Nontyphoidal Salmonella enterica (NTS) poses a major public health risk worldwide that is amplified by the existence of antimicrobial-resistant strains, especially those resistant to quinolones and extended-spectrum cephalosporins (ESC). Little is known on the dissemination of plasmids harboring the acquired genetic determinants that confer resistance to these antimicrobials across NTS serotypes from livestock in the United States.
KEYWORDS: AmpC, antimicrobial drug resistance, ESBL, Salmonella, fluoroquinolones, plasmids, swine
ABSTRACT
Nontyphoidal Salmonella enterica (NTS) poses a major public health risk worldwide that is amplified by the existence of antimicrobial-resistant strains, especially those resistant to quinolones and extended-spectrum cephalosporins (ESC). Little is known on the dissemination of plasmids harboring the acquired genetic determinants that confer resistance to these antimicrobials across NTS serotypes from livestock in the United States. NTS isolates (n = 183) from U.S. swine clinical cases retrieved during 2014 to 2016 were selected for sequencing based on their phenotypic resistance to enrofloxacin (quinolone) or ceftiofur (3rd-generation cephalosporin). De novo assemblies were used to identify chromosomal mutations and acquired antimicrobial resistance genes (AARGs). In addition, plasmids harboring AARGs were identified using short-read assemblies and characterized using a multistep approach that was validated by long-read sequencing. AARGs to quinolones [qnrB15, qnrB19, qnrB2, qnrD, qnrS1, qnrS2, and aac(6')Ib-cr] and ESC (blaCMY-2, blaCTX-M-1, blaCTX-M-27, and blaSHV-12) were distributed across serotypes and were harbored by several plasmids. In addition, chromosomal mutations associated with resistance to quinolones were identified in the target enzyme and efflux pump regulation genes. The predominant plasmid harboring the prevalent qnrB19 gene was distributed across serotypes. It was identical to a plasmid previously reported in S. enterica serovar Anatum from swine in the United States (GenBank accession number KY991369.1) and similar to Escherichia coli plasmids from humans in South America (GenBank accession numbers GQ374157.1 and JN979787.1). Our findings suggest that plasmids harboring AARGs encoding mechanisms of resistance to critically important antimicrobials are present in multiple NTS serotypes circulating in swine in the United States and can contribute to resistance expansion through horizontal transmission.
INTRODUCTION
Nontyphoidal Salmonella enterica (NTS) is a major foodborne pathogen (1). The impact of NTS is greater when strains become resistant to the antimicrobials used to treat clinical salmonellosis in humans (2). While most NTS infections are transient and do not require antibiotic treatment, the use of antimicrobials such as quinolones and extended-spectrum cephalosporins (ESC) is indicated in invasive infections (3).
Quinolones are bactericidal antibiotics that interfere with the uncoiling of bacterial DNA during replication by inhibiting the target enzymes DNA gyrase and topoisomerase IV. Three main genetic determinants of resistance to quinolones have been described in Salmonella (4): (i) chromosomal mutations or deletions in the target enzyme-encoding genes, (ii) chromosomal mutations or deletions in genes regulating efflux pumps, and (iii) acquired antimicrobial resistance genes (AARGs) harbored by plasmids. Cephalosporins are antibiotics whose bactericidal effect is mediated by alteration of the bacterial cell wall construction (5). Genetic resistance to cephalosporins in NTS is a result of plasmid-mediated AARGs (6).
Plasmids are transformable circular genetic elements of various sizes that play an important role in the dissemination of antimicrobial resistance (AMR) within and among bacterial species (7).
According to the National Antimicrobial Resistance Monitoring System (NARMS), the prevalences of resistance to ceftiofur (3rd-generation cephalosporin) in NTS isolates from poultry and swine sampled at the slaughterhouse have changed from 7.2% and 2.4%, respectively, in 2013 to 9.7% and 1.8% in 2015 (8). During the same period, no resistance to ciprofloxacin (a quinolone) was found in NTS from poultry (in which its use has been banned since 2005) (9), while ciprofloxacin resistance in NTS from swine remained low (it varied between 0.4% in 2013 and 0.2% in 2015) (8). The numbers of isolates that were resistant to ciprofloxacin and ceftriaxone (3rd-generation cephalosporin) in NTS isolates from human clinical samples in NARMS during 2014 were 9/2,172 (0.4%) and 51/2,172 (2.3%), respectively (10).
While AARGs which confer resistance to ESC were commonly found in NTS isolates from animals and environmental samples in the United States (11, 12), AARGs which confer resistance to quinolones have been reported only occasionally (13–15). However, these AARGs, along with chromosomal mutations in target enzymes and efflux pump regulation genes, were recently described in enrofloxacin-resistant S. enterica 4,[5],12:i:− strains isolated from Midwestern swine (16). Moreover, in contrast with NARMS reports of isolates from animals sampled at the slaughterhouse (8, 9), Hong et al. (17) found that enrofloxacin resistance has been increasing among swine clinical NTS isolates from the Midwest since 2008 and that certain serotypes had higher prevalences of resistance to enrofloxacin (a quinolone) and ceftiofur. Given the potential impact on public health from dissemination of resistance to such important antimicrobial families, we aimed to characterize the antimicrobial resistance-conferring determinants to quinolones and ESC in NTS serotypes from Midwestern swine and the plasmids contributing to their spread.
RESULTS
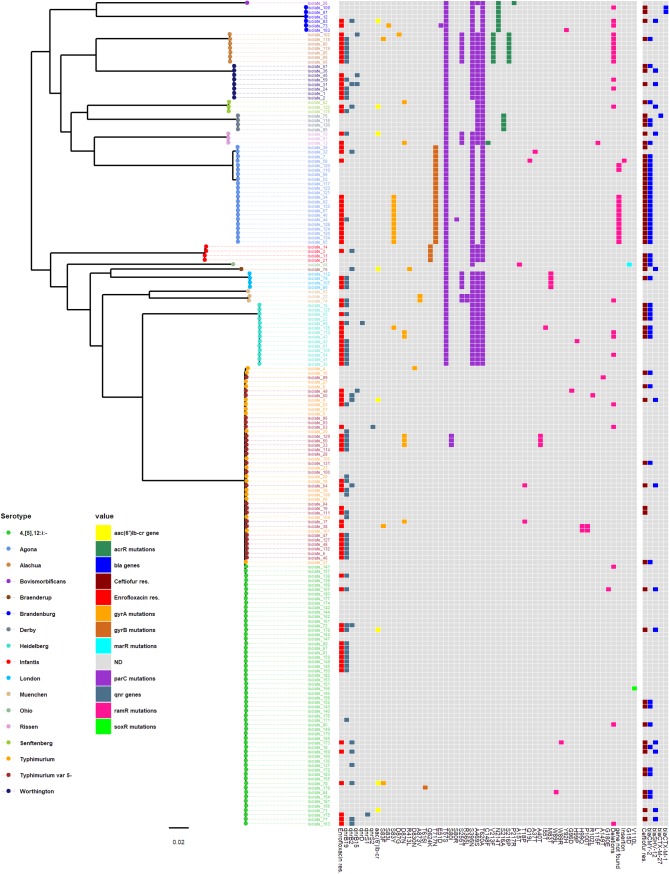
A maximum-likelihood tree was constructed using the core genome (3,252,309 bp, including 3,402 genes) from swine NTS isolates (between 1 and 39,174 pairwise single-nucleotide polymorphisms [SNPs], median = 26,716) (Fig. 1). The average number of within-serotype pairwise-SNP differences for serotypes represented by at least 10 isolates ranged from 18 to 399 pairwise SNPs. Overall, at least one potential resistance determinant was found in 87/89 and 65/68 of the isolates resistant to enrofloxacin and ceftiofur, respectively.
FIG 1.
Maximum-likelihood tree constructed using the core-genome alignment of nontyphoidal-Salmonella isolates collected from Midwestern swine during the years 2014 to 2016. Two S. Paratyphi type A outgroup strains (GenBank accession numbers SRR3033248 and SRR3277289) were used to root the tree (not included in the figure). The analysis included 122,201 variable sites in the alignment (i.e., SNPs). Tip colors indicate serotype. Data shown in heatmap include (i) resistance to enrofloxacin (MIC of ≥1 mg/liter), (ii) the presence of qnr and aac(6')Ib-cr genes, (iii) chromosomal mutations in target enzyme genes and genes involved in the regulation of efflux pumps, (iv) resistance to ceftiofur (MIC of ≥8 mg/liter), and (v) the presence of bla genes.
For well-represented serotypes (with at least 14 isolates resistant to either antimicrobial), phenotypic resistance to both enrofloxacin and ceftiofur (i.e., coresistance) was more frequent in S. enterica serovar Agona (13/22) and S. Heidelberg (5/14) than in S. 4,[5],12:i:− (5/26) and S. Typhimurium variant 5− (3/19) (see Table S4 in the supplemental material).
The AARGs to quinolones [qnrB19, qnrB2, qnrB15, qnrD, qnrS1, qnrS2, and aac(6')Ib-cr] and ESC (blaCMY-2, blaCTX-M-1, blaCTX-M-27, and blaSHV-12) were mostly detected among phenotypically resistant isolates (Fig. 1). For quinolones, multiple mutations in target enzyme genes (gyrA, gyrB, and parC), except parE, and in all efflux pump regulation genes (acrR, ramR, marR, and soxR), including base pair deletions of ramR, were found in isolates from multiple serotypes (Fig. 1, Table 1; Tables S2 and S3). In addition, ramR could not be detected in 13 S. Agona isolates (11 of which were enrofloxacin resistant) using the short-read assemblies, and only 35% of the gene was detected (100% identity) in one S. Agona isolate (isolate 44) following long-read assembly.
TABLE 1.
Potential quinolone resistance determinants identified in nontyphoidal-Salmonella serotypes, summarized by serotype and phenotypic resistance to enrofloxacin
| Salmonella serotype | Enrofloxacin resistanta | Total no. of isolates | No. of isolates with indicated potential resistance determinant |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Acquired gene |
Target enzyme geneb |
Efflux pump regulation geneb |
||||||||||
| Qnr | aac(6')Ib-cr | gyrA | gyrBc | parCc | parE | acrR | ramR | marR | soxR | |||
| Agona | No | 8 | 0 | 0 | 0 | 0 (8) | 0 (8) | 0 | 0 | 2d | 0 | 0 |
| Yes | 14 | 2 | 0 | 11 | 0 (14) | 1 (13) | 0 | 0 | 13d | 0 | 0 | |
| Alachua | Yes | 7 | 7 | 0 | 2 | 0 | 0 (7) | 0 | 7 | 7 | 0 | 0 |
| Bovismorbificans | No | 1 | 0 | 0 | 0 | 0 | 0 (1) | 0 | 1 | 0 | 0 | 0 |
| Braenderup | No | 1 | 1 | 1 | 1 | 0 | 0 (1) | 0 | 0 | 0 | 0 | 0 |
| Brandenburg | No | 4 | 0 | 0 | 0 | 0 | 0 (4) | 0 | 4 | 2 | 0 | 0 |
| Yes | 2 | 1 | 1 | 1 | 0 | 1 (1) | 0 | 2 | 2 | 0 | 0 | |
| Derby | No | 4 | 0 | 0 | 0 | 0 | 0 (4) | 0 | 4 | 0 | 0 | 0 |
| Heidelberg | No | 2 | 0 | 0 | 0 | 0 | 0 (2) | 0 | 0 | 0 | 0 | 0 |
| Yes | 12 | 9 | 0 | 3 | 0 | 0 (12) | 0 | 0 | 5 | 0 | 0 | |
| Infantis | No | 3 | 0 | 0 | 0 | 3 | 0 (3) | 0 | 0 | 0 | 0 | 0 |
| Yes | 1 | 1 | 0 | 0 | 1 | 0 (1) | 0 | 0 | 0 | 0 | 0 | |
| London | No | 1 | 0 | 0 | 0 | 0 | 0 (1) | 0 | 0 | 1 | 0 | 0 |
| Yes | 3 | 3 | 0 | 0 | 0 | 0 (3) | 0 | 0 | 3 | 0 | 0 | |
| 4,[5],12:i:- | No | 41 | 2 | 1 | 0 | 1 | 0 | 0 | 0 | 3 | 0 | 1 |
| Yes | 17 | 17 | 2 | 1 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | |
| Muenchen | No | 2 | 0 | 0 | 1 | 0 | 1 (1) | 0 | 0 | 0 | 0 | 0 |
| Yes | 1 | 1 | 0 | 1 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | |
| Ohio | No | 1 | 0 | 0 | 0 | 0 | 0 (1) | 0 | 0 | 1 | 1 | 0 |
| Rissen | No | 1 | 0 | 0 | 0 | 0 | 0 (1) | 0 | 0 | 0 | 0 | 0 |
| Yes | 2 | 1 | 1 | 1 | 0 | 0 (2) | 0 | 1 | 1 | 0 | 0 | |
| Senftenberg | No | 1 | 0 | 0 | 1 | 0 | 0 (1) | 0 | 0 | 0 | 0 | 0 |
| Yes | 2 | 2 | 1 | 0 | 0 | 0 (2) | 0 | 0 | 1 | 0 | 0 | |
| Typhimurium | No | 15 | 4 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| Yes | 4 | 4 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | |
| Typhimurium variant 5− | No | 7 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| Yes | 18 | 15 | 0 | 5 | 0 | 3 | 0 | 0 | 9 | 0 | 0 | |
| Worthington | No | 2 | 0 | 0 | 0 | 0 | 0 (2) | 0 | 0 | 0 | 0 | 0 |
| Yes | 6 | 6 | 0 | 0 | 0 | 0 (6) | 0 | 0 | 2 | 0 | 0 | |
A MIC of ≥1 mg/liter was used as the cutoff for phenotypic resistance to enrofloxacin.
Isolates in which nonsynonymous mutations were detected.
For gyrB and parC, the number of isolates harboring nonsynonymous mutations that were found regardless of resistance to enrofloxacin and were excluded from the analysis is indicated in brackets (see text and Tables S2 and S3 for more details).
Isolates in which ramR was not detected. These were regarded as nonsynonymous mutations.
The presence of qnr genes was significantly associated with enrofloxacin resistance in all isolates (P < 0.025, Pearson’s chi-square test) (Table 2) but not with high MICs (MIC of >2 mg/liter) among the resistant isolates (P > 0.025, Fisher’s exact test) (Table 3). Accordingly, in the well represented S. 4,[5],12:i:−, qnr genes were found in 17/17 and 2/41 of the enrofloxacin-resistant and -susceptible isolates, respectively. In contrast, qnr genes were rarely found (2/14) among S. Agona enrofloxacin-resistant isolates, while mutations in gyrA (11/14) and mutations and deletions in ramR (2/14 and 11/14, respectively) were common (Table 1). In addition, among isolates in which determinants of genetic resistance to quinolones were identified, 37/87 of the enrofloxacin-resistant isolates (MICs of ≥1 mg/liter) carried only AARGs [qnr with or without aac(6')Ib-cr genes] (Fig. 2). In 28 of these, qnrB19 was the sole identified gene encoding quinolone resistance.
TABLE 2.
Associations between phenotypic resistance to enrofloxacin and the presence of qnr genes or the total number of resistance determinants found among all nontyphoidal-Salmonella isolatesa
| Risk factor | No. of resistant isolates (MIC ≥ 1 mg/liter)/total no. of isolates (%) |
Odds ratio (95% CI)b | P valuec | |
|---|---|---|---|---|
| With the risk factor | Without the risk factor | |||
| Presence of qnr genes | 69/76 (90.78) | 20/107 (18.69) | 42.88 (17.14–107.26) | <0.001 |
| Total no. of resistance determinants higher than 2d | 17/18 (94.44) | 72/165 (43.64) | 21.96 (2.85–168.89) | <0.001 |
Univariable analyses for each risk factor were conducted with the Pearson’s chi-square test.
CI, confidence interval.
Statistically significant when P < 0.05/2 = 0.025 (the P value was adjusted to the number of tests; Bonferroni’s correction).
The total number of resistance determinants is the sum of the following: (i) qnr genes, (ii) aac(6')Ib-cr genes, and (iii) target enzyme and efflux pump regulation genes with at least one nonsynonymous mutation (please refer to the manuscript for further details). The maximal number of resistance determinants was five.
TABLE 3.
Associations between high MICs to enrofloxacin and the presence of qnr genes or the total number of resistance determinants found among enrofloxacin-resistant nontyphoidal-Salmonella isolatesa
| Risk factor | No. of isolates with high MICs (>2 mg/liter)/no. of resistant isolates (MIC ≥ 1 mg/liter) (%) |
Odds ratio (95% CI)b | P valuec | |
|---|---|---|---|---|
| With the risk factor | Without the risk factor | |||
| Presence of qnr genes | 21/69 (30.4) | 2/20 (10) | 3.94 (0.8–37.6) | 0.084d |
| Total no. of resistance determinants is higher than 2e | 13/17 (76.47) | 10/72 (13.89) | 20.15 (5.47–74.28) | <0.001 |
Univariable analyses for each risk factor were conducted with Pearson’s chi-square or Fisher’s exact tests.
CI, confidence interval.
Statistically significant when P < 0.05/2 = 0.025 (the P value was adjusted to the number of tests; Bonferroni’s correction).
Fisher’s exact test.
The total number of resistance determinants is the sum of the following: (i) qnr genes, (ii) aac(6')Ib-cr genes, and (iii) target enzyme and efflux pump regulation genes with at least one nonsynonymous mutation (please refer to the manuscript for further details). The maximal number of resistance determinants was five.
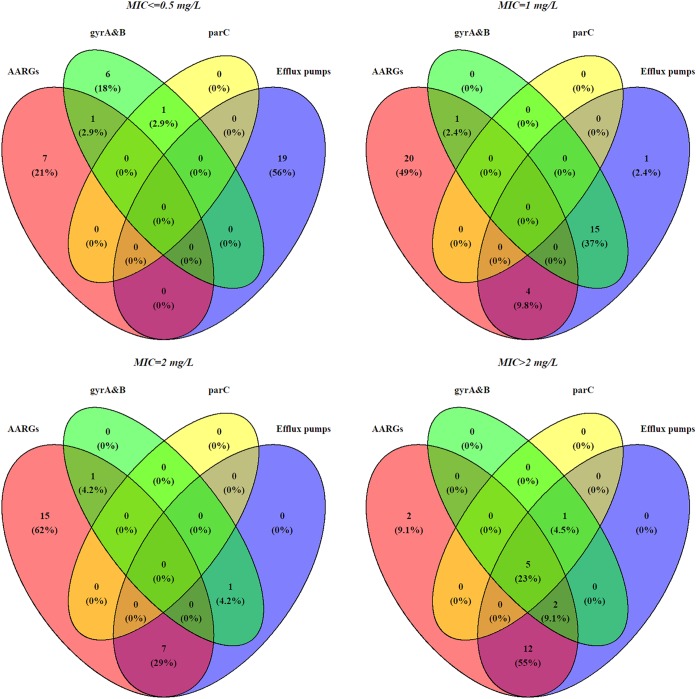
FIG 2.
Venn diagrams demonstrating the degree of overlap between enrofloxacin resistance determinants and different enrofloxacin MICs. Enrofloxacin resistance determinants were grouped as follows: (i) presence of at least one AARG [qnr and/or aac(6')Ib-cr] (red); (ii) at least one mutation in gyrA and/or gyrB target genes (green); (iii) at least one mutation in parC target gene (yellow); and (iv) at least one mutation (including deletions and insertions) in the efflux pump regulation genes (acrR, ramR, marR, and/or soxR) (blue).
Isolates with higher enrofloxacin MICs harbored multiple resistance determinants (Fig. 2); for example, only 2/32 (6.2%) isolates with MICs of ≤0.5 mg/liter harbored more than one resistance determinant, while 20/22 (90.9%) isolates with MICs of >2 mg/liter harbored two or more distinct resistance determinants. This was consistent with the significant association between enrofloxacin resistance (MIC of ≥1 mg/liter) and the presence of >2 resistance determinants that was found in all isolates (P < 0.025, Pearson’s chi-square test) (Table 2) and that remained significant when evaluated in resistant isolates while using a MIC of >2 mg/liter as a cutoff for high MICs (P < 0.025, Pearson’s chi-square test) (Table 3).
The presence of bla genes was significantly higher in ceftiofur-resistant isolates (65/68) than in susceptible isolates (1/115) (P < 0.001, Fisher’s exact test), and these genes were broadly distributed among serotypes (Fig. 1, Table 4). The AARGs blaCMY-2 (49/66), blaCTX-M-27 (1/66), and blaCTX-M-1 (2/66) were found only in isolates with high MICs (MIC of >8 mg/liter). The AARG blaSHV-12 (14/66) was found mainly in resistant isolates (MIC of 8 mg/liter) but also in one nonresistant isolate.
TABLE 4.
Presence of β-lactamase genes associated with resistance to ceftiofur, summarized by nontyphoidal-Salmonella serotype and resistance phenotype
| Nontyphoidal-Salmonella serotype | Ceftiofur resistanta | β-Lactamase gene(s) presentb (no. of genes detected) | No. of genes detected/no. of isolates (%) |
|---|---|---|---|
| Agona | No | 0/1 (0) | |
| Yes | blaCMY-2 (19), blaCMY-2-like (1) | 20/21 (95.24) | |
| Alachua | No | 0/6 (0) | |
| Yes | blaCMY-2 (1) | 1/1 (100) | |
| Bovismorbificans | No | 0/1 (0) | |
| Braenderup | Yes | blaSHV-12 (1) | 1/1 (100) |
| Brandenburg | No | 0/3 (0) | |
| Yes | blaCTX-M-1 (2), blaSHV-12 (1) | 3/3 (100) | |
| Derby | No | 0/1 (0) | |
| Yes | blaCMY-2 (1), blaCMY-2-like (1), blaCTX-M-27 (1) | 3/3 (100) | |
| Heidelberg | No | 0/7 (0) | |
| Yes | blaCMY-2 (7) | 7/7 (100) | |
| Infantis | No | 0/2 (0) | |
| Yes | blaCMY-2 (2) | 2/2 (100) | |
| London | No | 0/3 (0) | |
| Yes | blaCMY-2 (1) | 1/1 (100) | |
| 4,[5],12:i:- | No | 0/44 (0) | |
| Yes | blaCMY-2 (8), blaSHV-12 (6) | 14/14 (100) | |
| Muenchen | No | 0/3 (0) | |
| Ohio | Yes | blaCMY-2 (1) | 1/1 (100) |
| Rissen | No | 0/1 (0) | |
| Yes | blaCMY-2 (1), blaSHV-12 (1) | 2/2 (100) | |
| Senftenberg | No | blaSHV-12 (1) | 1/2 (50) |
| Yes | blaCMY-2 (1) | 1/1 (100) | |
| Typhimurium variant 5− | No | 0/21 (0) | |
| Yes | blaCMY-2 (1), blaSHV-12 (1) | 2/4 (50) | |
| Typhimurium | No | 0/15 (0) | |
| Yes | blaCMY-2 (3), blaSHV-12 (1) | 4/4 (100) | |
| Worthington | No | 0/5 (0) | |
| Yes | blaCMY-2 (1), blaSHV-12 (2) | 3/3 (100) |
A MIC of ≥8 mg/liter was used as the cutoff for phenotypic resistance.
blaTEM-1B and blaCARB-2 were not included in this list.
We were able to detect at least one plasmid group in the genome assemblies of isolates harboring all AARGs except for isolates harboring qnrS1 (Table 5; Table S5), and the plasmid groups identified were distributed across multiple serotypes (Fig. 3). Large plasmids (average size range, 53,880 to 324,077 bp) were detected for all bla, aac(6')Ib-cr, qnrB15, and qnrB2 genes. IncQ2 plasmids (average size, 7,748 bp) were detected for qnrS2, and small ColRNAI and Col3M plasmids were detected for qnrB19 and qnrD, respectively (Table 5).
TABLE 5.
General characteristics of the plasmid groups detected in short-read (Illumina) assemblies that were identified with the corresponding plasmids detected using long-read (Pac-Bio) assemblya
| AARG | Plasmid detected using Illumina reads |
Plasmid identified using hybrid assembly |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Plasmid groupb (no. of isolates) | Incompatibility group/colicin type | Size range (median) (bp) | Mobilityc | AMRc | Heavy metalsc | Isolate ID(s) | Size in bp [range (median)] | Circular | % identityd | % coverage [range (median)]e | |
| aac(6')Ib-cr | I (8) | IncHI2A, IncHI2, TrfAf | 318,782–328,945 (324,503) | + | + | + | 64, 76, 77g | 225,754–309,825 (307,579) | Yes | 99 | 59–94 (82) |
| qnrB15 | I (1) | IncN | 54,641 | + | + | − | |||||
| ND (3) | |||||||||||
| qnrB19 | I (11) | ColRNAI | 2,617–2,826 (2,699) | − | − | − | |||||
| II (39) | ColRNAIf | 3,071–3,082 (3,071) | − | − | − | 33, 44, 61, 69h | 3,071 (3,071) | Yes | 99 | 99–100 (99) | |
| III (1) | ColRNAI | 2,989 (2,989) | |||||||||
| ND (5) | |||||||||||
| qnrB2 | I (15) | IncHI2A, IncHI2, TrfAf | 261,310–339,962 (263,138) | + | + | + | 64, 76, 77g | 225,754–309,825 (307,579) | Yes | 99 | 62–99 (84) |
| ND (1) | |||||||||||
| qnrD | I (1) | Col3Mf | 2,682–2,683 (2,683) | − | − | − | 69i | 2,683 | Yes | 100 | 100 (100) |
| qnrS1 | ND (1) | ||||||||||
| qnrS2 | I (1) | IncQ2f | 7,748 | + | − | − | 63j | 7,555 | Yes | 100 | 99 (99) |
| blaCMY-2 | I (36) | IncA/C2f | 152,216–199,469 (166,161) | + | + | + | 44k | 179,765 | No | 99 | 80–98 (90) |
| II (8) | IncI1f | 99,184–109,170 (104,177) | NA | NA | NA | 116l | 98,867 | No | 99 | 99 (99) | |
| ND (5) | |||||||||||
| blaCTX-M-27 | I (1) | IncFIIf | 68,117–78,962 (73,540) | + | + | − | 75m | 68,088 | Yes | 99 | 99 (99) |
| blaCTX-M-1 | I (1) | IncN | 43,265–44,494 (43,880) | NA | − | NA | |||||
| blaSHV-12 | I (14) | IncHI2A, IncHI2, TrfAf | 314,137–328,945 (321,541) | + | + | + | 64, 76, 77g | 225,754–309,825 (307,579) | Yes | 99 | 59–94 (81.5) |
See Table S5 for detailed information and references.
Plasmid groups detected in the Illumina short-read genome assemblies of isolates harboring the AARGs; different plasmid groups are indicated by roman numerals. ND, not detected (isolates in which an AARG was found and yet a plasmid group was not detected).
Independent mobility of the plasmids, presence of multiple AMR genes, and presence of genes associated with resistance to heavy metals on the plasmids according to the literature (see Table S5 for details). NA, not available.
Minimal percent identity of the plasmid identified in the long-read (Pac-Bio) assembly with the plasmids identified in the short-read (Illumina) assembly, determined by megablast (through NCBI).
Percent coverage range and median value (when applicable) of the plasmid identified in the long-read (Pac-Bio) assembly and the plasmids identified in the short-read (Illumina) assembly, determined by megablast (through NCBI).
The same incompatibility group/colicin type was found in the plasmid detected using the long-read (Pac-Bio) assembly.
Isolates 64 and 77 harbored a plasmid carrying both blaSHV-12 and qnrB2 (accession numbers in GenBank of the plasmids detected using long-read assembly are MK191841 and MK191844, respectively). In isolate 76, aac(6')Ib-cr was also found on the same plasmid (the accession number in GenBank of the plasmid detected using long-read assembly is MK191835).
The plasmid accession numbers in GenBank are MK191837, MK191838, MK191839, and MK191842 for isolates 33, 44, 61, and 69, respectively.
The accession number in GenBank of the plasmid detected using long-read assembly is MK191843.
The accession number in GenBank of the plasmid detected using long-read assembly is MK191840.
The accession number in GenBank of the plasmid detected using long-read assembly is MK191845.
The accession number in GenBank of the plasmid detected using long-read assembly is MK191846.
The accession number in GenBank of the plasmid detected using long-read assembly is MK191836.
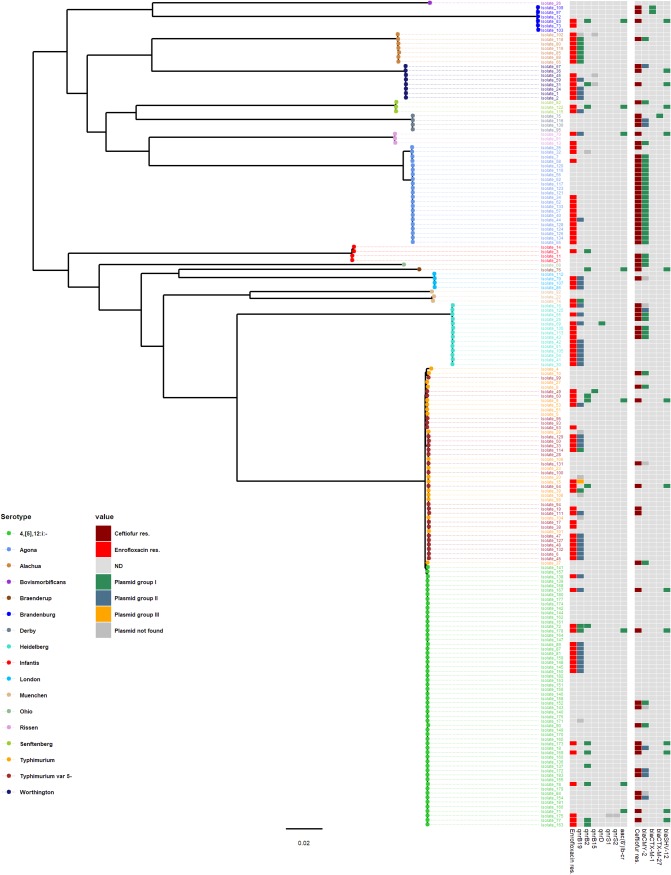
FIG 3.
Maximum-likelihood tree constructed using the core-genome alignment of nontyphoidal-Salmonella isolates collected from Midwestern swine during the years 2014 to 2016. Two S. Paratyphi type A outgroup strains (GenBank accession numbers SRR3033248 and SRR3277289) were used to root the tree (not included in the figure). The analysis included 122,201 variable sites in the alignment (i.e., SNPs). Tip colors indicate serotype. Data shown in the heatmap include (i) resistance to enrofloxacin (MIC of ≥1 mg/liter), (ii) the plasmid groups demonstrated in short-read assemblies for each qnr and aac(6')Ib-cr gene, (iii) resistance to ceftiofur (MIC of ≥8 mg/liter), and (v) the plasmid groups obtained in short-read assemblies for each bla gene.
Using long-read assemblies, we were able to identify the plasmids harboring AARGs in all 10 sequenced isolates. All plasmids detected belonged to the same incompatibility group/colicin type identified in the corresponding plasmid group identified by short-read assembly. Overall, the sequence identity range between plasmids whose sequences were assembled using short-read technology (Illumina) and long-read technology (Pacific Biosciences [Pac-Bio]) was 99% to 100%, while the coverage ranges varied between 99% to 100% and 52% to 100% for the small (<7,555 bp) and large (>68,117 bp) plasmids, respectively (Table 5; Table S5).
In the short-read assemblies, no more than one AARG was located on the same contig (data not shown). In addition, a common plasmid (GenBank accession number CP022696.1) was only found for aac(6')Ib-cr and blaSHV-12 (Table S5). However, using the long-read assemblies, we found large plasmids harboring both qnrB2 and blaSHV-12 in two isolates (isolates 69 and 77), plus aac(6')Ib-cr in another isolate (isolate 76). The long-read-assembly findings agreed with the significant collinearities observed only between these three AARGs (pairwise odds ratios ranged between 19 and 64).
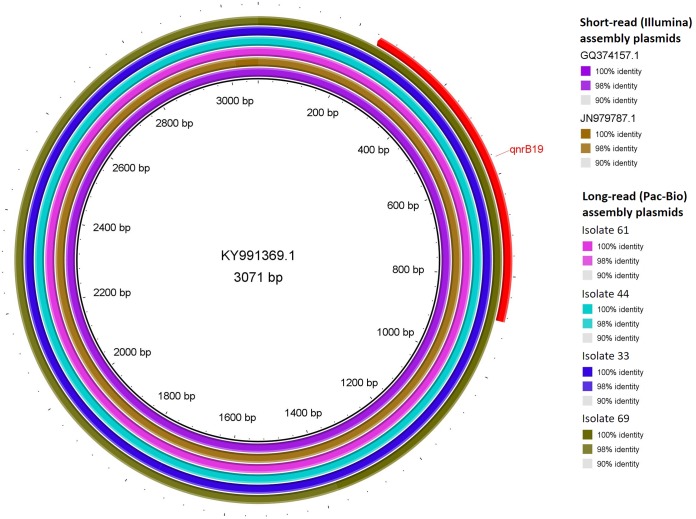
Circular plasmids harboring qnrB19 were found in four isolates (isolates 33, 44, 61, and 69) by using long-read assemblies. Three were identical and one was highly similar (one SNP difference) to a short-read-identified plasmid of group II whose GenBank accession number is KY991369.1 (Fig. 4, Table 5; Table S5). Similar plasmid groups were identified in 19 Salmonella isolates of swine origin harboring the qnrB19 gene in the FDA NARMS Now database (32). These were categorized as harboring plasmids of group I (n = 3) or group II (n = 15), while in one isolate, the plasmid could not be identified.
FIG 4.
A BLAST ring alignment of the two plasmids detected in short-read (Illumina) assemblies (GenBank accession numbers GQ374157.1 and JN979787.1) and four long-read (Pac-Bio) assembly plasmids (isolates 33, 44, 61, and 69) identified for the predominant group of plasmids harboring the qnrB19 gene (group II), with the short-read-assembly-identified plasmid with GenBank accession number KY991369.1 as a reference. The percentage of identity with the reference for each aligned sequence is indicated in the key. The location of the qnrB19 gene is indicated in red.
DISCUSSION
The whole-genome sequencing (WGS) analysis of NTS isolates from swine clinical samples revealed the presence of multiple plasmid-mediated genetic determinants conferring resistance to quinolones or ESC. Each of the AARGs detected was harbored on only a few plasmid groups that were distributed among serotypes, suggesting potential horizontal spread of resistance genes between and within serotypes. In addition, we found multiple known and novel mutations in target enzymes and efflux pump regulation genes. These novel mutations (which are likely to be identified more often given the increasing use of WGS) in the genes involved in the quinolone resistance mechanism may potentially lead to reduced susceptibility. This potential was demonstrated in isolate 58, which was resistant to enrofloxacin (MIC of 1 mg/liter) and did not harbor AARGs, and the only mutation that was found in this isolate was a nonsynonymous mutation in ramR. However, given the complexity of such mechanisms, further molecular studies are essential to determine the contribution of such novel mutations to phenotypic resistance.
As described before for NTS (4, 18), the mutations found here varied between serotypes, and target enzyme mutations were not restricted to the quinolone resistance-determining region (QRDR) only, as opposed to mutations described in Escherichia coli (4). Hopkins et al. (4) postulated that the mutations’ locations outside the QRDR in Salmonella suggest that these mutations confer resistance by different mechanisms than mutations within the QRDR. However, additional molecular studies are essential to support this theory.
The two antimicrobials studied here, ceftiofur and enrofloxacin, have been licensed in the United States for use in swine since 1992 and 2008, respectively (new animal drug application [NADA] application numbers 141-068 [enrofloxacin] and 140-338, 141-235, 141-288, and 200-420 [ceftiofur] [19]). According to NARMS reports, the prevalence of resistance to ESC in swine samples from the slaughterhouse has decreased and stabilized following the ban on extra-label use in livestock in 2012 (13) and the prevalence of resistance to quinolones remained low (<1%) between 2013 and 2015 (8). However, higher levels of resistance have been reported in clinical isolates from swine (17). Comparison of genotypic and phenotypic resistance in this study revealed significant associations between enrofloxacin resistance (MIC of ≥1 mg/liter) and the presence of qnr genes, as well as the number of resistance determinants. The presence of multiple resistance determinants was also significantly associated with high MICs (enrofloxacin MIC of >2 mg/liter) among enrofloxacin-resistant isolates, consistent with a cumulative effect of these genetic determinants. This has been described previously (4, 20), yet to our knowledge, not as part of a comprehensive comparison of all genetic determinants as conducted here. Resistance to ceftiofur was predominantly mediated by blaCMY-2, commonly found in farm animals in the United States (11, 12), and the presence of this gene has recently been linked with the occurrence of ESC-resistant S. Heidelberg strains in Europe (21). This AmpC β-lactamase confers extended resistance to cephalosporins similar to that provided by extended-spectrum β-lactamases (ESBL), but due to its additional resistance to clavulanic acid (6), it may constitute an even higher limitation upon options for medical treatment. The presence of this gene and the ESBL blaCTX genes, which are commonly found in Europe (22) but not in the United States (23), resulted in high MICs (ceftiofur MIC of >8 mg/liter).
Our multistep approach to detect and characterize plasmid groups using the short-read assemblies enabled the characterization of plasmid groups for almost all AARGs, which was further confirmed through long-read sequencing. Still, we had limited ability to identify the simultaneous presence of multiple AARGs on large (>200 kbp) plasmids, which is inevitable given the limitations imposed by the short length of the contigs assembled (24). Overall, the approach taken here could be a useful method to determine whether certain AARGs are distributed by similar plasmids (especially those smaller than 7,555 bp) in NTS across serotypes and/or host species (e.g., human and swine). This knowledge should advance the understanding of the dynamics behind the increasing prevalence of resistant strains and inform the potential design of mitigation measures.
The plasmid with GenBank accession number KY991369.1, present in all four isolates harboring qnrB19 and subjected to Pac-Bio sequencing, was first identified in the United States in an S. Anatum isolate from swine cecal samples in 2014 (14). This small plasmid is highly similar to plasmids in isolates from humans in Bolivia (2005) (25) and Argentina (2008) (26), and we have characterized it as part of the qnrB19-bearing plasmid group II. This group was identified in 39/57 and 15/19 of the isolates harboring qnrB19 in this study and in the NARMS Now data (32), respectively, suggesting it may be widely distributed in the United States. The presence of such a plasmid across multiple serotypes may have been attributed by the emergence of serotypes like S. 4,[5],12:i:− harboring qnrB19 genes (16). This finding is salient, given that our data and other recent studies (14, 16) suggest that qnrB19 alone may be sufficient to confer phenotypic resistance to quinolones, in contrast with previous reports (4, 20).
Among the plasmids harboring qnrB19 whose sequences were assembled using short-read technology (Illumina), the plasmids with GenBank accession numbers GQ374156.1 (25) and KU674895.1 (27) from plasmid group I and the plasmid with GenBank accession number GQ374157.1 (25) from plasmid group II demonstrated mobility via transformation in vitro. In addition, the plasmid with GenBank accession number FN428572.1 from plasmid group I lacked the mobilization system, and its horizontal spread was suggested to occur only through phage transduction, fusion with conjugative replicons, or transformation of naked DNA (28). These findings may suggest that the plasmids detected in Salmonella isolates from swine in this study are able to spread horizontally to other bacteria. However, further molecular studies for determination of the conjugation ability and plasmid transfer frequency are required for better evaluation of their potential for natural transmissibility and the risk for public health.
Coresistance to antimicrobials due to the presence of multiple AARGs in the same plasmid may underpin the persistence of resistance in a bacterial population even after eliminating the use of one of the antimicrobials (29). In addition, such multiresistant pathogens may lead to higher costs of treatment and to increased use of carbapenems or tigecycline in human clinical cases (3). In this study, the detection of large plasmids harboring qnrB2, blaSHV-12, and occasionally aac(6')Ib-cr indicates the possible spread of such coresistance in swine. This may impose an additional challenge for mitigating these resistant phenotypes. Interestingly, in S. Agona, in which phenotypic coresistance was abundant, these plasmids were not found and cooccurrence of AARGs against quinolones and ESC was rare, suggesting that other mechanisms not identified in this study may be involved in coresistance.
Higher prevalences of resistance to enrofloxacin and ceftiofur in serotypes 4,[5],12:i:− and Agona described previously in the Midwestern swine clinical samples (17) led us to hypothesize that the same genetic determinants could contribute to resistance in other serotypes. However, we found different resistance determinants in these serotypes: in S. 4,[5],12:i:−, resistance to ceftiofur was mediated by the presence of either blaCMY-2 or blaSHV-12, while resistance to enrofloxacin was mediated by the presence of qnrB2 and aac(6')Ib-cr (potentially in the same plasmid as blaSHV-12) or by the presence of qnrB19. In contrast, resistance to ceftiofur in S. Agona was associated mainly with blaCMY-2, while enrofloxacin resistance was mediated mainly by an S83Y mutation (a change of the amino acid Serine to Tyrosine in position 83) in gyrA previously described in Salmonella and associated with resistance (18). In addition, in the same S. Agona isolates, ramR was not detected in the short-read assemblies and a deletion of 375 bp in the gene sequence was identified in the long-read assembly. Akiyama and Khan (30) described a 315-bp deletion in ramR from S. Schwarzengrund isolates that resulted in the overexpression of ramA and led to reduced susceptibility to quinolones, and therefore, a similar effect could be hypothesized here.
Overall, these findings demonstrate that diverse determinants contribute to resistance to ESC and quinolones in NTS serotypes in swine in the United States, and the serotype-specific genotypes described here highlight the importance of serotype-specific AMR surveillance. Due to the importance of these antimicrobials in human medicine, the potential of transmission of such resistant strains to humans is of concern, especially given the presence of plasmids harboring AARGs that could spread horizontally and potentially be transmitted to other bacteria.
MATERIALS AND METHODS
Study population.
A subset of 183 NTS isolates comprising 17 serotypes recovered from Midwest swine clinical samples at the Minnesota Veterinary Diagnostic Laboratory during 2014 to 2016 were selected and sent for whole-genome sequencing (WGS) using Illumina platforms. Isolates were (i) resistant to either enrofloxacin (n = 56) or ceftiofur (n = 35) or both (n = 33) or (ii) susceptible to both (n = 59). Among these, the WGS information and resistance phenotypes of 48 S. 4,[5],12:i:− isolates were available from a previous study (16).
Clinical and Laboratory Standards Institute methodology (31) and clinical breakpoints for resistance were adopted (16) to determine phenotypic resistance to enrofloxacin (MIC of ≥1 mg/liter) and ceftiofur (MIC of ≥8 mg/liter). For this purpose, isolates with intermediate MICs (i.e., MICs above 0.25 and lower than 1 mg/liter and above 2 and lower than 8 mg/liter for enrofloxacin and ceftiofur, respectively) were regarded as susceptible.
In addition, the slaughterhouse findings of the U.S. Food and Drug Administration (FDA) NARMS Now surveillance program (32) were screened (using free-text search) to identify Salmonella isolates recovered from swine cecal samples between 2013 and 2015 that harbored the qnrB19 gene. Their raw reads were downloaded and subjected to the same analysis for plasmid characterization (see below).
Data analysis.
De novo-assembled contigs created using the SPAdes assembler (version 3.12.0) (33) were used to (i) perform core-genome alignment and phylogenetic analysis (see below), (ii) determine the presence of nonsynonymous mutations (herein simply “mutations”) in known target enzyme genes and genes involved in the regulation of efflux pumps (Table S1) using a local BLAST (version 2.4.0+) (34), and (iii) determine the presence of AARGs, plasmid replicons, and multilocus sequence types using the bacterial analysis pipeline (with the default settings, i.e., threshold cutoffs for gene detection were set to at least 90% identity and more than 60% coverage of the query sequence) at the Center for Genomic Epidemiology (CGE; https://cge.cbs.dtu.dk/services/).
In addition, the Salmonella In Silico Typing Resource (SISTR) platform (version 1.0.2) (35) was used to determine the serotypes (see the supplemental material for further details).
Core-genome alignment and phylogeny construction.
The de novo assemblies were annotated using Prokka (version 1.13.3) (36), and a core genome (including the outgroup strains; see below) was extracted using Roary (version 3.12.0) (37). The core-genome alignment was used for the construction of the maximum-likelihood trees using RAxML (version 8.2.10) (38) with the generalized time-reversible with gamma (GTR+Γ) substitution model. Trees were rooted using S. Paratyphi type A as an outgroup (GenBank accession numbers SRR3033248 and SRR3277289). Support for nodes on the trees was assessed using 5,000 bootstrap replicates. The packages ape (version 5.0) (39) and ggtree (version 1.10.5) (40) in R (version 3.4.3) (41) were used for visualization.
Identification of the plasmids harboring AARGs.
Following the detection of AARGs using the CGE website, local BLAST queries were used to identify the contigs containing the AARGs. These contigs were then BLASTed (online) against the NCBI repository (NCBI nt), and the first 10 matches (i.e., top matches sorted [ascending] by their E values) obtained for each contig were recorded (overall coverage and identity ranged from 6% to 100% and 90.89% to 100%, respectively). For each AARG, a matrix of contigs (each representing an isolate, as none of the isolates harbored more than a single copy of an AARG) and BLAST matches was generated. Then, the NCBI GenBank records from matches identified in at least 10% of the isolates were screened (using free-text search) to include only matches indicated as plasmids and in which the AARG in question or resistance to its antimicrobial family (quinolones/cephalosporins) were indicated. The sequences of these plasmids were then downloaded and used as a reference to align (using Bowtie2 version 2.3.4.1) (42) the raw reads of the isolates in which a given AARG was found. In these alignments, a conservative approach was taken and a base coverage depth of zero was assigned to locations with fewer than 8 bases aligned. The breadth of coverage percentile [breadth of coverage percentile = 100 × (reference genome length − number of positions with zero coverage)/reference genome length], the absolute breadth of coverage [absolute breadth of coverage = (breadth of coverage percentile × reference genome length)/100], and the number of single-nucleotide polymorphisms (SNPs) were then calculated for each alignment.
The plasmid references for which the alignments had the highest percent breadth of coverage (top two) or absolute breadth of coverage (top two) were selected out of the alignments that had at least 60% breadth of coverage (the same cutoff used as the default setting for gene detection in ResFinder [version 2.1]) (43) and fewer than 200 SNPs (arbitrarily selected to reduce the number of possible matches). The plasmids selected following this multistep filtering process were grouped based on similarity and were considered the most likely plasmids containing the AARG.
Pac-Bio sequencing.
Ten isolates were selected for Pacific Biosciences (Pac-Bio) long-read sequencing based on their plasmids identified in short-read assemblies. Hybrid assemblies of the Pac-Bio long reads with the Illumina short reads were created using Unicycler (version 0.4.4) (44). Bandage (version 0.8.1) (45) was used to visualize assemblies and to identify the plasmids harboring the AARGs in long-read assemblies. Pac-Bio-identified plasmids were compared with the plasmid groups identified in the previous step using NCBI nucleotide megablast, and the BLAST Ring Image Generator (BRIG version 0.95) (46) was used for alignment visualization.
Data summarization and statistical analysis.
Data were summarized using Microsoft Excel and R (version 3.43) (41). Associations between phenotypic enrofloxacin resistance and (i) the presence of qnr genes or (ii) the total number of potential resistance determinants (including AARGs and mutations) were evaluated in separate univariable analyses. The analyses were conducted in (i) all the isolates using a MIC of ≥1 mg/liter as a cutoff value for resistance and (ii) resistant isolates using a MIC of >2 mg/liter as a cutoff for high MICs. In addition, the association between the presence of bla genes and phenotypic resistance to ceftiofur was assessed. All associations were estimated in separate univariable analyses with Pearson’s chi-square or Fisher’s exact tests using the WinPEPI statistical package (47). A P value of <0.05 was considered to indicate statistical significance, and when necessary, it was adjusted for multiple comparisons using Bonferroni’s correction.
The relationships between the presence of resistance determinants and the MICs found for enrofloxacin were visualized (including only isolates in which resistance determinants to quinolones were identified [n = 121]) in Venn diagrams using the VennDiagram package (version 1.6.18) (48) in R. In addition, collinearity between AARG pairs was assessed following the method of Dohoo et al. (49).
For the purpose of this analysis, the chromosomal mutations detected (Table S3 in the supplemental material) were defined as potentially contributing to resistance to quinolones. However, the chromosomal mutations T717N in gyrB and T57S, S255T, S395N, A469S, and T620A in parC were found in multiple serotypes regardless of the enrofloxacin resistance phenotype and were excluded from the analyses (Fig. 1; supplemental material and Tables S2 and S3).
For further details on materials and methods, see the supplemental material.
Accession number(s).
The raw reads from Illumina and Pac-Bio sequencing were deposited at the NCBI under BioProject accession numbers PRJNA215333 and PRJNA505665. In addition, the plasmids harboring AARGs that were detected using the long-read assemblies were uploaded to GenBank under accession numbers MK191835 to MK191846.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Global Food Venture-MnDrive Initiative, the National Institute of Food and Agriculture (Animal Health Formula Fund project MIN-62-091) of the USDA, the Rapid Agricultural Response Fund (RARF), the Swine Disease Eradication Center (SDEC) at the University of Minnesota, and the GenomeTrakr project of the U.S. Food and Drug Administration (FDA). In addition, E.E. was supported by BARD, the United States-Israel Binational Agricultural Research and Development Fund, Vaadia-BARD Postdoctoral Fellowship award no. FI-565-17.
We thank Colette Friedenson for her help reviewing the literature.
The authors declare no conflict of interests.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.02602-18.
REFERENCES
- 1.Kirk MD, Pires SM, Black RE, Caipo M, Crump JA, Devleesschauwer B, Dopfer D, Fazil A, Fischer-Walker CL, Hald T, Hall AJ, Keddy KH, Lake RJ, Lanata CF, Torgerson PR, Havelaar AH, Angulo FJ. 2015. World Health Organization estimates of the global and regional disease burden of 22 foodborne bacterial, protozoal, and viral diseases, 2010: a data synthesis. PLoS Med 12:e1001921. doi: 10.1371/journal.pmed.1001921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Centers for Disease Control and Prevention. 2013. Antibiotic resistance threats in the United States, 2013. CDC, Atlanta, GA.
- 3.Crump JA, Sjolund-Karlsson M, Gordon MA, Parry CM. 2015. Epidemiology, clinical presentation, laboratory diagnosis, antimicrobial resistance, and antimicrobial management of invasive salmonella infections. Clin Microbiol Rev 28:901–937. doi: 10.1128/CMR.00002-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hopkins KL, Davies RH, Threlfall EJ. 2005. Mechanisms of quinolone resistance in Escherichia coli and Salmonella: recent developments. Int J Antimicrob Agents 25:358–373. doi: 10.1016/j.ijantimicag.2005.02.006. [DOI] [PubMed] [Google Scholar]
- 5.Caprile KA. 1988. The cephalosporin antimicrobial agents: a comprehensive review. J Vet Pharmacol Ther 11:1–32. doi: 10.1111/j.1365-2885.1988.tb00117.x. [DOI] [PubMed] [Google Scholar]
- 6.Jacoby GA. 2009. AmpC beta-lactamases. Clin Microbiol Rev 22:161–182. doi: 10.1128/CMR.00036-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carattoli A. 2009. Resistance plasmid families in Enterobacteriaceae. Antimicrob Agents Chemother 53:2227–2238. doi: 10.1128/AAC.01707-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.US FDA. 2017. NARMS Now: integrated data. US Food and Drug Administration, Department of Health and Human Services, Rockville, MD: https://www.fda.gov/AnimalVeterinary/SafetyHealth/AntimicrobialResistance/NationalAntimicrobialResistanceMonitoringSystem/ucm416741.htm. Accessed 12 January 2019. [Google Scholar]
- 9.US FDA. 2017. NARMS integrated report, 2015. US Food and Drug Administration, Department of Health and Human Services, Laurel, MD: https://www.fda.gov/AnimalVeterinary/SafetyHealth/AntimicrobialResistance/NationalAntimicrobialResistanceMonitoringSystem/ucm059103.htm. [Google Scholar]
- 10.Centers for Disease Control and Prevention. 2016. National Antimicrobial Resistance Monitoring System for enteric bacteria (NARMS): human isolates surveillance report for 2014 (final report). CDC, Atlanta, GA. [Google Scholar]
- 11.Alcaine SD, Sukhnanand SS, Warnick LD, Su WL, McGann P, McDonough P, Wiedmann M. 2005. Ceftiofur-resistant Salmonella strains isolated from dairy farms represent multiple widely distributed subtypes that evolved by independent horizontal gene transfer. Antimicrob Agents Chemother 49:4061–4067. doi: 10.1128/AAC.49.10.4061-4067.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao S, White DG, Friedman SL, Glenn A, Blickenstaff K, Ayers SL, Abbott JW, Hall-Robinson E, McDermott PF. 2008. Antimicrobial resistance in Salmonella enterica serovar Heidelberg isolates from retail meats, including poultry, from 2002 to 2006. Appl Environ Microbiol 74:6656–6662. doi: 10.1128/AEM.01249-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.US FDA. 2016. 2014–2015 National Antimicrobial Resistance Monitoring System (NARMS) retail meat interim report. US Food and Drug Administration, Silver Spring, MD: https://www.fda.gov/AnimalVeterinary/NewsEvents/CVMUpdates/ucm498038.htm. Accessed 3 October 2018. [Google Scholar]
- 14.Tyson GH, Tate HP, Zhao S, Li C, Dessai U, Simmons M, McDermott PF. 2017. Identification of plasmid-mediated quinolone resistance in Salmonella isolated from swine ceca and retail pork chops in the United States. Antimicrob Agents Chemother 61:e01318-17. doi: 10.1128/AAC.01318-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cummings KJ, Rodriguez-Rivera LD, Norman KN, Ohta N, Scott HM. 2016. Identification of a plasmid-mediated quinolone resistance gene in Salmonella isolates from Texas dairy farm environmental samples. Zoonoses Public Health doi: 10.1111/zph.12318. [DOI] [PubMed] [Google Scholar]
- 16.Elnekave E, Hong S, Mather AE, Boxrud D, Taylor AJ, Lappi V, Johnson TJ, Vannucci F, Davies P, Hedberg C, Perez A, Alvarez J. 2018. Salmonella enterica Serotype 4,[5],12:i:- in swine in the United States Midwest: an emerging multidrug-resistant clade. Clin Infect Dis 66:877–885. doi: 10.1093/cid/cix909. [DOI] [PubMed] [Google Scholar]
- 17.Hong S, Rovira A, Davies P, Ahlstrom C, Muellner P, Rendahl A, Olsen K, Bender JB, Wells S, Perez A, Alvarez J. 2016. Serotypes and antimicrobial resistance in Salmonella enterica recovered from clinical samples from cattle and swine in Minnesota, 2006 to 2015. PLoS One 11:e0168016. doi: 10.1371/journal.pone.0168016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eaves DJ, Randall L, Gray DT, Buckley A, Woodward MJ, White AP, Piddock LJ. 2004. Prevalence of mutations within the quinolone resistance-determining region of gyrA, gyrB, parC, and parE and association with antibiotic resistance in quinolone-resistant Salmonella enterica. Antimicrob Agents Chemother 48:4012–4015. doi: 10.1128/AAC.48.10.4012-4015.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.US FDA. 2008. Approved Animal Drug Products (Green Book). US Food and Drug Administration, Silver Spring, MD: https://www.fda.gov/AnimalVeterinary/Products/ApprovedAnimalDrugProducts/default.htm. Accessed 3 October 2018. [Google Scholar]
- 20.Hooper DC, Jacoby GA. 2015. Mechanisms of drug resistance: quinolone resistance. Ann N Y Acad Sci 1354:12–31. doi: 10.1111/nyas.12830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liakopoulos A, Geurts Y, Dierikx CM, Brouwer MS, Kant A, Wit B, Heymans R, van Pelt W, Mevius DJ. 2016. Extended-spectrum cephalosporin-resistant Salmonella enterica serovar Heidelberg strains, the Netherlands. Emerg Infect Dis 22:1257–1261. doi: 10.3201/eid2207.151377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rodriguez I, Barownick W, Helmuth R, Mendoza MC, Rodicio MR, Schroeter A, Guerra B. 2009. Extended-spectrum {beta}-lactamases and AmpC {beta}-lactamases in ceftiofur-resistant Salmonella enterica isolates from food and livestock obtained in Germany during 2003-07. J Antimicrob Chemother 64:301–309. doi: 10.1093/jac/dkp195. [DOI] [PubMed] [Google Scholar]
- 23.McDermott PF, Tyson GH, Kabera C, Chen Y, Li C, Folster JP, Ayers SL, Lam C, Tate HP, Zhao S. 2016. Whole-genome sequencing for detecting antimicrobial resistance in nontyphoidal Salmonella. Antimicrob Agents Chemother 60:5515–5520. doi: 10.1128/AAC.01030-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arredondo-Alonso S, Willems RJ, van Schaik W, Schürch AC. 2017. On the (im)possibility of reconstructing plasmids from whole-genome short-read sequencing data. Microb Genom 3:e000128. doi: 10.1099/mgen.0.000128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pallecchi L, Riccobono E, Sennati S, Mantella A, Bartalesi F, Trigoso C, Gotuzzo E, Bartoloni A, Rossolini GM. 2010. Characterization of small ColE-like plasmids mediating widespread dissemination of the qnrB19 gene in commensal enterobacteria. Antimicrob Agents Chemother 54:678–682. doi: 10.1128/AAC.01160-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tran T, Andres P, Petroni A, Soler-Bistue A, Albornoz E, Zorreguieta A, Reyes-Lamothe R, Sherratt DJ, Corso A, Tolmasky ME. 2012. Small plasmids harboring qnrB19: a model for plasmid evolution mediated by site-specific recombination at oriT and Xer sites. Antimicrob Agents Chemother 56:1821–1827. doi: 10.1128/AAC.06036-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fiegen U, Klein G, de Jong A, Kehrenberg C. 2017. Detection of a novel qnrB19-carrying plasmid variant mediating decreased fluoroquinolone susceptibility in Salmonella enterica serovar Hadar. Microb Drug Resist 23:280–284. doi: 10.1089/mdr.2016.0067. [DOI] [PubMed] [Google Scholar]
- 28.Hammerl JA, Beutlich J, Hertwig S, Mevius D, Threlfall EJ, Helmuth R, Guerra B. 2010. pSGI15, a small ColE-like qnrB19 plasmid of a Salmonella enterica serovar Typhimurium strain carrying Salmonella genomic island 1 (SGI1). J Antimicrob Chemother 65:173–175. doi: 10.1093/jac/dkp383. [DOI] [PubMed] [Google Scholar]
- 29.Canton R, Ruiz-Garbajosa P. 2011. Co-resistance: an opportunity for the bacteria and resistance genes. Curr Opin Pharmacol 11:477–485. doi: 10.1016/j.coph.2011.07.007. [DOI] [PubMed] [Google Scholar]
- 30.Akiyama T, Khan AA. 2012. Molecular characterization of strains of fluoroquinolone-resistant Salmonella enterica serovar Schwarzengrund carrying multidrug resistance isolated from imported foods. J Antimicrob Chemother 67:101–110. doi: 10.1093/jac/dkr414. [DOI] [PubMed] [Google Scholar]
- 31.CLSI. 2015. Performance standards for antimicrobial susceptibility testing; twenty-fifth informational supplement. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 32.US FDA. 2018. National Antimicrobial Resistance Monitoring System (NARMS) Now. US Food and Drug Administration, Department of Health and Human Services, Rockville, MD: https://www.fda.gov/animalveterinary/safetyhealth/antimicrobialresistance/nationalantimicrobialresistancemonitoringsystem/ucm570685.htm. Accessed 18 June 2018. [Google Scholar]
- 33.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 35.Yoshida CE, Kruczkiewicz P, Laing CR, Lingohr EJ, Gannon VP, Nash JH, Taboada EN. 2016. The Salmonella In Silico Typing Resource (SISTR): an open Web-accessible tool for rapidly typing and subtyping draft Salmonella genome assemblies. PLoS One 11:e0147101. doi: 10.1371/journal.pone.0147101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 37.Page AJ, Cummins CA, Hunt M, Wong VK, Reuter S, Holden MT, Fookes M, Falush D, Keane JA, Parkhill J. 2015. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 31:3691–3693. doi: 10.1093/bioinformatics/btv421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Paradis E, Claude J, Strimmer K. 2004. APE: Analyses of Phylogenetics and Evolution in R language. Bioinformatics 20:289–290. doi: 10.1093/bioinformatics/btg412. [DOI] [PubMed] [Google Scholar]
- 40.Yu G, Smith DK, Zhu H, Guan Y, Lam TT-Y. 2017. ggtree: an R package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol Evol 8:28–36. doi: 10.1111/2041-210X.12628. [DOI] [Google Scholar]
- 41.R Development Core Team. 2016. R: A language and environment for statistical computing. R Foundation, Vienna, Austria: https://www.R-project.org/. [Google Scholar]
- 42.Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, Aarestrup FM, Larsen MV. 2012. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wick RR, Judd LM, Gorrie CL, Holt KE. 2017. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol 13:e1005595. doi: 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wick RR, Schultz MB, Zobel J, Holt KE. 2015. Bandage: interactive visualization of de novo genome assemblies. Bioinformatics 31:3350–3352. doi: 10.1093/bioinformatics/btv383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Alikhan NF, Petty NK, Ben Zakour NL, Beatson SA. 2011. BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12:402. doi: 10.1186/1471-2164-12-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Abramson JH. 2011. WINPEPI updated: computer programs for epidemiologists, and their teaching potential. Epidemiol Perspect Innov 8:1. doi: 10.1186/1742-5573-8-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen H, Boutros PC. 2011. VennDiagram: a package for the generation of highly-customizable Venn and Euler diagrams in R. BMC Bioinformatics 12:35. doi: 10.1186/1471-2105-12-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dohoo IR, Martin SW, Stryhn H. 2012. Methods in epidemiologic research, p 359–400. UPEI, Charlottetown, Prince Edward Island, Canada. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.