, Tzelepi E. 2001. Multidrug-resistant strains of Neisseria gonorrhoeae in Greece. Antimicrob Agents Chemother 45:2651–2654. doi: 10.1128/AAC.45.9.2651-2654.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Johnson SR, Sandul AL, Parekh M, Wang SA, Knapp JS, Trees DL. 2003. Mutations causing in vitro resistance to azithromycin in Neisseria gonorrhoeae. Int J Antimicrob Agents 21:414–419. doi: 10.1016/S0924-8579(03)00039-6. [DOI] [PubMed] [Google Scholar]
- 34.Pham CD, Purfield AE, Fader R, Pascoe N, Lockhart SR. 2015. Development of a multilocus sequence typing system for medically relevant Bipolaris species. J Clin Microbiol 53:3239–3246. doi: 10.1128/JCM.01546-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Andrews S. 2010. FastQC: a quality control tool for high throughput sequence data. http://www.bioinformatics.babraham.ac.uk/projects/fastqc.
- 36.Krueger F. 2012. Trim Galore: a wrapper tool around Cutadapt and FastQC to consistently apply quality and adapter trimming to FastQ files. http://www.bioinformatics.babraham.ac.uk/projects/trim_galore.
- 37.Deatherage DE, Barrick JE. 2014. Identification of mutations in laboratory-evolved microbes from next-generation sequencing data using breseq. Methods Mol Biol 1151:165–188. doi: 10.1007/978-1-4939-0554-6_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cock PA, Antao T, Chang JT, Chapman BA, Cox CJ, Dalke A, Friedberg I, Hamelryck T, Kauff F, Wilczynski B., de Hoon MJL. 2009. Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics 25:1422–1423. doi: 10.1093/bioinformatics/btp163. [DOI] [PMC free article] [PubMed] [Google Scholar]