There is an urgent need for new therapies to overcome antimicrobial resistance especially in Gram-negative bacilli (GNB). Repurposing old U.S.
KEYWORDS: adjuvants, antibiotic resistance, Gram-negative bacteria
ABSTRACT
There is an urgent need for new therapies to overcome antimicrobial resistance especially in Gram-negative bacilli (GNB). Repurposing old U.S. Food and Drug Administration-approved drugs as complementary agents to existing antibiotics in a synergistic combination presents an attractive strategy. Here, we demonstrate that the anthelmintic drug niclosamide selectively synergized with the lipopeptide antibiotic colistin against colistin-susceptible but more importantly against colistin-resistant GNB, including clinical isolates that harbor the mcr-1 gene. Breakpoints for colistin susceptibility in resistant Gram-negative bacilli were reached in the presence of 1 μg/ml (3 μM) niclosamide. Reversal of colistin resistance was also observed in combinations of niclosamide and polymyxin B. Enhanced bacterial killing was evident for the combination, in comparison to colistin monotherapy, against resistant Pseudomonas aeruginosa, Acinetobacter baumannii, Klebsiella pneumoniae, Escherichia coli, and Enterobacter cloacae. Accumulating evidence in the literature, along with our results, strongly suggests the potential for the combination of niclosamide and colistin to treat colistin-resistant Gram-negative bacillary infections. Our finding is significant since colistin is an antibiotic of last resort for multidrug-resistant Gram-negative bacterial infections that are nonresponsive to conventional treatments. With the recent global dissemination of plasmid-encoded colistin resistance, the addition of niclosamide to colistin therapy may hold the key to overcome colistin resistance.
INTRODUCTION
The alarming threat of antimicrobial resistance has emerged as a worldwide problem, where the evolution of resistant microbes has seemingly outpaced the development of new antibiotics (1, 2).The rapid spread and dissemination of resistance-encoding genes among pathogens further exacerbate the problem, resulting in an ever-increasing rate of antimicrobial-resistant bacterial infections (3–5). Infections caused by Gram-negative bacilli (GNB) are arguably more difficult to treat since they are intrinsically resistant to many antibiotics and therefore often have limited options for therapy (6, 7).
Intrinsic resistance in GNB is comprised of their two protective (inner and outer) membranes and overexpressed efflux systems. These resistance mechanisms effectively prevent antibiotics from achieving an optimal intracellular concentration to elicit therapeutic activity. Moreover, adaptive resistance is common among GNB, in which altered genetic expression to confer fitness in the presence of either certain bacterial load threshold or suboptimal antibiotic concentration is observed (8–10). This coordinated intercellular bacterial response that involves the production, release, and group detection of signaling (autoinducer) molecules is called quorum sensing (QS) (11). Morphological changes, motility, biofilm formation, and secretion of virulence factors are among the bacterial processes orchestrated by QS. Quenching this communication network is an attractive strategy to attenuate pathogenicity of an infectious microbe (12, 13).
Recently, the U.S. Food and Drug Administration (FDA)-approved anthelmintic drug niclosamide (NIC) was discovered to inhibit QS in Pseudomonas aeruginosa by hindering the cell’s response and production of the autoinducers N-3-oxododecanoyl-homeserine lactone and N-butanoyl-homoserine lactone (14). The quenching of QS by niclosamide phenotypically impeded bacterial motility, biofilm formation, and secretion of the virulence factors elastase, pyocyanin, and rhamnolipids (14). More importantly, administration of niclosamide prior to injection of P. aeruginosa at lethal dose was shown to protect Galleria mellonella larvae from mortality (14). The antivirulence effects of niclosamide in P. aeruginosa, combined with its established safety profile, as an agent to treat parasitic worm infections (15) suggest that it could be potentially repurposed for GNB infections. However, it should be noted that the fundamental concept of anti-QS therapy, to disarm the pathogen of its ability to cause disease in vivo, has yet to gain clinical support (12, 16). We questioned whether niclosamide may be used in combination with an antibiotic to treat GNB infections. Here, we disclose the potent synergistic combination of niclosamide and the lipopeptide drug colistin (COL), but also polymyxin B, against clinically relevant multidrug-resistant (MDR) GNB. While it was previously reported that niclosamide can eradicate Gram-positive bacteria (such as methicillin-resistant Staphylococcus aureus and Enterococcus faecium) (17), we confirmed that niclosamide possessed poor antibacterial activity against GNB. However, niclosamide synergized with colistin to eradicate colistin-susceptible but, more importantly, colistin-resistant Gram-negative bacterial isolates, including two Escherichia coli clinical isolates (18) that harbor the mcr-1 gene. Furthermore, the combination demonstrated an enhanced bacterial killing against resistant GNB in comparison to colistin monotherapy. This finding is timely and relevant since the global dissemination of plasmid-encoded colistin resistance had been realized (19, 20) and there is an urgent need for new therapeutic strategies to combat resistance to this antibiotic of last resort.
RESULTS AND DISCUSSION
Niclosamide selectively synergized with colistin against P. aeruginosa.
We evaluated potential synergism of niclosamide with 31 clinically used antibiotics against wild-type P. aeruginosa PAO1 strain (Table 1 ). The antibiotics tested were chosen to represent a vast spectrum of antimicrobial classes, including surfactants and antiseptics, aminoglycosides, β-lactams and carbapenems, fluoroquinolones, and those that are active only against Gram-positive bacteria. Niclosamide by itself displayed very poor activity with a MIC of 512 μg/ml against PAO1. Of the 31 antibiotics evaluated, niclosamide displayed selective synergy with colistin against P. aeruginosa PAO1 with a fractional inhibitory concentration (FIC) index of 0.127. Other well-known surfactants (benzalkonium chloride, benzethonium chloride, and cetrimonium bromide) did not exhibit synergism with niclosamide. Colistin is considered an antibiotic of last resort for a multidrug-resistant (MDR) and extensively drug-resistant (XDR) Gram-negative bacterial infection that is nonresponsive to conventional antibiotic therapy. The recent realization of plasmid-encoded polymyxin resistance gene dissemination such as of the mcr-1 (19), mcr-2 (21), and mcr-3 (22), among other genes, greatly limits the therapeutic usage of colistin (20, 23). Therefore, our initial finding that niclosamide synergized with colistin against wild-type P. aeruginosa PAO1 warranted further studies.
TABLE 1.
Scan for synergistic combination of niclosamide and various antibiotics against wild-type P. aeruginosa PAO1
| Antibiotic | MICniclosamide [MICcombo], μg/ml | MICantibiotic [MICcombo], μg/ml | FIC index | Interpretation |
|---|---|---|---|---|
| Colistin | 512 [1] | 1 [0.125] | 0.1270 | Synergy |
| Benzalkonium chloride | 512 [0.125] | 64 [64] | 1.0002 | Indifferent |
| Benzethonium chloride | 512 [0.125] | 32 [32] | 1.0002 | Indifferent |
| Cetrimonium bromide | 512 [0.125] | 128 [128] | 1.0002 | Indifferent |
| Tobramycin | 512 [0.25] | 0.5 [0.25] | 0.5005 | Indifferent |
| Gentamicin | 512 [0.125] | 1 [1] | 1.0002 | Indifferent |
| Amikacin | 512 [32] | 1 [0.5] | 0.5625 | Indifferent |
| Streptomycin | 512 [32] | 16 [8] | 0.5625 | Indifferent |
| Kanamycin | 512 [0.5] | 64 [32] | 0.5010 | Indifferent |
| Minocycline | 512 [0.25] | 16 [8] | 0.5005 | Indifferent |
| Doxycycline | 512 [64] | 8 [4] | 0.6250 | Indifferent |
| Piperacillin | 512 [0.25] | 4 [2] | 0.5005 | Indifferent |
| Aztreonam | 512 [0.5] | 4 [2] | 0.5010 | Indifferent |
| Ceftazidime | 512 [64] | 2 [1] | 0.6250 | Indifferent |
| Cefotaxime | 512 [1] | 16 [8] | 0.5019 | Indifferent |
| Meropenem | 512 [1] | 1 [0.5] | 0.5019 | Indifferent |
| Doripenem | 512 [0.125] | 0.5 [0.5] | 1.0002 | Indifferent |
| Ciprofloxacin | 512 [0.125] | 0.125 [0.125] | 1.0002 | Indifferent |
| Levofloxacin | 512 [0.125] | 0.25 [0.25] | 1.0002 | Indifferent |
| Moxifloxacin | 512 [64] | 1 [0.5] | 0.6250 | Indifferent |
| Rifampicin | 512 [0.125] | 16 [8] | 0.5002 | Indifferent |
| Erythromycin | 512 [32] | 256 [128] | 0.5625 | Indifferent |
| Nitrofurantoin | 512 [8] | 512 [256] | 0.5156 | Indifferent |
| Vancomycin | 512 [0.125] | 512 [256] | 0.5002 | Indifferent |
| Chloramphenicol | 512 [0.125] | 32 [16] | 0.5002 | Indifferent |
| Trimethoprim | 512 [0.125] | 128 [64] | 0.5002 | Indifferent |
| Novobiocin | 512 [0.125] | 512 [256] | 0.5002 | Indifferent |
| Clindamycin | 512 [1] | 1,024 [512] | 0.5019 | Indifferent |
| Linezolid | 512 [0.125] | 1,024 [1,024] | 1.0002 | Indifferent |
| Fosfomycin | 512 [8] | 16 [8] | 0.5156 | Indifferent |
| Pleuromutilin | 512 [16] | 512 [256] | 0.5312 | Indifferent |
The combination of niclosamide and colistin was then assessed against an extended panel of colistin-susceptible P. aeruginosa isolates (Table 2). The panel included a wild-type strain, two efflux-deficient mutants (PAO200 and PAO750), and seven MDR/XDR clinical isolates. Synergy was observed in most P. aeruginosa strains except against the wild-type ATCC 27853 and the MDR PA259-96918 strains. Indifferent interaction of the combination against the two P. aeruginosa strains may be due to genetic and/or phenotypic variances. More importantly, the MIC of colistin was potentiated up to 64-fold against tested P. aeruginosa strains (Table 2) at a fixed concentration of 1 μg/ml (3 μM) niclosamide. These data suggest the prospective role of niclosamide as an adjuvant partner for colistin in treatment of P. aeruginosa bacterial infection.
TABLE 2.
Synergy of niclosamide with colistin against wild-type, efflux pump-deletion mutant and MDR/XDR colistin-susceptible P. aeruginosaa
| P. aeruginosa strain | MICNIC [MICcombo], μg/ml | MICCOL [MICcombo], μg/ml | FIC index | Absolute MICCOL,b μg/ml | Potentiation (fold)c |
|---|---|---|---|---|---|
| PAO1d | 512 [1] | 1 [0.125] | 0.127 | 0.125 | 8 |
| PAO200e | >512 [4] | 0.5 [0.062] | 0.125 < x < 0.133 | 0.125 | 4 |
| PAO750f | 2 [0.25] | 0.5 [0.125] | 0.375 | 0.062 | 8 |
| PA095 | 256 [0.125] | 0.25 [0.008] | 0.032 | 0.004 | 64 |
| PA259-96918 | 512 [1] | 0.25 [0.125] | 0.502 | 0.125 | 2 |
| PA260-97103 | 512 [0.5] | 0.5 [0.125] | 0.251 | 0.125 | 4 |
| PA262-101856 | 512 [0.5] | 2 [0.125] | 0.063 | 0.125 | 16 |
| PA264-104354 | 512 [4] | 2 [0.125] | 0.070 | 0.5 | 4 |
| 100036 | 512 [0.125] | 1 [0.125] | 0.125 | 0.125 | 8 |
| 101885 | 512 [0.5] | 1 [≤0.062] | 0.001 < x ≤ 0.063 | 0.125 | 8 |
| ATCC 27853d | >64 [4] | 0.5 [0.25] | 0.500 < x < 0.562 | 0.5 | None |
MICCOL, colistin MIC; MICNIC, niclosamide MIC.
MIC of colistin in the presence of 1 μg/ml (3 μM) niclosamide.
Degree of colistin potentiation in the presence of 1 μg/ml (3 μM) niclosamide.
Wild type.
The MexAB-OprM deletion strain.
The efflux-deficient strain that lacks five clinically relevant pumps (MexAB-OprM, MexCD-OprJ, MexEF-OprN, MexJK, and MexXY) and the outer membrane protein OpmH.
Efflux affected niclosamide activity against P. aeruginosa.
In comparison to wild-type P. aeruginosa PAO1 (MIC = 512 μg/ml), niclosamide’s MIC was reduced significantly to 2 μg/ml against the efflux-deficient PAO750 strain (Table 2), which lacked five clinically relevant efflux pumps and the outer membrane protein H (OpmH). The MIC of niclosamide was unchanged against PAO200, which only lacked the MexAB-OprM tripartite efflux system, in comparison to PAO1. These data strongly suggests that niclosamide is a substrate of efflux systems in P. aeruginosa but not the MexAB-OprM pump, and that negation of active efflux allows the accumulation of niclosamide to effectively elicit its antibacterial property. Niclosamide is known to possess potent antibacterial activity against Gram-positive bacteria; the associated mode of action of this antibacterial activity is still unclear (17).
Cooperative effect of niclosamide and colistin against P. aeruginosa resulted in synergy.
Colistin’s MIC did not change between wild-type P. aeruginosa PAO1 and the efflux-deficient P. aeruginosa strains (Table 2), confirming that it is a poor substrate of efflux pumps, as previously reported (24, 25). Suggestively, colistin may partly potentiate niclosamide’s antibacterial activity by disrupting efflux in P. aeruginosa, at least against wild-type and colistin-susceptible strains. The membrane-acting colistin may potentially block efflux in several ways: (i) by sequestering lipid composition surrounding the transmembrane domain of efflux pumps resulting in a conformational shift and inactivity of the domain and/or (ii) by disturbing the inner membrane organization that ultimately dissipate the proton motive force responsible to energize efflux systems. On this note, synergy between the two agents was expected to be null in efflux-deficient strains if the only purpose of colistin was to spoil active efflux of niclosamide. However, synergism of niclosamide and colistin was retained in efflux-deficient PAO750 strain (FIC index = 0.375), although it was less pronounced relative to wild-type PAO1 (FIC index = 0.127). This suggested that aside from disrupting active efflux, the well-known membrane permeabilizing properties of colistin also allowed an enhanced intracellular accumulation of niclosamide that resulted in synergy. We believe that colistin potentiates niclosamide in both ways, and yet these may only be applicable to strains that are phenotypically susceptible to colistin. It is also possible that niclosamide may induce an effect on P. aeruginosa that render them more susceptible to colistin.
A recent microarray analysis showed that niclosamide affected the transcription of 258 genes in P. aeruginosa, in which most of the downregulated genes were involved in QS and therefore may lead to attenuated virulence (14). About 40% of these niclosamide-controlled genes are still classified as hypothetical; hence, comprehensive details on the molecular effect of niclosamide may still not be realized. However, a closer look revealed that the downregulation of grxD (PA3533) transcription in P. aeruginosa by niclosamide may result in increased colistin susceptibility (14). The grxD gene encodes monothiol glutaredoxin (GrxD), to which nonfunctionality has been linked to enhanced colistin susceptibility in P. aeruginosa (26). Interestingly, increased expression of grxD does not result in colistin resistance (26). This mechanism of colistin potentiation by niclosamide through the regulation of genes that result in phenotypic switching may likely be responsible for the synergism between the two agents, especially against strains that are phenotypically resistant to colistin.
Synergism was retained across colistin-susceptible Gram-negative bacteria.
The synergistic combination of niclosamide and colistin was then evaluated against a panel of other colistin-susceptible Gram-negative bacilli, including Acinetobacter baumannii, Klebsiella pneumoniae, E. coli, and Enterobacter cloacae, as shown in Table 3. Niclosamide by itself possessed very poor activity against the four organisms. However, niclosamide strongly synergized with colistin against most of the tested strains, except K. pneumoniae 117493, E. coli 117255, and E. cloacae 117029. Niclosamide and colistin displayed synergy against all eight A. baumannii strains consisting of one wild-type and seven MDR/XDR clinical isolates (Table 3). Our data confirmed that the synergism between niclosamide and colistin was not Pseudomonas specific but was conserved in other Gram-negative bacilli.
TABLE 3.
Synergy of niclosamide with colistin against wild-type and MDR/XDR colistin-susceptible Gram-negative bacilli
| Organism | MICNIC [MICcombo], μg/ml | MICCOL [MICcombo], μg/ml | FIC index | Absolute MICCOL,a μg/ml | Potentiation (fold)b |
|---|---|---|---|---|---|
| A. baumannii AB027 | >256 [0.5] | 0.25 [0.031] | 0.125 < x < 0.127 | 0.031 | 8 |
| A. baumannii AB031 | >256 [2] | 0.25 [0.008] | 0.031 < x < 0.039 | 0.016 | 16 |
| A. baumannii LAC-4 | >256 [4] | 0.125 [0.008] | 0.062 < x < 0.078 | 0.016 | 8 |
| A. baumannii 87072 | >64 [0.5] | 0.5 [0.125] | 0.250 < x < 0.258 | 0.125 | 4 |
| A. baumannii 110193 | >256 [2] | 0.25 [0.031] | 0.125 < x < 0.133 | 0.062 | 4 |
| A. baumannii 119141 | >64 [2] | 0.5 [0.125] | 0.250 < x < 0.281 | 0.25 | 2 |
| A. baumannii 121621 | >64 [1] | 0.25 [0.062] | 0.250 < x < 0.266 | 0.062 | 4 |
| A. baumannii ATCC 17978c | >256 [4] | 0.25 [0.004] | 0.016 < x < 0.031 | 0.031 | 8 |
| K. pneumoniae 116381 | >256 [2] | 1 [0.031] | 0.031 < x < 0.039 | 0.125 | 8 |
| K. pneumoniae 117493 | >64 [1] | 0.25 [0.125] | 0.500 < x < 0.516 | 0.125 | 2 |
| E. coli 117255 | >64 [2] | 0.25 [0.125] | 0.500 < x < 0.531 | 0.25 | None |
| E. coli ATCC 25922c | >64 [0.25] | 0.5 [0.125] | 0.250 < x < 0.254 | 0.125 | 4 |
| E. cloacae 117029 | >256 [0.5] | 0.125 [0.008] | 0.062 < x < 0.064 | 0.008 | 16 |
| E. cloacae 117877 | >64 [0.25] | 0.25 [0.125] | 0.500 < x < 0.504 | 0.125 | 2 |
MIC of colistin in the presence of 1 μg/ml (3 μM) niclosamide.
Degree of colistin potentiation in the presence of 1 μg/ml (3 μM) niclosamide.
Wild type.
Niclosamide reversed colistin resistance against resistant Gram-negative bacteria.
The potential therapeutic utility of niclosamide in colistin therapy was further evaluated in vitro against MDR/XDR colistin-resistant Gram-negative bacterial isolates (Table 4). Biochemical alteration of the outer membrane leading to a net reduction of negative charge in lipopolysaccharide (LPS), via conjugation of positively charged biomolecules such as l-aminoarabinose, is the prevalent mechanism of colistin resistance (27). The reduction of anionicity in LPS prevents the cationic polymyxin to interact with the outer membrane and therefore results in a loss of antibacterial activity. We questioned whether niclosamide is able to revitalize colistin’s activity against colistin-resistant pathogens. Thirteen colistin-resistant clinical isolates were tested (Table 4), including P. aeruginosa, A. baumannii, K. pneumoniae, E. coli, and E. cloacae. Notably, the two E. coli isolates in the panel were known to harbor the mcr-1 gene plasmid that transcribe phosphoethanolamine transferase to confer colistin resistance (18). According to the Clinical and Laboratory Standards Institute (CLSI) (28), an MIC of ≤2 μg/ml against P. aeruginosa and A. baumannii denotes susceptibility to colistin, while no breakpoint is currently established against Enterobacteriaceae. For this article, we cautiously use the available colistin susceptibility breakpoint values against P. aeruginosa as an interpretative guide for Enterobacteriaceae. The MIC of colistin ranged from 4 to 1,024 μg/ml against the tested colistin-resistant panel. Niclosamide was found to significantly synergized with colistin against all thirteen colistin-resistant Gram-negative bacilli using a checkerboard assay (Table 4). More importantly, the MICs of colistin against all resistant strains were reduced below the susceptibility breakpoint in the presence of 1 μg/ml (3 μM) niclosamide. For instance, we observed a 2,048-fold MIC reduction of colistin in combination with 1 μg/ml (3 μM) niclosamide against MDR K. pneumoniae 113250 and 113254 strains, in which the MIC was reduced from 256 to 0.125 μg/ml for both. Similarly, we also observed that niclosamide synergized with polymyxin B against colistin-resistant Gram-negative bacilli (see Table S1 in the supplemental information). Our data clearly demonstrated that niclosamide was able to reverse colistin resistance even against those Gram-negative bacilli that harbors the mcr-1 gene (Table 4 and Table S1). Thus, these findings strongly suggest the use of niclosamide in combination with colistin to eradicate colistin-resistant Gram-negative pathogens.
TABLE 4.
Synergy of niclosamide with colistin against MDR/XDR colistin-resistant Gram-negative bacilli, including mcr-1 gene-positive E. coli strains
| Organism | MICNIC [MICcombo], μg/ml | MICCOL [MICcombo], μg/ml | FIC index | Absolute MICCOL,a μg/ml | Potentiation (fold)b |
|---|---|---|---|---|---|
| P. aeruginosa 91433 | 4 [0.125] | 4 [0.062] | 0.047 | ≤0.016 | ≥256 |
| P. aeruginosa 101243 | >256 [0.25] | 1024 [0.25] | 0.0002 < x < 0.001 | ≤0.062 | ≥16,384 |
| P. aeruginosa 114228 | >256 [16] | 4 [1] | 0.250 < x < 0.312 | 2 | 2 |
| A. baumannii 82479 | >64 [2] | >64 [1] | <0.047 | 2 | >32 |
| A. baumannii 92247 | >64 [8] | 8 [0.125] | 0.016 < x < 0.141 | 0.25 | 32 |
| K. pneumoniae 106931 | >64 [4] | 64 [0.25] | 0.004 < x < 0.066 | 1 | 64 |
| K. pneumoniae 110036 | >64 [8] | 8 [0.25] | 0.031 < x < 0.156 | 2 | 4 |
| K. pneumoniae 113250 | >256 [0.5] | 256 [0.25] | 0.001 < x < 0.003 | 0.125 | 2,048 |
| K. pneumoniae 113254 | >256 [0.5] | 256 [0.5] | 0.002 < x < 0.004 | 0.125 | 2,048 |
| E. coli 94393c | >64 [8] | 4 [0.25] | 0.062 < x < 0.187 | 1 | 4 |
| E. coli 94474c | >64 [8] | 16 [0.5] | 0.031 < x < 0.156 | 2 | 8 |
| E. cloacae 118564 | >64 [8] | >64 [0.125] | <0.127 | 0.5 | >128 |
| E. cloacae 121187 | >64 [4] | >64 [0.125] | <0.064 | 0.5 | >128 |
MIC of colistin in the presence of 1 μg/ml (3 μM) niclosamide.
Degree of colistin potentiation in the presence of 1 μg/ml (3 μM) niclosamide.
mcr-1 gene positive.
Dose effect of niclosamide on colistin potentiation.
The effect of different niclosamide doses to the antibacterial activity of colistin against Gram-negative bacilli was then evaluated. We fixed the niclosamide concentration at either 4 μg/ml (12 μM), 2 μg/ml (6 μM), 1 μg/ml (3 μM), 0.5 μg/ml (1.5 μM), or 0.25 μg/ml (0.75 μM) to explore the optimal adjuvant dosage in reducing the MIC of colistin. No pronounced differences were observed between the five niclosamide doses against most colistin-susceptible Gram-negative organisms (see Tables S2 and S3 in the supplemental information). However, 0.25 μg/ml (0.75 μM) niclosamide was not sufficient to reduce the MIC of colistin against five of eight colistin-susceptible MDR/XDR A. baumannii isolates (see Table S3 in the supplemental information). A defined effect of niclosamide doses to colistin’s antibacterial activity against colistin-resistant Gram-negative bacilli was observed (Table 5). The adjuvant potency of niclosamide declined on concentrations below 1 μg/ml (3 μM), with the exception against P. aeruginosa 114228 in which synergy was only achieved at a niclosamide concentration of 16 μg/ml (as shown in Table 4). For instance, the absolute MIC of colistin went up from 0.5 to 16 μg/ml against E. cloacae 118564 when the niclosamide dosage was changed from 1 to 0.5 μg/ml (Table 5). Therefore, our findings strongly suggest that ≥1 μg/ml of niclosamide is optimal in combination with colistin against Gram-negative pathogens.
TABLE 5.
Effect of different niclosamide doses on the MIC of colistin against MDR/XDR colistin-resistant Gram-negative bacilli
| Organism | MICCOL | Absolute MICCOL, μg/ml, at: |
||||
|---|---|---|---|---|---|---|
| 4 μg/ml (12 μM) niclosamide | 2 μg/ml (6 μM) niclosamide | 1 μg/ml (3 μM) niclosamide | 0.5 μg/ml (1.5 μM) niclosamide | 0.25 μg/ml (0.75 μM) niclosamide | ||
| P. aeruginosa 91433 | 4 | NAb | ≤0.016 | ≤0.016 | 0.031 | 0.031 |
| P. aeruginosa 101243 | 1024 | ≤0.062 | ≤0.062 | ≤0.062 | 0.125 | 0.25 |
| P. aeruginosa 114228 | 4 | 2 | 2 | 2 | 2 | 4 |
| A. baumannii 82479 | >64 | 1 | 1 | 2 | 8 | 32 |
| A. baumannii 92247 | 8 | 0.25 | 0.25 | 0.25 | 0.5 | 1 |
| K. pneumoniae 106931 | 64 | 0.25 | 0.25 | 1 | 2 | 16 |
| K. pneumoniae 110036 | 8 | 0.5 | 0.5 | 2 | 4 | 8 |
| K. pneumoniae 113250 | 256 | ≤0.062 | ≤0.062 | 0.125 | 0.25 | 2 |
| K. pneumoniae 113254 | 256 | 0.125 | 0.125 | 0.125 | 0.5 | 2 |
| E. coli 94393a | 4 | 0.5 | 0.5 | 1 | 2 | 2 |
| E. coli 94474a | 16 | 1 | 1 | 2 | 2 | 16 |
| E. cloacae 118564 | >64 | 0.25 | 0.25 | 0.5 | 16 | >64 |
| E. cloacae 121187 | >64 | 0.125 | 0.25 | 0.5 | 1 | 4 |
mcr-1 gene positive.
NA, not applicable (since the MICNIC is 4 μg/ml).
Combination of niclosamide and colistin enhanced bacterial killing in colistin-susceptible and colistin-resistant Gram-negative bacilli relative to colistin monotherapy.
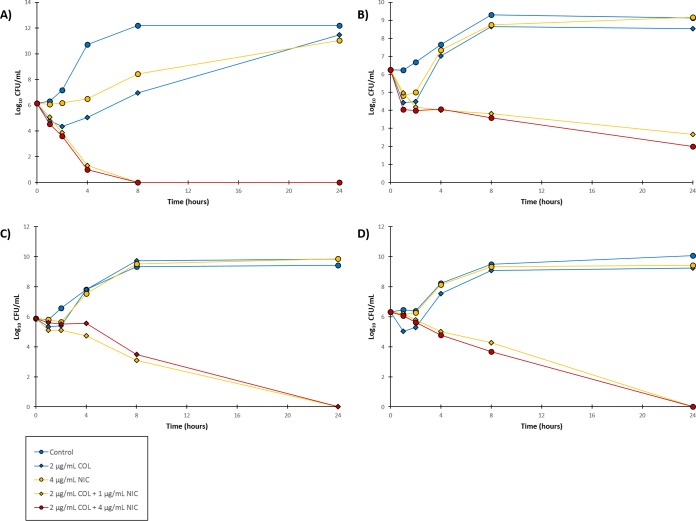
To further explore the degree of synergism, we looked at the bacterial killing induced by the synergistic combination of niclosamide and colistin against colistin-susceptible but more importantly colistin-resistant Gram-negative bacilli via time-kill kinetics assay. We purposely set the colistin concentration at the CLSI susceptibility breakpoint (2 μg/ml) and varied the concentration of niclosamide (1 or 4 μg/ml) since our initial data revealed that the optimal niclosamide concentration was ≥1 μg/ml. First, three clinical isolates of P. aeruginosa were tested (Fig. 1), two of which were colistin susceptible (PA262-11856 and PA264-104354) and one of which was colistin resistant (91433). Colistin at a 2 μg/ml concentration displayed bactericidal effects on both colistin-susceptible P. aeruginosa strains (Fig. 1A and B). However, significant bacterial growth was found after 24 h. While both colistin-susceptible strains had an MIC of 2 μg/ml for colistin, the concentration was clearly not enough to suppress bacterial regrowth. The addition of either 1 or 4 μg/ml niclosamide to 2 μg/ml colistin enhanced bacterial killing of the two colistin-susceptible strains (Fig. 1A and B). The combination lowered the bacterial load of P. aeruginosa PA262-11856 below the limit of detection by 8 h. However, P. aeruginosa PA262-11856 appeared to consist of colistin-heteroresistant subpopulation that niclosamide was not able to prevent regrowth since an increase in CFU was observed after 24 h (Fig. 1A). Other synergistic combinations of adjuvants with colistin against colistin-susceptible Gram-negative bacilli had also been reported to identify colistin-heteroresistant subpopulations (29). The colistin-susceptible PA264-104354 strain was eradicated completely by the combination of 2 μg/ml colistin and 4 μg/ml niclosamide after 24 h (Fig. 1B). More importantly, the combination of colistin (2 μg/ml) and niclosamide (at both 1 and 4 μg/ml) sterilized colistin-resistant P. aeruginosa 91433 after 24 h, while colistin monotherapy (2 μg/ml) resulted in visible bacterial growth after 8 h (Fig. 1C).
FIG 1.
Time-kill curves for the combination of niclosamide (NIC) and colistin (COL) against (A) colistin-susceptible P. aeruginosa PA262-101856, (B) colistin-susceptible P. aeruginosa PA264-104354 and (C) colistin-resistant P. aeruginosa 91433.
We then performed the same assay against other colistin-resistant Gram-negative bacilli (Fig. 2), including A. baumannii 92247, K. pneumoniae 113250, E. coli 94474, and E. cloacae 118564. In all tested colistin-resistant clinical isolates, significant bacterial killing was observed in comparison to colistin monotherapy. For instance, the combination of colistin (2 μg/ml) and niclosamide (at both 1 and 4 μg/ml) sterilized colistin-resistant A. baumannii 92247 after only 8 h (Fig. 2A), while a similar result was observed for colistin-resistant E. coli 94474 (Fig. 2C) and E. cloacae 118564 (Fig. 2D) after 24 h. Notably, our findings against E. coli 94474 were important, since this strain harbors the plasmid-mediated colistin resistance mcr-1 gene. Therefore, our data revealed that the synergistic combination of niclosamide and colistin augmented the bacterial killing of colistin-susceptible and more importantly colistin-resistant Gram-negative bacilli in comparison to colistin monotherapy.
FIG 2.
Time-kill curves for the combination of niclosamide (NIC) and colistin (COL) against (A) colistin- resistant A. baumannii 92247, (B) colistin-resistant K. pneumoniae 113250, (C) colistin-resistant E. coli 94474 and (D) colistin-resistant E. cloacae 118564.
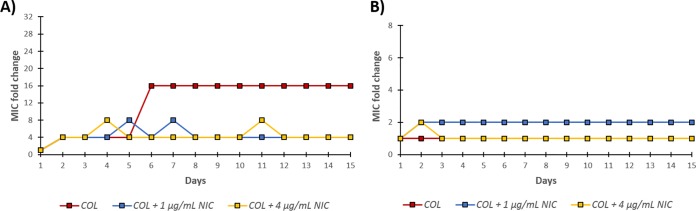
Addition of niclosamide to colistin therapy can suppress resistance development.
The emergence of resistance to either colistin monotherapy or combination of niclosamide and colistin was then assessed in Gram-negative bacilli in order to identify a strategy that would retain the MIC of colistin below its susceptibility breakpoint (≤2 μg/ml). We used the colistin-susceptible P. aeruginosa PA262-11856 (Fig. 3A and Table S4 in the supplemental information) and colistin-resistant E. coli 94474 (Fig. 3B and Table S5 in the supplemental information) for our assessment. We took an interest in P. aeruginosa PA262-11856 since our time-kill kinetic curve (Fig. 1A) indicated the presence of colistin-heteroresistant subpopulations. We wondered whether these heteroresistant subpopulations can accumulate further resistance after several passages. Serial passages of 15 days were performed by subjecting the organism to 1/2× the MIC of colistin and harvesting the surviving bacteria for the next generation. As shown in Fig. 3A, a 4-fold increase in the MIC of colistin (from 2 to 8 μg/ml) against P. aeruginosa PA262-11856 was observed in day 2 in both monotherapy and combination therapy. We believed that this increase in MIC was due to the colistin-heteroresistant subpopulation that was observed in our time-kill kinetics study. Colistin monotherapy elicited an additional 4-fold increase in the MIC of colistin on day 6 (from 8 to 32 μg/ml), which was a 16-fold increase relative to day 1 (see Table S4 in the supplemental information). However, the addition of either 1 or 4 μg/ml niclosamide prevented further increase in the MIC of colistin. The MIC of colistin in combination with either 1 or 4 μg/ml niclosamide remained below the CLSI susceptibility breakpoint (see Table S4 in the supplemental material), while the MIC of colistin in monotherapy increased to 32 μg/ml after 15 serial passages. This confirmed that the addition of niclosamide to colistin therapy against P. aeruginosa PA262-11856 suppressed the further emergence of resistance aside from the existing colistin-heteroresistant subpopulation.
FIG 3.
Emergence of bacterial resistance in panel A colistin-susceptible P. aeruginosa PA262-101856 and (B) colistin-resistant E. coli 94474 after 15 serial passages of ½ × MIC colistin (COL) with or without niclosamide (NIC). MIC fold change shown on the y axis is with respect to the MIC obtained on day 1. For the absolute MIC values, see Table S4 and S5 in the supplemental information.
No significant MIC fold change was observed in either colistin alone or with niclosamide (1 and 4 μg/ml) against the colistin-resistant E. coli 94474 after 15 serial passages (Fig. 3B). Since the strain was already colistin resistant, we were not surprised that the MIC of colistin did not further increase after a sub-MIC exposure. More importantly, the MIC of colistin (see Table S5 in the supplemental information) in the presence of 4 μg/ml niclosamide after 15 serial passages remained below the susceptibility breakpoint (MIC of 0.5 μg/ml). Indeed, our data revealed that the presence of niclosamide suppressed the development of colistin resistance.
Niclosamide and colistin are an advantageous combination therapy but with limitations.
An apparent advantage of this combination is that both are already FDA-approved therapeutic agents and that their pharmacological profiles are very well established. Toxicity is a concern with colistin usage due to its narrow therapeutic window, which required a plasma colistin concentration for therapeutic effect maybe in close range for the drug to exhibit nephrotoxicity depending on individuals (30–32). However, ≥1 μg/ml (≥3 μM) niclosamide significantly lowered the effective concentration of colistin (≤2 μg/ml) to eradicate both colistin-susceptible and colistin-resistant Gram-negative bacteria based in our in vitro data. Niclosamide was also reported to play a protective role in response to kidney injury. In a rat model of renal ischemia/reperfusion injury, treatment of niclosamide significantly improved renal function, as characterized by low levels of biomarkers (serum creatinine and blood urea nitrogen) that indicate kidney damage relative to the control (33). Moreover, niclosamide was reported to regulate the immune response by reducing the production of proinflammatory cytokines tumor necrosis factor alpha, interleukin-6 (IL-6), and HMGB1 and inducing the release of anti-inflammatory cytokine IL-10 relative to untreated subjects (33). Overall, the presence of niclosamide as an adjuvant in therapy may alleviate concerns regarding colistin’s toxicity toward the kidney.
Niclosamide, on the other hand, is known for its poor water solubility and bioavailability (34). Plasma concentration of niclosamide is restricted by its poor absorption in the intestinal tract (if administered via the oral route). For instance, a single oral dose of 2 g of niclosamide in adult humans to treat tapeworm infections was previously reported to result in a maximal serum concentration range of 0.25 to 6.0 μg/ml (0.76 to 18.35 μM); the variability is due to intraindividual differences (35). Interestingly, a recent report (36) contradicted previous pharmacokinetic data in which a 50-mg oral dose (administered three times daily) of niclosamide resulted in only a 35.7 to 88.3 ng/ml plasma concentration (n = 3). An increase in oral dose to 1,000 mg (administered three times daily) resulted in a 149 to 182 ng/ml plasma niclosamide concentration (n = 2) and yet elicited dose-limiting toxicities (36). However, caution is advised since the recent pharmacokinetic report was obtained in a phase 1 dose escalation study of niclosamide in castration-resistant prostate cancer patients who may had significantly weakened immune systems due to chemotherapy; moreover, that the study had a fairly limited (n = 5 total) number of participants (36).
Even though compounding evidence suggests that niclosamide is well tolerated through oral administration (37), the achievable niclosamide serum concentration is fairly limited. Unfortunately, niclosamide and other related salicylanilides are highly bound to serum proteins, which currently limits their systemic and intravenous applications (38). Another limitation of niclosamide is its reported cytotoxicity. Even though niclosamide was found to not rupture red blood cells (hemolysis) at a high concentration of 32 μg/ml, it was reported to elicit marked decreased cell viability (50% cytotoxic concentration ∼ 0.25 μg/ml of kidney HEK 293T/17 and liver HepG2 eukaryotic cell lines at 0.125 μg/ml) (39). A possible solution to these limitations is to develop a formulation (via encapsulation) that can effectively and selectively deliver niclosamide (and colistin) to the pathogen at the site of infection and prevent it from binding to plasma proteins. To avoid these niclosamide pharmacokinetic and toxicity issues, a more feasible approach is to instead develop this combination as a topical treatment for complicated infections caused by Gram-negative bacteria that may or may not be colistin resistant. This approach is similar to the commercially available topical ointment Polysporin, which consists of polymyxin B and bacitracin.
In conclusion, the anthelmintic drug niclosamide may serve as a potential adjuvant for colistin therapy. Aside from the reported inherent antivirulence activity of niclosamide by quenching QS in P. aeruginosa (14), we here disclosed its ability to selectively synergize with colistin against colistin-susceptible and colistin-resistant Gram-negative bacilli. Niclosamide significantly enhanced bacterial killing and suppressed the emergence of resistance to colistin monotherapy. These findings are important for the development of new therapeutic regimens to treat MDR and especially colistin-resistant GNB infections.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Most isolates were acquired from either the American Type Culture Collection (ATCC) or the Canadian Ward Surveillance (CANWARD) study (40). Bacterial strains from CANWARD were clinical specimens collected from patients suffering from a presumed infectious diseases admitted in a participating medical center across Canada. The two colistin-resistant E. coli strains from the CANWARD study had been reported to harbor the plasmid-mediated colistin resistance gene mcr-1, but not mcr-2 (18). The efflux-deficient P. aeruginosa strains PAO200 and PAO750 were obtained from previous studies (41, 42). PAO200 lacked the MexAB-OprM efflux system, while PAO750 lacked five clinically relevant pumps (MexAB-OprM, MexCD-OprJ, MexEF-OprN, MexJK, and MexXY) and the outer membrane protein OpmH. Prior to experiments, bacterial isolates were grown overnight in lysogeny broth (LB) on an incubator shaker at 37°C.
Antimicrobial susceptibility assay.
In vitro antibacterial activity of the tested agents were assessed by broth microdilution susceptibility testing according to the CLSI guidelines (28). Bacterial cultures grown overnight were then diluted in saline to achieve a 0.5 McFarland turbidity. Subsequently, the diluted bacterial culture was further diluted 1:50 in Mueller-Hinton broth (MHB) for inoculation to a final concentration of approximately 5 × 105 CFU/ml. The experiment was performed on a 96-well plates, where the tested agents were 2-fold serially diluted in MHB and incubated with equal volumes of bacterial inoculum at 37°C for 18 h. The MIC values for each tested agents were determined as the lowest concentration to inhibit visible bacterial growth in the form of turbidity, which was confirmed using an EMax Plus microplate reader (Molecular Devices) at a 590-nm wavelength. Wells with or without bacterial cells were used as positive or negative controls, respectively.
Checkerboard assay.
The assay was performed on a 96-well plate as previously described (43, 44). The antibiotic of interest was 2-fold serially diluted along the x axis, whereas niclosamide was 2-fold serially diluted along the y axis to create a matrix, where each well consists of a combination of both agents at different concentrations. Bacterial cultures grown overnight were then diluted in saline to 0.5 McFarland turbidity, followed by 1:50 further dilution in MHB and inoculation on each well to achieve a final concentration of approximately 5 × 105 CFU/ml. Wells comprising MHB with or without bacterial cells were used as positive or negative controls, respectively. The 96-well plates were then incubated at 37°C for 18 h and examined for visible turbidity, which was confirmed using an EMax Plus microplate reader (Molecular Devices) at a 590-nm wavelength. The fractional inhibitory concentration (FIC) of the antibiotic was calculated by dividing the MIC of antibiotic in the presence of niclosamide by the MIC of antibiotic alone. Similarly, the FIC of niclosamide was calculated by dividing the MIC of niclosamide in the presence of antibiotic by the MIC of niclosamide alone. The FIC index was the summation of both FIC values. FIC index values of ≤0.5, 0.5 < x < 4, and ≥4 were interpreted as synergistic, indifferent, and antagonistic, respectively.
Time-kill kinetics assay.
The kinetics of bacterial killing were determined as previously described (45), with minor modifications. A bacterial culture grown overnight was diluted in saline to 0.5 McFarland turbidity, followed by a further 1:50 dilution in LB. The cell culture was incubated at 37°C with either colistin, niclosamide, or combinations thereof at the desired concentrations to result in sets of culture tubes per designated time intervals. A 100-μl aliquot was obtained from each tubes at specified time intervals and plated on LB agar plates after serial dilution in phosphate-buffered saline (pH 7.2). Bacterial colonies were counted from the plates after incubation for 18 h at 37°C.
Emergence of resistance/serial passage assay.
The resistance development for colistin monotherapy or combination therapy of colistin and niclosamide was determined after a 15-day serial passage assay according to a reported protocol (46) with minor modifications. Briefly, the bacterial strains were subjected to an MIC assay on the first day with either colistin or combination of colistin and a fixed concentration of niclosamide (1 or 4 μg/ml). The bacterial cells growing at the highest concentration (1/2× the MIC) were harvested after 18 h of incubation at 37°C as the working inoculum for the subsequent experiment day; an additional MIC assay was then performed using this inoculum. This was repeated for the remaining duration of the assay (15 days in all); the MIC values for colistin were recorded and plotted as the MIC fold change compared to day 1 of the experiment.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by the Natural Sciences and Engineering Research Council of Canada NSERC-DG (2018-06047).
We thank M. Mulvey for providing the two E. coli isolates containing the mcr-1 plasmid.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.02574-18.
REFERENCES
- 1.Luepke KH, Mohr JF III.. 2017. The antibiotic pipeline: reviving research and development and speeding drugs to market. Expert Rev anti Infect Ther 15:425–433. doi: 10.1080/14787210.2017.1308251. [DOI] [PubMed] [Google Scholar]
- 2.Falagas ME, Mavroudis AD, Vardakas KZ. 2016. The antibiotic pipeline for multidrug-resistant gram negative bacteria: what can we expect? Expert Rev anti Infect Ther 14:747–763. doi: 10.1080/14787210.2016.1204911. [DOI] [PubMed] [Google Scholar]
- 3.MacVane SH. 2017. Antimicrobial resistance in the intensive care unit: a focus on Gram-negative bacterial infections. J Intensive Care Med 32:25–37. doi: 10.1177/0885066615619895. [DOI] [PubMed] [Google Scholar]
- 4.Prabaker K, Weinstein RA. 2011. Trends in antimicrobial resistance in intensive care units in the United States. Curr Opin Crit Care 17:472–479. doi: 10.1097/MCC.0b013e32834a4b03. [DOI] [PubMed] [Google Scholar]
- 5.Fraimow H, Nahra R. 2013. Resistant Gram-negative infections. Crit Care Clin 29:895–921. doi: 10.1016/j.ccc.2013.06.010. [DOI] [PubMed] [Google Scholar]
- 6.Blair JMA, Webber MA, Baylay AJ, Ogbolu DO, Piddock LJV. 2015. Molecular mechanisms of antibiotic resistance. Nat Rev Microbiol 13:42–51. doi: 10.1038/nrmicro3380. [DOI] [PubMed] [Google Scholar]
- 7.Domalaon R, Idowu T, Zhanel GG, Schweizer F. 2018. Antibiotic hybrids: the next generation of agents and adjuvants against Gram-negative pathogens? Clin Microbiol Rev 31:e00077-17. doi: 10.1128/CMR.00077-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schroeder M, Brooks BD, Brooks AE. 2017. The complex relationship between virulence and antibiotic resistance. Genes (Basel) 8:39. doi: 10.3390/genes8010039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Andersson DI, Hughes D. 2014. Microbiological effects of sublethal levels of antibiotics. Nat Rev Microbiol 12:465–478. doi: 10.1038/nrmicro3270. [DOI] [PubMed] [Google Scholar]
- 10.Fernandez L, Hancock REW. 2012. Adaptive and mutational resistance: role of porins and efflux pumps in drug resistance. Clin Microbiol Rev 25:661–681. doi: 10.1128/CMR.00043-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Papenfort K, Bassler BL. 2016. Quorum sensing signal-response systems in Gram-negative bacteria. Nat Rev Microbiol 14:576–588. doi: 10.1038/nrmicro.2016.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maura D, Ballok AE, Rahme LG. 2016. Considerations and caveats in anti-virulence drug development. Curr Opin Microbiol 33:41–46. doi: 10.1016/j.mib.2016.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rasko DA, Sperandio V. 2010. Anti-virulence strategies to combat bacteria-mediated disease. Nat Rev Drug Discov 9:117–128. doi: 10.1038/nrd3013. [DOI] [PubMed] [Google Scholar]
- 14.Imperi F, Massai F, Ramachandran Pillai C, Longo F, Zennaro E, Rampioni G, Visca P, Leoni L. 2013. New life for an old drug: the anthelmintic drug niclosamide inhibits Pseudomonas aeruginosa quorum sensing. Antimicrob Agents Chemother 57:996–1005. doi: 10.1128/AAC.01952-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brown D. 2015. Antibiotic resistance breakers: can repurposed drugs fill the antibiotic discovery void? Nat Rev Drug Discov 14:821–832. doi: 10.1038/nrd4675. [DOI] [PubMed] [Google Scholar]
- 16.Defoirdt T, Brackman G, Coenye T. 2013. Quorum sensing inhibitors: how strong is the evidence? Trends Microbiol 21:619–624. doi: 10.1016/j.tim.2013.09.006. [DOI] [PubMed] [Google Scholar]
- 17.Rajamuthiah R, Fuchs BB, Conery AL, Kim W, Jayamani E, Kwon B, Ausubel FM, Mylonakis E. 2015. Repurposing salicylanilide anthelmintic drugs to combat drug-resistant Staphylococcus aureus. PLoS One 10:e0124595. doi: 10.1371/journal.pone.0124595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Walkty A, Karlowsky JA, Adam HJ, Lagace-Wiens P, Baxter M, Mulvey MR, McCracken M, Poutanen SM, Roscoe D, Zhanel GG. 2016. Frequency of MCR-1-mediated colistin resistance among Escherichia coli clinical isolates obtained from patients in Canadian hospitals (CANWARD 2008-2015). C Open 4:E641–E645. doi: 10.9778/cmajo.20160080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu Y-Y, Wang Y, Walsh TR, Yi L-X, Zhang R, Spencer J, Doi Y, Tian G, Dong B, Huang X, Yu L-F, Gu D, Ren H, Chen X, Lv L, He D, Zhou H, Liang Z, Liu J-H, Shen J. 2016. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis 16:161–168. doi: 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- 20.Al-Tawfiq JA, Laxminarayan R, Mendelson M. 2017. How should we respond to the emergence of plasmid-mediated colistin resistance in humans and animals? Int J Infect Dis 54:77–84. doi: 10.1016/j.ijid.2016.11.415. [DOI] [PubMed] [Google Scholar]
- 21.Xavier BB, Lammens C, Ruhal R, Kumar-Singh S, Butaye P, Goossens H, Malhotra-Kumar S. 2016. Identification of a novel plasmid-mediated colistin-resistance gene, mcr-2, in Escherichia coli, Belgium, June 2016. Euro Surveill 21 Euro Surveill 21:30280. doi: 10.2807/1560-7917.ES.2016.21.27.30280. [DOI] [PubMed] [Google Scholar]
- 22.Yin W, Li H, Shen Y, Liu Z, Wang S, Shen Z, Zhang R, Walsh TR, Shen J, Wang Y. 2017. Novel plasmid-mediated colistin resistance gene mcr-3 in Escherichia coli. mBio 8:e00543-17. doi: 10.1128/mBio.00543-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schwarz S, Johnson AP. 2016. Transferable resistance to colistin: a new but old threat. J Antimicrob Chemother 71:2066–2070. doi: 10.1093/jac/dkw274. [DOI] [PubMed] [Google Scholar]
- 24.Masuda N, Sakagawa E, Ohya S, Gotoh N, Tsujimoto H, Nishino T. 2000. Substrate specificities of MexAB-OprM, MexCD-OprJ, and MexXY-oprM efflux pumps in Pseudomonas aeruginosa. Antimicrob Agents Chemother 44:3322–3327. doi: 10.1128/AAC.44.12.3322-3327.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Domalaon R, Berry L, Tays Q, Zhanel GG, Schweizer F. 2018. Development of dilipid polymyxins: investigation on the effect of hydrophobicity through its fatty acyl component. Bioorg Chem 80:639–648. doi: 10.1016/j.bioorg.2018.07.018. [DOI] [PubMed] [Google Scholar]
- 26.Romsang A, Leesukon P, Duangnkern J, Vattanaviboon P, Mongkolsuk S. 2015. Mutation of the gene encoding monothiol glutaredoxin (GrxD) in Pseudomonas aeruginosa increases its susceptibility to polymyxins. Int J Antimicrob Agents 45:314–318. doi: 10.1016/j.ijantimicag.2014.10.024. [DOI] [PubMed] [Google Scholar]
- 27.Baron S, Hadjadj L, Rolain J-M, Olaitan AO. 2016. Molecular mechanisms of polymyxin resistance: knowns and unknowns. Int J Antimicrob Agents 48:583–591. doi: 10.1016/j.ijantimicag.2016.06.023. [DOI] [PubMed] [Google Scholar]
- 28.The Clinical and Laboratory Standards Institute. 2016. Performance standards for antimicrobial susceptibility testing CLSI supplement M100S, 26th ed Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 29.Li J, Rayner CR, Nation RL, Owen RJ, Spelman D, Tan KE, Liolios L. 2006. Heteroresistance to colistin in multidrug-resistant Acinetobacter baumannii. Antimicrob Agents Chemother 50:2946–2950. doi: 10.1128/AAC.00103-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tran TB, Velkov T, Nation RL, Forrest A, Tsuji BT, Bergen PJ, Li J. 2016. Pharmacokinetics/pharmacodynamics of colistin and polymyxin B: are we there yet? Int J Antimicrob Agents 48:592–597. doi: 10.1016/j.ijantimicag.2016.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Forrest A, Garonzik SM, Thamlikitkul V, Giamarellos-Bourboulis EJ, Paterson DL, Li J, Silveira FP, Nation RL. 2017. Pharmacokinetic/toxicodynamic analysis of colistin-associated acute kidney injury in critically ill patients. Antimicrob Agents Chemother 61:e01367-17. doi: 10.1128/AAC.01367-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sorli L, Luque S, Grau S, Berenguer N, Segura C, Montero MM, Alvarez-Lerma F, Knobel H, Benito N, Horcajada JP. 2013. Trough colistin plasma level is an independent risk factor for nephrotoxicity: a prospective observational cohort study. BMC Infect Dis 13:380. doi: 10.1186/1471-2334-13-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang L-X, Zhao H-J, Sun D-L, Gao S-L, Zhang H-M, Ding X-G. 2017. Niclosamide attenuates inflammatory cytokines via the autophagy pathway leading to improved outcomes in renal ischemia/reperfusion injury. Mol Med Rep 16:1810–1816. doi: 10.3892/mmr.2017.6768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen W, Mook RAJ, Premont RT, Wang J. 2018. Niclosamide: Beyond an antihelminthic drug. Cell Signal 41:89–96. doi: 10.1016/j.cellsig.2017.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Andrews P, Thyssen J, Lorke D. 1982. The biology and toxicology of molluscicides, Bayluscide. Pharmacol Ther 19:245–295. doi: 10.1016/0163-7258(82)90064-X. [DOI] [PubMed] [Google Scholar]
- 36.Schweizer MT, Haugk K, McKiernan JS, Gulati R, Cheng HH, Maes JL, Dumpit RF, Nelson PS, Montgomery B, McCune JS, Plymate SR, Yu EY. 2018. A phase I study of niclosamide in combination with enzalutamide in men with castration-resistant prostate cancer. PLoS One 13:e0198389. doi: 10.1371/journal.pone.0198389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ofori-Adjei D, Dodoo ANO, Appiah-Danquah A, Couper M. 2008. A review of the safety of niclosamide, pyrantel, triclabendazole and oxamniquine. Int J Risk Saf Med 20:113–122. [Google Scholar]
- 38.Mohammed-Ali NA, Bogan JA. 1987. The pharmacodynamics of the flukicidal salicylanilides, rafoxanide, closantel and oxyclosanide. J Vet Pharmacol Ther 10:127–133. doi: 10.1111/j.1365-2885.1987.tb00089.x. [DOI] [PubMed] [Google Scholar]
- 39.Gooyit M, Janda KD. 2016. Reprofiled anthelmintics abate hypervirulent stationary-phase Clostridium difficile. Sci Rep 6:33642. doi: 10.1038/srep33642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhanel GG, Adam HJ, Baxter MR, Fuller J, Nichol KA, Denisuik AJ, Lagacé-Wiens PRS, Walkty A, Karlowsky JA, Schweizer F, Hoban DJ, Canadian Antimicrobial Resistance Alliance. 2013. Antimicrobial susceptibility of 22746 pathogens from Canadian hospitals: results of the CANWARD 2007-11 study. J Antimicrob Chemother 68:i7–i22. doi: 10.1093/jac/dkt022. [DOI] [PubMed] [Google Scholar]
- 41.Schweizer HP. 1998. Intrinsic resistance to inhibitors of fatty acid biosynthesis in Pseudomonas aeruginosa is due to efflux: application of a novel technique for generation of unmarked chromosomal mutations for the study of efflux systems. Antimicrob Agents Chemother 42:394–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kumar A, Chua K-L, Schweizer HP. 2006. Method for regulated expression of single-copy efflux pump genes in a surrogate Pseudomonas aeruginosa strain: identification of the BpeEF-OprC chloramphenicol and trimethoprim efflux pump of Burkholderia pseudomallei 1026b. Antimicrob Agents Chemother 50:3460–3463. doi: 10.1128/AAC.00440-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Domalaon R, Yang X, Lyu Y, Zhanel GG, Schweizer F. 2017. Polymyxin B3-tobramycin hybrids with Pseudomonas aeruginosa-selective antibacterial activity and strong potentiation of rifampicin, minocycline, and vancomycin. ACS Infect Dis 3:941–954. doi: 10.1021/acsinfecdis.7b00145. [DOI] [PubMed] [Google Scholar]
- 44.Domalaon R, Brizuela M, Eisner B, Findlay B, Zhanel GG, Schweizer F. 2018. Dilipid ultrashort cationic lipopeptides as adjuvants for chloramphenicol and other conventional antibiotics against Gram-negative bacteria. Amino Acids. doi: 10.1007/s00726-018-2673-9. [DOI] [PubMed] [Google Scholar]
- 45.Lyu Y, Yang X, Goswami S, Gorityala BK, Idowu T, Domalaon R, Zhanel GG, Shan A, Schweizer F. 2017. Amphiphilic tobramycin-lysine conjugates sensitize multidrug resistant Gram-negative bacteria to rifampicin and minocycline. J Med Chem 60:3684–3702. doi: 10.1021/acs.jmedchem.6b01742. [DOI] [PubMed] [Google Scholar]
- 46.Pollard JE, Snarr J, Chaudhary V, Jennings JD, Shaw H, Christiansen B, Wright J, Jia W, Bishop RE, Savage PB. 2012. In vitro evaluation of the potential for resistance development to ceragenin CSA-13. J Antimicrob Chemother 67:2665–2672. doi: 10.1093/jac/dks276. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.