Abstract
A series of imidazo[1,2-b]pyridazin-8-amine kinase inhibitors were discovered to allosterically inhibit the endoribonuclease function of the dual kinase-endoribonuclease inositol-requiring enzyme 1α (IRE1α), a key component of the unfolded protein response in mammalian cells and a potential drug target in multiple human diseases. Inhibitor optimization gave compounds with high kinome selectivity that prevented endoplasmic reticulum stress-induced IRE1α oligomerization and phosphorylation, and inhibited endoribonuclease activity in human cells. X-ray crystallography showed the inhibitors to bind to a previously unreported and unusually disordered conformation of the IRE1α kinase domain that would be incompatible with back-to-back dimerization of the IRE1α protein and activation of the endoribonuclease function. These findings increase the repertoire of known IRE1α protein conformations and can guide the discovery of highly selective ligands for the IRE1α kinase site that allosterically inhibit the endoribonuclease.
Introduction
The dual kinase-endoribonuclease inositol-requiring enzyme 1α (IRE1α; ERN1) is a central and conserved component of the unfolded protein response (UPR) activated by eukaryotic cells in reaction to endoplasmic reticulum (ER) stress, caused by an excess of misfolded proteins in the ER lumen.1,2 Activation of the UPR reduces protein translation and increases the protein-folding capacity. If this initial response does not alleviate ER stress, prolonged activation of the UPR signaling network leads to apoptosis. Thus, the balance of downstream anti- and proapoptotic signaling pathways activated by the UPR determines cell fate in the presence of acute or chronic proteotoxic stress.
IRE1α is an ER membrane resident protein with serine/threonine kinase and endoribonuclease (RNase) functions contained in its cytosolic domain.3 The accumulation of unfolded or misfolded proteins in the ER lumen triggers oligomerization of IRE1α and kinase autophosphorylation, resulting in activation of the RNase.4−6 The RNase specifically cleaves the messenger RNA (mRNA) coding for the unspliced transcription factor X-box protein 1 (XBP1u) to remove a 26 base pair intron, and subsequent ligation produces the mRNA of the active, spliced form (XBP1s) of the transcription factor.7 XBP1s-dependent transcription includes upregulation of ER-resident molecular chaperones required to relieve aberrant protein folding.8 In contrast, nonspecific cleavage of other mRNAs localized at the ER membrane by IRE1α RNase (termed regulated IRE1-dependent decay) is proposed to exert an apoptotic effect.9 Additionally, the interaction of TRAF2 with oligomerized phospho-IRE1α elicits signaling through the JNK/c-JUN axis, independent of IRE1α RNase activity.10 In mammalian cells, IRE1α functions with the transcriptional repression and regulation of apoptosis effected by the other UPR elements, ATF6 and PERK, to alleviate ER stress or to direct cell death.1,2
Dysregulation of IRE1α-XBP1s signaling is implicated in a number of human diseases, including cancer, diabetes, lipidemia, inflammatory disease, and neurodegeneration.1,11 Signaling through IRE1α-XBP1s has been proposed as important for the survival of myeloma cells, as well as breast and prostate cancers.12−14 Recently, in an ovarian cancer in vivo model, XBP1s function has been shown to suppress proper presentation of tumor-associated antigens by tumor-infiltrating dendritic cells, thus determining the degree of T-cell antitumor immune response and suggesting that modulation of XBP1 splicing in normal tissue may be an alternative approach for an anticancer therapy.15
There is a clear need for selective chemical tools to explore the consequences of IRE1α inhibition by different mechanisms in cancer and nontransformed cells and to study the IRE1α-XBP1s pathway in other human diseases. Given the diverse nature of IRE1α signaling outputs, inhibitors targeting the kinase or RNase functions, or both, may have different consequences for cell fate, which may further vary according to cell context. Reversible covalent binders targeting the RNase active site directly have been reported from a salicylaldehyde scaffold16−18 and have been shown to inhibit cancer cell growth.18−20 The pharmacology of inhibitors binding at the IRE1α kinase site is complicated by the possibility of allosteric inhibition or activation of the RNase function depending on the conformation of the kinase domain and oligomerization state that is stabilized.21 Allosteric inhibition of the RNase function has been associated with type II kinase site ligands that bind the DFG-out conformation of the kinase21 or other compounds that displace the αC-helix of the kinase from the active conformation.22−24 While a series of highly selective, kinase site binding, allosteric inhibitors of IRE1α RNase have shown no direct cytotoxic effect on a range of cancer cell lines,23 compounds with this profile remain of interest for other therapeutic approaches that target nonmalignant cells.25 In contrast to type II and αC-helix displacing scaffolds, reported type I IRE1α kinase site ligands promote dimerization and activate the RNase function.6,21 The identification of new activators and inhibitors of IRE1α RNase and understanding the allosteric mechanisms linking the kinase and RNase sites are therefore of continued importance for the development of new therapeutics.
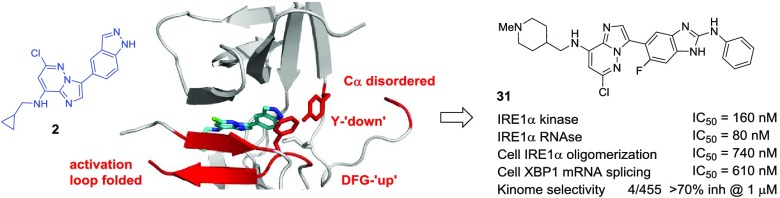
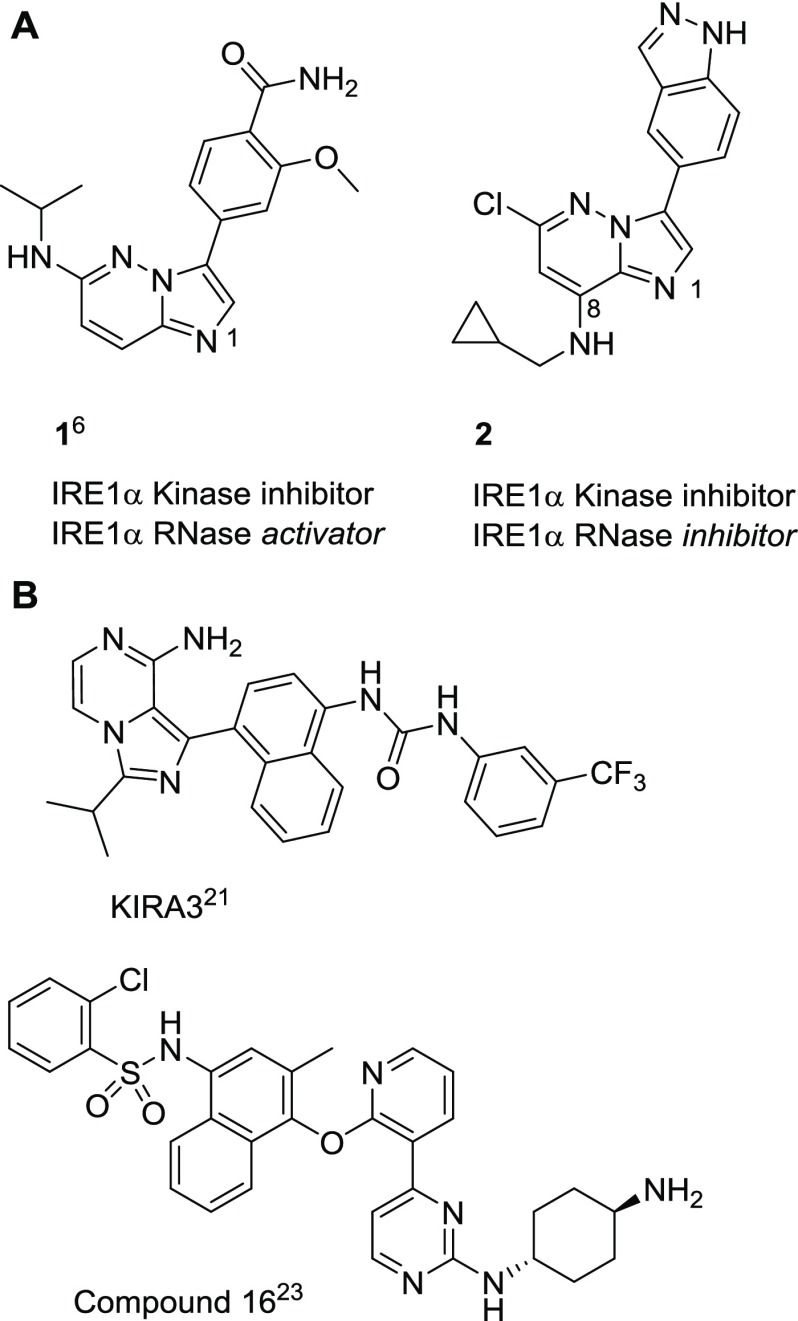
We previously described the type I imidazo[1,2-b]pyridazine kinase inhibitor 1 (Figure 1) that allosterically activates IRE1α endoribonuclease in vitro.6 While screening chemical scaffolds based on 1, we unexpectedly discovered the related type I imidazo[1,2-b]pyridazin-8-amine 2 (Figure 1A) to be an inhibitor of both the kinase and RNase functions of IRE1α. In this study, we show how 2 and more potent analogues bind to a previously undescribed inactive conformation of monomeric IRE1α involving major rearrangements of key secondary structure in the kinase domain, including disordering of the αC-helix and disruption of the hydrophobic spine of the protein, which together confer high IRE1α kinase binding selectivity and inhibition of both kinase and RNase functions. We confirm that these allosteric modulators disrupt all downstream signaling through IRE1α in human cells, thus validating targeting of this unusual binding mode as a new way to generate selective and cell active inhibitors of IRE1α.
Figure 1.
(A) Structures of the IRE1α RNase activator 1 and the structurally related RNase inhibitor 2. (B) Structures of selected published IRE1α kinase-RNase inhibitors.
Synthetic Chemistry
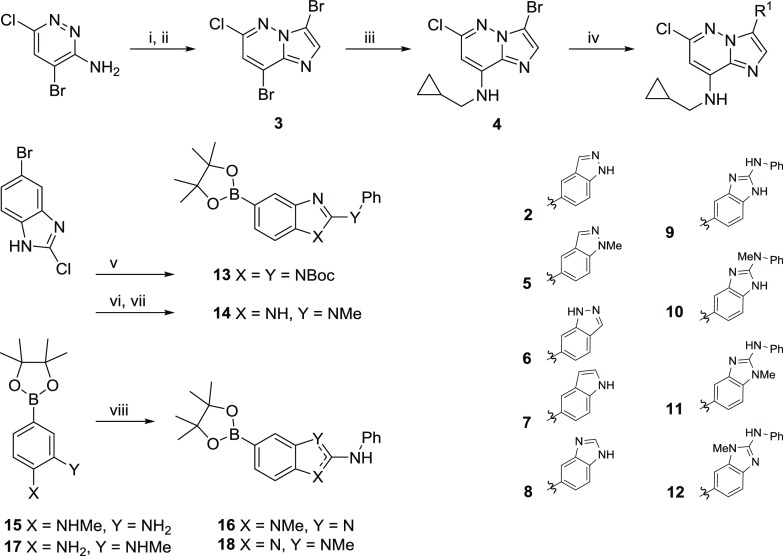
We established short synthetic routes to prepare analogues of 2 using the differential reactivity of the polyhalogenated imidazo[1,2-b]pyridazine 3 (Scheme 1). Selective displacement of the 8-bromo substituent introduced (cyclopropylmethyl)amine and was followed by Suzuki reactions of the 3-bromo group of 4 to give compounds 5–12. The 6-chloro substituent of 3 and 4 might be expected to undergo competing nucleophilic substitution or palladium insertion but proved notably stable under the conditions studied.26 To replace the indazol-5-yl substituent of 2 with 2-(N-phenyl)amino-1H-benzo[d]imidazole, the intermediate 13 was prepared from 5-bromo-2-chloro-1H-benzo[d]imidazole by displacement of the 2-chloro group with aniline and N-protection, before palladium-catalyzed installation of the boronate (Scheme 1). Compound 13 was obtained and used as a mixture of N-Boc 2-aminobenzimidazole regioisomers, with complete N-deprotection observed in the subsequent Suzuki reaction. 2-(Methylamino)benzimidazole 14 did not require N-protection for successful reaction. Other N-methylated 2-aminobenzimidazoles 16 and 18 were synthesized from the nitrophenyl boronates 15 and 17, respectively, by nitro group reduction and condensation with isothiocyanatobenzene.27
Scheme 1. Syntheses of Compounds 2–18.
Reagents and conditions: (i) 2-chloroacetaldehyde, EtOH, 50 °C, 21 h, 78%; (ii) N-bromosuccinimide, CH3Cl, 0 °C to room temperature (rt), 16 h, 84%; (iii) cyclopropylmethanamine, tetrahydrofuran (THF), rt, 3 h, 93%; (iv) heteroaryl boronate, Pd(OAc)2, 1,1′-bis(di-tert-butylphosphino)ferrocene, 2 M Na2CO3, dioxane, 120–135 °C, 15–22 h, 5–51%; (v) (a) PhNH2, 180 °C (microwave), 1 h; (b) (Boc)2O, 4-dimethylaminopyridine (DMAP), THF, rt, 16 h; (c) bis(pinacolato)diboron, KOAc, Pd(dppf)Cl2·CH2Cl2, 1,4-dioxane, 100 °C, 16 h, 53% over three steps; (vi) PhNHMe, 180 °C (microwave), 1 h, 86%; (vii) bis(pinacolato)diboron, KOAc, Pd(dppf)Cl2·CH2Cl2, 1,4-dioxane, 100 °C, 16 h, 36%; (viii) isothiocyanatobenzene, 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide (EDC), THF, 50 °C, 5 h, 17–45%.
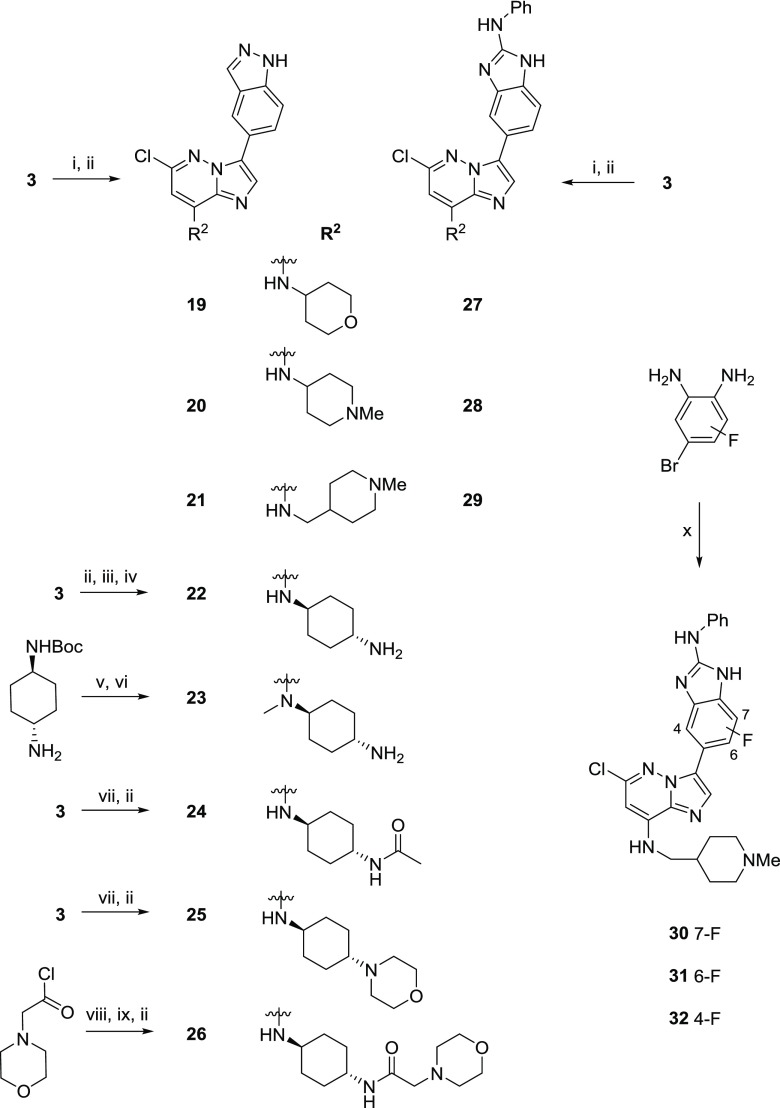
The synthesis was adapted to vary the 8-amino substituent of the imidazo[1,2-b]pyridazine by reacting a range of amines with substrate 3 to give 19–21 and 27–29 (Scheme 2). For the unsubstituted 1,4-diaminocyclohexanes 22 and 23, N-protection of the terminal amine, to ensure selectivity, and higher temperatures were required in the nucleophilic substitution step. The more elaborate 1,4-diaminocyclohexane substituents incorporated into 24–26 were made prior to reaction with 3. Fluorine substituents were introduced into the 2-(N-phenyl)amino-1H-benzo[d]imidazole group starting from the appropriate fluorinated 4-bromobenzene-1,2-diamines via condensation with isothiocyanatobenzene, and the route was telescoped to generate compounds 30–32.
Scheme 2. Syntheses of Compounds 19–32.
Reagents and conditions: (i) amine, THF, rt, 16 h, 60–97%; (ii) heteroaryl boronate, Pd(OAc)2, 1,1′-bis(di-tert-butylphosphino)ferrocene, 2 M Na2CO3, 1,4-dioxane, 130 °C, 16 h, 4–55%; (iii) tert-butyl ((1r,4r)-4-aminocyclohexyl)carbamate, THF, 50 °C, 16 h, quantitative; (iv) HCl, 1,4-dioxane, rt, 2 h, 99%; (v) (a) LiAlH4, THF, 0 °C to reflux, 4 h; (b) ethyl 1,3-dioxoisoindoline-2-carboxylate, Et3N, CH2Cl2, rt, 15 h; (c) 3, Et3N, 1,4-dioxane–dimethyl sulfoxide (DMSO), 100 °C, 22 h, 52% over three steps; (vi) 5-(4,4,5,5-tetramethyl-1,3,2-dioxaborolan-2-yl)-1H-indazole, Pd(OAc)2, 1,1′-bis(di-tert-butylphosphino)-ferrocene, 2 M Na2CO3, 1,4-dioxane, 130 °C, 21 h, then NH2NH2·H2O, 100 °C, 48 h, 34%; (vii) amine, THF, 50 °C, 16 h, 29–85%; (viii) (a) tert-butyl ((1r,4r)-4-aminocyclohexyl)carbamate, Et3N, CH2Cl2, rt, 20 h; (b) HCl–1,4-dioxane, MeOH, rt, 2 h, 37% over two steps; (ix) 3, Et3N, 1,4-dioxane-DMSO, 50 °C, 23 h, 58%; (x) (a) isothiocyanatobenzene, EDC, THF, 50 °C, 5 h; (b) (Boc)2O, DMAP, THF, rt, 16 h; (c) bis(pinacolato)diboron, KOAc, Pd(dppf)Cl2·CH2Cl2, 1,4-dioxane, 135 °C, 24 h; (d) 3-bromo-6-chloro-N-((1-methylpiperidin-4-yl)methyl)imidazo[1,2-b]pyridazin-8-amine, Pd(OAc)2, 1,1′-bis(di-tert-butylphosphino)-ferrocene, 2 M Na2CO3, 1,4-dioxane, 135 °C, 16 h, 2–5% over four steps.
Results
Compound 2 (Figure 1A and Table 1) was discovered through screening of type I kinase scaffolds related to the IRE1α kinase inhibitor-RNase activator 16. Unexpectedly, we determined that while inhibiting the kinase activity with similar potency to 1, compound 2 was an inhibitor of the RNase function in vitro, suggesting a different allosteric effect on the RNase compared to typical type I IRE1α kinase inhibitors.21 The small size of 2 compared to reported IRE1α kinase-RNase inhibitors from type II- or type I-extended scaffolds (e.g., KIRA3 and compound 16, respectively; Figure 1B) was also attractive as a potential starting point for inhibitor optimization.21,23,24 We hypothesized that the orientation of 2 within the IRE1α adenosine 5′-triphosphate (ATP) site would be similar to that determined for 1 (PDB 4Z7H).6 We anticipated that the key hinge-binding hydrogen bond from N-1 of the imidazo[1,2-b]pyridazine would be maintained and that a new hydrogen bond could be formed to the hinge region from the 8-amino group, replacing the CH···O interaction seen for 1. In this orientation, the indazole substituent would be directed toward the αC-helix, taking the place of the benzamide moiety in 1, while the cyclopropylmethyl group would be directed out toward solvent. The imidazo[1,2-b]pyridazin-8-amine hinge-binding group has recently been shown to adopt such a binding mode to the kinase, monopolar spindle 1 (MPS1).28,29
Table 1. In Vitro Binding and Inhibition of IRE1α Kinase and RNase Activities by 2, 5–12.
| no. | IRE1α S724 autophosphorylation IC50 (μM)a | IRE1α ATP-site binding IC50 (μM)b | IRE1α RNase activity IC50 (μM)c |
|---|---|---|---|
| 16 | 0.218 (±0.15)6 | 1.18 (±0.17) | activator EC50 0.1436 |
| 2 | 0.303 (±0.15) | 1.60 (±0.91) | 8.90 (±0.21) |
| 5 | >10 | >10 | n.d.d |
| 6 | >10 | >10 | n.d. |
| 7 | 4.06 (±0.74) | >10 | >10 |
| 8 | 0.89 (±0.14) | 46% @10 μMe | >10 |
| 9 | 0.81 (±0.4) | 0.84 (±0.42) | 2.67 (±1.89) |
| 10 | >10 | >10 | n.d. |
| 11 | >10 | >10 | n.d. |
| 12 | 2.7 (±1.1) | >10 | n.d. |
Inhibition of recombinant G547 IRE1α kinase-endoribonuclease extended (KEN) domain pS274 autophosphorylation measured in dissociation-enhanced lanthanide fluorescence immunoassay (DELFIA) format, mean (±standard deviation (SD)) for n ≥ 3.
Inhibition of ATP-site LanthaScreen tracer binding to recombinant dephosphorylated G547 IRE1α KEN domain, mean (±SD) for n ≥ 3.
Inhibition of G547 IRE1α-dependent cleavage of a fluorescence resonance energy transfer (FRET)-labeled stem-loop RNA containing the XBP1 cleavage site, mean (±SD) for n ≥ 3.
n.d. = not determined.
±4%, n = 4.
We used in vitro biochemical assays to measure compound binding and effects on IRE1α autophosphorylation and endoribonuclease activities, to investigate the structure–activity relationships around 2 and test the proposed binding model. Recombinant IRE1α protein corresponding to the cytoplasmic kinase-endoribonuclease extended (KEN) domain (G547 IRE1α) was produced separately in phosphorylated and dephosphorylated states, as confirmed by intact protein mass spectrometry (MS) (Figure S1).30 We used our previously described DELFIA format to determine the inhibition of recombinant IRE1α S274 autophosphorylation.31 To measure compound binding to IRE1α, we developed a LanthaScreen Eu Kinase binding FRET assay, which monitored the displacement of an ATP-site tracer. To determine the allosteric effect of the kinase inhibitors on IRE1α RNase activity, we used a FRET-based assay to measure the cleavage of a short stem-loop RNA sequence derived from the endogenous cleavage site in XBP1u mRNA (Figure S2).21,30
We initially investigated the role of the indazol-5-yl substituent in 2 (Table 1). The availability and correct positioning of hydrogen bond donor/acceptor functionality was critical, with neither the N-methyl (5) nor regioisomeric (6) analogues showing activity. Only weak inhibition of IRE1α autophosphorylation in the sensitive DELFIA format was observed for the indol-5-yl derivative (7). The benzimidazol-5-yl analogue (8) also inhibited IRE1α autophosphorylation but showed significantly weaker ATP-site binding in the LanthaScreen assay and no RNase inhibition. For the RNase assay, this may in part reflect the reduced potency of 8 compared to 2. The weak potency for ATP-site binding assay was unexpected, but based on the promising inhibition of IRE1α autophosphorylation, we pursued further substitution of the benzimidazol-5-yl scaffold.
We assumed that the indazol-5-yl substituent of 2 potentially interacted with the same region of the IRE1α active site as the benzamide of 1, in particular with residues such as D711 from the DFG motif or K599. Compound 1 also interacted with a water molecule that was in contact with E612 from the αC-helix.6 Therefore, we investigated 2-(N-phenyl)amino substitution of the benzimidazole 8. The 2-(N-phenyl)aminobenzimidazole motif is a known isostere for the diaryl urea motif more commonly found in type II kinase inhibitors, as it is capable of mimicking the hydrogen-bonding pattern made by type II urea inhibitors in the inactive DFG-out conformation of the kinase domain.32 The addition of aryl urea groups or their isosteres to convert type I kinase inhibitor scaffolds to type II molecules is a well-established approach.33 Gratifyingly, we observed that the 2-(N-phenyl)aminobenzimidazole 9 had similar kinase site binding affinity to 2, and improved on the inhibition of IRE1α RNase activity. Consistent with a role for the 2-(N-phenyl)aminoimidazole functionality in forming multiple hydrogen bonds, N-methylation at any position reduced inhibitory activity (see 10–12).
With two variations of the imidazo[1,2-b]pyridazin-8-amine core identified that led to RNase inhibition, the effects of varying the 8-amino group on potency and physicochemical properties were studied. The aqueous (aq) buffer solubilities of 2 (KSol = 0.8 μM) and 9 (KSol = 2.9 μM) were low, consistent with their measured lipophilicities (2, Log D7.4 = 3.4; 9, Log D7.4 = 4.1). We therefore sought to introduce polar or ionizable groups to improve solubility and hypothesized that the vector defined by the cyclopropylmethyl group should be directed toward solvent. Importantly, studies leading to the IRE1α inhibitor compound 16 (Figure 1B) found that appending cyclic amine or cyclohexylamine substituents directed out of the ATP-binding pocket was essential for achieving high potency.23 Our presumed binding mode of the core scaffolds 2 and 9 oriented the 8-amino substituent in a similar direction. We investigated a range of substituents, including cyclic amines analogous to those described in the development of compound 16, with selected examples highlighted in Table 2.
Table 2. In Vitro Binding and Inhibition of IRE1α Kinase and RNase Activities by 19–32 and Published Compound KIRA3.
| no. | IRE1α S724 autophosphorylation IC50 (μM)a | IRE1α ATP-site binding IC50 (μM)b | IRE1α RNase activity IC50 (μM)c |
|---|---|---|---|
| 19 | 0.19 (±0.11) | 0.25 (±0.07) | 0.48 (±0.19) |
| 20 | 0.77 (±0.13)d | 0.83 (±0.19) | 0.13e |
| 21 | 0.47 (±0.09) | 0.87 (±0.24) | 0.16e |
| 22 | 0.32 (±0.09) | 0.11 (±0.04) | 0.072 (±0.011) |
| 23 | >10 | >10 | n.d.f |
| 24 | 0.15 (±0.05) | 0.14 (±0.02) | 0.033e |
| 25 | 0.19 (±0.01)d | 0.11 (±0.01) | 0.122 (±0.026) |
| 26 | 0.24 (±0.09) | 0.087 (±0.022) | 0.027 (±0.007) |
| 27 | 0.20 (±0.05) | 2.0 (±0.94) | 0.51e |
| 28 | 0.13 (±0.06) | 0.67 (±0.23) | 0.18e |
| 29 | 0.13 (±0.03) | 0.39 (±0.11) | 0.09 (±0.05) |
| 30 | 0.15 (±0.02) | 0.33 (±0.04) | 0.10 (±0.05) |
| 31 | 0.16 (±0.06) | 0.27 (±0.17) | 0.08 (±0.05) |
| 32 | 0.16 (±0.05) | 0.48 (±0.05) | 0.17 (±0.09) |
| KIRA3 | 0.29 (±0.06) | 0.40 (±0.16) | 0.45 (±0.10) |
Inhibition of recombinant G547 IRE1α KEN domain pS274 autophosphorylation measured in DELFIA format, mean (±SD) for n ≥ 3.
Inhibition of ATP-site LanthaScreen tracer binding to recombinant dephosphorylated G547 IRE1α KEN, mean (±SD) for n ≥ 3.
Inhibition of G547 IRE1α-dependent cleavage of a FRET-labeled stem-loop RNA containing the XBP1 cleavage site, mean (±SD) for n ≥ 3.
n = 2.
Single determination.
n.d. = not determined.
Introduction of polar groups generally maintained the dual kinase-RNase inhibitory profile of the compounds while enhancing potency. Thus, replacement of the cyclopropylmethyl substituent by tetrahydropyran-4-yl resulted in submicromolar inhibitors of IRE1α RNase function, 19 and 27. Aqueous solubility remained low (19, KSol = 2.2 μM; 27, KSol = 2.4 μM), but was increased in the basic N-methylpiperidin-4-yl derivatives 20 (KSol = 58 μM, Log D7.4 = 1.8) and 28 (KSol = 41 μM, Log D7.4 = 2.7), which also showed improvements in RNase inhibition.
For the 3-(indazol-5-yl) substituted analogues, extension of the piperidine substituent had a negligible effect on compound affinity or potency (see 21). However, the trans-1,4-diaminocyclohexane analogue 22 showed enhanced binding and RNase inhibition. Notably, N-8-methylation to give 23 led to loss of activity, consistent with the assumed kinase hinge-binding role of the 8-amino substituent. Further substitution of the trans-1,4-diaminocyclohexane group was tolerated, exemplified by 24–26, and led to potent in vitro inhibitors of IRE1α RNase activity with acceptable aqueous solubility (e.g., 26, KSol = 80 μM, Log D7.4 = 1.8).
For the 3-(2-(N-phenyl)aminobenzimidazol-5-yl) substituted compounds, the (N-methylpiperidin-4-yl)methyl analogue 29 provided an enhancement in binding and RNase inhibitory potency without the need for further substitution. We examined fluorination of the benzimidazole group but found only marginal (<2-fold) gains in affinity and potency (see 30–32). The most potent RNase inhibitor 31 from this scaffold showed moderate solubility (KSol = 38 μM), reflecting the high lipophilicity (Log D7.4 = 3.7).
Cellular inhibition of IRE1α RNase function was measured for selected compounds using a bioluminescent reporter assay in HEK293 cells stably expressing an XBP1 luciferase fusion mRNA to quantify XBP1 mRNA splicing (Table 3). In the absence of ER stress, translation of the XBP1u mRNA terminates before the luciferase sequence, while exogenous induction of ER stress leads to IRE1α-dependent XBP1 mRNA splicing and results in tandem translation of the XBP1s-luciferase reporter protein. The imidazo[1,2-b]pyridazine 2 moderately inhibited tunicamycin-induced mRNA splicing. Enhancement of the in vitro potency and aqueous solubility of the scaffold, to give 22 and 26, was accompanied by increased cellular activity. Likewise, the most potent and soluble 3-(2-(N-phenyl)aminobenzimidazol-5-yl) substituted analogues 29 and 31 gave submicromolar inhibitors of IRE1α activity in cells. The optimization of in vitro potency in this series involved addition of basic groups, which may have impaired cell permeability and countered gains in potency for some of the more basic molecules. The cytotoxicity of compounds in the cell line was monitored to preclude interference in the assay, and no compound showed high cytotoxicity in the HEK293 cell line over the duration of the assay.
Table 3. Cellular Activities of Selected IRE1α Inhibitors.
| no. | XBP1 luciferase fusion mRNA splicing in HEK293 cells IC50 (μM)a | cytotoxicity in HEK293 reporter cells EC50 (μM)b |
|---|---|---|
| 2 | 4.2 (±1.7) | >49 |
| 22 | 2.9 (±0.87) | 28 (±6.9) |
| 26 | 1.12 (±0.24) | >49 |
| 29 | 0.89 (±0.1) | 12c |
| 31 | 0.71 (±0.33) | 9.0 (±1.6) |
| KIRA3 | 0.76c | >49 |
Inhibition of tunicamycin-induced IRE1α-dependent splicing of an XBP1u-luciferase mRNA reporter stably expressed in HEK293 cells, mean (±SD) for n ≥ 2.
Cytotoxicity in HEK293 cells stably expressing an XBP1u-luciferase mRNA reporter, measured using the Alamar Blue format, mean (±SD) for n ≥ 2.
Single determination.
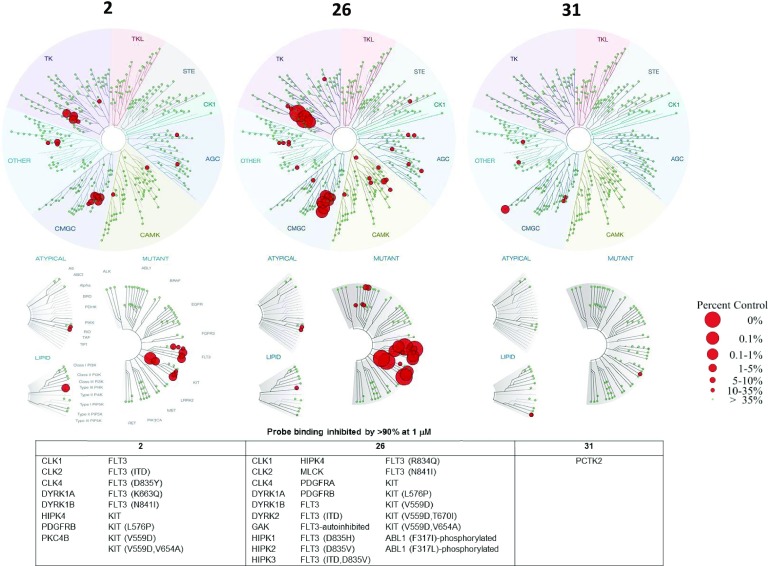
Compounds 2, 26, and 31 were profiled for broad kinase selectivity, as determined by inhibition of probe binding to recombinant human protein and lipid kinase domains at a concentration of 1 μM (KINOMEscan, Eurofins; Figure 2 and Table S1). Compound 2 gave >70% inhibition of 26/455 wild-type and mutant kinases tested. A distinct selectivity pattern was seen, with specific CMGC (CLK1, CLK2, CLK4, DYRK1A, DYRK1B, HIPK4) and TK (FLT3 wild type and mutants, KIT wild type and mutants, PDGFRB) subfamily members showing particularly high inhibition (>90%). The more potent derivative 26 maintained a very similar selectivity pattern, with additional kinases in the CMGC (DYRK2, HIPK1, HIPK2, HIPK3) and TK (PDGFRA) subfamilies related to those inhibited by 2 showing potent (>90%) inhibition. Overall, 54/455 wild-type and mutant kinases tested showed >70% inhibition, consistent with the increased potency of 26. In contrast, the 2-(N-phenyl)aminobenzimidazole 31 had a different selectivity pattern and very high selectivity with >70% inhibition of only 4/455 kinases. While two of the CMGC kinases inhibited by 2 and 26 (CLK1 and CLK4) were inhibited to a lesser extent by 31, the most potently inhibited kinase was another CGMC family member PCTK2 (97% inhibition at 1 μM test concentration). Interaction with wild-type and mutant TK subfamily kinases was minimal for 31. The inhibitors 26 and 31 were, respectively, 9- and 100-fold selective for IRE1α over the IRE1β isoform, which has 80% sequence identity in the kinase domain,34 in parallel LanthaScreen binding assays (Figure S3).
Figure 2.
Binding profiles of compounds 2, 26, and 31 tested at 1 μM concentrations of inhibitors against 455 wild-type and mutant human protein and lipid kinases (KINOMEscan, Eurofins) (see Table S1). Kinases where probe binding was inhibited by >90% at 1 μM test compound concentration are listed.
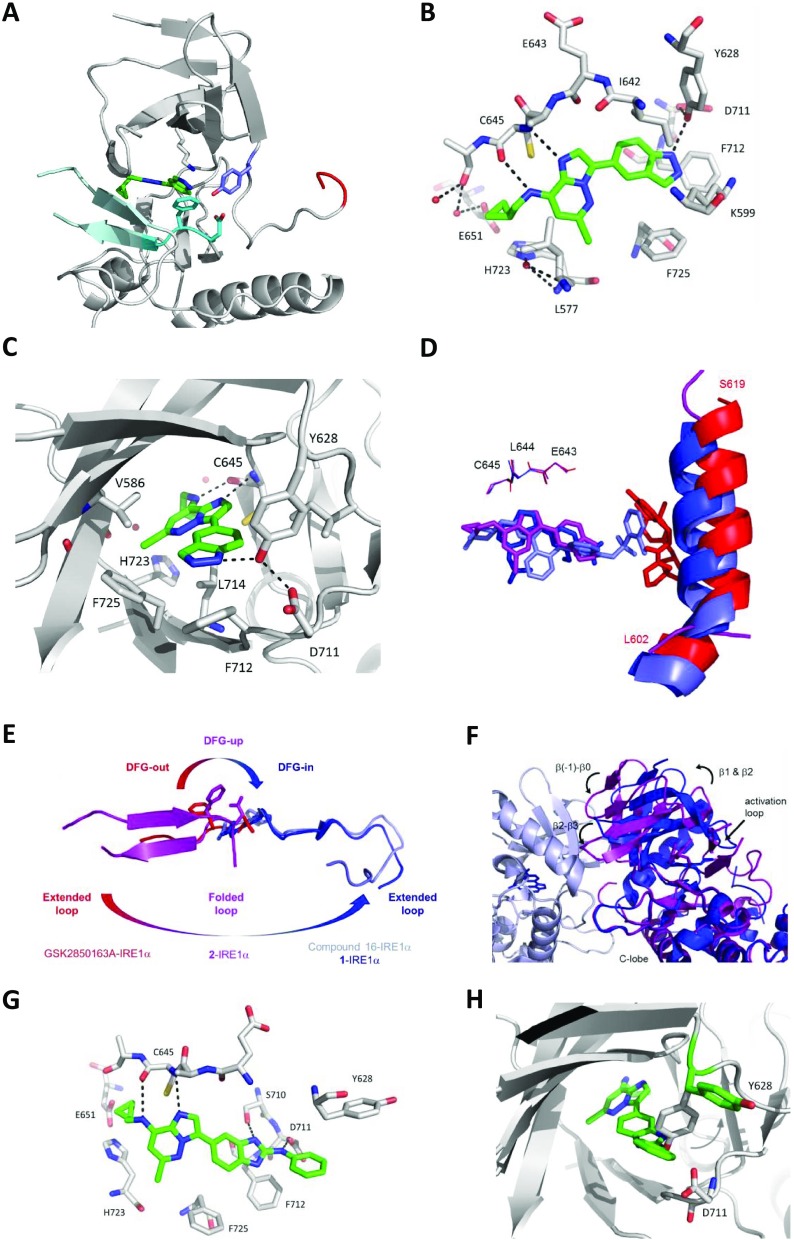
To better understand the origins of the potency and high selectivity of the inhibitors, we determined the X-ray structure of 2 bound to the cytosolic kinase-endoribonuclease (KEN) domain of IRE1α (PDB 6HX1; Figure 3A,B). The complex 2-IRE1α crystallized as a monomer in the unit cell. The kinase domain adopted an unusual conformation in which the αC-helix was remarkably disordered such that electron density was not modeled for most of this important secondary structural component. The DFG motif occupied an intermediate position between the canonical DFG-in and DFG-out conformations, which we term “DFG-up” for convenience. Thus, F712 was displaced from the “in” position that characterizes the active conformation of the regulatory hydrophobic spine of the kinase. A related intermediate conformation of the DFG motif has been observed in complexes of ligands bound to mutated Aurora A kinases.35,36 In the current structure, the residue Y628, also part of the hydrophobic spine in IRE1α, was directed down toward the DFG motif, forming hydrogen bonds to D711 (2.7 Å) and the bound ligand. This contrasted with the upward orientation of Y628 seen when the regulatory spine is intact and the IRE1α kinase domain is in the active conformation.6 The DFG-up, Y628-down conformation of 2-IRE1α was accompanied by an unusual folding of the activation loop in an antiparallel double β-strand across the face of the ATP site, forming a region of β-sheet with the P-loop that enclosed the bound ligand. The residue H723 in the activation loop appeared to anchor this folding, with direct hydrogen bonding to E651 (2.8 Å) in the kinase domain C-terminus and a water-mediated hydrogen bond observed between H723 and L577 in the P-loop. Folding of the activation loop across the active site is an autoinhibitory mechanism for some kinases. For example, structures of imidazo[1,2-b]pyridazines and other ligands bound to the kinase MPS1 show a single strand of the activation loop folded across the active site parallel to the P-loop, potentially blocking the interaction with substrate peptides.29 However, in these cases, the kinase adopts a DFG-in conformation on ligand binding and the αC-helix is preserved.
Figure 3.
Characterization of IRE1α inhibitor binding modes. (A) Part of the crystal structure (PDB 6HX1) of 2 (green sticks) bound to IRE1α KEN domain (gray image) showing the conformations of the DFG motif (cyan), activation loop segment (pale cyan), Y628 (blue), K599 (gray), and the observed part of the αC-helix (red). (B) Binding of 2 (green sticks) in the IRE1α ATP site (gray sticks) with key residues labeled. Hydrogen bonds are shown as black dotted lines and selected water molecules as red spheres. (C) Details of the interactions of 2 (green sticks) with the DFG motif and Y628 in the IRE1 ATP site (gray image) with key residues labeled. Hydrogen bonds are shown as black dotted lines and selected water molecules as red spheres. (D) Comparison of 2 (purple), 1 (blue; PDB 4Z7H), Compound 16 (light blue; PDB 4U6R), and GSK2850163 (red; PDB 4YZ9) binding to IRE1α KEN domain. Structures are overlaid on the hinge region (E643-L644-C645; lines), and the αC-helix (L602-S619; cartoon) is shown in the three structures in which it is present. (E) Schematic overlay showing the relationship between the DFG-out, DFG-up, and DFG-in conformations and the corresponding orientations of the observed activation loop segments (L714-S729) for GSK2850163-IRE1α (red), 2-IRE1 (purple), compound 16-IRE1α (light blue), and 1-IRE1α (blue). (F) Comparison of components of the N-terminal kinase domain dimerization interface for the DFG-in, active conformation of 1-IRE1α (blue and blue-white protomers) and DFG-up, and inactive conformation of 2-IRE1α (purple). Structures are aligned on the kinase C-lobe (shown) and endonuclease C-terminal domain (not shown). Bound ligands are rendered as sticks; (G) model of 9 (green sticks) bound to IRE1 (gray image) based on the conformation observed for 2-IRE1. Predicted hydrogen bonds are shown as black dotted lines. (H) Detail showing the movement of Y628 as positioned in 2-IRE1 (gray image) required to accommodate the modeled binding of 9 (green sticks).
Within the unusual kinase ATP-site conformation, the ligand 2 bound at the hinge region, with the imidazo[1,2-b]pyridazine ring sandwiched between L714 in the activation loop and V586 in the β2-strand, C-terminal to the P-loop (Figure 3B). Canonical hydrogen bonds were made from the imidazo[1,2-b]pyridazine N1 and 8-NH to the C645 NH (3.1 Å) and C=O (3.0 Å) functionalities, respectively. This closely resembled the binding modes described for 8-amino-imidazo[1,2-b]pyridazines in other kinases.28,29,37,38 An additional hydrogen bond (2.8 Å) was observed between the indazole NH and Y628, presumably contributing to stabilizing the tyrosine “down” conformation. The indazole occupied a hydrophobic pocket formed by the gatekeeper residue I642, F712 of the DFG motif, F725 from the activation loop, and the catalytic lysine K599. The 6-chloro substituent of 2 packed closely against the β-sheet surface formed by L577 and G578 of the P-loop and H723 and S724 of the activation loop. The cyclopropylmethyl side chain of 2 reached the edge of the ATP site and was oriented to provide a vector out into solvent.
The unusual binding mode seen for 2-IRE1α was independently reproduced with a close analogue, 33 (PDB 6HV0), in which the cyclopropylmethyl group was replaced by an isopropyl chain (Figure S4), supporting the likely biological relevance of the structure. Furthermore, the observed binding mode was consistent with the structure–activity relationships described above for the indazole-substituted 8-amino-imidazo[1,2-b]pyridazines, specifically the requirement for a hydrogen bond donor corresponding to the indazole N(1)–H (Table 1) and the tolerance of a wide range of substitution of the 8-amino group on the imidazo[1,2-b]pyridazine core (Table 2). Modeling of the more potent compound 26 into the conformation of IRE1α observed in the crystal structures reproduced this binding mode, with the suggestion of an additional hydrogen bond from the cyclohexylamine to E651 on the surface of the protein (Figure S5).
The unusual DFG-up inactive conformation of the kinase domain of 2- and 33-IRE1α differed considerably from other active and inactive kinase conformations reported for ligand-bound structures of IRE1α (Figure 3C,D).4,6,22,23 We previously described the IRE1α kinase inhibitor-RNase activator 1 that stabilized a classical DFG-in, active conformation of the kinase with the activation loop extended away from the active site, mimicking IRE1α autophosphorylation and activating the RNase function through competent dimerization of the endonuclease domain.6 In contrast, a large sulfonamide inhibitor (named compound 16), although it also bound to a DFG-in conformation of the kinase, was found to cause a shift of the αC-helix that was incompatible with the formation of the RNase domain dimer.23 A type III inhibitor of IRE1α, GSK2850163, has been described that occupied a pocket adjacent to the hinge region of the kinase and promoted a DFG-out kinase conformation, with the activation loop directed across the active site and the αC-helix further shifted and rotated away from the active conformation.22 In the latter two cases, although the active kinase conformation was disrupted, the αC-helix remained intact. For 2 and 33, our data showed a striking disruption of the αC-helix, as well as a distinctive refolding of the activation loop around the bound inhibitor. This conformation is incompatible with the back-to-back dimerization required to form an active endoribonuclease domain dimer (Figure 3E). The dimerization interface in the kinase domain is formed by the loops β(−1)−β(0) and β(2)−β(3) from each monomer.6 Compared to the active IRE1α dimer structure represented by 1-IRE1α, the binding of 2 and 33 shifted the position of the kinase domain relative to the endonuclease domain such that dimerization at the kinase domain would lead to steric clashes of the endoribonuclease domains.
We were unable to obtain a crystal structure of a 2-aminobenzimidazole-substituted analogue bound to IRE1α. Modeling of 9, the closest analogue to 2, into the structure of 2-IRE1 showed that alignment of the imidazo[1,2-b]pyridazine cores of the two inhibitors resulted in a clash between the 2-aminobenzimidazole substituent and residue Y628. When Y628 was allowed to move from the “down” orientation observed in 2-IRE1 to the “up” conformation observed in other IRE1α structures, this minimal change resulted in a satisfactory binding mode for the prototypical 2-(N-phenyl)aminobenzimidazole 9 (Figure 3F). The model retained binding of the imidazo[1,2-b]pyridazine at the hinge region and proposed a 180° rotation of the 2-aminobenzimidazole relative to the indazole substituent of 2 such that a new hydrogen bond was formed between the 2-amino group and D711 of the DFG motif. The terminal phenyl group occupied the space vacated by Y628 and packed against this residue now in its upward conformation. The introduction of a hydrogen bond to the DFG motif was consistent with precedent for other 2-aminobenzimidazole kinase inhibitors, where this functionality is often an isostere for urea-based inhibitors that bind to DFG-out conformations, as we had originally speculated in developing the series. Moreover, the modeled hydrogen bond to D711 was consistent with the observed structure–activity relationships, where methylation of the 2-amino group or imidazole nitrogens reduced activity (Table 1). The combination of multiple kinase conformational disruptions accompanying binding of the indazole and aminobenzimidazole scaffolds can account for the high IRE1α kinase selectivity of 26 and 31 (Figure 2). For 26, a discrete set of kinases in the CAMK and TK subfamilies were also inhibited. For this ligand, we cannot exclude the possibility that an alternative binding mode where the indazole contributes the donor–acceptor hinge-binding motif may be accessible in some kinases.39,40 Interestingly, while the majority of residues forming the binding site of 2 and 33 to IRE1α are identical in the sequence of the isoform IRE1β, the histidine H723 in IRE1α is replaced by a cysteine residue (C671) in IREβ (Figure S3B).34 Since hydrogen bonding of the imidazole side chain of H723 to E651 appeared important for the unusual activation loop fold in the structures 2 and 33 bound to IRE1α, we speculate that the absence of this residue may in part be involved in the observed selectivity of the compounds for IRE1α over IRE1β. The other major local difference (A646 in the hinge corresponds to R594 in IRE1β34) involves a side chain directed away from the binding site.
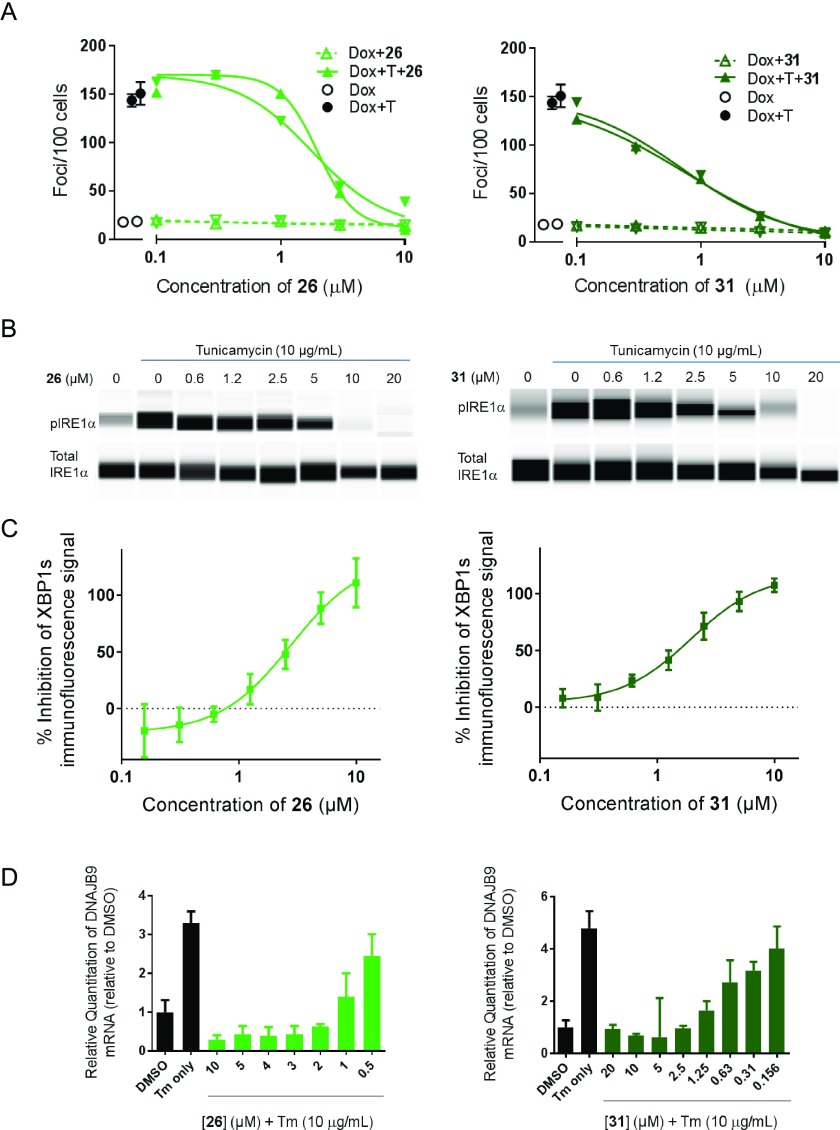
To confirm the functional consequences of the unusual binding modes of the inhibitors, we investigated the effects of 26 and 31 on ER stress-induced IRE1α signaling outputs in human cells. The effects of the compounds on IRE1α oligomerization were determined in HEK293 cells expressing doxycycline-inducible green fluorescent protein (GFP)-labeled IRE1α that forms quantifiable fluorescent foci in the ER membrane under stress conditions41 (Figure 4A). GFP-IRE1α foci produced upon treatment with tunicamycin were dose-dependently reduced by 26 (IC50 = 1.88 ± 0.09 μM) and 31 (IC50 = 0.74 ± 0.03 μM), indicating inhibition of IRE1α oligomer formation. No stimulation of GFP-IRE1α foci formation was observed for either compound on its own. Thus, the compounds are full inhibitors of oligomerization in cells and do not show partial activation of IRE1α through occupation of the nucleotide-binding site.
Figure 4.
Compounds 26 and 31 inhibit IRE1α oligomerization, IRE1α autophosphorylation, IRE1α RNase activity, XBP1s protein expression, and XBP1-dependent transcription in human cells. (A) Inhibition of tunicamycin-induced IRE1α oligomerization, measured by fluorescent foci formation in HEK293 cells stably transfected with doxycycline-inducible GFP-IRE1α after 5 h treatment with inhibitors (0–10 μM) in the absence or presence of tunicamycin (quantification of image fields from n = 2 experiments plotted separately). Dox = doxycycline, T = tunicamycin (10 μg/mL). (B) Inhibition of tunicamycin-induced pS724 IRE1α autophosphorylation as measured by capillary electrophoresis immunoassay (simple Western) relative to total IRE1α. Data shown for a single experiment representative of n = 3. (C) Inhibition of tunicamycin-induced XBP1s protein expression in H929 cells as measured by immunofluorescent assay (quantification of image fields from n > 3 experiments). (D) Inhibition of tunicamycin-induced XBP1s-dependent transcription of DNAJB9 mRNA as measured by real-time quantitative polymerase chain reaction (RT-qPCR). Data shown for a single experiment representative of n = 3. Tm = tunicamycin.
Compounds 26 and 31 inhibited both tunicamycin- and thapsigargin-induced IRE1α-dependent splicing of XBP1 luciferase fusion mRNA in HEK293 cells (Table 3 and Figure S6) with equivalent potencies (IC50 0.68–1.63 μM). In parallel, inhibition of tunicamycin-induced production of endogenous XBP1s mRNA was demonstrated in H929 myeloma cells using RT-qPCR at similar concentrations (Figure S7). The expression of the spliced transcription factor XBP1s following ER stress was measured by immunofluorescent staining in H929 myeloma cells (Figure 4C). Tunicamycin-induced expression of XBP1s protein was inhibited by 26 and 31 with similar potencies to the inhibition of IRE1α oligomerization and RNase activities in cells. The expected downstream effect on XBP1s-dependent transcription in H929 cells was confirmed by monitoring production of the mRNA coding for DNAJB9, an ER-resident molecular chaperone regulated by XBP1s8 (Figure 4D). Inhibition of IRE1α autophosphorylation in H929 cells was also observed on treatment with 26 and 31 (Figures 4B and S8), although full blockade appeared to require higher concentrations than the oligomerization and RNase functions. This is consistent with the proposed model for IRE1α activation, where conformationally productive oligomerization is the critical step in activating the endoribonuclease,4 while autophosphorylation contributes to stabilizing the active oligomers6 and XBP1-independent signaling to JNK through the binding of TRAF2.10 Previous studies by our group6 and others4 indicate that the trans-autophosphorylation of IRE1α proceeds through a face-to-face encounter of IRE1α protomers distinct from the back-to-back arrangement essential for activation of the RNase function.
Discussion and Conclusions
Through screening analogues of a type I IRE1α kinase inhibitor that activates the RNase function through binding to a classical DFG-in kinase conformation, we unexpectedly discovered a closely related series of imidazo[1,2-b]pyridazin-8-amine kinase inhibitors that also inhibited the RNase. This surprising finding challenged our initial assumption that type II or type III kinase inhibitors binding to a DFG-out conformation of the kinase would be needed to prevent IRE1α dimerization and activation of the RNase.
We increased the potency of the imidazo[1,2-b]pyridazin-8-amines using design hypotheses that assumed a type I binding mode of the core scaffold, leading to a series of indazoles, while further substitution generated 2-phenyl-benzimidazoles designed as potential type II inhibitors. However, crystal structures of the indazoles 2 and 33 bound to IRE1α showed the protein to adopt a highly unusual conformation, with the DFG motif in an intermediate “up” position and substantial disordering of the αC-helix. These structures and models derived from them accounted for the structure–activity relationships of the inhibitors, showing how a type I scaffold could promote a previously unreported conformation of the kinase domain that is incompatible with RNase domain back-to-back dimerization required for activation of the RNase function. The binding mode emphasized the role of the hydrophobic spine residue Y628 of IRE1α in compound binding and kinase conformational control.6,42
Other kinase signaling pathways have been observed to be modulated through transactivation of dimers controlled allosterically by kinase site occupation.43 Notably, ligand-promoted heterodimerization of inhibitor-bound BRAF with CRAF results in paradoxical activation of RAF signaling by some BRAF inhibitors.44,45 Kinase site inhibitors that can simultaneously suppress the catalytic function of kinase monomers and their dimerization with uninhibited partners are therefore a potentially useful therapeutic modality.46 Certain pseudokinases that lack intrinsic enzyme function also exploit dimerization to regulate signaling pathways and may be amenable to pharmacological intervention through nucleotide-binding-site ligands that prevent dimerization.47
The most advanced compound we studied showed nanomolar potency for biochemical inhibition of the kinase and RNase and, importantly, was highly selective for IRE1α over other kinases. It is possible that the high selectivity of 31 reflects binding to the unusual IRE1α kinase conformation incorporating multiple disruptions, as modeled for the analogue 9. Although elements of the conformational disruption of the IRE1α kinase domain have been seen separately in other contexts,22,23,28,29,35,36 to our knowledge, they have not been observed occurring simultaneously. Our findings thus increase the repertoire of conformations discovered for IRE1α and provide a means to achieve high specificity for IRE1α inhibition over other kinases, with reliable translation of kinase ATP-site occupation into RNase inhibition.
We confirmed that compounds engaging the newly discovered IRE1α conformation resulted in the inhibition of ER stress-induced signaling through IRE1α in multiple human cell types, resulting in inhibition of XBP1-dependent transcription. We determined substantial inhibition of the IRE1α signaling pathways to occur in cells at concentrations of 26 and 31 between ca. 1 and 2 μM in short-term assays. Selective IRE1α kinase-endoribonuclease inhibitors may have utility in situations where direct cytotoxicity is not the desired outcome of IRE1α-XBP1s inhibition, such as the modulation of XBP1s-dependent suppression of dendritic immune cell function. The identification of compounds 26 and 31 and their binding mode demonstrates a new means to achieve highly selective inhibition of IRE1α, which may be important in such contexts where the biology of untransformed host cells is targeted.48
Experimental Section
IRE1α Autophosphorylation Assay
Inhibition of IRE1α autophosphorylation was measured using a dissociation-enhanced lanthanide fluorescence immunoassay (DELFIA) as described previously.31 Briefly, dephosphorylated IRE1α G547-L977 (700 nM) was incubated for 25 min with 100 μM ATP in 15 μL of assay buffer (40 mM Tris (pH 7.5), 20 mM MgCl2, and 1 mM 1,4-dithiothreitol (DTT)). The assay was stopped by the addition of 40 mM ethylenediaminetetraacetic acid. Samples were transferred to a 384-well high-binding plate and incubated overnight at 4 °C. The plates were washed three times in 0.1% Tween-20, followed by blocking in 5% nonfat skimmed milk in phosphate-buffered saline (PBS) for 30 min at 37 °C. The plates were washed again before application of α-pS724 phospho-specific IRE1α primary antibody31 (160 pg/mL) in PBS to each well and incubated for 1.5 h at 37 °C. The plates were washed before addition of a europium-labeled α-rabbit secondary antibody (PerkinElmer Life Sciences). Another wash step was followed by the addition of DELFIA enhancement solution (PerkinElmer Life Sciences). Assay plates were read on an EnVision multimode plate reader (PerkinElmer Life Sciences).
IRE1α RNase Assay
Inhibition of IRE1α endoribonuclease activity was measured using a FRET de-repression assay monitoring cleavage of a 29-nucleotide stem-loop RNA containing the XBP1 cleavage site sequence and labeled with a fluorescence emitter (fluorescein amidite (FAM)) and a fluorescence quencher (Black Hole Quencher (BHQ)) at the 5′ and 3′ ends, respectively (Figure S2), as described previously.30,31 Briefly, varying volumes of compound in DMSO or DMSO alone were added to a low-volume 384-well plate (3676, Corning) to give final concentrations ranging from 100 μM to 0.313 nM using an Echo acoustic liquid dispenser (Labcyte, CA). Nonphosphorylated IRE1α G547-L977 was added to a final concentration of 200 nM. After incubating for 30 min at 30 °C, hairpin RNA XBP1 substrate mimic labeled with fluorescein and Black Hole Quencher (5′ FAM-GAACAAGAUAUCCGCA-GCAUAUACAGUUC-3′ BHQ, Eurofins MWG Operon, Germany) was added to 100 nM final concentration. After incubation for a further 15 min at 30 °C, the fluorescein fluorescence was measured on an EnVision multimode plate reader (PerkinElmer Life Sciences). Fluorescence in the presence of compound was expressed relative to that of DMSO alone (no compound).
IRE1α ATP-Site Binding Assay
Compound binding to IRE1α was measured using a LanthaScreen Eu Kinase binding FRET assay monitoring the displacement of an ATP-site tracer by inhibitors. Briefly, a master mix of (7.5 μL) 50 nM dephosphorylated IRE1α G547-L977, 2 nM biotinylated anti-his-tag antibody (PV6089, Thermo Fisher Scientific), 2 nM streptavidin–europium (PV5899, Thermo Fisher Scientific) diluted in assay buffer (PV3189, Thermo Fisher Scientific), and 1 mM 1,4-dithiothreitol (DTT) (D0632, Sigma-Aldrich) was incubated with 7.5 μL of 75 nM tracer (Tracer 236, Thermo Fisher Scientific) in assay buffer for 60 min at rt. During the incubation, the 384-well white low-volume plate (3674, Corning) was covered with foil. Assay plates were read on an EnVision multimode plate reader (PerkinElmer Life Sciences).
X-ray Crystallography of 2-IRE1α (PDB 6HX1)
Determination of the structure of 2-IRE1α was conducted by Proteros Biostructures GmbH (Martinsried, Germany). Expression, purification, and crystallization of IRE1α with compound were performed by analogy to previously reported protocols.6 Crystals were flash-frozen and data were collected at 100 K at the Swiss Light Source (PXI/X06SA, PILATUS 6 M detector; SLS, Villigen, Switzerland) using cryogenic conditions. Data were processed using the programs XDS and XSCALE.49 The structure of apo-IRE1α was solved by molecular replacement using a previously solved structure of IRE1α as a search model. Subsequent model building and refinement was performed according to standard protocols with the software packages CCP450 and COOT.51 For the calculation of the free R-factor, 5.4% of measured reflections were excluded from the refinement procedure (see Table S2). The ligand parameterization and generation of the corresponding library files were carried out with CORINA (Molecular Networks, GmbH). The water model was built with the “Find waters” algorithm of COOT by placing water molecules in peaks of the Fo–Fc map contoured at 3.0, followed by refinement with REFMAC552 and checking all waters with the validation tool of COOT. The criteria for the list of suspicious waters were: B-factor greater than 80 Å2, 2Fo–Fc map less than 1.2σ, and distance to the closest contact less than 2.3 Å or more than 3.5 Å. The suspicious water molecules and those in the ligand binding site (distance to ligand less than 10 Å) were checked manually. The occupancy of side chains in negative peaks in the Fo–Fc map (contoured at −3.0σ) was set to zero and subsequently to 0.5 if a positive peak occurred after the next refinement cycle.
In Silico Chemistry
The enzyme–inhibitor crystal structure of 2-IRE1α (PDB 6HX1) was prepared for modeling using Protein Preparation Wizard in Maestro,53 and all water molecules, apart from that interacting with residues L577 and H723, were removed. To propose predicted binding modes of the inhibitors, Glide (grid-based ligand docking with energetics)54 was used for the docking experiments. The receptor grid (model A; for 26 bound to IRE1α) was defined by a grid box of 25 × 25 × 25 Å3 with a default inner box (10 × 10 × 10 Å3) centered on the ligand 2 in 6HX1. For the “Y628-up” conformation (model B; for 9 bound to IRE1α), the side-chain orientation of residue Tyr628 in 6HX1 was flipped from the observed rotamer to the most probable rotamer without steric clashes according to the Schrödinger in-built rotamer library, energy-minimized, and a Glide receptor grid subsequently prepared as described above.
Ligands were prepared using LigPrep55 applying the OPLS_2005 force field with possible tautomeric and ionization states in the pH range 5.0–9.0 generated. Using Glide Extra Precision (XP) settings, flexible docking of ligands was conducted with the minimal constraints defined as satisfying at least one H-bonding interaction with either residue R643 or C645 (kinase hinge region interactions), analogous to the interactions observed in the structure of 2-IRE1α. Model A was used for the binding mode predictions of 26, whereas model B was applied to predict the binding mode of 9.
Hit compound 2 was analyzed for potential pan-assay interference behaviors using open access in silico tools (http://zinc15.docking.org/patterns/home), and no substructural alerts were returned. Specific inhibition of IRE1α kinase was demonstrated in two independent assay formats for 2 and active analogues (Tables 1 and 2).
General Synthetic Chemistry
Reactions were carried out under N2. Organic solutions were dried over MgSO4 or Na2SO4. Starting materials and solvents were purchased from commercial suppliers and were used without further purification. Reactions heated by microwave irradiation were carried out using a Biotage Initiator microwave reactor. Reactions in sealed tubes were carried out in pressure vessels with shield protection. Ion-exchange chromatography was performed using ISOLUTE Flash SCX-II (acidic) or Flash NH2 (basic) resin cartridges. Silica column chromatography was performed using Biotage SP1 or Isolera medium-pressure chromatography systems, using prepacked silica gel cartridges (normal phase, Biotage SNAP KP-Si; reverse phase, Biotage SNAP Ultra C18). Preparative high-performance liquid chromatography (HPLC) was carried out at rt using a 1200 Series Preparative HPLC (Agilent, Santa Clara) over a 15 min gradient elution from 60:40 to 0:100 water/MeOH (both modified with 0.1% formic acid) at flow rates of 5, 20, or 40 mL/min depending on the column size used. Standard injections of 500 μL to 2 mL (with needle wash) of the sample were made onto ACE 5 C18-PFP columns (5 μm, 250 × 10/250 × 21.2/250 × 30 mm2, Advanced Chromatography Technologies, Aberdeen, U.K.). UV–vis spectra were acquired at 254 nm on a 1200 Series Prep Scale diode array detector (Agilent, Santa Clara). NMR spectra were recorded on Bruker AMX500 or AV600 instruments using internal deuterium locks. Chemical shifts (δ) are reported relative to tetramethylsilane (δ = 0) and/or referenced to the solvent in which they were measured. Compounds were assessed for purity by tandem HPLC–MS. Combined HPLC–MS analyses were recorded using an Agilent 6210 time-of-flight (ToF) HPLC–MS with a Merck Purospher STAR column (RP-18e, 30 × 4 mm2) or a Waters Xevo G2QToF HPLC–MS with a Phenomenex Kinetex C18 column (30 × 2.1 mm2, 2.6 μm, 100 A). Analytical separation was carried out at 40 °C with UV detection at 254 nm, and ionization was by positive-ion electrospray. The mobile phase was a mixture of MeOH (solvent A) and water (solvent B), both containing formic acid at 0.1%. Gradient elution from 10:90 (A/B) to 90:10 (A/B) was carried out over 2 min (flow rate: Agilent, 3 mL/min; Xevo, 0.5 mL/min) or 4 min (flow rate: Agilent, 1.5 mL/min; Xevo, 0.3 mL/min). Biologically evaluated compounds gave >95% purity as determined by these methods (Table S2).
6-Chloro-N-(cyclopropylmethyl)-3-(1H-indazol-5-yl)imidazo[1,2-b]pyridazin-8-amine (2)
4-Bromo-6-chloropyridazin-3-amine (10.0 g, 48.1 mmol) and 2-chloroacetaldehyde (31.8 mL, 250 mmol; 50% w/w aqueous solution) in EtOH (67 mL) were stirred at 50 °C for 22 h. The mixture was cooled to rt, concentrated, and partitioned between CH2Cl2 (300 mL) and saturated aqueous NaHCO3 (200 mL). The aqueous phase was extracted with CH2Cl2 (150 mL). The combined organic layers were dried, filtered, and evaporated to dryness to give 8-bromo-6-chloroimidazo[1,2-b]pyridazine (11.2 g, quantitative) that was taken to the next step without further purification. 1H NMR (500 MHz, CDCl3) δ 8.01 (d, J = 1.2 Hz, 1H), 7.84 (d, J = 1.1 Hz, 1H), 7.40 (s, 1H); 13C NMR (126 MHz, CDCl3) δ 145.9, 137.0, 134.8, 124.2, 121.2, 118.8; liquid chromatography mass spectrometry (LCMS) (2 min) electrospray ionization (ESI), (m/z) tR = 1.04 min, m/z (ESI+) 233 (M + H+). N-Bromosuccinimide (5.17 g, 29.0 mmol) was added to a solution of 8-bromo-6-chloroimidazo[1,2-b]pyridazine (4.50 g, 19.4 mmol) in CHCl3 (59 mL) at 0 °C. The mixture was warmed to rt and stirred for 18 h. The mixture was partitioned between saturated aqueous NaHCO3 (150 mL) and CH2Cl2 (100 mL × 2). The combined organic layers were dried, filtered, and evaporated to dryness. Silica column chromatography, eluting with 30% EtOAc–hexanes, gave 3,8-dibromo-6-chloroimidazo[1,2-b]pyridazine (3) (5.05 g, 84%). 1H NMR (500 MHz, CDCl3) δ 7.85 (s, 1H), 7.46 (s, 1H); 13C NMR (126 MHz, CDCl3) δ 147.0, 137.5, 135.3, 124.3, 121.4, 103.2; LCMS (2.0 min) tR = 1.36 min, m/z (ESI+) 311 (M + H+). Cyclopropanemethylamine (4.82 mL, 56.2 mmol) was added to a stirred solution of 3 (3.50 g, 11.24 mmol) in THF (56 mL) at rt. After 3 h, the mixture was evaporated and partitioned between saturated aqueous NaHCO3 (200 mL) and CH2Cl2 (100 mL × 2). The combined organic layers were washed with brine (100 mL), dried, and evaporated to dryness. Silica column chromatography, eluting with 15–20% EtOAc–hexanes, gave 3-bromo-6-chloro-N-(cyclopropylmethyl)imidazo[1,2-b]pyridazin-8-amine (4) (3.20 g, 93%). 1H NMR (500 MHz, CDCl3) δ 7.49 (s, 1H), 6.04 (s, 1H), 6.00 (br t, J = 5.3 Hz, 1H), 3.17 (dd, J = 5.2, 7.1 Hz, 2H), 1.20–1.17 (m, 1H), 0.69–0.66 (m, 2H), 0.36–0.33 (m, 2H); 13C NMR (126 MHz, CDCl3) δ 149.5, 142.9, 133.1, 130.9, 101.1, 92.2, 47.8, 10.0, 3.8 (2C); LCMS (2.0 min) tR = 1.63 min, m/z (ESI+) 303 (M + H+). N2 was bubbled for 10 min through a mixture of 4 (0.096 g, 0.318 mmol), 5-(4,4,5,5-tetramethyl-1,3,2-dioxaborolan-2-yl)-1H-indazole (0.121 g, 0.350 mmol), and aq Na2CO3 (2 M; 0.637 mL, 1.27 mmol) in 1,4-dioxane (1.59 mL). Pd(OAc)2 (7.1 mg, 0.032 mmol) and 1,1′-bis(di-tert-butylphosphino)ferrocene (0.017 g, 0.032 mmol) were added and the mixture was heated at 135 °C in a sealed tube for 22 h. The mixture was cooled and purified by ion-exchange chromatography on acidic resin (5 g), eluting with 2 M NH3 in MeOH. The fractions containing product were concentrated and purified by silica column chromatography, eluting with 5% MeOH–CH2Cl2, to give 2 (0.047 g, 44%). 1H NMR (500 MHz, DMSO-d6) δ 13.18 (s, 1H), 8.51 (t, J = 1.1 Hz, 1H), 8.19 (t, J = 1.2 Hz, 1H), 8.07–7.79 (m, 3H), 7.66 (dt, J = 8.9, 0.9 Hz, 1H), 6.34 (s, 1H), 3.32 (s, 2H), 1.23–1.14 (m, 1H), 0.51–0.45 (m, 2H), 0.35–0.27 (m, 2H); 13C NMR (126 MHz, DMSO-d6) δ 147.6, 143.7, 139.1, 134.2, 132.9, 129.0, 128.8, 125.5, 122.9, 120.8, 118.4, 110.4, 90.6, 46.1, 10.2, 3.4 (2C); LCMS (2 min) tR = 1.50 min, m/z (ESI+) 339 (M + H+); high-resolution mass spectrometry (HRMS) m/z calcd for C17H16N635Cl (M + H) 339.1125, found 338.1133.
6-Chloro-N-(cyclopropylmethyl)-3-(1-methyl-1H-indazol-5-yl)imidazo[1,2-b]pyridazin-8-amine (5)
A mixture of 1-methyl-5-(4,4,5,5-tetramethyl-1,3,2-dioxaborolan-2-yl)-1H-indazole (0.086 g, 0.333 mmol), 4 (0.084 g, 0.278 mmol), aqueous Na2CO3 (2 M; 0.56 mL, 1.11 mmol), and Pd(OAc)2 (6.23 mg, 0.028 mmol), and 1,1′-bis(di-tert-butylphosphino)ferrocene (0.013 g, 0.028 mmol) in 1,4-dioxane (1.4 mL) was heated at 120 °C in a sealed tube for 22 h. The mixture was cooled, absorbed onto silica gel, and purified by silica column chromatography, eluting with 70% EtOAc–hexanes, to give 5 (0.046 g, 47%). 1H NMR (500 MHz, DMSO-d6) δ 8.52 (dd, J = 1.6, 0.8 Hz, 1H), 8.17 (d, J = 1.0 Hz, 1H), 8.02–7.93 (m, 3H), 7.76 (dt, J = 9.0, 0.9 Hz, 1H), 6.34 (s, 1H), 4.09 (s, 3H), 3.31–3.17 (m, 2H), 1.21–1.15 (m, 1H), 0.51–0.44 (m, 2H), 0.35–0.26 (m, 2H); 13C NMR (126 MHz, DMSO-d6) δ 148.1, 144.1, 139.3, 133.5, 133.4, 129.5, 129.1, 125.9, 124.0, 121.4, 118.9, 110.5, 91.1, 46.5, 35.9, 10.7, 3.9 (2C); LCMS (2 min) tR = 1.56 min, m/z (ESI+) 353 (M + H+); HRMS m/z calcd for C18H18N635Cl (M + H) 353.1281, found 353.1275.
6-Chloro-N-(cyclopropylmethyl)-3-(1H-indazol-6-yl)imidazo[1,2-b]pyridazin-8-amine (6)
N2 was bubbled for 10 min through a mixture of 4 (0.073 g, 0.242 mmol), 6-(4,4,5,5-tetramethyl-1,3,2-dioxaborolan-2-yl)-1H-indazole (0.065 g, 0.266 mmol), and aqueous Na2CO3 (2 M; 0.48 mL, 0.968 mmol) in 1,4-dioxane (1.21 mL). Pd(OAc)2 (5.4 mg, 0.024 mmol) and 1,1′-bis(di-tert-butylphosphino)ferrocene (0.013 g, 0.024 mmol) were added and the mixture was heated at 135 °C in a sealed tube for 15 h. The mixture was cooled and purified by ion-exchange chromatography on acidic resin (5 g), eluting with 2 M NH3 in MeOH. The fractions containing product were concentrated and purified by silica column chromatography, eluting with 5% MeOH–CH2Cl2, to give 6 (0.042 g, 51%). 1H NMR (500 MHz, DMSO-d6) δ 13.32–13.16 (m, 1H), 8.49 (q, J = 1.1 Hz, 1H), 8.12–8.08 (m, 2H), 8.03 (t, J = 6.1 Hz, 1H), 7.86 (dd, J = 8.5, 0.9 Hz, 1H), 7.71 (dt, J = 8.5, 1.0 Hz, 1H), 6.40 (d, J = 1.5 Hz, 1H), 3.34 (s, 2H), 1.24–1.14 (m, 1H), 0.53–0.45 (m, 2H), 0.35–0.28 (m, 2H); 13C NMR (126 MHz, DMSO-d6) δ 147.7, 143.7, 140.0, 139.9, 133.5, 130.0, 128.3, 126.0, 121.9, 120.8, 119.5, 107.0, 90.9, 46.1, 10.2, 3.4 (2C); LCMS (4 min), tR = 3.21 min, m/z (ESI+) 339 (M + H+); HRMS m/z calcd for C17H15N635Cl (M + H) 339.1119, found 339.1115.
6-Chloro-N-(cyclopropylmethyl)-3-(1H-indol-5-yl)imidazo[1,2-b]pyridazin-8-amine (7)
Prepared from 4 and (1H-indol-5-yl)boronic acid using the method described for 6 to give 7 (50%). 1H NMR (500 MHz, DMSO-d6) δ 11.23 (s, 1H), 8.29–8.20 (m, 1H), 7.88 (t, J = 6.1 Hz, 1H), 7.84 (s, 1H), 7.68 (dd, J = 1.7, 8.5 Hz, 1H), 7.50 (dt, J = 0.9, 8.6 Hz, 1H), 7.40 (t, J = 2.8 Hz, 1H), 6.53 (ddd, J = 0.9, 1.9, 3.1 Hz, 1H), 6.30 (s, 1H), 3.25 (s, 2H), 1.19 (dd, J = 2.0, 4.9 Hz, 1H), 0.52–0.45 (m, 2H), 0.34–0.28 (m, 2H); 13C NMR (126 MHz, DMSO-d6) δ 147.4, 143.6, 135.4, 132.5, 129.9, 128.4, 127.6, 126.2, 120.5, 119.3, 118.6, 111.6, 101.6, 90.3, 46.1, 10.3, 3.4 (2C); LCMS (2 min) tR = 1.52 min, m/z (ESI+) 338 (M + H+); HRMS m/z calcd for C18H16N535Cl (M + H) 338.1172, found 338.1161.
6-Chloro-N-(cyclopropylmethyl)-3-(1H-benzo[d]imidazol-5-yl)imidazo[1,2-b]pyridazin-8-amine (8)
Prepared from 4 and 6-(4,4,5,5-tetramethyl-1,3,2-dioxaborolan-2-yl)-1H-benzo[d]imidazole using the method described for 6 with purification by preparative HPLC to give 8 (5%). 1H NMR (500 MHz, DMSO-d6) δ 12.60 (d, J = 36.4 Hz, 1H), 8.39 (d, J = 8.9 Hz, 1H), 8.28 (s, 1H), 7.95 (d, J = 15.4 Hz, 2H), 7.88–7.60 (m, 2H), 6.34 (s, 1H), 3.25 (s, 2H), 1.23–1.14 (m, 1H), 0.52–0.45 (m, 2H), 0.35–0.28 (m, 2H); 13C NMR (126 MHz, DMSO-d6) δ 148.0, 144.1, 143.4, 133.4, 129.5 (2C), 122.7, 121.6, 91.0, 46.5, 10.7, 3.9 (2C); LCMS (2 min) tR = 1.19 min, m/z (ESI+) 339 (M + H+); HRMS m/z calcd for C17H15N635Cl (M + H) 339.1125, found 339.1127.
6-Chloro-N-(cyclopropylmethyl)-3-(2-(phenylamino)-1H-benzo[d]imidazol-5-yl)imidazo[1,2-b]pyridazin-8-amine (9)
A mixture of 5-bromo-2-chloro-1H-benzo[d]imidazole (5.00 g, 21.6 mmol) and aniline (10.1 mL, 110 mmol) was heated by microwave irradiation at 180 °C for 60 min. The mixture was cooled, diluted with EtOAc (60 mL), and washed with water (60 mL) and brine (60 mL). The organic layer was dried and concentrated. Silica chromatography, eluting with 0–2% MeOH–CH2Cl2, gave 5-bromo-N-phenyl-1H-benzo[d]imidazol-2-amine (4.67 g, 75%). 1H NMR (500 MHz, DMSO-d6) δ 11.02 (s, 1H), 9.54 (s, 1H), 7.76–7.69 (m, 2H), 7.46 (s, 1H), 7.35–7.27 (m, 2H), 7.24 (d, J = 8.2 Hz, 1H), 7.11 (dd, J = 2.0, 8.4 Hz, 1H), 6.98–6.91 (m, 1H); 13C NMR (126 MHz, DMSO-d6) δ 151.4, 140.5, 128.8 (2C), 122.9, 122.2, 120.9, 118.1, 117.9, 117.3 (2C), 112.1, 111.0; LCMS (2 min) tR = 0.95 min, m/z (ESI+) 288 and 290 (M + H+); HRMS m/z calcd for C13H10N379Br (M + H) 288.0136, found 288.0148. DMAP (0.4 g, 3.24 mmol) and di-tert-butyl-dicarbonate (8.84 g, 40.5 mmol) were added to a solution of 5-bromo-N-phenyl-1H-benzo[d]imidazol-2-amine (4.67 g, 16.2 mmol) in dry THF (160 mL). The mixture was stirred at rt for 16 h and then concentrated. The residue was dissolved in EtOAc (100 mL) and washed with water (100 mL) and brine (100 mL). The organic layer was dried and concentrated. Silica chromatography, eluting with 0–2% MeOH-CH2Cl2, gave tert-butyl 5-bromo-2-((tert-butoxycarbonyl)(phenyl)amino)-1H-benzo[d]imidazole-1-carboxylate (7.9 g, quantitative) as a mixture of three di-Boc-protected isomers, which was used directly in the next step. LCMS (2 min) tR = 1.69, 1.87, 1.91 min; m/z (ESI+) 488 and 490 (M + H+). A portion of the material (5.4 g, 11.1 mmol) was dissolved in dry 1,4-dioxane (110 mL) and stirred at rt. Bis(pinacolato)diboron (3.37 g, 13.3 mmol), 1,1′-bis(diphenylphosphino)ferrocene-palladium(II) dichloride (0.81 g, 1.11 mmol), and KOAc (2.17 g, 22.1 mmol) were added and the mixture was heated at 100 °C for 15 h. The mixture was cooled, filtered through celite, and concentrated. The residue was dissolved in EtOAc (50 mL) and washed with water (50 mL) and brine (50 mL). The organic layer was dried and concentrated. Silica chromatography, eluting with 0–10% EtOAc–cyclohexane, gave tert-butyl 2-((tert-butoxycarbonyl)(phenyl)amino)-5-(4,4,5,5-tetramethyl-1,3,2-dioxaborolan-2-yl)-1H-benzo[d]imidazole-1-carboxylate 13 (4.40 g, 74%) as a mixture of three di-Boc protected isomers, which was used directly in the next step. LCMS (2 min) tR = 1.70 min, 1.78 min, 1.82 min; m/z (ESI+) 536 (M + H+). The mixture of isomers of 13 was reacted with 4 using the method described for 6 to give 9 (33%). 1H NMR (500 MHz, DMSO-d6) δ 11.14 and 11.01 (2 × s, 1H, NH); 9.53 and 9.50 (2 × s, 1H, NH), 8.06 (dd, J = 1.7, 11.6 Hz, 1H), 7.93–7.84 (m, 2H), 7.82–7.74 (m, 2H), 7.62 (td, J = 1.8, 7.9 Hz, 1H), 7.41 (dd, J = 8.2, 28.3 Hz, 1H), 7.33 (td, J = 2.9, 8.0 Hz, 2H), 6.94 (td, J = 1.2, 7.2 Hz, 1H), 6.31 (s, 1H), 3.25 (s, 2H), 1.23–1.15 (m, 1H), 0.53–0.45 (m, 2H), 0.35–0.29 (m, 2H); 13C NMR (126 MHz, DMSO-d6) δ 151.4, 151.2, 147.5, 143.6, 143.2, 143.1, 140.7, 132.9, 132.7, 132.6, 129.7 (2C), 128.8, 128.5, 120.9, 120.7, 120.0, 119.9, 119.0, 117.2 (2C), 117.1, 115.8, 114.2, 109.5, 107.7, 90.3, 46.1, 10.2, 3.4 (2C); LCMS (2 min) tR = 1.30 min, m/z (ESI+) 430 (M + H+); HRMS m/z calcd for C23H20N735Cl (M + H) 430.1547, found 430.1537.
6-Chloro-N-(cyclopropylmethyl)-3-(2-(methyl(phenyl)amino)-1H-benzo[d]imidazol-5-yl)imidazo[1,2-b]pyridazin-8-amine (10)
A mixture of 5-bromo-2-chloro-1H-benzo[d]imidazole (0.700 g, 3.02 mmol) and N-methylaniline (0.98 mL, 9.07 mmol) in EtOH (7 mL) was heated under microwave irradiation at 180 °C for 30 min. The mixture was cooled and concentrated. Silica chromatography, eluting with 10% EtOAc–cyclohexane and then with 50% EtOH–EtOAc, gave 5-bromo-N-methyl-N-phenyl-1H-benzo[d]imidazol-2-amine (0.800 g, 88%). 1H NMR (500 MHz, DMSO-d6) δ 12.85 (br s, 1H), 7.54 (d, J = 3.1 Hz, 4H), 7.45 (d, J = 1.6 Hz, 1H), 7.41 (tt, J = 2.7, 6.0 Hz, 1H), 7.30–7.23 (m, 2H), 3.56 (s, 3H); 13C NMR (126 MHz, DMSO-d6) δ 152.1, 142.4, 130.1 (2C), 127.3, 125.4 (2C), 124.5, 114.5, 113.6, 113.3 (2 quaternary C not observed; NCH3 obscured by solvent but detected in heteronuclear single quantum coherence NMR); LCMS (2 min) tR = 0.94 min, m/z (ESI+) 302 and 304 (M + H+); HRMS m/z calcd for C14H12N379Br (M + H) 302.0293, found 302.0300. A mixture of 5-bromo-N-methyl-N-phenyl-1H-benzo[d]imidazol-2-amine (0.800 g, 2.65 mmol), bis(pinacolato)diboron (0.690 g, 2.73 mmol), 1,1′-bis(diphenylphosphino)ferrocene-palladium(II) dichloride (0.190 g, 0.260 mmol), and KOAc (0.520 g, 5.30 mmol) in dry 1,4-dioxane (32 mL) was heated at 100 °C for 15 h. The mixture was cooled, filtered through celite, and concentrated. Ion-exchange chromatography on acidic resin, eluting with 2 M NH3 in MeOH, gave N-methyl-N-phenyl-5-(4,4,5,5-tetramethyl-1,3,2-dioxaborolan-2-yl)-1H-benzo[d]imidazol-2-amine (14) (0.29 g, 31%), which was used in the next step without further purification. LCMS (2 min) tR = 1.03 min, m/z (ESI+) 350 (M + H+). The crude product 14 was reacted with 4 using the method described for 6 to give 10 (24%). 1H NMR (500 MHz, DMSO-d6) δ 11.45 (s, 1H), 8.01 (d, J = 1.6 Hz, 1H), 7.90 (t, J = 6.1 Hz, 1H), 7.84 (s, 1H), 7.57 (dd, J = 1.7, 8.2 Hz, 1H), 7.49–7.42 (m, 4H), 7.32 (d, J = 8.1 Hz, 1H), 7.27–7.20 (m, 1H), 6.30 (s, 1H), 3.34 (s, 3H), 3.24 (s, 2H), 1.24–1.11 (m, 1H), 0.52–0.43 (m, 2H), 0.35–0.26 (m, 2H); 13C NMR (126 MHz, DMSO-d6) δ 154.8, 147.4, 144.7, 143.6, 132.6, 129.6, 129.4 (2C), 128.4, 124.8, 123.9 (2C), 119.5, 90.3, 46.1, 38.8, 10.2, 3.4 (2C), 5 quaternary C not observed; LCMS (2 min) tR = 1.25 min, m/z (ESI+) 444 (M + H+); HRMS m/z calcd for C24H22N735Cl (M + H) 444.1703, found 444.1703.
6-Chloro-N-(cyclopropylmethyl)-3-(1-methyl-2-(phenylamino)-1H-benzo[d]imidazol-5-yl)imidazo[1,2-b]pyridazin-8-amine (11)
Isothiocyanatobenzene (0.20 mL, 0.23 mmol) and EDC (0.56 g, 2.9 mmol) were added to a stirred solution of N1-methyl-4-(4,4,5,5-tetramethyl-1,3,2-dioxaborolan-2-yl)benzene-1,2-diamine (15)56 in dry THF (16 mL) at rt. The mixture was heated to 50 °C for 16 h. The mixture was concentrated and the residue was partitioned between water (30 mL) and EtOAc (30 mL). The organic layer was washed with brine (30 mL), dried, and concentrated. Ion-exchange chromatography on acidic resin, eluting with 2 M NH3–MeOH, gave 1-methyl-N-phenyl-5-(4,4,5,5-tetramethyl-1,3,2-dioxaborolan-2-yl)-1H-benzo[d]imidazol-2-amine (16) (0.25 g, 45%). 1H NMR (500 MHz, DMSO-d6) δ 8.94 (s, 1H), 7.82–7.90 (m, 2H), 7.65 (d, J = 0.9 Hz, 1H), 7.41 (dd, J = 1.1, 7.9 Hz, 1H), 7.29–7.35 (m, 3H), 6.96 (tt, J = 1.2, 7.4 Hz, 1H), 3.72 (s, 3H), 1.30 (s, 12H); 13C NMR (126 MHz, DMSO-d6) δ 150.8, 141.4, 140.8, 136.8, 128.6 (2C), 126.3, 122.3, 121.1, 120.1, 117.9 (2C), 107.7, 83.2 (2C), 29.1, 24.7 (4C); LCMS (2 min) tR = 1.10 min, m/z (ESI+) 350 (M + H+); HRMS m/z calcd for C20H25BN3O2 (M + H) 350.2043, found 350.2035. The product 16 was combined with 4 using the method described for 6 with purification by preparative HPLC to give 11 (15%). 1H NMR (500 MHz, DMSO-d6) δ 8.99 (s, 1H), 8.12 (dd, J = 0.5, 1.6 Hz, 1H), 7.94–7.87 (m, 4H), 7.71 (dd, J = 1.6, 8.3 Hz, 1H), 7.42 (d, J = 8.2 Hz, 1H), 7.38–7.30 (m, 2H), 6.97 (tt, J = 1.1, 7.3 Hz, 1H), 6.32 (s, 1H), 3.76 (s, 3H), 3.25 (s, 1H), 1.29–1.13 (m, 2H), 0.52–0.43 (m, 2H), 0.35–0.28 (m, 2H); 13C NMR (126 MHz, DMSO-d6) δ 151.1, 147.5, 143.7, 141.8, 140.8, 134.0, 132.7, 129.5, 128.7, 128.6 (2C), 121.3, 121.2, 118.9, 118.0 (2C), 114.3, 108.2, 90.4, 46.1, 29.2, 10.2, 3.4 (2C); LCMS (2 min) tR = 1.41 min, (ESI+) 444 (M + H+); HRMS m/z calcd for C24H23N735Cl (M + H) 444.1703, found 444.1706.
6-Chloro-N-(cyclopropylmethyl)-3-(1-methyl-2-(phenylamino)-1H-benzo[d]imidazol-6-yl)imidazo[1,2-b]pyridazin-8-amine (12)
Isothiocyanatobenzene was combined with N1-methyl-5-(4,4,5,5-tetramethyl-1,3,2-dioxaborolan-2-yl)benzene-1,2-diamine (17)53 using the method described for 16 to give 1-methyl-N-phenyl-6-(4,4,5,5-tetramethyl-1,3,2-dioxaborolan-2-yl)-1H-benzo[d]imidazol-2-amine (18) (17%). 1H NMR (500 MHz, DMSO-d6) δ 9.00 (s, 1H), 7.81–7.93 (m, 2H), 7.56 (d, J = 1.0 Hz, 1H), 7.42 (dd, J = 1.1, 7.8 Hz, 1H), 7.28–7.38 (m, 3H), 6.97 (tt, J = 1.1, 7.3 Hz, 1H), 3.73 (s, 3H), 1.31 (s, 12H); LCMS (2 min) tR = 1.25 min, 350 (M + H+). The product 18 was combined with 4 using the method described for 6 to give 12 (38%). 1H NMR (500 MHz, DMSO-d6) δ 9.02 (s, 1H), 7.97 (d, J = 1.6 Hz, 1H), 7.96–7.91 (m, 2H), 7.91–7.87 (m, 2H), 7.81 (dd, J = 1.7, 8.3 Hz, 1H), 7.50–7.46 (m, 1H), 7.38–7.30 (m, 2H), 6.97 (tt, J = 1.1, 7.2 Hz, 1H), 6.32 (s, 1H), 3.77 (s, 3H), 3.25 (s, 2H), 1.26–1.13 (m, 1H), 0.52–0.45 (m, 2H), 0.35–0.28 (m, 2H); 13C NMR (126 MHz, DMSO-d6) δ 151.3, 147.5, 143.6, 141.5, 140.7, 134.4, 132.7, 129.5, 128.7, 128.6 (2C), 121.3, 120.2, 120.0, 118.1 (2C), 115.9, 106.3, 90.4, 46.1, 29.1, 10.2, 3.5 (2C); LCMS (2 min) tR = 1.41 min, m/z (ESI+) 444 (M + H+); HRMS m/z calcd for C24H23N735Cl (M + H) 444.1703, found 444.1704.
6-Chloro-3-(1H-indazol-5-yl)-N-(tetrahydro-2H-pyran-4-yl)imidazo[1,2-b]pyridazin-8-amine (19)
A mixture of 3 (150 mg, 0.482 mmol) and tetrahydro-2H-pyran-4-amine (73 mg, 1.5 equiv, 0.723 mmol) in THF (1.2 mL) was stirred at rt for 20 h. The mixture was diluted with THF (3 mL), and polystyrene–benzaldehyde resin (0.50 g, 0.723 mmol) was added. The suspension was gently stirred for 18 h at rt and then purified by ion-exchange chromatography on acidic resin (5 g), eluting with MeOH, 20% MeOH–CH2Cl2, and then 1:1 CH2Cl2/7 M NH3 in MeOH. The basic fractions were concentrated and dried in vacuo to give 3-bromo-6-chloro-N-(tetrahydro-2H-pyran-4-yl)imidazo[1,2-b]pyridazin-8-amine (144 mg, 90%), which was used directly in the next step. A sample of the crude material (60 mg, 0.18 mmol) was added to a mixture of 5-(4,4,5,5-tetramethyl-1,3,2-dioxaborolan-2-yl)-1H-indazole (53 mg, 0.216 mmol), 1,1′-bis(di-tert-butylphosphino)ferrocene (8.54 mg, 0.018 mmol), Pd(OAc)2 (4.04 mg, 0.018 mmol), and aq Na2CO3 (2 M; 0.360 mL, 0.720 mmol) in 1,4-dioxane (1.5 mL). The mixture was heated in a sealed tube at 130 °C for 18 h and then cooled and purified by ion-exchange chromatography on acidic resin (2 g), eluting with MeOH and then with 1:1 CH2Cl2/7 M NH3 in MeOH. The basic fractions were concentrated. Preparative HPLC, followed by preparative thin-layer chromatography, eluting with 1% NH3 (aq) in 7% MeOH–CH2Cl2, gave 19 (2.90 mg, 4%). 1H NMR (500 MHz, MeOH-d4) δ 8.54 (dd, J = 0.9, 1.6 Hz, 1H), 8.14 (d, J = 1.0 Hz, 1H), 7.95 (dd, J = 1.6, 8.8 Hz, 1H), 7.81 (s, 1H), 7.63 (dt, J = 1.0, 8.9 Hz, 1H), 6.34 (s, 1H), 4.01 (dt, J = 3.7, 11.9 Hz, 2H), 3.84 (d, J = 10.4 Hz, 1H), 3.60 (td, J = 2.2, 11.6 Hz, 2H), 2.06 (ddt, J = 2.3, 4.6, 12.2 Hz, 2H), 1.69 (dtd, J = 4.3, 10.8, 13.2 Hz, 2H); 13C NMR (126 MHz, MeOH-d4) δ 149.9, 143.4, 135.6, 134.5, 132.4, 131.1, 129.5, 127.6, 124.4, 122.6, 120.5, 111.3, 93.2, 67.4, 49.6 (2C), 33.3 (2C); LCMS (2 min) tR = 1.35 min, m/z (ESI+) 369 (M + H+); HRMS m/z calcd for C18H18N6O35Cl (M + H) 369.1225, found 369.1216.
6-Chloro-3-(1H-indazol-5-yl)-N-(1-methylpiperidin-4-yl)imidazo[1,2-b]pyridazin-8-amine (20)
Prepared from 3, 1-methylpiperidin-4-amine, and 5-(4,4,5,5-tetramethyl-1,3,2-dioxaborolan-2-yl)-1H-indazole using the method described for 19 to give 20 (11% over two steps). 1H NMR (600 MHz, DMSO-d6) δ 13.2 (s, 1H), 8.51 (s, 1H), 8.20 (s, 1H), 8.00 (s, 1H), 7.98 (s, 1H), 7.74 (br d, J = 8.5 Hz, 1H), 7.67 (br d, J = 8.8 Hz, 1H), 6.40 (s, 1H), 3.66 (br s, 1H), 2.95–2.93 (m, 2H), 2.33 (br s, 5H), 1.93–1.90 (m, 2H), 1.82–1.76 (m, 2H); 13C NMR (151 MHz, DMSO-d6) δ 148.1, 143.3, 139.6, 134.6, 133.3, 129.4, 129.3, 125.9, 123.4, 121.2, 118.9, 110.9, 91.4, 55.4, 54.2, 45.4, 34.2, 31.8 (2C); LCMS (2 min) tR = 0.94 min, m/z (ESI+) 382 (M + H+); HRMS m/z calcd for C19H21N735Cl (M + H) 382.1547, found 382.1542.
6-Chloro-3-(1H-indazol-5-yl)-N-((1-methylpiperidin-4-yl)methyl)imidazo[1,2-b]pyridazin-8-amine (21)
Prepared from 3, (1-methylpiperidin-4-yl)methanamine, and 5-(4,4,5,5-tetramethyl-1,3,2-dioxaborolan-2-yl)-1H-indazole using the method described for 19 to give 21 (10% over two steps). 1H NMR (500 MHz, MeOH-d4) δ 8.53 (t, J = 0.7 Hz, 1H), 8.14 (d, J = 0.9 Hz, 1H), 7.94 (dd, J = 1.6 and 8.8 Hz, 1H), 7.78 (s, 1H), 7.62 (d, J = 8.8 Hz, 1H), 6.20 (s, 1H), 3.23 (d, J = 7.0 Hz, 2H), 2.94 (br d, J = 12 Hz, 2H), 2.31 (s, 3H), 2.10–2.05 (m, 2H), 1.86 (br d, J = 13 Hz, 2H), 1.74–1.70 (m, 1H), 1.42–1.34 (m, 2H); 13C NMR (126 MHz, Methanol-d4) δ 148.4, 143.4, 139.6, 134.1, 133.0, 129.5, 128.1, 126.1, 122.9, 121.2, 118.9, 110.0, 91.4, 54.8, 44.8, 34.5, 29.2; LCMS (2 min) tR = 1.09 min, m/z (ESI+) 396 (M + H+); HRMS m/z calcd for C20H23N735Cl (M + H) 396.1703, found 396.1707.
(1r,4r)-N1-(6-Chloro-3-(1H-indazol-5-yl)imidazo[1,2-b]pyridazin-8-yl)cyclohexane-1,4-diamine (22)
tert-Butyl ((1r,4r)-4-aminocyclohexyl)carbamate (0.929 g, 4.34 mmol) was added to a stirred solution of 3 (0.450 g, 1.45 mmol) in dimethylformamide (10 mL) at rt. The reaction was heated at 50 °C for 16 h, then cooled and partitioned between Na2CO3 aq and EtOAc. The organic layer was washed with brine, dried, and concentrated. Silica chromatography, eluting with 0–40% EtOAc–cyclohexane, gave tert-butyl ((1r,4r)-4-((3-bromo-6-chloroimidazo[1,2-b]pyridazin-8-yl)amino)cyclohexyl)-carbamate (0.643 g, 100%). 1H NMR (500 MHz, CDCl3) δ 7.48 (s, 1H), 6.16 (s, 1H), 5.97 (d, J = 7.7 Hz, 1H), 4.08 (d, J = 13.4 Hz, 1H), 3.79 (s, 1H), 3.56 (s, 1H), 3.11 (ddd, J = 13.1, 9.6, 3.4 Hz, 1H), 3.03 (dd, J = 13.3, 8.5 Hz, 1H), 2.16–2.09 (m, 1H), 1.84 (d, J = 14.3 Hz, 1H), 1.75–1.64 (m, 2H), 1.48 (s, 9H); LCMS (2 min) tR = 1.57 min m/z, (ESI+) 446 (M + H+). A mixture of tert-butyl ((1r,4r)-4-((3-bromo-6-chloroimidazo[1,2-b]pyridazin-8-yl)amino)cyclohexyl)-carbamate (0.100 g, 0.225 mmol), 5-(4,4,5,5-tetramethyl-1,3,2-dioxaborolan-2-yl)-1H-indazole (0.055 g, 0.225 mmol), Pd(OAc)2 (5.05 mg, 0.022 mmol), 1,1′-bis(di-t-butylphosphino)ferrocene (0.011 g, 0.022 mmol), and Na2CO3 aq (2 M; 0.45 mL, 0.900 mmol) in 1,4-dioxane (2 mL) was heated at 130 °C in a sealed tube for 24 h. The mixture was cooled, filtered through celite, and absorbed onto silica. Chromatography, eluting with 0–50% EtOAc–cyclohexane, gave tert-butyl ((1r,4r)-4-((6-chloro-3-(1H-indazol-5-yl)imidazo[1,2-b]pyridazin-8-yl)amino)cyclohexyl)carbamate (0.060 g, 55%). 1H NMR (500 MHz, CDCl3) δ 8.50 (s, 1H), 8.22 (s, 1H), 7.94 (d, J = 8.7 Hz, 1H), 7.74 (s, 1H), 7.61 (d, J = 8.8 Hz, 1H), 6.18 (d, J = 8.0 Hz, 1H), 6.04 (d, J = 7.0 Hz, 1H), 4.54–4.47 (m, 1H), 3.54 (s, 1H), 3.47–3.37 (m, 1H), 2.52–2.12 (m, 4H), 1.48 (s, 9H), 1.37–1.24 (m, 4H); LCMS (2 min) tR = 1.56 min m/z, (ESI+) 482 (M + H+). tert-Butyl ((1r,4r)-4-((6-chloro-3-(1H-indazol-5-yl)imidazo[1,2-b]pyridazin-8-yl)amino)cyclohexyl)carbamate (30 mg, 0.062 mmol) was dissolved in HCl–1,4-dioxane (4 M; 1 mL) and stirred at rt for 2 h. The mixture was concentrated. Ion-exchange chromatography on acidic resin, eluting with 2 M NH3–MeOH, followed by preparative HPLC gave 22 (24 mg, 99%). 1H NMR (500 MHz, DMSO-d6) δ 8.50 (t, J = 1.2 Hz, 1H), 8.19 (d, J = 1.0 Hz, 1H), 7.96–7.89 (m, 2H), 7.69–7.62 (m, 1H), 7.55 (s, 1H), 6.34 (s, 1H), 3.52 (s, 1H), 3.17 (br s, 3H), 2.57 (tt, J = 10.5, 3.6 Hz, 1H), 1.99–1.71 (m, 4H), 1.50 (q, J = 13.3, 12.7 Hz, 2H), 1.25–1.15 (m, 2H); 13C NMR (126 MHz, DMSO-d6) δ 165.6, 147.7, 142.78, 139.3, 134.0, 132.8, 128.9, 125.4, 122.9, 120.8, 118.4, 110.6, 90.9, 49.7, 48.5, 29.5 (2C), 29.3 (2C); LCMS (2 min) tR = 1.11 min, m/z, (ESI+), 382 (M + H+); HRMS m/z calcd for C19H20N735Cl (M + H) 382.1547, found 382.1538.
(1r,4r)-N1-(6-Chloro-3-(1H-indazol-5-yl)imidazo[1,2-b]pyridazin-8-yl)-N1-methylcyclohexane-1,4-diamine (23)
LiAlH4 in THF (1 M; 25 mL, 25.0 mmol) was added to a solution of tert-butyl ((1r,4r)-4-aminocyclohexyl)carbamate (1.30 g, 6.07 mmol) in THF (5 mL) at 0 °C. The mixture was heated to reflux for 4 h, forming a thick white suspension. The reaction was cooled, diluted with Et2O (80 mL), and water (0.95 mL) was slowly added dropwise, followed by 15% w/v NaOH aq (0.95 mL). After stirring for 1 h, water (2.85 mL) was added dropwise. The mixture was filtered through celite, washed with Et2O, and the filtrate was concentrated to give crude (1r,4r)-N1-methylcyclohexane-1,4-diamine (0.597 g). A solution of the crude amine, Et3N (1.3 mL, 9.31 mmol), and ethyl 1,3-dioxoisoindoline-2-carboxylate (0.928 g, 4.23 mmol) in CH2Cl2 (42 mL) was stirred at rt for 15 h. Saturated (NH4)2CO3 aq (50 mL) was added, and the mixture was extracted with CH2Cl2 (3 × 30 mL). The combined organic layers were dried and concentrated to give crude 2-((1r,4r)-4-(methylamino)cyclohexyl)isoindoline-1,3-dione (0.99 g). A mixture of the crude amine, 3 (0.918 g, 2.95 mmol), and Et3N (0.82 mL, 5.90 mmol) in 1,4-dioxane (3.7 mL) and DMSO (3.7 mL) was heated at 100 °C for 21 h. The mixture was cooled and diluted with CH2Cl2 (15 mL). The solid was collected and washed with dichloromethane (DCM, 5 mL) to give 2-((1r,4r)-4-((3-bromo-6-chloroimidazo[1,2-b]pyridazin-8-yl)(methyl)amino)cyclohexyl)isoindoline-1,3-dione (1.06 g, 52% over three steps). 1H NMR (500 MHz, DMSO-d6) δ 7.88–7.82 (m, 4H), 7.80 (s, 1H), 6.26 (s, 1H), 5.76 (br s, 1H), 4.18–4.08 (m, 1H), 3.10 (br s, 3H), 2.39–2.25 (m, 2H), 1.97–1.77 (m, 6H); LCMS (4 min) tR = 3.53 min, m/z (ESI+) 490 (M + H+). A mixture of 2-((1r,4r)-4-((3-bromo-6-chloroimidazo[1,2-b]pyridazin-8-yl)(methyl)amino)cyclohexyl)-isoindoline-1,3-dione (0.210 g, 0.430 mmol), 5-(4,4,5,5-tetramethyl-1,3,2-dioxaborolan-2-yl)-1H-indazole (0.126 g, 0.516 mmol), Pd(OAc)2 (9.6 mg, 0.043 mmol), 1,1′-bis(di-tert-butylphosphino)ferrocene (0.020 g, 0.043 mmol), and Na2CO3 aq (2 M; 0.86 mL, 1.72 mmol) in 1,4-dioxane (2.1 mL) was heated at 130 °C for 21 h. The reaction mixture was cooled and NH2NH2·H2O (0.5 mL) was added. The mixture was heated at 100 °C for 48 h. The mixture was cooled and purified by ion-exchange chromatography on acidic resin, eluting with 2 M NH3 in MeOH. Reversed-phase silica chromatography, eluting with 10–90% MeOH–water containing 0.1% HCO2H, followed by normal phase chromatography, eluting with 2–20% EtOH–CH2Cl2, gave 23 (0.058 g, 34%). 1H NMR (600 MHz, DMSO-d6) δ 13.20 (s, 1H), 8.47–8.44 (m, 1H), 8.20 (d, J = 0.9 Hz, 1H), 7.96 (s, 1H), 7.90 (dd, J = 8.7, 1.6 Hz, 1H), 7.67 (d, J = 8.7 Hz, 1H), 6.19 (s, 1H), 5.70 (br s, 1H), 3.07 (br s, 3H), 2.57 (tt, J = 11.1, 3.9 Hz, 1H), 1.88–1.81 (m, 2H), 1.76–1.70 (m, 4H), 1.62 (br s, 2H), 1.32–1.18 (m, 2H); 13C NMR (151 MHz, DMSO-d6) δ 147.6, 145.1, 139.7, 134.5, 133.7, 129.3, 128.9, 126.3, 123.3, 121.2, 119.4, 110.9, 94.1, 57.9, 50.0, 35.8 (2C), 32.7, 28.9 (2C); LCMS (4 min) tR = 2.26 min, m/z (ESI+) 396 (M + H+); HRMS m/z calcd for C20H22N735Cl (M + H) 396.1703, found 396.1700.
N-(trans-4-((6-Chloro-3-(1H-indazol-5-yl)imidazo[1,2-b]pyridazin-8-yl)amino)cyclohexyl)acetamide (24)
Prepared from 3, N-((1r,4r)-4-aminocyclohexyl)acetamide, and 5-(4,4,5,5-tetramethyl-1,3,2-dioxaborolan-2-yl)-1H-indazole using the methods described for 19, except that the first step was conducted at 50 °C, to give 24 (35% over two steps). 1H NMR (500 MHz, DMSO-d6) δ 13.18 (s, 1H), 8.50 (dt, J = 0.8, 1.6 Hz, 1H), 8.19 (t, J = 1.3 Hz, 1H), 7.96–7.90 (m, 2H), 7.80 (d, J = 7.7 Hz, 1H), 7.65 (dt, J = 0.9, 8.7 Hz, 1H), 7.61 (d, J = 8.8 Hz, 1H), 6.42 (s, 1H), 3.60 (s, 1H), 3.50 (dtt, J = 4.1, 8.1, 11.9 Hz, 1H), 1.93 (t, J = 8.2 Hz, 2H), 1.79 (s, 5H), 1.56 (d, J = 12.8 Hz, 2H), 1.41–1.27 (m, 2H); 13C NMR (126 MHz, DMSO-d6) δ 168.3, 147.7, 142.8, 139.1, 134.2, 132.9, 128.9, 128.8, 125.5, 122.9, 120.8, 118.4, 110.4, 90.7, 50.1, 47.2, 30.9 (2C), 30.3 (2C), 22.8; LCMS (2 min), tR = 1.32 min, m/z (ESI+) 424 (M + H+); HRMS m/z calcd for C21H23N7O35Cl (M + H) 424.1647, found 424.1635.
6-Chloro-3-(1H-indazol-5-yl)-N-(trans-4-morpholinocyclohexyl)imidazo[1,2-b]pyridazin-8-amine, Formate Salt (25)
Prepared from 3, (1r,4r)-4-morpholinocyclohexan-1-amine, and 5-(4,4,5,5-tetramethyl-1,3,2-dioxaborolan-2-yl)-1H-indazole using the methods described for 19, except that the first step was conducted at 50 °C, to give 25 (4% over two steps), isolated as the formate salt from preparative HPLC. 1H NMR (500 MHz, MeOH-d4) δ 8.56–8.52 (m, 1H), 8.36 (s, 1H), 8.14 (d, J = 1.0 Hz, 1H), 7.94 (dd, J = 1.6, 8.8 Hz, 1H), 7.79 (s, 1H, formate), 7.63 (d, J = 8.6 Hz, 1H), 6.26 (s, 1H), 3.88 (t, J = 4.7 Hz, 4H), 3.55–3.51 (m, 1H), 3.13 (t, J = 4.6 Hz, 4H), 3.00 (ddd, J = 3.3, 8.4, 12.1 Hz, 1H), 2.28–2.15 (m, 4H), 1.63 (qd, J = 3.2, 12.6 Hz, 2H), 1.46 (qd, J = 2.9, 13.1 Hz, 2H); 13C NMR (126 MHz, MeOH-d4) δ 167.8 (formate), 149.8, 143.5, 141.1, 135.5, 134.4, 131.0, 129.6, 127.5, 124.4, 122.6, 120.4, 111.4, 93.1, 65.9 (2C), 65.4, 51.4, 50.4 (2C), 31.4 (2C), 26.6 (2C); LCMS (2 min), tR = 1.00 min, m/z (ESI+) 452 (M + H+); HRMS m/z calcd for C23H27N7O35Cl (M + H) 452.1966, found 452.1961.
2-(((1r,4r)-4-((6-Chloro-3-(1H-indazol-5-yl)imidazo[1,2-b]pyridazin-8-yl)amino)cyclohexyl)amino)-1-morpholinoethan-1-one (26)
A solution of 2-chloro-1-morpholinoethanone (0.66 mL, 5.05 mmol), tert-butyl ((1r,4r)-4-aminocyclohexyl)carbamate (902 mg, 4.21 mmol), and Et3N (2.1 mL, 15.1 mmol) in CH2Cl2 (35 mL) was stirred at rt for 20 h. The mixture was partitioned between water (50 mL) and CH2Cl2 (30 mL × 2). The combined organic layers were dried and concentrated. The resulting crude material was dissolved in MeOH (42 mL), and HCl in 1,4-dioxane (4 M; 42 mL, 168 mmol) was added. After 2 h at rt, the mixture was concentrated and purified by ion-exchange chromatography on acidic resin, eluting with 2 M NH3 in MeOH, to give 2-(((1r,4r)-4-aminocyclohexyl)amino)-1-morpholinoethan-1-one (0.375 g, 37%). 1H NMR (500 MHz, CDCl3) δ 3.71–3.59 (m, 7H), 3.49–3.36 (m, 4H), 2.72–2.58 (m, 3H), 2.42–2.33 (m, 1H), 1.94–1.78 (m, 4H), 1.24–1.04 (m, 4H). A sample of the material (0.370 g, 1.55 mmol), 3 (0.440 g, 1.41 mmol), and Et3N (0.39 mL, 2.83 mmol) in 1,4-dioxane (1.8 mL) and DMSO (1.8 mL) was heated at 50 °C for 23 h. The mixture was cooled and purified by ion-exchange chromatography on acidic resin, eluting with 2 M NH3 in MeOH, and then by silica chromatography, eluting with 2–20% EtOH–CH2Cl2, to give 2-(((1r,4r)-4-((3-bromo-6-chloroimidazo[1,2-b]pyridazin-8-yl)amino)cyclohexyl)amino)-1-morpholinoethan-1-one (0.38 g, 58%). 1H NMR (500 MHz, CDCl3) δ 7.44 (s, 1H), 6.04 (s, 1H), 5.80 (d, J = 8.1 Hz, 1H), 3.75–3.62 (m, 7H), 3.47 (s, 2H), 3.43–3.40 (m, 3H), 2.56–2.47 (m, 1H), 2.23–2.15 (m, 2H), 2.10–2.01 (m, 2H), 1.43–1.29 (m, 4H); LCMS (2 min) tR = 0.97 min, m/z (ESI+) 473 (M + H+). A sample of the material (0.365 g, 0.774 mmol), 5-(4,4,5,5-tetramethyl-1,3,2-dioxaborolan-2-yl)-1H-indazole (0.227 g, 0.928 mmol), Pd(OAc)2 (17.0 mg, 0.077 mmol), 1,1′-bis(di-tert-butylphosphino)ferrocene (37.0 mg, 0.077 mmol), and Na2CO3 aq (2 M; 1.55 mL, 3.09 mmol) were mixed in 1,4-dioxane (3.9 mL) and heated at 135 °C for 19 h. The mixture was cooled and purified by ion-exchange chromatography on acidic resin, eluting with 2 M NH3 in MeOH, then by silica chromatography, eluting with 2–20% MeOH-CH2Cl2, and then by reversed-phase chromatography, eluting with 10–70% MeOH–water with 0.1% formic acid, to give 26 (0.069 g, 18%). 1H NMR (500 MHz, MeOH-d4) δ 8.49 (dd, J = 1.6, 0.9 Hz, 1H), 8.11 (d, J = 1.0 Hz, 1H), 7.89 (dd, J = 8.8, 1.6 Hz, 1H), 7.72 (s, 1H), 7.59 (d, J = 8.7 Hz, 1H), 6.17 (s, 1H), 3.67–3.63 (m, 4H), 3.59–3.55 (m, 2H), 3.46 (s, 2H), 3.45–3.42 (m, 3H), 2.50–2.41 (m, 1H), 2.13–2.06 (m, 2H), 2.02–1.96 (m, 2H), 1.40–1.24 (m, 4H); 13C NMR (126 MHz, MeOH-d4) δ 171.2, 149.8, 143.5, 141.0, 135.5, 134.4, 130.9, 129.4, 127.5, 124.4, 122.6, 120.3, 111.3, 92.9, 67.7, 67.6, 57.2, 52.2, 48.0, 46.1, 43.4, 32.0 (2C), 31.6 (2C); LCMS (2 min) tR = 1.05 min, m/z (ESI+) 509 (M + H+); HRMS m/z calcd for C25H30N8O235Cl (M + H) 509.2180, found 509.2187.
6-Chloro-3-(2-(phenylamino)-1H-benzo[d]imidazol-5-yl)-N-(tetrahydro-2H-pyran-4-yl)imidazo[1,2-b]pyridazin-8-amine (27)
Prepared from 4, tetrahydro-2H-pyran-4-amine, and 13 using the method described for 19 to give 27 (14% over two steps). 1H NMR (500 MHz, DMSO-d6) δ 11.13 and 11.01 (2 × s, 1H, NH); 9.53 and 9.50 (2 × s, 1H, NH), 8.06 (dd, J = 1.7, 12.9 Hz, 1H), 7.87 (d, J = 11.0 Hz, 1H), 7.82–7.75 (m, 2H), 7.71 (dd, J = 6.7, 8.7 Hz, 1H), 7.64–7.58 (m, 1H), 7.41 (dd, J = 8.3, 28.4 Hz, 1H), 7.36–7.29 (m, 2H), 6.94 (tt, J = 1.2, 7.3 Hz, 1H), 6.43 (s, 1H), 3.83–3.96 (m, 2H), 3.45 (td, J = 2.0, 11.7 Hz, 2H), 3.33 (s, 1H), 1.92–1.79 (m, 2H), 1.78–1.64 (m, 2H); 13C NMR (126 MHz, DMSO-d6) δ 151.4, 151.2, 147.5, 143.2, 143.1, 142.6, 140.7, 132.9, 132.6, 129.7, 128.8 (2C), 128.5, 120.9, 120.7, 120.0, 117.2 (2C), 117.1, 115.8, 114.2, 109.5, 107.7, 90.6, 66.0 (2C), 48.2, 31.8 (2C); LCMS (4 min) tR = 2.60 min, m/z (ESI+) 460 (M + H+); HRMS m/z calcd for C24H22N7O35Cl (M + H) 460.1653, found 460.1651.
6-Chloro-N-(1-methylpiperidin-4-yl)-3-(2-(phenylamino)-1H-benzo[d]imidazol-5-yl)imidazo[1,2-b]pyridazin-8-amine (28)
Prepared from 3, 1-methylpiperidin-4-amine, and 13 using the method described for 19 to give 28 (5% over two steps). 1H NMR (500 MHz, DMSO-d6) δ 11.14 and 11.02 (2 × s, 1H, NH), 9.54 and 9.51 (2 × s, 1H, NH), 8.11–8.10 (m, 1H), 7.86 (d, J = 10.6 Hz, 1H), 7.78 (d, J = 7.9 Hz, 2H), 7.65–7.57 (m, 2H), 7.41 (dd, J = 8.3, 28.5 Hz, 1H), 7.36–7.28 (m, 2H), 6.97–6.90 (m, 1H), 6.35 (s, 1H), 3.57 (d, J = 2.3 Hz, 1H), 2.80 (d, J = 11.1 Hz, 2H), 2.21 (s, 3H), 2.08 (t, J = 11.7 Hz, 2H), 1.86 (d, J = 12.2 Hz, 2H), 1.80–1.67 (m, 2H); 13C NMR (126 MHz, DMSO-d6) δ 151.4, 151.2, 147.5, 143.2, 143.1, 142.8, 140.7, 132.9, 132.6, 129.7, 128.8 (2C), 128.4, 120.9, 120.7, 119.9, 119.0, 117.2 (2C), 117.1, 115.8, 114.2, 109.5, 107.7, 90.6, 54.1 (2C), 48.8, 45.8, 30.7 (2C); LCMS (2 min) tR = 0.86 min, m/z (ESI+) 473 (M + H+); HRMS m/z calcd for C25H25N835Cl (M + H) 473.1969, found 473.1961.
6-Chloro-N-((1-methylpiperidin-4-yl)methyl)-3-(2-(phenylamino)-1H-benzo[d]imidazol-5-yl)imidazo[1,2-b]pyridazin-8-amine (29)
Prepared from 3, (1-methylpiperidin-4-yl)methanamine, and 13 using the method described for 19 to give 29 (13% over two steps). 1H NMR (500 MHz, DMSO-d6) δ 11.19 and 11.07 (2 × s, 1H, NH), 9.57 and 9.54 (2 × s, 1H, NH), 8.05 (d, J = 13.4 Hz, 1H), 7.93 (s, 1H), 7.85 (d, J = 9.2 Hz, 1H), 7.79 (d, J = 8.0 Hz, 2H), 7.61 (d, J = 8.2 Hz, 1H), 7.40 (dd, J = 8.2, 9.4 Hz, 1H), 7.32 (t, J = 7.8 Hz, 2H), 6.93 (tt, J = 1.2, 7.3 Hz, 1H), 6.30 (s, 1H), 3.23 (s, 2H), 2.79 (d, J = 11.2 Hz, 2H), 2.17 (s, 3H), 1.96–1.81 (m, 2H), 1.74–1.60 (m, 3H), 1.32–1.16 (m, 2H); 13C NMR (126 MHz, DMSO-d6) δ 151.4, 151.3, 147.5, 143.9, 143.2, 143.1, 140.8, 132.9, 132.6, 129.7, 128.8 (2C), 128.4, 120.7, 119.9, 119.0, 117.2 (2C), 115.8, 114.2, 109.5, 107.7, 90.3, 54.9 (2C), 47.2, 45.9, 34.2, 29.4 (2C); LCMS (2 min) tR = 1.03 min, m/z (ESI+) 487 (M + H+); HRMS m/z calcd for C26H27N835Cl (M + H) 487.2125, found 487.2130.
6-Chloro-3-(7-fluoro-2-(phenylamino)-1H-benzo[d]imidazol-5-yl)-N-((1-methylpiperidin-4-yl)methyl)imidazo[1,2-b]pyridazin-8-amine (30)
Prepared from 5-bromo-3-fluorobenzene-1,2-diamine and 3-bromo-6-chloro-N-((1-methylpiperidin-4-yl)methyl)imidazo[1,2-b]pyridazin-8-amine using the method described for 31 to give 30 (5% over 4 steps). 1H NMR (500 MHz, DMSO-d6) δ 11.67 (s, 1H), 9.82 (s, 1H), 7.97 (t, J = 6.5 Hz, 1H), 7.94 (s, 2H), 7.83–7.72 (m, 2H), 7.54 (d, J = 12.2 Hz, 1H), 7.39–7.29 (m, 2H), 6.96 (tt, J = 1.2, 7.3 Hz, 1H), 6.33 (s, 1H), 3.24 (s, 2H), 2.80 (d, J = 11.2 Hz, 2H), 1.88 (t, J = 11.5 Hz, 2H), 2.18 (s, 3H), 1.77–1.61 (m, 3H), 1.32–1.17 (m, 2H); 13C NMR (151 MHz, DMSO-d6) δ 152.0, 148.1, 144.3, 140.9, 133.4, 129.5, 129.3 (2C), 128.9, 121.5, 120.8, 117.9 (2C), 105.8 (d, JC–F = 16.6 Hz), 104.5, 91.0, 55.5 (2C), 47.8, 46.6, 34.8, 30.0 (2C) (3 quaternary C not observed); LCMS (2 min) tR = 1.13 min, m/z (ES+) 505 (M + H+); HRMS m/z calcd for C26H26N8F35Cl (M + H) 505.2031, found 505.2036.
6-Chloro-3-(6-fluoro-2-(phenylamino)-1H-benzo[d]imidazol-5-yl)-N-((1-methylpiperidin-4-yl)methyl)imidazo[1,2-b]pyridazin-8-amine (31)
A mixture of (1-methylpiperidin-4-yl)methanamine (1.24 g, 9.64 mmol), 3 (2.00 g, 6.42 mmol), and Et3N (1.79 mL, 12.9 mmol) in 1,4-dioxane (4 mL) and DMSO (4 mL) was stirred at 50 °C for 18 h. The mixture was cooled, diluted with EtOAc, and the organic layer was washed sequentially with water and brine. The organic layer was dried and concentrated. Ion-exchange chromatography on acidic resin, eluting with 2 M NH3 in MeOH, followed by silica chromatography, eluting with 0–5% MeOH–CH2Cl2 and then with 5–10% MeOH–DCM + 0.2 M NH3, gave 3-bromo-6-chloro-N-((1-methylpiperidin-4-yl)methyl)imidazo[1,2-b]pyridazin-8-amine (2.00 g, 87%), which was used directly in subsequent steps. LCMS (2 min) tR = 0.84 min, m/z (ESI+) 360 (M + H+). Separately, isothiocyanatobenzene (1.22 mL, 10.2 mmol) and EDC (3.37 g, 17.5 mmol) were added to a stirred solution of 4-bromo-5-fluorobenzene-1,2-diamine (2.00 g, 9.75 mmol) in dry THF (100 mL) at rt. The mixture was heated at 50 °C for 4 h, cooled, and concentrated. The residue was partitioned between water (100 mL) and EtOAc (100 mL). The organic layer was washed with brine (100 mL), dried, and concentrated. Ion-exchange chromatography on acidic resin, eluting with MeOH, gave 5-bromo-6-fluoro-N-phenyl-1H-benzo[d]imidazol-2-amine that was used directly in the next step (2.53 g, 85%). 1H NMR (500 MHz, DMSO-d6) δ 11.10 and 10.99 (2 × s, 1H, NH), 9.61 and 9.58 (2 × s, 1H, NH), 7.74–7.60 (m, 2H), 7.60–7.45 (m, 1H), 7.35–7.23 (m, 3H), 6.95 (t, J = 7.3 Hz, 1H); 13C NMR (126 MHz, DMSO-d6) δ 154.6, 152.5, 152.0, 143.4, 140.3, 130.4, 128.8 (2C), 121.15, 121.1, 118.5, 117.4 (2C), 117.3, 112.3, 103.2 (d, J = 26.0 Hz), 97.8 (d, J = 26.0 Hz), (1 quaternary C not observed); LCMS (2 min) tR = 1.01 min, m/z (ESI+) 306 (M + H+); HRMS m/z calcd for C13H9N3F79Br (M + H) 306.0042, found 306.0047. A mixture of 5-bromo-6-fluoro-N-phenyl-1H-benzo[d]imidazol-2-amine (2.53 g, 8.26 mmol), (Boc)2O (4.50 g, 20.7 mmol), and DMAP (0.20 g, 1.65 mmol) in THF (80 mL) was stirred at rt for 16 h. The mixture was concentrated, and the residue was partitioned between EtOAc (100 mL) and water (100 mL). The organic layer was washed with brine (100 mL), dried, and concentrated. Silica chromatography, eluting with 0–10% EtOAc–cyclohexane, gave the product as a mixture of three different di-Boc-protected positional isomers (1.94 g), which was used as such in the next step. LCMS (2 min) tR = 1.68, 1.79, and 1.92 min, m/z (ESI+) 506 (M + H+). This material (1.94 g) was stirred at rt in 1,4-dioxane (42 mL), and bis(pinacolato)diboron (1.17 g, 4.59 mmol), PdCl2dppf (0.28 g, 0.38 mmol), and KOAc (0.75 g, 7.65 mmol) were added. The mixture was heated at 120 °C for 24 h, then cooled and filtered through celite. The filtrate was concentrated and dissolved in THF (80 mL). DMAP (0.093 g, 0.77 mmol) and (Boc)2O (1.67 g, 7.66 mmol) were added, and the mixture was stirred for 6 h at rt. Solvent was removed by evaporation, and the residue was partitioned between water (100 mL) and EtOAc (100 mL). The organic layer was washed with brine (100 mL), dried, and concentrated. Silica chromatography, eluting with 0–50% EtOAc-cyclohexane, gave the product as a mixture of three different di-Boc-protected positional isomers (1.08 g), which was used as such in the next step. LCMS (2 min) tR = 1.67, 1.78, and 1.81 min, m/z (ESI+) 554 (M + H+). A mixture of this material (1.08 g), 3-bromo-6-chloro-N-((1-methylpiperidin-4-yl)methyl)imidazo[1,2-b]pyridazin-8-amine (0.63 g, 1.76 mmol) and Na2CO3 aq (2 M; 3.51 mL, 7.03 mmol) in dry 1,4-dioxane (18 mL) was purged with N2 for 10 min. Pd(OAc)2 (0.079 g, 0.35 mmol) and 1,1′-bis(di-tert-butylphosphino)ferrocene (0.17 g, 0.35 mmol) were added and the tube was sealed and stirred at 135 °C for 15 h. The mixture was cooled, filtered through celite pad, and concentrated. The residue was purified by silica chromatography, eluting with 0–5% MeOH–CH2Cl2 and then with 5–10% MeOH–CH2Cl2 + 1% 0.2 M NH3 aq, to give 31 (100 mg, 3% over three steps). 1H NMR (500 MHz, DMSO-d6) δ 11.14 (s, 1H), 9.62 (s, 1H), 8.00 (d, J = 6.5 Hz, 1H), 7.82 (d, J = 6.6 Hz, 1H), 7.79–7.74 (m, 2H), 7.70 (d, J = 2.5 Hz, 1H), 7.35–7.31 (m, 2H), 7.28 (d, J = 11.1 Hz, 1H), 6.98–6.92 (m, 1H), 6.31 (s, 1H), 3.23 (s, 2H), 2.75 (d, J = 10.8 Hz, 2H), 2.13 (s, 3H), 1.87–1.73 (m, 2H), 1.73–1.59 (m, 3H), 1.31–1.16 (m, 2H); 13C NMR (151 MHz, DMSO-d6) δ 154.1 (d, JC–F = 257.2 Hz), 148.1, 144.3, 140.9, 132.9, 131.0, 129.8, 129.3 (2C), 124.5, 121.5, 117.9 (2C), 110.0, 102.8, 91.1, 55.5 (2C), 47.8, 46.6, 34.8, 30.1 (2C) (3 quaternary C not observed); LCMS (2 min) tR = 0.91 min, m/z (ESI+) 505 (M + H+); ESI m/z calcd for C26H26N8F35Cl (M + H) 505.2031, found 505.2036.
6-Chloro-3-(4-fluoro-2-(phenylamino)-1H-benzo[d]imidazol-5-yl)-N-((1-methylpiperidin-4-yl)methyl)imidazo[1,2-b]pyridazin-8-amine (32)
Prepared from 4-bromo-3-fluorobenzene-1,2-diamine and 3-bromo-6-chloro-N-((1-methylpiperidin-4-yl)methyl)imidazo[1,2-b]pyridazin-8-amine using the method described for 31 to give 32 (2% over four steps). 1H NMR (500 MHz, DMSO-d6) δ 11.38 (s, 1H), 9.62 (s, 1H), 7.99 (t, J = 6.3 Hz, 1H), 7.81–7.74 (m, 2H), 7.69 (d, J = 1.4 Hz, 1H), 7.38 (t, J = 7.3 Hz, 1H), 7.36–7.32 (m, 2H), 7.25 (d, J = 8.2 Hz, 1H), 6.96 (tt, J = 1.2, 7.3 Hz, 1H), 6.29 (s, 1H), 3.23 (s, 2H), 2.79–2.68 (m, 2H), 2.13 (s, 3H), 1.86–1.74 (m, 2H), 1.73–1.60 (m, 3H), 1.30–1.16 (m, 2H); 13C NMR (151 MHz, DMSO-d6) δ 151.9, 148.1, 144.3, 140.9, 133.0, 130.7, 129.3 (2C), 124.8, 122.0, 121.4, 117.8 (2C), 107.9, 106.2, 91.1, 55.4 (2C), 47.7, 46.4, 34.7, 29.9 (2C) (3 quaternary C not observed); LCMS (2 min) tR = 1.14 min, m/z (ESI+) 505 (M + H+); HRMS m/z calcd for C26H26N8F35Cl (M + H) 505.2031, found 505.2013.
Acknowledgments
This work was supported by Cancer Research UK grants C309/A11566 (to the Cancer Therapeutics Unit, ICR), C20826/A12 (to F.E.D.), C24461/A13231 and C24461/A12772 (to R.B.), Janssen Biotech Inc., and The Institute of Cancer Research. The authors thank Dr. Sebastian Naud for the original synthesis of 2; Dr. Amin Mirza, Dr. Maggie Liu, and Meirion Richards for assistance in the structural characterization of compounds; Prof. Peter Walter for providing GFP-IRE1α HEK293 cells; Miri Jwa for details of the GFP-IRE1α oligomerization assay; Dr. Martin Augustin (Proteros Biostructures GmbH) for crystallography data; and Drs. Olivia Rossanese, Julian Blagg, Matthias Versele, and Ian Stansfield for helpful discussions and their interest in this work.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.jmedchem.8b01721.
Mass spectrometry characterization of IRE1α proteins; schematic of the fluorescence-labeled stem-loop RNA used to measure inhibition of IRE1α RNase activity; selectivity of compounds 26 and 31 for IRE1α vs IRE1β kinase inhibition; kinase selectivity profiles of compounds 2, 26, and 31; chemical structure and IRE1α inhibition data for 33; crystal structure of 33-IRE1α; model of 26 bound to IRE1α; microscopic images showing inhibition of stress-induced GFP-IRE1α foci formation in HEK293 cells by 26 and 31; titrations of inhibition of stress-induced splicing of XBP1-Luc reporter mRNA in HEK293 cells by 26 and 31; titrations of inhibition of stress-induced XBP1s mRNA expression in NCI-H929 cells by 26 and 31; quantification of inhibition of pS724 IRE1α relative to total IRE1α using capillary electrophoresis immunoassay in H929 cells by 26 and 31; microscopic images showing inhibition of stress-induced expression of XBP1s in NCI-H929 cells by 26 and 31; titrations of the inhibition of IRE1α autophosphorylation, IRE1α ATP-site binding, and IRE1α RNase by compounds 2, 26, and 31 in in vitro assays; selectivity profiles of 2, 26, and 31 tested against 455 human kinases at 1000 nM; HPLC purities of compounds 2, 5–12, and 19–33; experimental method for the generation of an HEK293TN XBP1 luciferase reporter cell line; experimental methods to determine inhibition of an XBP1 luciferase reporter in HEK293TN cells and assess compound cytotoxicity to HEK293TN cells; experimental method for the quantification of XBP1s expression in NCI-H929 cells by immunofluorescence; experimental method for the quantification of XBP1s and DNAJB9 mRNA expression in NCI-H929 cells by qPCR; experimental method for the quantification of the inhibition of IRE1α-GFP oligomerization in T-Rex293-IRE1-3F6HGFP cells; experimental methods for the quantification of total IRE1α and pIRE1α expression in NCI-H929 cells; experimental method for the determination of the crystal structure of 33-IRE1α; summary of crystallographic data for 2-IRE1α and 33-IRE1α; experimental methods for the synthesis of 33; copies of 1H and 13C NMR spectra of compounds 2, 5–12, and 19–33; copies of HPLC traces for compounds 2, 5–12, and 19–33; and molecular formula strings for compounds 2, 5–12, and 19–33 (PDF)
Molecular formula strings (CSV)
Accession Codes
PDB ID Codes: 2-IRE1α, 6HX1; 33-IRE1α, 6HV0.
Author Present Address
⊥ UAMS Myeloma Institute, 4018 W Capitol Avenue, Little Rock, Arkansas 72205, United States (F.E.D.).
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
The authors declare the following competing financial interest(s): All authors who are, or have been, employed by The Institute of Cancer Research are subject to a Rewards to Inventors Scheme which may reward contributors to a program that is subsequently licensed. The Institute of Cancer Research has a commercial interest in the development of inhibitors of IRE1-alpha.
Supplementary Material
References
- Hetz C.; Papa F. R. The unfolded protein response and cell fate control. Mol. Cell 2018, 69, 169–181. 10.1016/j.molcel.2017.06.017. [DOI] [PubMed] [Google Scholar]
- Corazzari M.; Gagliardi M.; Fimia G. M.; Piacentini M. Endoplasmic reticulum stress, unfolded protein response and cancer cell fate. Front. Oncol. 2017, 7, 78. 10.3389/fonc.2017.00078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K. P.; Dey M.; Neculai D.; Cao C.; Dever T. E.; Sicheri F. Structure of the dual enzyme Ire1 reveals the basis for catalysis and regulation in nonconventional RNA splicing. Cell 2008, 132, 89–100. 10.1016/j.cell.2007.10.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korennykh A. V.; Egea P. F.; Korostelev A. A.; Finer-Moore J.; Zhang C.; Shokat K. M.; Stroud R. M.; Walter P. The unfolded protein response signals through high-order assembly of Ire1. Nature 2009, 457, 687–693. 10.1038/nature07661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali M. M.; Bagratuni T.; Davenport E. L.; Nowak P. R.; Silva-Santisteban M. C.; Hardcastle A.; McAndrews C.; Rowlands M. G.; Morgan G. J.; Aherne W.; Collins I.; Davies F. E.; Pearl L. H. Structure of the Ire1 autophosphorylation complex and implications for the unfolded protein response. EMBO J. 2011, 30, 894–905. 10.1038/emboj.2011.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi A.; Newbatt Y.; McAndrew P. C.; Stubbs M.; Burke R.; Richards M. W.; Bhatia C.; Caldwell J. J.; McHardy T.; Collins I.; Bayliss R. Molecular mechanisms of human IRE1 activation through dimerization and ligand binding. Oncotarget 2015, 6, 13019–13035. 10.18632/oncotarget.3864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida H.; Matsui T.; Yamamoto A.; Okada T.; Mori K. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell 2001, 107, 881–891. 10.1016/S0092-8674(01)00611-0. [DOI] [PubMed] [Google Scholar]
- Lee A. H.; Iwakoshi N. N.; Glimcher L. H. XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Mol. Cell Biol. 2003, 23, 7448–7459. 10.1128/MCB.23.21.7448-7459.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollien J.; Weissman J. S. Decay of endoplasmic reticulum-localized mRNAs during the unfolded protein response. Science 2006, 313, 104–107. 10.1126/science.1129631. [DOI] [PubMed] [Google Scholar]
- Urano F.; Wang X.; Bertolotti A.; Zhang Y.; Chung P.; Harding H. P.; Ron D. Coupling of stress in the ER to activation of JNK protein kinases by transmembrane protein kinase IRE1. Science 2000, 287, 664–666. 10.1126/science.287.5453.664. [DOI] [PubMed] [Google Scholar]
- Wang M.; Kaufman R. J. Protein misfolding in the endoplasmic reticulum as a conduit to human disease. Nature 2016, 529, 326–335. 10.1038/nature17041. [DOI] [PubMed] [Google Scholar]
- Carrasco D. R.; Sukhdeo K.; Protopopova M.; Sinha R.; Enos M.; Carrasco D. E.; Zheng M.; Mani M.; Henderson J.; Pinkus G. S.; Munshi N.; Horner J.; Ivanova E. V.; Protopopov A.; Anderson K. C.; Tonon G.; DePinho R. A. The differentiation and stress response factor XBP-1 drives multiple myeloma pathogenesis. Cancer Cell 2007, 11, 349–360. 10.1016/j.ccr.2007.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X.; Iliopoulos D.; Zhang Q.; Tang Q.; Greenblatt M. B.; Hatziapostolou M.; Lim E.; Tam W. L.; Ni M.; Chen Y.; Mai J.; Shen H.; Hu D. Z.; Adoro S.; Hu B.; Song M.; Tan C.; Landis M. D.; Ferrari M.; Shin S. J.; Brown M.; Chang J. C.; Liu X. S.; Glimcher L. H. XBP1 promotes triple-negative breast cancer by controlling the HIF1α pathway. Nature 2014, 508, 103–107. 10.1038/nature13119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storm M.; Sheng X.; Arnoldussen Y. J.; Saatcioglu F. Prostate cancer and the unfolded protein response. Oncotarget 2016, 7, 54051–54066. 10.18632/oncotarget.9912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubillos-Ruiz J. R.; Silberman P. C.; Rutkowski M. R.; Chopra S.; Perales-Puchalt A.; Song M.; Zhang S.; Bettigole S. E.; Gupta D.; Holcomb K.; Ellenson L. H.; Caputo T.; Lee A. H.; Conejo-Garcia J. R.; Glimcher L. H. ER stress sensor XBP1 controls anti-tumor immunity by disrupting dendritic cell homeostasis. Cell 2015, 161, 1527–1538. 10.1016/j.cell.2015.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkmann K.; Lucas J. L.; Vuga D.; Wang X.; Brumm D.; Stiles C.; Kriebel D.; Der-Sarkissian A.; Krishnan K.; Schweitzer C.; Liu Z.; Malyankar U. M.; Chiovitti D.; Canny M.; Durocher D.; Sicheri F.; Patterson J. B. Potent and selective inhibitors of the inositol-requiring enzyme 1 endoribonuclease. J. Biol. Chem. 2011, 286, 12743–12755. 10.1074/jbc.M110.199737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross B. C.; Bond P. J.; Sadowski P. G.; Jha B. K.; Zak J.; Goodman J. M.; Silverman R. H.; Neubert T. A.; Baxendale I. R.; Ron D.; Harding H. P. The molecular basis for selective inhibition of unconventional mRNA splicing by an IRE1-binding small molecule. Proc. Natl. Acad. Sci. U.S.A. 2012, 109, E869–E878. 10.1073/pnas.1115623109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ming J.; Ruan S.; Wang M.; Ye D.; Fan N.; Meng Q.; Tian B.; Huang T. A novel chemical, STF-083010, reverses tamoxifen-related drug resistance in breast cancer by inhibiting IRE1/XBP1. Oncotarget 2015, 6, 40692–40703. 10.18632/oncotarget.5827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papandreou I.; Denko N. C.; Olson M.; Van Melckebeke H.; Lust S.; Tam A.; Solow-Cordero D. E.; Bouley D. M.; Offner F.; Niwa M.; Koong A. C. Identification of an Ire1alpha endonuclease specific inhibitor with cytotoxic activity against human multiple myeloma. Blood 2011, 117, 1311–1314. 10.1182/blood-2010-08-303099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao N.; Cao J.; Xu L.; Tang Q.; Dobrolecki L. E.; Lv X.; Talukdar M.; Lu Y.; Wang X.; Hu D. Z.; Shi Q.; Xiang Y.; Wang Y.; Liu X.; Bu W.; Jiang Y.; Li M.; Gong Y.; Sun Z.; Ying H.; Yuan B.; Lin X.; Feng X. H.; Hartig S. M.; Li F.; Shen H.; Chen Y.; Han L.; Zeng Q.; Patterson J. B.; Kaipparettu B. A.; Putluri N.; Sicheri F.; Rosen J. M.; Lewis M. T.; Chen X. Pharmacological targeting of MYC-regulated IRE1/XBP1 pathway suppresses MYC-driven breast cancer. J. Clin. Invest. 2018, 128, 1283–1299. 10.1172/JCI95873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L.; Perera B. G.; Hari S. B.; Bhhatarai B.; Backes B. J.; Seeliger M. A.; Schürer S. C.; Oakes S. A.; Papa F. R.; Maly D. J. Divergent allosteric control of the IRE1α endoribonuclease using kinase inhibitors. Nat. Chem. Biol. 2012, 8, 982–989. 10.1038/nchembio.1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Concha N. O.; Smallwood A.; Bonnette W.; Totoritis R.; Zhang G.; Federowicz K.; Yang J.; Qi H.; Chen S.; Campobasso N.; Choudhry A. E.; Shuster L. E.; Evans K. A.; Ralph J.; Sweitzer S.; Heerding D. A.; Buser C. A.; Su D. S.; DeYoung M. P. Long-range inhibitor-induced conformational regulation of human IRE1α endoribonuclease activity. Mol. Pharmacol. 2015, 88, 1011–1023. 10.1124/mol.115.100917. [DOI] [PubMed] [Google Scholar]
- Harrington P. E.; Biswas K.; Malwitz D.; Tasker A. S.; Mohr C.; Andrews K. L.; Dellamaggiore K.; Kendall R.; Beckmann H.; Jaeckel P.; Materna-Reichelt S.; Allen J. R.; Lipford J. R. Unfolded protein response in cancer: IRE1α inhibition by selective kinase ligands does not impair tumor cell viability. ACS Med. Chem. Lett. 2015, 6, 68–72. 10.1021/ml500315b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman H. C.; Tong M.; Wang L.; Meza-Acevedo R.; Gobillot T. A.; Lebedev I.; Gliedt M. J.; Hari S. B.; Mitra A. K.; Backes B. J.; Papa F. R.; Seeliger M. A.; Maly D. J. Structural and functional analysis of the allosteric inhibition of IRE1α with ATP-competitive ligands. ACS Chem. Biol. 2016, 11, 2195–2205. 10.1021/acschembio.5b00940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita S.; Villalta S. A.; Feldman H. C.; Register A. C.; Rosenthal W.; Hoffmann-Petersen I. T.; Mehdizadeh M.; Ghosh R.; Wang L.; Colon-Negron K.; Meza-Acevedo R.; Backes B. J.; Maly D. J.; Bluestone J. A.; Papa F. R. Targeting ABL-IRE1α signaling spares ER-stressed pancreatic β cells to reverse autoimmune diabetes. Cell Metab. 2017, 25, 883–897. 10.1016/j.cmet.2017.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaccaro W.; Chen Z.; Dodd D. S.; Huynh T. N.; Lin J.; Liu C.; Mussari C. P.; Tokarski J. S.; Tortolani D. R.; Wrobleski S. T.; Lin S.. Preparation of Fused Heterocyclic Compounds Useful as Kinase Modulators. US20080045536, 2008.
- Clark R. F.; Ba-Maung N. Y.; Erickson S. A.; Fidanze S. D.; Mantei R. A.; Sheppard G. S.; Sorensen B. K.; Wang G. T.; Wang J.; Bell R. L.. Preparation of Pyrimidine Inhibitors of Kinase Activity. International Application No. WO2010138575, 2010.
- Kusakabe K.; Ide N.; Daigo Y.; Itoh T.; Yamamoto T.; Hashizume H.; Nozu K.; Yoshida H.; Tadano G.; Tagashira S.; Higashino K.; Okano Y.; Sato Y.; Inoue M.; Iguchi M.; Kanazawa T.; Ishioka Y.; Dohi K.; Kido Y.; Sakamoto S.; Ando S.; Maeda M.; Higaki M.; Baba Y.; Nakamura Y. Discovery of imidazo[1,2-b]pyridazine derivatives: selective and orally available Mps1 (TTK) kinase inhibitors exhibiting remarkable antiproliferative activity. J. Med. Chem. 2015, 58, 1760–1775. 10.1021/jm501599u. [DOI] [PubMed] [Google Scholar]
- Uitdehaag J. C. M.; de Man J.; Willemsen-Seegers N.; Prinsen M. B. W.; Libouban M. A. A.; Sterrenburg J. G.; de Wit J. J. P.; de Vetter J. R. F.; de Roos J. A. D. M.; Buijsman R. C.; Zaman G. J. R. Target residence time-guided optimization on TTK kinase results in inhibitors with potent anti-proliferative activity. J. Mol. Biol. 2017, 429, 2211–2230. 10.1016/j.jmb.2017.05.014. [DOI] [PubMed] [Google Scholar]
- Itzhak D.; Bright M.; McAndrew P.; Mirza A.; Newbatt Y.; Strover J.; Widya M.; Thompson A.; Morgan G.; Collins I.; Davies F. Multiple autophosphorylations significantly enhance the endoribonuclease activity of human inositol requiring enzyme 1α. BMC Biochem. 2014, 15, 3. 10.1186/1471-2091-15-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newbatt Y.; Hardcastle A.; McAndrew P. C.; Strover J. A.; Mirza A.; Morgan G. J.; Burke R.; Davies F. E.; Collins I.; van Montfort R. L. Identification of autophosphorylation inhibitors of the inositol-requiring enzyme 1 alpha (IRE1α) by high-throughput screening using a DELFIA assay. J. Biomol. Screen. 2013, 18, 298–308. 10.1177/1087057112465647. [DOI] [PubMed] [Google Scholar]
- Potashman M. H.; Bready J.; Coxon A.; DeMelfi T. M. Jr.; DiPietro L.; Doerr N.; Elbaum D.; Estrada J.; Gallant P.; Germain J.; Gu Y.; Harmange J. C.; Kaufman S. A.; Kendall R.; Kim J. L.; Kumar G. N.; Long A. M.; Neervannan S.; Patel V. F.; Polverino A.; Rose P.; van der Plas S.; Whittington D.; Zanon R.; Zhao H. Design, synthesis, and evaluation of orally active benzimidazoles and benzoxazoles as vascular endothelial growth factor-2 receptor tyrosine kinase inhibitors. J. Med. Chem. 2007, 50, 4351–4373. 10.1021/jm070034i. [DOI] [PubMed] [Google Scholar]
- Liu Y.; Gray N. S. Rational design of inhibitors that bind to inactive kinase conformations. Nat. Chem. Biol. 2006, 2, 358–364. 10.1038/nchembio799. [DOI] [PubMed] [Google Scholar]
- Iwawaki T.; Hosoda A.; Okuda T.; Kamigori Y.; Nomura-Furuwatari C.; Kimata Y.; Tsuru A.; Kohno K. Translational control by the ER transmembrane kinase/ribonuclease IRE1 under ER stress. Nat. Cell Biol. 2001, 3, 158–164. 10.1038/35055065. [DOI] [PubMed] [Google Scholar]
- Dodson C. A.; Kosmopoulou M.; Richards M. W.; Atrash B.; Bavetsias V.; Blagg J.; Bayliss R. Crystal structure of an Aurora-A mutant that mimics Aurora-B bound to MLN8054: insights into selectivity and drug design. Biochem. J. 2010, 427, 19–28. 10.1042/BJ20091530. [DOI] [PubMed] [Google Scholar]; Erratum in:; Biochem. J. 2010, 427, 551. 10.1042/BJ4270551
- Aliagas-Martin I.; Burdick D.; Corson L.; Dotson J.; Drummond J.; Fields C.; Huang O. W.; Hunsaker T.; Kleinheinz T.; Krueger E.; Liang J.; Moffat J.; Phillips G.; Pulk R.; Rawson T. E.; Ultsch M.; Walker L.; Wiesmann C.; Zhang B.; Zhu B. Y.; Cochran A. G. A class of 2,4-bisanilinopyrimidine Aurora A inhibitors with unusually high selectivity against Aurora B. J. Med. Chem. 2009, 52, 3300–3307. 10.1021/jm9000314. [DOI] [PubMed] [Google Scholar]
- Moslin R.; Gardner D.; Santella J.; Zhang Y.; Duncia J. V.; Liu C.; Lin J.; Tokarski J. S.; Strnad J.; Pedicord D.; Chen J.; Blat Y.; Zupa-Fernandez A.; Cheng L.; Sun H.; Chaudhry C.; Huang C.; D’Arienzo C.; Sack J. S.; Muckelbauer J. K.; Chang C.; Tredup J.; Xie D.; Aranibar N.; Burke J. R.; Carter P. H.; Weinstein D. S. Identification of imidazo[1,2-b]pyridazine TYK2 pseudokinase ligands as potent and selective allosteric inhibitors of TYK2 signalling. MedChemComm 2017, 8, 700–712. 10.1039/C6MD00560H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas M. C.; Goldstein D. M.; Hermann J. C.; Kuglstatter A.; Liu W.; Luk K. C.; Padilla F.; Slade M.; Villasenor A. G.; Wanner J.; Xie W.; Zhang X.; Liao C. Rational design of highly selective spleen tyrosine kinase inhibitors. J. Med. Chem. 2012, 55, 10414–10423. 10.1021/jm301367c. [DOI] [PubMed] [Google Scholar]
- Smith C. R.; Dougan D. R.; Komandla M.; Kanouni T.; Knight B.; Lawson J. D.; Sabat M.; Taylor E. R.; Vu P.; Wyrick C. Fragment-based discovery of a small molecule inhibitor of Bruton’s tyrosine kinase. J. Med. Chem. 2015, 58, 5437–5444. 10.1021/acs.jmedchem.5b00734. [DOI] [PubMed] [Google Scholar]
- Miller R. M.; Paavilainen V. O.; Krishnan S.; Serafimova I. M.; Taunton J. Electrophilic fragment-based design of reversible covalent kinase inhibitors. J. Am. Chem. Soc. 2013, 135, 5298–5301. 10.1021/ja401221b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H.; Korennykh A. V.; Behrman S. L.; Walter P. Mammalian endoplasmic reticulum stress sensor IRE1 signals by dynamic clustering. Proc. Natl. Acad. Sci. U.S.A. 2010, 107, 16113–16118. 10.1073/pnas.1010580107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayliss R.; Haq T.; Yeoh S. The Ys and wherefores of protein kinase autoinhibition. Biochim. Biophys. Acta 2015, 1854, 1586–1594. 10.1016/j.bbapap.2015.04.025. [DOI] [PubMed] [Google Scholar]
- Lavoie H.; Li J. J.; Thevakumaran N.; Therrien M.; Sicheri F. Dimerization-induced allostery in protein kinase regulation. Trends Biochem. Sci. 2014, 39, 475–486. 10.1016/j.tibs.2014.08.004. [DOI] [PubMed] [Google Scholar]
- Hatzivassiliou G.; Song K.; Yen I.; Brandhuber B. J.; Anderson D. J.; Alvarado R.; Ludlam M. J.; Stokoe D.; Gloor S. L.; Vigers G.; Morales T.; Aliagas I.; Liu B.; Sideris S.; Hoeflich K. P.; Jaiswal B. S.; Seshagiri S.; Koeppen H.; Belvin M.; Friedman L. S.; Malek S. RAF inhibitors prime wild-type RAF to activate the MAPK pathway and enhance growth. Nature 2010, 464, 431–435. 10.1038/nature08833. [DOI] [PubMed] [Google Scholar]
- Poulikakos P. I.; Zhang C.; Bollag G.; Shokat K. M.; Rosen N. RAF inhibitors transactivate RAF dimers and ERK signalling in cells with wild-type BRAF. Nature 2010, 464, 427–430. 10.1038/nature08902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora R.; Di Michele M.; Stes E.; Vandermarliere E.; Martens L.; Gevaert K.; Van Heerde E.; Linders J. T.; Brehmer D.; Jacoby E.; Bonnet P. Structural investigation of B-Raf paradox breaker and inducer inhibitors. J. Med. Chem. 2015, 58, 1818–1831. 10.1021/jm501667n. [DOI] [PubMed] [Google Scholar]
- Byrne D. P.; Foulkes D. M.; Eyers P. A. Pseudokinases: update on their functions and evaluation as new drug targets. Future Med. Chem. 2017, 9, 245–265. 10.4155/fmc-2016-0207. [DOI] [PubMed] [Google Scholar]
- Song M.; Sandoval T. A.; Chae C. S.; Chopra S.; Tan C.; Rutkowski M. R.; Raundhal M.; Chaurio R. A.; Payne K. K.; Konrad C.; Bettigole S. E.; Shin H. R.; Crowley M. J. P.; Cerliani J. P.; Kossenkov A. V.; Motorykin I.; Zhang S.; Manfredi G.; Zamarin D.; Holcomb K.; Rodriguez P. C.; Rabinovich G. A.; Conejo-Garcia J. R.; Glimcher L. H.; Cubillos-Ruiz J. R. IRE1α-XBP1 controls T cell function in ovarian cancer by regulating mitochondrial activity. Nature 2018, 562, 423–428. 10.1038/s41586-018-0597-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W. XDS. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2010, 66, 125–132. 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winn M. D.; Ballard C. C.; Cowtan K. D.; Dodson E. J.; Emsley P.; Evans P. R.; Keegan R. M.; Krissinel E. B.; Leslie A. G.; McCoy A.; McNicholas S. J.; Murshudov G. N.; Pannu N. S.; Potterton E. A.; Powell H. R.; Read R. J.; Vagin A.; Wilson K. S. Overview of the CCP4 suite and current developments. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2011, 67, 235–242. 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P.; Lohkamp B.; Scott W. G.; Cowtan K. Features and development of Coot. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2010, 66, 486–501. 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov G. N.; Vagin A. A.; Dodson E. J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr., Sect. D: Biol. Crystallogr. 1997, 53, 240–255. 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Maestro, version 9.3; Schrödinger, LLC: New York, 2012.
- Glide, version 5.8; Schrödinger, LLC: New York, 2012.
- LigPrep, version 2.5; Schrödinger, LLC: New York, 2011.
- Zhegalova N. G.; Gonzales G.; Berezin M. Y. Synthesis of nitric oxide probes with fluorescence lifetime sensitivity. Org. Biomol. Chem. 2013, 11, 8228–8234. 10.1039/c3ob41498a. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.