Summary
Stereotyped synaptic connections define the neural circuits of the brain. In vertebrates, stimulus-independent activity contributes to neural circuit formation. It is unknown whether this type of activity is a general feature of nervous system development. Here, we report patterned, stimulus-independent neural activity in the Drosophila visual system during synaptogenesis. Using in vivo calcium, voltage, and glutamate imaging, we found that all neurons participate in this spontaneous activity, which is characterized by brain-wide periodic active and silent phases. Glia are active in a complementary pattern. Each of the 15 examined of the over 100 specific neuron types in the fly visual system exhibited a unique activity signature. The activity of neurons that are synaptic partners in the adult was highly correlated during development. We propose that this cell type-specific activity coordinates the development of the functional circuitry of the adult brain.
eTOC blurb
Akin & Bajar et al. report that stimulus-independent neural activity occurs in cell type-specific patterns during synapse formation in Drosophila. Their findings suggest that neural activity may be a fundamental feature of brain development.
Introduction
Synaptic connections between neurons determine how neural circuits process information. Understanding how the specificity of these connections is established is a central challenge in neurobiology. In vertebrates, cell autonomous genetic programs and neural activity—both evoked and spontaneous—contribute to synapse development. Spontaneous activity has been observed throughout the developing central nervous system (CNS) (reviewed in Blankenship and Feller, 2009). Retinal waves are the best characterized examples of spontaneous activity (Galli and Maffei, 1988; Meister et al., 1991; reviewed in Ackman and Crair, 2014). In mammals, retinal waves occur between the completion of axon guidance and eye opening, and the activity propagates from the retina to higher-order visual centers (Ackman et al., 2012). Significant progress has been made toward characterizing the organizing principles of spontaneous activity in the developing vertebrate brain, and the precise developmental role of this activity is an area of active interest.
By contrast to vertebrates, brain development in invertebrates has been thought to occur largely independent of activity. In the Drosophila visual system, photoreceptor neurons can develop the wild-type complement of synapses in a stimulus-independent manner (Hiesinger et al., 2006). However, the existence and significance of spontaneous activity during invertebrate brain development remains an open question.
Some of the most detailed understanding of brain development in the fly comes from the visual system. Visual information from the compound eye is topographically relayed to the optic neuropils—the lamina, medulla, and the lobula complex, each organized into layers and columns. Over 100 different neuronal cell types form precise synaptic connections, typically with several different cell types as revealed by 3D EM reconstructions (Takemura et al., 2013).
Visual system development in the fly takes place during the last stage of larval development and the ensuing 100 hours of metamorphosis, or pupal development. In the first 50 hours after puparium formation (hAPF), a dynamic self-assembly process mediated by cell surface molecules brings most of the cell types of the visual system to where they belong in the adult brain, ready for synaptogenesis (Hadjieconomou et al., 2011). Over the remaining 50 hours of pupal development, synapse assembly proceeds in parallel with the upregulation of genes involved in neural activity and new sets of cell recognition molecules (Chen et al., 2014; Zhang et al., 2016), and vast networks comprising distant cells are assembled through specific synaptic connections and compatible gene expression profiles (e.g. matching neurotransmitter systems and receptors). Little is known about the molecules and mechanisms that coordinate this period of brain development.
Here we report the discovery of stimulus-independent neural activity in the developing Drosophila CNS and its initial characterization in the visual system. We find that the visual system as a whole, all 15 of the individual neuronal cell types examined, as well as astrocytes, participate in patterned, stimulus-independent neural activity (PSINA), during the late stages of circuit assembly.
Results
Patterned neuronal and glial activity in the developing fly brain
To assess whether neural activity contributes to visual system development in Drosophila, we used an in vivo 2-photon imaging protocol (Akin and Zipursky, 2016; Langen et al., 2015). We expressed the genetically-encoded calcium indicator (GECI) GCaMP6s (Chen et al., 2013) throughout the central nervous system using a pan-neuronal GAL4 driver. Between 40 and 50 hAPF, the optic lobe is largely inactive, aside from sporadic, isolated activity with no clear spatial or temporal coordination (Movie S1). Starting shortly before 50 hAPF, a subset of neuronal processes begins to exhibit periodic bouts of increased fluorescence. By 55 hAPF, processes in all optic neuropils, as well as fibers originating from the central brain, participate in regular 12–15 minute long cycles comprising active and silent phases (Figures 1A–1B, Movie S1).
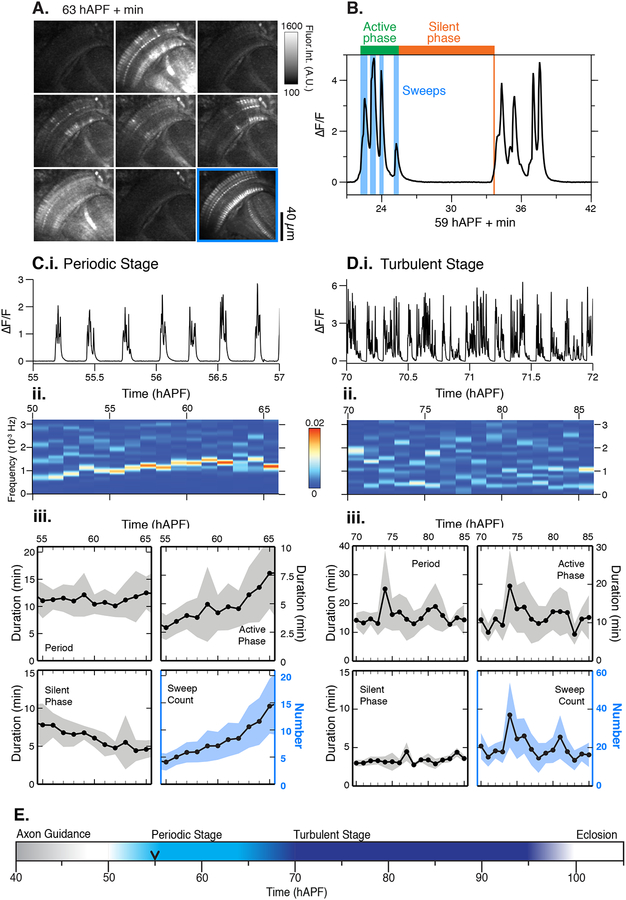
Figure 1. Patterned stimulus independent neural activity (PSINA) in the developing visual system.
A. Micrograph montage showing a single cycle at 63 hAPF; framed panel (lower right) is the average intensity projection through the active phase. B. Representative cycle. C.i. Representative trace during the periodic stage (55–65 hAPF). C.ii. Frequency analysis (Fourier transform) between 50–65 hAPF; C.iii. Average traces of cycle metrics in the periodic stage (n = 54 columns from 6 flies). Shaded area, standard deviation. D.i. Representative trace during the turbulent stage (70 hAPF to eclosion); D.ii. Frequency analysis (Fourier transform) between 70–85 hAPF; D.iii. Average traces of cycle metrics in the turbulent stage (n = 46 columns from 4 flies). Shaded area, standard deviation; E. Summary of spontaneous activity stages during pupal development. Black arrowhead marks the time point after which 100% of columns participate in each cycle. See Table S1 for genotypes.
The active phase of each cycle comprises several distinct bouts of activity, which we term sweeps (Figure 1B). Between 55 and 65 hAPF, or the periodic stage, the cycle period remains roughly constant (~0.001–0.002 Hz), the number of sweeps per cycle and the duration of the active phase increase, and the silent phase decreases (Figure 1C). There were no significant differences between periodicity, sweeps per cycle, active and silent phase durations between different animals, consistent with the notion that the mechanisms underlying these metrics are stereotyped (Figure S1A).
To study the activity beyond 65 hAPF, we moved our expression system to the cn,bw genetic background (Thimann and Beadle, 1937) to eliminate retinal pigmentation which allows for imaging through eclosion. We found no significant differences in periodic stage metrics between cn,bw flies and control flies (Figure S1A). By 70 hAPF, the simple temporal pattern is replaced with multiple frequencies reflecting cycles with variable periods (Figure 1D, Movie S1). Compared to the periodic stage, during the, turbulent stage individual cycles exhibit higher sweeps per cycle, and, on average, longer active and shorter silent phases (Figure 1D). The turbulent stage persists until the final hour of pupal development, after which the activity dissipates before eclosion (Figure S1B). Thus, pupal activity is divided into an early periodic stage and a later turbulent stage, and continues until the final hour of development (Figure 1E).
We next asked whether activity was present beyond the visual system. Recently, a detailed study of motoneuron development showed that peripheral neurons exhibit periodic bouts of activity, starting at 48 hAPF, which grow stronger as development proceeds (Constance et al., 2018). In pupae, the pan-neuronally driven GCaMP6s was detectable using a widefield epifluorescence microscope, making it possible to image the whole CNS of multiple animals simultaneously. We observed cycles of activity that matched our observations from the 2-photon setup in the optic lobes and the central brain (Figure 2A, Movie S2). We also saw that the pattern of activity is the same in both sexes (Figure S2A).
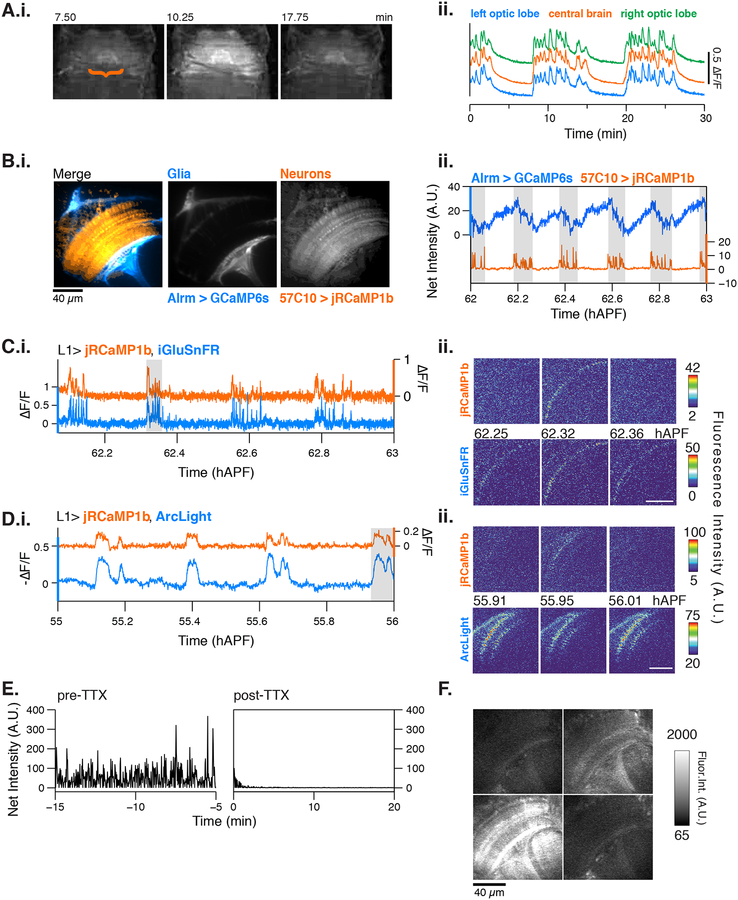
Figure 2. Characterization of PSINA.
A.i. Representative epifluorescence images of a single cycle in an intact pupa expressing pan-neuronal GCaMP6s.. A.ii. Average traces from ROIs encircling the left optic lobe (blue), central brain (orange), and right optic lobe (green) between 58–60 hAPF. B.i. Representative micrograph showing astrocytes expressing GCaMP6s (blue) and pan-neuronal expression of jRCaMP1b (orange). Scale bar, 40 μm. B.ii. Representative trace comparing glial (blue) and neuronal activity from (orange) between 62–63 hAPF. Active phases of the neuronal cycles are shaded in gray. C. Representative traces (i.) and micrographs (ii.) from L1 neurons expressing jRCaMP1b (orange, top) and iGluSnFr (blue, bottom). Note that iGluSnFr reports more sweeps than jRCaMP1b; we suspect that the L1-expressed glutamate sensor’s responds to neurotransmitter released by L1 itself, neighboring cells or both. D. Representative traces (i.) and micrographs (ii.) from L1 neurons expressing jRCaMP1b (orange, top) and ArcLight (blue, bottom). E. Representative traces of activity as reported by panneuronal GCaMP6s before (left) and after (right) addition of 1μM tetrodotoxin. F. Micrographs of norpAnull mutant flies expressing pan-neuronal GCaMP6s shows that visual stimuli are not required for activity. See Table S1 for genotypes.
Given the broad domain of the activity, we also examined glia using orthogonal expression systems to express GCaMP6s in astrocytes and jRCaMP1b in all neurons (Figures 2B, S2B–S2C). Before 55 hAPF, there is no significant correlation between the glial GECI signal and neuronal activity (Figure S2B). This changes during the periodic stage, when the glia exhibit tonic oscillations alongside the neuronal cycles (Figures 2B and S2C). The glial cycles are phase-shifted with respect to those of neurons: When neurons are active, astrocytes show a progressive loss in GECI signal, which is rebuilt during the neuronal silent phase (Figures 2B, S2D–S2E).
To establish whether the oscillatory activity ceases in the adult, we used a head-fixed cranial window preparation of adult flies (Aptekar et al., 2015; Seelig et al., 2010). We observed stimulus-independent activity in newly eclosed (1 hr old) flies as well as 1- and 5-day old adults, all of which also had intact, robust responses to visual stimuli (Figure S3A). Stimulus-independent activity in the adult did not engage the entire optic lobe, exhibited fewer sweeps in a given cycle, oscillated at a higher frequency (Figure S3B), and diminished with age. These differences suggest the involvement of different mechanisms in generating the pupal and adult stimulus-independent activities.
Activity correlates with changes in membrane voltage and neurotransmitter release and is independent of visual stimulus
To assess whether the pupal calcium activity is due to neuronal excitation, we examined the relationship of the pupal GECI signal to neurotransmitter release and changes in membrane potential by co-expressing the red-shifted GECI, jRCaMP1b (Dana et al., 2016), with either the genetically encoded glutamate sensor iGluSnFR (Marvin et al., 2013) or the genetically encoded voltage indicator ArcLight (Cao et al., 2013; Jin et al., 2012). Pan-neuronal co-expression of both indicator pairings revealed glutamate release and membrane voltage dynamics that correlated with the GECI signal (Figures S4A–B). All neuronal cell types we studied (see below) also display the same GECI activity pattern we describe for pan-neuronal expression. As such, we co-expressed the indicators in a single cell type, the L1 lamina monopolar neuron, a glutamatergic first-order interneuron (Gao et al., 2008). We observed strong correlation between the GECI signal and both the iGluSnFR-reported glutamate release (Figure 2C, Movie S3) and ArcLight-reported drops in membrane voltage (Figures 2D, Movie S4) at the level of individual sweeps. Indeed, cross-correlograms between GECI signals and iGluSnFR or ArcLight showed a sharp peak at a lag time of 0 s, and both cross-correlograms and auto-correlograms showed subsequent peaks at lag times of 12–15 mins, reflecting the shared active and silent phases between indicators (Figure S4C).
To pharmacologically manipulate neural activity, we used the head-fixed cranial window preparation in late stage pupae (90–95 hAPF). We found that calcium activity is severely attenuated with the administration of tetrodotoxin, a voltage-gated sodium channel blocker that inhibits action potentials (Figures 2E and S4D). Together, these results indicate that the GECI signal observed during pupal development reflects neuronal electrical activity.
Next, we assessed the contribution of visual stimulus to the activity in three classic vision mutants: hdc (histidine carboxylase, required for the synthesis of the photoreceptor neurotransmitter histamine (Burg et al., 1993)), ort (histamine receptor expressed by the post-synaptic partners of photoreceptors (Gengs et al., 2002)), and norpA (phospholipase C essential to phototransduction (Bloomquist et al., 1988)). In all mutants, the activity was still present during pupal development (Figure 2F, norpAnull shown, Figure S5A). In hdc and ort null animals, the frequency of the periodic stage cycles was comparable to wildtype (Figure S5B), suggesting that histamine signaling in visual processing, or more broadly in the CNS, is not required to drive the activity or establish its rhythm. By contrast, with NorpA we found a dose dependent-decrease in the frequency of the cycles (Figure S4A–S5B). The null allele used in this work, norpA36 (Pearn et al., 1996), removes both the retina-specific isoform and a more broadly expressed second isoform of the gene (Kim et al., 1995). We confirmed that retinal function was indeed abolished by testing norpA36 animals for optomotor response to widefield stimulus and closed-loop bar fixation (Figure S5C–E). The dose-dependent changes to the period with NorpA may reflect its function as a non-specialized phospholipase C acting broadly in the developing CNS, or in a subset of cells, to regulate some aspect of intracellular signaling or Ca2+ homeostasis. These results are consistent with the reported timing for the onset of photoreceptor light response at 82 hAPF, which is ~27 hours after the onset of activity (Hardie et al., 1993). As the the pupal neuronal activity is independent of visual stimuli, we refer to this phenomenon observed in the development of the Drosophila CNS as patterned, stimulus-independent neuronal activity, or PSINA (pronounced ‘see-na’) in the remainder of the text.
Cell type-specific dynamics of PSINA
We assessed PSINA in specific neuronal types in the visual system with 2-photon imaging of GCaMP6s, in 15 cell types, representing some of the major visual system classes (i.e. photoreceptors (R7, R8), lamina monopolar neurons (L1, L3, L5), medulla intrinsic neurons (Mi1, Mi4), distal medulla neurons (Dm3, Dm4, Dm9), transmedullary neurons (Tm3, Tm4, Tm9), and T neurons (T4, T5)) (Figure 3A). During the periodic stage, the temporal pattern of PSINA in all neurons followed the pan-neuronal archetype; as a group, cells of a type cycled through active and silent phases lasting 12–15 minutes, starting around 50–55 hAPF and gradually increasing the duration and the sweep complement of the active phase over time (Figure 3B).
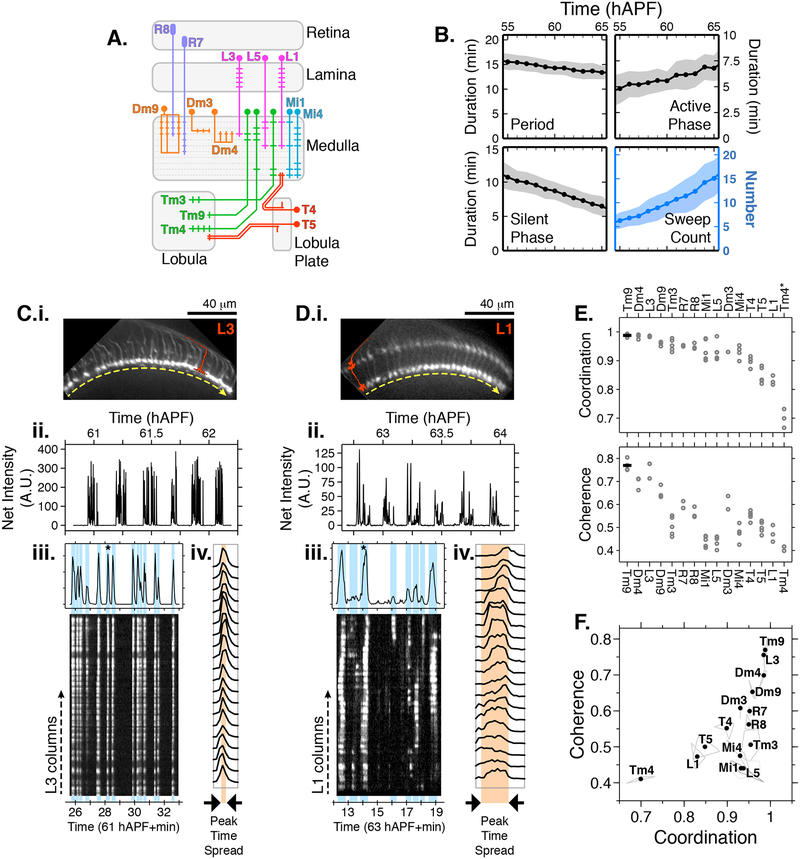
Figure 3. Cell type specific PSINA dynamics.
A. Schematic of visual system cell types described in Figures 3 and 4. B. Cycle metrics in the periodic stage, averaged over 15 cell types and 55 time series (i.e. flies). Shaded areas, standard deviation. C. PSINA dynamics in L3 cells. C.i. Average intensity projection of GCaMP6s expressing L3 processes in the M3 layer of the medulla neuropil. Single L3 schematically shown in red. Dashed yellow arrow sits below the thin profile through M3 used to generate the kymograph in (iii); direction matches the layout of the columns in the kymograph. C.ii. Average net fluorescence intensity along the profile described in (i). Active phase with gray background shown in greater detail in (iii). C.iii. Plot shows expanded view of an active phase with sweeps highlighted in light blue. Star marks the sweep expanded into individual column traces in (iv). Kymograph of net fluorescence derived from the profile described in (i). C.iv. Plot of fluorescence change in individual medulla columns in the star marked sweep in (iii). D. Same as (C) for an L1 time series. Kymograph generated from a thin profile through the L1 processes in M5 (i.e. layer just above the yellow line). E. Coordination (top) and coherence (bottom) values calculated for different cell types. Round gray markers are individual time series, black bars are the average for each cell type. Data from 2–6 flies shown for each cell type. Metrics for each time series calculated over 55–65 hAPF, using an average of 41+/−9 cycles and 10–20 columns per cycle. *The outlier coordination value of Tm4 is due to sparse labeling of this cell type with the driver used. F. Scatter plot of coordination v. coherence. Vertices of light gray polygons, individual time series; black dots, average for each cell type. See Table S1 for genotypes used in this figure.
While the broad temporal characteristics of PSINA are shared, how the activity propagates across the repeated columnar array of a cell type varies. For example, nearly all L3s participate in every sweep of an active phase while in L1s, fractional participation can change between sweeps (Figures 3C–3D, Movie S5). Further, during a sweep, L3s reach peak intensity within a narrower time window compared to L1s (Figures 3C–3D). To compare PSINA dynamics between repeated observations of the same cell type and across different cell types, we defined two scalar metrics, coordination and coherence. Coordination is the average of the fraction of columns that participate in each sweep. Coherence is the largest fraction of columns that peak within the same time point, averaged over all sweeps. Distinct observations of PSINA in L3s all yield comparably high coordination and coherence values in contrast to L1 (Figure 3E). For the 15 cell types we studied, coordination and coherence values from separate observations cluster around means characteristic to each cell type, independent of the specific drivers used for GECI expression or transgenic constitution of the animals (Figures 3F and S6A, Movie S6). For most cell types, coordination and coherence are roughly constant during the periodic stage, between 55–65 hAPF (Figure S6B). In the few that do show changes, we observe loss of coordination that is attributable to loss of image quality as developing retinal pigmentation degrades the observed GECI signal, particularly with weaker cell type-specific drivers. A notable exception is L1; here, despite the loss of net signal, both metrics increase over time (Figure S6B), consistent with evolving PSINA dynamics in L1 (Figures S6C–S6D).
We also analyzed PSINA dynamics using a generalized approach to measure correlated activity, the spike time tiling coefficient (STTC, Cutts and Eglen, 2014) (Figure S7), which has been used to study the distance-dependent correlation of propagating activity, such as retinal waves (e.g. Xu et al., 2016). With STTC, we again found that different cell types cluster around characteristic values, both for STTC at the shortest distance value (4.5 μm) and for a measure of STTC decay at a fixed distance (36 μm) (Figure S7C–S7D). While differences in both sampling rate and the physical scales involved confound direct quantitative comparisons to STTC analyses of retinal waves (Cutts and Eglen, 2014), this analysis points to a diversity of wave-like propagation patterns, some of which are qualitatively similar to the dynamics seen in the vertebrate retina. While there are some similarities in how coordination-coherence and STTC rank each cell type, such as the highly synchronous set of Tm9, L3, and Dm4, these approaches are not directly comparable since the former analyzes individual sweeps and the latter full active phases of each cycle.
In summary, we find that the fine spatiotemporal structure of PSINA is cell type-specific, stereotyped and can be dynamic over the course of development.
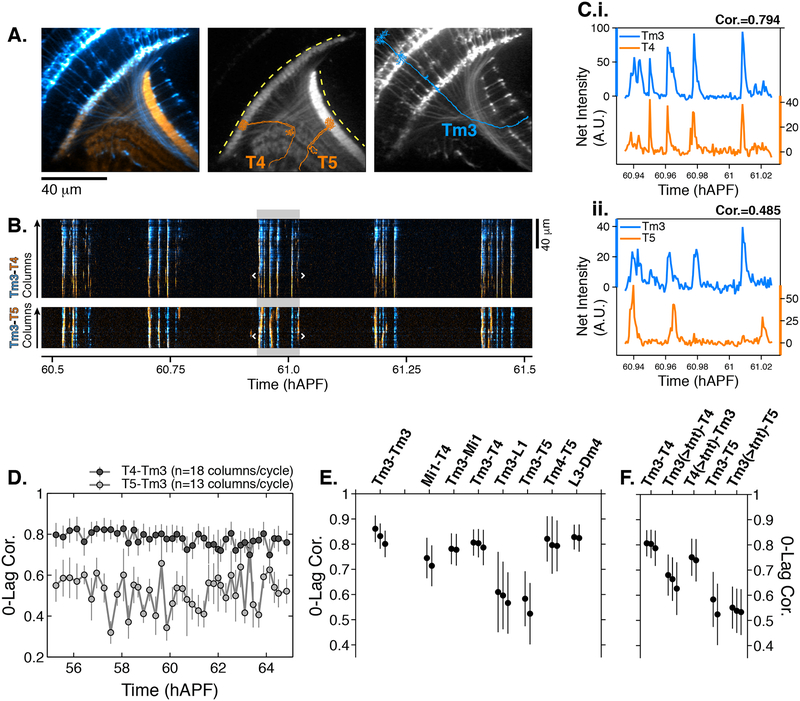
The two approaches used to analyze cell type specific PSINA patterns rely on ensemble metrics that do not preserve cell or column specific timing information and cannot inform on correlated activity between two cell types. To measure this directly, we imaged pairs of neurons expressing red and green GECIs (Figure 4). For example, we compared the activity in Tm3 transmedullary neurons, with processes in both the medulla and the lobula neuropils, to the medulla-resident dendrites of T4s and the lobula-resident dendrites of T5s (Figures 4A–4D, Movie S7). Between 55–65 hAPF, the Tm3-T4 activity was highly correlated (0.8+/−0.06, n=3) while the Tm3-T5 correlation was significantly lower (0.55+/−0.1, n=2) (Figures 4D–4E, S8A–S8B). The results were the same when these measurements were repeated with the opposite cell type and color pairing (Figure S8C). Notably, in the adult, Tm3 and T4 are synaptic partners in the ON-motion circuit (reviewed in Silies et al., 2014) while T5, which is part of the OFF-motion circuit, is not a synaptic partner with Tm3.
Figure 4. Synaptic release is required for correlated PSINA activity.
A. Average intensity projection images of GCaMP6s expressing Tm3 (blue) and RCaMP1b expressing T4–5 (orange) cells. Single Tm3, T4, and T5 projections are schematically shown. Dashed yellow arcs in center panel abut the thin profiles through M9–10 and the lobula used to generate the kymographs in (B). B. Tm3-T4 (top) and Tm3-T5 (bottom) kymographs of net fluorescence derived from the profiles described in (A). Columns between the white brackets in active phase with gray background were used to generate the plots in (C). C. Tm3-T4 (i) and Tm3-T5 (ii) net fluorescence intensity along the columns marked in (B). D. 0-Lag cross correlation values between 55–65 hAPF for Tm3-T4 (dark gray) and Tm3-T5 (light gray) for the time series used in (A-C). Markers are the average correlation value for 10–20 columns per cycle, gray vertical lines are standard deviation. E. 0-lag correlation values for pairs of cell types, averaged over 55–65 hAPF. Black markers and vertical lines are the average and standard deviation for each time series. Data from 2–3 flies shown per pair. 43+/−10 cycles with 15+/−3 columns per cycle used for each fly. The Tm3-Tm3 pair represents the highest correlation we expect to observe for a perfect match given the signal-to-noise statistics of the data (see Figures S6A–S6B). F. TNT expression in Tm3 reduces Tm3-T4 correlation but has no effect on Tm3-T5 correlation. Data statistics as in (E). Unperturbed pairs reproduced from (E) for ease of comparison. See Table S1 for genotypes used in this figure.
Downstream of photoreceptors, L1 provides the principal input to the ON-motion circuit, with Tm3 and Mi1 as its major post-synaptic partners, which then synapse with T4 (Figure 3A). Activities of the Mi1-Tm3 and Mi1-T4 pairs are also well correlated while L1-Tm3 has lower correlation (Figure 4E). As discussed above, PSINA dynamics in L1 evolve through pupal development, and may converge with a presumptive ON-motion PSINA channel during the turbulent stage. Alternatively, if PSINA is propagated through some form of synaptic coupling, the low L1-Tm3 correlation may report that L1 is an inhibitory synaptic partner at this stage of development.
Returning to T5, we found that its activity is highly correlated with Tm4, an OFF-motion circuit input into T5 in the adult (Shinomiya et al., 2014) (Figure 4E). Finally, we observed highly correlated activity between a pair of high coordination-coherence cells, L3 and Dm4 (Figure 4E), which are also synaptic partners in the adult. Together, these results confirm the presence of multiple distinct channels of PSINA activity.
Correlated activity patterns between many adult synaptic partners ~45 hours before the end of development hinted at an early form of synaptic pairing. To address this, we expressed tetanus toxin (TNT) in one cell type of a pair and measured the correlation of the PSINA activities as before. Driving TNT expression in Tm3s reduced the correlation of the Tm3-T4 pair significantly while the Tm3-T5 value was unaffected (Figure 4F). By contrast, T4 expression of TNT had no effect on the Tm3-T4 correlation (Figure 4F), indicating that the correlated PSINA activity in Tm3 and T4 is dependent on synaptic release from Tm3 and that PSINA propagation is achieved through some form of synaptic coupling.
Discussion
We report the discovery and initial characterization of PSINA in the developing fly visual system. We observe three distinct stages of PSINA: a periodic stage (55 to 65 hAPF), a turbulent stage (70 hAPF to the final hour of pupal development), and an adult stage that persists alongside mature stimulus responses through at least five days after eclosion. In the periodic stage, which coincides with the onset of synaptogenesis in the fly CNS (Chen et al., 2014; Muthukumar et al., 2014), each of the 15 neuronal cell types analyzed exhibited stereotyped and distinct activity patterns. Many adult synaptic partners had correlated activity, which depended on synaptic release.
Distinct periodic calcium dynamics were also seen in astrocytes. Astrocytes in the developing adult brain elaborate processes which infiltrate the neuropil during synapse formation (Muthukumar et al., 2014; Richier et al., 2017). Ablating astrocytes significantly reduces the total synapse count in the brain, supporting a role for these cells in regulating synaptogenesis (Muthukumar et al., 2014). Our finding that astrocytes exhibit cycles of calcium activity alongside PSINA raises the possibility that astrocytes and PSINA contribute to the formation, specificity, or maturation of synapses within the visual system.
Work on retinal waves supports a role for stimulus-independent activity in synaptic development. In the vertebrate visual system, retinal ganglion cell (RGC) projections target the lateral geniculate nucleus (LGN) and the superior colliculus (SC), where they create retinotopic maps of the visual field and segregate based on the eye of origin. Retinotopy and eye-specific segregation in the LGN and SC are refined during cholinergic retinal waves. Early work in the field established that pharmacological manipulation of spontaneous activity in the cat retina disrupts the organization of RGC projections in the LGN (Shatz and Stryker, 1988; Sretavan et al., 1988). Later studies, using progressively more refined methods, have shown that disrupting the cholinergic circuit of starburst amacrine cells and RGCs largely eliminates retinal waves and leads to defects in the refinement of retinotopy and eye-specific segregation of RGC projections (Bansal et al., 2000; Burbridge et al., 2014; McLaughlin et al., 2003; and others).
The diversity of cell types in the vertebrate retina—as many as 30 for RGCs alone (Sanes and Masland, 2015)—is comparable to the fly visual system. Whether there is cell type-specific texture to the retinal waves similar to PSINA described here is not known, although broad classes of RGCs and cone bipolar cells have been shown to exhibit temporally offset firing patterns (Akrouh and Kerschensteiner, 2013; Kerschensteiner and Wong, 2008). With improving genetic handles for distinct retinal cell types and ongoing efforts at describing the high resolution connectome, it will be possible to explore cell type-specific patterns and the contribution of retinal waves to retinal circuitry (Seung and Sümbül, 2014).
In Drosophila, peristaltic contractions of body wall muscles have recently been appreciated as part of broad neuronal activity during embryonic development (Baines and Bate, 1998; Vonhoff and Keshishian, 2016). This activity is similar to PSINA observed during pupal development with respect to periodicity and timing relative to synapse formation and refinement. Preventing motor neurons from participating in this neuronal activity, or disrupting calcium-dependent intracellular signaling results in ectopic synapses (Carrillo et al., 2010; Vonhoff and Keshishian, 2016).
Based on previous studies in the fly visual system, and of the role of spontaneous activity in other systems, we propose a general conceptual framework for the role of PSINA in regulating the assembly of the adult connectome. Our results suggest that some adult synaptic pairings are already established by 55 hAPF, soon after the first pre-synapses can be detected and co-incident with the onset of PSINA. The global coordination of PSINA indicates that an early connectome, one that must be built through largely contact-dependent interactions of over 100 different neuronal cell types, is present at this time. While the level of organization achieved through such activity-independent mechanisms is astonishing, the early connectome may still be an approximation of what is required in the adult.
PSINA, by orchestrating cellular communication at temporal and spatial scales inaccessible to other signaling mechanisms, may act to refine this draft connectome to complete the self-assembly of the brain. The diversity of spatiotemporal patterns in PSINA presents many opportunities to establish circuit-specific topographic integration. Sweeps of activity repeatedly coursing through the brain through different ‘channels’ could link distinct sets of neurons to direct coordinated morphological changes and sculpt cell-cell contacts, strengthen synapses with correct targets while weakening incorrect pairings, and control transcription programs that direct circuit refinement (Lee et al., 2017; Tyssowski et al., 2018). PSINA may act as a ‘dress rehearsal’ for neural networks, preparing for ‘opening night’ at the completion of development. Individual cells know their own lines, with whom they interact, and their respective positions on the stage; however, repeated practice of each scene ensures that each of the cast can perform as part of a whole ensemble.
We find it remarkable that as a process that engages most, if not all, of the CNS, PSINA is the collective output of the genetically hardwired developmental programs of individual neurons. Thus, the organizing principles, the driving forces, and the functional significance of PSINA at the level of circuits, cells, and molecules should be discoverable through genetic analysis. Undertaking this effort in the fly visual system, where structures analogous to the vertebrate retinal plexiforms, LGN, and SC (Sanes and Zipursky, 2010) are compactly organized in a single microscopic field of view and for which the EM-derived connectome is available, may yield valuable insights into whether and how PSINA affects synaptic specificity and circuit maturation. We expect that the ever-expanding genetic toolkit of Drosophila, complemented with improvements in genomic/transcriptomic analysis and imaging technology, will offer a robust experimental track toward understanding PSINA’s contribution to brain development and function.
STAR METHODS
Key Resources Table
The table highlights the genetically modified organisms and strains, cell lines, reagents, software, and source data essential to reproduce results presented in the manuscript. Depending on the nature of the study, this may include standard laboratory materials (i.e., food chow for metabolism studies), but the Table is not meant to be comprehensive list of all materials and resources used (e.g., essential chemicals such as SDS, sucrose, or standard culture media don’t need to be listed in the Table). Items in the Table must also be reported in the Method Details section within the context of their use. The number of primers and RNA sequences that may be listed in the Table is restricted to no more than ten each. If there are more than ten primers or RNA sequences to report, please provide this information as a supplementary document and reference this file (e.g., See Table S1 for XX) in the Key Resources Table.
Please note that ALL references cited in the Key Resources Table must be included in the References list. Please report the information as follows:
REAGENT or RESOURCE: Provide full descriptive name of the item so that it can be identified and linked with its description in the manuscript (e.g., provide version number for software, host source for antibody, strain name). In the Experimental Models section, please include all models used in the paper and describe each line/strain as: model organism: name used for strain/line in paper: genotype. (i.e., Mouse: OXTRfl/fl: B6.129(SJL)-Oxtrtm1.1Wsy/J). In the Biological Samples section, please list all samples obtained from commercial sources or biological repositories. Please note that software mentioned in the Methods Details or Data and Software Availability section needs to be also included in the table. See the sample Table at the end of this document for examples of how to report reagents.
SOURCE: Report the company, manufacturer, or individual that provided the item or where the item can obtained (e.g., stock center or repository). For materials distributed by Addgene, please cite the article describing the plasmid and include “Addgene” as part of the identifier. If an item is from another lab, please include the name of the principal investigator and a citation if it has been previously published. If the material is being reported for the first time in the current paper, please indicate as “this paper.” For software, please provide the company name if it is commercially available or cite the paper in which it has been initially described.
- IDENTIFIER: Include catalog numbers (entered in the column as “Cat#” followed by the number, e.g., Cat#3879S). Where available, please include unique entities such as RRIDs, Model Organism Database numbers, accession numbers, and PDB or CAS IDs. For antibodies, if applicable and available, please also include the lot number or clone identity. For software or data resources, please include the URL where the resource can be downloaded. Please ensure accuracy of the identifiers, as they are essential for generation of hyperlinks to external sources when available. Please see the Elsevier list of Data Repositories with automated bidirectional linking for details. When listing more than one identifier for the same item, use semicolons to separate them (e.g. Cat#3879S; RRID: AB_2255011). If an identifier is not available, please enter “N/A” in the column.
- A NOTE ABOUT RRIDs: We highly recommend using RRIDs as the identifier (in particular for antibodies and organisms, but also for software tools and databases). For more details on how to obtain or generate an RRID for existing or newly generated resources, please visit the RII or search for RRIDs.
Please use the empty table that follows to organize the information in the sections defined by the subheading, skipping sections not relevant to your study. Please do not add subheadings. To add a row, place the cursor at the end of the row above where you would like to add the row, just outside the right border of the table. Then press the ENTER key to add the row. Please delete empty rows. Each entry must be on a separate row; do not list multiple items in a single table cell. Please see the sample table at the end of this document for examples of how reagents should be cited.
TABLE FOR AUTHOR TO COMPLETE
Please upload the completed table as a separate document. Please do not add subheadings to the Key Resources Table. If you wish to make an entry that does not fall into one of the subheadings below, please contact your handling editor. (NOTE: For authors publishing in Current Biology, please note that references within the KRT should be in numbered style, rather than Harvard.)
Contact for Reagent and Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by Orkun Akin (akin.orkun@gmail.com).
Experimental Model and Subject Details
Flies were reared at 18°C or 25°C on standard cornmeal/molasses medium. Pupal development was staged with respect to white pre-pupa formation (0 hAPF) or head eversion (12 hAPF). The specific developmental age studied in each experiment is indicated in the figures and figure legends. Female flies were used for all experiments except those involoving hemizygous NorpA males shown in Figure S5B–E; see Table S1 for the genotypes used in all figures. The GAL4/UAS and LexA/LexAop expression systems (Brand and Perrimon, 1993; Lai and Lee, 2006) were used to drive cell type-specific transgene expression; complete genotypes used in each experiment can be found in Table S1.
Method Details
2P Imaging of the Developing Visual System
Pupae were prepared for imaging as previously described (Akin and Zipursky, 2016). Briefly, the cuticle around the heads were removed with fine forceps and the animals were attached eye-down on a coverslip coated with a thin layer of embryo glue. A water reservoir on the objective side of the coverglass provided sufficient immersion medium to last through the hours-long imaging sessions; another reservoir below the pupae kept the animals from dehydrating.
Time-lapse imaging of the visual system was carried out on a custom-built 2P microscope (Akin and Zipursky, 2016) equipped with a 20x water immersion objective (Zeiss, W Plan-Apochromat 10x/1.0 DIC) and 2 GaAsP detectors (Hamamatsu). Over the 2–24 hr imaging sessions, the pupae were kept at 25°C using an objective heater system (Bioptechs). A tunable Ti:Sapphire pulsed laser (Chameleon Ultra II, Coherent) was used as the light source. Green fluors were excited at 940 or 970 nm with ~30 mW under-the-objective power; 1020 nm at ~60 mW was used for red fluors and two-color imaging. Animals imaged under these conditions developed normally and eclosed on schedule. To observe a thicker cross-section of the visual system than possible with a single optical slice, we used the maximum intensity projection of three successive images taken 2 μm apart in the z-axis as the frame for an individual time point. Thus, the effective sampling rate of these time series was 0.4 Hz (2.5s per frame).
Wide-field Imaging
Pupae were staged for head eversion and reared at 25°C. At 58–60 hAPF, pupae were affixed to a Sylgard 184 Silicone Elastomer plate (Dow Corning) with double-stick adhesive tape (3M). Images were acquired with a SteREO Discovery.V8 stereomicroscope (Zeiss) with illumination provided by an X-Cite Series 120PC light source (Excelitas) and captured on a Vixia HF R20 1/4.85 inch CMOS camera (Canon). Images were acquired at 30 Hz. Time series were processed with Fiji (ImageJ) (Schindelin et al., 2012) and analyzed using MATLAB (Mathworks, Natick, MA, USA).
Adult Functional Imaging
Calcium imaging was performed as previously described (Keleş and Frye, 2017). Briefly, a single fly was anesthetized at 4°C and placed into a chemically etched metal shim attached to a custom 3D-printed holder. Holder design was based on (Weir et al., 2016); details can be found at http://ptweir.github.io/flyHolder/. The head capsule and thorax were glued to the metal shim using a UV-curable glue (www.esslinger.com). Legs and the antennae were immobilized using beeswax applied with a heated metal probe (Waxelectric-1, Renfert). The head capsule was bathed in saline (103mM NaCl, 3mM KCl, 1.5mM CaCl2, 4mM MgCl2, 26mM NaHCO3, 1mM NaH2PO4, 10mM trehalose, 10mM glucose, 5mM TES, 2mM sucrose) and a small window was opened using fine forceps (Dumont, #5SF). Muscles and fat covering the optic lobe were cleared before placing the fly under a 2P microscope (3i, Denver, CO). Neurons expressing GCaMP6s were imaged at 920 nm using a Ti:Sapphire pulse laser (Chameleon Vision, Coherent). Images were acquired at 10 Hz.
An arena of 48 8×8 LED matrices (470 nm, Adafruit) was used to deliver the visual stimulus. Three layers of blue filter (Rosco no. 59 Indigo) were placed between the screen and the fly to eliminate leakage of the LED light into the PMTs. The screen extended ±108° along the azimuth and ± 72° in elevation. Each LED pixel corresponded to a coverage of 2.2° on the retina equator. However, the projection of each pixel on the retina was variable due to the difference between the curvature of the eye and that of the screen. Visual stimulus consisted of a wide-field grating with a spatial frequency of 35° and presented at a temporal frequency of 0.62 Hz in both directions (ipsi-to-contra and contra-to-ipsi) along the horizontal axis. The presentation order of the visual stimuli was randomized to prevent sensory adaptation. Each experimental condition was tested three to four times per animal.
Tetrodotoxin Treatment
Pupal development was staged for white pre-pupa formation and reared at 25°C. Between 90–95 hAPF, the pupal case was removed with fine forceps. These late pupae were prepared for imaging following the protocol described above for adult functional imaging. Viability was verified by leg or trachea movement. Neurons expressing GCaMP6s were imaged at 920 nm using a Ti:Sapphire pulsed laser (Chameleon Vision, Coherent). Images were acquired at 10 Hz.
Tetrodotoxin at 1 μM final concentration was mixed into the saline solution after 40 minutes of imaging and the fly was observed for another 20 minutes after the application of the drug. Viability was confirmed before and after tetrodotoxin administration, and the data were excluded from analysis if the animal did not survive the experiment.
Visual Flight Simulator
Flies were cold anesthetized at 4°C, tethered to tungsten pins using UV activated glue, and allowed to recover for 1–2 hours in a small, humidified acrylic container with a red desk lamp providing heat. This recovery regime improves flight performance consistency. The majority of the experiments were performed in the afternoon when flies are most active.
A visual flight simulator composed of 32×96 cylindrical green (570 nm) LEDs was used to deliver visual stimuli (Reiser and Dickinson, 2008). The arena covered ± 180° along the azimuth and ± 60° in elevation. Single flies were positioned in the center of the arena and illuminated from above with an 880 nm infrared LED. The shadow cast by the wings was detected with an optical sensor. Signal form this sensor was analyzed by an instrument called the wingbeat analyzer (JFI Electronics Laboratory, University of. Chicago, Chicago, IL, USA) that calculates left and right wing beat amplitudes (WBA). The difference in the left and right WBA is proportional to the fly’s steering effort in the yaw axis.
For bar fixation closed-loop experiments, a dark bar that is 120° in height and 30° in width was presented to the flies. Positional displacement of the bar in the yaw axis was coupled to the steering effort of the fly, allowing the animal to have the active control of the bar position. Each fly was tested for closed-loop fixation behavior for four minutes. To test open-loop optomotor responses, flies were presented with wide-field gratings with a spatial frequency of 30° and a temporal frequency of 3 Hz for four seconds.
Quantification and Statistical Analysis
The number and type (i.e. flies/time series, visual system columns, or activity cycles) of replicates are indicated in the figure legends.
Analysis of Pupal Imaging Data
Pre-processing:
Processing and analysis of image data were carried out with custom scripts written in MATLAB (Mathworks, Natick, MA, USA). Fiji (ImageJ) was used for some user-assisted tasks and figure preparation. Time series were processed in blocks corresponding to ~6 hours of observation (~9000 frames). In the pre-processing step of reducing lateral motion, the general approach of maximizing the cross correlation of individual frames to a reference image was modified to meet the specific challenges of developmental imaging. First, a series of reference images were generated as averages of pools of high signal frames distributed across each block. After ~55 hAPF, the optic lobes begin to twitch with a period similar to that of PSINA. These fast movements can introduce significant blur into the pool-averaged reference images. To reduce this blur, 300 random subsets of each pool were tested to find the sharpest average reference image. Sequential registration of this series of reference images to each other produced a stabilized representation of the visual system which continues to move and grow over the course of observation (Akin and Zipursky, 2016; Langen et al., 2015). In a second step, the registration of the reference series was refined to minimize the movement of a user defined region of interest (ROI). These internally registered reference images then served as local registration targets for nearby frames of the full block. Finally, the block was corrected for any rotational motion of the ROI.
Signal and Feature Extraction:
Per frame pixel averages of masked regions were used to define raw signal (F) traces from the image time series. Time-dependent fluorescence baseline (Fo) was estimated using a moving window approach and used to calculate the net signal (F-Fo, Figures 2, 3 and 4) and change-in-signal ((F-Fo)/Fo, Figures 1 and 2) traces. User-defined, static masks were used for pan-neuronal and glial expression experiments. For cell type-specific experiments, dynamic masks, corresponding to the active columns in each cycle, were defined automatically from the kymograph representation of the time series. Briefly, kymographs were generated as concatenated line profiles from user-defined, segmented arcs of 7–9 pixel (3–4 μm) thickness, drawn across a single layer of the medulla or lobula neuropil. Baseline subtracted, net signal kymographs were used in all subsequent analysis. Projecting along the spatial dimension of the kymographs yielded one dimensional net signal traces, which were used to identify the limits of PSINA cycles. Within each active phase, sweeps were defined by ordering intensity peaks with respect to their amplitudes, and, from the largest peak on down, marking the continuous time spans with net signal intensity greater than 75% of peak value; lesser peaks present in the sweep of a larger one were removed from the ordered peak list used in sweep identification. Dynamic masks were based on peaks in the activity profiles of PSINA cycles, produced for each active phase by projecting along the temporal dimension of the kymographs. The width of each mask was determined by testing the spatial neighborhood of each peak for correlated net intensity changes in the time domain. The maximum number of dynamic masks identified in each cycle was set to 20.
Frequency Analysis:
Analysis was implemented in MATLAB, following the guidelines of Uhlen (Uhlén, 2004). Change-in-signal (ΔF/F) traces were processed using a 2-hour sliding window which traversed the time series in 1-hour steps. After filtering with a Hanning window to reduce spectral leakage, each 2-hour block was transformed with the FFT algorithm to obtain non-parametric power spectrum density estimates. The fidelity of the power spectrum density estimate was confirmed by applying the inverse transform on the highest-power peak and comparing the resultant signal to the raw data. One-sided power spectrum density estimates were plotted for each 2-hour block in Figure 1 and Figure S2.
PSINA Dynamics:
For each cycle, unit signal-to-noise (S2N) value was defined as twice the standard deviation of the net signal trace in the silent phase. A dynamically masked column was considered to participate in a given sweep if it had a net intensity peak greater than or equal to 1.0 S2N within the sweep limits. This scoring scheme was the basis of the definition of the coordination metric. For coherence, the largest fraction of columns that reach peak intensity at the same time point within each sweep was calculated. To ensure consistent comparisons across different cell types, only high participation (≥90%) sweeps were considered for the coherence metric.
STTC analysis was carried out according to Cutts and Eglen (Cutts and Eglen, 2014). For each time series, the active phase of each cycle was treated as a separate recording. The net intensity traces from dynamically masked columns were converted to spike trains using a hard threshold of four times the standard deviation of the net trace in the preceding silent phase. STTC values were calculated for a range of Δts; the value of 5 s marked a decrease in the slope of STTC v. Δt and yielded the greatest dynamic range across different cell types and therefore was chosen for the analysis shown in Figures S8B–S8D. Distances between columns were binned in increments of 4.5 μm, the average column width in the optic neuropils during development. As a measure of how STTC decays over distance for a cell type, we report the ratio of the STTC value at 36 μm to that at 4.5 μm; the 36 μm mark was chosen to ensure that all cell types could be compared using this metric, including ones with processes in the lobula plate (e.g. T5), which is only partially visible in our setup.
Correlation Analysis:
For the analysis of two-color neuronal imaging experiments, two separate kymographs were generated using the same segmented arc. Dynamic masks were derived from the average activity profile of these two kymographs to ensure that the masks captured columns active in both channels. Cycle limits were determined using the brighter channel. For each cycle, masks with a maximum S2N value of at least 1.0 in both channels were used to calculate pairwise 0-lag cross-correlation. Cycles with fewer than 10 masks above the signal quality threshold were excluded in the calculation of time series ensemble statistics (i.e. mean and standard deviation.)
For correlation analysis of GECI combined with voltage or glutamate imaging, one ROI was generated using a segmented arc over the first layer of the medulla. Mean population traces were generated for both indicators; single column analysis was not possible due to low S2N. MATLAB functions for normalized cross-correlation and auto-correlation were used to compare lags between traces. The same approach was used for two-color calcium imaging of neurons and glia; however, separate ROIs were used for neuronal signals (medulla) and glia (inner optic chiasm).
Analysis of Adult Calcium Imaging Data
Images were pre-processed to correct for lateral motion using the registration algorithm described above. To find active pixels in the lobula, we defined a mask excluding other neuropils (medulla and lobula plate). For every pixel in this mask, the mean value and standard deviation were calculated for the full time series; the test value for each pixel was defined as the product of these metrics. Pixels with test values greater than or equal to twice the mean value of all pixels in the mask were used in analysis. In our experience, this thresholding approach enriches for active pixels over background and shot noise in the selected mask. The frame average of active pixels were used to produce the signal trace for the time series. Repeated observations were averaged for each fly and a single average trace per experiment was generated.
Analysis of Visual Fixation Behavior
Behavioral data from the visual display and the wing beat analyzer was collected with a Digidata 1440A digitizer (Molecular Devices, San Jose, CA, USA) sampled at 1 kHz. Data were processed using custom written scripts in MATLAB (Mathworks, Natick, MA, USA). Briefly, the first 100 milliseconds of the trials were removed and the first data point of the remaining signal was subtracted from the entire trial to set the initial WBA to zero. ΔWBA was calculated by subtracting left from right WBA. Flies which stopped flying during the experiments were excluded from further analysis. Trials for the same experimental conditions were averaged and calculated for all animals. No statistical tests were conducted to pre-determine the sample size. To analyze closed-loop fixation data, the bar position was binned into 96 positions around the visual azimuth and bar histograms for each fly was calculated. Data were then averaged across the animals for the time bar spent at each position.
Data and Software Availability
Scripts developed by the authors and used in this study are available upon request.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Bacterial and Virus Strains | ||
| Biological Samples | ||
| Chemicals, Peptides, and Recombinant Proteins | ||
| Tetrodotoxin | Sigma-Aldrich | 554412; CAS 18660–81–6 |
| Critical Commercial Assays | ||
| Deposited Data | ||
| Experimental Models: Cell Lines | ||
| Experimental Models: Organisms/Strains | ||
| Drosophila: 13XLexAop2-IVS-Syn21-OpGCaMP6s-p10 (su(Hw)attP5) | Rubin Lab, JRC, HHMI | N/A |
| Drosophila: 13XLexAop2-IVS-Syn21-OpGCaMP6s-p10 (su(Hw)attP8) | Rubin Lab, JRC, HHMI | N/A |
| Drosophila: 20XUAS-iGluSnFR.A184V (attP2) | Bloomington | 59611 |
| Drosophila: 20XUAS-IVS-NES-jRCaMP1b-p10 (VK00005) | Bloomington | 63793 |
| Drosophila: 20XUAS-IVS-Syn21-opGCaMP6s-p10 (su(Hw)attP1) | Rubin Lab, JRC, HHMI | N/A |
| Drosophila: 20XUAS-IVS-Syn21-opGCaMP6s-p10 (su(Hw)attP5) | Rubin Lab, JRC, HHMI | N/A |
| Drosophila: 20XUAS-IVS-Syn21-opGCaMP6s-p10 (su(Hw)attP8) | Rubin Lab, JRC, HHMI | N/A |
| Drosophila: 6–60–GAL4 (Chr 3) |
Ulrike Heberlein, JRC, HHMI; Nern et al., 2008 | N/A |
| Drosophila: alrm-Gal4 (Chr 2) | Bloomington | 67031 |
| Drosophila: alrm-Gal4 (Chr 3) | Bloomington | 67032 |
| Drosophila: brp(RSRT.Stop)V5–2A–LexA–VP16 (VK00003b) | This study (see Chen et al., 2014) | N/A |
| Drosophila: cn[1], bw[1] | Bloomington | 264 |
| Drosophila: Dm3-Gal4 [R25F07-GAL4 (attP2)] | Bloomington | 49130 |
| Drosophila: Dm4-Gal4 [R75F06-GAL4 (attP2)] | Bloomington | 39901 |
| Drosophila: Dm4-LexA [R75F06-LexA (attP40)] | Bloomington | 54100 |
| Drosophila: Dm9–1–Gal4 [VT025981.GAL4 (attP2)] | VDRC | 205425 |
| Drosophila: Dm9–2–Gal4 [R56G04-GAL4 (attP2)] | Bloomington | 39161 |
| Drosophila: Hdc[JK910] | Bloomington | 64203 |
| Drosophila: Hdc[MB07212] | Bloomington | 25260 |
| Drosophila: L3–1–Gal4 [R29F12-GAL4 (attP2)] | Bloomington | 49495 |
| Drosophila: L3–2–Gal4 [R9D03-GAL4 (attP2)] | Bloomington | 40726 |
| Drosophila: Mi1-LexA [R89C04-LexA (attp40)] | Bloomington | 54382 |
| Drosophila: Mi1-SplitGal4 [w1118; R55C05-p65ADZp (attP40); R71D01-ZpGdbd (attP2)] | Strother et al., 2017 (available from http://splitgal4.janelia.org) |
JRC_SS00955 |
| Drosophila: Mi4-Gal4 [R49B06-GAL4 (attP2)] | Bloomington | 50409 |
| Drosophila: Mi4-LexA [R49B06-LexA (attP40)] | Bloomington | 52707 |
| Drosophila: ort[1] | Bloomington | 1133 |
| Drosophila: PanLamina-1-FLP1 [27G05-FLP1 (su(Hw)attP5)] | This study (see Pecot et al., 2013) | N/A |
| Drosophila: PanLamina-2-FLP1 [27G05-FLP1 (Chr X)] | Tadros et al., 2016 | N/A |
| Drosophila: PanNeuronal-Gal4 [R57C10-Gal4 (attP2)] | Bloomington | 39171 |
| Drosophila: PanNeuronal-LexA [R57C10-lexA (attP40)] | Bloomington | 52817 |
| Drosophila: R7-GAL4 [sevEP-GAL4.B (Chr 2)] | Bloomington | 5793 |
| Drosophila: sens-R.pest (su(Hw)attP8) | This study (see Akin and Zipursky, 2016) | N/A |
| Drosophila: svp-Gal4 [svpNP0724(87B5)] | Kyoto DGGR | 103727 |
| Drosophila: T4/T5-Gal4 [R23G12-GAL4 (attP2)] | Bloomington | 49044 |
| Drosophila: T4/T5-LexA [R23G12-LexA (su(Hw)attP8)] | Bloomington | 65044 |
| Drosophila: Tm3-Gal4 [R13E12-GAL4 (attP2)] | Bloomington | 48569 |
| Drosophila: Tm3-LexA [R13E12-LexA (attP40)] | Bloomington | 48569 |
| Drosophila: Tm4-LexA [R35H01-LexA (attp40)] | Bloomington | 52459 |
| Drosophila: Tm9–1–Gal4 [R24C08-GAL4 (attP2)] | Bloomington | 48050 |
| Drosophila: Tm9–2–Gal4 [R25F10-GAL4 (attP2)] | Bloomington | 49132 |
| Drosophila: tubP-(FRT.GAL80) (attP40) | Rubin Lab, JRC, HHMI | N/A |
| Drosophila: UAS-ArcLight (attP2) | Bloomington | 51056 |
| Drosophila: UAS-CD-tdTOM (VK00033) | Bloomington | 35837 |
| Drosophila: UAS-TeTxLC.tnt (Chr 2) | Bloomington | 28838 |
| Drosophila: UAS-TeTxLC.tnt (Chr X) | Bloomington | 28996 |
| Drosophila: w[*] norpA[36] | Bloomington | 9048 |
| Oligonucleotides | ||
| Recombinant DNA | ||
| Software and Algorithms | ||
| Other | ||
Highlights.
Stimulus-independent neural activity occurs in the developing Drosophila visual system
Cell types exhibit unique activity patterns within a common oscillatory structure
Activity patterns reflect adult connectivity in the ON and OFF motion circuits
Acknowledgments
We thank Gerald M. Rubin, Barrett Pfeiffer, David J. Anderson, Yon-il Jung, Douglas Kim, Vivek Jayaraman, and Yi Sun for GCaMP6s and jRCaMP1b flies, and for technical advice. For assistance and advice with preliminary experiments, we thank Na Ji, Anderson Chen, and Rongwen Lu. For discussing unpublished results and providing feedback on our findings, we thank Claude Desplan, Ben Jiwon Choi, Chris Doe, Marla Feller, Haig Keshishian, Robin Hiesinger, and Kristin Scott. We thank members of the Zipursky Lab for their support and insights. We apologize to all whose contributions could not be cited directly due to space constraints. This work was supported by NIH T32-GM008032 and T32-MH073526 to BTB, NIH R01-EY026031 to MAF, and Howard Hughes Medical Institute. SLZ is an investigator of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests
The authors declare no competing interests.
Supplemental Information
Supplemental Information includes seven figures, one table, and seven movies.
References
- Ackman JB, and Crair MC (2014). Role of emergent neural activity in visual map development. Curr. Opin. Neurobiol 24, 166–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ackman JB, Burbridge TJ, and Crair MC (2012). Retinal waves coordinate patterned activity throughout the developing visual system. Nature 490, 219–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akin O, and Zipursky SL (2016). Frazzled promotes growth cone attachment at the source of a Netrin gradient in the Drosophila visual system. eLife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akrouh A, and Kerschensteiner D (2013). Intersecting circuits generate precisely patterned retinal waves. Neuron 79, 322–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aptekar JW, Keles MF, Mongeau J-M, Lu PM, Frye MA, and Shoemaker PA (2014). Method and software for using m-sequences to characterize parallel components of higher-order visual tracking behavior in Drosophila. Front. Neural Circuits 8, 130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baines RA, and Bate M (1998). Electrophysiological Development of Central Neurons in the Drosophila Embryo. J. Neurosci 18, 4673–4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bansal A, Singer JH, Hwang BJ, Xu W, Beaudet A, and Feller MB (2000). Mice lacking specific nicotinic acetylcholine receptor subunits exhibit dramatically altered spontaneous activity patterns and reveal a limited role for retinal waves in forming ON and OFF circuits in the inner retina. J. Neurosci. Off. J. Soc. Neurosci 20, 7672–7681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blankenship AG, and Feller MB (2009). Mechanisms underlyingspontaneous patterned activityin developing neural circuits. Nat. Rev. Neurosci 11, 18–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloomquist BT, Shortridge RD, Schneuwly S, Perdew M, Montell C, Steller H, Rubin G, and Pak WL (1988). Isolation of a putative phospholipase C gene of Drosophila, norpA, and its role in phototransduction. Cell 54, 723–733. [DOI] [PubMed] [Google Scholar]
- Brand AH, and Perrimon N (1993). Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118, 401–415. [DOI] [PubMed] [Google Scholar]
- Burbridge TJ, Xu H-P, Ackman JB, Ge X, Zhang Y, Ye M-J, Zhou ZJ, Xu J, Contractor A, and Crair MC (2014). Visual Circuit Development Requires Patterned Activity Mediated by Retinal Acetylcholine Receptors. Neuron 84, 1049–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burg MG, Sarthy PV, Koliantz G, and Pak WL (1993). Genetic and molecular identification of a Drosophila histidine decarboxylase gene required in photoreceptor transmitter synthesis. EMBO J. 12, 911–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao G, Platisa J, Pieribone VA, Raccuglia D, Kunst M, and Nitabach MN (2013). Genetically targeted optical electrophysiology in intact neural circuits. Cell 154, 904–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrillo RA, Olsen DP, Yoon KS, and Keshishian H (2010). Presynaptic Activity and CaMKII Modulate Retrograde Semaphorin Signaling and Synaptic Refinement. Neuron 68, 32–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T-W, Wardill TJ, Sun Y, Pulver SR, Renninger SL, Baohan A, Schreiter ER, Kerr RA, Orger MB, Jayaraman V, et al. (2013). Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature 499, 295–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Akin O, Nern A, Tsui CYK, Pecot MY, and Zipursky SL (2014). NeuroResource. Neuron 81, 280–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constance WD, Mukherjee A, Fisher YE, Pop S, Blanc E, Toyama Y, and Williams DW (2018). Neurexin and Neuroligin-based adhesion complexes drive axonal arborisation growth independent of synaptic activity. eLife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutts CS, and Eglen SJ (2014). Detecting pairwise correlations in spike trains: an objective comparison of methods and application to the study of retinal waves. J. Neurosci. Off. J. Soc. Neurosci 34, 14288–14303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dana H, Mohar B, Sun Y, Narayan S, Gordus A, Hasseman JP, Tsegaye G, Holt GT, Hu A, Walpita D, et al. (2016). Sensitive red protein calcium indicators for imaging neural activity. eLife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galli L, and Maffei L (1988). Spontaneous impulse activity of rat retinal ganglion cells in prenatal life. Science 242, 90–91. [DOI] [PubMed] [Google Scholar]
- Gao S, Takemura S, Ting C-Y, Huang S, Lu Z, Luan H, Rister J, Thum AS, Yang M, Hong S-T, et al. (2008). The Neural Substrate of Spectral Preference in Drosophila. Neuron 60, 328–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gengs C, Leung H-T, Skingsley DR, Iovchev MI, Yin Z, Semenov EP, Burg MG, Hardie RC, and Pak WL (2002). The target of Drosophila photoreceptor synaptic transmission is a histamine-gated chloride channel encoded by ort (hclA). J. Biol. Chem 277, 42113–42120. [DOI] [PubMed] [Google Scholar]
- Hadjieconomou D, Timofeev K, and Salecker I (2011). A step-by-step guide to visual circuit assembly in Drosophila. Curr. Opin. Neurobiol 21, 76–84. [DOI] [PubMed] [Google Scholar]
- Hardie RC, Peretz A, Pollock JA, and Minke B (1993). Ca2+ limits the development of the light response in Drosophila photoreceptors. Proc. Biol. Sci 252, 223–229. [DOI] [PubMed] [Google Scholar]
- Hiesinger PR, Zhai RG, Zhou Y, Koh T-W, Mehta SQ, Schulze KL, Cao Y, Verstreken P, Clandinin TR, Fischbach K-F, et al. (2006). Activity-Independent Prespecification of Synaptic Partners in the Visual Map of Drosophila. Curr. Biol 16, 1835–1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin L, Han Z, Platisa J, Wooltorton JRA, Cohen LB, and Pieribone VA (2012). Single action potentials and subthreshold electrical events imaged in neurons with a fluorescent protein voltage probe. Neuron 75, 779–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keleş MF, and Frye MA (2017). Object-Detecting Neurons in Drosophila. Curr. Biol 27, 680–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerschensteiner D, and Wong ROL (2008). A precisely timed asynchronous pattern of ON and OFF retinal ganglion cell activity during propagation of retinal waves. Neuron 58, 851–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, McKay RR, Miller K, and Shortridge RD (1995). Multiple Subtypes of Phospholipase C Are Encoded by the norpA Gene of Drosophila melanogaster. J. Biol. Chem 270, 14376–14382. [DOI] [PubMed] [Google Scholar]
- Lai S-L, and Lee T (2006). Genetic mosaic with dual binary transcriptional systems in Drosophila. Nat. Neurosci 9, 703–709. [DOI] [PubMed] [Google Scholar]
- Langen M, Agi E, Altschuler DJ, Wu LF, Altschuler SJ, and Hiesinger PR (2015). The Developmental Rules of Neural Superposition in Drosophila. Cell 162, 120–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee PR, Cohen JE, Iacobas DA, Iacobas S, and Fields RD (2017). Gene networks activated by specific patterns of action potentials in dorsal root ganglia neurons. Sci. Rep 7, 43765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marvin JS, Borghuis BG, Tian L, Cichon J, Harnett MT, Akerboom J, Gordus A, Renninger SL, Chen T-W, Bargmann CI, et al. (2013). An optimized fluorescent probe for visualizing glutamate neurotransmission. Nat. Methods 10, 162–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin T, Torborg CL, Feller MB, and O’Leary DDM (2003). Retinotopic map refinement requires spontaneous retinal waves during a brief critical period of development. Neuron 40, 1147–1160. [DOI] [PubMed] [Google Scholar]
- Meister M, Wong RO, Baylor DA, and Shatz CJ (1991). Synchronous bursts of action potentials in ganglion cells of the developing mammalian retina. Science 252, 939–943. [DOI] [PubMed] [Google Scholar]
- Muthukumar AK, Stork T, and Freeman MR (2014). Activity-dependent regulation of astrocyte GAT levels during synaptogenesis. Nat. Publ. Group 17, 1340–1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearn MT, Randall LL, Shortridge RD, Burg MG, and Pak WL (1996). Molecular, Biochemical, and Electrophysiological Characterization of Drosophila norpA Mutants. J. Biol. Chem 271, 4937–4945. [DOI] [PubMed] [Google Scholar]
- Reiser MB, and Dickinson MH (2008). A modular display system for insect behavioral neuroscience. J. Neurosci. Methods 167, 127–139. [DOI] [PubMed] [Google Scholar]
- Richier B, de Miguel Vijandi C, Mackensen S, and Salecker I (2017). Lapsyn controls branch extension and positioning of astrocyte-like glia in the Drosophila optic lobe. Nat. Commun 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanes JR, and Masland RH (2015). The Types of Retinal Ganglion Cells: Current Status and Implications for Neuronal Classification. Annu. Rev. Neurosci 38, 221–246. [DOI] [PubMed] [Google Scholar]
- Sanes JR, and Zipursky SL (2010). Design Principles of Insect and Vertebrate Visual Systems. Neuron 66, 15–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seelig JD, Chiappe ME, Lott GK, Dutta A, Osborne JE, Reiser MB, and Jayaraman V (2010). Two-photon calcium imaging from head-fixed Drosophila during optomotor walking behavior. Nat. Methods 7, 535–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seung HS, and Sümbül U (2014). Neuronal cell types and connectivity: lessons from the retina. Neuron 83, 1262–1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shatz CJ, and Stryker MP (1988). Prenatal tetrodotoxin infusion blocks segregation of retinogeniculate afferents. Science 242, 87–89. [DOI] [PubMed] [Google Scholar]
- Shinomiya K, Karuppudurai T, Lin T-Y, Lu Z, Lee C-H, and Meinertzhagen IA (2014). Candidate Neural Substrates for Off-Edge Motion Detection in Drosophila. Curr. Biol 24, 1062–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silies M, Gohl DM, and Clandinin TR (2014). Motion-detecting circuits in flies: coming into view. Annu. Rev. Neurosci 37, 307–327. [DOI] [PubMed] [Google Scholar]
- Sretavan DW, Shatz CJ, and Stryker MP (1988). Modification of retinal ganglion cell axon morphology by prenatal infusion of tetrodotoxin. Nature 336, 468–471. [DOI] [PubMed] [Google Scholar]
- Takemura S, Bharioke A, Lu Z, Nern A, Vitaladevuni S, Rivlin PK, Katz WT, Olbris DJ, Plaza SM, Winston P, et al. (2013). A visual motion detection circuit suggested by Drosophila connectomics. Nature 500, 175–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimann KV, and Beadle GW (1937). Development of the Eye Colors in Drosophila: Extraction of the Diffusible Substances Concerned. Proc. Natl. Acad. Sci. U. S. A 23, 143–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyssowski KM, DeStefino NR, Cho J-H, Dunn CJ, Poston RG, Carty CE, Jones RD, Chang SM, Romeo P, Wurzelmann MK, et al. (2018). Different Neuronal Activity Patterns Induce Different Gene Expression Programs. Neuron 98, 530–546.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlén P (2004). Spectral analysis of calcium oscillations. Sci. STKE 2004, pl15. [DOI] [PubMed] [Google Scholar]
- Vonhoff F, and Keshishian H (2016). Cyclic nucleotide signaling is required during synaptic refinement at the Drosophila neuromuscular junction. Dev. Neurobiol 77, 39–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir PT, Henze MJ, Bleul C, Baumann-Klausener F, Labhart T, and Dickinson MH (2016). Anatomical Reconstruction and Functional Imaging Reveal an Ordered Array of Skylight Polarization Detectors in Drosophila. J. Neurosci 36, 5397–5404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu HP, Burbridge TJ, Ye M, Chen M, Ge X, Zhou ZJ, and Crair MC (2016). Retinal Wave Patterns Are Governed by Mutual Excitation among Starburst Amacrine Cells and Drive the Refinement and Maintenance of Visual Circuits. J. Neurosci 36, 3871–3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang KX, Tan L, Pellegrini M, Zipursky SL, and McEwen JM (2016). Rapid Changes in the Translatome during the Conversion of Growth Cones to Synaptic Terminals. CellReports 14, 1258–1271. [DOI] [PubMed] [Google Scholar]
- Zheng J, Lee S, and Zhou ZJ (2006). A transient network of intrinsically bursting starburst cells underlies the generation of retinal waves. Nat. Neurosci 9, 363–371. [DOI] [PubMed] [Google Scholar]
- Zheng J-J, Lee S, and Zhou ZJ (2004). A developmental switch in the excitability and function of the starburst network in the mammalian retina. Neuron 44, 851–864. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Scripts developed by the authors and used in this study are available upon request.