Rifapentine is a rifamycin used to treat tuberculosis. As is the case for rifampin, plasma exposures of rifapentine are associated with the treatment response.
KEYWORDS: AADAC, SLCO1B1, pharmacogenetics, population pharmacokinetics, rifapentine, tuberculosis
ABSTRACT
Rifapentine is a rifamycin used to treat tuberculosis. As is the case for rifampin, plasma exposures of rifapentine are associated with the treatment response. While concomitant food intake and HIV infection explain part of the pharmacokinetic variability associated with rifapentine, few studies have evaluated the contribution of genetic polymorphisms. We evaluated the effects of functionally significant polymorphisms of the genes encoding OATP1B1, the pregnane X receptor (PXR), constitutive androstane (CAR), and arylacetamide deacetylase (AADAC) on rifapentine exposure. Two studies evaluating novel regimens among southern African patients with drug-susceptible pulmonary tuberculosis were included in this analysis. In the RIFAQUIN study, rifapentine was administered in the continuation phase of antituberculosis treatment in 1,200-mg-once-weekly or 900-mg-twice-weekly doses. In the Daily RPE study, 450 or 600 mg was given daily during the intensive phase of treatment. Nonlinear mixed-effects modeling was used to describe the pharmacokinetics of rifapentine and to identify significant covariates. A total of 1,144 drug concentration measurements from 326 patients were included in the analysis. Pharmacogenetic information was available for 162 patients. A one-compartment model with first-order elimination and transit compartment absorption described the data well. In a typical patient (body weight, 56 kg; fat-free mass, 45 kg), the values of clearance and volume of distribution were 1.33 liters/h and 25 liters, respectively. Patients carrying the AA variant (65.4%) of AADAC rs1803155 were found to have a 10.4% lower clearance. HIV-infected patients had a 21.9% lower bioavailability. Once-weekly doses of 1,200 mg were associated with a reduced clearance (13.2%) compared to that achieved with more frequently administered doses. Bioavailability was 23.3% lower among patients participating in the Daily RPE study than in those participating in the RIFAQUIN study. This is the first study to report the effect of AADAC rs1803155AA on rifapentine clearance. The observed increase in exposure is modest and unlikely to be of clinical relevance. The difference in bioavailability between the two studies is probably related to the differences in food intake concomitant with the dose. HIV-coinfected patients had lower rifapentine exposures.
INTRODUCTION
Rifamycins play a key role in the multidrug treatment of tuberculosis. Their sterilizing activity is exposure dependent (1–3). Rifapentine was approved by the Food and Drug Administration (FDA) in 1998 for the treatment of pulmonary tuberculosis (3, 4). Rifapentine pharmacokinetics are influenced by age, weight, dosing pattern, human immunodeficiency virus (HIV) infection, and sex (5, 6). Rifapentine is less rapidly absorbed than rifampin, with peak plasma concentrations being reached within 5 h. Concomitant food intake markedly increases its absorption; the extent of rifapentine absorption increased by 33 to 86% when given with meals (7). Rifapentine has a half-life of approximately 12 h in humans (8, 9). With its long half-life and excellent sterilizing activity, rifapentine is an attractive alternative to rifampin and is increasingly used to treat active tuberculosis and latent infection. However, there is marked interpatient variability in rifamycin pharmacokinetics (10). The primary metabolic pathways for rifapentine involve deacetylation to the primary enzymatic metabolite, 25-desacetyl rifapentine, which is mediated by human arylacetamide deacetylase (AADAC), and nonenzymatic hydrolysis, resulting in the formation of the secondary metabolites 3-formyl rifapentine and 3-formyldesacetyl rifapentine (11). The protein binding of rifapentine is estimated to be about 98% (3, 12). Like other rifamycins, rifapentine induces its own metabolism (9).
Previously published data indicate that single nucleotide polymorphisms (SNPs) in the solute carrier organic anion transporter 1B1 (SLCO1B1) gene encoding the OATP1B1 transmembrane receptor affect rifampin concentrations (13, 14). The SLCO1B1 rs4149032 C > T polymorphism, found in 70% of South Africans with tuberculosis living in Cape Town, was associated with 20% and 28% reductions in rifampin bioavailability in heterozygotes and homozygotes, respectively (14). Rifamycins are also substrates of the drug efflux pump P glycoprotein, coded for by the polymorphic ABCB1 gene (15), and are metabolized mainly by polymorphic human arylacetamide deacetylase (AADAC) (16). Human rifamycin exposures are also modulated by the pregnane X receptor (PXR) and constitutive androstane (CAR) nuclear receptors (17). Since the development of resistance to rifamycins and their bactericidal effects are related to rifamycin concentrations, SNPs substantially influencing rifamycin concentrations may be of therapeutic importance. Little is known about the pharmacogenetic correlates of rifapentine pharmacokinetics, which may potentially help in finding the optimal dose of rifapentine. Therefore, the aim of this study was to determine the effect of polymorphisms of SLCO1B1, PXR, CAR, and AADAC on rifapentine pharmacokinetics.
RESULTS
A total of 326 patients were included in the study and contributed a total of 1,151 concentrations-time points. Only 7 concentrations were below the lower limit of quantification (LLOQ) and were omitted from the analysis. The median body weight and the median age of the study participants were 56 kg and 32 years, respectively. All demographic characteristics are summarized in Table 1.
TABLE 1.
Demographic and clinical characteristics of patients
| Demographic or clinical characteristic | Values for patients from the following study: |
||||
|---|---|---|---|---|---|
| Daily RPE, 450 mg (n = 44) | Daily RPE, 600 mg (n = 41) | RIFAQUIN, 900 mg (n = 116) | RIFAQUIN, 1,200 mg (n = 125) | Overall (n = 326) | |
| No. of samples for PKa analysis | 166 | 130 | 416 | 432 | 1,144 |
| No. of males/no. of females | 33/11 | 32/9 | 72/44 | 81/44 | 218/108 |
| No. (%) of HIV-positive patients | 6 (13.6) | 7 (17.1) | 30 (25.9) | 16(12.8) | 59(18.1) |
| Median (range) age (yr) | 29 (19–61) | 29 (18–63) | 31 (19–64) | 34 (19– 80) | 32 (18–80) |
| Median (range) wt (kg) | 55 (45–79) | 55 (45–94) | 55 (38–77) | 57 (38– 78) | 56 (38–94) |
| Median (range) FFM (kg) | 47 (32–58) | 47 (32–56) | 45 (27–62) | 45 (27–60) | 45 (27–62) |
PK, pharmacokinetic.
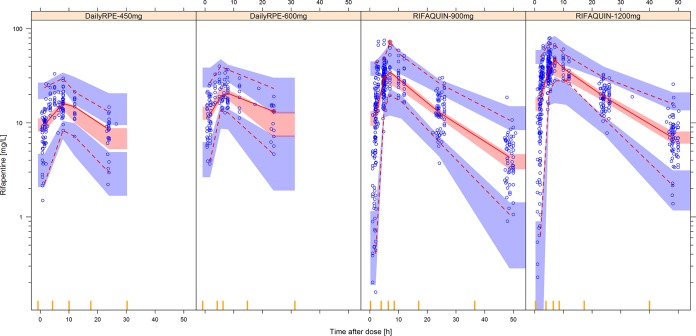
The population pharmacokinetics of rifapentine were well described by a one-compartment model with first-order elimination and transit compartment absorption. Fat-free mass (FFM) was found to be the best size descriptor for clearance (change in the NONMEM objective function value [ΔOFV], 93 points [P < 0.001] when including FFM for allometric scaling on clearance and 23 points better than when using body weight), and total body weight was found to be the best size descriptor for the volume of distribution (ΔOFV, 20; P < 0.001). The absorption of rifapentine was described using a series of transit compartments, which significantly improved the model with respect to the use of simple first-order absorption (ΔOFV, 421; P < 0.001). In a typical patient (FFM, 46 kg; weight, 56 kg), the values of clearance and volume of distribution were 1.33 liters/h and 25 liters, respectively. Final parameter estimates (shown in Table 2) were in agreement with the previously published results (6, 18), and a visual predictive check (VPC) of the final model is shown in Fig. 1.
TABLE 2.
Final parameter estimates for rifapentine population pharmacokinetic modela
| Parameter | Estimate |
Variability |
||
|---|---|---|---|---|
| Value | 95% CIc | % CV | 95% CI | |
| CLb (liters/h) | 1.33 | 1.14, 1.54 | 23.0 (IIV) | 17.7, 28.6 |
| Vb (liters) | 25 | 21.9, 28.4 | 12.8 (IIV) | 8.8, 17.4 |
| ka (h−1) | 0.814 | 0.568, 1.26 | 48.9 (IOV) | 36.4, 59.8 |
| MTT (h) | 1.47 | 1.20, 1.78 | 37.4 (IOV) | 28.3, 48.6 |
| NN | 10.2 | 6.70, 14.0 | ||
| F | 1 (fixed) | 20.3 (IOV) | 14.9, 26.4 | |
| Proportional residual error (%) | 9.56 | 7.09, 13.2 | ||
| Additive residual error (mg/liter) | 0.247 | 0.143, 0.401 | ||
| Effect of HIV+ on F (%) | −21.9 | −33.2, −6.64 | ||
| Effect of group on 1,200-mg dose in RIFAQUIN study on CL (%) | −13.2 | −22.8, −4.36 | ||
| Effect of Daily RPE study on F (%) | −23.3 | −35.6, −9.25 | ||
| AADAC rs1803155 (AA) effect on CL (%) | −10.4 | −17.3, −3.53 | ||
CL, oral clearance; V, apparent volume of distribution in the central compartment; ka, first-order absorption rate constant; MTT, absorption mean transit time; NN, number of hypothetical transit compartments; F, oral bioavailability; HIV+, human immunodeficiency virus positivity; AADAC, arylacetamide deacetylase gene; IIV, interindividual variability; IOV, interoccasion variability; CV, coefficient of variation; CI, confidence interval.
The typical values of clearance and volume of distribution reported for a patient with a body weight of 56 kg and FFM of 46 kg.
The 95% confidence interval of parameter estimates was obtained with sampling importance resampling (SIR; n = 1,000) of the final model.
FIG 1.
Visual predictive check (VPC) for the final rifapentine population pharmacokinetic model in log scale, stratified according to the different dose groups in the analysis. The lower, middle, and upper solid lines are the 2.5th, 50th, and 97.5th percentiles of the observed plasma concentration, respectively. The shaded areas are the 95% confidence intervals for the same percentiles, obtained from resimulations of the same trial.
Of 326 patients, pharmacogenetic data were available for 162 (49.7%), all of whom were enrolled from South African sites. The distribution of genotype and allele frequencies are presented in Table 3. SLCO1B1 rs2306283 and AADAC rs1803155 variant alleles were found in 82% of patients, whereas the NR1I2 rs2472677 and NR1I2 rs1523130 variant alleles existed at a low overall frequency of 33.5% and 16.4%, respectively. In keeping with our previous findings among South Africans in Cape Town (14), the SLCO1B1 rs4149032 variant allele frequency was found to be 0.75 (Table 3).
TABLE 3.
Observed genotype and allele frequency of single nucleotide polymorphisms in the studya
| Genotype | Genotype (frequencyb) | Allele (frequency) | |||
|---|---|---|---|---|---|
| SLCO1B1 A > G rs2306283 | AA (8, 4.94) | AG (43, 26.5) | GG (111, 68.5) | A (0.18) | G (0.82) |
| SLCO1B1 C > T rs4149032 | CC (15, 9.26) | CT (52, 32.1) | TT (95, 58.6) | C (0.25) | T (0.75) |
| NR1I2 C > T rs2472677c | CC (71, 44.1) | CT (72, 44.7) | TT (18, 11.2) | C (0.67) | T (0.34) |
| NR1I2 T > C rs1523130 | TT (116, 71.6) | CT (39, 24.1) | CC (7, 4.3) | T (0.84) | C (0.16) |
| AADAC G > A rs1803155 | GG (3, 1.85) | GA (53, 32.7) | AA (106, 65.4) | G (0.18) | A (0.82) |
Data are for 162 patients, unless indicated otherwise.
The data in parentheses represent the number, percent, of patients.
Data are available only for 161 patients.
After screening and inclusion of genetic information (and imputation of the missing genotype with a mixture model), patients homozygous for the AADAC rs1803155 AA polymorphism were found to have a 10.4% lower clearance of rifapentine than subjects that were rs1803155 GG or GA (ΔOFV, 6.2; P = 0.013). Initially, the three categories of rs1803155 (AA, GA, GG) were analyzed as separate groups to estimate the respective effects of GA and GG. However, the estimated effects were similar for GG and GA, and when combined, the model goodness of fit (GOF) was not affected. Using the principle of parsimony, we decided to use the simpler model, as the effects of GG and GA were not statistically significant. The other pharmacogenetic variants did not affect the pharmacokinetic parameters.
Patients infected with HIV were found to have a 21.9% lower bioavailability (ΔOFV, 42; P < 0.001). The patients who were treated with high 1,200-mg doses of rifapentine tended to have clearance reduced by 13.2% compared to the clearance for the patients in the other dose groups (ΔOFV, 17; P < 0.001). The pharmacokinetic differences between the two studies were explored, and it was found that the bioavailability of rifapentine in the Daily RPE study was 23.3% lower than that in the RIFAQUIN study (ΔOFV, 59; P < 0.001). The pharmacogenetic covariates other than the AADAC rs1803155 polymorphism did not have significant effects on the pharmacokinetic parameters.
DISCUSSION
The present study is the first to investigate the influence of various plausible physiologically relevant candidate gene polymorphisms on rifapentine pharmacokinetics. We developed a population pharmacokinetic model of rifapentine which was consistent with that developed in previous studies and tested the effect of genotype information on the pharmacokinetic parameters. We showed that the AADAC rs1803155 polymorphism is associated with rifapentine clearance. Subjects carrying the AA genotype had a 10.4% lower clearance than those carrying AG or GG, thus leading to increased rifapentine exposure. The low clearance due to this polymorphism is consistent with the findings of previous studies reporting the decreased activity of AADAC due to the presence of the variant allele (19). The majority of patients in our study had the AADAC rs1803155 AA variant allele, which occurred at a frequency of 0.82, and 65% were homozygous for the single nucleotide polymorphism, which could, in part, account for the relatively high rifapentine exposures described. The polymorphism occurs at lower frequencies of 0.50 to 0.64 in European American, African American, Korean, and Japanese populations (19). Another study identified lower rifapentine concentrations in black Africans, but the influence of pharmacogenetic factors, which might account for the difference in the genotype frequencies between the populations, was not explored (20), whereas Sloan et al., who explored the influence of AADAC gene polymorphisms on rifampin pharmacokinetics in Malawian patients, did not identify a significant relationship (21).The prevalence of variant genotypes is different between African ethnic groups and may be the reason for this contrasting effect. As only 3 of 162 patients had rs1803155 GG, no meaningful separate estimate of clearance for this genotype could be obtained. In further attempts to explain the variability in rifapentine pharmacokinetics, we explored the effects of several polymorphisms of drug transporters and transcriptional regulators. The choice of polymorphisms was based on those previously described to affect drug disposition and also previous pharmacogenetic studies conducted with rifampin. Interestingly, we could not detect the effect of the SLCO1B1 rs4149032 polymorphism on the pharmacokinetics of rifapentine, even with a carrier-no carrier approach. The frequency of SLCO1B1 in our cohort was 0.75, which is in agreement with previous findings in South African patients from the Cape Town region. Similarly, we did not find a statistically significant effect associated with SLCO1B1 rs2306283, which existed in our study population at a frequency of 0.82. SLCO1B1 polymorphisms have been reported to be associated with low rifampin levels (13, 14), and the lack of an effect on rifapentine may suggest differences in the absorption, distribution, metabolism, and excretion of the two drugs. It may be that this transporter does not play a major role in the pharmacokinetics of rifapentine or that the variant allele is associated with greater induction by rifampin. We did not observe an effect due to polymorphisms of the transcriptional regulators. This could be due to the activation of PXR or CAR by rifapentine, which may have overridden any constitutive effects.
Additionally, we found that HIV-infected patients have a lower bioavailability of rifapentine. While the association of HIV infection with antituberculosis drug exposures is inconsistent, our findings for rifapentine are consistent with those from recent studies (22–24). The data available were not sufficient to identify potential interactions of rifapentine with the various antiretroviral drugs prescribed concomitantly.
Patients in the higher-dose group (1,200 mg given once weekly) had increased exposure in the current study, contrary to the findings of Savic et al., who described a decrease in the bioavailability of rifapentine with increased dose (6). The reduced dosing frequency in this group may have led to reduced autoinduction and, thus, increased exposure.
Previous reports demonstrated that exposure to rifamycins is reduced in males due to a higher FFM/body weight ratio (25). The study by Langdon et al. described a 35% reduction in the clearance of 25-desacetyl rifapentine among females (5). In the present analysis, as allometric scaling with FFM accounted for the variability associated with sex, we did not observe any outstanding effects of sex. There was a difference in bioavailability between the two studies included in this analysis. This may be due to differences in food intake with the dose. Rifapentine absorption is strongly enhanced when it is administered with food (7). The finding that the Daily RPE study had a lower bioavailability may arise from the fact that meals with the dose were not standardized, in contrast to the RIFAQUIN study, where a standard meal was provided throughout the study.
To conclude, our study is the first to show that the AADAC rs1803155 (AA) genotype is associated with lower rifapentine clearance, leading to increased rifapentine exposure. This effect should be confirmed in a larger independent analysis. The pharmacogenetic association was modest compared to the study effect, which is likely linked to differences in the pattern of food use across the studies and highlights the importance of food intake recommendations both when the drug is used in a programmatic setting and when its pharmacokinetics are investigated. Additionally, we found that rifapentine exposure was lower in HIV-infected patients, a finding that is consistent with the findings of previous studies and that warrants further investigation to assess whether dose adjustment strategies should be considered. Lastly, patients dosed with 1,200-mg-once-weekly doses had lower clearance, possibly as a result of less pronounced autoinduction.
MATERIALS AND METHODS
Study population.
This analysis was performed on patients diagnosed with pulmonary tuberculosis from two clinical studies: the phase III RIFAQUIN study (registration number ISRCTN44153044) (26) and the two-stage activity-safety study of daily rifapentine (27), referred to here as the Daily RPE study (ClinicalTrials.gov registration number NCT00814671). A subset of participants from these studies provided their consent for us to assess the effect of genetic polymorphisms of nuclear receptors, drug-metabolizing enzymes, and drug transporters on the pharmacokinetics of rifapentine.
The RIFAQUIN study included two experimental arms in which patients were dosed with daily moxifloxacin, rifampin, pyrazinamide, and ethambutol for 2 months, followed by a continuation phase with either 4 months of 1,200 mg rifapentine once weekly together with 400 mg moxifloxacin or 2 months of 400 mg moxifloxacin twice weekly with 900 mg rifapentine. The RIFAQUIN study was conducted at sites in the Western Cape and Gauteng regions of South Africa and in Harare, Zimbabwe. The doses of rifapentine and moxifloxacin were taken with 240 ml of water 15 min after a light meal of 2 hard-boiled eggs with bread. During the 4th month of treatment, blood samples were drawn for determination of plasma rifapentine concentrations. The pharmacokinetic assessment involved rich sampling (with samples drawn at pre-dose and 1, 2, 3, 5, 7, 10, 12, 26, and 50 h after dosing) or sparse sampling (with samples drawn at about 2, 5, and 24 or 48 h after dosing).
The Daily RPE study was open label and had two experimental arms. Patients with pulmonary tuberculosis were randomized to 450 or 600 mg rifapentine daily, which replaced 600 mg rifampin during the intensive phase of standard therapy. The study participants were recruited in the Western Cape, South Africa. The patients were advised to take the required rifapentine dose with food, but no standardized meal was provided during the study, and no accurate details about food intake with the dose were recorded. Pharmacokinetic sampling was performed at approximately 1 month after starting therapy, and samples were obtained with either intensive sampling (with samples drawn at pre-dose and at 0.75, 1.5, 3.5, 5, 12, and 24 h after dosing) or sparse sampling (with samples drawn 0.5 to 2 h and 5 to 8 h after dosing). Separate written informed consent for the pharmacogenetic study was obtained from participants retrospectively. The pharmacogenetic study was reviewed and approved by the Research Ethics Committee of the University of Cape Town and the University of the Witwatersrand.
Drug determination.
Plasma rifapentine concentrations were determined with a validated liquid chromatography-tandem mass spectrometry (LC-MS/MS) assay developed in the Division of Clinical Pharmacology, University of Cape Town. Samples were processed with a protein precipitation extraction method using rifaximin as the internal standard, followed by high-performance liquid chromatography with MS/MS detection using an AB Sciex API 3200 instrument. The analyte and internal standard were monitored at mass transitions of the protonated precursor ions m/z 877.3 and m/z 786.3 to the product ions m/z 845.4 and m/z 754.1 for rifapentine and rifaximin, respectively. The calibration curves fit quadratic (weighted by 1/concentration) regressions over the range of 0.156 to 40.0 mg/liter for rifapentine. The accuracies for the rifapentine assay were 103.9%, 102.8%, and 97.5% at the low, medium, and high quality control levels, respectively, during interbatch validation. The lower limit of quantification (LLOQ) was 0.156 mg/liter.
SNP genotyping.
Genomic DNA was extracted from 200 μl whole blood using a QIAamp DNA minikit (Qiagen, Inc., Valencia, CA) in accordance with the manufacturer’s protocol. DNA was quantified spectrophotometrically using a NanoDrop spectrophotometer (Thermo Fisher Scientific Inc., Wilmington, DE) before storage at −20°C. Genotyping was performed by real-time PCR on a DNA Engine Chromo4 system (Bio-Rad Laboratories, Inc., Hercules, CA). The PCR protocol involved an initial denaturation step at 95°C for 15 min, followed by 50 cycles of amplification at 95°C for 15 s and a final annealing at 60°C for 1 min. TaqMan genotyping master mix and assays for SLCO1B1 rs2306283 (SNP identifier C_1901697_20), SLCO1B1 rs4149032 (C_1901709_10), NR1I2 rs2472677 (C_26079845_10), NR1I2 rs1523130 (C_9152783_20), and AADAC rs1803155 (C_8911003_1_) were obtained from Thermo Fisher Scientific (Waltham, MA). Allelic discrimination plots and genotype assignments were performed using Opticon Monitor (version 3.1) software from Bio-Rad Laboratories.
Pharmacokinetic analysis.
Rifapentine plasma concentration-time data were analyzed using a nonlinear mixed-effects model implemented in NONMEM (version 7.4.2) software (28). The execution of runs was through the Perl-speaks-NONMEM, Pirana, and graphical diagnostics were created using Xpose (version 4.6.0) and R software (29, 30). Estimation of typical population pharmacokinetic parameters, along with their random interindividual variability (IIV) and interoccasion variability (IOV), was performed using a first-order conditional estimation method with the ε-η interaction (FOCE INTER). A lognormal distribution was assumed for IIV and IOV, and a combined additive and proportional model for the residual unexplained variability (RUV) was evaluated. Various structural models including a one- or two-compartment distribution with first-order elimination and first-order absorption with or without a lag time or transit compartment absorption were tested (31). The influence of genetic polymorphisms on the rifapentine pharmacokinetics for patients with an unknown genotype was identified using mixture modeling (32). The effect of the genotype was first tested using the method EXTRA, which not only estimates the association only for patients with available genetic information but also estimates an additional covariate effect for patients with the unknown genotype. Subsequently, the MIX method to impute values using mixture modeling was applied to include the patient with unknown genotype to strengthen the robustness of the findings (32). Model selection was based on changes in the NONMEM objective function value (ΔOFV) and visual inspection of conditional weighted residuals (CWRES) versus time, visual predictive checks (33), and basic goodness-of-fit (GOF) plots. During model development, physiological plausibility and the precision of the parameter estimates were also considered. The model parameters of the final model were evaluated for their precision using the sampling importance resampling (SIR) method (34).
Allometric scaling was applied to clearance (CL) and the volume of distribution (V) to adjust for the effect of body size, as described by Anderson and Holford (35). Fat-free mass (FFM) and fat mass (FAT) were tested as alternative size predictors instead of total body weight through allometric scaling (35, 36). After the inclusion of allometric scaling, potential demographic, study site-specific, and pharmacogenetic covariates were screened by inspecting parameter-versus-covariate plots and then tested in the model using drops in the objective function value (which was assumed to be χ2 distributed and, thus, in which a 3.84-point drop was considered significant at a P value of <0.05 for the inclusion of a single parameter) while scrutinizing the physiological plausibility of the effect (37).
ACKNOWLEDGMENTS
Computations were performed using facilities provided by the University of Cape Town's ICTS High Performance Computing team (http://hpc.uct.ac.za).
The study was supported by the European and Developing Countries Clinical Trials Partnership (CT.2004.32011.002), the U.S. Food and Drug Administration Orphan Products Program (R01FD003524), and the Wellcome Trust (WT081199/Z/06/Z). The drug assays were supported in part by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health (UM1 AI068634, UM1 AI068636, UM1 AI106701, and U01 AI068632), the Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD), and the National Institute of Mental Health (AI068632). H.M.M., and A.O. are funded by the Wellcome Trust (grants 206379/Z/17/Z and 204776/Z/16/Z, respectively), and A.O. is an affiliate of the African Academy of Sciences. P.D. is supported by the South African National Research Foundation (IFR170227223728).
REFERENCES
- 1.Gumbo T, Louie A, Deziel MR, Liu W, Parsons LM, Salfinger M, Drusano GL. 2007. Concentration-dependent Mycobacterium tuberculosis killing, and prevention of resistance by rifampin. Antimicrob Agents Chemother 51:3781–3788. doi: 10.1128/AAC.01533-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rosenthal IM, Williams K, Tyagi S, Peloquin CA, Vernon AA, Bishai WR, Grosset JH, Nuermberger EL. 2006. Potent twice-weekly rifapentine-containing regimens in murine tuberculosis. Am J Respir Crit Care Med 174:94–101. doi: 10.1164/rccm.200602-280OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sirgel FA, Fourie PB, Donald PR, Padayatchi N, Rustomjee R, Levin J, Roscigno G, Norman J, McIlleron H, Mitchison DA. 2005. The early bactericidal activities of rifampin and rifapentine in pulmonary tuberculosis. Am J Respir Crit Care Med 172:128–135. doi: 10.1164/rccm.200411-1557OC. [DOI] [PubMed] [Google Scholar]
- 4.Dorman SE, Savic RM, Goldberg S, Stout JE, Schluger N, Muzanyi G, Johnson JL, Nahid P, Hecker EJ, Heilig CM, Bozeman L, Feng PI, Moro RN, MacKenzie W, Dooley KE, Nuermberger EL, Vernon A, Weiner M, Tuberculosis Trials Consortium. 2015. Daily rifapentine for treatment of pulmonary tuberculosis. A randomized, dose-ranging trial. Am J Respir Crit Care Med 191:333–343. doi: 10.1164/rccm.201410-1843OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Langdon G, Wilkins JJ, Smith PJ, McIlleron H. 2004. Consecutive-dose pharmacokinetics of rifapentine in patients diagnosed with pulmonary tuberculosis. Int J Tuberc Lung Dis 8:862–867. [PubMed] [Google Scholar]
- 6.Savic RM, Lu Y, Bliven-Sizemore E, Weiner M, Nuermberger E, Burman W, Dorman SE, Dooley KE. 2014. Population pharmacokinetics of rifapentine and desacetyl rifapentine in healthy volunteers: nonlinearities in clearance and bioavailability. Antimicrob Agents Chemother 58:3035–3042. doi: 10.1128/AAC.01918-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zvada SP, Van Der Walt J-S, Smith PJ, Fourie PB, Roscigno G, Mitchison D, Simonsson USH, McIlleron HM. 2010. Effects of four different meal types on the population pharmacokinetics of single-dose rifapentine in healthy male volunteers. Antimicrob Agents Chemother 54:3390–3394. doi: 10.1128/AAC.00345-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burman WJ, Gallicano K, Peloquin C. 2001. Comparative pharmacokinetics and pharmacodynamics of the rifamycin antibacterials. Clin Pharmacokinet 40:327–341. doi: 10.2165/00003088-200140050-00002. [DOI] [PubMed] [Google Scholar]
- 9.Keung A, Eller MG, McKenzie KA, Weir SJ. 1999. Single and multiple dose pharmacokinetics of rifapentine in man: part II. Int J Tuberc Lung Dis 3:437–444. [PubMed] [Google Scholar]
- 10.Wilkins JJ, Savic RM, Karlsson MO, Langdon G, McIlleron H, Pillai G, Smith PJ, Simonsson USH. 2008. Population pharmacokinetics of rifampin in pulmonary tuberculosis patients, including a semimechanistic model to describe variable absorption. Antimicrob Agents Chemother 52:2138–2148. doi: 10.1128/AAC.00461-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nakajima A, Fukami T, Kobayashi Y, Watanabe A, Nakajima M, Yokoi T. 2011. Human arylacetamide deacetylase is responsible for deacetylation of rifamycins: rifampicin, rifabutin, and rifapentine. Biochem Pharmacol 82:1747–1756. doi: 10.1016/j.bcp.2011.08.003. [DOI] [PubMed] [Google Scholar]
- 12.Weiner M, Bock N, Peloquin CA, Burman WJ, Khan A, Vernon A, Zhao Z, Weis S, Sterling TR, Hayden K, Goldberg S. 2004. Pharmacokinetics of rifapentine at 600, 900, and 1,200 mg during once-weekly tuberculosis therapy. Am J Respir Crit Care Med 169:1191–1197. doi: 10.1164/rccm.200311-1612OC. [DOI] [PubMed] [Google Scholar]
- 13.Weiner M, Peloquin C, Burman W, Luo C-C, Engle M, Prihoda TJ, Mac Kenzie WR, Bliven-Sizemore E, Johnson JL, Vernon A. 2010. Effects of tuberculosis, race, and human gene SLCO1B1 polymorphisms on rifampin concentrations. Antimicrob Agents Chemother 54:4192–4200. doi: 10.1128/AAC.00353-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chigutsa E, Visser ME, Swart EC, Denti P, Pushpakom S, Egan D, Holford NHG, Smith PJ, Maartens G, Owen A, McIlleron H. 2011. The SLCO1B1 rs4149032 polymorphism is highly prevalent in South Africans and is associated with reduced rifampin concentrations: dosing implications. Antimicrob Agents Chemother 55:4122–4127. doi: 10.1128/AAC.01833-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schuetz EG, Schinkel AH, Relling MV, Schuetz JD. 1996. P-glycoprotein: a major determinant of rifampicin-inducible expression of cytochrome P4503A in mice and humans. Proc Natl Acad Sci U S A 93:4001–4005. doi: 10.1073/pnas.93.9.4001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reith K, Keung A, Toren PC, Cheng L, Eller MG, Weir SJ. 1998. Disposition and metabolism of 14C-rifapentine in healthy volunteers. Drug Metab Dispos 26:732–738. [PubMed] [Google Scholar]
- 17.Maglich JM, Stoltz CM, Goodwin B, Hawkins-Brown D, Moore JT, Kliewer SA. 2002. Nuclear pregnane X receptor and constitutive androstane receptor regulate overlapping but distinct sets of genes involved in xenobiotic detoxification. Mol Pharmacol 62:638–646. doi: 10.1124/mol.62.3.638. [DOI] [PubMed] [Google Scholar]
- 18.Langdon G, Wilkins J, Mcfadyen L, Smith P, Simonsson USH, McIlleron H. 2005. Population pharmacokinetics of rifapentine and its primary desacetyl metabolite in South African tuberculosis patients. Antimicrob Agents Chemother 49:4429–4436. doi: 10.1128/AAC.49.11.4429-4436.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shimizu M, Fukami T, Kobayashi Y, Takamiya M, Aoki Y, Nakajima M, Yokoi T. 2012. A novel polymorphic allele of human arylacetamide deacetylase leads to decreased enzyme activity. Drug Metab Dispos 40:1183–1190. doi: 10.1124/dmd.112.044883. [DOI] [PubMed] [Google Scholar]
- 20.Egelund EF, Weiner M, Singh RP, Prihoda TJ, Gelfond JAL, Derendorf H, Mac Kenzie WR, Peloquin CA. 2014. Protein binding of rifapentine and its 25-desacetyl metabolite in patients with pulmonary tuberculosis. Antimicrob Agents Chemother 58:4904–4910. doi: 10.1128/AAC.01730-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sloan DJ, McCallum AD, Schipani A, Egan D, Mwandumba HC, Ward SA, Waterhouse D, Banda G, Allain TJ, Owen A, Khoo SH, Davies GR. 2017. Genetic determinants of the pharmacokinetic variability of rifampin in Malawian adults with pulmonary tuberculosis. Antimicrob Agents Chemother 61:e00210-17. doi: 10.1128/AAC.00210-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gengiah TN, Botha JH, Soowamber D, Naidoo K, Abdool Karim SS. 2014. Low rifampicin concentrations in tuberculosis patients with HIV infection. J Infect Dev Ctries 8:987–993. doi: 10.3855/jidc.4696. [DOI] [PubMed] [Google Scholar]
- 23.Jeremiah K, Denti P, Chigutsa E, Faurholt-Jepsen D, PrayGod G, Range N, Castel S, Wiesner L, Hagen CM, Christiansen M, Changalucha J, McIlleron H, Friis H, Andersen AB. 2014. Nutritional supplementation increases rifampicin exposure among tuberculosis patients coinfected with HIV. Antimicrob Agents Chemother 58:3468–3474. doi: 10.1128/AAC.02307-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Savic RM, Weiner M, MacKenzie WR, Engle M, Whitworth WC, Johnson JL, Nsubuga P, Nahid P, Nguyen NV, Peloquin CA, Dooley KE, Dorman SE. 2017. Defining the optimal dose of rifapentine for pulmonary tuberculosis: exposure–response relations from two phase II clinical trials. Clin Pharmacol Ther 102:321–331. doi: 10.1002/cpt.634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chirehwa MT, Rustomjee R, Mthiyane T, Onyebujoh P, Smith P, McIlleron H, Denti P. 2015. Model-based evaluation of higher doses of rifampin using a semimechanistic model incorporating autoinduction and saturation of hepatic extraction. Antimicrob Agents Chemother 60:487–494. doi: 10.1128/AAC.01830-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jindani A, Harrison TS, Nunn AJ, Phillips PPJ, Churchyard GJ, Charalambous S, Hatherill M, Geldenhuys H, McIlleron HM, Zvada SP, Mungofa S, Shah NA, Zizhou S, Magweta L, Shepherd J, Nyirenda S, van Dijk JH, Clouting HE, Coleman D, Bateson ALE, McHugh TD, Butcher PD, Mitchison DA. 2014. High-dose rifapentine with moxifloxacin for pulmonary tuberculosis. N Engl J Med 371:1599–1608. doi: 10.1056/NEJMoa1314210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dawson R, Narunsky K, Carman D, Gupte N, Whitelaw A, Efron A, Barnes GL, Hoffman J, Chaisson RE, McIlleron H, Dorman SE. 2015. Two-stage activity-safety study of daily rifapentine during intensive phase treatment of pulmonary tuberculosis. Int J Tuberc Lung Dis 19:780–786. doi: 10.5588/ijtld.14.0868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Beal S, Sheiner L, Boeckmann A, Bauer R. 2009. NONMEM user’s guides. (1989-2009). ICON Development Solutions, Ellicott City, MD. [Google Scholar]
- 29.Keizer RJ, Karlsson MO, Hooker A. 2013. Modeling and simulation workbench for NONMEM: tutorial on Pirana, PsN, and Xpose. CPT Pharmacometrics Syst Pharmacol 2:e50. doi: 10.1038/psp.2013.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.R Core Team. 2013. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- 31.Savic RM, Jonker DM, Kerbusch T, Karlsson MO. 2007. Implementation of a transit compartment model for describing drug absorption in pharmacokinetic studies. J Pharmacokinet Pharmacodyn 34:711–726. doi: 10.1007/s10928-007-9066-0. [DOI] [PubMed] [Google Scholar]
- 32.Keizer RJ, Zandvliet AS, Beijnen JH, Schellens JHM, Huitema ADR. 2012. Performance of methods for handling missing categorical covariate data in population pharmacokinetic analyses. AAPS J 14:601–611. doi: 10.1208/s12248-012-9373-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Karlsson MO, Holford NHG. 2008. A tutorial on visual predictive checks, abstr 1434. Abstr Ann Meet Population Approach Group Europe. https://www.page-meeting.org/?abstract=1434. [Google Scholar]
- 34.Dosne AG, Bergstrand M, Harling K, Karlsson MO. 2016. Improving the estimation of parameter uncertainty distributions in nonlinear mixed effects models using sampling importance resampling. J Pharmacokinet Pharmacodyn 43:583–596. doi: 10.1007/s10928-016-9487-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Anderson BJ, Holford NHG. 2008. Mechanism-based concepts of size and maturity in pharmacokinetics. Annu Rev Pharmacol Toxicol 48:303–332. doi: 10.1146/annurev.pharmtox.48.113006.094708. [DOI] [PubMed] [Google Scholar]
- 36.Janmahasatian S, Duffull SB, Ash S, Ward LC, Byrne NM, Green B. 2005. Quantification of lean bodyweight. Clin Pharmacokinet 44:1051–1065. doi: 10.2165/00003088-200544100-00004. [DOI] [PubMed] [Google Scholar]
- 37.Wählby U, Jonsson EN, Karlsson MO. 2002. Comparison of stepwise covariate model building strategies in population pharmacokinetic-pharmacodynamic analysis. AAPS Pharm Sci 4:E27. doi: 10.1208/ps040427. [DOI] [PMC free article] [PubMed] [Google Scholar]