Abstract
Background
Osteoarthritis (OA) of the knee is a common disease that is associated with chronic pain. This study aimed to identify and investigate the functional role of biomarkers associated with long noncoding RNA (lncRNA) in the progression of OA of the knee by lncRNA-associated competing endogenous RNA (ceRNA) integrated network analysis.
Material/Methods
High-quality microRNA (miRNA)-lncRNA and miRNA-mRNA interactions and lncRNA and mRNA expression profiles for patients with OA of the knee with mild and severe pain were obtained from the Gene Expression Omnibus (GEO) database (GSE99662). A three-step computational method was used to construct the lncRNA-associated ceRNA interaction network in OA by integrating miRNA-lncRNA/mRNA interactions and lncRNA/mRNA expression profiles in patients with OA with mild and severe pain.
Results
A total of 1,870 dysregulated lncRNA-mRNA interactions were obtained in the lncRNA-associated ceRNA network in OA, including 476 gain and 1,394 loss interactions, covering 131 lncRNAs and 1,251 mRNAs. Characterization of the lncRNA-associated ceRNA network in OA indicated that lncRNAs had roles in the network. Further differential expression analysis identified eight lncRNA biomarkers, which could distinguish between patients with OA with mild pain and severe pain. These lncRNA-associated interactions showed significantly different co-expression patterns in samples from patients with OA of the knee associated with mild pain.
Conclusions
Integrated network analysis of lncRNA-associated ceRNA identified eight lncRNA molecular biomarkers associated with the progression of OA of the knee.
MeSH Keywords: Biological Markers; Osteoarthritis; RNA, Long Noncoding
Background
Primary osteoarthritis (OA) of the knee is a common chronic disease with increasing incidence and prevalence [1–3]. Effective therapies remain to be discovered to prevent the progression of OA and reduce its severity, and OA is predicted to become an increasing economic burden as the population ages [1,4,5]. Chronic pain from OA is a common public health problem that has a detrimental impact on patient health and function [6]. Chronic pain also reduces the quality of life in patients with OA. Therefore, an accurate assessment of the severity of OA is clinically important [7,8].
Currently, several studies have been published on the association between the expression of long noncoding RNA (lncRNA) and the progression of OA. The lncRNA has a length of more than 200 nucleotides and has widespread biological functions, including regulation of transcriptional patterns and protein activity [9,10]. Recent studies have shown that some lncRNAs are involved in the pathogenesis of OA [11–16]. Dudek et al. reported that lncRNA H19 was overexpressed in chondrocytes and was regulated by SOX9 [17]. Large-scale RNA sequencing studies in patients with OA of the knee with severe pain and mild pain showed that several lncRNAs were differentially expressed, which indicates that these dysregulated lncRNAs may be involved in the progression of OA [1,18]. However, the molecular mechanisms of the role of dysregulated lncRNAs in the progression of OA remain poorly understood and further functional studies are needed to determine these roles.
In 2011, the hypothesis was proposed that competing endogenous RNAs (ceRNAs) could act as miRNA sponges through their miRNA response elements (MREs) and regulate their expression [19]. Several studies have shown that lncRNAs can act as ceRNAs and have important roles in malignancy and chronic diseases, including OA [20–24]. Shen et al. found that the lncRNA-SNHG5 could function as a ceRNA to competitively sponge miR-26a to regulate the expression of mRNA SOX2, and had a role in OA [23]. Also, Li et al. identified the modulating effect of lncRNA X-inactive specific transcript (XIST) in OA [24]. These authors showed that lncRNA-XIST served as a ceRNA for miR-211 to counteract miR-211-mediated CXCR4 suppression and modulate chondrocyte proliferation and apoptosis through downstream mitogen-activated protein kinase (MAPK) signaling [24]. These studies have demonstrated the importance of lncRNAs as ceRNAs in OA. However, the identification of genome-wide expression of lncRNA associated with ceRNA-ceRNA interactions in OA remains unknown.
Therefore, this study aimed to identify and investigate the functional role of biomarkers associated with lncRNA in the progression of OA of the knee by lncRNA-associated ceRNA integrated network analysis.
Material and Methods
Data collection
Human microRNA- long noncoding RNA (miRNA-lncRNA) and miRNA-mRNA target data were collected from StarBase [25,26]. High-quality miRNA-lncRNA and miRNA-mRNA interactions were validated by experiments performed in triplicate that included crosslinking immunoprecipitation (CLIP) and high-throughput sequencing. The miRNA-mRNA interactions were predicted by more than two computing methods as the final dataset. In total, 18,482 and 69,7637 interactions were identified for miRNA-lncRNA and miRNA-mRNA, respectively. Also, lncRNA and mRNA expression profiles for patients with osteoarthritis (OA) of the knee and with mild and severe pain were obtained from the Gene Expression Omnibus (GEO) database (accession GSE99662) [1].
Construction of the integrated network analysis of lncRNA-associated ceRNA
A three-step computational method was used to integrate miRNA-lncRNA/mRNA interactions and lncRNA/mRNA expression profiles in patients with OA with mild and severe pain.
The lncRNA and mRNA pairs were identified that were regulated by same miRNAs. The hypergeometric test was used to determine whether a given lncRNA and mRNA pair significantly shared miRNAs. The Bonferroni correction was performed to control the multiple hypotheses. The lncRNA and mRNA pairs with a false discovery rate (FDR) <0.01 were identified as candidate ceRNA-ceRNA interactions.
Pearson’s correlation coefficients between each lncRNA-mRNA pair in the context of mild pain and severe pain due to OA were identified. Then, the Pearson’s correlation coefficients (PCCs) were defined to measure the lncRNA-mRNA correlation difference between patients with OA of the knee associated with mild pain and severe pain as dPCC=PCCsevere-pain–PCCmild-pain. The lncRNA-mRNA pairs with dPCC >0.5 and significantly positive PCCsevere-pain were considered as gain ceRNA interactions, while lncRNA-mRNA pairs with dPCC <−0.5 and significantly positive PCCmild-pain were considered as loss ceRNA interactions. Assembling all the gain and loss of the ceRNA-ceRNA interactions, a lncRNA-associated ceRNA network in OA was constructed (Supplementary Table 1, available on request from authors). Each node represented a lncRNA or a mRNA, and two nodes were connected if they were dysregulated in OA associated with severe pain.
Network visualization
The networks were visualized by Cytoscape version 3.3.0 [27], including the lncRNA-associated ceRNA network in OA and the differential expression associated subnetwork.
Identification of differential expression of mRNA and lncRNA
Student’s t-test was used to evaluate the significance of differential expression of RNAs between samples of OA associated with mild pain and severe pain. Considering the small sample sizes, the RNAs were selected with P <0.2 as the differentially expressed RNAs [28,29]. Also, the edges were extracted that were related to differentially expressed RNAs and the differential expression subnetwork was constructed of the lncRNA-associated ceRNA network in OA (Supplementary Table 2, available on request from authors).
Results
Construction and characterization of the long noncoding RNA (lncRNA)-associated competing endogenous RNA (ceRNA) network in osteoarthritis (OA)
From the matched lncRNA and mRNA expression profiles for patients with osteoarthritis (OA) and the experimentally validated miRNA-lncRNA/mRNA target interactions, the lncRNA-associated ceRNA network was constructed in OA. The steps taken to construct the network were as described in the Methods. Following the hypergeometric test and Bonferroni correction test to identify the co-regulated lncRNA-mRNA pairs, 40,298 target relationships between 278 lncRNAs and 4,138 mRNAs were identified.
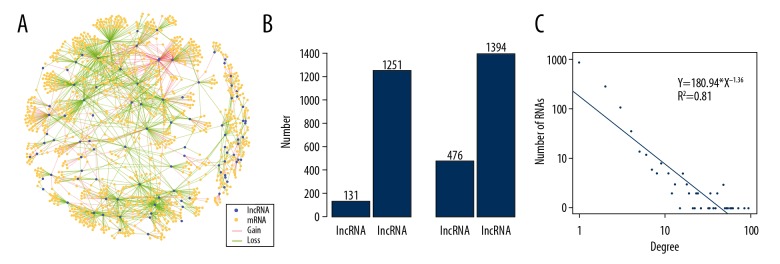
Each pair of lncRNA-mRNAs was found to share at least four miRNAs. The Pearsons’ correlation coefficients (PCCs) were calculated and the significance of each lncRNA-mRNA pair in patients with OA of the knee with mild pain and severe pain was determined. The lncRNA-mRNA pairs with R>0 and P<0.05 were considered as the lncRNA-associated ceRNA network in OA of the knee. The dPCC was defined to measure the different correlation between each lncRNA-mRNA pair in patients with OA of the knee associated with mild pain and severe pain. Combining the dPCC and the p-value using Pearson’s correlation of significance, there were 1,870 dysregulated lncRNA-mRNA interactions identified and the network was constructed of 131 lncRNAs and 1,251 mRNAs (Figure 1A, 1B).
Figure 1.
Dysregulated long noncoding RNA (lncRNA)-associated competing endogenous RNA (ceRNA) network in osteoarthritis (OA) of the knee. (A) Global view of the lncRNA-associated ceRNA network in osteoarthritis. The purple and orange nodes represent lncRNA and mRNA respectively. Red and green edges represent gain and loss interactions in samples from patients with severe osteoarthritis (OA) of the knee compared with samples from patients with mild OA of the knee. (B) Statistics of the nodes and edges of the lncRNA-associated ceRNA network in OA of the knee. (C) The degree of distribution of the lncRNA-associated ceRNA network in OA of the knee.
The lncRNA-associated ceRNA network in OA of the knee included a quarter of the gain ceRNA-ceRNA interactions and three-quarters of the loss of ceRNA-ceRNA interactions (Figure 1B). This result suggested that there were relatively more ceRNA-ceRNA that lost their consistent expression patterns in samples from patients with severe pain associated with OA of the knee. To explore the structure and features of the ceRNA-ceRNA interaction network in OA, the degree of distribution analysis of the nodes in the network was performed. As a result, most nodes had relatively few interactions while a small proportion of nodes had several interactions, which fits with a power law distribution, indicating that the network was scale-free and different from randomly generated networks (Figure 1C).
The lncRNA had central roles in the integrated network of lncRNA-associated ceRNA in OA
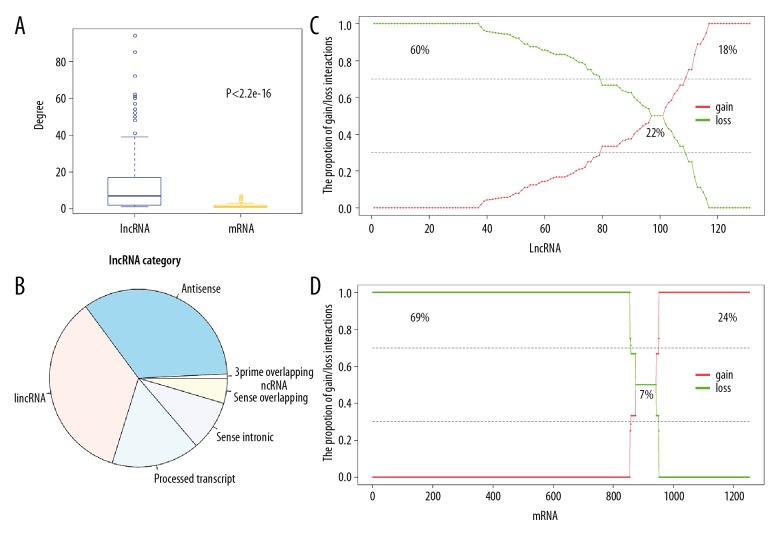
To characterize the roles of lncRNAs and mRNAs in the ceRNA-ceRNA interaction network in OA of the knee, the network degree of each lncRNA and mRNA were calculated. The lncRNAs were found to have more dysregulated neighbors than mRNAs (P<2.2e-16) (Figure 2A). One lncRNA was linked to a maximum of 94 mRNAs while one mRNA was dysregulated with a maximum of seven lncRNAs. This result indicated that lncRNAs tended to be hub nodes and lncRNAs had centralized roles in the integrated network of lncRNA-associated ceRNA in OA. The categories of lncRNAs in the dysregulated network were analyzed and the results showed that a variety of lncRNAs tended to be dysregulated in OA, especially antisense lncRNA and lncRNA (Figure 2B).
Figure 2.
Characteristics of the long noncoding RNA (lncRNA) and mRNAs in the competing endogenous RNA (ceRNA) network interaction in osteoarthritis (OA). (A) The degree of lncRNAs and mRNAs. (B) The category of lncRNAs in the ceRNA-ceRNA network interaction in osteoarthritis (OA). (C) The proportion of gain and loss interactions for each lncRNA in the ceRNA-ceRNA network interaction in OA. (D) The proportion of gain and loss interactions for each mRNA in the ceRNA-ceRNA network interaction in OA.
The gain and loss interactions for each lncRNA were analyzed. For each lncRNA, their proportion of gain and loss interactions were analyzed in all interactions with mRNAs. There were 60% lncRNAs that lost their interactions and 18% lncRNAs had mostly gain interactions with associated mRNAs in patients with OA of the knee that was associated with severe pain (Figure 2C). Also, the gain and loss interactions for each mRNA were calculated. As a result, 69% and 24% of mRNAs were found to be related to loss and gain interactions with lncRNAs in patients with OA of the knee and severe pain (Figure 2D). Only 22% of lncRNAs and 7% of mRNAs showed nondirectional interactions with their neighbors (30–70% gain and 70–30% loss (Figures 2C, 2D).
The differential expression pattern of lncRNAs in the dysregulated ceRNA network
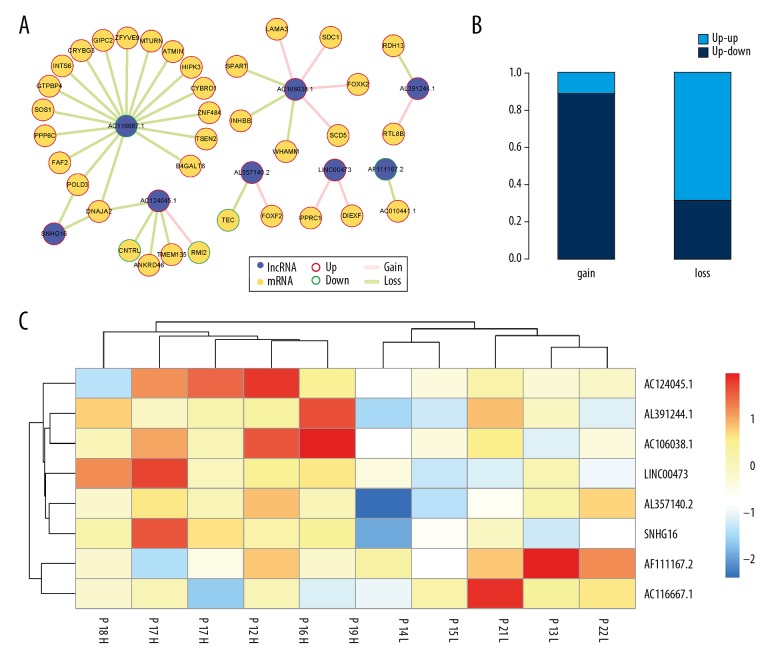
Previous studies have shown that dysregulated ceRNAs contributed to the abnormal transcriptome [30]. Therefore, the expression pattern of lncRNAs and mRNAs in the ceRNA-ceRNA interaction network in OA of the knee were investigated. The upregulated and down-regulated lncRNAs and mRNAs were extracted in samples from patients with OA of the knee associated with severe pain and compared with samples from patients with OA associated with mild pain and identified 16 and 180 differential expression lncRNAs and mRNAs, respectively. Also, the edges were extracted that linked the differentially expressed RNAs and we constructed the differential expression subnetwork with 38 edges, covering eight lncRNAs and 35 mRNAs (Figure 3A). Next, we analyzed the differential expression direction of dysregulated lncRNA-associated ceRNA pairs. As a result, 89% of the gain of ceRNA pairs showed the same dysregulated expression direction, while only 31% of the loss of ceRNA pairs had the same dysregulated expression direction (Figure 3B). This finding was consistent with previous studies [31], and suggested that ceRNA pairs with enhanced co-regulation in samples from patients with OA of the knee associated with severe pain had a consistent expression pattern.
Figure 3.
The eight long noncoding RNAs (lncRNAs) identified as possible biomarkers of osteoarthritis (OA). (A) The differential expression subnetwork of the competing endogenous RNA (ceRNA) network in osteoarthritis. The purple and orange nodes represent lncRNAs and mRNAs, respectively. Red and green edges represent gain and loss interactions. (B) The differential expression pattern of ceRNA pairs in the subnetwork. (C) The unsupervised hierarchical clustering heatmap of samples from patients with OA of the knee associated with mild pain and severe pain based on the expression profile of eight lncRNA biomarkers.
To test whether the eight lncRNAs with a dysregulated expression pattern could distinguish between samples from patients with OA associated with severe pain from samples from patients with OA associated with mild pain, the unsupervised hierarchical clustering for five OA with mild pain and five OA with severe pain with the expression profiles of the eight lncRNAs were performed. The result showed that samples from patients with OA associated with mild pain and samples from patients with OA associated with severe pain were separately classified into two clusters (Figure 3C), suggesting that the eight dysregulated lncRNAs could be biomarkers in prediction of pain intensity in patients with OA of the knee.
SNHG16 and LINC00473 affected OA by dysregulated ceRNA interactions
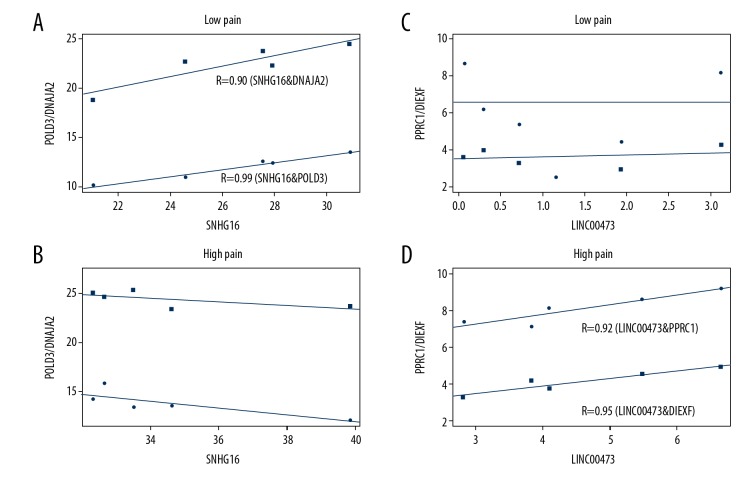
In the differential expression subnetwork, eight dysregulated lncRNAs were identified as candidate biomarkers for OA of the knee, including AC124045.1, AL391244.1, AC106038.1, LINC00473, AL357140.2, SNHG16, AF111167.2, and AC116667.1. Zhu et al. previously showed that SNHG16 was highly expressed in osteosarcoma [32]. In this study, SNHG16 was upregulated in samples from patients with OA of the knee associated with severe pain, suggesting that SNHG16 could promote the severity of OA. Also, the findings suggested that not only the abnormal expression pattern of SNHG16 but also the loss of associated ceRNA interactions (SNHG16 and POLD3, SNHG16 and DNAJA2) could accelerate the severity of OA of the knee (Figure 4A, 4B). However, Zhang et al. found that ZBTB7A could inhibit cisplatin-included apoptosis by repressing LINC00473 expression [33]. We found that LINC00473 was upregulated in patients with severe pain associated with OA and the associated ceRNA interactions were enhanced (Figure 4C, 4D), which showed that LINC00473 and the associated dysregulated ceRNAs might be potential biomarkers for OA.
Figure 4.
The differences in co-expression patterns of long noncoding RNAs (lncRNAs) as biomarkers associated with competing endogenous RNA (ceRNA) pairs. The co-expression patterns of SNHG16 and POLD3/DNAJA2 in patients with osteoarthritis (OA) of the knee associated with mild pain (A) and severe pain (B), and the co-expression patterns of LINC00473 and PPRC1/DIEXF in patients with osteoarthritis (OA) of the knee associated with mild pain (C) and severe pain (D).
Discussion
In this study, a long noncoding RNA (lncRNA)-associated competing endogenous RNA (ceRNA) integrated network analysis was performed in patients with osteoarthritis (OA) of the knee. Gain and loss interactions were obtained. Gain interactions represented the enhanced ceRNA-ceRNA interactions in samples from patients with OA associated with severe pain compared with samples from patients with OA associated with mild pain, while the loss of interactions represented the weakened ceRNA-ceRNA interactions. Analysis of the dysregulated ceRNA network showed that lncRNAs played central roles in the network and most of the lncRNAs or mRNAs were related to one type of dysregulated interaction, gain or loss. Integrating the differential expression analysis of lncRNAs identified eight lncRNA biomarkers. Further analysis showed the efficient prediction of the pain level in OA patients based on the expression profiles of eight lncRNAs with an abnormal expression pattern, indicating that these lncRNAs might have potential as biomarkers for OA. The findings of this study showed novel candidate biomarkers associated with the progression of OA of the knee and identified their specific functional mechanism in promoting OA as dysregulated ceRNAs.
Most previous molecular studies have focused on the aberrant expression pattern of single RNA molecules [1,34]. In the present study, we investigated the difference between samples from patients with OA of the knee associated with mild pain and severe pain by evaluating the competitive interactions between ceRNAs instead of single lncRNAs or mRNAs. Only 12% of lncRNAs and 14% of mRNAs in the ceRNA-ceRNA interaction network in OA of the knee showed differential expression. Also, only 2% of the edges in the ceRNA-ceRNA interaction network in OA contained two differentially expressed RNAs. These results suggested that not only abnormal expression but also the abnormal interaction with other RNAs could influence the progression of OA [31]. Calculating the percentage of gain and loss interactions for each lncRNA and mRNA showed that most lncRNAs or mRNAs tended to enhance or weaken the interactions with their neighbors. The dysregulated interactions between the lncRNA and mRNA could be caused by the abnormal expression of the related miRNAs, the abnormal regulation of miRNAs to the lncRNA or mRNA, and the abnormal length of the 3′UTR of lncRNA or mRNA [35,36]. Further studies are needed to explain these findings and to understand the molecular mechanism of the dysregulation of lncRNA-associated ceRNA interactions in OA of the knee.
Conclusions
By integrating the microRNA (miRNA), long noncoding RNA (lncRNA), and mRNA target interactions and expression of lncRNAs and mRNAs in patients with osteoarthritis (OA) of the knee associated with mild pain and severe pain, it was possible to construct a lncRNA-associated competing endogenous RNA (ceRNA) integrated network. Analysis of the network characteristics identified eight lncRNA biomarkers. The findings support the need for further studies on the potential role of lncRNA biomarkers in OA.
Footnotes
Source of support: This study was supported by the Natural Science Foundation of China (81171692)
Conflict of interest
None.
References
- 1.Bratus-Neuenschwander A, Castro-Giner F, Frank-Bertoncelj M, et al. Pain-associated transcriptome changes in synovium of knee osteoarthritis patients. Genes. 2018;9(7) doi: 10.3390/genes9070338. pii: E338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pereira D, Ramos E, Branco J. Osteoarthritis. Acta Med Port. 2015;28(1):99–106. doi: 10.20344/amp.5477. [DOI] [PubMed] [Google Scholar]
- 3.Flemming DJ, Gustas-French CN. Rapidly progressive osteoarthritis: A review of the clinical and radiologic presentation. Curr Rheumatol Rep. 2017;19(7):42. doi: 10.1007/s11926-017-0665-5. [DOI] [PubMed] [Google Scholar]
- 4.March LM, Bachmeier CJ. Economics of osteoarthritis: A global perspective. Baillieres Clin Rheumatol. 1997;11(4):817–34. doi: 10.1016/s0950-3579(97)80011-8. [DOI] [PubMed] [Google Scholar]
- 5.Running and osteoarthritis: Does recreational or competitive running increase the risk? J Orthop Sports Phys Ther. 2017;47(6):391. doi: 10.2519/jospt.2017.0505. [DOI] [PubMed] [Google Scholar]
- 6.White DK, Master H. Patient-reported measures of physical function in knee osteoarthritis. Rheum Dis Clin North Am. 2016;42(2):239–52. doi: 10.1016/j.rdc.2016.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Creamer P. Osteoarthritis pain and its treatment. Curr Opin Rheumatol. 2000;12(5):450–55. doi: 10.1097/00002281-200009000-00019. [DOI] [PubMed] [Google Scholar]
- 8.Sinusas K. Osteoarthritis: Diagnosis and treatment. Am Fam Physician. 2012;85(1):49–56. [PubMed] [Google Scholar]
- 9.Fu M, Huang G, Zhang Z, et al. Expression profile of long noncoding RNAs in cartilage from knee osteoarthritis patients. Osteoarthritis Cartilage. 2015;23(3):423–32. doi: 10.1016/j.joca.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 10.Cen X, Huang XQ, Sun WT, et al. Long noncoding RNAs: A new regulatory code in osteoarthritis. Am J Transl Res. 2017;9(11):4747–55. [PMC free article] [PubMed] [Google Scholar]
- 11.Li Y, Li S, Luo Y, et al. LncRNA PVT1 regulates chondrocyte apoptosis in osteoarthritis by acting as a sponge for miR-488-3p. DNA Cell Biol. 2017;36(7):571–80. doi: 10.1089/dna.2017.3678. [DOI] [PubMed] [Google Scholar]
- 12.Jiang SD, Lu J, Deng ZH, et al. Long noncoding RNAs in osteoarthritis. Joint Bone Spine. 2017;84(5):553–56. doi: 10.1016/j.jbspin.2016.09.006. [DOI] [PubMed] [Google Scholar]
- 13.Xing D, Liang JQ, Li Y, et al. Identification of long noncoding RNA associated with osteoarthritis in humans. Orthop Surg. 2014;6(4):288–93. doi: 10.1111/os.12147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen WK, Yu XH, Yang W, et al. lncRNAs: Novel players in intervertebral disc degeneration and osteoarthritis. Cell Prolif. 2017;50(1) doi: 10.1111/cpr.12313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fathollahi A, Aslani S, Jamshidi A, Mahmoudi M. Epigenetics in osteoarthritis: Novel spotlight. J Cell Physiol. 2019 doi: 10.1002/jcp.28020. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 16.Lu Z, Luo M, Huang Y. lncRNA-CIR regulates cell apoptosis of chondrocytes in osteoarthritis. J Cell Biochem. 2018 doi: 10.1002/jcb.27997. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 17.Dudek KA, Lafont JE, Martinez-Sanchez A, Murphy CL. Type II collagen expression is regulated by tissue-specific miR-675 in human articular chondrocytes. J Biol Chem. 2010;285(32):24381–87. doi: 10.1074/jbc.M110.111328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Park S, Lee M, Chun CH, Jin EJ. The lncRNA, nespas, is associated with osteoarthritis progression and serves as a potential new prognostic biomarker. Cartilage. 2017 doi: 10.1177/1947603517725566. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Salmena L, Poliseno L, Tay Y, et al. A ceRNA hypothesis: The Rosetta Stone of a hidden RNA language? Cell. 2011;146(3):353–58. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cesana M, Cacchiarelli D, Legnini I, et al. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147(2):358–69. doi: 10.1016/j.cell.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu Y, Lin L, Zou R, et al. MSC-derived exosomes promote proliferation and inhibit apoptosis of chondrocytes via lncRNA-KLF3-AS1/miR-206/GIT1 axis in osteoarthritis. Cell Cycle. 2018;17(21–22):2411–22. doi: 10.1080/15384101.2018.1526603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fan X, Yuan J, Xie J, et al. Long non-protein coding RNA DANCR functions as a competing endogenous RNA to regulate osteoarthritis progression via miR-577/SphK2 axis. Biochem Biophys Res Commun. 2018;500(3):658–64. doi: 10.1016/j.bbrc.2018.04.130. [DOI] [PubMed] [Google Scholar]
- 23.Shen H, Wang Y, Shi W, et al. LncRNA SNHG5/miR-26a/SOX2 signal axis enhances proliferation of chondrocyte in osteoarthritis. Acta Biochim Biophys Sin (Shanghai) 2018;50(2):191–98. doi: 10.1093/abbs/gmx141. [DOI] [PubMed] [Google Scholar]
- 24.Li L, Lv G, Wang B, Kuang L. The role of lncRNA XIST/miR-211 axis in modulating the proliferation and apoptosis of osteoarthritis chondrocytes through CXCR4 and MAPK signaling. Biochem Biophys Res Commun. 2018;503(4):2555–62. doi: 10.1016/j.bbrc.2018.07.015. [DOI] [PubMed] [Google Scholar]
- 25.Yang JH, Li JH, Shao P, et al. StarBase: A database for exploring microRNA-mRNA interaction maps from Argonaute CLIP-Seq and Degradome-Seq data. Nucleic Acids Res. 2011;39:D202–9. doi: 10.1093/nar/gkq1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li JH, Liu S, Zhou H, et al. starBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42:D92–97. doi: 10.1093/nar/gkt1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shannon P, Markiel A, Ozier O, et al. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Birati EY, Hanff TC, Maldonado D, et al. Predicting long term outcome in patients treated with continuous flow left ventricular assist device: The Penn-Columbia risk score. J Am Heart Assoc. 2018;7(6) doi: 10.1161/JAHA.117.006408. pii: e006408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Okumura LM, Antunes VD, Aguiar KS, et al. Tyrosine kinase inhibitors in patients with chronic myelogenous leukemia: Defining the role of social risk factors and non-adherence to treatment. Pharm Pract (Granada) 2015;13(2):559. doi: 10.18549/pharmpract.2015.02.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li M, Wang J, Liu D, Huang H. High throughput sequencing reveals differentially expressed lncRNAs and circRNAs, and their associated functional network, in human hypertrophic scars. Mol Med Rep. 2018;18(6):5669–82. doi: 10.3892/mmr.2018.9557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shao T, Wu A, Chen J, et al. Identification of module biomarkers from the dysregulated ceRNA-ceRNA interaction network in lung adenocarcinoma. Mol Biosyst. 2015;11(11):3048–58. doi: 10.1039/c5mb00364d. [DOI] [PubMed] [Google Scholar]
- 32.Zhu C, Cheng D, Qiu X, et al. Long noncoding RNA SNHG16 promotes cell proliferation by sponging microRNA-205 and upregulating ZEB1 expression in osteosarcoma. Cell Physiol Biochem. 2018;51(1):429–40. doi: 10.1159/000495239. [DOI] [PubMed] [Google Scholar]
- 33.Zhang L, Wang Y, Li X, et al. ZBTB7A enhances osteosarcoma chemoresistance by transcriptionally repressing lncRNALINC00473-IL24 activity. Neoplasia. 2017;19(11):908–18. doi: 10.1016/j.neo.2017.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu B, Wu P, Mei L, et al. [Differential expression of exosomal miRNAs in osteoblasts in osteoarthritis]. Zhong Nan Da Xue Xue Bao Yi Xue Ban. 2018;43(12):1294–300. doi: 10.11817/j.issn.1672-7347.2018.12.003. [in Chinese] [DOI] [PubMed] [Google Scholar]
- 35.Park HJ, Ji P, Kim S, et al. 3′ UTR shortening represses tumor-suppressor genes in trans by disrupting ceRNA crosstalk. Nat Genet. 2018;50(6):783–89. doi: 10.1038/s41588-018-0118-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li L, Wang D, Xue M, et al. 3′UTR shortening identifies high-risk cancers with targeted dysregulation of the ceRNA network. Sci Rep. 2014;4:5406. doi: 10.1038/srep05406. [DOI] [PMC free article] [PubMed] [Google Scholar]