Vancomycin-resistant Enterococcus (VRE) is a leading cause of hospital-acquired infection, with limited treatment options. Resistance to one of the few remaining drugs, daptomycin, is a growing clinical problem and has previously been described in this hospital.
KEYWORDS: Enterococcus, antimicrobial resistance, antimicrobial stewardship, daptomycin, evolution
ABSTRACT
Vancomycin-resistant Enterococcus (VRE) is a leading cause of hospital-acquired infection, with limited treatment options. Resistance to one of the few remaining drugs, daptomycin, is a growing clinical problem and has previously been described in this hospital. In response to increasing resistance, an antimicrobial stewardship intervention was implemented to reduce hospital-wide use of daptomycin. To assess the impact of the intervention, daptomycin prescribing patterns and clinically reported culture results from vancomycin-resistant Enterococcus faecium (VREfm) bloodstream infections (BSIs) from 2011 through 2017 were retrospectively extracted and the impact of the intervention was estimated using interrupted time series analysis (ITS). We corrected for a change in MIC determination methodology by retesting 262 isolates using Etest and broth microdilution. Hospital-wide and within-patient resistance patterns of corrected daptomycin MICs are reported. Our data show that daptomycin prescriptions decreased from an average of 287 days of therapy/month preintervention to 151 days of therapy/month postintervention. Concurrently, the proportion of patients experiencing an increase in daptomycin MIC during an infection declined from 14.6% (7/48 patients) in 2014 to 1.9% (1/54 patients) in 2017. Hospital-wide resistance to daptomycin also decreased in the postintervention period, but this was not maintained. This study shows that an antimicrobial stewardship-guided intervention reduced daptomycin use and improved individual level outcomes but had only transient impact on the hospital-level trend.
INTRODUCTION
Antimicrobial resistance poses an increasing threat to public health, with the use of antimicrobials being the leading cause of increased resistance (1). Managing resistance evolution in an acute care setting, where the use of antibiotics is crucial to patient survival, presents an important challenge. Evolution of resistance within patients in response to treatment is easily observed and well-documented (2, 3). Understanding the many additional factors that contribute to population-wide resistance, such as transmission dynamics (4–6), fitness of resistance mutations (7), and collateral resistance or sensitivity (8), is more complex. In response to the increased threat posed by antimicrobial resistance, antimicrobial stewardship practices have been put forward as one part of the solution (9, 10). Many studies have demonstrated the ability of antimicrobial stewardship initiatives to successfully decrease the use of targeted antimicrobials within hospitals (11). Furthermore, antimicrobial stewardship interventions can reduce resistance within hospitals (12–15).
Vancomycin-resistant Enterococcus faecium (VREfm) is an important cause of health care-associated infections (HAI), being the second most common cause of multidrug-resistant HAI (16, 17). Resistance to multiple classes of drugs has resulted in limited treatment options, with daptomycin often being the preferred treatment (18–20). In 2011, routine daptomycin MIC determination of all VREfm bloodstream infection (BSI) isolates was introduced at Michigan Medicine, a 1,000-bed tertiary hospital. Over the subsequent 4 years, a continual increase in daptomycin resistance both within patients and hospital wide was observed (2). Based on these findings, the adult antimicrobial stewardship program implemented an intervention to reduce daptomycin use in the hospital. Here, we assess the impact of an intervention on the use of daptomycin and on the evolution of resistance within patient and hospital wide. Complicating the analysis is a change in testing methodology that occurred simultaneously with the intervention. This study also addresses the importance of considering changes in testing methodology when investigating long-term trends in resistance and how to correct for these changes.
RESULTS
Impact of interventions on drug use.
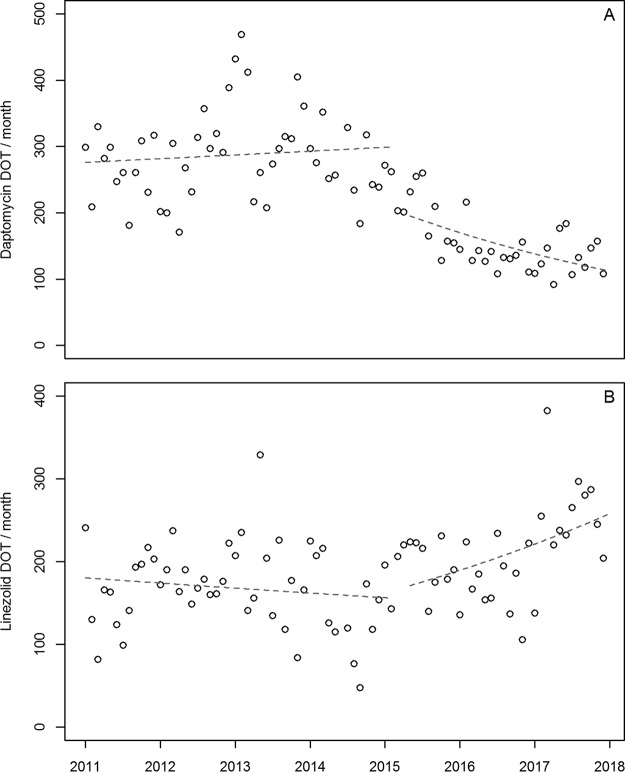
Following the intervention, daptomycin use in the hospital reduced from a mean of 287 days of therapy (DOT)/month to 151 DOT/month (Fig. 1A). Despite this large drop, it is difficult to interpret the main effect of the intervention due to a significant interaction with time (relative risk [RR] = 0.98 [95% CI: 0.97, 0.99], P = 0.003). Following the intervention, the use of linezolid, which is used to treat similar infections as daptomycin, increased from a mean of 168 DOT/month to 210 DOT/month (Fig. 1B). There was a significant interaction due to a steady increase in linezolid use during the postintervention period (RR = 1.02 [1.00, 1.03], P = 0.014). There was no significant change in vancomycin use following the intervention (RR = 0.94 [0.86, 1.03], P = 0.156) (see Fig. S2 in the supplemental material). Patient days were only reliably available from 2013 onwards, and therefore, we have reported DOT/month rather than DOT/1,000 patient days. From 2013 to 2017, monthly DOT was highly correlated with DOT/1,000 patient days (r2 = 0.99) and, therefore, DOT/month is likely to be highly representative of DOT/1,000 patient days.
FIG 1.
Total days of therapy (DOT) per month in the hospital for daptomycin (A) and linezolid (B). Dotted lines show the fit results from the interrupted time series analysis.
The proportion of patients with VREfm BSIs that were treated with daptomycin dropped after the intervention (Table 1). There was a reduction (from 0.83 to 0.34) in the proportion of patients receiving daptomycin therapy in the early period (<4 days), likely before the MIC results were available, a reduction (from 0.66 to 0.29) in daptomycin treatment later, when the MICs would be known, and a reduction in the median length of treatment by 3 days.
TABLE 1.
Treatment of patients with VREfm BSIsa
| Period | Initial isolate MIC | Early therapy |
Nonempirical therapy |
||
|---|---|---|---|---|---|
| Proportion of patients (n) | Median DOT (IQR) | Proportion of patients (n) | Median DOTs (IQR) | ||
| Preintervention | <4 μg/ml | 0.84 (56/67) | 3 (2–4) | 0.78 (52/67) | 9 (6–12) |
| 4 μg/ml | 0.83 (99/119) | 3 (2–4) | 0.71 (85/119) | 7 (4–11) | |
| >4 μg/ml | 0.82 (23/28) | 2 (1–3) | 0.18 (5/28) | 2 (1–8) | |
| All | 0.83 (178/214) | 3 (2–4) | 0.66 (142/214) | 8 (4–11) | |
| Postintervention | <4 μg/ml | 0.40 (21/53) | 3 (2–4) | 0.38 (20/53) | 4.5 (2–8.5) |
| 4 μg/ml | 0.33 (15/46) | 3 (2–4) | 0.28 (13/46) | 6 (2–10) | |
| >4 μg/ml | 0.14 (2/14) | 4 (4–4) | 0.00 (0/14) | - | |
| All | 0.34 (38/113) | 3 (2–4) | 0.29 (33/113) | 5 (2–10) | |
Early therapy includes any days of therapy (DOTs) of daptomycin between the date of the initial BSI isolate and up to 4 days following the initial isolate date. Nonempirical therapy includes any DOTs given greater than 4 days after the initial isolate and up to 30-days post-initial isolate. Median DOT reflect only those patients who received therapy. IQR, interquartile range. Bold data are for all initial isolates for each period.
Daptomycin MIC of VREfm BSI.
From 2011 to 2017, 338 patients had VREfm bloodstream infections. Daptomycin MICs of the initial isolates from 216 of these infections were originally determined by Etest and 122 by the Trek system. Based on clinically reported daptomycin MICs, resistance to daptomycin increased in the hospital from 2011 through 2014, followed by a sharp decline in 2015, and then an increase through 2016 and 2017 (Fig. 2).
FIG 2.
Average daptomycin MIC for VREfm initial isolates by quarter. Open circles are calculated from original clinical MICs, and closed circles are calculated from corrected values. Means prior to February 2015 are only shown as original values, as a correction for these values was not required. Error bars represent 95% confidence interval (CI) of the mean, and the numbers are the number of infections in each quarter. The gray circles show each initial isolate per patient as susceptible or nonsusceptible. Gray line is a local polynomial regression (LOESS) of the daptomycin susceptibility of initial isolates, showing the moving average of the proportion of initial isolates that are daptomycin nonsusceptible. Highlighted panel indicates the study intervention period.
MIC test method comparisons.
MICs by broth microdilution (BMD) were highly repeatable, with 99.7% of repeat testing results being within a single 2-fold dilution of the sample mode, and 77.2% of tests being in agreement with the mode (see Fig. S3 in the supplemental material). Six of the 40 samples returned the same MIC on all eight tests, while for one sample the replicate tests were divided equally between two MICs. These results match the variation expected by chance (see Fig. S4 in the supplemental material). This variation was not associated with the MIC of the sample (χ2 = 13.55, degrees of freedom [df] = 9, P = 0.14).
The BMD retest MICs had a 97.0% essential agreement (within 2-fold) with samples originally tested on the Trek system and a 98.4% essential agreement with samples originally tested by Etest (Fig. 3). Despite good overall agreement between the two original test methods and the BMD results, the original results for samples originally tested on Trek were more likely to be lower when tested by BMD (χ2 = 9.76, df = 1, P < 0.001), while isolates originally tested with Etest showed no significant skew (χ2 = 1.00, df = 1, P = 0.32). This difference indicates that a correction is required in order to directly compare the Trek and Etest result data sets.
FIG 3.
Comparison of original test results with repeat BMD results. (A and B) Show the outcomes of repeat testing based on original reported daptomycin MIC. (C and D) Show the fold difference between original test results and repeat test results as a proportion of the tested isolates. Negative fold changes occur when the original testing method is lower than the reference BMD, while positive values indicate that the original testing method is higher than the reference BMD. (A and C) Represent isolates that were originally tested by Trek (n = 199), and (B and D) show isolates that were originally tested on the Etest system (n = 63).
Adjusting for testing method.
The relationship between the initial Trek and retest BMD MICs fit a linear model where the slope was not significantly different to 1 (t = 0.03, df = 1, P = 0.98), and so the Trek data set was corrected by a single offset for all values (equation 1).
| (1) |
The relationship between the initial Etest and retest BMD MICs was linear with neither the slope or intercept being significantly different to 1 (slope: t = −0.79, df = 1, P = 0.43; intercept: t = 0.44, df = 1, P = 0.66). Therefore, no correction was required for values initially tested by Etest.
Hospital trend.
Applying the correction did not change the original trend; however it reduced the magnitude of the decline in 2015 due to the upward shift of all MIC data following the testing methodology change and, therefore, following the intervention (Fig. 2). Following the intervention, there was a significant drop in mean MIC (interrupted time series [ITS]: β = −0.84 [−1.25, −0.42], P < 0.001) (see Fig. S5 in the supplemental material); however, due to the continued increase in MIC throughout the postintervention period, there was no overall difference in mean daptomycin MICs before (3.61 μg/ml) and after (3.73 μg/ml) the intervention.
Correcting the trend using Etest as the reference method rather than BMD resulted in the application of different correction factors, but this did not change the qualitative results (see Fig. S6 in the supplemental material).
Relationship of daptomycin use to daptomycin MICs.
Prior to the intervention, mean daptomycin MIC (by quarter) and mean DOT/quarter were positively correlated (R2 = 0.404, P = 0.008) (Fig. 4). Following the intervention, the relationship between daptomycin DOT and daptomycin MICs remained significantly correlated (R2 = 0.465, P = 0.030), but the direction of the relationship changed, with an increase in MIC now associated with a decrease in DOT (Fig. 4).
FIG 4.
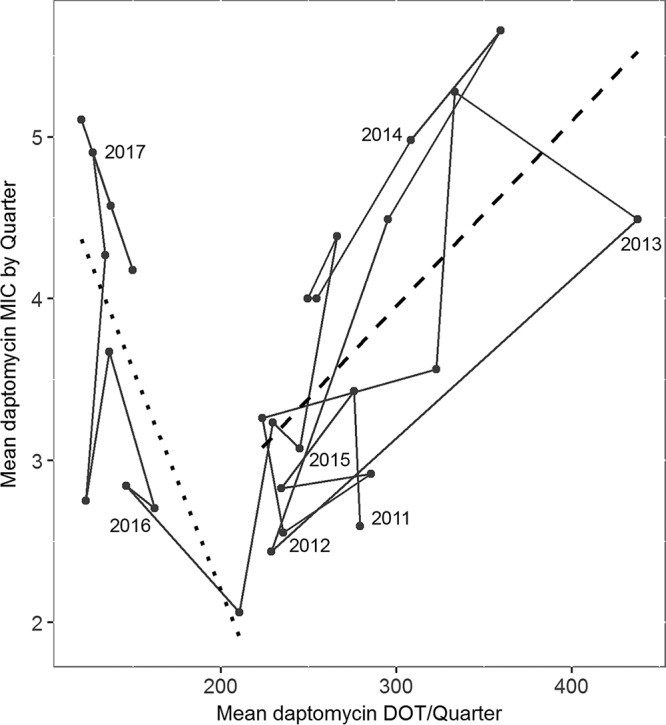
Relationship of mean daptomycin DOT to mean daptomycin MIC. Each point is the mean of three months (one quarter), and points are connected through time. The first quarter of each year is indicated by the year text. The dashed line is a linear regression through all points preintervention, and the dotted line is a linear regression through the postintervention points. Quarters that contained any time period within the intervention period were excluded from the regressions.
Within-patient evolution.
The impact of reduced daptomycin prescribing for VREfm BSIs can also be seen when looking at within-patient evolution of resistance. The majority of BSIs lasted less than 3 days (the window for retesting drug susceptibilities), although infections ranged from <3 to 27 days. Increases in MIC were observed between 3 and 27 days after the initial isolate (median, 6 days). Patients where an increase in MIC was observed received a median of 4 doses of daptomycin prior to the increase (range: 0 to 16 doses). From 2011 through 2014 the proportion of patients where a subsequent VREfm isolate (within 30 days of the initial isolate) had a higher MIC than the initial isolate rose from 3.6% (2/55) to 14.6% (7/48). Following the intervention and reduced daptomycin prescribing, this proportion dropped consistently each year to 1.9% (1/53) in 2017 (Fig. 5). Prior to 2015 all patients with an increase in MIC received at least one dose of daptomycin between the date of the initial isolate and the date of the observed MIC increase. However, in 2015 one of the four patients with MIC increases had received no daptomycin treatment, and the one patient in each of 2016 and 2017 with an MIC increase had received no daptomycin.
FIG 5.
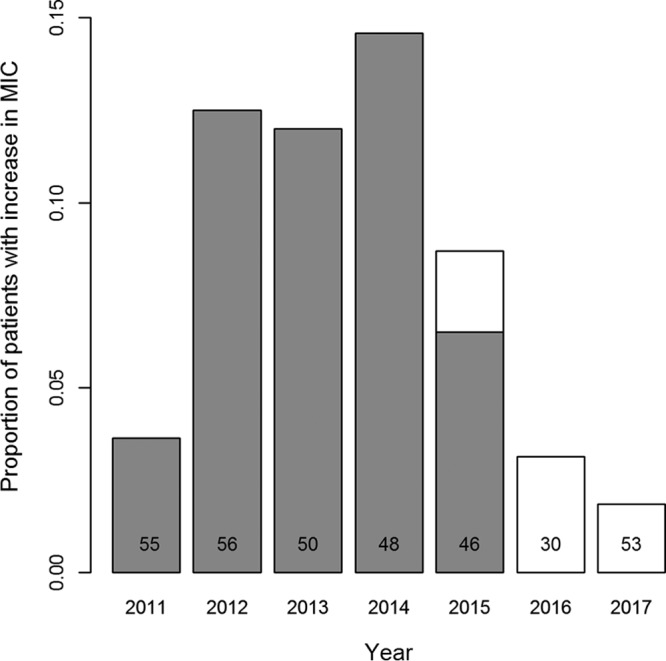
Within-patient evolution. Proportion of patient with VREfm BSI where the daptomycin MIC of the infection increases during the infection (within 30 days of the initial isolate). Numbers indicate the number of VREfm BSI infections each year. Gray bars indicate patients who had received daptomycin treatment between the date of initial infection and the date of MIC increase, and white bars indicate patients who received no daptomycin treatment.
DISCUSSION
Following the intervention, daptomycin use was reduced hospital-wide and as treatment for VREfm BSIs. There was a concurrent increase in the use of linezolid, an alternate treatment for VREfm infections. Importantly, for VREfm bloodstream infections ultimately identified as daptomycin nonsusceptible, the intervention resulted in a reduction in the proportion of patients receiving daptomycin treatment in the time before MIC data were available.
Comparative testing demonstrated a need to correct for testing methods to accurately describe MIC change over time, but this correction did not qualitatively change the trend in resistance (Fig. 2). Consistent with previous studies (21–23), testing methods show significant differences, with Etest returning a higher mean MIC than the Trek method (23). While previous studies showed BMD to give lower MICs on average than Etest (24), our study found the BMD method to be more consistent with Etest than Trek. While it has been suggested that BMD is not highly reproducible across laboratories and brands of media (25), BMD in our hands was highly repeatable (Fig. S3 and S4) and, therefore, an appropriate reference method for this study.
The proportion of patients who had a within-patient increase in MIC during an infection decreased following the intervention. This change likely reflects the reduced number of patients receiving treatment with daptomycin, which more than halved following the intervention (Table 1). While all patients who experienced increases in resistance prior to 2015 were treated with daptomycin, in 2016 and 2017 none of the patients showing within-patient increases received daptomycin. The 2016 to 2017 within-patient increases are consistent with the sampling error seen in the repeated testing (Fig. S4).
Daptomycin resistance in the hospital increased steadily from the beginning of the study period in January 2011 until 2015. During this preintervention period, mean daptomycin MICs were correlated with daptomycin use in the hospital. Following the intervention, there was a transient drop in hospital-wide resistance, but this was followed by an increase in daptomycin resistance that was no longer correlated with daptomycin use (Fig. 4). Indeed, the average MIC at the end of our time series was as high as it had been when daptomycin use was at its peak. This dissolution of the relationship between daptomycin use and resistance suggests a shift in the drivers of evolution in this setting. While one possibility is that the preintervention correlation is spurious, we argue that this is unlikely, as the correlation exists across several increases and decreases in daptomycin use that are separated in time, the correlation is consistent with the increase in within-host evolution of resistance (Fig. 4), and the interrupted time series shows a decrease in MIC associated with the intervention (Fig. S5).
Some potential drivers of the dissolution of the relationship between resistance and drug use deserve consideration. First, the postintervention increase could result from more resistant strains being introduced into the hospital. Given that daptomycin is not generally used in community settings, this suggests that selection for resistance may be occurring at other hospitals or care facilities in the area. The price of daptomycin fell around this time as generics became available, so it is possible that there was increased use in other health care settings. Second, the 2016 to 2017 increase could be due to indirect selection for daptomycin resistance via selection on a correlated trait. This could occur if, for example, resistance to other antibiotics, disinfectants, or antimicrobial peptides confer cross-resistance to daptomycin (26–31), or if adaption to some other environmental novelty also confers increased resistance (32). Third, hitchhiking is also a possibility, where daptomycin resistance alleles could be carried to high frequency either due to stochastic dynamics or because they happen to be present in a clone that for unrelated reasons had a fitness advantage in the hospital setting (33). While there are no known overlaps in resistance mutations between linezolid and daptomycin (34, 35), coresistance is common in some settings (36). Thus, the increased linezolid use could drive daptomycin resistance; however, an increase in linezolid resistance was not observed in this time period. Fourth, resistance mutations with a lower cost of resistance may have emerged (7, 37, 38), or circulating hospital strains may have acquired compensatory mutations (39, 40). Disconnects between drug use and MIC dynamics at a population level have been seen in other pathogen systems (41–43), where, like here, selection at the individual level and at the population level differed. Fifth, it is possible that changes in daptomycin utilization not captured in our study could impact the evolution of resistance, such as the doses used or the nature of the underlying infection. Determining which of these drivers may be acting is important, as they suggest different approaches to address the continued resistance trend. Future work combining genome sequencing of archived strains and epidemiological methods may be able to distinguish between importation from outside the hospital and within-hospital spread, as well as a shift in the genetic determinants of resistance.
Limitations of this study include the retrospective observational design, which restricts the ability to identify causal relationships, and being a single center study, so caution should be taken with the generalization of these findings. Despite this, our study highlights the complex interactions of antimicrobials and resistance in a hospital setting and demonstrates the importance of looking at not only antimicrobial use but also the impact on resistance when assessing the outcome of antimicrobial use interventions. While minimizing the use of antimicrobials is important for reducing within-host selection for resistance and likely plays a significant role in hospital-wide levels of resistance, our study suggests that simply substituting one front-line drug for another need not restore sensitivity to the first.
MATERIALS AND METHODS
Study strategy.
A retrospective observational cohort study was conducted at Michigan Medicine to determine the effect of antibiotic policy change on antibiotic use and hospital-wide levels of daptomycin resistance in VREfm bloodstream infections. The study was approved by the University of Michigan Institutional Review Board (identification [ID] no. HUM00102282), which determined that informed consent was not required, as all samples were collected for patient treatment purposes.
Data sets.
Four data sets of daptomycin MICs and two daptomycin use data sets were used for this study, as outlined in Table 2 and described below. Daptomycin, linezolid, and vancomycin use data were calculated as days of therapy (DOT) per month across the entire hospital population and were extracted from pharmacy billing databases. Individual-level clinical data were extracted retrospectively from the electronic medical record system.
TABLE 2.
Data sets used in this study
| Data set | Use | Samples included | Data source | No. of isolates, test results, patients, or doses |
|---|---|---|---|---|
| Daptomycin resistance trend | To investigate the change in daptomycin resistance over time | All VREfm BSI isolates 2011–2017 (initial isolates only) | Clinical reports: 2011–Feb 2015 by Etest, Feb 2015–2017 by Trek system | 338 isolates |
| Testing method correction | To determine the relationship between MICs generated by the two different testing methods used in the hospital to allow for a direct comparison of MICs over time | Enterococcus BSIs Sept 2013–Sept 2016 | Clinical reports: repeat testing by BMD and Etest | 262 isolates, 524 test results |
| Correction assay repeatability | To assess the repeatability of the reference assays | Representative (by species, vancR, and MIC) subset of testing method correction samples | Repeat testing by BMD and Etest | 40 isolates, 320 test results |
| Within-patient change in resistance | To determine proportions of patients where daptomycin resistance increases during the infection | All VREfm BSI isolates 2009–Jan 2018 | Clinical reports | 574 isolates, 338 patients |
| Days of therapy | To determine changes in overall drug doses used in the hospital | Monthly days of therapy of daptomycin, linezolid, and IV vancomycin between Jan 2011 and Dec 2017 | Hospital electronic medical records | |
| Daptomycin use by patient | To investigate direct impact of the intervention on treatment of VREfm BSI infections | All doses of daptomycin administered to patients with VREfm between Jan 2011–Jan 2017 | Hospital electronic medical records | 3,534 doses, 254 patients |
The daptomycin resistance trend data set (Table 2) includes all VREfm BSI initial isolates between January 2011 and December 2017. This data set contains two subsets; the first consists of MIC results from samples originally tested by Etest (bioMérieux) that were collected prior to the testing switch date of February 15, 2015. The second data set collected on or after the switch date contained MIC results for samples originally tested with the Trek system (Thermo Fisher).
The testing method correction data set (Table 2) was used to determine the relationship of MICs produced by the Etest and Trek methods. This correction allows the impact of the change in MIC testing method to be separated from the impact of the antimicrobial stewardship intervention. This data set consists of 262 stored Enterococcus isolates from 235 patients that were retested by both broth microdilution (BMD) and Etest. Daptomycin MICs for 63 of the isolates were originally tested by Etest and 199 isolates with the Trek system. This sample set included all stored Enterococcus BSI initial isolates between September 2013 and September 2016. Prior to January 2016, isolates were stored in a haphazard fashion. Starting January 2016, all Enterococcus BSI isolates were systematically stored. From September 2013 to December 2015 the sample set included 51% of the initial isolates tested in the hospital, and from January 2016 to September 2016, this included all the initial isolates tested in the hospital. Additionally, all stored daptomycin nonsusceptible Enterococcus (DNSE) BSI isolates for the same period were included, even if they were not the initial isolate for that patient, to improve representation of higher MIC values in the data set. For six patients, two different species of Enterococcus were isolated from their initial sample, and both species were included as initial isolates.
The correction assay repeatability data set (Table 2) contained 40 samples that were representative of the full correction data set based on species, vancomycin susceptibility, and daptomycin MIC (see Fig. S1 in the supplemental material). These samples were tested a total of eight times each to assess the repeatability of BMD.
Finally, within-patient change in daptomycin resistance was investigated for all patients with an initial VREfm BSI isolate between January 2011 and December 2017. A within-patient increase was defined as at least a 2-fold increase in daptomycin MIC within 30 days of the initial isolate. Isolates are routinely tested for drug susceptibilities every 3 days.
Intervention.
The adult antimicrobial stewardship program (ASP) at Michigan Medicine consists of 3 infectious disease physicians, 3 infectious disease pharmacists, and 1 infectious disease pharmacy resident. In response to rising daptomycin nonsusceptibility among VREfm isolated from bloodstream infections (2), the ASP launched an initiative to reduce hospital-wide daptomycin use. Over three months (February 2015 to April 2015), the ASP led educational efforts to implement this initiative. Two 1-hour lectures were provided to the Department of Infectious Diseases during a faculty conference and business meeting. Targeted education was provided to the transplant infectious disease service because the patient population they manage has relatively higher rates of VRE infection. An existing institutional policy was continued, requiring prescribing physicians to seek preauthorization for the use of restricted antimicrobial agents, including daptomycin, between the hours of 7 a.m. and 11 p.m., 7 days per week. Although no prior approval was required overnight, a pharmacist member of the ASP conducted daily prospective audits and feedback of all restricted antimicrobial agents Monday through Friday to ensure appropriate use and dosing, facilitate de-escalation, and optimize duration of therapy. The ASP used these procedures as opportunities to reduce the use of daptomycin hospital-wide and preferred the use of linezolid instead of daptomycin for the empirical and definitive treatment of VRE infections when not contraindicated.
Susceptibility testing methodology.
Susceptibility testing for the testing method correction was performed by the broth microdilution method using frozen reference panels (Thermo Fisher Scientific) and also by Etest (bioMérieux). Both methods were preformed according to CLSI M7 guidelines (44). MIC testing plates contained 2-fold dilutions of daptomycin with final concentrations ranging from 0.125 μg/ml to 128 μg/ml. Enterococcus faecalis ATCC 29212 and Staphylococcus aureus ATCC 29213 were included for quality control with each batch of testing.
Data analysis.
For the testing method correction, general linear models allowing linear, quadratic, and cubic terms were fitted by original test type to determine the relationship of original daptomycin MICs to the MIC for each reference method. Nonsignificant terms (P > 0.05) were removed from the models in a stepwise fashion in order to determine the simplest model for each relationship. The models were then applied to all VREfm initial bloodstream isolates from 2011 to 2017 based on their original testing method to correct for the change in testing methods. Analyses were generated using SAS software, version 9.4, of the SAS System for Windows.
Time series data were analyzed using interaction model interrupted time series (ITS). For the DOT outcomes, we used multiplicative quasi-Poisson models, and the MIC model was regressed using the normal distribution. Any autocorrelation in the time series was corrected for with heteroskedasticity-consistent estimation of the covariance matrix with the function vcovHAC in the R package sandwich (45, 46). The models consisted of terms for time and study period and an interaction between the two. If the interaction terms had no statistical significance, then simpler main effects models were used in order to preserve model parsimony. Analyses were performed with R (R Core Team, 2017).
Supplementary Material
ACKNOWLEDGMENTS
We thank E. Hanks for statistical advice; A. Smith for data extraction; N. Fischer, J. Tolles, C. Brenke, and M. Forstchen for assistance with retesting correction samples; E. Hansen and V. Morley for discussion; and two anonymous reviewers for their comments.
This work was supported by the National Institutes of Health (grant number K08 AI119182 to R.J.W.). and by Pennsylvania State University.
C.L.K., C.Y., V.M., and R.J.W. have no potential conflicts. A.F.R. and T.S.P. report grants from Merck, outside the submitted work. D.W.N. reports personal fees from NaviDx LLC, outside the submitted work.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.01800-18.
REFERENCES
- 1.Centers for Disease Control and Prevention. 2013. Antibiotic resistance threats in the United States, 2013. US Department of Health and Human Services, Washington DC: https://www.cdc.gov/drugresistance/pdf/ar-threats-2013-508.pdf. [Google Scholar]
- 2.Woods RJ, Patel TS, Nagel JL, Newton DW, Read AF. 2018. Institution-wide and within-patient evolution of daptomycin susceptibility in vancomycin-resistant Enterococcus faecium bloodstream infections. Infect Control Hosp Epidemiol 39:226–228. doi: 10.1017/ice.2017.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arias CA, Panesso D, McGrath DM, Qin X, Mojica MF, Miller C, Diaz L, Tran TT, Rincon S, Barbu EM, Reyes J, Roh JH, Lobos E, Sodergren E, Pasqualini R, Arap W, Quinn JP, Shamoo Y, Murray BE, Weinstock GM. 2011. Genetic basis for in vivo daptomycin resistance in enterococci. N Engl J Med 365:892–900. doi: 10.1056/NEJMoa1011138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lipsitch M, Bergstrom CT, Levin BR. 2000. The epidemiology of antibiotic resistance in hospitals: paradoxes and prescriptions. Proc Natl Acad Sci U S A 97:1938–1943. doi: 10.1073/pnas.97.4.1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kleef E, van Luangasanatip N, Bonten MJ, Cooper BS. 2017. Why sensitive bacteria are resistant to hospital infection control. Wellcome Open Res 2:16. doi: 10.12688/wellcomeopenres.11033.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cooper BS, Medley GF, Stone SP, Kibbler CC, Cookson BD, Roberts J. a, Duckworth G, Lai R, Ebrahim S. 2004. Methicillin-resistant Staphylococcus aureus in hospitals and the community: stealth dynamics and control catastrophes. Proc Natl Acad Sci U S A 101:10223–10228. doi: 10.1073/pnas.0401324101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Andersson DI, Hughes D. 2010. Antibiotic resistance and its cost: is it possible to reverse resistance? Nat Rev Microbiol 8:260–271. doi: 10.1038/nrmicro2319. [DOI] [PubMed] [Google Scholar]
- 8.Baym M, Stone LK, Kishony R. 2016. Multidrug evolutionary strategies to reverse antibiotic resistance. Science 351:aad3292. doi: 10.1126/science.aad3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barlam TF, Cosgrove SE, Abbo LM, Macdougall C, Schuetz AN, Septimus EJ, Srinivasan A, Dellit TH, Falck-Ytter YT, Fishman NO, Hamilton CW, Jenkins TC, Lipsett PA, Malani PN, May LS, Moran GJ, Neuhauser MM, Newland JG, Ohl CA, Samore MH, Seo SK, Trivedi KK. 2016. Implementing an antibiotic stewardship program: guidelines by the Infectious Diseases Society of America and the Society for Healthcare Epidemiology of America. Clin Infect Dis 62:e51–e77. doi: 10.1093/cid/ciw118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Siegel JD, Rhinehart E, Jackson M, Chiarello L. 2007. Management of multidrug-resistant organisms in healthcare settings, 2006. Am J Infect Control 35:S165–S193. doi: 10.1016/j.ajic.2007.10.006. [DOI] [PubMed] [Google Scholar]
- 11.Davey P, Marwick CA, Scott CL, Charani E, McNeil K, Brown E, Gould IM, Ramsay CR, Michie S. 2017. Interventions to improve antibiotic prescribing practices for hospital inpatients. Cochrane Database Syst Rev 2:CD003543. doi: 10.1002/14651858.CD003543.pub4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Del Arco A, Tortajada B, de la Torre J, Olalla J, Prada JL, Fernández F, Rivas F, García-Alegría J, Faus V, Montiel N. 2015. The impact of an antimicrobial stewardship programme on the use of antimicrobials and the evolution of drug resistance. Eur J Clin Microbiol Infect Dis 34:247–251. doi: 10.1007/s10096-014-2225-5. [DOI] [PubMed] [Google Scholar]
- 13.Timbrook TT, Hurst JM, Bosso JA. 2016. Impact of an antimicrobial stewardship program on antimicrobial utilization, bacterial susceptibilities, and financial expenditures at an academic medical center. Hosp Pharm 51:703–711. doi: 10.1310/hpj5109-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Charbonneau P, Parienti J, Thibon P, Ramakers M, Daubin C, Du Cheyron D, Lebouvier G, Le Coutour X, Leclercq R; French Fluoroquinolone Free (3F) Study Group. 2006. Fluoroquinolone use and methicillin-resistant Staphylococcus aureus isolation rates in hospitalized patients: a quasi experimental study. Clin Infect Dis 42:778–784. doi: 10.1086/500319. [DOI] [PubMed] [Google Scholar]
- 15.Fridkin SK, Lawton R, Edwards JR, Tenover FC, McGowan JE, Gaynes RP. 2002. Monitoring antimicrobial use and resistance: comparison with a national benchmark on reducing vancomycin use and vancomycin-resistant enterococci. Emerg Infect Dis 8:702–707. doi: 10.3201/eid0807.010465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arias CA, Murray BE. 2012. The rise of the Enterococcus: beyond vancomycin resistance. Nat Rev Microbiol 10:266–278. doi: 10.1038/nrmicro2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sievert DM, Ricks P, Ms JRE, Schneider A, Patel J, Srinivasan A, Kallen A, Limbago B, Fridkin; National Healthcare Safety Network Team and Participating NHSN Facilities. 2013. Antimicrobial-resistant pathogens associated with healthcare- associated infections: summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2009–2010. Infect Control Hosp Epidemiol 34:1–14. doi: 10.1086/668770. [DOI] [PubMed] [Google Scholar]
- 18.O’Driscoll T, Crank CW. 2015. Vancomycin-resistant enterococcal infections: epidemiology, clinical manifestations, and optimal management. Infect Drug Resist 8:217–230. doi: 10.2147/IDR.S54125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Britt NS, Potter EM, Patel N, Steed ME. Comparison of the effectiveness and safety of linezolid and daptomycin in vancomycin-resistant enterococcal bloodstream infection: a national cohort study of veterans affairs patients. Clin Infect Dis 61:871–878. doi: 10.1093/cid/civ444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arias CA, Contreras GA, Murray BE. 2010. Management of multidrug-resistant enterococcal infections. Clin Microbiol Infect 16:555–562. doi: 10.1111/j.1469-0691.2010.03214.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Riedel S, Neoh KM, Eisinger SW, Dam LM, Tekle T, Carroll KC. 2014. Comparison of commercial antimicrobial susceptibility test methods for testing of staphylococcus aureus and enterococci against vancomycin, daptomycin, and linezolid. J Clin Microbiol 52:2216–2222. doi: 10.1128/JCM.00957-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kirn TJ, Onyeaso E, Syed M, Weinstein MP. 2014. Systematic evaluation of commercial susceptibility testing methods for determining the in vitro activity of daptomycin versus Staphylococcus aureus and Enterococci. J Clin Microbiol 52:1877–1882. doi: 10.1128/JCM.03439-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bryant KA, Roberts AL, Rupp ME, Anderson JR, Lyden ER, Fey PD, Van Schooneveld TC. 2013. Susceptibility of enterococci to daptomycin is dependent upon testing methodology. Diagn Microbiol Infect Dis 76:497–501. doi: 10.1016/j.diagmicrobio.2013.04.019. [DOI] [PubMed] [Google Scholar]
- 24.Shukla BS, Shelburne S, Reyes K, Kamboj M, Lewis JD, Rincon SL, Reyes J, Carvajal LP, Panesso D, Sifri CD, Zervos MJ, Pamer EG, Tran TT, Adachi J, Munita JM, Hasbun R, Arias CA. 2016. Influence of minimum inhibitory concentration in clinical outcomes of Enterococcus faecium bacteremia treated with daptomycin: is it time to change the breakpoint? Clin Infect Dis 62:1514–1520. doi: 10.1093/cid/ciw173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Campeau SA, Schuetz AN, Kohner P, Arias CA, Hemarajata P, Dien Bard J, Humphries RM. 2018. Variability of daptomycin minimum inhibitory concentrations for Enterococcus faecium when measured by reference broth microdilution and gradient diffusion tests. Antimicrob Agents Chemother 62:e00745-18. doi: 10.1128/AAC.00745-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Howden BP, McEvoy CRE, Allen DL, Chua K, Gao W, Harrison PF, Bell J, Coombs G, Bennett-Wood V, Porter JL, Robins-Browne R, Davies JK, Seemann T, Stinear TP. 2011. Evolution of multidrug resistance during Staphylococcus aureus infection involves mutation of the essential two component regulator WalKR. PLoS Pathog 7:e1002359. doi: 10.1371/journal.ppat.1002359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bhardwaj P, Hans A, Ruikar K, Guan Z. 2018. Reduced chlorhexidine and daptomycin susceptibility in vancomycin-resistant Enterococcus faecium after serial chlorhexidine exposure. Antimicrob Agents Chemother 62:1–12. doi: 10.1128/AAC.01235-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mwangi MM, Wu SW, Zhou Y, Sieradzki K, Lencastre H, De Richardson P, Bruce D, Rubin E, Myers E, Siggia ED, Tomasz A. 2007. Tracking the in vivo evolution of multidrug resistance in Staphylococcus aureus by whole-genome sequencing. Proc Natl Acad Sci U S A 104:9451–9456. doi: 10.1073/pnas.0609839104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pillai SK, Wennersten C, Venkataraman L, Eliopoulos GM, Moellering RC, Karchmer AW. 2009. Development of reduced vancomycin susceptibility in methicillin‐susceptible Staphylococcus aureus. Clin Infect Dis 49:1169–1174. doi: 10.1086/605636. [DOI] [PubMed] [Google Scholar]
- 30.Mishra NN, McKinnell J, Yeaman MR, Rubio A, Nast CC, Chen L, Kreiswirth BN, Bayer AS. 2011. In vitro cross-resistance to daptomycin and host defense cationic antimicrobial peptides in clinical methicillin-resistant Staphylococcus aureus isolates. Antimicrob Agents Chemother 55:4012–4018. doi: 10.1128/AAC.00223-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen C, Huang Y, Chiu C. 2015. Multiple pathways of cross-resistance to glycopeptides and daptomycin in persistent MRSA bacteraemia. J Antimicrob Chemother 70:2965–2972. doi: 10.1093/jac/dkv225. [DOI] [PubMed] [Google Scholar]
- 32.Knöppel A, Näsvall J, Andersson DI. 2017. Evolution of antibiotic resistance without antibiotic exposure. Antimicrob Agents Chemother 61:e01495–e01417. doi: 10.1128/AAC.01495-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barton NH. 2000. Genetic hitchhiking. Philos Trans R Soc Lond B Biol Sci 355:1553–1562. doi: 10.1098/rstb.2000.0716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Munita JM, Panesso D, Diaz L, Tran TT, Reyes J, Wanger A, Murray BE, Arias CA. 2012. Correlation between mutations in liaFSR of enterococcus faecium and MIC of daptomycin: Revisiting daptomycin breakpoints. Antimicrob Agents Chemother 56:4354–4359. doi: 10.1128/AAC.00509-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Diaz L, Tran TT, Munita JM, Miller WR, Rincon S, Carvajal LP, Wollam A, Reyes J, Panesso D, Rojas NL, Shamoo Y, Murray BE, Weinstock GM, Arias CA. 2014. Whole-genome analyses of Enterococcus faecium isolates with diverse daptomycin MICs. Antimicrob Agents Chemother 58:4527–4534. doi: 10.1128/AAC.02686-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Greene MH, Harris BD, Nesbitt WJ, Watson ML, Wright PW, Talbot TR, Nelson GE. 2018. Risk factors and outcomes associated with acquisition of daptomycin and linezolid-nonsusceptible vancomycin-resistant Enterococcus. Open Forum Infect Dis 5:ofy185. doi: 10.1093/ofid/ofy185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guillard T, Pons S, Roux D, Pier GB, Skurnik D. 2016. Antibiotic resistance and virulence: Understanding the link and its consequences for prophylaxis and therapy. Bioessays 38:682–693. doi: 10.1002/bies.201500180. [DOI] [PubMed] [Google Scholar]
- 38.Melnyk AH, Wong A, Kassen R. 2015. The fitness costs of antibiotic resistance mutations. Evol Appl 8:273–283. doi: 10.1111/eva.12196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kunz AN, Begum AA, Wu H, D'Ambrozio JA, Robinson JM, Shafer WM, Bash MC, Jerse AE. 2012. Impact of fluoroquinolone resistance mutations on gonococcal fitness and in vivo selection for compensatory mutations. J Infect Dis 205:1821–1829. doi: 10.1093/infdis/jis277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li Q-J, Jiao W, Yin Q, Xu F, Li J-Q, Sun L, Xiao J, Li Y, Mokrousov I, Huang H, Shen A. 2016. Compensatory mutations of rifampicin resistance are associated with transmission of multidrug-resistant Mycobacterium tuberculosis Beijing genotype strains in China. Antimicrob Agents Chemother 60:2807–2812. doi: 10.1128/AAC.02358-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Enne VI, Livermore DM, Stephens P, Hall LMC. 2001. Persistence of sulphonamide resistance in Escherichia coli in the UK despite national prescribing restriction. Lancet 357:1325–1328. doi: 10.1016/S0140-6736(00)04519-0. [DOI] [PubMed] [Google Scholar]
- 42.Harbarth S, Harris AD, Carmeli Y, Samore MH. 2001. Parallel analysis of individual and aggregated data on antibiotic exposure and resistance in gram-negative bacilli. Clin Infect Dis 33:1462–1468. doi: 10.1086/322677. [DOI] [PubMed] [Google Scholar]
- 43.Donnan PT, Wei L, Steinke DT, Phillips G, Clarke R, Noone A, Sullivan FM, MacDonald TM, Davey PG. 2004. Presence of bacteriuria caused by trimethoprim resistant bacteria in patients prescribed antibiotics: multilevel model with practice and individual patient data. BMJ 328:1297–1290. doi: 10.1136/bmj.328.7451.1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Clinical and Laboratory Standards Institute. 2015. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard, 10th ed Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 45.Zeileis A. 2004. Econometric computing with HC and HAC covariance matrix estimators. J Stat Softw 11:1–17. [Google Scholar]
- 46.Zeileis A. 2006. Object-oriented computation of sandwich estimators. J Stat Softw 16:1–16. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.