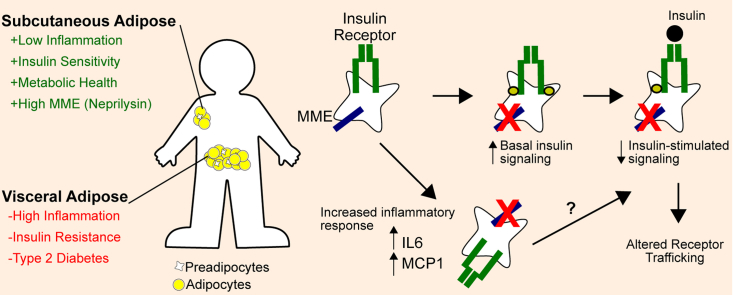
Abstract
Objective
Accumulation of visceral white adipose tissue (WAT) associates with insulin resistance, adipose tissue inflammation, and metabolic syndrome, whereas accumulation of subcutaneous WAT may be protective. We aimed to identify molecular mechanisms that might provide mechanistic insights underlying the phenotypic differences in these tissues. Membrane Metallo-Endopeptidase (MME/Neprislyin) is an extracellular, membrane-bound protease enriched in subcutaneous WAT that can target degradation of a variety of peptides, including insulin, IL6, and β-amyloids. We hypothesized that MME contributes to adipose depot-specific metabolic properties.
Methods
We performed RNA sequencing on human subcutaneous and visceral preadipocytes and array gene expression profiling in murine subcutaneous and visceral preadipocytes. We conducted several insulin signaling and inflammatory response experiments on different cellular states of MME expression.
Results
MME in white preadipocytes is expressed at a higher level in subcutaneous compared to visceral WAT and favors insulin signaling and a low inflammatory response. Thus, knockdown of MME in subcutaneous preadipocytes increased the inflammatory response to substance P and amyloid β aggregates. This associated with increased basal insulin signaling and decreased insulin-stimulated signaling. Moreover, MME differentially regulates the internalization and turnover of the α/β subunits of the insulin receptor.
Conclusion
MME is a novel regulator of the insulin receptor in adipose tissue. Given the clinical significance of both chronic inflammation and insulin sensitivity in metabolic disease, these results show a potentially new target to increase insulin sensitivity and decrease inflammatory susceptibility.
Keywords: Insulin receptor, Adipose, Neprilysin, Visceral, Insulin signaling, Inflammation
Graphical abstract
Highlights
-
•
Preadipocytes show depot-specific gene expression differences, one of which is the membrane metallo-endopeptidase MME.
-
•
MME/Neprilysin perturbations affect insulin signaling and inflammatory response in preadipocytes.
-
•
MME knockdown differentially affects insulin receptor subunit expression and/or turnover.
1. Introduction
Adipose is a complex tissue with an important role in energy homeostasis, endocrine function, and the regulation of immune response. It has a high degree of plasticity and is capable of expanding, contracting, and remodeling to meet a wide range of metabolic challenges [1]. Adipose tissue is primarily composed of adipocytes and adipocyte progenitors but also contains perivascular cells, endothelial cells, and myriad immune cells. Adipocytes are the main adipose constituent by mass and serve the primary role for energy storage and endocrine function. Adipocyte progenitor cells, including mesenchymal stem cells and committed preadipocytes, represent a small mass of tissue but can comprise as much as 50% of adipose tissue by cell number and are the primary source for new adipocytes [2].
Adipose tissue can be divided into at least two metabolically-distinct types: brown adipose tissue (BAT) and white adipose tissue (WAT). BAT is primarily an energy expending tissue, whose human relevance has recently become an active area of research [3], [4]. White adipose tissue, in contrast, is primarily an energy storing tissue. White adipose tissue has been shown to have regional variation based on anatomical location in humans [5], [6]. Some of the most striking differences in WAT are observed between (abdominal) subcutaneous and visceral white adipose tissue. Accumulation of visceral adipose tissue, i.e. central obesity, has been associated with insulin resistance, metabolic syndrome, and diabetes mellitus [6], [7], [8]. By contrast, accumulation of subcutaneous adipose tissue has been associated with metabolically beneficial characteristics, including increased insulin sensitivity and decreased inflammation [7], [9]. White adipose tissue from other regions, such as gluteal adipose tissue, perirenal fat, and bone marrow, also have different properties, including differences in cytokine response and proliferation rates [10], [11]. This regional variation within white adipose tissue stresses the need to understand the underlying mechanisms accounting for differences in white adipose depots in order to develop targeted therapies for diabetes, lipodystrophy, and related metabolic complications.
Several molecular differences between visceral and subcutaneous adipose tissue have been described. One of the most demarcating differences between adipose depots is the signature of developmental genes, including Hox, Shox, and T-box genes [12], [13]. Lineage tracing studies have revealed key developmental signatures in adipocyte development, such as the Myf5 lineage marking brown adipocytes and a subset of white adipocytes across different fat depots [14], [15]. The mesodermal developmental gene TBX15 has also been shown to mark a subset of white adipocytes which have a higher glycolytic rate [16]. Traditionally, surface markers have been used to establish region specificity and to distinguish adipocyte-lineage cells from other adipose-resident cells, although there is disagreement over the exact panel of surface marker expression in the adipocyte lineage [17], [18]. More extensive reviews on this topic are provided elsewhere [2], [19], [20].
Membrane metallo-endopeptidase (MME/Neprilysin/CD10/CALLA) is a membrane-bound protein with a distinct extracellular protease domain. MME was first isolated from rabbit kidney and described as a thermolysin-like enzyme [21]. Since then, MME has been shown to be well-conserved across different species from C. elegans to mammals [22]. MME is a zinc metalloprotease and shares substrates and structural similarity with several related extracellular proteases, including Endothelin Converting Enzyme 1 (ECE1), Phosphate-regulating neutral endopeptidase X-linked (PHEX), and Kell blood group antigen (KEL) [23], [24], [25]. MME is also expressed in the brain, where the MME knockout mouse has been shown to have an increase in amyloidβ peptides, suggesting that MME may play a role in protection from Alzheimer's disease [26]. The whole-body MME knockout (MMEKO) mouse was created in 1995 and was described as a septic shock model because it showed hypersensitivity to treatment with different cytokines [27].
In the context of metabolism, the MMEKO mouse develops age-related obesity. This is thought to be mediated through hyperphagia [28], although the exact mechanism is unclear. Interestingly, whole-body knockdown or overexpression of the Drosophila MME homolog NEP4 decreases larval food intake and decreases the levels of circulating insulin-like peptide DILP1 [29]. In humans, MME is also found in plasma, and circulating levels of MME positively correlate with BMI and HOMA-IR [30]. Additionally, MME mutants have been associated with Charcot-Marie-Tooth disease [31], emphasizing that MME is expressed in a variety of tissue types, including adipose, brain, and lymphatic tissue [26], [27], [28].
MME has been shown to target a variety of small peptides including amyloidβ, insulin B-chain, and several neuropeptides [25], [32]. Additionally, the MME intracellular domain is known interact with PTEN, suggesting it potentially could modify signaling pathways active via the PI3K/Akt pathway [33], [34], [35]. Both adipocytes and preadipocytes express MME, and preadipocytes have been shown to secrete exosome-bound MME, which can be endocytosed by non-adipose cell types such as neuronal cells in vitro [36].
In the present study, we sought to determine the cellular role of MME in distinguishing subcutaneous from visceral preadipocytes and its role in insulin sensitivity and inflammatory response in adipose tissue.
2. Materials and methods
2.1. Human cell culture
Primary cell cultures (4 abdominal subcutaneous, 4 omental, and 4 mesenteric) were obtained from the Kirkland lab [37]. Primary cells for subcutaneous, omental, and mesenteric preadipocytes were grown to 90% confluency in 10-cm plates. Primary cell cultures were maintained for no more than 10 passages. Human neck-derived immortalized white preadipocyte cell lines were obtained from the Tseng lab and cultured as previously described [38], [39]. Cells were maintained in DMEM-H, 10% FBS, 1% Pen-Strep (100 U/mL), and 0.2% normocin (100 μg/mL). Cells were passaged at 90% confluency by a 1:5 dilution.
2.2. RNA sequencing
Total RNA was harvested from a 100% confluent 10-cm plate with 1 mL Trizol and stored at −80 °C. Samples were thawed, homogenized with a 25-gauge needle, and 200 μL chloroform was added. The samples were vortexed (20 s), centrifuged (15,000 g, 30 min), and the aqueous phase was isolated for purification. The aqueous phase was combined with 1 volume of isopropanol, centrifuged (15,000 g, 60 min), and washed with 70% ethanol. The resulting RNA pellet was air dried for 1 h and resuspended in 30 μL RNase-free water. RNA quality was assessed on an Agilent 2100 Bioanalyzer to ensure RIN values were greater than 8.0.
For library prep, we used the NEBNext Ultra Directional Library Prep Kit (New England Biolabs). Briefly, 500 ng of RNA in 50 μL water was combined with 15 μL NEBNext Oligo d(T)25 magnetic beads (pre-washed twice with RNA binding buffer) and incubated at 65 °C for 5 min. The beads were washed twice with 200 μL wash buffer and eluted with 200 μL elution buffer. The binding-washing was repeated once more. The final elution was in 15 μL first strand synthesis buffer and incubated at 94 °C for 15 min. Then 10 μL of the supernatant was used for cDNA synthesis reaction, which was purified using Agencourt AMPure XP beads. End-repair was performed with NEBNext end repair reaction buffer and purified with AMPure XP beads. Library enrichment and multiplexing was performed using the NEBNext High-Fidelty PCR Master Mix (14 cycles of PCR) and NEBNext Multiplex Oligos.
The cDNA libraries were multiplexed on the Illumina HiSeq 2000 and 2500. Raw 50bp paired-end reads were aligned via the Spliced Transcripts to Alignment Reference (STAR 2.3.0e) [40] to the human genome build hg19 and annotation file gencode v19. The non-default and non-directory parameters for building the reference genome in STAR were: STAR --runMode genomeGenerate --sjdbOverhang 49 --runThreadN 8, and the parameters for alignment were STAR --readFilesCommand bzcat --outFilterMismatchNmax 6 --outFilterIntronMotifs RemoveNoncanonicalUnannotated. Raw counts from the alignment files were extracted using HT-seq (v0.5.4) with the following parameters: htseq-count -i gene_id -q -m union -s reverse. Exploratory and differential gene expression analyses were carried out in R (v3.03) and other packages including DESeq2 [41], EBimage [42], and ggplot2 [43]. The data are available on GEO under the accession GSE117353.
2.3. Murine inguinal and epididymal cell culture and microarray
Wild-type C57BL/6 male mice were maintained on chow diet. Mice aged 49–63 days were sacrificed and the inguinal and epididymal adipose depots were surgically extracted. The fat pads were minced, incubated with 5 mL digestion buffer (DMEM-H, 1%BSA, 1 mg/mL collagenase), and incubated on a shaking water bath for 30 min at 37 °C. The solution was filtered through a 150–250 μm mesh-syringe, combined with 10 mL 1%-BSA DMEM-H and centrifuged (800 g, 5 min, 4 °C). The supernatant was removed, the pellet was washed with 10 mL FACS buffer (PBS, 2% Fetal Calf Serum) and centrifuged again (800 g, 5 min, 4 °C). The pellet was resuspended in 1 mL erythrocyte lysis buffer (Ammonium-Chloride-Potassium lysis buffer) and incubated on ice for 5 min. FACS buffer was added to bring the volume to 10 mL and centrifuged (800 g, 5 min, 4 °C). The supernatant was aspirated, and the pellet was resuspended in 300 μL Blocking buffer (PBS, 2% Fetal Calf Serum, 1:150 Fc Block (BD Biosciences)) and centrifuged (800 g, 5 min, 4 °C). The pellet was resuspended in 100 μL of the antibody mix (CD45-, TER119-, CD31, CD34+, Sca1+) and incubated for 15 min on ice. Blocking buffer was added to bring the volume to 250 μL followed by centrifugation (800 g, 5 min, 4 °C). The pellet was resuspended in 200 μL FACS buffer, submitted to FACS, and plated onto 6-well plates. Total RNA was harvested by Trizol (as for the human cells above) and submitted to the Joslin Diabetes Center Genomics Core for microarray hybridization to generate gene expression profiles. The data are available on GEO under the accession GSE117353.
2.4. Quantitative PCR
RNA for qPCR was extracted via the Qiagen RNAeasy mini kit (Qiagen). Complementary DNA synthesis was performed with 200 ng/sample of RNA via the High Capacity cDNA Reverse Transcription kit (Applied Biosystems). Quantitative PCR was performed via the SYBR Green PCR Master Mix (Bio-Rad) on the CFX384 Real-Time PCR Detection System according to the manufacturer's protocol. For relative qPCR, expression was normalized to the reference gene HPRT1. For absolute qPCR, synthetic templates were created as a single gBlock (IDT) and used to calculate copy number.
The sequences for primers or gBlock are as follows: HPRT1 (F:5′-TGAAAAGGACCCCACGAAG, R:5′-AAGCAGATGGCCACAGAACTAG),CCL2 (F:5′-CATAGCAGCCACCTTCATTCC, R:5′-TCTGCACTGAGATCTTCCTATTGG), TNFα (F:5′-TCAGAGGGCCTGTACCTCAT, R: 5′-GGAGGTTGACCTTGGTCTGG), IL6 (F:5′-GGTACATCCTCGACGGCATCT, R:5′-GTGCCTCTTTGCTGCTTTCAC), MME exon 2 (F: 5′-GCTATTGATAGCAGAGGTGGAGA, 5′-GTTGCTACTGGCCACCCATA), MME exon 10 (F: 5′-TGTGGCCAGATTGATTCGTCA, R:5′-TGGCAATTTCTTTTTCCAATTCCA), MME exon 20 (F:5′-GAGATGTGCAAGTGGCGAAG, R:5′-GTGACCCCTCAGCAGATCC), MME exon 21 (F:5′-TCTCGACAATACCCGTTGGC, R:5′-GAGTGGCAGAATAGGGCTGG), and the synthetic template for absolute qPCR (5′-GGGGTCTCGACAATACCCGTTGGCAAGGAGTCTGCCTCCATGCTGCAGTGTTCGAGTGGATTGTAGGTGCAAGATGGAAAGGATTGTAGGTGCAAGCTGTCCAGAGAAAAGAGTCCTTGTTCCAGCCCTATTCTGCCACTCTTTTTGAGATGTGCAAGTGGCGAAGCTTGACCGAGAGCAGGCTGGAGCAGCCGCCCAACTCCTGGCGCGGGATCTGCTGAGGGGTCACTTTTTTGTGGCCAGATTGATTCGTCAGGAAGAAAGATTGCCCATCGATGAAAACCAGCTTGCTTTGGAAATGAATAAAGTTATGGAATTGGAAAAAGAAATTGCCATTTTTGCTATTGATAGCAGAGGTGGAGAACCTCTACTCAAACTGTTACCAGACATATATGGGTGGCCAGTAGCAACTTTTTTGAAAAGGACCCCACGAAGTGTTGGATATAAGCCAGACTTTGTTGGATTTGAAATTCCAGACAAGTTTGTTGTAGGATATGCCCTTGACTATAATGAATACTTCAGGGATTTGAATCATGTTTGTGTCATTAGTGAAACTGGAAAAGCAAAATACAAAGCCTAAGATGAGAGTTCAAGTTGAGTTTGGAAACATCTGGAGTCCTATTGACATCGCCAGTAAAATTATCAATGTTCTAGTTCTGTGGCCATCTGCTTGGGG).
2.5. MME perturbations and insulin signaling
Immortalized human neck derived white preadipocytes, generated as previously described [38], were transfected with MME or non-targeting control (GFP) siRNA using Lipofectamine 3000 (Thermo Fisher) for 24 or 48 h. For the pharmacological inhibition of MME experiments, preadipocytes were incubated for 24 or 48 h with the MME inhibitor omapatrilat (1 μM). For overexpression, the plasmids pCMV-GFP (Addgene #12337), pCMV-MME (Addgene #12338) or pCMV-MMEX (Addgene #12340) were transduced for 48 h with Lipofectamine 3000. The preadipocytes were then serum-starved overnight (24 h) and stimulated with or without 100 nM insulin for 20 min. Cell lysates were collected on ice with cell lysis buffer (1X RIPA, 0.1% SDS, 1X protease and phosphatase inhibitor cocktails (Bimake)). Lysates were centrifuged (10,000 g, 10 min), and supernatants were used for immunoblotting.
2.6. Immunoblotting
Purified protein lysates were quantified with a bicinchoninic acid (BCA) assay (ThermoFisher). For SDS-PAGE, 20 μg/sample of prepared protein (cell lysate with 1X LDS sample buffer) were run on a 4–12% Bis-Tris polyacrylamide gel at 120 V for 60 min in 1X running buffer (2.5 mM MOPS, 2.5 mM Tris base, 0.005% SDS, 50 μM EDTA, pH 7.7). Protein was transferred to PVDF membrane at 30 V for 960 min in 1X transfer buffer (1.25 mM Bicine, 1.25 mM Bis-Tris, 50 μM EDTA, pH 7.2). Membranes were blocked in Starting Block T20 (ThermoFisher). Primary antibodies were used at a 1:2000 dilution (MME (human: Santa Cruz, mouse: Abcam), IRα (Santa Cruz), IRβ (Cell Signaling), pIR/pIGF1R (Cell Signaling), IRS1 (Millipore), pIRS1 (Cell Signaling), pan-AKT (Cell Signaling), pAKT (Cell Signaling), ERK1/2 (Cell Signaling), pERK1/2 (Cell Signaling), Actin (Santa Cruz), GAPDH (Santa Cruz)). Secondary antibodies were used at a 1:10,000 dilution (anti-mouse (Bio-Rad) or anti-rabbit (Bio-Rad)). Signal was detected by Pico or Femto Chemiluminescent Substrate (ThermoFisher).
2.7. MMEKO mice and GTT/ITT
MMEKO mice, originally created on a C57BL/6 background, were kindly provided by Bao Lu [27] to create a colony at Joslin Diabetes Center. MMEKO mice were bred with WT C57BL/6 mice from Charles River Laboratories, and F2 homozygous male KO mice were used for subsequent experiments. MMEKO and WT mice were maintained on chow diet (22% fat by weight). For glucose tolerance testing, 20-week-old mice were starved for 16 h before intraperitoneal injection of 10 μg/gBW (micrograms per gram body weight) glucose. For insulin tolerance testing, the same cohort at 22-weeks old was starved for 4 h and then injected intraperitoneally with 0.1 mU/gBW insulin.
2.8. Isolation of preadipocytes from MMEKO and WT mice
MMEKO and WT mice aged 12 weeks were sacrificed with isoflurane. The inguinal and epididymal adipose tissue depots were surgically removed, minced, and placed in 500 μL digestion buffer (DMEM-H, 2% BSA, 1.5 mg/mL collagenase). The solutions were placed in a shaking water bath (37 °C, 90 cycles/min) until cells were dispersed, but for no more than 40 min. Then, 20 mL growth media (DMEM-H, 10% FBS, 1% Penicillin-Streptomycin, 0.2% Normocin) was added, and the suspension was filtered through a 100-μm cell strainer and centrifuged (1500 rpm, 5 min). The supernatant was aspirated, and the stromal vascular pellet containing preadipocytes was re-suspended in 2 mL growth media before being plated into a single well of a 6-well plate.
2.9. Insulin receptor internalization
Immortalized human neck-derived white preadipocytes [38], [39] were seeded at 50,000 cells/well in 6-well plates. Transient overexpression or knockdown MME was performed as described above. Cells were serum-starved overnight (24 h) and then stimulated with 100 nM insulin for 0, 30 min, or 120 min. Cells were washed with ice-cold 1X PBS and incubated with Sulfo-NHS-Biotin (0.3 mg/mL in PBS) for 30 min at 4 °C to label cell surface proteins. The biotinylation reaction was quenched with 4 °C 100 mM glycine. Cells were washed with ice-cold 1X PBS and lysed in a buffer containing 1X RIPA, 0.1% SDS, 1X protease and phosphatase inhibitor cocktails (Bimake). Lysates were centrifuged (10,000 g, 10 min), and the supernatant was used for streptavidin pulldown.
Purified protein lysates were quantified with a bicinchoninic acid (BCA) assay (ThermoFisher). For each sample, 100 μg of protein was combined with 10 μL of magnetic streptavidin beads and incubated on a rotating rack at 4 °C overnight. The bead pellets were washed with 600 μL 1X RIPA, 600 μL 1X RIPA +500 mM NaCl, and 600 μL 1X RIPA in series. The bead pellets were eluted with 40 μL 1X LDS buffer incubated at 96 °C for 5 min. The immunoprecipitates and total protein were used for SDS-PAGE and western blot as described above.
2.10. Subcellular fractionation
Cell fractionation in immortalized human neck-derived white preadipocytes was performed using a subcellular fractionation kit (Thermo Fisher #78840). Briefly, MME overexpression in 6-well plates was performed as described above. Lysis buffers CEB, MEB, NEB, and PEB were prepared as described in the manufacturer's protocol. Cells were scrapped as a single layer, centrifuged (5 min, 500 g), washed with ice-cold 1X PBS, and lysed with 200 μL/sample ice-cold CEB (incubated at 4 °C for 10 min on a rotary shaker). Pellets were centrifuged (5 min, 500 g), supernatant collected (cytoplasmic fraction), and 200 μL/sample ice-cold MEB was added (vortexed for 5 s and incubated at 4 °C for 10 min). Pellets were centrifuged (5 min, 3000 g), supernatant collected (membrane fraction), and 100 μL/sample ice-cold NEB was added (vortexed for 15 s and incubated at 4 °C for 30 min). Pellets were centrifuged (5 min, 5000 g), supernatant collected (soluble nuclear fraction), and 100 μL/sample ice-cold chromatin-NEB was added (vortexed for 15 s, incubated at 37 °C for 5 min, and vortexed again for 15 s). Pellets were centrifuged (16,000 g, 5 min), supernatant collected (chromatin fraction), and 100 μL/sample of ice-cold PEB was added (vortexed for 15 s and incubated at room temp for 10 min). Pellets were centrifuged (16,000 g, 5 min), and the supernatant collected (cytoskeletal fraction). Isolates were stored at −80 °C. Note that since each of these fractions are isolated in series, residual protein carryover is expected in downstream protein fractions.
2.11. Insulin receptor MME co-immunoprecipitation
Immortalized murine brown IR/IGFR double-knockout preadipocytes [44] were seeded at 50,000 cells/well in 6-well plates. Preadipocytes were then transiently transfected with FLAG-tagged insulin receptor (pCMV-IR-3XFLAG), MME (pCSC-SP-PW-Nep, Addgene #12338), or a combination of the two for 48 h before insulin signaling. For insulin signaling, cells were serum-starved for 3 h and incubated with or without insulin for 15 min and harvested with 1 mL/well co-ip lysis buffer (20 mM Hepes (pH 7.4), 150 mM NaCl, 50 mM KF, 50 mM β-glycerolphosphate, 2 mM EGTA (pH8.0), 1 mM Na3VO4, 1% Triton X-100, 10% glycerol, and 1 × protease inhibitor cocktail (Calbiochem)). Lysates were centrifuged (10,000 g, 10 min) and the supernatant was used for immunoprecipitation.
Anti-FLAG M2 magnetic beads were washed twice with co-ip lysis buffer prior to use. For each sample, 600 μg protein lysate was combined with 20 μL washed M2 magnetic beads and incubated overnight on a rotating rack at 4 °C. The bead pellets were washed three times each with 1 mL co-ip lysis buffer, 1 mL co-ip lysis buffer +500 mM NaCl, and 1 mL co-ip lysis buffer. The bead pellets were then eluted with 40 μL 1X LDS (Lithium Dodecyl Sulfate) running buffer incubated at 96 °C for 5 min. The immunoprecipitates and total protein were used for SDS-PAGE and western blot as described above.
2.12. MME inflammation assays
Immortalized human neck-derived white preadipocytes were seeded at 50,000 cells/well in 6-well plates and MME knockdown was performed as described above for 24-hours. For inflammatory response in primary stromal vascular cells, subcutaneous and omental preadipocytes were seeded at 30,000 cells/well in 24-well plates. Cells were serum-starved for 24 h and then treated with IL1β (5 ng/μL), Substance P (100 nM), Amyloidβ aggregates (20 μM), and/or Omapatrilat (1 μM) for 24 h. Amyloidβ aggregates were generated by incubating Amyloidβ peptide (fragment 25–35; 40 μM) for 7 days at 37 °C. Cell lysis and RNA extraction was performed with the Qiagen RNAeasy mini kit (Qiagen), and qPCR was performed as described above.
2.13. Human protein-protein interactome between MME and INSR
The human protein-protein interaction network pulled from BioGRID (Biological General Repository for Interaction Datasets) was used to determine the shortest paths between insulin receptor (INSR) and MME. Briefly, the R package igraph was used to analyze the protein-protein interactome (PPI). The PPI (Biogrid-ALL-3.4.145) was loaded and the shortest paths between INSR and MME were determined with Djikstra's algorithm via the function all_shortest_paths. Normalized, pre-processed RNA-seq data from subcutaneous adipose tissue was taken from ENCODE [45].
2.14. MME activity assay
Immortalized human neck-derived white preadipocytes were seeded at 50,000 cells/well in 6-well plates. Cells were treated with either vehicle (DMSO) or Omapatrilat (1 μM) for 24 h. MME activity was assayed with a Neprilysin activity assay kit (BioVision). Briefly, pelleted cells were lysed in 400 μL assay buffer and incubated on ice for 10 min. Samples were centrifuged (12,000 g, 10 min, 4 °C) and 10 μL/sample of supernatant loaded into the 96-well assay plate. Assay buffer was added to a final volume of 90 μL/well. Each well received 10 μL of prepared substrate solution. Fluorescence was measured at 430 nm in kinetic mode at 37 C for 1 h. MME activity was determined with a prepared absolute standard curve.
3. Results
3.1. Preadipocytes show depot-specific gene expression patterns
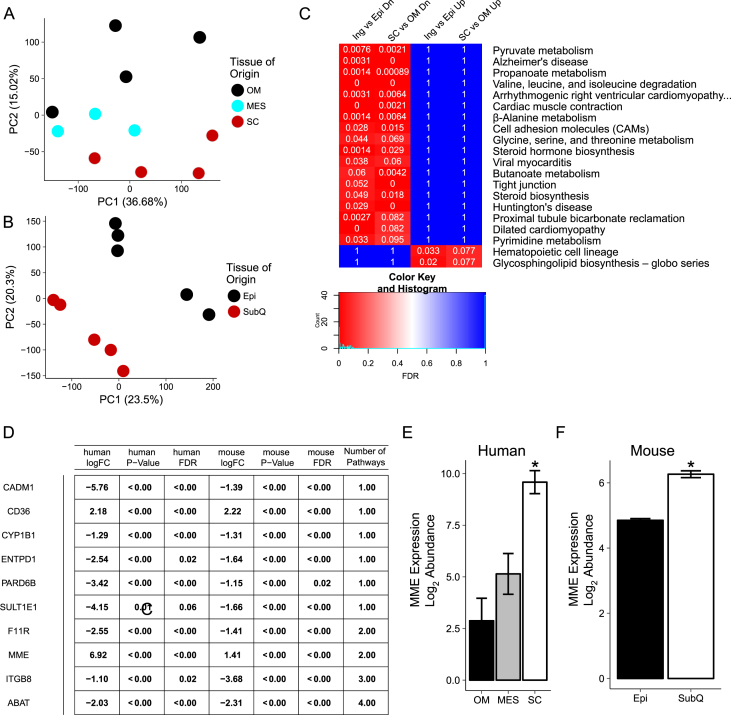
RNA-seq was performed on human preadipocytes from subcutaneous and two visceral depots: omental and mesenteric fat. These were compared to the two standard murine depots used to represent subcutaneous and visceral fat, namely the inguinal and epididymal, respectively. Principal component analysis of the data showed that the gene expression profiles cluster by depot of origin in both human (Figure 1A) and murine (Figure 1B) preadipocytes. Gene set enrichment analysis [46], [47] of differentially expressed genes between subcutaneous and visceral preadipocytes (represented by subcutaneous vs omental fat in humans and inguinal vs epididymal in the mouse) showed a set of 20 significant pathways which showed conserved differential expression across species (Figure 1C).
Figure 1.
RNA-Seq of preadipocytes revealed depot-specific signatures and MME as a highly differentially expressed gene between visceral and subcutaneous preadipocytes. Human subcutaneous, omental, and mesenteric preadipocyte primary cultures were grown to confluency, harvested for RNA, and subjected to RNA-seq to generate gene expression profiles. For mouse gene expression profiles, inguinal and epididymal preadipocyte primary cultures were grown to confluency, harvested for RNA, and subjected microarray analysis. (A) Principal component analysis of human subcutaneous, mesenteric, and omental gene expression profiles shows clustering by depot. (B) Principal component analysis of inguinal and epididymal gene expression profiles shows clustering by depot. (C) Differential gene expression followed by Gene Set Enrichment Analysis (GSEA) revealed a set of pathways differentially expressed between subcutaneous and visceral preadipocytes in human and mouse tissue. SC = abdominal subcutaneous. MES = Mesenteric. OM = Omental. Epi = Epididymal. SubQ = Inguinal. N = 3–5. (D) The 20 significant pathways were analyzed to reveal genes found in multiple pathways. A total of four genes (ABAT, ITGB8, MME, and F11R) were significant in multiple pathways. (E) RNA-seq expression of Membrane Metallo-Endopeptidase (MME) in subcutaneous and visceral preadipocytes (Omental = OM, Mesenteric = MES). (F) Microarray expression of MME in mouse subcutaneous (SubQ) and visceral preadipocytes (Epi). Asterisks indicate p ≤ 0.05 by moderated t-test (limma). N = 3–5. Bars indicate mean ± s.e.m.
The majority of these pathways were related to metabolism of different substrates and were lower in expression in rodent inguinal vs epididymal fat, as well as human subcutaneous vs omental fat (Figure 1C). These included pathways of branched-chained amino metabolism, short-chain fatty acids metabolism (e.g. butanoate metabolism), and pyrimidine metabolism. They also included several disease-defined pathways (e.g. Alzheimer's disease and Huntington's disease), which reflected differences in expression of genes in the mitochondrial electron transport chain. Interestingly, several pathways were annotated as being cardiac-related (e.g. dilated cardiomyopathy and viral myocarditis); these represented differences in genes involved in calcium signaling and regulation of the extracellular matrix. Differences in cell surface markers (e.g. hematopoietic cell lineage) and in glycosphingolipid biosynthesis were up in subcutaneous preadipocytes of both mice and humans. The former has been previously described [48], [49], [50].
3.2. Membrane metallo-endopeptidase marks subcutaneous preadipocytes but not visceral preadipocytes
In order to narrow the list of genes contributing to these differences, we sought to determine the genes common across at least two pathways. A total of 10 significant genes in the inter-depot comparisons were found in overlapping pathways, although only 4 genes (F11R, MME, ITGB8, and ABAT) were in common across statistically significant pathways (Figure 1D). Of these 4 genes, MME was the only gene significantly higher in subcutaneous preadipocytes compared to visceral preadipocytes in both human (Figure 1E) and mouse (Figure 1F), and was found in two significant pathways: Alzheimer's disease and Hematopoietic cell lineage.
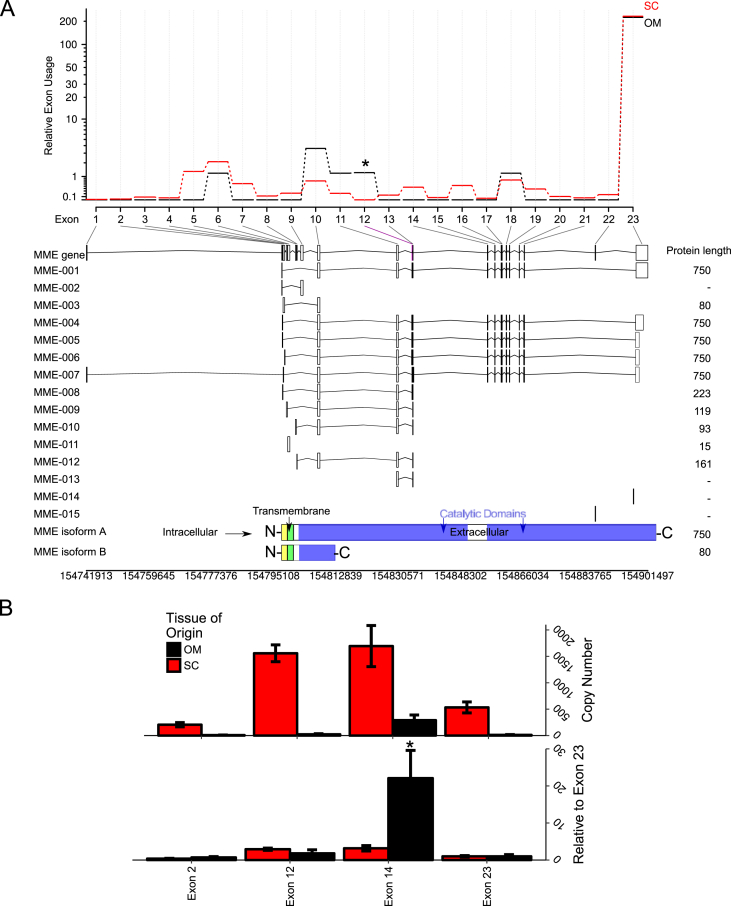
The MME gene encodes a 750 amino acid (AA) protein product with three distinct domains: an N-terminal 28-AA intracellular domain, a 23-AA transmembrane domain, and a 699-AA extracellular domain ending at the C-terminus of the protein [33] (see structure at bottom of Figure 2A). Previous research has reported that MME has tissue-specific alternative splicing in the 5'-UTR for the same 750-AA protein isoform [51], [52]. Additionally, the annotation of the human genome suggested a putative isoform with a shorter 80-AA protein having a truncated extracellular domain (MME isoform B, RefSeq NP_001341573.1) [53], [54]. Several other truncated transcripts from the MME locus have been reported, although not all have been shown to produce a protein product [55], [56].
Figure 2.
RNA-seq reveals differential exon usage in MME between subcutaneous and omental preadipocytes. Exon-level count tables were generated by HTSeq for omental and subcutaneous RNA seq gene expression profiles. Differential exon usage was performed with DEXSeq. (A) The relative exon usage map shows at least one differentially-expressed exon in MME between subcutaneous (red) and omental (black) preadipocytes. The transcript table of the detected exons (i.e. at least 1 unique read mapped to the region) showed at least one exon (exon 12) was comparatively higher in omental preadipocytes than in subcutaneous preadipocytes after normalizing to the average expression of every other exon. (B) Absolute qPCR was performed with exon-specific primer sets and copy number determined by a standard generated with synthetic template (see methods). Exon 14 (ENSE00001019488) was relatively higher in omental preadipocytes rather than exon 12. Asterisk in (A) indicates p ≤ 0.05 by a likelihood ratio test. Asterisks in (B) indicate p ≤ 0.05 by Student's t-test. N = 3–4. Bars indicate mean ± s.e.m.
To determine whether subcutaneous or omental preadipocytes had preferential exon expression in the MME gene, we subjected our RNA seq data to differential exon usage (DEXSeq) [57], [58]. Differential exon splicing showed subcutaneous preadipocytes have less expression of exon 12 (ENSE00000934033) compared to every other exon (exons 1–11 and exons 13–23) within the MME gene (Figure 2A). Exon 12 codes for a 32 amino acid segment in the first catalytic domain (amino acid position 79–483). Interestingly, loss of exon 12 produces an in-frame deletion. Thus the translated product would be a 718 amino acid protein product with an intact catalytic domain. Absolute qPCR using synthetic standards showed the exon-specific regulation of MME between subcutaneous and omental preadipocytes (Figure 2B), although the significant exon was exon 14 (ENSE00000934033). These two exons are found in the same extracellular N-terminal peptidase domain. These results suggest that the depot-specific regulation of exons 12 and 14 may influence the expression of truncated MME protein isoforms.
3.3. MME regulates the inflammatory response of white preadipocytes
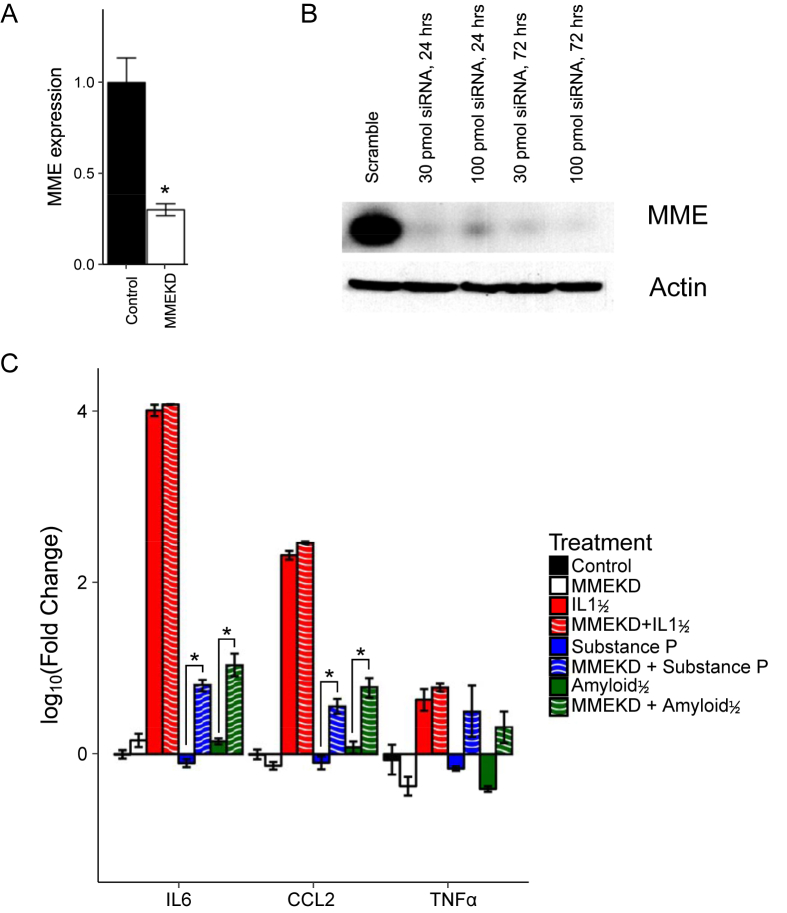
The extracellular region of MME is known to function as a protease for a variety of substrates including several pro-inflammatory cytokines [25], [32]. To determine whether reducing MME expression in subcutaneous preadipocytes would reduce the response to pro-inflammatory cytokines, we transiently knocked down MME in immortalized human neck (HN) white subcutaneous preadipocytes using RNAi. The expression of MME mRNA was reduced by 60% as assessed by qPCR (Figure 3A). This produced a 90% reduction of MME protein expression even at the lowest siRNA amount (30 pmol) as determined by western blotting (Figure 3B). To determine if this affected the cellular response to peptides that MME has been shown to degrade, we exposed the cells to substance P or amyloidβ aggregates. Consistent with a role for MME in response to these stimuli, MME knockdown enhanced the ability of both substance P and amyloidβ aggregates to stimulate IL6 and CCL2 mRNA expression (Figure 3C). TNF-α showed similar trends of a knockdown-mediated increase in mRNA expression in response to these inflammatory mediators, but this was not statistically significant (Figure 3C). Interestingly, MME knockdown cells exposed to IL1β, a pro-inflammatory cytokine postulated (but not proven) to be a target of MME [33], [59], did not have higher expression of CCL2 or IL6 compared to controls. These results suggest that MME in subcutaneous preadipocytes serves to limit the pro-inflammatory response to its degradation targets substance P and amyloidβ.
Figure 3.
Knockdown of MME increases the expression of pro-inflammatory markers IL6 and CCL2 in subcutaneous white preadipocytes. Immortalized human neck white subcutaneous preadipocytes were transiently transfected with siRNA targeting MME and harvested for RNA or protein. (A) qPCR of transfected cells shows a significant reduction of MME mRNA after 72 h post-transfection of 100 pmol siRNA targeting MME. (B) Western blot on protein lysates collected at 24-hrs and 72-hrs after transfection showed efficient knockdown of MME protein by 24 h. (C) Transient transfection with MME siRNA was performed for 24 h followed by serum-starvation for 24 h. Then, treatment with IL1β (5 ng/μL), Substance P (100 nM), or Amyloidβ aggregates (20 μM) was performed for 24 h before RNA extraction and qPCR of IL6, CCL2, and TNFα. Knockdown of MME increases expression of pro-inflammatory markers IL6 and CCL2 in response to substance P and Amyloidβ aggregates. Asterisks indicate p ≤ 0.05 by Student's t-test. N = 3. Bars indicate mean ± s.e.m. qPCR expression calculated by deltadeltaCT to the reference gene: HPRT.
To further explore the role of MME in the inflammatory response, we compared primary human abdominal subcutaneous and omental (visceral) preadipocytes to stimulation of IL1β, substance P, or amyloidβ in the absence or presence of the MME inhibitor Omapatrilat [60]. Omapatrilat alone did not significantly affect the mRNA expression of inflammatory markers CCL2 and IL6 (Supplementary Figure 1). Compared to subcutaneous preadipocytes, the omental preadipocytes showed a trend of increased mRNA expression of CCL2 and IL6 in response to IL1β (Supplementary Figure 1). Substance P did not show any statistically significant changes in CCL2 and IL6 mRNA expression with or without Omapatrilat (Supplementary Figure 1). Interestingly, amyloidβ with Omapatrilat decreased the mRNA expression of CCL2 and IL6 relative to amyloidβ alone in subcutaneous preadipocytes (Supplementary Figure 1). This decrease in inflammatory response may be related to the anti-inflammatory effects Omapatrilat previously shown in vivo [61].
3.4. MME perturbs insulin signaling in white preadipocytes and regulates the expression of the insulin receptor subunits
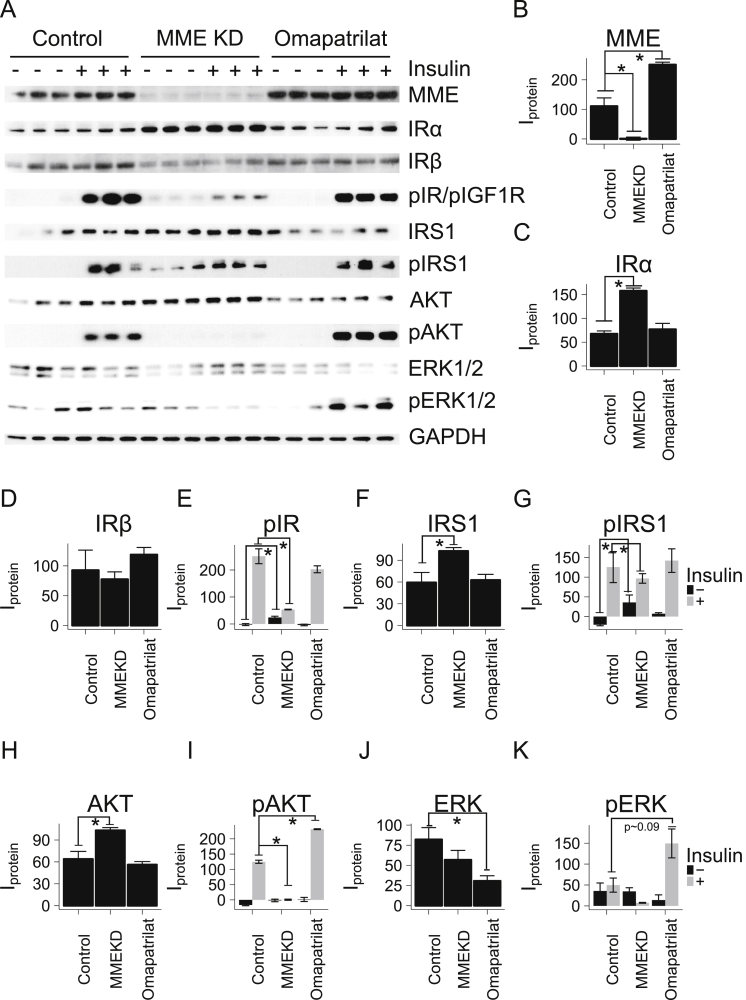
MME is known to have crosstalk with insulin and IGF1 signaling by regulating signaling through PTEN/PI3K/AKT in a Casein Kinase 2 (CSNK2) dependent manner, as demonstrated in in HEK293 cells [34]. However, the effects of MME on the insulin receptor tyrosine kinase have not been studied. To determine the effects on insulin receptor and downstream insulin signaling, immortalized HN subcutaneous white preadipocytes were transduced with MME siRNA for 48-hours before insulin signaling experiments (Figure 4A). Protein levels of MME were reduced by 99% (p < 0.05) in the knockdown cells (Figure 4B). This resulted in a 2-fold increase in the levels of insulin receptor α (IRα) subunit as determined by western blotting (Figure 4C). Interestingly, the level of Insulin receptor β (IRβ) subunit showed a trend to decrease with MME knockdown (Figure 4D). This was paralleled by an even more marked decrease in insulin stimulation of insulin receptor phosphorylation (pIR) at the Tyr1150/1151 site in the receptor kinase domain in the knockdown (Figure 4E). Total IRS1 and AKT protein levels were slightly increased after insulin stimulation, while phosphorylation of IRS1 at Tyr612 and AKT at Ser473 were decreased by 30% and 99% in the MME knockdown (Figure 4F–I). Total ERK and ERK phosphorylation in response to insulin stimulation were unchanged in the knockdown (Figure 4J–K).
Figure 4.
Knockdown of MME perturbs insulin signaling and increases IRα but not IRβ. Immortalized human neck white subcutaneous preadipocytes were transiently transfected with siRNA targeting MME, non-targeting siRNA, or Omapatrilat for 48 h. Cells were starved for 24 h before a 20-minute insulin stimulation (100 nM) followed by protein isolation and western blot. (A) Western blot of knockdown of MME or treatment with Omapatrilat in immortalized subcutaneous preadipocytes. (B–K) Densitometric analysis of protein levels. The levels of unphosphorylated proteins were grouped by treatment (Control, MME KD, or Ompatrilat). The levels of phosphorylated proteins were grouped by treatment and insulin stimulation (black = no insulin, grey = 20 min after 100 nM insulin). Omapatrilat = dual ACE/MME pharmacological inhibitor. Asterisks indicate p ≤ 0.05 by Student's t-test. N = 3 or 6. Bars indicate mean ± s.e.m.
To determine the contribution of the enzymatic activity of MME to these changes in insulin signaling, immortalized HN subcutaneous white preadipocytes were treated with Omapatrilat, an MME enzymatic inhibitor (Supplementary Figure S6), for 48 h before insulin stimulation. Interestingly, Omapatrilat treatment resulted in a ∼2-fold increase in MME protein levels (p < 0.05) (Figure 4B), but there was no change in protein levels of the insulin receptor α- or β-subunits (Figure 4C–D). Similarly, there was no change in the protein levels of the downstream proteins IRS1, AKT, and ERK (Figure 4F,H,J). Likewise, insulin-stimulated phosphorylation the insulin receptor and IRS1 were unchanged (Figure 4E,G). However, phosphorylation of AKT and ERK was slightly increased compared to untreated insulin-stimulated control (Figure 4I,K).
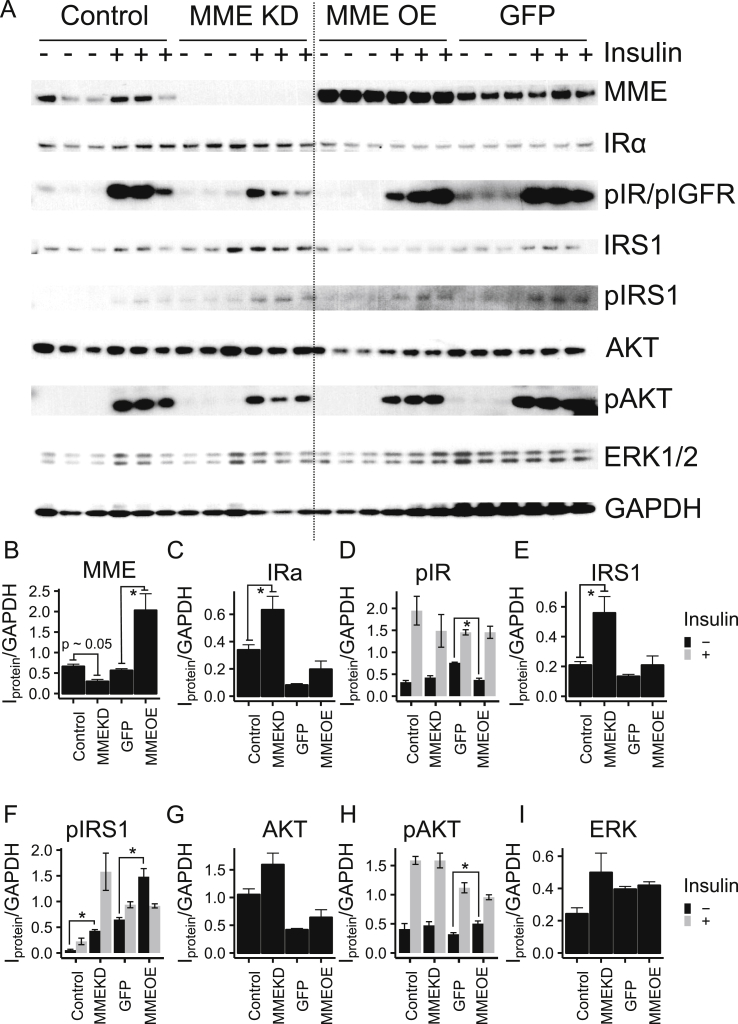
To further explore the role of MME on insulin signaling, immortalized HN subcutaneous white preadipocytes were transiently transduced with either MME (full length 750 amino acid protein) or GFP driven by CMV promoter for 48-hours before insulin signaling (Figure 5A). Knockdown with siRNA was also performed in parallel to confirm reproducibility. Again, knockdown of MME increased the IR α-subunit expression (1.4-fold) but also showed a trend to decrease both pIR and AKT phosphorylation (Figure 5A). Overexpressing MME, on the other hand, did not affect IRα protein levels (Figure 5B–C). This was paralleled by a decrease in basal pIR levels (Figure 5D). Total IRS1, AKT, and ERK levels were unchanged in the overexpression, while the phosphorylation of both IRS1 and AKT increased in the MME overexpression compared to the control GFP overexpression without insulin stimulation (Figure 5E–I). Taken together, these results demonstrate that MME is involved in insulin signaling, and that MME can regulate insulin signaling at the level of the insulin receptor, at least in part, by modulating the levels of IR protein and altering the ratio of α and β subunits.
Figure 5.
Overexpression of MME does not show a reciprocal perturbation on insulin signaling compared to the MME knockdown. Immortalized human neck white subcutaneous preadipocytes were transiently transduced with either MME (full-length 750 amino acid protein) or GFP driven by CMV promoter for 48 h before insulin signaling. Knockdown was performed by a transient transduction of siRNA targeting MME or GFP 48 h before insulin signaling. Cells were starved for 24 h before a 20-minute insulin stimulation followed by protein isolation and western blot. (A) Western bot of knockdown and overexpression of MME showed marked changes in the insulin signaling cascade. (B–I) Densitometric analysis of protein levels. The levels of unphosphorylated proteins were grouped by treatment (Control, MME KD, MME OE, or GFP). The levels of phosphorylated proteins were grouped by treatment and insulin stimulation (black = no insulin, grey = 20 min after 100 μM insulin). Asterisks indicate p ≤ 0.05 by 2-way repeated measures ANOVA followed by a Tukey's test. N = 3 or 6. Bars indicate mean ± s.e.m.
3.5. MMEKO preadipocytes have impaired insulin signaling
MMEKO mice were previously found to have an age-dependent obesity phenotype, with relatively young mice (age < 15 weeks) having a very mild to no phenotype and old mice (age > 36 weeks) having marked obesity under chow diet [28], [30]. In our colony, at 20 weeks, MMEKO mice on chow diet showed no difference in body weight compared to C57BL/6 controls (Supplementary Figure 2A). A glucose tolerance test (GTT) at that time showed no difference from control (Supplementary Figure 2B). However, an insulin tolerance test (ITT) showed a more marked reduction in circulating blood glucose in response to an i.p. injection of insulin in the MMEKO (Supplementary Figure 2C), indicating increased insulin sensitivity in MMEKO mice compared to controls.
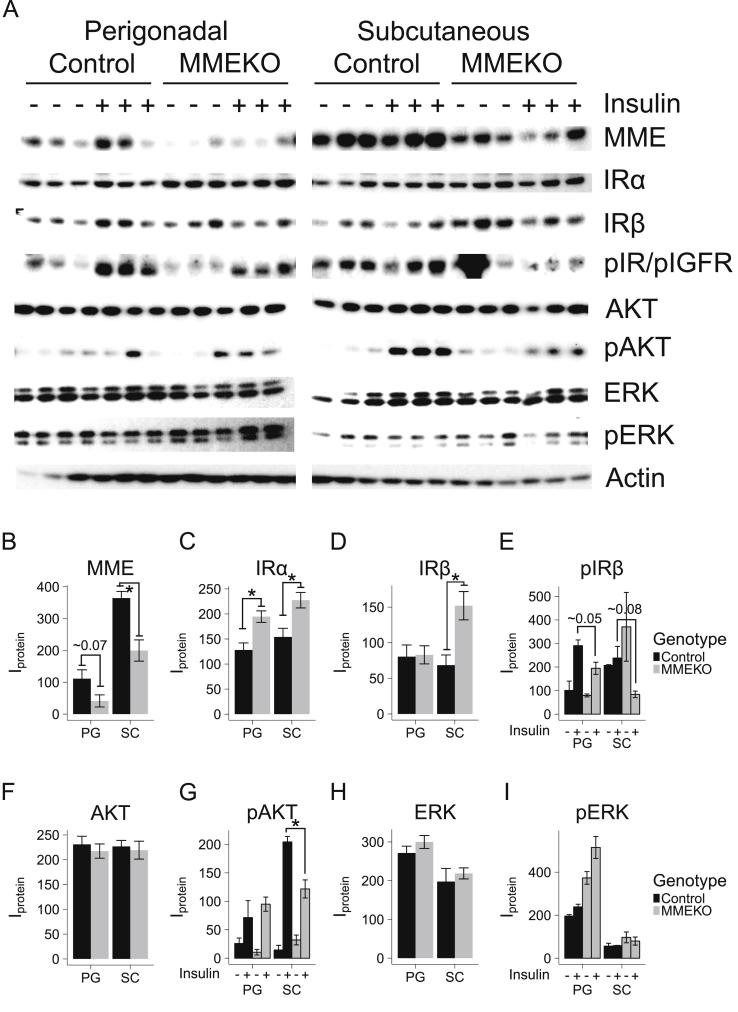
To investigate whether there were any early cellular changes in insulin response, subcutaneous (inguinal) and visceral (epididymal) preadipocytes of 12-week old control and MMEKO mice were subjected to in vitro insulin treatment (Figure 6A). As expected, the levels of MME were markedly lower in the MMEKO mice and epididymal preadipocytes compared to WT and subcutaneous preadipocytes (Figure 6B). Consistent with our in vitro knockdown studies, the levels of IRα were increased in the MMEKO preadipocytes, although in the subcutaneous preadipocytes (but not perigonadal preadipocytes) IRβ levels were also increased (Figure 6C–D). Again, this was also met with a reduction in insulin-stimulated pIR and pAKT in MMEKO preadipocytes, especially in the subcutaneous WAT depot (Figure 6E–G). As before, levels of total ERK and pERK in the MMEKO preadipocytes compared to WT were largely unchanged (Figure 6H–I).
Figure 6.
Subcutaneous and perigonadal MMEKO preadipocytes show impaired insulin signaling. MMEKO and control (wild-type C57BL/6) mice aged 12 weeks were sacrificed for in vitro insulin signaling. The stromal vascular cells (preadipocytes) from the subcutaneous (inguinal) and visceral (perigonadal) adipose depots were isolated. Cells were serum-starved for 4 h before a 20-minute insulin stimulation followed by protein isolation and western blot. (A) Western blot of protein of subcutaneous and perigonadal MMEKO preadipocytes showed impaired response among different insulin-signaling proteins. (B–I) Densitometric analysis of protein levels. The levels of unphosphorylated proteins were grouped by adipose depot (PG = Perigondal, SC = Subcutaneous) and genotype (black = Control, grey = MMEKO). The levels of phosphorylated proteins were grouped by adipose depot, genotype, and insulin stimulation (– = no insulin, + = 20 min after 100 μM insulin). Asterisks indicate p ≤ 0.05 by Student's t-test. N = 3. Bars indicate mean ± s.e.m.
3.6. MME may regulate the internalization and cellular localization of the insulin receptor
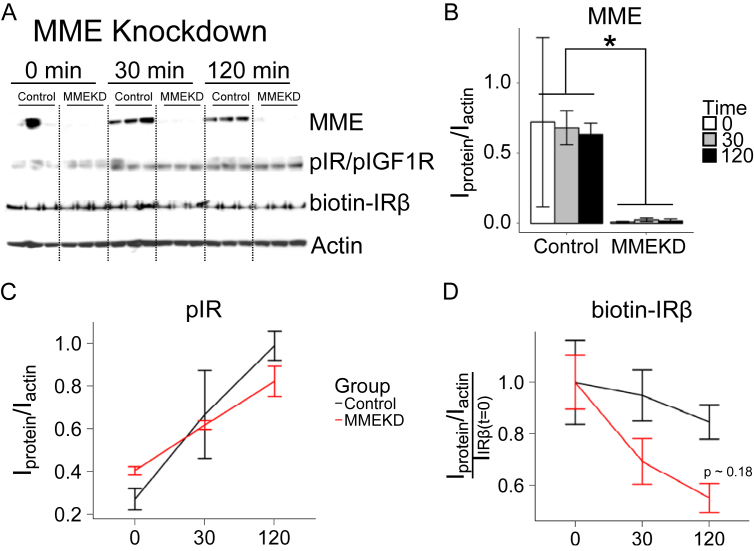
Insulin receptor internalization is one of the major methods to modulate insulin signaling and can lead to either recycling or degradation of the insulin receptor [62]. Thus, we examined the acute internalization of the insulin receptor after insulin stimulation in knockdown or overexpression of MME protein levels. Total surface proteins in immortalized HN subcutaneous white preadipocytes were labeled with biotin after insulin stimulation. Immunoblotting was used to assess the internalization of the insulin receptor at 0, 30, and 120 min post insulin stimulation in control and knockdown cells (Figure 7A). The level of MME protein was decreased by 99% in the knockdown (Figure 7B). Insulin receptor phosphorylation (pIR) increased as a function of time in both the knockdown and control (Figure 7C). As before, the MME knockdown showed a trend of higher basal pIR (0 min) and lower insulin-stimulated pIR (120 min) (Figure 7C). The levels of biotinylated insulin receptor (biotin-IRβ) showed a decreasing trend in the MME knockdown at 30 min and 120 min (Figure 7D). These results suggest that a reduction in MME expression caused an increase in insulin receptor internalization.
Figure 7.
Knockdown of MME may increase the rate of internalization of the insulin receptor. Immortalized human neck white subcutaneous preadipocytes were transiently transduced with either siRNA targeting MME (MMEKD) or non-targeting siRNA (Control) for 48 h before insulin signaling and receptor internalization experiments. Cells were starved for 24 h before a 0, 30-min, or 120-minute insulin stimulation followed by surface-receptor labeling (biotinylation), protein isolation, and Western blot. (A) Western blot of MME, pIR, biotin-IRβ, and actin in the MME knockdown. (B–C) Densitometric analysis of protein levels. Proteins were grouped by treatment (Control = black, MMEKD = red) and time (0, 30, or 120 min after 100 μM insulin stimulation). Asterisks indicate p ≤ 0.05 by 2-way ANOVA followed by a Tukey's test. N = 3. Bars indicate mean ± s.e.m.
To complement the MME knockdown, insulin receptor internalization was assessed after the overexpression of MME or GFP control (Supplementary Figure 3A). The overexpression of MME showed the most robust changes at 0 min and 120 min compared to the GFP control (Supplementary Figure 3B). As in the knockdown, insulin receptor phosphorylation showed a time-dependent increase with no statistically-significant difference in the MME and GFP overexpressing cells at any time points (Supplementary Figure 3C). Interestingly, the levels of biotin-IRβ showed that the MME overexpressing cells had a similar time-dependent pattern as the MME knockdown cells (Supplementary Figure 3D).
To determine whether MME overexpression may affect insulin receptor subcellular localization, we performed subcellular fractionation in immortalized HN subcutaneous white preadipocytes overexpressing GFP, MME, or a catalytically-deficient MME (MMEX). These proteins were overexpressed and the cell lysates fractionated into cytosplasmic, membrane, nuclear, chromatin, and cytoskeletal protein fractions using a kit and differential centrifugation. As expected, cells overexpressing WT MME or MMEX showed the highest expression of MME protein in the membrane fraction, with lesser amounts in the cytoplasmic and nuclear fraction (Supplementary Figure 4). The levels of IR α-subunit were highest in the membrane fraction, although expression could be detected in the cytoplasmic and nuclear fraction as well. The overexpression of either MME or MMEX as compared to GFP overexpression did not significantly affect IRα localization in any subcellular fraction (Supplementary Figure 4). Thus, changing the level of MME does not significantly alter insulin receptor subcellular localization.
3.7. MME and insulin receptor do not co-immunoprecipitate
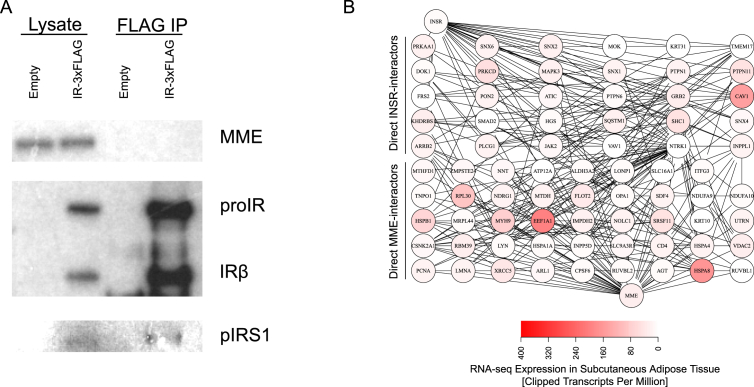
To determine whether MME may directly interact with the insulin receptor, we performed immunoprecipitation of MME and Insulin Receptor (IR) FLAG tagged on the C-terminus of the IRβ subunit transduced into immortalized murine brown IR/IGF1R double knockout preadipocytes [44]. Under normal growth (in media containing 10% fetal bovine serum), MME was not detected in the immunoprecipitate of FLAG-tagged IR, although phosphorylated IRS1 was detected as a positive control (Figure 8A). Similarly, in cells which had been serum-starved followed by 15-min insulin stimulation, MME could not be detected in the immunoprecipitate of the FLAG-tagged IR (Supplementary Figure 5). These results suggest that MME and the insulin receptor do not have a direct protein-protein interaction.
Figure 8.
MME does not form a protein complex with the insulin receptor. Immortalized IR/IGFR double-knockout murine brown preadipocytes were transiently transduced with either pCMV-MME alone or pCMV-MME and pCMV-IR-3XFLAG for 48 h before protein isolation and immunoprecipitation followed by western blot. (A) Western blot of protein lysate and the immunoprecipitation of IR-FLAG in IR/IGFR double-knockout brown preadipocytes in serum-fed media. N = 2 (B) Protein-protein interaction network between MME and INSR. The shortest paths in between MME (bottom node) and INSR (top node) was determined in the PPI as defined in BioGRID. RNA-seq from subcutaneous adipose tissue from ENCODE was plotted on the nodes. One possible path is INSR-CAV1-FLOT2-MME, which are proteins found on endocytic vesicles.
To assess whether MME and the insulin receptor might have any more indirect interaction, we analyzed the human protein-protein interactome (PPI) as defined in the Biological General Repository for Interaction Datasets (BioGRID) [63]. Again, the human PPI showed no direct interaction between MME and the insulin receptor (INSR). The calculated shortest paths showed that there may be at least two intermediate proteins in between MME and INSR (Figure 8B). For example, one possible path (i.e. protein complex) between MME and INSR would be through Caveolin 1 (CAV1) and Flotillin 2 (FLOT2), both of which are involved in endocytosis. Taken together, these results suggest that instead of a direct interaction, MME may affect insulin receptor by a protein complex involving at least two intermediate proteins.
4. Discussion
Adipose tissue shows remarkable regional differences in multiple phenotypic characteristics including inflammatory response, insulin sensitivity, and metabolic behavior. Here, we show that MME is among the most significant differentially expressed genes between subcutaneous and visceral preadipocytes, both in humans and mice. More importantly, we provide novel evidence that MME modulates the inflammatory response and insulin signaling cascade in white preadipocytes. In particular, reducing MME expression increases the expression of pro-inflammatory genes in subcutaneous white preadipocytes. Similarly, we find that perturbations in MME expression differentially regulate the insulin receptor subunits. Thus, knockdown of MME increases the expression of IRα subunit but not IRβ subunit. This is associated with an increase in basal and decrease in insulin-stimulated IR signaling. These results suggest that MME is not only a marker of subcutaneous versus visceral preadipocytes but also a regulator of adipose insulin signaling and inflammatory response.
MME has been shown to have at least three distinct transcripts in humans producing the same 750-bp protein product [64]. The most recent human genome build (GrCh38) has up to 18 different transcripts (coding, non-coding, and predicted) from the MME locus. In rats, these mRNA species have been shown to have tissue-specific expression driven by different promoters resulting in unique 5' untranslated regions [52]. Truncated transcripts of MME have been reported in previous studies [55], [56]. A truncated form of MME missing exon 16 was identified in human lung tissue and reported to have significantly impaired enzymatic activity [56]. There are also differences in glycosylation of MME in different tissues resulting in tissue-specific differences in molecular weight of MME ranging from 85 to 110 kDa [65]. Our data show differential exon-usage in MME between human subcutaneous and omental preadipocytes in the extracellular N-terminal peptidase domain (exons 12 and 14), suggesting distinct tissue-specific regulation of MME isoforms.
In terms of its protease activity, MME has a broad range of substrates being able to target glucagon, bradykinin, GLP1, and several other classes of circulating small molecules [25]. MME has been shown to target free insulin B-chain [32], although whether MME could target and degrade the insulin receptor is unknown. Our data suggest that MME is affecting some aspect of INSR trafficking such that the alpha subunit accumulates, whereas the beta subunit does not. Several factors can contribute to this observation. One is that the alpha subunit has a longer half-life than the beta subunit in the presence of insulin [66]. Another is that the glycosylation state of the receptor affects not only western blot detection but also the processing and degradation of the receptor [67]. Lastly, previous research has shown that glucose deprivation causes aberrant glycosylation of the insulin proreceptor that results in an accumulation of the beta subunit but not the alpha subunit [68].
MME may be part of a larger class of extracellular proteases important in regulating insulin signaling. For example, β-secretase 2 (BACE2), another extracellular protease that shares at least one common substrate with MME (amyloid precursor protein), has been shown to regulate insulin receptor (IRβ) internalization in pancreatic β-cells [69]. Furthermore, mice with functionally-inactive BACE2 show increased β-cell mass and increased circulating insulin levels [70]. Interestingly, the closely related homolog BACE1 does not affect insulin receptor internalization [69]. Thus, the MME-dependent regulation of insulin signaling may share a mechanism similar to BACE2, where proteolytic cleavage of INSR is modulated.
The effects of MME on insulin signaling are potentially responsible for its documented effects on adipogenesis. For example, MME overexpression has been shown to potentiate adipogenesis by activating the AKT-PI3K pathway [35]. Preadipocytes sorted by MME expression predicts adipogenic capacity: preadipocytes with low MME expression have lower lipid accumulation after adipogenesis and vice versa [50]. Furthermore, depot-specific differences in MME expression are present in both the preadipocytes and adipocytes [50]. Mice with knockout of MME do not show defects in adipose tissue development, at least up to 20 weeks of age [30]. However, MMEKO mice on chow diet develop obesity, which is evident around 36 weeks of age [28]. In 20 week-old MMEKO, we observed that a reduction in MME expression increased basal insulin receptor phosphorylation and decreased insulin-stimulated receptor phosphorylation. It is possible that this altered insulin response already at 20 weeks of age contributed to the consequent development of obesity with age.
Elevated circulating levels of soluble MME are associated with increased BMI and HOMA-IR [30]. Also, pharmacological treatment of rats with MME inhibitors has been associated with increased insulin sensitivity [71], consistent with our data in which the MMEKO mice are more insulin sensitive than their littermate controls as assessed by an ITT with a low insulin dose (0.1 mU/gBW). This may be due to increased circulating MME-degraded hormones such as bradykinin [71], [72]. MME has also been shown to be secreted on exosomes from adipose-derived stem cells [36]. Thomou et al. have shown that exosomes released from adipose tissue may regulate gene expression in distal tissues [73]; while this activity has been shown to be related to exosomal miRNAs, this may also be related to exosomal-bound MME. In effect, subcutaneous-adipose-derived circulating MME may have distal regulatory effects on peripheral tissue. Thus, MME may have a distinct cellular role to regulate insulin signaling at the level of the insulin receptor and a somewhat distinct systemic effect to regulate whole-body insulin sensitivity by degrading several small peptide hormones. Determining how to modify each of these hierarchical roles will be crucial in therapeutic interventions for adipose-associated diseases mediated through MME.
In regards to regional variation in metabolic behavior of adipose tissue, subcutaneous adipose tissue transplantation has been shown to reprogram visceral adipose tissue to have subcutaneous-like phenotypic behavior, whereas visceral-to-subcutaneous transplantation does not promote a more detrimental phenotype [7], [74]. MME, as a membrane-bound protease, can modify the local milieu in a substrate-specific manner in an adipose transplantation, whereas the absence of MME would not worsen the depot. Thus, MME may also be involved in adipose tissue reprogramming by degrading the appropriate hormones to promote a metabolically-beneficial phenotype.
Although we have focused on MME as one of the central differences between subcutaneous and visceral preadipocytes, the gene expression in both human and mouse datasets showed several other distinct differences. For example, another differentially expressed gene, 4-aminobutyrate aminotransferase (ABAT), has been shown to be differentially expressed between brown and white adipocytes [75] and may be involved in the maintenance of mitochondria [76], which are much higher in brown than in white adipocytes. Thus, ABAT may also be involved in the metabolic differences within different types of white adipocytes, such as between subcutaneous and visceral adipocytes. Similarly, the remaining differentially expressed genes, Integrin beta 8 (ITGB8) and Junction Adhesion Molecule A (F11R), may play an as-of-yet unidentified role in the phenotypic differences between depot-specific adipocytes.
In summary, we have shown that MME has distinct roles in white preadipocytes. Functionally in a systemic context, MME can target a broad class of inflammatory cytokines, thus its high expression in subcutaneous preadipocytes may serve to decrease inflammatory responses while this protective effect may not occur in visceral preadipocytes due to low expression of MME. More importantly, we have shown that MME can regulate receptor trafficking, suggesting a cellular role in insulin signaling. Thus, MME in preadipocytes and adipose tissue may serve as a potential therapeutic target to modulate insulin sensitivity and inflammatory response in an adipose depot-specific manner.
Author contributions
A.K.R., S.D., S.K., M.S. and W.C.analyzed the results. A.K.R., S.D. S.K. and C.R.K. wrote the paper. A.K.R and S.D. performed the RNA sequencing. A.K.R. and W.C. performed the insulin signaling and receptor internalization. A.K.R and M.S. performed the adipose tissue extraction and murine experiments. A.K.R. assembled the figures.
Acknowledgements
We would like to thank James Kirkland for providing the human preadipocyte stromal vascular isolates; Bao Lu and Craig Gerard for providing breeding pairs of the MMEKO mice. A.K.R. was supported by NIH grant T32 DK007260-37. This work was supported by P30DK036836, R01DK0835659 and R37031036.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.molmet.2019.01.006.
Conflict of interest
The authors declare no competing financial interests.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
Supplementary Figure 1. Pharmacological inhibition of MME does not significantly affect the expression of pro-inflammatory markers CCL2 and IL6 in subcutaneous or visceral preadipocytes. Confluent cultures of primary human abdominal subcutaneous and omental preadipocytes were serum-starved for 24 h. Then, treatment with IL1β (5 ng/μL), Substance P (100 nM), Amyloidβ aggregates (20 μM), or Omapatrilat (1 μM) was performed for 24 h before RNA harvest and qPCR of CCL2 and IL6. Asterisks indicate p ≤ 0.05 by Student's t-test. N = 3. Bars indicate mean ± s.e.m. qPCR expression calculated by deltadeltaCT to the reference gene: HPRT.
Supplementary Figure 2. 20-week old MMEKO mice are more insulin sensitive compared to control mice. (A) MMEKO mice (red), compared to control littermates (black), showed no difference in weight at 20 weeks. (B) A GTT performed on 20-week old mice showed a slight increase in glucose levels at 15 min in MMEKO mice. (C) An ITT performed on the same cohort at 22-weeks old showed a marked reduction in glucose levels in MMEKO mice. Asterisks indicate p ≤ 0.05 by Student's t-test. N = 3. Bars indicate mean ± s.e.m.
Supplementary Figure 3. Overexpression of MME may affect the rate of internalization of the insulin receptor. Immortalized human neck white subcutaneous preadipocytes were transiently transduced with either pCMV-MME (MMEOE) or pCMV-GFP (GFP) for 48 h before insulin signaling and receptor internalization experiments. Cells were starved for 24 h before a 0, 30-min, or 120-minute insulin stimulation followed by surface-receptor labeling (biotinylation), protein isolation, and western blot. (A) Western blot of MME, pIR, biotin-IRβ, and actin in the MME overexpression. (B-D) Densitometric analysis of protein levels. Proteins were grouped by treatment (GFP = black, MMEOE = red) and time (0, 30, or 120 min after 100 μM insulin stimulation). Asterisks indicate p ≤ 0.05 by 2-way ANOVA followed by a Tukey's test. N = 3. Bars indicate mean ± s.e.m.
Supplementary Figure 4. MME overexpression does not affect insulin receptor cellular localization. Immortalized human neck white subcutaneous preadipocytes were transiently transduced with either pCMV-GFP, pCMV-MME, or pCMV-MMEX for 48 h before subcellular protein fractionation and western blot. Wild-type MME and catalytically-deficient MME (MMEX) were overexpressed as expected and primarily localized to the membrane. Overexpression of MME or MMEX did not affect IRα localization. Cyto = Cytoplasm, Mem = Membrane, Nuc = Nuclear, Chrom = Chromatin, CytoS = Cytoskeleton. N = 1.
Supplementary Figure 5. MME does not form a protein complex with the insulin receptor following insulin stimulation. Immortalized IR/IGFR double-knockout murine brown preadipocytes were transiently transduced with either pCMV-MME alone or pCMV-MME and pCMV-IR-3XFLAG for 48 h before insulin signaling, protein isolation, and immunoprecipitation followed by western blot. Western blot of protein lysate and the immunoprecipitation of IR-FLAG in IR/IGFR double-knockout brown preadipocytes after serum-starvation followed by 100 μM insulin stimulation. N = 2.
Supplementary Figure 6. Omapatrilat inhibits MME enzymatic activity. Immortalized human white preadipocytes were treated vehicle (DMSO) or Omapatrilat (1 μM) for 24 h before cell lysis. Cell lysis and MME activity measurements were performed with a MME activity assay kit. N = 3.
References
- 1.Hyvönen M.T., Spalding K.L. Maintenance of white adipose tissue in man. The International Journal of Biochemistry & Cell Biology. 2014;56:123–132. doi: 10.1016/j.biocel.2014.09.013. [DOI] [PubMed] [Google Scholar]
- 2.Tchkonia T., Thomou T., Zhu Y., Karagiannides I., Pothoulakis C., Jensen M.D. Mechanisms and metabolic implications of regional differences among fat depots. Cell Metabolism. 2013;17(5):644–656. doi: 10.1016/j.cmet.2013.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cypess A.M., White A.P., Vernochet C., Schulz T.J., Xue R., Sass C.A. Anatomical localization, gene expression profiling and functional characterization of adult human neck brown fat. Nature Medicine. 2013;19(5):635–639. doi: 10.1038/nm.3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shinoda K., Luijten I.H.N., Hasegawa Y., Hong H., Sonne S.B., Kim M. Genetic and functional characterization of clonally derived adult human brown adipocytes. Nature Medicine. 2015;21(4):389–394. doi: 10.1038/nm.3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tchoukalova Y.D., Votruba S.B., Tchkonia T., Giorgadze N., Kirkland J.L., Jensen M.D. Regional differences in cellular mechanisms of adipose tissue gain with overfeeding. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(42):18226–18231. doi: 10.1073/pnas.1005259107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ibrahim M.M. Subcutaneous and visceral adipose tissue: structural and functional differences. Obesity Reviews an Official Journal of the International Association for the Study of Obesity. 2010;11(1):11–18. doi: 10.1111/j.1467-789X.2009.00623.x. [DOI] [PubMed] [Google Scholar]
- 7.Tran T.T., Kahn C.R. Transplantation of adipose tissue and stem cells: role in metabolism and disease. Nature Reviews Endocrinology. 2010;6(4):195–213. doi: 10.1038/nrendo.2010.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kelley D.E., Thaete F.L., Troost F., Huwe T., Goodpaster B.H. Subdivisions of subcutaneous abdominal adipose tissue and insulin resistance. American Journal of Physiology Endocrinology and Metabolism. 2000;278(5):E941–E948. doi: 10.1152/ajpendo.2000.278.5.E941. [DOI] [PubMed] [Google Scholar]
- 9.Kim J.Y., Van De Wall E., Laplante M., Azzara A., Trujillo M.E., Hofmann S.M. Obesity-associated improvements in metabolic profile through expansion of adipose tissue. Journal of Clinical Investigation. 2007;117(9):2621–2637. doi: 10.1172/JCI31021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karastergiou K., Fried S.K., Xie H., Lee M.J., Divoux A., Rosencrantz M. a. Distinct developmental signatures of human abdominal and gluteal subcutaneous adipose tissue depots. Journal of Clinical Endocrinology & Metabolism. 2013;98(1):362–371. doi: 10.1210/jc.2012-2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cawthorn W.P., Scheller E.L., Learman B.S., Parlee S.D., Simon B.R., Mori H. Bone marrow adipose tissue is an endocrine organ that contributes to increased circulating adiponectin during caloric restriction. Cell Metabolism. 2014;20(2):368–375. doi: 10.1016/j.cmet.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gesta S., Blüher M., Yamamoto Y., Norris A.W., Berndt J., Kralisch S. Evidence for a role of developmental genes in the origin of obesity and body fat distribution. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(17):6676–6681. doi: 10.1073/pnas.0601752103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yamamoto Y., Gesta S., Lee K.Y., Tran T.T., Saadatirad P., Kahn C.R. Adipose depots possess unique developmental gene signatures. Obesity (Silver Spring, Md.) 2010;18(5):872–878. doi: 10.1038/oby.2009.512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sanchez-Gurmaches J., Guertin D. a. Adipocyte lineages: tracing back the origins of fat. Biochimica et Biophysica Acta. 2013 doi: 10.1016/j.bbadis.2013.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sanchez-Gurmaches J., Hung C.-M., Sparks C.A., Tang Y., Li H., Guertin D.A. PTEN loss in the Myf5 lineage redistributes body fat and reveals subsets of white adipocytes that arise from Myf5 precursors. Cell Metabolism. 2012;16(3):348–362. doi: 10.1016/j.cmet.2012.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee K.Y., Sharma R., Gase G., Ussar S., Li Y., Welch L. Tbx15 defines a glycolytic subpopulation and white adipocyte heterogeneity. Diabetes. 2017;66(11):2822–2829. doi: 10.2337/db17-0218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cawthorn W.P., Scheller E.L., Macdougald O.A. Adipose tissue stem cells meet preadipocyte commitment : going back to the future. Journal of Lipid Research. 2012;53:227–246. doi: 10.1194/jlr.R021089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ussar S., Lee K.Y., Dankel S.N., Boucher J., Haering M., Kleinridders A. ASC-1 , PAT2 , and P2RX5 are cell surface markers for white, beige and brown adipocytes. Science Translational Medicine. 2014;6(247) doi: 10.1126/scitranslmed.3008490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schoettl T., Fischer I.P., Ussar S. Heterogeneity of adipose tissue in development and metabolic function. Journal of Experimental Biology. 2018;221(Suppl. 1):jeb162958. doi: 10.1242/jeb.162958. [DOI] [PubMed] [Google Scholar]
- 20.Gesta S., Tseng Y.-H., Kahn C.R. Developmental origin of fat: tracking obesity to its source. Cell. 2007;131(2):242–256. doi: 10.1016/j.cell.2007.10.004. [DOI] [PubMed] [Google Scholar]
- 21.Kerr M.A., Kenny A.J. The purification and specificity of a neutral endopeptidase from rabbit kidney brush border. Biochemical Journal. 1974;137(3):477–488. doi: 10.1042/bj1370477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bland N.D., Pinney J.W., Thomas J.E., Turner A.J., Isaac R.E. Bioinformatic analysis of the neprilysin (M13) family of peptidases reveals complex evolutionary and functional relationships. BMC Evolutionary Biology. 2008;8(1):1–10. doi: 10.1186/1471-2148-8-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson G.D., Stevenson T., Ahn K. Hydrolysis of peptide hormones by endothelin-converting. Biochemistry. 1999;274(7):4053–4058. doi: 10.1074/jbc.274.7.4053. [DOI] [PubMed] [Google Scholar]
- 24.Whyteside A.R., Turner A.J. Human neprilysin-2 (NEP2) and NEP display distinct subcellular localisations and substrate preferences. FEBS Letters. 2008;582(16):2382–2386. doi: 10.1016/j.febslet.2008.05.046. [DOI] [PubMed] [Google Scholar]
- 25.Webster C.I., Burrell M., Olsson L.L., Fowler S.B., Digby S., Sandercock A. Engineering neprilysin activity and specificity to create a novel therapeutic for alzheimer's disease. PLoS One. 2014;9(8) doi: 10.1371/journal.pone.0104001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yasojima K., McGeer E.G., McGeer P.L. Relationship between β amyloid peptide generating molecules and neprilysin in Alzheimer disease and normal brain. Brain Research. 2001;919(1):115–121. doi: 10.1016/s0006-8993(01)03008-6. [DOI] [PubMed] [Google Scholar]
- 27.Lu B.B., Gerard N.P., Kolakowski L.F., Bozza M., Gerard C., Sue I. 1995. Neutral endopeptidase modulation o f septic shock. David Zurakowski, Oretta Finco,* Michael C. Carroll,* 181(June) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Becker M., Siems W.E., Kluge R., Gembardt F., Schultheiss H.P., Schirner M. New function for an old enzyme: NEP deficient mice develop late-onset obesity. PLoS One. 2010;5(9):1–8. doi: 10.1371/journal.pone.0012793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hallier B., Schiemann R., Cordes E., Vitos-faleato J., Heinisch J., Malmendal A. Drosophila neprilysins control insulin signaling and food intake via cleavage of regulatory peptides. ELife. 2016:1–22. doi: 10.7554/eLife.19430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Standeven K.F., Hess K., Carter A.M., Rice G.I., Cordell P.A., Balmforth A.J. Neprilysin, obesity and the metabolic syndrome. International Journal of Obesity. 2011;35(8):1031–1040. doi: 10.1038/ijo.2010.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Higuchi Y., Hashiguchi A., Yuan J., Yoshimura A., Mitsui J., Ishiura H. Mutations in MME cause an autosomal-recessive Charcot-Marie-Tooth disease type 2. Annals of Neurology. 2016:1–14. doi: 10.1002/ana.24612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sexton T., Hitchcook L.J., Rodgers D.W., Bradley L.H., Hersh L.B. Active site mutations change the cleavage specificity of neprilysin. PLoS One. 2012;7(2) doi: 10.1371/journal.pone.0032343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maguer-Satta V., Besançon R., Bachelard-Cascales E. Concise review: neutral endopeptidase (CD10): a multifaceted environment actor in stem cells, physiological mechanisms, and cancer. Stem Cells. 2011;29(3):389–396. doi: 10.1002/stem.592. [DOI] [PubMed] [Google Scholar]
- 34.Siepmann M., Kumar S., Mayer G., Walter J. Casein Kinase 2 dependent phosphorylation of neprilysin regulates receptor tyrosine kinase signaling to Akt. PLoS One. 2010;5(10):1–9. doi: 10.1371/journal.pone.0013134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim J., Han D., Byun S.-H., Kwon M., Cho S.-J., Koh Y.H. Neprilysin facilitates adipogenesis through potentiation of the phosphatidylinositol 3-kinase (PI3K) signaling pathway. Molecular and Cellular Biochemistry. 2017;0(0):0. doi: 10.1007/s11010-017-2948-6. [DOI] [PubMed] [Google Scholar]
- 36.Katsuda T., Tsuchiya R., Kosaka N., Yoshioka Y., Takagaki K., Oki K. Human adipose tissue-derived mesenchymal stem cells secrete functional neprilysin-bound exosomes. Scientific Reports. 2013:1–11. doi: 10.1038/srep01197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tchkonia T., Lenburg M., Thomou T., Giorgadze N., Frampton G., Pirtskhalava T. Identification of depot-specific human fat cell progenitors through distinct expression profiles and developmental gene patterns. American Journal of Physiology Endocrinology and Metabolism. 2007:298–307. doi: 10.1152/ajpendo.00202.2006. [DOI] [PubMed] [Google Scholar]
- 38.Xue R., Lynes M.D., Dreyfuss J.M., Shamsi F., Schulz T.J., Zhang H. Clonal analyses and gene profiling identify genetic biomarkers of the thermogenic potential of human brown and white preadipocytes. Nature Medicine. 2015;21(7):760–768. doi: 10.1038/nm.3881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kriszt R., Arai S., Itoh H., Lee M.H., Goralczyk A.G., Ang X.M. Optical visualisation of thermogenesis in stimulated single-cell brown adipocytes. Scientific Reports. 2017;7(1):1–14. doi: 10.1038/s41598-017-00291-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S. STAR: ultrafast universal RNA-seq aligner. Bioinformatics (Oxford England) 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology. 2014;15(12):1–21. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pau G., Fuchs F., Sklyar O., Boutros M., Huber W. EBImage-an R package for image processing with applications to cellular phenotypes. Bioinformatics. 2010;26(7):979–981. doi: 10.1093/bioinformatics/btq046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wichkam H. 2nd ed. Springer; 2016. ggplot2: elegant graphics for data analysis. [Google Scholar]
- 44.Boucher J., Tseng Y.H., Kahn C.R. Insulin and insulin-like growth factor-1 receptors act as ligand-specific amplitude modulators of a common pathway regulating gene transcription. Journal of Biological Chemistry. 2010;285(22):17235–17245. doi: 10.1074/jbc.M110.118620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lin S., Lin Y., Nery J.R., Urich M.A., Breschi A., Davis C.A. Comparison of the transcriptional landscapes between human and mouse tissues. Proceedings of the National Academy of Sciences. 2014;111(48):17224–17229. doi: 10.1073/pnas.1413624111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Subramanian A., Subramanian A., Tamayo P., Tamayo P., Mootha V.K., Mootha V.K. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Varemo L., Nielsen J., Nookaew I. Enriching the gene set analysis of genome-wide data by incorporating directionality of gene expression and combining. Statistical Hypotheses and Methods. 2013;41(8):4378–4391. doi: 10.1093/nar/gkt111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.MacKenzie S.M., Huda S.S., Sattar N., Fraser R., Connell J.M.C., Davies E. Depot-specific steroidogenic gene transcription in human adipose tissue. Clinical Endocrinology. 2008;69(6):848–854. doi: 10.1111/j.1365-2265.2008.03262.x. [DOI] [PubMed] [Google Scholar]
- 49.Li H., Zimmerlin L., Marra K.G., Donnenberg V.S., Donnenberg A.D., Rubin J.P. Adipogenic potential of adipose stem cell subpopulations. Plastic and Reconstructive Surgery. 2011;128(3):663–672. doi: 10.1097/PRS.0b013e318221db33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ong W.K., Tan C.S., Chan K.L., Goesantoso G.G., Chan X.H.D., Chan E. Identification of specific cell-surface markers of adipose-derived stem cells from subcutaneous and visceral fat depots. Stem Cell Reports. 2014;2(2):171–179. doi: 10.1016/j.stemcr.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.D'Adamio L., Shipp M.A., Masteller E.L., Reinherz E.L. Organization of the gene encoding common acute lymphoblastic leukemia antigen (neutral endopeptidase 24.11): multiple miniexons and separate 5' untranslated regions. Proceedings of the National Academy of Sciences of the United States of America. 1989;86(18):7103–7107. doi: 10.1073/pnas.86.18.7103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li C., Booze R.M., Hersh L.B. Tissue-specific expression of rat neutral endopeptidase (Neprilysin) mRNAs. Journal of Biological Chemistry. 1995:5723–5728. doi: 10.1074/jbc.270.11.5723. [DOI] [PubMed] [Google Scholar]
- 53.Muzny D.M., Scherer S.E., Kaul R., Wang J., Yu J., Sudbrak R. The DNA sequence, annotation and analysis of human chromosome 3. Nature. 2006;440(7088):1194–1198. doi: 10.1038/nature04728. [DOI] [PubMed] [Google Scholar]
- 54.Strausberg R.L., Feingold E.A., Grouse L.H., Derge J.G., Klausner R.D., Collins F.S. Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(26):16899–16903. doi: 10.1073/pnas.242603899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Llorens-Cortes C., Giros B., Schwartz J.C. A novel potential metallopeptidase derived from the enkephalinase gene by alternative splicing. Journal of Neurochemistry. 1990;55(6):2146–2148. doi: 10.1111/j.1471-4159.1990.tb05810.x. [DOI] [PubMed] [Google Scholar]
- 56.Iijima H., Gerard N.P., Squassoni C., Ewig J., Face D., Drazen J.M. Exon 16 del: a novel form of human neutral endopeptidase (CALLA) American Journal of Physiology – Lung Cellular and Molecular Physiology. 1992;262(6 Pt 1):725–729. doi: 10.1152/ajplung.1992.262.6.L725. [DOI] [PubMed] [Google Scholar]
- 57.Anders S., Reyes A., Huber W. Detecting diferential usage of exons from RNA-seq data. Genome Research. 2012;22(10):2008–2017. doi: 10.1101/gr.133744.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W. Limma powers differential expression analyses for RNA sequencing and microarray studies. Nucleic Acids Research. 2015;43(7) doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Van Der Velden V.H.J., Hulsmann A.R. Peptidases: structure, function and modulation of peptide-mediated effects in the human lung. Clinical and Experimental Allergy. 1999;29(4):445–456. doi: 10.1046/j.1365-2222.1999.00462.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Robl J.A., Sun C.-Q., Stevenson J., Ryono D.E., Simpkins L.M., Cimarusti M.P. Dual metalloprotease inhibitors: mercaptoacetyl-based fused heterocyclic dipeptide. Mimetics as Inhibitors of Angiotensin Converting Enzyme and Neutral Endopeptidase. 1997;6(5):1570–1577. doi: 10.1021/jm970041e. [DOI] [PubMed] [Google Scholar]
- 61.Pu Q., Amiri F., Gannon P., Schiffrin E.L. Dual angiotensin-converting enzyme/neutral endopeptidase inhibition on cardiac and renal fibrosis and inflammation in DOCA-salt hypertensive rats. Journal of Hypertension. 2005;23(2):401–409. doi: 10.1097/00004872-200502000-00023. [DOI] [PubMed] [Google Scholar]
- 62.Bergeron J.J.M., Di Guglielmo G.M., Dahan S., Dominguez M., Posner B.I. Spatial and temporal regulation of receptor tyrosine kinase activation and intracellular signal transduction. Annual Review of Biochemistry. 2016:1–25. doi: 10.1146/annurev-biochem-060815-014659. (March) [DOI] [PubMed] [Google Scholar]
- 63.Chatr-Aryamontri A., Oughtred R., Boucher L., Rust J., Chang C., Kolas N.K. The BioGRID interaction database: 2017 update. Nucleic Acids Research. 2017;45(D1):D369–D379. doi: 10.1093/nar/gkw1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shipp M.A., Richardson N.E., Sayre P.H., Brown N.R., Masteller E.L., Clayton L.K. Molecular cloning of the common acute lymphoblastic leukemia antigen (CALLA) identifies a type II integral membrane protein. Proceedings of the National Academy of Sciences of the United States of America. 1988;85(13):4819–4823. doi: 10.1073/pnas.85.13.4819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Relton J.M., Gee N.S., Matsas R., Turner A.J., Kenny A.J. Purification of endopeptidase-24.11 (‘enkephalinase’) from pig brain by immunoadsorbent chromatography. Biochemical Journal. 1983;215(3):519–523. doi: 10.1042/bj2150519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kasuga M., Kahn C.R., Hedo J.A., Van Obberghen E., Yamada K.M. Insulin-induced receptor loss in cultured human lymphocytes is due to accelerated receptor degradation. Proceedings of the National Academy of Sciences of the United States of America. 1981;78(11):6917–6921. doi: 10.1073/pnas.78.11.6917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hwang J.B., Frost S.C. Effect of alternative glycosylation on insulin receptor processing. Journal of Biological Chemistry. 1999;274(32):22813–22820. doi: 10.1074/jbc.274.32.22813. [DOI] [PubMed] [Google Scholar]
- 68.Hwang J.B., Hernandez J., Leduc R., Frost S.C. Alternative glycosylation of the insulin receptor prevents oligomerization and acquisition of insulin-dependent tyrosine kinase activity. Biochimica et Biophysica Acta Molecular Cell Research. 2000;1499(1–2):74–84. doi: 10.1016/s0167-4889(00)00109-9. [DOI] [PubMed] [Google Scholar]
- 69.Casas S., Casini P., Piquer S., Altirriba J., Soty M., Cadavez L. 2010. BACE2 plays a role in the insulin receptor trafficking in pancreatic β -cells; pp. 1087–1095. [DOI] [PubMed] [Google Scholar]
- 70.Esterházy D., Stützer I., Wang H., Rechsteiner M.P., Beauchamp J., Döbeli H. Bace2 is a β cell-enriched protease that regulates pancreatic β cell function and mass. Cell Metabolism. 2011;14(3):365–377. doi: 10.1016/j.cmet.2011.06.018. [DOI] [PubMed] [Google Scholar]
- 71.Arbin V., Claperon N., Fournie-Zaluski M.C., Roques B.P., Peyroux J. Effects of dual angiotensin-converting enzyme and neutral endopeptidase 24-11 chronic inhibition by mixanpril on insulin sensitivity in lean and obese Zucker rats. Journal of Cardiovascular Pharmacology. 2003;41(2):254–264. doi: 10.1097/00005344-200302000-00015. [DOI] [PubMed] [Google Scholar]
- 72.Wang C.H., Leung N., Lapointe N., Szeto L., Uffelman K.D., Giacca A. Vasopeptidase inhibitor omapatrilat induces profound insulin sensitization and increases myocardial glucose uptake in Zucker fatty rats: studies comparing a vasopeptidase inhibitor, angiotensin-converting enzyme inhibitor, and angiotensin II type I recept. Circulation. 2003;107(14):1923–1929. doi: 10.1161/01.CIR.0000062646.09566.CC. [DOI] [PubMed] [Google Scholar]
- 73.Thomou T., Mori M.A., Dreyfuss J.M., Konishi M., Sakaguchi M., Wolfrum C. Adipose-derived circulating miRNAs regulate gene expression in other tissues. Nature. 2017;542(7642):450–455. doi: 10.1038/nature21365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tran T.T., Yamamoto Y., Gesta S., Kahn C.R. Beneficial effects of subcutaneous fat transplantation on metabolism. Cell Metabolism. 2008;7(5):410–420. doi: 10.1016/j.cmet.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ramirez A.K., Lynes M.D., Shamsi F., Xue R., Tseng Y.-H., Kahn C.R. Integrating extracellular flux measurements and genome-scale modeling reveals differences between Brown and white adipocytes. Cell Reports. 2017;21(11):3040–3048. doi: 10.1016/j.celrep.2017.11.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Besse A., Wu P., Bruni F., Donti T., Graham B.H., Craigen W.J. The GABA transaminase, ABAT, is essential for mitochondrial nucleoside metabolism. Cell Metabolism. 2015;21(3):417–427. doi: 10.1016/j.cmet.2015.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 6. Omapatrilat inhibits MME enzymatic activity. Immortalized human white preadipocytes were treated vehicle (DMSO) or Omapatrilat (1 μM) for 24 h before cell lysis. Cell lysis and MME activity measurements were performed with a MME activity assay kit. N = 3.