HCV genotype 6 (GT-6) is found predominantly in East and Southeast Asia. Clinical studies have focused on patients infected with hepatitis C virus (HCV) GT-6a, where high sustained virologic response (SVR) rates to direct-acting antivirals (DAAs) have been achieved.
KEYWORDS: HCV, NS3, NS5A, NS5B, genotype 6, ombitasvir, polymorphism, resistance, sofosbuvir, velpatasvir
ABSTRACT
HCV genotype 6 (GT-6) is found predominantly in East and Southeast Asia. Clinical studies have focused on patients infected with hepatitis C virus (HCV) GT-6a, where high sustained virologic response (SVR) rates to direct-acting antivirals (DAAs) have been achieved. However, GT-6 is highly diverse, with 29 reported subtypes. We explored the diversity of GT-6 polymorphisms at residues associated with DAA resistance, their impact on DAA in vitro potency when evaluated in a GT-6a consensus replicon, and their association with specific GT-6 subtypes. GT-6 sequences from 25 patient-derived samples and 105 sequences from the U.S. HCV database were compared, and substitutions at resistance-associated residue positions were phenotyped against different DAAs. Preexisting resistance-associated substitutions (RASs) to NS3 protease (A156V and D168E) and NS5B nucleotide (L159F and S282C) inhibitors were rare (<4%). Preexisting RASs to NS5A inhibitors were common, especially at L28 (A/F/G/M/T/V) and R30 (E/N/S). In vitro susceptibilities of NS5A-L28A and -L28T were dramatically reduced against all tested NS5A drugs (90% effective concentration [EC90] range, 119 to 2,032 nM) compared with susceptibilities against a GT-6a consensus replicon (EC90 range, 0.1 to 19 nM). These L28 RASs preexisted in combination with R30S (EC90 [L28A-R30S] of ≥720 nM or EC90 [L28T-R30S] of ≥128 nM against tested DAAs) or as L28T-L31I (EC90 [tested DAAs] of >5,000 nM) and were detected in evaluated GT-6b and -6f sequences. NS5A-L28A-R30A, observed in GT-6r, did not replicate. In conclusion, HCV GT-6b, GT-6f, and GT-6r sequences harbored highly resistant RASs to all evaluated NS5A drugs. Therefore, monitoring SVR in patients infected with these GT-6 subtypes treated with NS5A drug-containing regimens is suggested to confirm any association between noted NS5A polymorphisms and treatment failure.
INTRODUCTION
The hepatitis C virus (HCV) is genetically diverse, with seven recognized genotypes (GTs), most of which have many subtypes (1). Among the seven genotypes of HCV, GT-6 exhibits the greatest diversity and therefore is thought to represent the oldest HCV lineage (2, 3). GT-6 has now been classified into 29 subtypes, 6a to 6xf (1). In addition, a number of lineages that remain unassigned to a subtype have been identified based on the complete sequence. Furthermore, many novel variants have been reported by sequencing partial genomic regions (4, 5). Geographically, HCV GT-6 isolates are mainly detected in residents of Southeast Asia, suggesting the ancestral region of origin (6–10). In Southeast Asia, the prevalence of GT-6 HCV is approximately 31%, whereas it is 0.1% in Europe (11). A wide spectrum of GT-6 isolates have been detected in North America (primarily Canada and the United States), accounting for approximately 1.8% of total HCV infections; almost all GT-6 isolates were sampled from Asian immigrants (12, 13).
Since the introduction of direct-acting antivirals (DAAs) for treatment of chronic HCV infection, sustained virologic response (SVR) rates have markedly improved for infected patients (14). However, most HCV therapy studies have focused on the predominant genotypes in North America, eastern Asia, and Europe (GT-1 and GT-3). Recently, several DAA-based regimens have been evaluated in patients with a HCV GT-6 infection (15, 16). In general, these DAA-based regimens have demonstrated high SVR in patients with an HCV GT-6 infection.
With HCV GT-1 infection, there is extensive evidence that GT-1 subtype (1a or 1b) influences the efficacy and barrier to resistance of some DAAs and DAA-based HCV therapeutic regimens (17–21). However, limited information has been reported regarding GT-6 subtype effects on the efficacy and resistance profile of DAA-based therapeutic regimens.
An NS5A inhibitor is a component of most approved DAA regimens. Daclatasvir (DCV) was the first (22) and has been evaluated in multiple clinical studies, and it demonstrated robust antiviral responses and a good safety and tolerability profile in combination with other DAA agents targeting the HCV NS3 protease (asunaprevir [ASV], BMS-650032, and simeprevir [SMV]) (23–28) and/or the NS5B polymerase (sofosbuvir [SOF], beclabuvir [BCV], and BMS-791325) (26–31). DCV-based regimens have mostly been evaluated in patients infected with HCV GT-1 through -4 and only in a very limited number of patients with GT-6 HCV (29, 30, 32, 33).
The objective of this study was to explore the resistance profiles of approved drugs targeting three HCV GT-6 proteins: NS3 protease, NS5A, and NS5B polymerase. We investigated the distribution of naturally occurring drug resistance-associated substitutions (RASs) in HCV GT-6 sequences, the in vitro effect of these RASs on drug sensitivity, and their potential significance by GT-6 subtype for treatment recommendations.
(A subset of the NS5A results from this analysis was presented at the Asian Pacific Digestive Week, 23 to 26 September 2017 [34].)
RESULTS
Subtyping of patient-derived sequences.
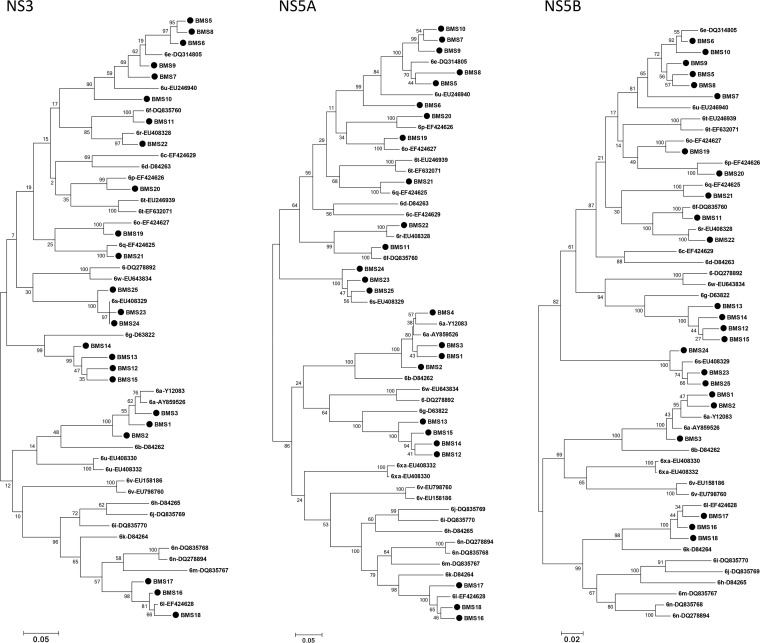
The GT-6 subtypes of 25 patient-derived sequences were determined based on segregation of their sequences from those of the usHCVdb with confirmed GT-6 subtypes (Fig. 1). Most isolates clustered with GT-6a, -6e, -6f, -6g, -6l, -6o, -6p, -6q, -6r, -6s, or -6u (see Table S3 in the supplemental material).
FIG 1.
Subtyping of GT-6 patient-derived NS3, NS5A, and NS5B sequences using phylogenetic analyses. Twenty-four NS3, 25 NS5A, and 24 NS5B patient-derived sequences from patients infected with HCV GT-6 and 30 prototype sequences of assigned subtypes (1) from the Los Alamos HCV Sequence Database (usHCVdb) (47) were analyzed phylogenetically. Closed circles represent patient-derived sequences. Sequences downloaded from the usHCVdb are labeled with GT-6 subtypes and accession numbers.
Baseline polymorphisms at residue positions previously associated with drug resistance.
Baseline polymorphisms at NS5A, NS3, and NS5B residue positions where substitutions have been previously shown to emerge in patients and confer resistance to DCV, ASV, and BCV, respectively, against HCV GT-1 (21, 35, 36) were identified by comparison with a GT-6a consensus sequence (derived from 21 GT-6a sequences; Table S2).
Of 130 available GT-6 NS5A sequences, many polymorphisms were detected at residue positions associated with NS5A drug resistance (33, 35, 36). Polymorphisms at L28, R30, and T58 were common, reference polymorphisms were most frequently detected at L31, A92, and T93, and P29 and P32 were conserved (Table 1).
TABLE 1.
Prevalence of HCV GT NS3, NS5A, and NS5B polymorphisms at clinically relevant residue positions associated with drug resistance
| Polymorphisms (n) ina: | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NS3 sequences (n = 104) | NS5A sequences (n = 130) | NS5B sequencesb (n = 104) | ||||||||||||||
| V36 | Y56 | K80 | A156 | D168 | I170 | L28 | R30 | L31 | T58 | T93 | L159 | S282 | L392 | V421 | V494 | P495 |
| V (78) | Y (93) | Q (81) | A (103) | D (102) | V (70) | V (52) | S (58) | L (126) | P (65) | T (109) | L (100) | S (102) | L (96) | V (103) | A (77) | P (103) |
| L (20) | F (11) | K (22) | V (1) | E (2) | I (33) | L (36) | R (38) | I (2) | T(46) | S (18) | F (3) | C (1) | I (8) | A (1) | V (25) | L (1) |
| I (6) | L (1) | A (1) | F (14) | A (28) | M (2) | G (5) | I (2) | ? (1) | ? (1) | M (2) | ||||||
| T (13) | N (4) | S (6) | ? (1) | |||||||||||||
| A (6) | E (1) | A (4) | ||||||||||||||
| M (6) | T (1) | ? (1) | ||||||||||||||
| ? (2) | L (2) | |||||||||||||||
| G (1) | L/Pc (1) | |||||||||||||||
The first row depicts the GT-6a consensus sequence (determined from alignment of 24 NS3, 36 NS5A, and 24 NS5B sequences). Underlining indicates polymorphisms previously shown to be associated with DAA resistance (21, 33, 35–37, 48); ?, undefined polymorphism. Only GT-6a reference polymorphisms were observed at NS3 (T54, V55, and R155), NS5A (P29 and P32), and NS5B (V321 and A499) resistance-associated residue positions. NS5A-A92P was detected in 1/130 NS5A sequences.
BCV-associated RASs include L392I, A421V, V494A, L495L/S, and A499T; SOF-associated RASs were found at L159F, S282T, and V321I.
One patient-derived NS5A sequence had a mixture of polymorphisms detected at T58. Each of these polymorphisms (L58 and P58) represented ≥15% of the viral population.
Of 104 available GT-6 NS3 sequences, RASs (A156V and D168E) associated with moderate to high-level resistance in GT-1 (35) were infrequently detected (1/104 and 2/104 sequences, respectively; Table 1), while the K80 RAS, associated with minimal to low-level resistance in GT-1, was more frequently detected (22/104).
Of the 104 available GT-6 NS5B sequences, polymorphisms associated with BCV (thumb-1 inhibitor) resistance were common, especially with respect to the low-level RAS V421 (21) or the V494A RAS (37), while polymorphisms associated with SOF (nucleoside inhibitor) resistance at L159 (3/104) or S282 (1/104) were rarely observed.
HCV GT-6 NS5A resistance-associated substitutions by HCV GT-6 subtype.
The frequency and combination of NS5A polymorphisms at residue positions L28, R30, L31, T58, and T93 are summarized by GT-6 subtype in Table 2. Of the 130 NS5A sequences, approximately half represented three GT-6 subtypes: GT-6a (n = 36), GT-6e (n = 10), and GT-6f (n = 16). The majority (24/36) of GT-6a sequences matched the GT-6a-cons sequence at residue positions associated with NS5A drug resistance, while the remaining sequences harbored either L28F (n = 10), L28V (n = 1), or the dual combination L28F-T93I. None of the other evaluated GT-6 subtypes had a similar polymorphism sequence at the five noted residue positions.
TABLE 2.
Prevalence of HCV GT-6 subtypes by NS5A polymorphisms and country
| GT-6 subtype | No. of NS5A sequences | Countrya | No. of NS5A sequences with noted polymorphism combinations | NS5A polymorphismsc |
||||
|---|---|---|---|---|---|---|---|---|
| L28 | R30 | L31 | T58 | T93 | ||||
| 6a | 36 | AU (n = 1), CN (n = 16), HK (n = 15), VN (n = 1), ND (n = 3) | 24 | |||||
| 10 | F | |||||||
| 1 | F | I | ||||||
| 1 | V | |||||||
| 6b | 2 | TH | 2 | T | I | S | ||
| 6cb | 1 | TH | 1 | V | A | G | ||
| 6d | 1 | VN | 1 | V | T | S | I | |
| 6e | 10 | AU (n = 2), CN (n = 10), US (n = 2), VN (n = 2), ND (n = 3) | 5 | V | S | P | ||
| 1 | V | A | M | L | ||||
| 4 | M | S | P | |||||
| 6f | 16 | TH (n = 15), ND (n = 1) | 10 | T | S | P | ||
| 1 | T | S | L | S | ||||
| 2 | A | S | P | |||||
| 2 | A | S | S | |||||
| 1 | A | S | L | |||||
| 6g | 6 | HK (n = 1), ID (n = 1), TW (n = 4) | 1 | M | N | P | ||
| 1 | M | S | P | |||||
| 1 | N | P | ||||||
| 1 | N | L/P | ||||||
| 1 | N | S | ||||||
| 1 | S | P | ||||||
| 6h | 1 | VN | 1 | V | A | P | ||
| 6i | 3 | TH | 2 | V | A | P | ||
| 1 | ? | A | A | |||||
| 6j | 2 | TH | 1 | V | A | ? | ||
| 1 | V | A | P | ? | ||||
| 6k | 5 | CN (n = 4), VN (n = 1) | 5 | V | A | P | ||
| 6l | 7 | AU (n = 1), US (n = 1), VN (n = 3), ND (n = 3) | 6 | V | A | P | ||
| 1 | V | E | P | |||||
| 6m | 4 | TH | 4 | V | S | S | ||
| 6n | 6 | CN (n = 2), MY (n = 1), TH (n = 3) | 6 | V | S | S | ||
| 6o | 4 | CA (n = 1), US (n = 1), VN (n = 1), ND (n = 1) | 2 | A | A | |||
| 2 | A | A | S | |||||
| 6p | 3 | CA (n = 1), US (n = 1), VN (n = 1) | 2 | V | S | P | ||
| 1 | M | S | P | |||||
| 6q | 2 | AU (n = 1), CA (n = 1) | 2 | V | S | P | ||
| 6r | 2 | AU (n = 1), CA (n = 1) | 1 | A | A | P | ||
| 1 | G | A | P | |||||
| 6s | 4 | AU (n = 3), CA (n = 1) | 4 | S | P | |||
| 6t | 4 | VN | 4 | V | S | G | ||
| 6u | 4 | CN (n = 3), VN (n = 1) | 3 | V | S | P | ||
| 1 | V | S | P | S | ||||
| 6v | 4 | CN | 4 | V | S | P | S | |
| 6w | 3 | CN (n = 1), TW (n = 2) | 3 | F | A | P | ||
Country codes: AU, Australia; CA, Canada; CN, Mainland China; HK, Hong Kong; ID, Indonesia; MY, Malaysia; ND, not determined; TH, Thailand; TW, Taiwan; US, United States; VN, Vietnam.
NS5A-A92P was detected in the single HCV GT-6c sequence.
?, undefined polymorphism.
Of 10 evaluated HCV GT-6e NS5A sequences, the most frequently detected combination of polymorphisms included R30S with either L28V (N = 5) or L28M (N = 4). These dual combinations of polymorphisms were also detected in other GT-6 subtypes representing g/m/n/p/q/t/u/v (n = 28) (Table 2). In 15 NS5A sequences from GT-6 subtypes m/n/u/v, L28V-R30S was also combined with T93S. There was a single GT-6e sequence with an L31M polymorphism combined with L28V-R30S. NS5A-L31M was not detected in any other GT-6 sequences.
Of 16 evaluated GT-6f NS5A sequences, polymorphism L28A (N = 5) or L28T (N = 11) was detected in combination with R30S (Table 2). NS5A-L28A was also detected in one GT-6r sequence combined with R30A, while L28T was detected in combination with L31I-T58S in both evaluated GT-6b sequences (Table 2). Another NS5A L28 polymorphism of potential interest (L28G) was detected in one GT-6r sequence in combination with R30S.
In vitro susceptibility of HCV GT-6 NS5A RASs.
There were numerous permutations of GT-6 polymorphism combinations detected at NS5A residue positions previously shown to be associated with drug resistance (Table 2). We therefore focused on examining the susceptibilities of single substitutions previously shown to modulate NS5A inhibitor activity (underlined polymorphisms in Table 1) and compared these potencies to that of a GT-6a-cons replicon. A summary of the susceptibilities of replicating HCV GT-6a NS5A replicon variants to inhibition by different drugs is shown in Table 3. Against the GT-6a-cons replicon, LDV and ombitasvir (OMV) had similar 90% effective concentration (EC90) values (EC90 [LDV], 14 nM; EC90 [OMV], 19 nM]), while DCV was ≥20-fold more potent (EC90, 0.7 nM) and VEL was >100-fold more potent (EC90, 0.1 nM).
TABLE 3.
Susceptibility of HCV GT-6 NS5A polymorphisms at residue positions associated with NS5A drug resistance
| GT-6a NS5A rep bkgda | RASb | ECs (±SD)c [nM] for: |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| DCV |
LDV |
OMV |
VEL |
||||||
| EC50 | EC90 | EC50 | EC90 | EC50 | EC90 | EC50 | EC90 | ||
| Cons | Cons | 0.30 (0.02) | 0.70 (0.04) | 4.3 (0.2) | 14 (1) | 8.1 (0.8) | 19 (1) | 0.05 (0.01) | 0.10 (0.01) |
| Cons | L28A | 296 (20) | 735 (41) | 677 (31) | 989 (142) | 1,720 (107) | 2,032 (131) | 100 (8) | 331 (24) |
| Cons | L28F | 0.65 (0.04) | 1.4 (0.1) | 38 (2) | 91 (6) | 13 (1) | 34 (3) | 0.09 (0.01) | 0.20 (0.03) |
| Cons | L28M | 9.3 (0.8) | 22 (1) | 152 (5) | 250 (22) | 21 (1) | 39 (2) | 0.60 (0.07) | 1.5 (0.2) |
| Cons | L28T | 224 (13) | 432 (32) | 560 (3) | 620 (60) | 1,483 (49) | 1,683 (21) | 54 (1) | 119 (4) |
| Cons | L28V | 1.9 (0.1) | 4.5 (0.1) | 31 (2) | 75 (4) | 81 (3) | 177 (11) | 0.55 (0.02) | 1.3 (0.1) |
| Cons | R30A | 0.40 (0.02) | 0.90 (0.02) | 1.5 (0.1) | 5.3 (0.2) | 0.0020 (0.0001) | 0.009 (0.001) | 0.0100 (0.0004) | 0.040 (0.002) |
| Cons | L31I | 6.4 (0.0) | 30 (1) | 334 (22) | 891 (141) | 108 (7) | 365 (15) | 1.0 (0.2) | 3.2 (0.3) |
| Cons | T93I | 1.2 (0.1) | 2.7 (0.1) | 21 (2) | 45 (1) | 29 (3) | 55 (12) | 0.08 (0.00) | 0.25 (0.02) |
| Cons | T93S | 1.1 (0.0) | 2.7 (0.2) | 25 (2) | 68 (5) | 46 (5) | 90 (7) | 0.15 (0.01) | 0.35 (0.02) |
| Cons | L28M, R30S | 47 (4) | 218 (22) | 188 (17) | 481 (33) | 0.25 (0.02) | 0.75 (0.06) | 0.30 (0.02) | 0.65 (0.20) |
| Cons | L28T, R30S | 735 (72) | 1,081 (101) | 876 (226) | 1,113 (279) | 1,746 (16) | 2,056 (16) | 63 (5) | 128 (39) |
| Cons | L28V, R30A | 0.55 (0.07) | 1.4 (0.2) | 1.8 (0.1) | 4.7 (0.2) | 0.085 (0.01) | 0.25 (0.01) | 0.05 (0.00) | 0.18 (0.01) |
| Cons | L28V, R30S | 8.7 (0.7) | 20 (2) | 62 (1) | 76 (2) | 9.5 (0.3) | 25 (1) | 0.25 (0.01) | 0.85 (0.01) |
| Cons | L28V, R30T | 4.8 (0.3) | 14 (1) | 38 (3) | 115 (3) | 2.9 (0.2) | 11 (0) | 0.09 (0.01) | 0.20 (0.02) |
| Cons | L28T, L31I | 2349 (128) | >5,000 | >5,000 | >5,000 | >5,000 | >5,000 | 42,221 (750) | >5,000 |
| Cons | L28V, R30S, T93S | 91 (13) | 254 (48) | 296 (31) | 618 (75) | 174 (27) | 418 (26) | 3.9 (0.4) | 12 (1) |
| JFH-1 | Cons | 0.10 (0.01) | 0.20 (0.01) | 2.7 (0.3) | 5.6 (0.1) | 5.2 (1.3) | 7.0 (0.2) | 0.006 (0.004) | 0.02 (0.01) |
| JFH-1 | L31M | 85 (3) | 137 (5) | 398 (97) | 544 (29) | 26 (2) | 43 (1) | 5.2 (0.5) | 8.3 (0.6) |
| JFH-1 | T58A | 3.6 (0.3) | 6.3 (0.2) | 33 (1) | 52 (1) | 74 (5) | 124 (19) | 0.023 (0.001) | 0.050 (0.003) |
| JFH-1 | T58S | 3.5 (0.1) | 6.1 (0.1) | 41 (13) | 56 (5) | 44 (8) | 70 (1) | 0.020 (0.000) | 0.05 (0.01) |
| Con1 | Cons | 0.088 (0.005) | 0.39 (0.07) | 0.60 (0.06) | 3.2 (0.2) | 1.4 (0.1) | 9.0 (1.0) | 0.014 (0.001) | 0.043 (0.005) |
| Con1 | R30E | 1.8 (0.0) | 9.6 (0.8) | 4.2 (0.8) | 29 (5) | 0.31 (0.05) | 1.6 (0.2) | 0.018 (0.001) | 0.075 (0.012) |
| Con1 | R30N | 0.35 (0.02) | 1.7 (0.1) | 0.53 (0.03) | 4.3 (0.2) | 0.003 (0.001) | 0.013 (0.002) | 0.009 (0.001) | 0.027 (0.002) |
| Con1 | R30S | 0.33 (0.03) | 1.6 (0.1) | 0.51 (0.04) | 3.7 (0.3) | 0.003 (0.001) | 0.012 (0.001) | 0.009 (0.001) | 0.024 (0.003) |
| Con1 | L28A R30S | 1,139 (155) | 3,269 (486) | 1,263 (43) | 2,648 (265) | 1,121 (123) | 1,630 (59) | 183 (60) | 720 (114) |
| Con1 | L28M, R30N | 20 (3) | 181 (20) | 74 (4) | 370 (27) | 0.024 (0.006) | 0.096 (0.016) | 0.61 (0.02) | 2.7 (0.1) |
GT-6a NS5A consensus (Cons) sequence was introduced into GT-2a (JFH-1) and GT-1b (Con1) replicon constructs in case GT-6a NS5A substitutions did not result in viable replicons in the GT-6a (NS3-5B) consensus replicon background (rep bkgd).
Certain GT-6 NS5A replicon constructs harboring various substitutions and combinations of substitutions did not replicate in our replicon system. NS5A-A92P conferred no change in NS5A drug potencies compared with that of the HCV GT-6a-cons replicon (data not shown).
EC, effective concentration; SD, standard deviation.
The potency of all drugs was modulated by different NS5A-L28 substitutions with a consistent potency order: VEL > DCV > OMV ≅ LDV (Table 3). NS5A substitutions L28A and L28T conferred high resistance to all four drugs; EC90 values ranged from 119 to 2,032 nM depending on the drug.
Against NS5A-R30 substitutions, VEL maintained its potency, the R30E substitution conferred low-level resistance to LDV (9-fold resistance), and R30E/N/S substitutions conferred low-level resistance to DCV (4- to 25-fold resistance); however, DCV was still slightly more potent than LDV against all GT-6a NS5A-R30 replicon variants (DCV EC90 range, 0.9 to 9.6 nM; LDV EC90 range, 3.7 to 29 nM) (Table 2). OMV potency was enhanced dramatically when R30 was changed to any other substitution, making OMV slightly more potent (EC90, 0.009 to 0.013 nM) than VEL (EC90, 0.024 to 0.040 nM) except against R30E (OMV EC90, 1.6 nM; VEL EC90, 0.075 nM).
Against NS5A-L31 substitutions, all drugs conferred resistance, although the level of resistance depended on the substitution: DCV potency was impacted by L31M (EC90, 137 nM) more so than L31I (EC90, 30 nM) and vice versa for OMV (EC90 [L31M], 43 nM; EC90 [L31I], 365 nM). Irrespective of L31 substitutions, LDV and VEL potencies were in the high- or low-nanomolar range, respectively.
Fold resistance to NS5A-T93 (T93I or T93S) or -T58 (T58A or T58S) substitutions were similar for the four drugs compared with EC90 values reported against GT-6a-cons NS5A. Relative potency order in each case was VEL > DCV > OMV ≅ LDV (Table 3).
The susceptibility of tested HCV GT-6a NS5A replicon variants harboring two or three substitution combinations detected in non-GT-6a subtypes to inhibition by NS5A drugs is shown in Table 3. A combination of either L28A or L28T with R30S conferred high resistance to all four drugs (EC90 range, 128 to 3,269 nM), as did NS5A substitutions L28T and L31I (EC90 values, >5,000 nM).
Not all NS5A-L28 substitution combinations impacted the potency of NS5A drugs; these included L28V with R30A, R30S, or R30T (Table 3). NS5A-L28M combined with R30S only produced increased resistance to DCV compared with the single L28M substitution (EC90 [L28M-R30S], 218 nM; EC90 [L28M], 22 nM). For LDV and VEL, potencies against L28M-R30S (EC90 values of 481 nM and 0.65 nM, respectively) were comparable to those of L28M alone (EC90 values of 620 nM and 1.5 nM, respectively), and OMV was significantly less resistant (EC90 [L28M-R30S], 0.75 nM; EC90 [L28M], 39 nM). NS5A-R30N appeared to rescue the negative potency effects of L28M on OMV to a greater extent than R30S (EC90 [L28M-R30N], 0.096 nM; EC90 [L28M+R30S], 0.75 nM), although OMV potencies against the single substitutions of R30N and R30S were similar (Table 3).
HCV GT-6a replicons harboring NS5A-T93S combined with L28V-R30S similarly increased fold resistance to each tested drug compared to the level for L28V-R30S alone (8- to 17-fold increase depending on drug) (Table 3).
In vitro susceptibility of NS3 RASs and their association with HCV GT-6 subtype.
Assessment of RASs by GT-6 subtype from 104 NS3 sequences indicated that K80 preexisted only in GT-6a sequences, where it was the most frequently detected polymorphism (22/24; Table S4), while K80Q predominated in other GT-6 subtypes. NS3-A156V was detected in a single GT-6l sequence, while D168E was detected in 1/24 GT-6a and 1/6 GT-6g sequences; K80 was also detected with D168E in the GT-6a sequence.
A summary of the susceptibilities of HCV GT-6a replicons harboring noted NS3 polymorphisms to inhibition by different NS3 inhibitors is provided in Table S5. Against the HCV GT-6a-cons replicon, ASV and SMV had similar potencies (EC90 values of 130 nM and 96 nM, respectively) but were noticeably less potent than the tested next-generation NS3 inhibitors, such as grazoprevir (GZV) and BMS-986144 (EC90 values of 8.1 nM and 15 nM, respectively). Against the D168E substitution, the potency variation between early- and next-generation inhibitors was also large (EC90 values of 513 to 1,583 nM versus 21 to 22 nM, respectively). The NS3-K80 polymorphism exerted a greater effect on SMV potency than the other NS3 inhibitors in comparisons of GT-6a-cons replicons with K80 versus Q80 (15-fold EC90 difference). A potency difference was also observed for K80-D168E versus K80Q-D168E (Table S5).
In vitro susceptibility of NS5B RASs and their association with HCV GT-6 subtype.
Assessment of RASs by GT-6 subtype from 104 NS5B sequences indicated that the thumb-1 signature RAS P495L was detected in 1/24 GT-6a sequences (Table S6). The L392I RAS was detected in 3/6 GT-6g sequences and combined with V494A in 4/10 GT-6e and 1/2 GT-6q sequences. NS5B-V494A preexisted in most GT-6 subtypes except for GT-6a and GT-6g, where it was detected in 4/24 and 2/6 NS5B sequences, respectively. The SOF-associated RAS L159F was only detected in 3/3 GT-6s sequences, while S282C was detected in a GT-6w sequence.
A summary of the susceptibilities of HCV GT-6a replicons harboring noted NS5B polymorphisms to inhibition by different NS5B inhibitors is provided in Table S7. Significantly reduced potencies were observed against the tested RASs known to impact thumb-1 inhibitors (L392I, V494A, and P495L), while potencies for SOF and a palm site inhibitor (BMS-929075) were comparable against GT-6a-cons and different thumb-1 variant replicons.
DISCUSSION
There is a greater understanding of SVR rates to DAA treatment in patients infected with HCV genotypes 1 to 3, while clinical data for patients infected with other genotypes, such as GT-6, is limited (15). Nevertheless, SVR rates after DAA treatment for the few treated patients infected with GT-6 have been promising (15, 32). Most patients in these clinical studies were infected with HCV GT-6a or GT-6e. Given the diversity and very limited number of patients infected with other GT-6 subtypes in these studies, any requirement for differentiating treatment options by GT-6 subtype is unknown.
Examination of patient-derived and usHCVdb GT-6 NS5A, NS3, and NS5B sequences for preexisting RASs that could impact drugs targeting these HCV proteins indicated a high level of NS5A heterogeneity compared with the levels for NS3 and NS5B. Signature NS3 RASs (A156V and D168E) were detected as the predominant viral species in approximately 3% of evaluated sequences. The SMV-associated secondary RAS in GT-1a, Q80K (35), was the predominant polymorphism in GT-6a, although it was not detected in other GT-6 subtypes. In vitro GT-6a data suggest that HCV GT-6-infected patients with preexisting D168E with or without K80 may have reduced SVR with certain ASV- and SMV-containing treatments, while patients treated with next-generation NS3 inhibitors, such as GZV, would be expected to respond.
Signature NS5B RASs either targeting nucleoside (at S282) or thumb-1 inhibitors (at P495) were detected as the predominant viral species in only 1% of evaluated sequences. NS5B-S282 substitutions are rarely detected by next-generation sequencing at treatment failure, let alone pretreatment. NS5B-S282C was detected by Sanger sequencing in a GT-6w sequence from Taiwan. This substitution has been observed in GT-2 sequences by next-generation sequencing (38). Evaluation of GT-6a NS5B-S282C on SOF in vitro potency was not possible using our replicon model. NS5B-L159F, a secondary SOF RAS, was only detected in GT-6s sequences from Australia. The implication of this finding is unknown. The thumb-1 RAS NS5B-V494A was the only polymorphism detected in all GT-6 subtypes except for GT-6a and GT-6g. This RAS conferred high resistance to BCV when tested in the GT-6a-cons replicon. NS5B-V494A, however, is not always associated with high resistance (37). The other RAS shown to confer high BCV resistance (NS5B-L392I) was detected in half of the GT-6e and GT-6g isolates, a single GT-6q isolate, and in certain cases in combination with V494A, suggesting that BCV is not effective against these GT-6 subtypes.
More than 80% of the GT-6 NS5A sequences had polymorphisms at residue positions associated with drug resistance compared with the GT-6a-cons sequence. Residue positions L28, R30, and T58 were highly polymorphic, while L31, A92, and T93 were the predominant species representing >80% to 99% of sequences. The only conserved resistance-associated residue positions were P29 and P32.
Of the observed GT-6 NS5A-L28 polymorphisms, HCV GT-6a-cons replicons harboring L28A or L28T were shown to confer very high resistance to all tested NS5A drugs. These L28 polymorphisms were detected in 15% of studied GT-6 NS5A sequences, of which the majority (17/19) also harbored R30A or R30S polymorphisms. This polymorphism combination was detected in all tested GT-6f isolates from Thailand and a GT-6r isolate from Australia. NS5A-L28G was observed in combination with R30A in one GT-6r sequence from Canada. Susceptibility evaluation of L28G or L28G-R30A in our GT-6a replicon models was not possible, since these constructs did not replicate. In patients infected with GT-1a, glycine at residue 28 emerged during treatment failure (21). Unpublished susceptibility data for GT-1a replicons harboring glycine versus alanine or threonine at residue position 28 indicated that a glycine variant conferred the highest level of resistance to DCV, with fold resistance of 54,812 versus 3,181 or 364, respectively, compared with the DCV EC90 value against a GT-1a (H77) reference replicon. It is therefore possible L28G also confers high resistance in the context of the GT-6 sequence.
A noteworthy observation from our study was the interplay between NS5A substitutions at L28 and R30 on in vitro OMV potency compared with those of the other tested NS5A drugs. In general, OMV potency was inferior against substitutions at L28 than those of DCV and VEL. OMV EC90 values were ≥19 nM against GT-6a-cons replicons harboring L28 (control) or harboring L28F or L28V substitution. Interestingly, OMV potency was dramatically enhanced to low-picomolar levels against GT-6a NS5A replicons harboring substitutions at R30 (A/N/S). Enhanced OMV potency was also maintained in HCV GT-6a NS5A replicons harboring various L28M/V-R30A/N/S substitution combinations compared with its potency against L28 substitutions alone. This is pertinent given that HCV GT-6 NS5A-R30 polymorphisms preexisted in combination with L28 polymorphisms in 60% of our evaluated sequences. Nevertheless, R30S could not rescue the loss of OMV potency conferred by L28A or L28T.
The NS5A-L31I polymorphism was only detected in combination with L28T in two GT-6b sequences from Thailand. This combination of substitutions was the most deleterious of tested substitutions in our in vitro HCV GT-6a-cons replicon assay with the abrogation of NS5A drug activity (EC90 of >5,000 nM).
The frequently observed baseline NS5A RAS, a histidine (H) at position 93, whose preexistence has led to reduced treatment response rates against HCV GT-1 and GT-3 infection (28, 39), was not detected in evaluated GT-6 NS5A sequences. NS5A-Y93S, however, was detected in 14% of evaluated sequences. Unlike GT-1a or GT-3 replicons harboring NS5A-Y93H (35, 40), fold drug resistance conferred by a GT-6a replicon harboring Y93S was low (approximately 4- to 5-fold against tested drugs). This frequently observed substitution, by itself, would not be expected to impact treatment response to NS5A drug-containing regimens.
An integration of HCV GT-6 clinical data recently described by Boyd et al. (15) showed very high SVR rates in 90 patients treated with different DAA regimens, including LDV/SOF and VEL/SOF. The majority of these patients were infected with HCV GT-6 subtypes representing -6a or -6e and treated with VEL/SOF. From the lack of highly resistant NS5A polymorphisms detected in our evaluated GT-6a sequences, excellent SVR rates would be anticipated. The GT-6 subtypes for HCV-infected patients treated with VEL/SOF were described by Hezode et al. (41). Four of these patients were infected with GT-6 subtypes representing -6m, -6n, and -6v, although subtype-specific NS5A polymorphisms were not described. However, Hezode et al. reported that VEL-specific GT-6 NS5A RASs L28M/V, T93S, and L28M/V-T93S preexisted in their patient-derived GT-6 NS5A sequences. In our analyses, all 14 GT-6 NS5A sequences representing GT-6m, -6n, and -6v harbored the L28V-T93S polymorphism combination with R30S, a polymorphism that confers no resistance to VEL, and when evaluated in the context of our GT-6a-cons replicon, it resulted in a VEL EC90 potency in the low-nanomolar range. This level of VEL potency in our replicon model may therefore correlate with a positive clinical outcome. In contrast, we observed significant potency reductions for VEL and the other tested drugs against various HCV GT-6a NS5A-L28A or -L28T replicon variants. These RASs were detected in patients infected with HCV GT-6 isolates representing subtypes -6b, -6f, and -6r and may impact treatment response to NS5A drug-containing regimens. In the Hezode analysis, none of the VEL/SOF-treated patients were infected with GT-6b, -6f, or -6r (41).
We tested the susceptibility of GT-6 NS5A substitutions to inhibition by four approved NS5A drugs but not to elbasvir and pibrentasvir (PIB). Elbasvir is not approved for the treatment of HCV GT-6 (42). PIB, which has been studied and approved for patients infected with HCV GT-6 (42), may be a substitute for VEL, although direct comparisons in the clinical trial setting have not been performed. Pham et al. (43) have studied VEL and PIB using an in vitro HCV GT-6a replicon model and showed that while VEL-resistant NS5A-L31V was susceptible to PIB inhibition, both were highly resistant to the L28S-L31V combination. Since GT-6b harbors NS5A-L28T-L31I, PIB may also confer resistance to this subtype.
A limitation of our study is that the susceptibilities of noted HCV GT-6 polymorphisms were tested by introduction into a HCV GT-6a-cons replicon. For respective NS3 or NS5B drugs targeting various HCV genotype subtypes harboring a RAS, in vitro susceptibilities and fold resistance changes conferred by the specific RAS have been fairly comparable irrespective of genotype and/or subtype (35). This is not always the case for drugs targeting NS5A (35). Establishing viable HCV GT-6 replicons representing NS5A subtypes other than GT-6a has been a challenge. Nevertheless, our susceptibility analyses of 1-, 2-, and 3-RAS combinations observed in evaluated HCV GT-6 subtype sequences still provides insight into how each drug interacts with the NS5A protein and the relative fold resistance for each drug. Although additional NS5A polymorphisms observed in certain HCV GT-6 subtype sequences may further influence NS5A drug susceptibilities, certain combinations of preexisting GT-6 NS5A RASs were shown to significantly impact drug susceptibility.
For patients infected with HCV GT-6, differentiation of subtype using the most commonly available commercial HCV genotyping assays, such as the VERSANT HCV Genotype 2.0 Line probe assay (Siemens Healthcare, Germany), is limited. In addition, the Abbott RealTime HCV genotype PCR assay (Abbott Molecular, USA) is not recommended for HCV GT-6 identification. The lack of reliable HCV genotyping assays could be an issue in Asian countries such as China and Thailand, where HCV GT-6 subtypes are more diverse and prominent. Thus, for GT-6 patients who fail currently approved DAA treatments, it may be important to confirm the GT-6 subtype and explore whether there is an association with SVR rates and various regimens. In the absence of a reliable commercial assay, HCV NS5A sequencing and phylogenetic analysis could be employed to identify the NS5A RAS and GT-6 subtype and facilitate retreatment options.
In conclusion, in vitro susceptibilities of GT-6a replicons harboring combinations of NS5A RASs that naturally occur in GT-6 subtypes -6b, -6f, and -6r were substantially reduced for the evaluated NS5A drugs. Given these findings, we suggest monitoring SVR rates to NS5A drug-containing regimens in patients infected with these HCV GT-6 subtypes to confirm any association between preexisting NS5A-L28A, -L28T, and possibly -L28G polymorphism combinations with treatment failure.
MATERIALS AND METHODS
Clinical samples.
Twenty-five plasma samples were obtained from patients who were infected with GT-6 HCV. Six were baseline samples from patients who enrolled in clinical studies assessing the safety and efficacy of DCV-based regimens, of which one was from ALLY-1 (AI444-215; ClinicalTrials.gov NCT02032875) (29), one was from UNITY-2 (AI443-113; ClinicalTrials.gov NCT01973049) (30), and four were from UNITY-4 (AI443-123; ClinicalTrials.gov NCT02170727) (33). Nine samples were obtained from Monash Health and Monash University (Melbourne, Australia), nine were purchased from Boca Biolistics (Pompano Beach, FL), and one was purchased from SeraCare Life Sciences (Milford, MA) and previously reported (44).
Genotypic analysis of clinical samples.
Viral RNA purification, cDNA synthesis, and PCR amplification were performed as previously described (19). The NS3, NS5A, and NS5B coding regions were amplified, population-based sequencing (threshold of ≥15% for reporting polymorphisms at each residue position) was performed using the BigDye Terminator v3.1 cycle sequencing kit (Applied Biosystems, Foster City, CA), and genotype-specific primers were selected from the list in Table S1 in the supplemental material. A GT-6a consensus sequence was generated for each target gene from each sample. Resulting sequences were compared to a GT-6a consensus strain (Fig. S1).
HCV GT-6 subtype distribution and prevalence of NS3, NS5A, and NS5B RASs were analyzed using 24 patient-derived NS3 sequences, 25 patient-derived NS5A sequences, and 24 patient-derived NS5B sequences with either 80 (for NS3 and NS5B) or 105 NS5A sequences from the usHCVdb. Accession numbers of the 80 NS3, 105 NS5A, and 80 NS5B database sequences are provided in Table S2.
Phenotypic analysis.
To evaluate the susceptibility of HCV GT-6 variants to NS3, NS5A, and NS5B inhibitors, a pGT-6a-cons replicon was constructed (Fig. S1), and substitutions of interest were introduced into this construct by site-directed mutagenesis and confirmed by sequence analysis. For GT-6 NS5A variants, if replication was not observed in the GT-6a-cons replicon, substitutions were also introduced into hybrid GT-6a NS5A constructs in the Con1 or JFH-1 replicon background (44, 45).
The susceptibility of cell lines harboring replicons with HCV GT-6 variants to inhibition by DAAs was evaluated using an HCV replicon luciferase-based assay as described previously (45). The 50% and 90% effective concentrations (EC50 and EC90) were calculated as the concentration of inhibitor required for a 50% and 90% reduction in luciferase activity. When testing the susceptibility of HCV GT-6a replicon variants to DAA inhibition, control assays employing replicons representing HCV GT-1b Con1 (44) or HCV GT-2a JFH-1 (45) and/or HCV GT-6-cons were performed in parallel.
Phylogenetic analysis.
Phylogenetic analyses were performed on NS3, NS5A, and NS5B sequences by aligning the sequences using MEGA, version 6, software (46) with the ClustalW algorithm. A maximum-likelihood tree was created based on the alignments and drawn using MEGA, version 6, software. The NS3, NS5A, and NS5B analyses used 24 patient-derived NS3 sequences (encoding amino acids 1 to 181), 25 patient-derived NS5A sequences (encoding amino acids 1 to 359), or 24 patient-derived NS5B sequences (encoding amino acids 72 to 549), respectively, each with HCV GT-6 infection (Table S3), and 30 prototype GT-6 sequences of assigned subtypes (1) from the Los Alamos HCV Sequence Database (usHCVdb) (47) (Table S2). Subtyping of patient-drived GT-6 sequences was based on the phylogenetic grouping of patient-derived sequences using usHCVdb sequences with confirmed GT-6 subtypes.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dennis Hernandez for his work on HCV GT-6a NS3 constructs and Bernie Kienzle for her support with sequencing. Fiona McPhee is an employee of Bristol-Meyers Squibb. This work was funded by Bristol-Myers Squibb.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.02205-18.
REFERENCES
- 1.Smith DB, Bukh J, Kuiken C, Muerhoff AS, Rice CM, Stapleton JT, Simmonds P. 2017. HCV classification. International Committee on Taxonomy of Viruses. https://talk.ictvonline.org/ictv_wikis/flaviviridae/w/sg_flavi/56/hcv-classification.
- 2.Salemi M, Vandamme AM. 2002. Hepatitis C virus evolutionary patterns studied through analysis of full-genome sequences. J Mol Evol 54:62–70. doi: 10.1007/s00239-001-0018-9. [DOI] [PubMed] [Google Scholar]
- 3.Wang H, Yuan Z, Barnes E, Yuan M, Li C, Fu Y, Xia X, Li G, Newton PN, Vongsouvath M, Klenerman P, Pybus OG, Murphy D, Abe K, Lu L. 2013. Eight novel hepatitis C virus genomes reveal the changing taxonomic structure of genotype 6. J Gen Virol 94:76–80. doi: 10.1099/vir.0.047506-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li C, Pham VH, Abe K, Lu L. 2014. Nine additional complete genome sequences of HCV genotype 6 from Vietnam including new subtypes 6xb and 6xc. Virology 468-470:172–177. doi: 10.1016/j.virol.2014.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li C, Barnes E, Newton PN, Fu Y, Vongsouvath M, Klenerman P, Okamoto H, Abe K, Pybus OG, Lu L. 2015. An expanded taxonomy of hepatitis C virus genotype 6: characterization of 22 new full-length viral genomes. Virology 476:355–363. doi: 10.1016/j.virol.2014.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bernier L, Willems B, Delage G, Murphy DG. 1996. Identification of numerous hepatitis C virus genotypes in Montreal, Canada. J Clin Microbiol 34:2815–2818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pham VH, Nguyen HD, Ho PT, Banh DV, Pham HL, Pham PH, Lu L, Abe K. 2011. Very high prevalence of hepatitis C virus genotype 6 variants in southern Vietnam: large-scale survey based on sequence determination. Jpn J Infect Dis 64:537–539. [PMC free article] [PubMed] [Google Scholar]
- 8.Pybus OG, Barnes E, Taggart R, Lemey P, Markov PV, Rasachak B, Syhavong B, Phetsouvanah R, Sheridan I, Humphreys IS, Lu L, Newton PN, Klenerman P. 2009. Genetic history of hepatitis C virus in East Asia. J Virol 83:1071–1082. doi: 10.1128/JVI.01501-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shinji T, Kyaw YY, Gokan K, Tanaka Y, Ochi K, Kusano N, Mizushima T, Fujioka S, Shiraha H, Lwin AA, Shiratori Y, Mizokami M, Khin M, Miyahara M, Okada S, Koide N. 2004. Analysis of HCV genotypes from blood donors shows three new HCV type 6 subgroups exist in Myanmar. Acta Med Okayama 58:135–142. doi: 10.18926/AMO/32110. [DOI] [PubMed] [Google Scholar]
- 10.Stuyver L, Wyseur A, van Arnhem W, Hernandez F, Maertens G. 1996. Second-generation line probe assay for hepatitis C virus genotyping. J Clin Microbiol 34:2259–2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Petruzziello A, Marigliano S, Loquercio G, Cozzolino A, Cacciapuoti C. 2016. Global epidemiology of hepatitis C virus infection: an up-date of the distribution and circulation of hepatitis C virus genotypes. World J Gastroenterol 22:7824–7840. doi: 10.3748/wjg.v22.i34.7824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Murphy DG, Willems B, Deschenes M, Hilzenrat N, Mousseau R, Sabbah S. 2007. Use of sequence analysis of the NS5B region for routine genotyping of hepatitis C virus with reference to C/E1 and 5' untranslated region sequences. J Clin Microbiol 45:1102–1112. doi: 10.1128/JCM.02366-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nainan OV, Alter MJ, Kruszon-Moran D, Gao FX, Xia G, McQuillan G, Margolis HS. 2006. Hepatitis C virus genotypes and viral concentrations in participants of a general population survey in the United States. Gastroenterology 131:478–484. doi: 10.1053/j.gastro.2006.06.007. [DOI] [PubMed] [Google Scholar]
- 14.Feeney ER, Chung RT. 2014. Antiviral treatment of hepatitis C. BMJ 348:g3308. doi: 10.1136/bmj.g3308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boyd SD, Harrington P, Komatsu TE, Naeger LK, Chan-Tack K, Murray J, Birnkrant D, Struble K. 2018. HCV genotype 4, 5 and 6: distribution of viral subtypes and sustained virologic response rates in clinical trials of approved direct-acting antiviral regimens. J Viral Hepat 25:969–975. doi: 10.1111/jvh.12896. [DOI] [PubMed] [Google Scholar]
- 16.Brown A, Hezode C, Zuckerman E, Foster GR, Zekry A, Roberts SK, Lahser F, Durkan C, Badshah C, Zhang B, Robertson M, Wahl J, Barr E, Haber B, C-SCAPE Study Investigators. 2018. Efficacy and safety of 12 weeks of elbasvir +/− grazoprevir +/− ribavirin in participants with hepatitis C virus genotype 2, 4, 5 or 6 infection: the C-SCAPE study. J Viral Hepat 25:457–464. doi: 10.1111/jvh.12801. [DOI] [PubMed] [Google Scholar]
- 17.Fridell RA, Wang C, Sun JH, O'Boyle DR Jr, Nower P, Valera L, Qiu D, Roberts S, Huang X, Kienzle B, Bifano M, Nettles RE, Gao M. 2011. Genotypic and phenotypic analysis of variants resistant to hepatitis C virus nonstructural protein 5A replication complex inhibitor BMS-790052 in humans: in vitro and in vivo correlations. Hepatology 54:1924–1935. doi: 10.1002/hep.24594. [DOI] [PubMed] [Google Scholar]
- 18.Poveda E, Wyles DL, Mena A, Pedreira JD, Castro-Iglesias A, Cachay E. 2014. Update on hepatitis C virus resistance to direct-acting antiviral agents. Antiviral Res 108:181–191. doi: 10.1016/j.antiviral.2014.05.015. [DOI] [PubMed] [Google Scholar]
- 19.McPhee F, Hernandez D, Yu F, Ueland J, Monikowski A, Carifa A, Falk P, Wang C, Fridell R, Eley T, Zhou N, Gardiner D. 2013. Resistance analysis of hepatitis C virus genotype 1 prior treatment null responders receiving daclatasvir and asunaprevir. Hepatology 58:902–911. doi: 10.1002/hep.26388. [DOI] [PubMed] [Google Scholar]
- 20.Nguyen LT, Gray E, Dean J, Carr M, Connell J, De Gascun C, Nguyen LA, O’Leary A, Bergin C, Hall W, Norris S. 2015. Baseline prevalence and emergence of protease inhibitor resistance mutations following treatment in chronic HCV genotype-1-infected individuals. Antivir Ther 20:865–869. doi: 10.3851/IMP2964. [DOI] [PubMed] [Google Scholar]
- 21.McPhee F, Hernandez D, Zhou N, Ueland J, Yu F, Vellucci V, Huang X, Wang X, Ishikawa H, Karino Y, Kumada H. 2018. Pooled analysis of HCV genotype 1 resistance-associated substitutions in NS5A, NS3 and NS5B pre-and post-treatment with 12 weeks of daclatasvir, asunaprevir and beclabuvir. Antivir Ther 23:53–66. doi: 10.3851/IMP3177. [DOI] [PubMed] [Google Scholar]
- 22.Gao M, Nettles RE, Belema M, Snyder LB, Nguyen VN, Fridell RA, Serrano-Wu MH, Langley DR, Sun J-H, O’Boyle DR Jr, Lemm JA, Wang C, Knipe JO, Chien C, Colonno RJ, Grasela DM, Meanwell NA, Hamann LG. 2010. Chemical genetics strategy identifies an HCV NS5A inhibitor with a potent clinical effect. Nature 465:96–100. doi: 10.1038/nature08960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kumada H, Suzuki Y, Ikeda K, Toyota J, Karino Y, Chayama K, Kawakami Y, Ido A, Yamamoto K, Takaguchi K, Izumi N, Koike K, Takehara T, Kawada N, Sata M, Miyagoshi H, Eley T, McPhee F, Damokosh A, Ishikawa H, Hughes E. 2014. Daclatasvir plus asunaprevir for chronic HCV genotype 1b infection. Hepatology 59:2083–2091. doi: 10.1002/hep.27113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Manns M, Pol S, Jacobson IM, Marcellin P, Gordon SC, Peng CY, Chang TT, Everson GT, Heo J, Gerken G, Yoffe B, Towner WJ, Bourliere M, Metivier S, Chu CJ, Sievert W, Bronowicki JP, Thabut D, Lee YJ, Kao JH, McPhee F, Kopit J, Mendez P, Linaberry M, Hughes E, Noviello S, HALLMARK-DUAL Study Team. 2014. All-oral daclatasvir plus asunaprevir for hepatitis C virus genotype 1b: a multinational, phase 3, multicohort study. Lancet 384:1597–1605. doi: 10.1016/S0140-6736(14)61059-X. [DOI] [PubMed] [Google Scholar]
- 25.Zeuzem S, Hezode C, Bronowicki JP, Loustaud-Ratti V, Gea F, Buti M, Olveira A, Banyai T, Al-Assi MT, Petersen J, Thabut D, Gadano A, Pruitt R, Makara M, Bourliere M, Pol S, Beumont-Mauviel M, Ouwerkerk-Mahadevan S, Picchio G, Bifano M, McPhee F, Boparai N, Cheung K, Hughes EA, Noviello S, LEAGUE-1 Study Team. 2016. Daclatasvir plus simeprevir with or without ribavirin for the treatment of chronic hepatitis C virus genotype 1 infection. J Hepatol 64:292–300. doi: 10.1016/j.jhep.2015.09.024. [DOI] [PubMed] [Google Scholar]
- 26.Sulkowski MS, Gardiner DF, Rodriguez-Torres M, Reddy KR, Hassanein T, Jacobson I, Lawitz E, Lok AS, Hinestrosa F, Thuluvath PJ, Schwartz H, Nelson DR, Everson GT, Eley T, Wind-Rotolo M, Huang SP, Gao M, Hernandez D, McPhee F, Sherman D, Hindes R, Symonds W, Pasquinelli C, Grasela DM, AI444040 Study Group. 2014. Daclatasvir plus sofosbuvir for previously treated or untreated chronic HCV infection. N Engl J Med 370:211–221. doi: 10.1056/NEJMoa1306218. [DOI] [PubMed] [Google Scholar]
- 27.Wyles DL, Ruane PJ, Sulkowski MS, Dieterich D, Luetkemeyer A, Morgan TR, Sherman KE, Dretler R, Fishbein D, Gathe JC Jr, Henn S, Hinestrosa F, Huynh C, McDonald C, Mills A, Overton ET, Ramgopal M, Rashbaum B, Ray G, Scarsella A, Yozviak J, McPhee F, Liu Z, Hughes E, Yin PD, Noviello S, Ackerman P, ALLY-2 Investigators. 2015. Daclatasvir plus sofosbuvir for HCV in patients coinfected with HIV-1. N Engl J Med 373:714–725. doi: 10.1056/NEJMoa1503153. [DOI] [PubMed] [Google Scholar]
- 28.Nelson DR, Cooper JN, Lalezari JP, Lawitz E, Pockros PJ, Gitlin N, Freilich BF, Younes ZH, Harlan W, Ghalib R, Oguchi G, Thuluvath PJ, Ortiz-Lasanta G, Rabinovitz M, Bernstein D, Bennett M, Hawkins T, Ravendhran N, Sheikh AM, Varunok P, Kowdley KV, Hennicken D, McPhee F, Rana K, Hughes EA, ALLY-3 Study Team. 2015. All-oral 12-week treatment with daclatasvir plus sofosbuvir in patients with hepatitis C virus genotype 3 infection: ALLY-3 phase III study. Hepatology 61:1127–1135. doi: 10.1002/hep.27726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Poordad F, Schiff ER, Vierling JM, Landis C, Fontana RJ, Yang R, McPhee F, Hughes EA, Noviello S, Swenson ES. 2016. Daclatasvir with sofosbuvir and ribavirin for hepatitis C virus infection with advanced cirrhosis or post-liver transplantation recurrence. Hepatology 63:1493–1505. doi: 10.1002/hep.28446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Muir AJ, Poordad F, Lalezari J, Everson G, Dore GJ, Herring R, Sheikh A, Kwo P, Hezode C, Pockros PJ, Tran A, Yozviak J, Reau N, Ramji A, Stuart K, Thompson AJ, Vierling J, Freilich B, Cooper J, Ghesquiere W, Yang R, McPhee F, Hughes EA, Swenson ES, Yin PD. 2015. Daclatasvir in combination with asunaprevir and beclabuvir for hepatitis C virus genotype 1 infection with compensated cirrhosis. JAMA 313:1736–1744. doi: 10.1001/jama.2015.3868. [DOI] [PubMed] [Google Scholar]
- 31.Toyota J, Karino Y, Suzuki F, Ikeda F, Ido A, Tanaka K, Takaguchi K, Naganuma A, Tomita E, Chayama K, Fujiyama S, Inada Y, Yoshiji H, Watanabe H, Ishikawa H, Hu W, McPhee F, Linaberry M, Yin PD, Swenson ES, Kumada H. 2017. Daclatasvir/asunaprevir/beclabuvir fixed-dose combination in Japanese patients with HCV genotype 1 infection. J Gastroenterol 52:385–395. doi: 10.1007/s00535-016-1245-6. [DOI] [PubMed] [Google Scholar]
- 32.Hezode C, Aberge LA, Chas J, Conti F, Cotte L, Tateo M, Alric L, Vergniol J. 2018. Sustained virologic response to daclatasvir and sofosbuvir, with or without ribavirin, among patients in the French ATU programme infected with HCV genotype 4, 5 and 6. J Hepatol 64:S755. doi: 10.1016/S0168-8278(16)01471-9. [DOI] [Google Scholar]
- 33.Kao JH, Yu ML, Peng CY, Heo J, Chu CJ, Chang TT, Lee YJ, Hu TH, Yoon KT, Paik SW, Lim YS, Ahn SH, Isakov V, McPhee F, Hu W, Scott Swenson E, Yin PD, Treitel M. 2017. Daclatasvir/asunaprevir/beclabuvir, all-oral, fixed-dose combination for patients with chronic hepatitis C virus genotype 1. J Gastroenterol Hepatol 32:1998–2005. doi: 10.1111/jgh.13796. [DOI] [PubMed] [Google Scholar]
- 34.McPhee F, Ueland J, Vellucci V, Bowden S, Sievert W, Zhou N. 2017. J Gastroenterol Hepatol 32(Suppl S3):165. [Google Scholar]
- 35.Lontok E, Harrington P, Howe A, Kieffer T, Lennerstrand J, Lenz O, McPhee F, Mo H, Parkin N, Pilot-Matias T, Miller V. 2015. Hepatitis C virus drug resistance-associated substitutions: state of the art summary. Hepatology 62:1623–1632. doi: 10.1002/hep.27934. [DOI] [PubMed] [Google Scholar]
- 36.McPhee F, Hernandez D, Zhou N, Yu F, Ueland J, Monikowski A, Chayama K, Toyota J, Izumi N, Yokosuka O, Kawada N, Osaki Y, Hughes EA, Watanabe H, Ishikawa H, Kumada H. 2014. Virological escape in HCV genotype-1-infected patients receiving daclatasvir plus ribavirin and peginterferon alfa-2a or alfa-2b. Antivir Ther 19:479–490. doi: 10.3851/IMP2729. [DOI] [PubMed] [Google Scholar]
- 37.Sims KD, Lemm J, Eley T, Liu M, Berglind A, Sherman D, Lawitz E, Vutikullird AB, Tebas P, Gao M, Pasquinelli C, Grasela DM. 2014. Randomized, placebo-controlled, single-ascending-dose study of BMS-791325, a hepatitis C virus (HCV) NS5B polymerase inhibitor, in HCV genotype 1 infection. Antimicrob Agents Chemother 58:3496–3503. doi: 10.1128/AAC.02579-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ji H, Kozak RA, Biondi MJ, Pilon R, Vallee D, Liang BB, La D, Kim J, Van Domselaar G, Leonard L, Sandstrom P, Brooks J. 2015. Next generation sequencing of the hepatitis C virus NS5B gene reveals potential novel S282 drug resistance mutations. Virology 477:1–9. doi: 10.1016/j.virol.2014.12.037. [DOI] [PubMed] [Google Scholar]
- 39.McPhee F, Suzuki Y, Toyota J, Karino Y, Chayama K, Kawakami Y, Yu ML, Ahn SH, Ishikawa H, Bhore R, Zhou N, Hernandez D, Mendez P, Kumada H. 2015. High sustained virologic response to daclatasvir plus asunaprevir in elderly and cirrhotic patients with hepatitis C virus genotype 1b without baseline NS5A polymorphisms. Adv Ther 32:637–649. doi: 10.1007/s12325-015-0221-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hernandez D, Zhou N, Ueland J, Monikowski A, McPhee F. 2013. Natural prevalence of NS5A polymorphisms in subjects infected with hepatitis C virus genotype 3 and their effects on the antiviral activity of NS5A inhibitors. J Clin Virol 57:13–18. doi: 10.1016/j.jcv.2012.12.020. [DOI] [PubMed] [Google Scholar]
- 41.Hezode C, Reau N, Svarovskaia ES, Doehle BP, Shanmugam R, Dvory-Sobol H, Hedskog C, McNally J, Osinusi A, Brainard DM, Miller MD, Mo H, Roberts SK, O'Leary JG, Shafran SD, Zeuzem S. 2018. Resistance analysis in patients with genotype 1-6 HCV infection treated with sofosbuvir/velpatasvir in the phase III studies. J Hepatol 68:895–903. doi: 10.1016/j.jhep.2017.11.032. [DOI] [PubMed] [Google Scholar]
- 42.Boyd SD, Tracy L, Komatsu TE, Harrington PR, Viswanathan P, Murray J, Sherwat A. 2017. US FDA perspective on elbasvir/grazoprevir treatment for patients with chronic hepatitis C virus genotype 1 or 4 infection. Clin Drug Investig 37:317–326. doi: 10.1007/s40261-017-0492-5. [DOI] [PubMed] [Google Scholar]
- 43.Pham LV, Ramirez S, Gottwein JM, Fahnoe U, Li YP, Pedersen J, Bukh J. 2018. HCV genotype 6a escape from and resistance to velpatasvir, pibrentasvir, and sofosbuvir in robust infectious cell culture models. Gastroenterology 154:2194–2208. doi: 10.1053/j.gastro.2018.02.017. [DOI] [PubMed] [Google Scholar]
- 44.Wang C, Jia L, O'Boyle DR Jr, Sun JH, Rigat K, Valera L, Nower P, Huang X, Kienzle B, Roberts S, Gao M, Fridell RA. 2014. Comparison of daclatasvir resistance barriers on NS5A from hepatitis C virus genotypes 1 to 6: implications for cross-genotype activity. Antimicrob Agents Chemother 58:5155–5163. doi: 10.1128/AAC.02788-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fridell RA, Qiu D, Wang C, Valera L, Gao M. 2010. Resistance analysis of the hepatitis C virus NS5A inhibitor BMS-790052 in an in vitro replicon system. Antimicrob Agents Chemother 54:3641–3650. doi: 10.1128/AAC.00556-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kuiken C, Yusim K, Boykin L, Richardson R. 2005. The Los Alamos hepatitis C sequence database. Bioinformatics 21:379–384. doi: 10.1093/bioinformatics/bth485. [DOI] [PubMed] [Google Scholar]
- 48.Zhou N, Hernandez D, Ueland J, Yang X, Yu F, Sims K, Yin PD, McPhee F. 2016. NS5A sequence heterogeneity and mechanisms of daclatasvir resistance in hepatitis C virus genotype 4 infection. J Infect Dis 213:206–215. doi: 10.1093/infdis/jiv379. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.