Abstract
Epidermal growth factor receptor (EGFR) genotyping is critical for treatment guidelines such as the use of tyrosine kinase inhibitors in lung adenocarcinoma. Conventional identification of EGFR genotype requires biopsy and sequence testing which is invasive and may suffer from the difficulty of accessing tissue samples. Here, we propose a deep learning model to predict EGFR mutation status in lung adenocarcinoma using non-invasive computed tomography (CT).
We retrospectively collected data from 844 lung adenocarcinoma patients with pre-operative CT images, EGFR mutation and clinical information from two hospitals. An end-to-end deep learning model was proposed to predict the EGFR mutation status by CT scanning.
By training in 14 926 CT images, the deep learning model achieved encouraging predictive performance in both the primary cohort (n=603; AUC 0.85, 95% CI 0.83–0.88) and the independent validation cohort (n=241; AUC 0.81, 95% CI 0.79–0.83), which showed significant improvement over previous studies using hand-crafted CT features or clinical characteristics (p<0.001). The deep learning score demonstrated significant differences in EGFR-mutant and EGFR-wild type tumours (p<0.001).
Since CT is routinely used in lung cancer diagnosis, the deep learning model provides a non-invasive and easy-to-use method for EGFR mutation status prediction.
Short abstract
Deep learning provides a noninvasive method for EGFR mutation prediction (AUC 0.81) in lung adenocarcinoma, which shows significant improvement over using hand-crafted CT features or clinical characteristics http://ow.ly/LtDJ30nhc5Q
Introduction
Lung adenocarcinoma is a common histological type of lung cancer and the discovery of epidermal growth factor receptor (EGFR) mutations has revolutionised its treatment [1, 2]. In first-line treatment, detecting an EGFR mutation is critical since EGFR tyrosine kinase inhibitors can target specific mutations within the EGFR gene, and have resulted in improved outcomes in EGFR-mutant lung adenocarcinoma patients [3, 4]. Mutational sequencing of biopsies has become the gold standard of EGFR mutation detection. However, biopsy testing for measuring EGFR status probably suffers from having to locate tissue regions because of the extensive heterogeneity of lung tumours [5, 6]. In addition, biopsy testing raises a potential risk of cancer metastasis [7]. Furthermore, repeated tumour sampling, difficulty of accessing tissue samples, poor DNA quality [8] and the relative high costs can limit the applicability of mutational sequencing [9]. In these situations, a non-invasive and easy-to-use method for predicting EGFR mutation status is necessary.
Computed tomography (CT) as a routinely used technique in cancer diagnosis provides a non-invasive way to analyse lung cancer [10–12]. Recent studies revealed that features extracted from lung cancer CT images were related to gene expression patterns [13–16] and showed predictive power on EGFR profiles [17–19]. Although image assessment cannot replace biopsies, image-driven studies can provide additional information that is complementary to biopsies [5, 9]. For example, CT imaging provides a complete scope of a tumour and its microenvironment, enabling us to predict EGFR mutation status by considering intra-tumour heterogeneity. In addition, predicting EGFR-mutation status by CT imaging helps us to choose the most suspicious tumour for biopsy if multiple tumours present in a patient. Furthermore, CT imaging is non-invasive and easy to acquire throughout the course of treatment.
Early findings demonstrated that CT semantic features and quantitative “radiomic” features showed predictive value to EGFR mutation status [9]. However, these methods can only reflect generalised image features which lack specificity to EGFR mutation. In addition, the radiomics methods based on feature engineering rely on precise tumour boundary annotation, which requires human labelling efforts. Since radiomic features are computed only inside the tumour area, the microenvironment and tumour-attached tissues are ignored. In contrast, advanced artificial intelligence models can overcome these problems through a self-learning strategy such as deep learning methods [20–22]. Benefiting from a strong feature-learning ability, deep learning models have shown human expert-level performance in classification of skin cancer [23], diagnosis of eye diseases [24] and prediction of non-invasive liver fibrosis [25]. Moreover, deep learning models present a promising performance in assisting lung cancer analysis [26–29]. Compared with feature engineering-based radiomic methods, deep learning-based radiomics do not require precise tumour boundary annotation and learn features automatically from image data [30]. Furthermore, deep learning-based radiomics can extract features that are adaptive to specific clinical outcomes, while feature engineering-based radiomics can only describe general features that may lack specificity for outcome prediction.
In this study, we proposed a deep learning model to mine CT image information that is related to EGFR mutation status. Our method is an end-to-end pipeline that requires only the manually selected tumour region in a CT image without precise tumour boundary segmentation or human-defined features, which is different to conventional radiomic methods based on feature engineering. The proposed model can learn EGFR mutation-related features from CT images automatically and predicts the probability of the tumour being EGFR-mutant. Furthermore, the deep learning model can discover suspicious tumour subregions that are strongly related to EGFR mutation status, aiming to rapidly facilitate clinicians' treatment decision-making for patients. To evaluate the performance of the deep learning model, we collected a large dataset from two independent hospitals (844 patients) and provided independent validation results of the proposed deep learning model.
Material and methods
Patients
The institutional review board of Tianjin Medical University (Tianjin, China) and Shanghai Pulmonary Hospital (Shanghai, China) approved this retrospective study and waived the need to obtain informed consent from the patients. Patients who meet the following inclusion criteria were collected into this study. 1) Histologically confirmed primary lung adenocarcinoma; 2) pathological examination of tumour specimens carried out with proven records of EGFR mutation status; and 3) pre-operative contrast-enhanced CT data obtained. Patients were excluded if 1) clinical data including age, sex and stage was missing; 2) pre-operative treatment was received; or 3) the duration between CT examination and subsequent surgery exceeded 1 month. Finally, 844 patients from two hospitals were used for this study. We allocated the patients into a primary cohort and an independent validation cohort according to the hospital. The primary cohort included 603 patients from Shanghai Pulmonary Hospital between January 2013 and July 2014. The validation cohort included 241 patients from Tianjin Medical University between January 2013 and February 2014. The primary and validation cohorts were used for developing and validating the deep learning model, respectively. CT scanning parameters and detailed descriptions about the datasets are presented in the supplementary methods.
With regard to molecular profiles, tumour specimens were obtained using surgical resection. EGFR mutations were identified on four tyrosine kinase domains (exons 18–21), which are frequently mutated in lung cancer. The mutation status was determined using an amplification refractory mutation system with a human EGFR gene mutations detection kit (Beijing ACCB Biotech Ltd, Beijing, China). If any exon mutation was detected, the tumour was identified as EGFR-mutant; otherwise, the tumour was identified as EGFR-wild type. In this study, we therefore focused on predicting these binary outcomes (EGFR-mutant and EGFR-wild type) for patients with lung adenocarcinoma.
Development of the deep learning model
Deep learning is a hierarchical neural network that aims at learning the abstract mapping between raw data to the desired label. The computational units in the deep learning model are defined as layers and they are integrated to simulate the analysis process of human brain. The main computational formulas are convolution, pooling, activation and batch normalisation. The terms of the computational process in building the deep learning model are defined in the supplementary methods.
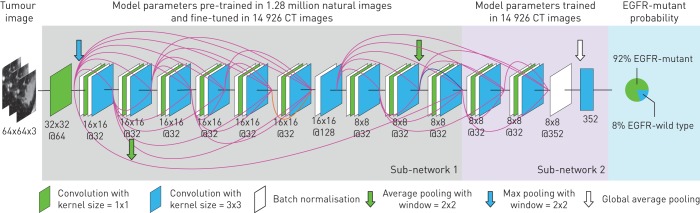
Figure 1 illustrates the pipeline of the EGFR mutation status prediction. For applying the deep learning model, a cubic region of interest (ROI) containing the entire tumour was manually selected (by J. Shi and Y. Liu) according to the following rule: the ROI should include the full tumour region, including the edges of tumours. This rule is easy to use in practice since we do not require the tumour to be precisely in the centre of the ROI (supplementary figure S1 illustrates several ROIs selected by users). Afterwards, the ROI was resized to 64×64 pixels by third-order spline interpolation in each CT slice, and fed into the deep learning model. Through a sequential activation of convolution and pooling layers, the deep learning model gave an EGFR-mutant probability for the image. To make a robust prediction, all the CT slices of the tumour were fed into the deep learning model, and the average probability is treated as the EGFR-mutant probability for the tumour. Specifically, all the adjacent three CT slices were combined as a three-channel image and were fed into the deep learning model for prediction (supplementary figure S2).
FIGURE 1.
Illustration of the deep learning model. This model is composed of convolutional layers with kernel size 3×3 and 1×1, batch normalisation and pooling layers. Sub-network 1 shares the same structure with the first 20 layers in DenseNet [31], which was pre-trained using 1.28 million natural images. Sub-network 2 was trained in the epidermal growth factor receptor (EGFR) mutation dataset, aiming at capturing the association between image features to EGFR mutation labels. When we feed a tumour into the deep learning model, it predicts the probability of the tumour being EGFR-mutant. CT: computed tomography.
During model training, we used transfer learning to train the first 20 convolutional layers (sub-network 1 in figure 1) by 1.28 million natural images from the ImageNet dataset [31]. This transfer learning technique has shown good performance in disease diagnosis since it enlarged the training data [23, 32]. Afterwards, the last four convolutional layers (sub-network 2 in figure 1) were trained using 14 926 CT images from lung adenocarcinoma tumours in the primary cohort. Details about building the model are presented in the supplementary methods.
Given the CT image of tumour, the deep learning model predicts a probability of the tumour being EGFR-mutant directly without any pre- or post-processing or image segmentation. The deep learning model generated using the primary cohort of this study is available at http://radiomics.net.cn/post/110. Part of the CT images from the validation cohort can be downloaded as examples for testing the deep learning model.
Visualisation of the deep learning model
Due to the end-to-end manner of deep learning, the inference process of the deep learning model is not intuitive for users. To further understand the prediction process of the deep learning model, we used visualisation techniques to analyse features learned by the model. The most important component of the deep learning model is the convolutional layer. Therefore, we visualised convolutional layers from two perspectives to understand the inference process of the deep learning model: 1) visualising the feature patterns extracted by convolutional layer; and 2) visualising the response of each convolutional layer to different tumours.
A convolutional layer consists of multiple convolutional filters where each convolutional filter extracts different features. Through a filter-visualising algorithm [33, 34], we can visualise the feature pattern extracted by a convolutional filter, and we define this feature pattern as a deep learning feature (supplementary methods).
To further explore the meaning of the deep learning features, we observed the response of each convolutional filter to different tumours. Given a tumour image, each convolutional filter in the deep learning model generates a response map indicating the corresponding feature patterns in the tumour. The average value of the response map is defined as response value. A good convolutional filter should have different response values between EGFR-mutant and EGFR-wild type tumours. Therefore, visualising the response values for a convolutional filter in different tumour groups can help us evaluate the performance of the convolutional filter.
Statistical analysis
Statistical analysis was performed using SPSS Statistics 21 (IBM, Armonk, NY, USA). The independent-samples t-test was adopted to assess the significance of the mean value on ages between the patients in EGFR-mutant and EGFR-wild type groups. The same statistical analysis was performed to assess the difference of deep learning score between the EGFR-mutant and EGFR-wild type groups. The Chi-squared test was used to evaluate the difference of categorical variables such as sex and tumour stage in all the cohorts. In addition, we used the DeLong test to evaluate the difference of the receiver operating characteristic (ROC) curves between various models. A p-value <0.05 was treated as significant. Our implementation of the deep learning model used the Keras toolkit and Python 2.7 (Python Software Foundation; www.python.org/).
Results
Clinical characteristics of patients
The clinical characteristics of patients are presented in table 1. There was no significant difference between the primary and validation cohorts in terms of age and sex (p=0.083 for age, p=0.321 for sex). The tumour stage showed statistical differences between the two cohorts, probably because of regional differences, since patients in the two cohorts are from two different cities in China. To eliminate this difference, we performed a stratified analysis in the two cohorts to validate the robustness of the deep learning model. Clinical characteristics such as age, sex and stage illustrated difference between EGFR-mutant and EGFR-wild type patients; therefore, these characteristics were used to build a clinical model for comparison to the deep learning model.
TABLE 1.
Clinical characteristics of patients in the primary and validation cohorts
| Primary cohort | p-value | Validation cohort | p-value | |||
| EGFR-wild type | EGFR-mutant | EGFR-wild type | EGFR-mutant | |||
| Subjects n | 603 | 241 | ||||
| Age years | 59.50±9.72 | 61.36±8.96 | 0.016 | 59.59±8.83 | 59.21±7.28 | 0.716 |
| Sex | <0.001 | <0.001 | ||||
| Female | 99 (39.76) | 206 (58.19) | 52 (42.62) | 79 (66.39) | ||
| Male | 150 (60.24) | 148 (41.81) | 70 (57.38) | 40 (33.61) | ||
| Stage | 0.047 | 0.017 | ||||
| I | 181 (72.69) | 240 (67.80) | 50 (40.98) | 65 (54.62) | ||
| II | 27 (10.84) | 27 (7.63) | 22 (18.03) | 8 (6.72) | ||
| III | 36 (14.46) | 69 (19.49) | 43 (35.25) | 35 (29.41) | ||
| IV | 5 (2.01) | 18 (5.08) | 7 (5.74) | 11 (9.24) | ||
| EGFR mutation | 249 (41.29) | 354 (58.71) | 122 (50.62) | 119 (49.38) | ||
Data are presented as mean±sd, or n (%), unless otherwise stated. EGFR: epidermal growth factor receptor.
Diagnostic validation of the deep learning model
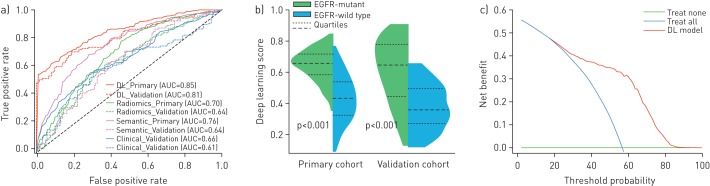
Table 2 lists the predictive performance of the deep learning model where we used area under the ROC curve (AUC), accuracy, sensitivity and specificity as main measurements. In our study, all the results were measured for tumour-level predictions, which are equivalent to reflect subject-level evaluations, since each patient only has one tumour. In the primary cohort, the deep learning model showed good predictive performance by five-fold cross-validation (AUC 0.85, 95% CI 0.83–0.88). This performance was further confirmed in the independent validation cohort (AUC 0.81, 95% CI 0.79–0.83). The close AUC between the primary and validation cohorts indicated that the deep learning model generalised well on predicting EGFR mutation status of unseen new patients. Benefiting from transfer learning with 1.28 million natural images, the deep learning model did not suffer from over-fitting. The ROC curves of the deep learning model in the two cohorts are presented in figure 2a. Moreover, the deep learning score revealed a significant difference between EGFR-mutant and EGFR-wild type groups in the two cohorts (p<0.001 in both the primary and validation cohorts; figure 2b).
TABLE 2.
Predictive performance of various methods in the primary and validation cohorts
| AUC (95% CI) | Accuracy % (95% CI) | Sensitivity % (95% CI) | Specificity % (95% CI) | |
| Clinical model | ||||
| Primary | 0.66 (0.62–0.70) | 61.60 (57.90–65.15) | 64.39 (59.75–68.90) | 56.75 (50.65–62.68) |
| Validation | 0.61 (0.58–0.64) | 61.83 (58.88–64.88) | 56.30 (52.41–60.41) | 67.21 (63.20–71.20) |
| Semantic model | ||||
| Primary | 0.76 (0.72–0.80) | 64.77 (61.31–68.22) | 71.49 (67.86–75.09) | 61.22 (57.45–65.12) |
| Validation | 0.64 (0.61–0.67) | 62.24 (59.94–64.72) | 63.03 (59.61–66.60) | 61.48 (58.22–64.92) |
| Radiomics model | ||||
| Primary | 0.70 (0.66–0.74) | 66.27 (62.96–69.83) | 85.05 (81.81–88.46) | 40.98 (35.82–46.34) |
| Validation | 0.64 (0.61–0.67) | 61.47 (58.69–64.69) | 64.04 (60.34–68.34) | 58.97 (55.10–63.10) |
| DL model | ||||
| Primary | 0.85 (0.83–0.88) | 77.02 (74.02–79.97) | 76.83 (73.17–80.49) | 79.03 (74.26–83.61) |
| Validation | 0.81 (0.79–0.83) | 73.86 (71.82–75.82) | 72.27 (69.27–75.27) | 75.41 (72.32–78.32) |
Data are presented as % (95% CI). All the results in the primary cohort were evaluated by five-fold cross-validation. Bold type represents the best performance. AUC: area under the receiver operating characteristic curve.
FIGURE 2.
Predictive performance of the deep learning model. a) Receiver operating characteristic curves of the deep learning (DL) model, radiomics model, semantic model and clinical model in the primary/validation cohorts. b) DL score between epidermal growth factor receptor (EGFR)-mutant and EGFR-wild type groups in the primary and validation cohorts. c) Decision curve of the DL model. The green line represents the benefit of treating all the patients as EGFR-wild type, and the blue line represents the benefit of treating all the patients as EGFR-mutant. The red line shows the benefit of using the DL model.
In addition, we performed a stratified analysis to validate the diagnostic performance of the deep learning model concerning tumour stage. Supplementary table S1 and supplementary figure S3 indicate that the deep learning model achieved good results in all the tumour stages. Moreover, the deep learning score showed a significant difference between EGFR-mutant and EGFR-wild type groups, regardless of tumour stages.
Figure 2c plots the decision curve of the deep learning model. This curve shows that if the threshold probability of a patient or doctor is >10%, using the deep learning model to predict EGFR mutation status in lung adenocarcinoma adds more benefit than either the treat-all-patients scheme or the treat-none scheme [35]. This highlights the clinical use of the deep learning model.
Comparison between the deep learning model and other methods
In early studies, clinical characteristics, semantic features [17, 36] and quantitative “radiomic” features [9] were used for EGFR mutation status prediction. Therefore, we built a clinical model, a semantic model and a radiomics model as comparison to the proposed deep learning model. The clinical model involved sex, stage and age as features, and used a support vector machine (SVM) with radius-basis kernel for EGFR mutation prediction. The semantic model used 16 semantic features reported in the previous study and a multivariate logistic regression (details in supplementary methods and supplementary table S4) [17]. The radiomics model extracted 1108 features by the PyRadiomics toolkit [37] and selected eight features using recursive feature elimination (RFE). Finally, a random forest containing 100 trees was built for EGFR mutation prediction in the radiomics model.
The quantitative performance in table 2 and the ROC curves in figure 2a indicate that the deep learning model had better performance than the clinical model, with significant difference (AUC 0.66, 95% CI 0.62–0.70 in the primary cohort, p<0.0001; AUC 0.61, 95% CI 0.58–0.64 in the validation cohort, p<0.0001). In addition, a significant improvement over the semantic model was observed in the two cohorts (AUC 0.76, 95% CI 0.72–0.80 in the primary cohort, p<0.0001; AUC 0.64, 95% CI 0.61–0.67 in the validation cohort, p<0.0001). Similar improvement over the radiomics model was confirmed in the two cohorts (AUC 0.70, 95% CI 0.66–0.74 in the primary cohort, p<0.0001; AUC 0.64, 95% CI 0.61–0.67 in the validation cohort, p=0.0002).
Suspicious tumour area discovery
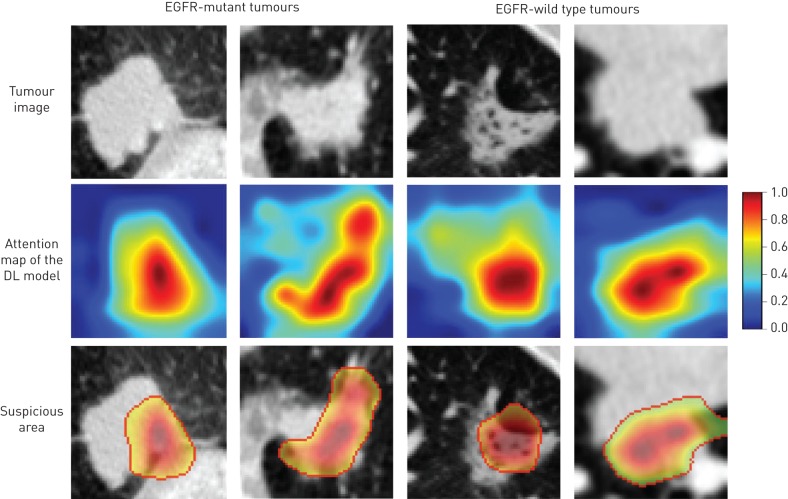
Since deep learning is an end-to-end prediction model that learns abstract mappings between tumour image and EGFR mutation status directly, it is important to explain the predicting process such that we can estimate how reliable the prediction is. We used a deep learning visualisation method [33, 34] to find the tumour region that was most related to EGFR mutation status (supplementary methods). This important region was defined as the suspicious area in our study. When the deep learning model predicts an EGFR mutation status, it tells clinicians which area draws the attention of the model at the same time.
Figure 3 depicts the suspicious areas found by the deep learning model. For a lung adenocarcinoma tumour, the deep learning model generated an attention map indicating the importance of each part in the tumour; we used 0.5 as the cut-off value to reserve the high-response area (suspicious tumour area). These areas were more important than other regions of tumour since they drew the attention of the deep learning model. As shown in the bottom row in figure 3, the suspicious areas found by the deep learning model varied in different tumours. For example, the suspicious area in figure 3a was the tissue between tumour and pleura, whereas the suspicious area in figure 3b was the tumour edge. Based on these observations, the deep learning model interpreted these two tumours as EGFR-mutant. In contrast, the deep learning model focused on the cavitary area in figure 3c and predicted it to be EGFR-wild type. Since the deep learning model required only raw CT image of tumours as input without any tumour segmentation, some normal tissues can be fed into the model. However, the model was capable of finding suspicious areas inside tumours instead of being disturbed by normal tissues. Figure 3d illustrates a tumour adjacent to the mediastinum. In this case, the ROI for the deep learning model included some normal tissues outside the tumour. However, the deep learning model found a suspicious area inside the tumour instead of the normal tissues. The suspicious tumour area was inferred to be strongly related to EGFR mutation status by the deep learning model. Therefore, it can potentially provide a biopsy position for clinicians to avoid false negative diagnoses caused by intra-tumour hetrogeneity. The difference between the suspicious tumour area and other tumour areas may be further explained by combining positron emission tomography–CT data.
FIGURE 3.
Suspicious tumour area discovery. We used 0.5 as cut-off value to acquire the suspicious areas according to the attention map of the deep learning (DL) model. EGFR: epidermal growth factor receptor.
Deep learning feature analysis
The advantage of deep learning mainly comes from its automatic feature-learning ability. By learning from 14 926 tumour images, the deep learning model detects features that are strongly associated with EGFR mutation status.
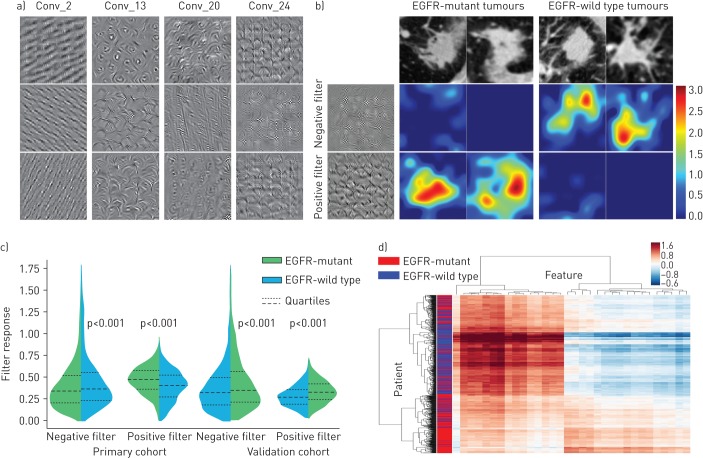
For a better understanding of the deep learning feature, we visualised several convolutional filters in the deep learning model (figure 4a). The shallow convolutional layer learned low-level simple features such as horizontal and diagonal edges (Conv_2). A deeper convolutional layer learned more complex features such as tumour shape. For instance, the filters in layer Conv_13 had strong response to circle or arch shapes, because most tumours contain circular or arch-shaped structures. When going deeper, the features became more abstract and were gradually related to EGFR mutation status (Conv_20, Conv_24). In supplementary figure S4, we compared the convolutional filters before training and after transfer learning (trained in CT data). This figure indicates that the convolutional filters learned various feature patterns that are different with their initial status. Furthermore, transfer learning makes the filters more specific to CT data, especially in deeper network layers.
FIGURE 4.
Deep learning feature analysis. a) Convolutional filters (Conv_) from the 2nd, 13th, 20th and 24th layers of the deep learning model. Each convolutional layer includes hundreds of filters, and only the first three filters are illustrated in each layer. b) Response of the negative filter and the positive filter in epidermal growth factor receptor (EGFR)-mutant/-wild type tumours. The positive filter has strong response to EGFR-mutant tumours and the negative filter has strong response to EGFR-wild type tumours. All the tumour images are from the validation cohort. c) Response value of the positive and the negative filters in the two cohorts. d) Unsupervised clustering of lung adenocarcinoma patients (n=844) on the vertical axis and deep learning feature expression (feature dimension=32, the Conv_24 layer) on the horizontal axis.
To further demonstrate the association between the deep learning features and EGFR mutation status, we extracted two convolutional filters from the last convolutional layer (the positive and negative filters). These two filters captured different texture patterns (the first column in figure 4b) responding to EGFR-mutant and EGFR-wild type tumours. When we fed EGFR-wild type tumours to the deep learning model, the negative filter generated a strong response, while the positive filter was nearly shut down. Similarly, when we fed EGFR-mutant tumours to the deep learning model, the negative filter was depressed, but the positive filter was strongly activated. As depicted in figure 4c, the response of the positive/negative filters on EGFR-mutant and EGFR-wild type tumours were significantly different in all the cohorts (p<0.001). In figure 4d, the clustering map of deep learning features from the last convolutional layer (Conv_24) in the whole dataset (844 patients) is illustrated. The deep learning features showed obvious clusters that had different responses to EGFR-mutant and EGFR-wild type patients. Meanwhile, tumours of different EGFR mutation status (EGFR-mutant/-wild type) can be roughly separated (vertical axis in figure 4d).
To compare the importance of the deep learning features and the radiomic features, we combined the 32 deep learning features from the Conv_24 layer with the 1108 radiomic features, and used RFE to select the important features. In this step, the RFE used linear SVM and five-fold cross-validation to determine the optimal feature amount using the primary cohort, which is consistent with the RFE settings in building the radiomics model. Finally, 11 features were selected, including eight deep learning features and three radiomic features. This indicates that the deep learning features showed stronger association with EGFR mutation status than radiomic features. In addition, we calculated the univariate AUC for all the deep learning features and the radiomic features. As illustrated in supplementary figure S5, many of the deep learning features have higher AUCs than the radiomic features.
Discussion
In this study, we proposed a deep learning model using non-invasive CT images to predict EGFR mutation status for patients with lung adenocarcinoma. We trained the deep learning model in 14 926 CT images from the primary cohort (603 patients), and validated its performance in an independent validation cohort from another hospital (241 patients). The deep learning model showed encouraging results in the primary cohort (AUC 0.85, 95% CI 0.83–0.88) and achieved strong performance in the independent validation cohort (AUC 0.81, 95% CI 0.79–0.83). The deep learning model revealed that there was a significant association between high-dimensional CT image features and EGFR genotype. Our analysis provides an alternative method to non-invasively assess EGFR information for patients, and offers a great supplement to biopsy. Meanwhile, our model can discover the suspicious tumour area that dominates the prediction of EGFR mutation status. This analysis offered visual interpretation to clinicians about understanding the prediction outcomes in CT data. Moreover, the deep learning model requires only the raw tumour image as input and predicts the EGFR mutation status directly without further human assistance, is easy to use and very fast.
Previous studies used clinical factors [8] and radiomics based on feature engineering [9, 17, 18] to predict EGFR mutation status. For example, clinical factors such as age, sex, tumour stage and predominant subtype were used to build a nomogram for EGFR mutation status prediction [8]. In this study, the clinical factors achieved AUC 0.64 in a validation cohort including 464 Asian patients. The clinical model is interpretable, since clinical factors are widely used and the nomogram represents an intuitive linear model. However, clinical features such as stage and predominant subtype require invasive biopsy. In addition, clinical features only reflect few tumour information in pathological level. By contrast, radiomic methods used CT images to quantify tumour information at the macroscopic level, and built the relationship between tumour image and EGFR mutation status. Compared with clinical factors, radiomic analysis provides quantitative features to mine high-dimensional information associated with EGFR genotype. In a cohort including 353 patients, the radiomic method achieved AUC 0.69 by using hand-crafted CT image features [9]. Despite the advantages of the radiomic method, the hand-crafted feature requires time-consuming tumour boundary segmentation and may lack specificity to EGFR genotype. Consequently, we proposed a deep learning method to learn EGFR-related tumour features automatically and avoid complex tumour boundary segmentation. Furthermore, the deep learning method only requires a user-defined ROI of the tumour instead of four complex procedures in radiomics based on feature engineering (tumour boundary segmentation, feature extraction, feature selection and model building).
Advantages of deep learning
Previous studies suggested that CT-based semantic features [18, 19] and quantitative radiomic features [9, 17] reflected EGFR mutation status. However, they can only reflect low-order visual features or simple high-order features. There are abstract features that can probably be associated with EGFR mutation status; however, they are difficult to represent using hand-crafted feature engineering. In these situations, deep learning demonstrates its advantage since it can mine abstract features that are difficult to formulise but are important for identifying EGFR mutation status.
Compared with previously reported hand-crafted features, the deep learning model has the following advantages. 1) Through a hierarchical neural network structure, the deep learning model extracts multi-level features from visual characteristics to abstract mappings that are directly related to EGFR information; 2) the deep learning model does not require time-consuming tumour boundary annotation, which is a big advantage over hand-crafted feature engineering. Moreover, the microenvironment of tumours and the relationship between tumours and attached tissues (pleura traction, etc.) are considered in the deep learning model; 3) the deep learning model is fast and easy to use, requires only the raw CT image as input and predicts the EGFR mutation status directly without further human input.
Clinical utility of the deep learning model
The deep learning model provides potential clinical utility from the following perspectives. 1) The proposed deep learning model provides a non-invasive method to predict EGFR mutation status, which can be used easily in routine CT diagnosis. 2) If the biopsy result of a tumour shows EGFR-wild type, the result may include false negatives because of intra-tumour heterogeneity. At this time, the deep learning model can be seen as an alternative validation tool. If the deep learning model predicts the tumour to be EGFR-mutant, clinicians may need to re-biopsy tissues [38]. 3) The deep learning model only requires routinely used CT images, without adding cost. Therefore, this model can be used multiple times throughout the course of treatment [9]. 4) Most importantly, although we studied only adenocarcinoma, the deep learning model shows predictive value in other histological types. This enables the deep learning model to be used directly in CT scans of lung cancer without identifying histological types. To validate this hypothesis, we additionally collected 125 patients with other lung cancer histological types from Shanghai Pulmonary Hospital between January 2013 and July 2014 (clinical characteristics described in supplementary table S2). Quantitative results in supplementary table S3 indicate that the deep learning model can achieve AUC 0.77 (95% CI 0.73–0.81) in other histological types of lung cancer. Consequently, even without knowing the histological type of a lung cancer, the deep learning model can achieve AUC 0.81 in adenocarcinoma and AUC 0.77 in other histological types.
Despite the encouraging performance of the deep learning model, this study has several limitations. First, we only examined patients in an Asian population. However, EGFR mutation rate can be affected by race. In future work, populations from multiple sources will be necessary to test whether the deep learning model can be generalised to other populations. Second, although the deep learning model shows better performance than clinical, semantic and radiomics models, the combination of these models is unclear. The predictive performance may be improved if we combine these models together. Third, our study only focused on EGFR mutation status. The relationship between EGFR mutation and other genetic mutations (e.g. ROS-1, ALK) can be explored in future work.
Supplementary material
Please note: supplementary material is not edited by the Editorial Office, and is uploaded as it has been supplied by the author.
Supplementary material ERJ-00986-2018_Supplement (567.6KB, pdf)
Footnotes
This article has supplementary material available from erj.ersjournals.com
Author contributions: D. Dong, J. Shi, Y. Liu and Z. Ye collected the clinical dataset. Z. Liu, K. Wang and Y. Zhu processed and analysed the data. H. Zhou provided statistical analysis. S. Wang, D. Yu and M. Zhou built the deep learning model and wrote the article. O. Gevaert and J. Tian conceived the project and edited the article.
Conflict of interest: S. Wang has nothing to disclose.
Conflict of interest: J. Shi has nothing to disclose.
Conflict of interest: Z. Ye has nothing to disclose.
Conflict of interest: D. Dong has nothing to disclose.
Conflict of interest: D. Yu has nothing to disclose.
Conflict of interest: M. Zhou has nothing to disclose.
Conflict of interest: Y. Liu has nothing to disclose.
Conflict of interest: O. Gevaert has nothing to disclose.
Conflict of interest: K. Wang has nothing to disclose.
Conflict of interest: Y. Zhu has nothing to disclose.
Conflict of interest: H. Zhou has nothing to disclose.
Conflict of interest: Z. Liu has nothing to disclose.
Conflict of interest: J. Tian has nothing to disclose.
Support statement: This work was supported by the National Key R&D Programme of China (2017YFA0205200, 2017YFC1308700, 2017YFC1309100, and 2016YFC010380), National Natural Science Foundation of China (81227901, 81771924, 81501616, 61231004, 81671851, 81527805), the Beijing Municipal Science and Technology Commission (Z171100000117023, Z161100002616022), the Beijing Natural Science Foundation (L182061), the Bureau of International Cooperation of Chinese Academy of Sciences (173211KYSB20160053), the Instrument Developing Project of the Chinese Academy of Sciences (YZ201502) and the Youth Innovation Promotion Association CAS (2017175). O. Gevaert was supported by the National Institute of Biomedical Imaging and Bioengineering of the National Institutes of Health under award number R01EB020527. Funding information for this article has been deposited with the Crossref Funder Registry.
References
- 1.Sequist LV, Yang JC, Yamamoto N, et al. Phase III study of afatinib or cisplatin plus pemetrexed in patients with metastatic lung adenocarcinoma with EGFR mutations. J Clin Oncol 2013; 31: 3327–3334. [DOI] [PubMed] [Google Scholar]
- 2.Maemondo M, Inoue A, Kobayashi K, et al. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med 2010; 362: 2380–2388. [DOI] [PubMed] [Google Scholar]
- 3.Li T, Kung H-J, Mack PC, et al. Genotyping and genomic profiling of non-small-cell lung cancer: implications for current and future therapies. J Clin Oncol 2013; 31: 1039–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou C, Wu Y-L, Chen G, et al. Erlotinib versus chemotherapy as first-line treatment for patients with advanced EGFR mutation-positive non-small-cell lung cancer (OPTIMAL, CTONG-0802): a multicentre, open-label, randomised, phase 3 study. Lancet Oncol 2011; 12: 735–742. [DOI] [PubMed] [Google Scholar]
- 5.Itakura H, Achrol AS, Mitchell LA, et al. Magnetic resonance image features identify glioblastoma phenotypic subtypes with distinct molecular pathway activities. Sci Transl Med 2015; 7: 303ra138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sacher AG, Dahlberg SE, Heng J, et al. Association between younger age and targetable genomic alterations and prognosis in non-small-cell lung cancer. JAMA Oncol 2016; 2: 313–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Loughran C, Keeling C. Seeding of tumour cells following breast biopsy: a literature review. Br J Radiol 2011; 84: 869–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Girard N, Sima CS, Jackman DM, et al. Nomogram to predict the presence of EGFR activating mutation in lung adenocarcinoma. Eur Respir J 2012; 39: 366–372. [DOI] [PubMed] [Google Scholar]
- 9.Rios Velazquez E, Parmar C, Liu Y, et al. Somatic mutations drive distinct imaging phenotypes in lung cancer. Cancer Res 2017; 77: 3922–3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lambin P, Leijenaar RT, Deist TM, et al. Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol 2017; 14: 749–762. [DOI] [PubMed] [Google Scholar]
- 11.Gillies RJ, Kinahan PE, Hricak H. Radiomics: images are more than pictures, they are data. Radiology 2016; 278: 563–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kauczor H-U, Heussel CP, von Stackelberg O. Time to take CT screening to the next level? Eur Respir J 2017; 49: 1700064. [DOI] [PubMed] [Google Scholar]
- 13.Aerts HJ, Velazquez ER, Leijenaar RT, et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun 2014; 5: 4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karlo CA, Di Paolo PL, Chaim J, et al. Radiogenomics of clear cell renal cell carcinoma: associations between CT imaging features and mutations. Radiology 2014; 270: 464–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gevaert O, Xu J, Hoang CD, et al. Non-small cell lung cancer: identifying prognostic imaging biomarkers by leveraging public gene expression microarray data – methods and preliminary results. Radiology 2012; 264: 387–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou M, Leung A, Echegaray S, et al. Non-small cell lung cancer radiogenomics map identifies relationships between molecular and imaging phenotypes with prognostic implications. Radiology 2018; 286: 307–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu Y, Kim J, Qu F, et al. CT features associated with epidermal growth factor receptor mutation status in patients with lung adenocarcinoma. Radiology 2016; 280: 271–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yano M, Sasaki H, Kobayashi Y, et al. Epidermal growth factor receptor gene mutation and computed tomographic findings in peripheral pulmonary adenocarcinoma. J Thorac Oncol 2006; 1: 413–416. [PubMed] [Google Scholar]
- 19.Zhou J, Zheng J, Yu Z, et al. Comparative analysis of clinicoradiologic characteristics of lung adenocarcinomas with ALK rearrangements or EGFR mutations. Eur Radiol 2015; 25: 1257–1266. [DOI] [PubMed] [Google Scholar]
- 20.LeCun Y, Bengio Y, Hinton G. Deep learning. Nature 2015; 521: 436–444. [DOI] [PubMed] [Google Scholar]
- 21.Silver D, Schrittwieser J, Simonyan K, et al. Mastering the game of Go without human knowledge. Nature 2017; 550: 354–359. [DOI] [PubMed] [Google Scholar]
- 22.Hazlett HC, Gu H, Munsell BC, et al. Early brain development in infants at high risk for autism spectrum disorder. Nature 2017; 542: 348–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Esteva A, Kuprel B, Novoa RA, et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature 2017; 542: 115–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ting DSW, Cheung CY-L, Lim G, et al. Development and validation of a deep learning system for diabetic retinopathy and related eye diseases using retinal images from multiethnic populations with diabetes. JAMA 2017; 318: 2211–2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang K, Lu X, Zhou H, et al. Deep learning radiomics of shear wave elastography significantly improved diagnostic performance for assessing liver fibrosis in chronic hepatitis B: a prospective multicentre study. Gut 2018: gutjnl-2018–316204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lakhani P, Sundaram B. Deep learning at chest radiography: automated classification of pulmonary tuberculosis by using convolutional neural networks. Radiology 2017; 284: 574–582. [DOI] [PubMed] [Google Scholar]
- 27.Wang S, Zhou M, Liu Z, et al. Central focused convolutional neural networks: developing a data-driven model for lung nodule segmentation. Med Image Anal 2017; 40: 172–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shen W, Zhou M, Yang F, et al. Multi-crop convolutional neural networks for lung nodule malignancy suspiciousness classification. Pattern Recognit 2017; 61: 663–673. [Google Scholar]
- 29.Wang S, Liu Z, Chen X, et al. Unsupervised deep learning features for lung cancer overall survival analysis. In: 2018 40th Annual International Conference of the IEEE Engineering in Medicine and Biology Society, IEEE, 2018; p. 2583–2586. [DOI] [PubMed] [Google Scholar]
- 30.Wang S, Liu Z, Rong Y, et al. Deep learning provides a new computed tomography-based prognostic biomarker for recurrence prediction in high-grade serous ovarian cancer. Radiother Oncol 2018: S0167–8140. [DOI] [PubMed] [Google Scholar]
- 31.Huang G, Liu Z, Weinberger KQ, et al. Densely connected convolutional networks. In: Proceedings of the IEEE conference on computer vision and pattern recognition; 2017, IEEE, 2017; p. 3. [Google Scholar]
- 32.Kermany DS, Goldbaum M, Cai W, et al. Identifying medical diagnoses and treatable diseases by image-based deep learning. Cell 2018; 172: 1122–1131. [DOI] [PubMed] [Google Scholar]
- 33.Selvaraju RR, Cogswell M, Das A, et al. Grad-CAM: visual explanations from deep networks via gradient-based localization. In: 2017 IEEE International Conference on Computer Vision, IEEE, 2017; pp. 618–626. [Google Scholar]
- 34.Kotikalapudi R. keras-vis. https://github.com/raghakot/keras-vis, 2017.
- 35.Huang Y-Q, Liang C-H, He L, et al. Development and validation of a radiomics nomogram for preoperative prediction of lymph node metastasis in colorectal cancer. J Clin Oncol 2016; 34: 2157–2164. [DOI] [PubMed] [Google Scholar]
- 36.Gevaert O, Echegaray S, Khuong A, et al. Predictive radiogenomics modeling of EGFR mutation status in lung cancer. Sci Rep 2017; 7: 41674 10.1038/srep41674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van Griethuysen JJ, Fedorov A, Parmar C, et al. Computational radiomics system to decode the radiographic phenotype. Cancer Res 2017; 77: e104–e107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu Y, Kim J, Balagurunathan Y, et al. Radiomic features are associated with EGFR mutation status in lung adenocarcinomas. Clin Lung Cancer 2016; 17: 441–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Please note: supplementary material is not edited by the Editorial Office, and is uploaded as it has been supplied by the author.
Supplementary material ERJ-00986-2018_Supplement (567.6KB, pdf)