Abstract
Pt nanoparticles play an essential role in a wide variety of catalytic reactions. The activity of the particles strongly depends on their three-dimensional (3D) structure and exposed facets, as well as on the reactive environment. High-resolution electron microscopy has often been used to characterize nanoparticle catalysts but unfortunately most observations so far have been either performed in vacuum and/or using conventional (2D) in situ microscopy. The latter however does not provide direct 3D morphological information. We have implemented a quantitative methodology to measure variations of the 3D atomic structure of nanoparticles under the flow of a selected gas. We were thereby able to quantify refaceting of Pt nanoparticles with atomic resolution during various oxidation–reduction cycles. In a H2 environment, a more faceted surface morphology of the particles was observed with {100} and {111} planes being dominant. On the other hand, in O2 the percentage of {100} and {111} facets decreased and a significant increase of higher order facets was found, resulting in a more rounded morphology. This methodology opens up new opportunities toward in situ characterization of catalytic nanoparticles because for the first time it enables one to directly measure 3D morphology variations at the atomic scale in a specific gaseous reaction environment.
Keywords: in situ STEM, Pt nanoparticles, 3D imaging, catalysis, convolutional neural networks, quantitative ADF STEM
It is widely accepted that the catalytic activity of metal nanoparticles (NPs) is highly sensitive to their three-dimensional (3D) shape and atomic surface structure.1−5 The rational design of nanocatalysts with optimized functional properties therefore strongly depends on the availability of advanced quantitative 3D characterization techniques. Electron tomography currently enables one to analyze the structure and composition of nanostructures in 3D, even with atomic resolution.6−10 Unfortunately, all of these measurements are mostly performed at room temperature and in ultrahigh vacuum, which are conditions that are completely irrelevant for the use of NPs in real applications. Measuring the 3D structure of NPs under static conditions is furthermore not sufficient to understand their structural evolution and connected changes of their properties when being exposed to various reaction environments. Therefore, we decided to study Pt NPs, a model catalyst for numerous gas phase reactions.11−14 Such reactions often occur in the presence of H2 and O2 and it is known that surface energies will usually vary through adsorption of the H2 or O2 molecules, leading to (de)stabilization of certain facets, which in turn affects the catalytic behavior and activity of the NPs.15−18 Morphological changes of metallic NPs during oxidation–reduction cycles have been previously investigated by in situ high-resolution transmission electron microscopy (HRTEM), but these experiments are typically limited by providing only 2D projections of 3D objects.12,15,19−22 For example, Figure S1 shows a 2D HRTEM image acquired from a 10 nm Pt NP. From this image, it is not possible to decide which of the two presented models predicts the correct structure, due to the lack of thickness information along the viewing direction. A quantitative 3D description of the atomic structure and morphology, including the occurrence of surface facets, is therefore far from straightforward based on 2D projection images only. Moreover, experimentally imaging the evolution path of the 3D structure and facet distribution of catalytic NPs in a changing environment is technically very challenging but would become an essential tool toward design and control over the catalyst structure and performance. Although electron tomography experiments can be performed using an environmental TEM,23 the pressures that can be obtained are still too low to be representative of the actual environment during catalytic reactions.24,25 Much higher pressures can be reached using environmental sample holders but the tilting range of these holders is currently too limited for electron tomography. We present here an innovative methodology that allowed us to quantify refaceting of Pt NPs during reduction and oxidation reactions with atomic resolution in 3D.
Pt NPs with average size of 9.7 ± 0.3 nm were obtained by reduction of K2PtCl4 with sodium borohydride in the presence of tetradecyltrimethylammonium bromide, at 50 °C.26 More synthetic details are provided in the Supporting Information. Although Pt particles that are of interest for catalysis are often even smaller, the selectivity and catalytic activity of Pt NPs of this size is exploited in various gas phase reactions.11 High-angle annular dark-field scanning TEM (HAADF-STEM) images of the Pt particles in ultrahigh vacuum and at room temperature are presented in Figure S2. The same Pt NPs were investigated by HAADF-STEM under realistic thermal and gaseous conditions using a gas and heating cell holder.
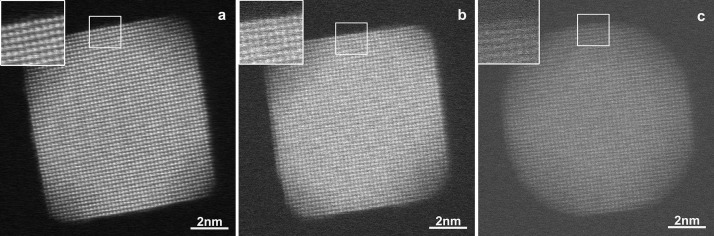
We have previously shown that the number of atoms in an atomic column within a NP can be counted with single atom sensitivity from HAADF-STEM images with a sufficiently large signal-to-noise ratio (SNR).10,27,28 Applying a similar approach for HAADF-STEM images acquired in the gas environment of a dedicated holder is unfortunately far from straightforward because of several technical and more fundamental reasons. One of the main problems is that the NPs are placed in between two SiN windows, each having a thickness of 30 nm. This necessarily lowers the quality of HAADF-STEM images and the effect will become even more pronounced in a gaseous environment. This issue is illustrated in Figure 1, where an image of a Pt NP on a 25 nm thick SiN bottom support is compared to an image of a similar Pt NP in a gas cell in vacuum, as well as in a gas environment.
Figure 1.
HAADF-STEM images of Pt NPs (a) at room temperature and in ultrahigh vacuum on a 25 nm thick SiN support, (b) at room temperature and in ultrahigh vacuum in a gas cell, and (c) at 300 °C and in 5% H2/Ar environment in a gas cell. The quality is drastically reduced when imaging is performed in a gas cell.
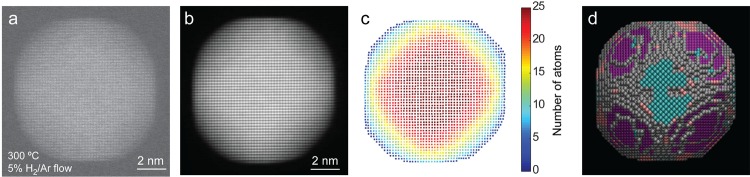
An additional problem commonly observed when imaging NPs by HAADF-STEM is related to particle rotation while scanning, which will cause image distortions and hamper a reliable quantification.29 This effect is demonstrated in Figure S3, which shows HAADF-STEM images from the same Pt NP using increasing acquisition times of 1, 2, 4, and 8 s. It is clear that distortions become more dominant for images acquired with longer acquisition times. To minimize particle rotation and to improve the SNR, we acquired multiple consecutive images at high speed (acquisition of 2k × 2k images with a scan time of 1 s for each frame). As a consequence of the high acquisition speed, effects of kHz–MHz frequency distortions are unfortunately very strong and often highly correlated along the fast-scan direction. An example of a HAADF-STEM image, acquired from a Pt NP at 300 °C and a 1 bar of 5% H2 in Ar environment, is presented in Figure 2a. Frame average procedures based on rigid30 and nonrigid registration have been reported,31−33 but they are not properly designed to correct for high speed distortions. Therefore, we developed and applied a novel methodology based on deep convolutional neural networks (CNNs). This approach has become state-of-the-art because of its ability to learn from data during a training process.34 By training the neural network, it implicitly learns how to detect the presence of distortions and to correct for these, regardless of the level and combination of distortions present in the image that is used as an input. Our network was trained using simulated HAADF-STEM images, in the presence and absence of different types of distortions that can be expected for these images. Next, five HAADF-STEM images individually corrected using the CNN method served as an input for rigid and nonrigid averaging procedures, the result of which is presented in Figure 2b. From these corrected images, it now becomes possible to reliably count the number of atoms in each of the atomic columns.10,27,28 The counting results, displayed in Figure 2c, are used to generate a 3D starting configuration by positioning the atoms in each atomic column parallel to the beam direction and symmetrically around a central plane. By means of molecular dynamics simulations that employ the embedded atom method potential, a relaxed 3D model for the structure of the NP is obtained.35−38Figure 2d presents the final 3D model obtained for the particle in Figure 1a–c. Further technical details on these procedures are presented in Sections 3–6 of the Supporting Information.
Figure 2.
Structural characterization of Pt nanoparticles at 300 °C and under a continuous 5% H2 in Ar flow. (a) HAADF-STEM image (acquired within a frame time of 1 s) of a Pt NP at 300 °C and in a continuous 5% H2 in Ar flow. (b) Averaged and corrected high-resolution HAADF-STEM image (based on five frames) of the same Pt particle. (c) Number of atoms in every atomic column. (d) Final 3D model, the atoms are presented in different colors, according to the type of surface facet: blue = {100}, pink = {110}, purple = {111}, gray = higher index.
To demonstrate the reliability of our methodology, we performed a high resolution electron tomography experiment for one of the Pt NPs in vacuum. First, we acquired a tilt series of HAADF-STEM images over ±70° with a tilt increment of 2°. After applying the approach explained above to correct for distortions (Figure 2b), the resulting images corresponding to different tilt angles, for example, Figure 3a, were aligned with respect to each other. The tilt series was used as an input for the conventional simultaneous iterative reconstruction technique (SIRT) algorithm. The 3D visualizations of the resulting reconstruction are presented in Figure 3b,c. Next, the HAADF-STEM image in Figure 3a was used for atom counting and relaxation with the result presented in Figures 3d,e. The comparison between the 3D reconstructions obtained using conventional high resolution electron tomography and the methodology based on atom counting and relaxation shows very good agreement. This methodology can therefore be considered as a reliable approach to investigate 3D changes of NPs in the presence of a gaseous environment of a gas cell holder which cannot tilt over a tilt range sufficient for conventional high resolution tomography.
Figure 3.
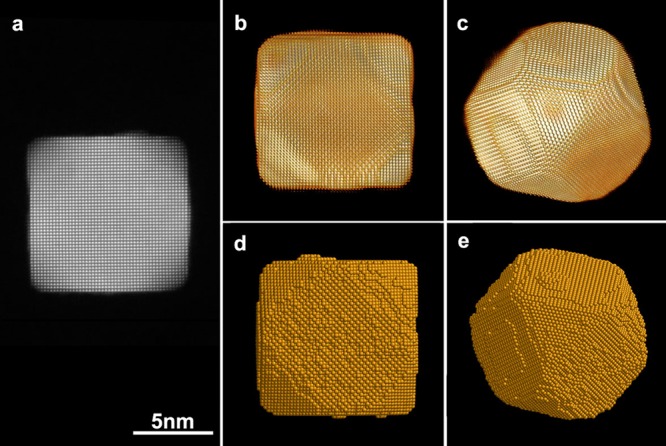
Comparison of the 3D shapes obtained by the reconstruction of a high resolution tomography series and by atom counting, for the same Pt NP. (a) High-resolution HAADF-STEM image of a Pt NP which was used for conventional high-resolution electron tomography and for atom counting and relaxation. (b,c) Three-dimensional visualization of the reconstructed volume obtained by conventional high-resolution tomography, along different viewing directions. (d,e) Three-dimensional models of the same Pt NP obtained using atom counting and relaxation.
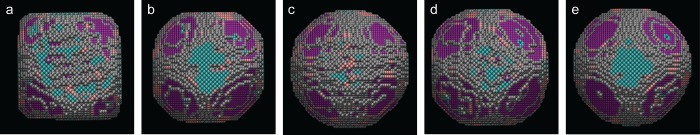
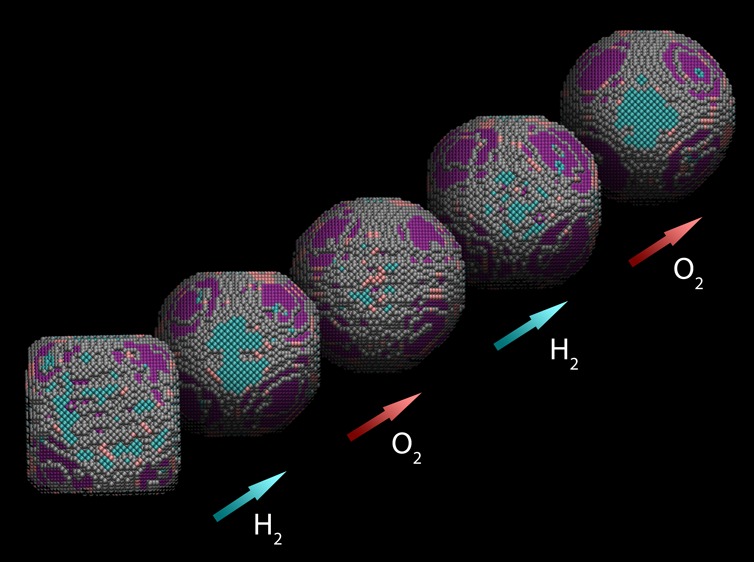
To demonstrate the impact and the possibilities of this methodology, we investigated the morphological stability of the Pt NPs under different environmental conditions. In Figure 4, we show the 3D structures obtained using the approach described above for a particle in vacuum (Figure 4a) and for a similar particle in different gas environments (Figure 4b–e). Figure S2 illustrates the high monodispersity of the Pt NPs. We therefore consider the two selected particles to be representative of the sample. The in situ experiment was performed at 300 °C and the gas flow was varied from vacuum to a 1 bar of 5% H2 in Ar flow (Figure 4b), and subsequently to a 1 bar O2 environment (Figure 4c), corresponding to conditions which are similar to those typically used during reduction and oxidation reactions. To investigate the behavior of the NP during continuous reducing-oxidizing environments, we switched once more the gas flow to 5% H2 in Ar (Figure 4d) and subsequently to O2 (Figure 4e). Two-dimensional HAADF-STEM images, counting results, and additional viewing directions of the obtained 3D structures for a single Pt particle in different gaseous environments are presented in Figure S4. It should be noted that for the different steps of the experiment in gas flow, the average total number of atoms in the structure remained constant and was measured with an error of 0.9%. This demonstrates that there is no removal of atoms. Moreover, shape changes, such as those shown in Figure 4b–e, only occur when changing the gas environment and are not related to knock on surface displacements of atoms due to the electron beam. Indeed, when comparing Figure 4a to Figure 4b–e, it is clear that the gas influences the particle morphology. In vacuum, the particle shape corresponds to a truncated cube, whereas in H2, the shape resembles a truncated octahedron (Figure 4b,d). The morphology is found to become more rounded in an O2 environment (Figure 4c,e). Such information is critical to reveal the behavior of the catalyst particle in a real working environment. It is interesting to note that when heating the particle to 300 °C in vacuum, its morphology does not change significantly (Figure S5). The changes observed in Figure 4 are therefore due to changes in the gaseous environment.
Figure 4.
Structural evolution of Pt NPs under different environmental conditions. Morphology of a Pt NP in vacuum (a) and in different gaseous environments, that is, (b,d) in 5% H2 in Ar; (c,e) in O2, all at 300 °C. The atoms are presented in different colors, according to the type of surface facet: blue = {100}, pink = {110}, purple = {111}, gray = higher index.
It is known that the morphology of Pt nanoparticles can change during catalytic reactions due to the presence of adsorbates.15 Both H2 and O2 can absorb on the surface of the particles and in this manner the free energy is decreased. Through ab initio calculations it was shown that for Pd and Rh nanoparticles higher oxygen pressures tend to reduce the differences in the surface energies between different facets yielding rounder crystals with more atomically rough surfaces.39 Although these calculations were performed for Pd and Rh, it is likely that also Pt nanoparticles follow similar behavior.
In this work, we were able to go beyond a qualitative interpretation of the 3D structure, as we succeeded in quantifying the number of 7-, 8-, and 9-coordinated surface atoms. On the basis of these values, we can extract the occurrence of different types of surface facets, which is illustrated by the color codes in Figure 4. Additionally, Figure 5 shows the occurrence of different surface facets as a function of changes in the gas flow with time. The percentage of the different types of surface facets is determined within an error of less than 1.7%. Approximately 7% of the surface atoms correspond to {110} facets, independent of the environment. The more faceted appearance of the particle in H2 is reflected by the obvious presence of {100} and {111} planes. In O2, we observe a decrease of the percentage of these facets and a significant increase of higher order facets, corresponding to a more rounded morphology. It is both interesting and important to note that the H2–O2–H2 morphological changes are reversible, but a small hysteresis effect is observed for the final change to O2. Indeed, for the morphology presented in Figure 4e, the decrease in the percentage of {100} and {111} facets and the increase of higher order facets is less pronounced in comparison to the previous cycle (Figure 4c). Because this effect will certainly influence the properties of the NPs,12 these quantitative insights are of high relevance toward understanding, for example, catalytic deactivation.
Figure 5.
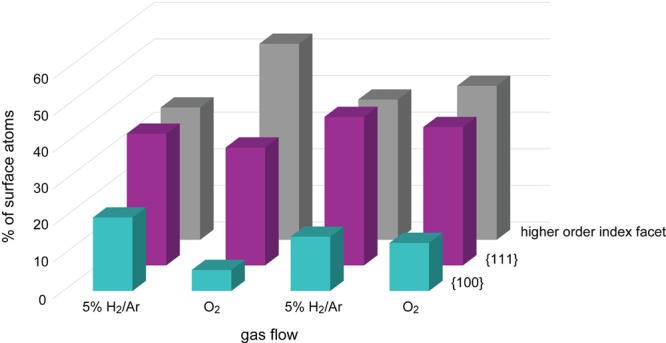
Occurrence of different surface facets as a function of the gas flow in time. Note that a smaller, but constant presence of {110} facets was found for the different gaseous environments (not shown in this graph).
Morphology changes during reduction and oxidation reactions were previously described by in situ TEM. However, given that the reported images invariably correspond to 2D projections of 3D objects, the interpretation of the particle morphology remains challenging. The methodology that we present here enables us to go beyond these limitations, because we are able to directly describe dynamic changes in the 3D atomic structure resulting from different gaseous environments in a quantitative manner. Moreover, it is clear from our experiments that the morphology of the NPs cannot be represented by a simple geometrical model, e.g. such as those in Figure S1. Indeed, the actual 3D structure of the NPs is far more complex and the particles contain a significant percentage of high index surface facets, which is of crucial importance for catalysis.
In conclusion, our methodology enables the direct quantification of real 3D morphological changes at an atomic scale in gaseous environments. The information that we extract from these experiments can be used directly to propose realistic input models for density functional theory calculations, enabling one to unambiguously link the properties of the nanocatalysts in operation to their 3D structure.
Acknowledgments
This work was supported by the European Research Council (Grant 335078 COLOURATOM to S.B. and Grant 770887 PICOMETRICS to S.V.A.). The authors acknowledge funding from the European Commission Grant (EUSMI 731019 to S.B., L.M.L.-M., and Q.X. and MUMMERING 765604 to S.B. and Q.X.). The authors gratefully acknowledge funding from the Research Foundation Flanders (FWO, Belgium) through project fundings (G.0368.15N, G.0369.15N, and G.0267.18N), postdoctoral grants to T.A. and A.D.B, and an FWO [PEGASUS]2 Marie Sklodowska-Curie fellowship to Y.Z. (12U4917N). L.M.L.-M. acknowledges funding from the Spanish Ministerio de Economía y Competitividad (Grant MAT2017-86659-R). We gratefully acknowledge the support of NVIDIA Corporation with the donation of the Titan X Pascal GPU used for this research.
Glossary
ABBREVIATIONS
- 3D
three-dimensional
- NPs
nanoparticles
- HR
high resolution
- HRTEM
HR transmission electron microscopy
- HAADF-STEM
high-angle annular dark-field scanning transmission electron microscopy
- CNNs
convolutional neural networks
- SNR
signal-to-noise ratio
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.nanolett.8b04303.
Synthesis, HRTEM image of Pt NPs, experimental details of the HAADF-STEM observations, HAADF-STEM image restoration method based on a deep convolutional neural network and nonrigid registration, atom-counting, and energy minimization (PDF)
Author Contributions
T.A. and I.L. contributed equally. The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
The authors declare no competing financial interest.
Supplementary Material
References
- Hansen P. L.; Wagner J. B.; Helveg S.; Rostrup-Nielsen J. R.; Clausen B. S.; Topsøe H. Science 2002, 295, 2053–2055. 10.1126/science.1069325. [DOI] [PubMed] [Google Scholar]
- Tao F.; Dag S.; Wang L. W.; Liu Z.; Butcher D. R.; Bluhm H.; Salmeron M.; Somorjai G. A. Science 2010, 327, 850–853. 10.1126/science.1182122. [DOI] [PubMed] [Google Scholar]
- Tian N.; Zhou Z. Y.; Sun S. G.; Ding Y.; Zhong L. W. Science 2007, 316, 732–735. 10.1126/science.1140484. [DOI] [PubMed] [Google Scholar]
- Honkala K.; Hellman A.; Remediakis I. N.; Logadottir A.; Carlsson A.; Dahl S.; Christensen C. H.; Nørskov J. K. Science 2005, 307, 555–558. 10.1126/science.1106435. [DOI] [PubMed] [Google Scholar]
- Barron H.; Barnard A. S. Catal. Sci. Technol. 2015, 5, 2848–2855. 10.1039/C5CY00123D. [DOI] [Google Scholar]
- Goris B.; Bals S.; Van den Broek W.; Carbó-Argibay E.; Gómez-Graña S.; Liz-Marzán L. M.; Van Tendeloo G. Nat. Mater. 2012, 11, 930–935. 10.1038/nmat3462. [DOI] [PubMed] [Google Scholar]
- Bals S.; Casavola M.; Van Huis M. A.; Van Aert S.; Batenburg K. J.; Van Tendeloo G.; Vanmaekelbergh D. Nano Lett. 2011, 11, 3420–3424. 10.1021/nl201826e. [DOI] [PubMed] [Google Scholar]
- Goris B.; De Backer A.; Van Aert S.; Gómez-Graña S.; Liz-Marzán L. M.; Van Tendeloo G.; Bals S. Nano Lett. 2013, 13, 4236–4241. 10.1021/nl401945b. [DOI] [PubMed] [Google Scholar]
- Scott M. C.; Chen C.-C.; Mecklenburg M.; Zhu C.; Xu R.; Ercius P.; Dahmen U.; Regan B. C.; Miao J. Nature 2012, 483, 444–447. 10.1038/nature10934. [DOI] [PubMed] [Google Scholar]
- Van Aert S.; Batenburg K. J.; Rossell M. D.; Erni R.; Van Tendeloo G. Nature 2011, 470, 374–377. 10.1038/nature09741. [DOI] [PubMed] [Google Scholar]
- Grass M. E.; Rioux R. M.; Somorjai G. A. Catal. Lett. 2009, 128, 1–8. 10.1007/s10562-008-9754-4. [DOI] [Google Scholar]
- Vendelbo S. B.; Elkjær C. F.; Falsig H.; Puspitasari I.; Dona P.; Mele L.; Morana B.; Nelissen B. J.; Van Rijn R.; Creemer J. F.; et al. Nat. Mater. 2014, 13, 884–890. 10.1038/nmat4033. [DOI] [PubMed] [Google Scholar]
- El-Nakat J. H.; Dance I. G.; Fisher K. J.; Willett G. D. Polyhedron 1993, 12, 2477–2487. 10.1016/S0277-5387(00)83073-9. [DOI] [Google Scholar]
- Roobol S.; Onderwaater W.; van Spronsen M.; Carlà F.; Balmes O.; Navarro V.; Vendelbo S. B.; Kooyman P. J.; Elkjaer C.; Helveg S.; et al. Phys. Chem. Chem. Phys. 2017, 19, 8485–8495. 10.1039/C6CP08041C. [DOI] [PubMed] [Google Scholar]
- Cabié M.; Giorgio S.; Henry C. R.; Axet M. R.; Philippot K.; Chaudret B. J. Phys. Chem. C 2010, 114, 2160–2163. 10.1021/jp906721g. [DOI] [Google Scholar]
- Shi A.-C.; Masel R. I. J. Catal. 1989, 120, 421–431. 10.1016/0021-9517(89)90282-0. [DOI] [Google Scholar]
- Rupprechter G.; Freund H. Top. Catal. 2000, 14, 3–14. 10.1023/A:1009094613850. [DOI] [Google Scholar]
- Ramachandran A. S.; Anderson S. L.; Datye A. K. Ultramicroscopy 1993, 51, 282–297. 10.1016/0304-3991(93)90154-P. [DOI] [Google Scholar]
- Yoshida H.; Kuwauchi Y.; Jinschek J. R.; Sun K.; Tanaka S.; Kohyama M.; Shimada S.; Haruta M.; Takeda S. Science 2012, 335, 317–319. 10.1126/science.1213194. [DOI] [PubMed] [Google Scholar]
- Giorgio S.; Cabie M.; Henry C. R. Gold Bull. 2008, 41, 167–173. 10.1007/BF03216594. [DOI] [Google Scholar]
- Fujita T.; Guan P.; McKenna K.; Lang X.; Hirata A.; Zhang L.; Tokunaga T.; Arai S.; Yamamoto Y.; Tanaka N.; et al. Nat. Mater. 2012, 11, 775–780. 10.1038/nmat3391. [DOI] [PubMed] [Google Scholar]
- Harris P. J. F. Nature 1986, 323, 792–794. 10.1038/323792a0. [DOI] [Google Scholar]
- Roiban L.; Li S.; Aouine M.; Tuel A.; Farrusseng D.; Epicier T. J. Microsc. 2018, 269, 117–126. 10.1111/jmi.12557. [DOI] [PubMed] [Google Scholar]
- Creemer J. F.; Helveg S.; Hoveling G. H.; Ullmann S.; Molenbroek A. M.; Sarro P. M.; Zandbergen H. W. Ultramicroscopy 2008, 108, 993–998. 10.1016/j.ultramic.2008.04.014. [DOI] [PubMed] [Google Scholar]
- Allard L. F.; Overbury S. H.; Bigelow W. C.; Katz M. B.; Nackashi D. P.; Damiano J. Microsc. Microanal. 2012, 18, 656–666. 10.1017/S1431927612001249. [DOI] [PubMed] [Google Scholar]
- Lee H.; Habas S. E.; Kweskin S.; Butcher D.; Somorjai G. A.; Yang P. Angew. Chem., Int. Ed. 2006, 45, 7824–7828. 10.1002/anie.200603068. [DOI] [PubMed] [Google Scholar]
- Van Aert S.; De Backer A.; Martinez G. T.; Goris B.; Bals S.; Van Tendeloo G.; Rosenauer A. Phys. Rev. B: Condens. Matter Mater. Phys. 2013, 87, 64107. 10.1103/PhysRevB.87.064107. [DOI] [Google Scholar]
- De Backer A.; Martinez G. T.; Rosenauer A.; Van Aert S. Ultramicroscopy 2013, 134, 23–33. 10.1016/j.ultramic.2013.05.003. [DOI] [PubMed] [Google Scholar]
- Li Z. Y.; Young N. P.; Di Vece M.; Palomba S.; Palmer R. E.; Bleloch A. L.; Curley B. C.; Johnston R. L.; Jiang J.; Yuan J. Nature 2008, 451, 46–48. 10.1038/nature06470. [DOI] [PubMed] [Google Scholar]
- Reddy B. S.; Chatterji B. N. IEEE Trans. Image Process. 1996, 5, 1266–1271. 10.1109/83.506761. [DOI] [PubMed] [Google Scholar]
- Jones L.; Yang H.; Pennycook T. J.; Marshall M. S. J.; Van Aert S.; Browning N. D.; Castell M. R.; Nellist P. D. Adv. Struct. Chem. Imaging 2015, 1, 8. 10.1186/s40679-015-0008-4. [DOI] [Google Scholar]
- Wang H.; Dong L.; O’Daniel J.; Mohan R.; Garden A. S.; Kian Ang K.; Kuban D. A.; Bonnen M.; Chang J. Y.; Cheung R. Phys. Med. Biol. 2005, 50, 2887–2905. 10.1088/0031-9155/50/12/011. [DOI] [PubMed] [Google Scholar]
- Cachier P.; Pennec X.; Ayache N.. Technol. Rep. 1999, No. RR-3706, INRIA, 1–25. [Google Scholar]
- Krizhevsky A.; Sutskever I.; Geoffrey E. H. Adv. Neural Inf. Process. Syst. 25 2012, 1, 1097–1105. [Google Scholar]
- Geuchies J. J.; Van Overbeek C.; Evers W. H.; Goris B.; De Backer A.; Gantapara A. P.; Rabouw F. T.; Hilhorst J.; Peters J. L.; Konovalov O.; et al. Nat. Mater. 2016, 15, 1248–1254. 10.1038/nmat4746. [DOI] [PubMed] [Google Scholar]
- De Backer A.; Jones L.; Lobato I.; Altantzis T.; Goris B.; Nellist P. D.; Bals S.; Van Aert S. Nanoscale 2017, 9, 8791–8798. 10.1039/C7NR02656K. [DOI] [PubMed] [Google Scholar]
- Peters J. L.; Van Den Bos K. H. W.; Van Aert S.; Goris B.; Bals S.; Vanmaekelbergh D. Chem. Mater. 2017, 29, 4122–4128. 10.1021/acs.chemmater.7b01103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones L.; Macarthur K. E.; Fauske V. T.; Van Helvoort A. T. J.; Nellist P. D. Nano Lett. 2014, 14, 6336–6341. 10.1021/nl502762m. [DOI] [PubMed] [Google Scholar]
- Mittendorfer F.; Seriani N.; Dubay O.; Kresse G. Phys. Rev. B: Condens. Matter Mater. Phys. 2007, 76, 233413. 10.1103/PhysRevB.76.233413. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.