Abstract
Background
Long noncoding RNAs (lncRNAs) play important roles in cancer development and therapeutic resistance. However, the role of small nucleolar RNA host gene 16 (SNHG16) in the development of hepatocellular carcinoma (HCC) remains largely unknown.
Material/Methods
In situ hybridization (ISH) staining was performed to detect the expression level of SNHG16 in HCC tissues and adjacent non-cancerous tissues. Quantitative real-time polymerase chain reaction (qRT-PCR) was used to detect the level of SNHG16 in HCC samples, adjacent non-cancerous tissues and HCC cell lines. Transwell assay was performed to investigate the migration and invasion ability of HCC cells. Cell viability assays were performed to determine the ability of proliferation and sorafenib resistance of HCC cells.
Results
We found that SNHG16 was upregulated in HCC tissues and cell lines and that it was negatively correlated with survival time in HCC patients. Univariate and multivariate analyses revealed that SNHG16 was a significant and independent predictor for the overall survival of HCC patients. Furthermore, downregulation of SNHG16 inhibited proliferation, migration, invasion, and sorafenib resistance in hepatocellular carcinoma cell lines.
Conclusions
Our findings revealed that lncRNA SNHG16 could be used as an oncogene to predict the outcome of hepatocellular carcinoma.
MeSH Keywords: Carcinoma, Hepatocellular; Prognosis; RNA, Long Noncoding
Background
Hepatocellular carcinoma (HCC) is one of the most lethal human malignancies in China. A report on the global surveillance of cancer survival trends showed that HCC accounts for 12.5% of the 18 most prevalent cancers and that the 5-year survival rate after HCC diagnosis is less than 15% in China [1]. Many strategies have been explored to treat HCC in the past, including surgical resection, radiotherapy, chemotherapy, and biotherapy. However, improvements in HCC prognosis have been limited [2]. The molecular carcinogenesis of HCC remains unclear and it is widely accepted that alterations in gene expression can lead to the abnormal proliferation of cancer cells. Therefore, novel molecules need to be identified to better diagnose and treat HCC.
In recent years, long noncoding RNAs (lncRNAs) have been identified and have been shown to participate in numerous signalling pathways as auxiliary regulators of proteins. lncRNAs have been shown to target several chromatin modification complexes involved in gene regulation [3]. In addition, several other studies have demonstrated that active chromatin states are associated with lncRNAs [4]. The associations between lncRNAs and gene regulation in HCC have been widely studied, and many lncRNAs, such as UFC1 and PVT1, have been used to predict HCC outcome. Mechanistically, these lncRNAs can directly sponge microRNAs or interact with mRNA stabilizing proteins to disrupt gene regulation [5,6].
lncRNA SNHG16, located at chromosome 17q25.1, was originally identified as an oncogene in neuroblastoma [7]. Ectopic expression of SNHG16 has been shown to induce transformation of NIH3T3 cells, while knockdown of endogenous SNHG16 significantly inhibits cell growth [7]. Recently, several studies investigated the oncogenic effects of SNHG16 in a variety of tumors, such as acute lymphoblastic leukaemia, non-small cell lung cancer, and hemangioma [8–10]. In addition, lncRNA SNHG16 is also involved in the LPS-induced inflammatory pathway via the regulation of miR-15a/16 [11]. However, the relationship between HCC prognosis and lncRNA SNHG16 remains unclear.
In this study, we analysed the clinicopathologic and prognostic significance of lncRNA SNHG16 in HCC patients. We also downregulated the expression of lncRNA SNHG16 in 2 HCC cell lines to detect its function in vitro. Our results indicated that lncRNA SNHG16 expression could potentially be used for HCC prognosis in the future.
Material and Methods
Patients and specimens
In this study, 2 independent cohorts were enrolled. In cohort 1, fresh HCC tissues and adjacent non-cancerous samples were collected from 10 patients who had been pathologically diagnosed with HCC and undergone a hepatectomy between April 2016 and December 2016 at the First Affiliated Hospital of the Anhui Medical University (Hefei, China). In cohort 2, formalin-fixed, paraffin-embedded samples from 61 HCC patients who underwent initial surgical resection between February 2008 to December 2012 were collected randomly from the Department of Pathology at the First Affiliated Hospital of the Anhui Medical University. Informed consent was obtained from all patients and the study was approved by the Biomedical Ethics Committee of Anhui Medical University. Pathological staging was performed according to the international staging system [12].
In situ hybridization (ISH) staining
ISH was performed according to the manufacturer’s protocol (ISH kit, Boster Bio-Engineering Company, Wuhan, China). ISH staining was scored by 2 independent pathologists as negative (−), weakly positive (+), or strongly positive (++).
Follow-up
In cohort 2, each patient was scheduled for an examination every 3 months for the first 2 years and at 6-month intervals thereafter during the follow-up period (5 years). Testing included blood analysis, ultrasound examination, and computed tomography.
Cell culture and transfection
One normal liver cell line (HL-7702) and 4 classic HCC cell lines (SK-Hep-1, Huh7, Hep3B, and HepG2) were purchased from the Cell Bank of the Type Culture Collection (Chinese Academy of Sciences, Shanghai, China) and maintained in Dulbecco’s modified Eagle’s medium (DMEM) (Gibco, Grand Island, NY) supplemented with 10% fetal bovine serum (Gibco), 2 mmol/l L-glutamine (HyClone, Logan, UT), and penicillin (50 U/ml)/streptomycin (50 μg/ml) (HyClone) at 37°C in a 5% CO2 humidified chamber.
RNA interference
The siRNA of human SNHG16 and a scrambled negative control siRNA were purchased from Genepharma (Shanghai, China). Transfection was performed using Lipofectamine RNAiMAX reagent (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions.
Quantitative RT-PCR
Total RNA from the cell lines and fresh tissues was extracted with TRIzol reagent (Invitrogen) according to the manufacture’s protocol. Next, 2 μg of total RNA was reverse transcribed using a First Strand cDNA kit (Invitrogen) to synthesize the cDNA template. qRT-PCR was performed on a 7500 Fast Real-Time PCR system (Applied Biosystem, Rotkreuz, Switzerland) using a SYBR Green PCR kit (Takara, Dalian, China). The primer sequences were as follows:
SNHG16 F: 5′-GCAGAATGCCATGGTTTCCC-3′;
R: 5′-GGACAGCTGGCAAGAGACTT-3′
Cell viability assay
Cell viability assays were performed using the Cell Counting Kit-8 (CCK-8, Dojindo, Japan) according to the manufacturer’s protocol. For treatment with SNHG16 siRNA and the scrambled negative control, cells were plated 48 h after transfection and incubated for 1–3 days. In the sorafenib resistance assay, transfected cells were treated with sorafenib (dose range, 0–32 μM) for 72 h, and then cell inhibition was measured using the CCK-8 assay.
Cell migration and invasion assay
Cell migration and invasion assays were performed in Transwell chambers (Corning Costar, Cambridge, MA), with the cell invasion assays carried out in a chamber that contained a Matrigel (BD Biosciences, San Jose, CA)-precoated membrane. HepG2 cells were transfected with SNHG16 siRNA or a scrambled negative control siRNA and were then serum-starved for 24 h before conducting the assays. For the migration assay, 1×105 cells were plated into the top chamber of a Transwell device (Corning Costar) with 0.1% bovine serum albumin (BSA). For the invasion assay, cells were plated on filters coated with 20 μg/cm2 of reconstituted Matrigel basement membranes. The bottom chamber was filled with complete medium. After incubation at 37°C for 24 h, the cells that had not migrated or invaded from the upper membrane surface were removed using cotton swabs. Cells adhered on the lower membrane surface of the filters were fixed in 100% methanol and stained with 0.5% crystal violet at room temperature. Cell numbers were quantitated by counting the cells in 5 random fields (original magnification, ×200).
Statistical analysis
The chi-squared test was used to examine the association between lncRNA SNHG16 expression and various patient clinicopathologic factors. Survival time comparisons are shown using Kaplan-Meier curves, and significant differences were compared with a log-rank test. Prognostic value was determined by multivariate Cox regression analysis. P values <0.05 were considered statistically significant. All statistical analyses were performed using SPSS 19.0 software (Abbott Laboratories, North Chicago, IL).
Results
SNHG16 is upregulated in HCC tissues and cell lines
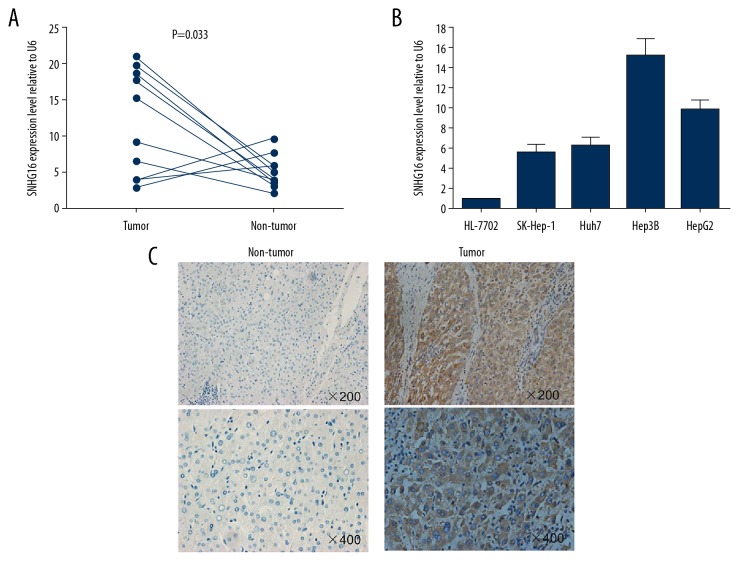
We investigated SNHG16 expression in HCC tissues and cell lines. The expression of SNHG16 was detected in 10 paired HCC samples and adjacent non-cancerous tissues by qRT-PCR (Cohort 1). SNHG16 expression levels were significantly upregulated in cancerous tissues compared to non-tumor tissues (P=0.033, Figure 1A). Furthermore, all 4 HCC cell lines (SK-Hep-1, Huh7, Hep3B, and HepG2) expressed higher levels of SNHG16 compared to the normal hepatocyte cell line HL-7702 (Figure 1B). To further validate its dysregulated expression, we investigated the levels of SNHG16 in an additional 61 paired paraffin-embedded samples by ISH (Cohort 2). Samples with negative or weakly positive SNHG16 levels were defined as having low expression, while strongly positive samples were defined as having high expression, and 35 out of 61 (57.4%) cases exhibited high expression of SNHG16 (Figure 1C). Together, these results indicate that SNHG16 is upregulated in HCC.
Figure 1.
SNHG16 overexpression in HCC tissues and cell lines. (A) Analysis of SNHG16 level in 10 paired HCC samples and adjacent non-cancerous tissues by qRT-PCR (Cohort 1). (B) Analysis of SNHG16 in HCC cell lines and a normal hepatocyte cell line HL-7702. (C) Analysis of SNHG16 expression in HCC tissues by ISH assays (Cohort 2).
Correlation between SNHG16 and clinicopathologic factors in patients with HCC
To investigate whether SNHG16 is associated with HCC progression, we analysed the correlation between SNHG16 expression and clinicopathologic parameters in 61 HCC patients (Cohort 2). As shown in Table 1, statistical analysis revealed a positive correlation between SNHG16 expression and tumor size (P=0.031), AFP level (P<0.001), portal vein tumor thrombus (PVTT) (P=0.007), and metastasis (P=0.007). However, no statistically significant correlations were found between SNGH16 and age, sex, pathologic grade, and liver cirrhosis (P>0.05).
Table 1.
Correlation between SNHG16 expression and HCC clinicopathologic features.
| Characteristic | SNHG16 expression level | P-value | |
|---|---|---|---|
| Low N=26 (42.6%) | High N=35 (57.4%) | ||
| Age | 0.149 | ||
| ≤55 | 16 (61.5%) | 15 (42.9%) | |
| >55 | 10 (38.5%) | 20 (57.1%) | |
| Sex | 0.186 | ||
| Male | 16 (61.5%) | 27 (77.1%) | |
| Female | 10 (38.5%) | 8 (22.9) | |
| Tumor size (cm) | 0.031 | ||
| ≤5 | 21 (80.8%) | 19 (54.3%) | |
| >5 | 5 (19.2%) | 16 (45.7%) | |
| Pathologic grade | 0.096 | ||
| I–II | 16 (61.5%) | 14 (40.0%) | |
| III–IV | 10 (38.5%) | 21 (60.0%) | |
| AFP level (ng/ml) | <0.001 | ||
| ≤500 | 23 (88.5%) | 15 (42.9%) | |
| >500 | 3 (11.5%) | 20 (57.1%) | |
| Liver cirrhosis | 0.856 | ||
| With | 15 (57.7%) | 21 (60.0%) | |
| Without | 11 (42.3%) | 14 (40.0%) | |
| PVTT | 0.007 | ||
| No | 25 (96.2%) | 24 (68.6%) | |
| Yes | 1 (3.8%) | 11 (31.4%) | |
| Metastasis | 0.007 | ||
| No | 25 (96.2%) | 24 (68.6%) | |
| Yes | 1 (3.8%) | 11 (31.4%) | |
PTVV – portal vein tumor thrombus.
SNHG16 expression is associated with poor survival of HCC patients
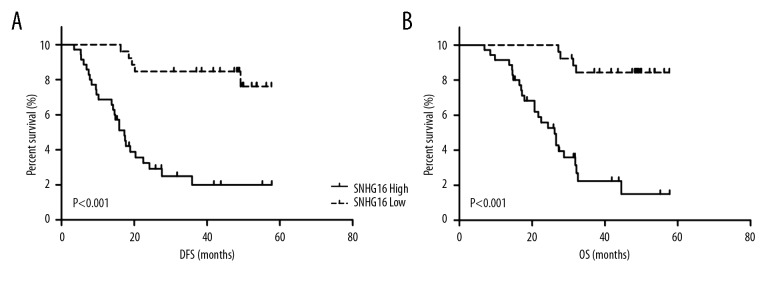
To investigate the relationship between SNGH16 and HCC patient prognosis, Kaplan-Meier analysis and log-rank tests were performed to evaluate the association between SNHG16 expression and overall survival (OS) or disease-free survival (DFS) of HCC patients. The results indicated that patients with higher SNHG16 expression have worse DFS (P<0.001) and OS (P<0.001) compared to patients with lower SNHG16 expression (Figure 2). To further evaluate whether the level of SNHG16 could be identified as a prognostic factor for HCC patients, univariate and multivariate analysis was performed. Univariate analysis demonstrated that tumor size (P<0.001), AFP level (P<0.001), PVTT (P<0.001), metastasis (P<0.001), and SNHG16 expression (P<0.001) were significantly correlated with OS in HCC patients (Table 2). Multivariate analysis indicated that SNHG16 expression (HR=4.985, 95% CI=1.451–17.132, P=0.011), AFP level (HR=7.078, 95% CI=2.465–20.326, P<0.001), PVTT (HR=12.082, 95% CI=3.660–39.890, P<0.001) and metastasis (HR=6.179, 95% CI=2.188–17.444, P=0.001) were significant independent predictors for patients with HCC (Table 2).
Figure 2.
Kaplan-Meier curves of (A) DFS and (B) OS according to the level of lncRNA SNHG16 expression.
Table 2.
Univariate and multivariate analysis of OS by Cox regression analysis.
| Variables | Univariate analysis | Multivariate analysis | ||
|---|---|---|---|---|
| HR (95% CI) | P | HR (95% CI) | P | |
| Age | 1.809 (0.863–3.796) | 0.117 | ||
| Sex | 0.845 (0.374–1.909) | 0.685 | ||
| Tumor size | 4.871 (2.300–10.320) | <0.001 | ||
| Pathologic grade | 1.013 (0.489–2.099) | 0.973 | ||
| AFP level | 13.411 (5.170–34.786) | <0.001 | 7.078 (2.465–20.326) | <0.001 |
| Liver cirrhosis | 1.059 (0.505–2.219) | 0.880 | ||
| PVTT | 10.133 (4.132–24.848) | <0.001 | 12.082 (3.660–39.890) | <0.001 |
| Metastasis | 8.468 (3.768–19.028) | <0.001 | 6.179 (2.188–17.444) | 0.001 |
| SNHG16 | 10.238 (3.493–30.007) | <0.001 | 4.985 (1.451–17.132) | 0.011 |
PTVV – portal vein tumor thrombus.
Knockdown of SNHG16 inhibits proliferation, migration, and invasion and reverses sorafenib resistance in HCC cells
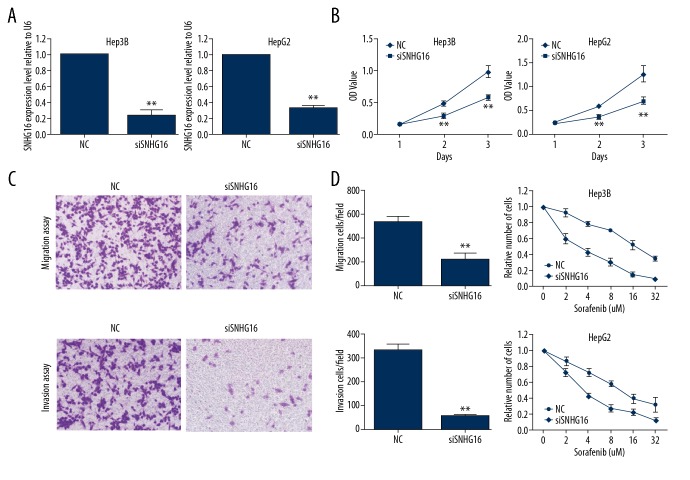
Subsequently, to investigate the biological role of SNHG16 in HCC progression and therapy resistance, we used siRNA to downregulate the expression of SNHG16 in Hep3B and HepG2 cell lines. As shown in Figure 3A, the expression of SNHG16 was effectively silenced using siRNA (P<0.01). The CCK-8 assay indicated that SNHG16 knockdown significantly decreased cellular proliferation in Hep3B and HepG2 cell lines (Figure 3B). In addition, knockdown of SNHG16 expression reduced the migration and invasion ability of HCC cells (Figure 3C). Moreover, the sorafenib resistance assay indicated that knockdown of SNHG16 expression significantly reverses sorafenib resistance in both Hep3B and HepG2 cell lines (Figure 3D). Collectively, these data show that knockdown of SNHG16 expression significantly inhibits proliferation, migration, invasion, and sorafenib resistance in HCC cells.
Figure 3.
Knockdown of SNHG16 inhibits proliferation, migration, and invasion and reverses sorafenib resistance in HCC cells. (A) Hep3B and HepG2 cells were transfected with SNHG16 siRNA or a scrambled negative control. The transfection efficiency was detected by qRT-PCR. ** P<0.01. (B) Hep3B and HepG2 cells were transfected with SNHG16 siRNA or a scrambled negative control. Cell growth was detected at days 1–3 after transfection. The data are presented as the mean ±SD. ** P<0.01. (C) HepG2 cells were transfected with SNHG16 siRNA or a scrambled negative control. Cell migration and invasion abilities were analysed by Transwell migration assay (above) and invasion assay (below) (magnification: ×200). ** P<0.01. (D) Hep3B and HepG2 cells were transfected with SNHG16 siRNA or a scrambled negative control. The sensitivity to sorafenib was detected by CCK-8 assay.
Discussion
In this study, we detected the expression of SNHG16 in HCC tissues and cell lines and then analysed the prognostic value of SNHG16 in patients with HCC. Our results revealed that SNHG16 is upregulated in HCC tissues and cell lines. The SNHG16 expression level was correlated with tumor size, AFP level, PVTT, and metastasis. Further statistical analysis indicated that high expression of SNHG16 is associated with worse prognosis in HCC patients. Multivariate analysis indicated that SNHG16 could be an independent prognosticator for HCC patients. Furthermore, suppression of SNHG16 significantly inhibited proliferation, migration, invasion, and sorafenib resistance in HCC cells in vitro. Collectively, our results show that SNHG16 may play an important role in the development and progression of HCC.
It is well established that lncRNAs play crucial roles in a large variety of cellular processes and that dysregulated lncRNA expression is probably pervasive in human cancers [13]. For example, one of the most well-studied lncRNAs, HOTAIR, promotes cancer metastasis by reprograming the chromatin state [14]. HULC is upregulated in HCC tissues compared to adjacent non-cancerous tissues, and ectopic expression of HULC facilitates HCC progression by promoting cell viability, invasion, and vascularization [15,16].
Recently, accumulating evidence demonstrated that lncRNAs can serve as biomarkers and prognosticators for many cancers, including HCC [17,18]. For example, overexpression of UFC1 is correlated with tumor size, clinical stage, and patient outcomes [5]. These data indicate that lncRNAs may play crucial roles in HCC carcinogenesis and progression. In this study, we demonstrated that SNHG16 is upregulated in HCC and that it correlates with a worse prognosis in HCC patients. Importantly, SNHG16 has been reported to be a potential oncogene in several carcinomas, including bladder [19], breast [20], neuroblastoma [7], and non-small cell lung cancer [9], which is in line with results of the present study. In breast cancer, SNHG16 is highly expressed and promotes cell growth and invasiveness by sponging miR-98 [20]. A similar analysis also reported that SNHG16 overexpression predicted poor survival in patients with esophageal squamous cell carcinoma [21]. However, the underlying mechanisms by which SNHG16 promotes cancer progression, including in HCC, remain largely unknown. Numerous studies have demonstrated that lncRNA regulatory mechanisms strongly depend on the location of the lncRNA [22]. lncRNAs located in the cytoplasm may serve as partners with miRNA, mRNA, or proteins to regulate target genes in a post-transcriptional pathway [23]. In the present study, ISH indicated that SNHG16 is located in the cytoplasm (Figure 1C), where it may function as a sponge for miRNAs or integrate with proteins to regulate downstream molecules. The detailed mechanisms of SNHG16 in HCC progression and sorafenib resistance will be investigated in the future.
Conclusions
In summary, our results revealed significant associations between SNHG16 expression and various clinicopathologic characteristics and prognosis in patients with HCC. Moreover, survival analysis showed that the lncRNA SNHG16 is an independent prognostic factor for DFS and OS in HCC. These findings suggest that SNHG16 could be used as a pathological marker to identify individuals with poor outcomes and providing a reference for clinical therapy in the future.
Footnotes
Source of support: This work was supported by a grant from the National Nature and Science Foundation of China (No. 81872047)
Conflict of interest
None.
References
- 1.Allemani C, Matsuda T, Di Carlo V, et al. Global surveillance of trends in cancer survival 2000–14 (CONCORD-3): Analysis of individual records for 37 513 025 patients diagnosed with one of 18 cancers from 322 population-based registries in 71 countries. Lancet. 2018;391(10125):1023–75. doi: 10.1016/S0140-6736(17)33326-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhao C, Jin M, Le RH, et al. Poor adherence to hepatocellular carcinoma surveillance: A systematic review and meta-analysis of a complex issue. Liver Int. 2018;38(3):503–14. doi: 10.1111/liv.13555. [DOI] [PubMed] [Google Scholar]
- 3.Lin XQ, Huang ZM, Chen X, et al. XIST Induced by JPX suppresses hepatocellular carcinoma by sponging miR-155-5p. Yonsei Med J. 2018;59(7):816–26. doi: 10.3349/ymj.2018.59.7.816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Malek R, Gajula RP, Williams RD, et al. TWIST1-WDR5-hottip regulates Hoxa9 chromatin to facilitate prostate cancer metastasis. Cancer Res. 2017;77(12):3181–93. doi: 10.1158/0008-5472.CAN-16-2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cao C, Sun J, Zhang D, et al. The long intergenic noncoding RNA UFC1, a target of MicroRNA 34a, interacts with the mRNA stabilizing protein HuR to increase levels of beta-catenin in HCC cells. Gastroenterology. 2015;148(2):415–26. doi: 10.1053/j.gastro.2014.10.012. [DOI] [PubMed] [Google Scholar]
- 6.Zhang Y, Mo WJ, Wang X, et al. Microarraybased bioinformatics analysis of the prospective target gene network of key miRNAs influenced by long noncoding RNA PVT1 in HCC. Oncol Rep. 2018;40(1):226–40. doi: 10.3892/or.2018.6410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu M, Ohira M, Li Y, et al. High expression of ncRAN, a novel non-coding RNA mapped to chromosome 17q25.1, is associated with poor prognosis in neuroblastoma. Int J Oncol. 2009;34(4):931–38. doi: 10.3892/ijo_00000219. [DOI] [PubMed] [Google Scholar]
- 8.Yang T, Jin X, Lan J, et al. Long non-coding RNA SNHG16 has tumor suppressing effect in acute lymphoblastic leukemia by inverse interaction on hsa-miR-124-3p. IUBMB Life. 2019;71(1):134–42. doi: 10.1002/iub.1947. [DOI] [PubMed] [Google Scholar]
- 9.Han W, Du X, Liu M, et al. Increased expression of long non-coding RNA SNHG16 correlates with tumor progression and poor prognosis in non-small cell lung cancer. Int J Biol Macromol. 2019;121:270–78. doi: 10.1016/j.ijbiomac.2018.10.004. [DOI] [PubMed] [Google Scholar]
- 10.Zhao W, Fu H, Zhang S, et al. LncRNA SNHG16 drives proliferation, migration, and invasion of hemangioma endothelial cell through modulation of miR-520d-3p/STAT3 axis. Cancer Med. 2018 doi: 10.1002/cam4.1562. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang W, Lou C, Gao J, et al. LncRNA SNHG16 reverses the effects of miR-15a/16 on LPS-induced inflammatory pathway. Biomed Pharmacother. 2018;106:1661–67. doi: 10.1016/j.biopha.2018.07.105. [DOI] [PubMed] [Google Scholar]
- 12.Wittekind C. [2010 TNM system: on the 7th edition of TNM classification of malignant tumors]. Pathologe. 2010;31(5):331–32. doi: 10.1007/s00292-010-1349-3. [in German] [DOI] [PubMed] [Google Scholar]
- 13.Sahu A, Singhal U, Chinnaiyan AM. Long noncoding RNAs in cancer: From function to translation. Trends Cancer. 2015;1(2):93–109. doi: 10.1016/j.trecan.2015.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gupta RA, Shah N, Wang KC, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464(7291):1071–76. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cui M, Xiao Z, Wang Y, et al. Long noncoding RNA HULC modulates abnormal lipid metabolism in hepatoma cells through an miR-9-mediated RXRA signaling pathway. Cancer Res. 2015;75(5):846–57. doi: 10.1158/0008-5472.CAN-14-1192. [DOI] [PubMed] [Google Scholar]
- 16.Li SP, Xu HX, Yu Y, et al. LncRNA HULC enhances epithelial-mesenchymal transition to promote tumorigenesis and metastasis of hepatocellular carcinoma via the miR-200a-3p/ZEB1 signaling pathway. Oncotarget. 2016;7(27):42431–46. doi: 10.18632/oncotarget.9883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee YR, Kim G, Tak WY, et al. Circulating exosomal non-coding RNAs as prognostic biomarkers in human hepatocellular carcinoma. Int J Cancer. 2019;144(6):1444–52. doi: 10.1002/ijc.31931. [DOI] [PubMed] [Google Scholar]
- 18.Wu D, Yang B, Chen J, et al. Upregulation of long non-coding RNA RAB1A-2 induces FGF1 expression worsening lung cancer prognosis. Cancer Lett. 2018;438:116–25. doi: 10.1016/j.canlet.2018.09.016. [DOI] [PubMed] [Google Scholar]
- 19.Feng F, Chen A, Huang J, et al. Long noncoding RNA SNHG16 contributes to the development of bladder cancer via regulating miR-98/STAT3/Wnt/beta-catenin pathway axis. J Cell Biochem. 2018;119(11):9408–18. doi: 10.1002/jcb.27257. [DOI] [PubMed] [Google Scholar]
- 20.Cai C, Huo Q, Wang X, et al. SNHG16 contributes to breast cancer cell migration by competitively binding miR-98 with E2F5. BiochemBiophys Res Commun. 2017;485(2):272–78. doi: 10.1016/j.bbrc.2017.02.094. [DOI] [PubMed] [Google Scholar]
- 21.Han GH, Lu KJ, Wang P, et al. LncRNA SNHG16 predicts poor prognosis in ESCC and promotes cell proliferation and invasion by regulating Wnt/beta-catenin signaling pathway. Eur Rev Med Pharmacol Sci. 2018;22(12):3795–803. doi: 10.26355/eurrev_201806_15262. [DOI] [PubMed] [Google Scholar]
- 22.Wang XQ, Crutchley JL, Dostie J. Shaping the genome with non-coding RNAs. Curr Genomics. 2011;12(5):307–21. doi: 10.2174/138920211796429772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Geisler S, Coller J. RNA in unexpected places: Long non-coding RNA functions in diverse cellular contexts. Nat Rev Mol Cell Biol. 2013;14(11):699–712. doi: 10.1038/nrm3679. [DOI] [PMC free article] [PubMed] [Google Scholar]