Abstract
Objective
Soybean seeds are an important source of vegetable proteins for both food and industry worldwide. Conglycinins (7S) and glycinins (11S), which are two major families of storage proteins encoded by a small family of genes, account for about 70% of total soy seed protein. Mutant alleles of these genes are often necessary in certain breeding programs, as the relative abundance of these protein subunits affect amino acid composition and soy food properties. In this study, we set out to test the efficiency of the CRISPR/Cas9 system in editing soybean storage protein genes using Agrobacterium rhizogenes-mediated hairy root transformation system.
Results
We designed and tested sgRNAs to target nine different major storage protein genes and detected DNA mutations in three storage protein genes in soybean hairy roots, at a ratio ranging from 3.8 to 43.7%. Our work provides a useful resource for future soybean breeders to engineer/develop varieties with mutations in seed storage proteins.
Electronic supplementary material
The online version of this article (10.1186/s13104-019-4207-2) contains supplementary material, which is available to authorized users.
Keywords: Seed storage proteins, Soybean, CRISPR/Cas9, Genome editing
Introduction
Just as seeds are an important foundation of Agriculture, soybean seeds are an important source of vegetable proteins for both food and industry world-wide. Two major families of storage proteins, conglycinins (7S) and glycinins (11S), account for about 70% of total soy seed protein [1]. Both quantity and quality of storage proteins, in soybean seeds, are major biochemical components influencing the quality of tofu and other soy food products [2, 3]. Soybean breeders have developed a series of mutant soybean genotypes differing in seed storage glycinin and conglycinin subunit composition. These mutant lines encompass a wide range of genetic variability available to breeders to improve soy protein functional properties for specific end uses [4–6]. Conventionally, breeders have to repeatedly introgress the mutations into elite soybean cultivars by conducting genetic crosses and rounds of selection over several generations and years. This is a long and labour-intensive process, which has been a major limiting factor for the timely delivery of quality soybean varieties, in an effort to cope with a continuously changing agriculture environment. Even though new plant breeding techniques have been constantly sought after by the plant genetics and genomics research community, it seems that the CRISPR-Cas9 system (the Clustered Regularly Interspaced Short Palindromic Repeat (CRISPR)/CRISPR-associated 9 (Cas9)) is revolutionizing our breeding practices [7–9].
The CRISPR/Cas9 system has emerged as a robust technology for efficient genome editing [10, 11], and has been successfully applied in many major crops, including soybean [12–16]. In this study, we set out to test the efficiency of the CRISPR/Cas9 system in editing soybean storage protein genes using Agrobacterium rhizogenes-mediated hairy root transformation system. Since stable transgenic soybean plants require a relatively long time (approximately 9 months) to develop, we opted to use soybean hairy roots as a model system which only takes about 3 weeks. Therefore, assessing the efficiency of sgRNAs in generating InDels at target sites in hairy roots, prior to whole-plant transformation, could solve the labor-intensive problem of the traditional transformation techniques. More importantly, soybean hairy roots are true soybean tissue and thus, making them ideal for purposes of quick testing of the genome editing efficiency of the sgRNAs and method optimization.
Main text
Materials and methods
Plant materials and growth conditions
Wild-type soybean line “Harosoy 63” was gifted from Dr. Sangeeta Dhaubhadel. “Harosoy 63” was registered in 1964 [17]. All pots and trays were sterilized with 10% bleach solution. Seeds were sterilized with H2O2/ethanol (10% of 30% H2O2, 75% of 95% ethanol, 15% sterile distilled water) for 2 min by inverting gently. Washing solution was decanted and seeds were rinsed 5–6 times with excess amount of sterile distilled water. Pots were filled with sterilized vermiculite and sterilized seeds were placed 1–2 cm deep in vermiculite.
SgRNA Design and Construction of sgRNA: Cas9 Expression
We used EnsemblPlants (http://plants.ensembl.org/Glycine_max/Gene/Summary?g=GLYMA_19G254600;r=19:49937807-49940339;t=KRG97147;db=core) to obtain the gene sequences for all nine conglycinin and glycinin genes. To design sgRNA, we employed CRISPR-PLANT (https://www.genome.arizona.edu/crispr/CRISPRsearch.html). Sequences of all sgRNAs are listed in Table 1. We used pZG23C05 vector (Cas9/gRNA construction kit for dicots, Bar resistant from ZGene Biotechnology Inc.) and followed the manufacturer’s protocol to construct our gRNA target sequences into Cas9/gRNA plasmid. Plants and bacteria transformed by pZG23C05 should be resistant to Basta and Kanamycin, respectively.
Table 1.
Summary of mutations generated for each sgRNA
| Genes | ID for sgRNA | gRNA-PAMa | No. hairy roots genotyped | No. hairy roots w. mutations detected | % mutation |
|---|---|---|---|---|---|
| Glyma.20g148400 | g1 | 5′-CCTTCTGATGAGGTGGGCGTGGG-3′ | 17 | 1 | 5.8 |
| Glyma.20g146200 | g1 | 5′-GGACAATCCGGTAGTCTCGAAGG-3′ | 46 | 0 | |
| g2 | 5′-ACAGAAGCAGAAACAGGAAGAGG-3′ | 37 | 0 | ||
| g3 | 5′-GGATTCTCTGGGCATCGCCACCC-3′ | 42 | 0 | ||
| Glyma.10g246300 | g1 | 5′-TCTCGCTATTGCAACTTCGGAGG-3′ | 27 | 0 | |
| g2 | 5′-CAGTGTTGTGGATATGAACGAGG-3′ | 32 | 0 | ||
| g3 | 5′-AGAAGAAGAAGACCAAGACGAGG-3′ | 21 | 0 | ||
| Glyma.20g148200 | g1 | 5′-GGATTCTCTGGGCATCGCCAGGG-3′ | 75 | 0 | |
| g2 | 5′-ACTCTCTTTGAGAACCAAAACGG-3′ | 63 | 0 | ||
| g3 | 5′-GGACAATCCGGTAGTCTCGAAGG-3′ | 28 | 0 | ||
| Glyma.10g037100 | g1 | 5′-CATCTTACTCACCTTATCCCCGG-3′ | 99 | 0 | |
| g2 | 5′-GACGTACTCGTGATTCCTCCGG-3′ | 54 | 0 | ||
| g3 | 5′-CTCCATTCGCGGCTCTTGCGAGG-3′ | 42 | 0 | ||
| Glyma.03g163500 | g1 | 5′-GATAACCGTATAGAGTCAGAAGG-3′ | 26 | 1 | 3.8 |
| Glyma.19g164900 | g1 | 5′-GATAACCGTATAGAGTCAGAAGG-3′ | 32 | 14 | 43.7 |
| Glyma.13g123500 | g1 | 5′-GACCACCGCGTTGAGTCCGAAGG-3′ | 38 | 0 | |
| g2 | 5′-GACGTACTAGTGATTCCTCCTGG-3′ | 64 | 0 | ||
| g3 | 5′-TCGGGCCTGCTTGGTCGCTGTGG-3′ | 48 | 0 | ||
| Glyma.19g164800 | g1 | 5′-GACAACCTCATCGAATCCCAAGG-3′ | 49 | 0 | |
| g2 | 5′-AGCGTGGCCTACGTGACGAGTGG-3′ | 76 | 0 | ||
| g3 | 5′-TCGGTGTTCAGCGGCGCTGTTGG-3′ | 49 | 0 |
aThe PAM sequences are highlighted in bold
Hairy root transformation using A. rhizogenes K599
The plasmid vector used for expressing Cas9 and single-guide RNA was mobilized into A. rhizogenes K599 via electroporation. Hairy root transformation was performed following [18]. The 5-day-old seedlings with unopened cotyledons were selected for inoculation. A syringe needle was used to deliver the bacterial culture by stabbing at the cotyledonary node and/or at the hypocotyl proximal to the cotyledon. The puncture site was covered with sterile moist vermiculite. The whole tray was covered with a clear dome and left in a growth cabinet at 28 °C/12 h light (200 µmol/m2s light intensity), 25 °C/12 h dark and 80% humidity.
Evaluation of the ratio of transgenic hairy roots
To evaluate the ratio of transgenic hairy roots, a Green Florescence Protein (GFP) expressing vector pB7WG2D [19] was used. Hairy roots grown to a length of 5–6 cm were labelled with numbers and screened using a dissecting fluorescence microscope (Nikon SMZ1500), and the transgenic roots showed bright GFP fluorescence labeling while non-transgenic roots remained dark.
Screening for mutations induced by the CRISPR/Cas9 system
Genomic DNA was extracted from hairy roots by adding 400 µl extraction buffer (3% CTAB, 4% B-Me EtOH, 2 M NaCl, 5% PVP, 100 mM Tris–HCl, 25 mM EDTA) and incubating for 45 min at 70 °C water bath. 400 µl of chloroform was added and quickly vortexed. The samples were spun at 14,000 rpm for 10 min. About 400 µl of the supernatant was aspirated into a new centrifuge tube and 400 µl of isopropanol was added to precipitate the DNA at − 20 °C for 30 min. The DNA was pelleted by spinning at 14,000 rpm for 20 min. DNA pellets were washed 2 times with 750 µl of 75% ethanol, spinning at 14,000 rpm each time. DNA pellets were dissolved with 50 µl of sterile distilled water. For PCR, 5 µl of DNA was used to amplify the genomic fragments containing the sgRNA targeting sites. PCR primers were designed to amplify a 400–800 bp amplicon containing the target sequence, which was cloned into pGEM®-T Easy vector (Promega). Bacterial colony PCR was conducted and positive clones were picked for sequencing. Primer sequences used are listed in Additional file 1: Table S1.
Results and discussion
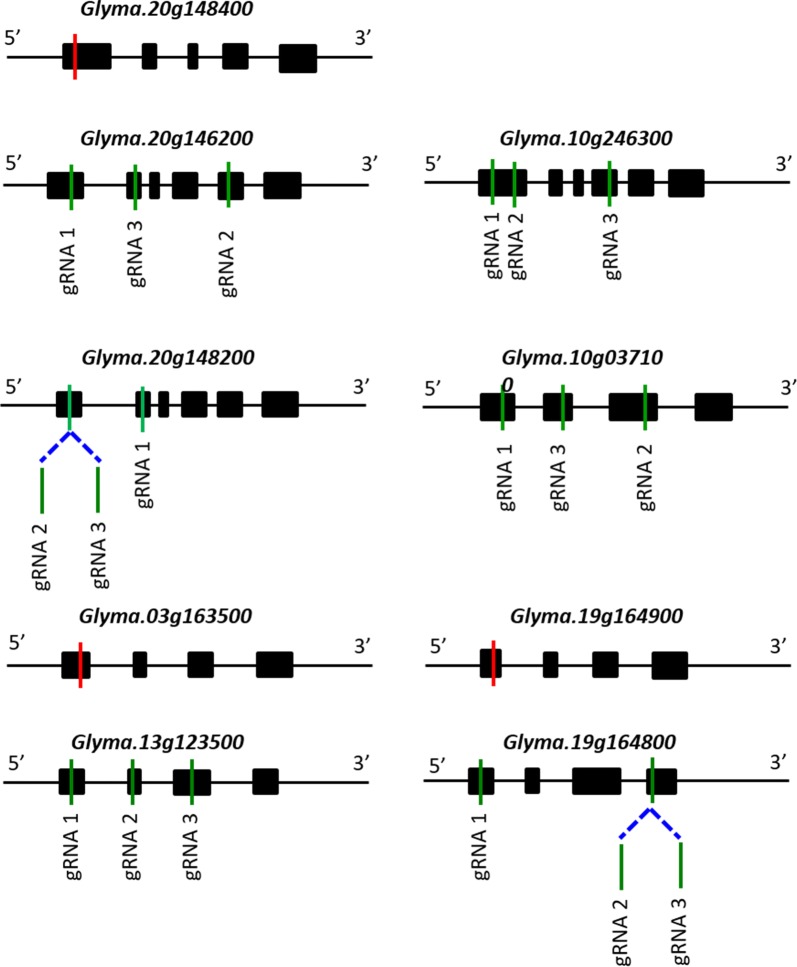
To explore whether the CRISPR/Cas9 system can generate mutations in soybean genes encoding seed storage proteins, we designed one sgRNA for each of the nine seed storage protein genes. Since our goal was to find sgRNAs that could disrupt the coding sequences, we tried to choose the sgRNA that targets the first exon of each gene. And when the first chosen sgRNA failed to cause any mutation, we continued to test additional up to three sgRNAs for each gene. Schematic presentations showing the positions of the sgRNAs in each gene are depicted in Fig. 1. The AGI numbers of the nine soybean storage protein genes are Glyma.20g148400, Glyma.20g146200, Glyma.10g246300, Glyma.20g148200, Glyma.10g037100, Glyma.03g163500, Glyma.19g164900, Glyma.13g123500, and Glyma.19g164800. The sgRNAs (driven by the AtU6 promoter) were individually cloned into the pZG23C05 vector carrying Basta (driven by the 35S promoter) and Cas9 (driven by the Ubi promoter) expression cassettes (Additional file 2: Figure S1).
Fig. 1.
Schematic representation of nine storage protein genes and sgRNAs used in this study. Black boxes and lines represent exons and non-coding regions, respectively. Red and green vertical lines indicate sgRNAs that caused mutation and failed to make mutation, respectively
The constructs were introduced into Agrobacterium rhizogenes K599, whose bacterial cultures were then used to inoculate at the junction site between the cotyledon and hypocotyl of 5-day-old soybean seedling to induce hairy roots (Additional file 3: Figure S2). About 10–15 days after inoculation, hairy roots could be observed to emerge from the puncture sites. Hairy roots were harvested individually at day 20 and genotyped for rapid evaluation of gene editing. Previous studies have shown that hairy roots induced by Agrobacterium rhizogenes may not necessarily be transgenic. Thus, in order to estimate the percentage of transgenic hairy roots in our transformation system, we used a construct that expresses Green Florescent Protein (GFP) under the control of the 35S promoter. Positive transgenic roots can easily be distinguished from non-transgenic ones by checking the GFP signal under a microscope (Additional file 4: Figure S3). The ratio of GFP positive hairy roots differed substantially (0–80%) in each plant (Additional file 5: Table S2). However, among the 385 hairy roots generated from 54 soybean plants, 112 hairy roots were positive transgenic roots showing strong GFP signal. The average ratio of transgenic roots, in our hairy root transformation system, is therefore 29.1%.
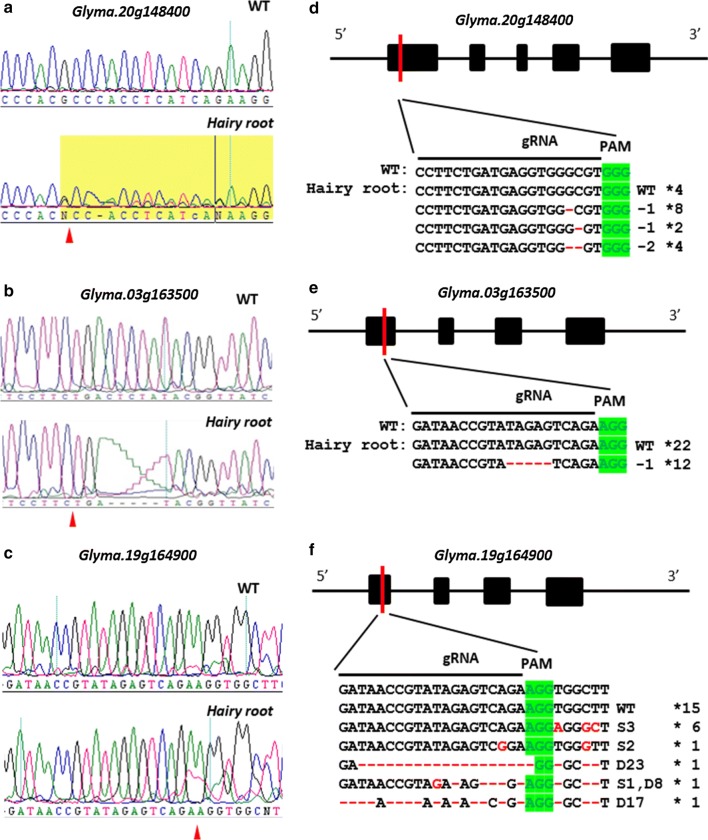
In order to quickly evaluate gene editing efficacy of the sgRNAs in soybean storage protein genes, we directly sanger sequenced PCR products amplified from genomic DNAs of individual hairy roots. PCR primers were designed to warrant detection of the targeting sites of sgRNAs in the PCR products. In this analysis, we used at least 8 plants or 17 hairy roots for each gene. We reasoned that, if the editing efficacy is relatively high, the chromatograph of the sequencing results should show “mixed” peaks at the edited nucleotide(s). From the data collected, we found gene editing events in 3 out of 9 genes. First, we found that 1 out of 17 hairy roots showing gene editing by the sgRNA (5′-CCTTCTGATGAGGTGGGCGT-3′) which was predicted to target the first exon of gene Glyma.20g148400 (Fig. 2a and Table 1). Second, for the sgRNA (5′-GATAACCGTATAGAGTCAGA-3′) that is predicted to target the first exon of both Glyma.03g163500 and Glyma.19g164900, we indeed observed editing of both genes. In Glyma.03g163500, editing by this sgRNA was identified in 1 hairy root (26 roots tested), and editing in Glyma.19g164900 was detected in 14 out of 32 hairy roots examined (Fig. 2b, c, and Table 1). These data suggest that two identical target sites in two different soybean storage protein genes could be simultaneously mutated by only one customized sgRNA. This might be beneficial for the application of CRIPSR/Cas9 to soybean when disruption of two genes is required at the same time. In terms of the editing efficiency, gene editing events were detected at about 5.8% of 17 hairy roots for Glyma.20g148400, 3.8% for Glyma.03g163500, and 43.7% for Glyma.19g164900. However, since the average ratio of transgenic hairy roots is not 100% as estimated above (Additional file 5: Table S2), the actual ratio of gene editing for these three genes could potentially be higher. For the remaining six sgRNAs which did not produce any editing, we designed and tested two more sgRNAs for each of them (Table 1). Despite our extensive screening of a large number of hairy roots, we were not able to detect signs of gene editing based on PCR-sanger sequencing. It has been previously reported that some genomic regions are more difficult to be edited by CRISPR/Cas9 [20–22], however, the precise reason(s) for this observation remain(s) largely unclear.
Fig. 2.
CRISPR/Cas9-mediated disruption of soybean storage protein genes in hairy roots. DNA sequencing peaks showing successful gene editing in target regions of Glyma20g28650/Glyma20g28660 (a), Glyma030g32030 (b), and Glyma19g34780 (c). Sequencing result from WT served as the negative control. Red triangles point to the putative cutting sites by Cas9. Cloning and Sequencing results of mutant alleles of Glyma20g28650/Glyma20g28660 (d), Glyma030g32030 (e), and Glyma19g34780 (f). The top row is the schematic representation of genomic locus. Black boxes and black lines represent exons and UTRs, respectively. Red vertical line indicates the position of sgRNAs. Letters D and S indicate the number of nucleotides deleted and substituted, respectively. The asterisks indicate the numbers of independent clones sequenced
To investigate whether the editing events found in these 3 genes caused frame shifts, we cloned and sequenced the PCR fragments. Several different types of mutations were detected (Fig. 2d–f). For gene Glyma.20g148400, we observed one to two nucleotides deletions (Fig. 2d). In gene Glyma.03g163500, a 5-bp DNA deletion (missing TAGAG) was detected (Fig. 2e). And lastly, in gene Glyma.19g164900, we found five different types of InDels, some of which contain small deletion of up to 23 nucleotides (Fig. 2f). These PCR-sequencing results clearly demonstrate that our sgRNA constructs could cause mutations that would disrupt the reading frames of the seed storage protein genes. Since these target sites are all located upstream in the coding regions of the genes, these mutant alleles, when recapitulated in stable soybean transgenic plants, would be considered as null alleles, i.e., the storage protein subunits they encode would not be expressed/detected.
In conclusion, we have demonstrated that CRISPR/Cas9 system could mutate seed storage protein genes in soybean hairy roots. Different sgRNAs produced different types of mutations, and even the same sgRNA at two identical target sites but in two different loci could produce different InDels. Regarding those storage protein genes that were difficult candidates for editing by CRISPR, editing events might still be happening, but possibly with too low efficiency to be detected by sanger sequencing of the PCR products. This observation also strongly suggests that testing the effectiveness of sgRNAs at target sites, in soybean hairy roots, before generating transgenic plants can be time saving, less labor intensive and more cost effective. Our results confirm that the CRISPR system is a simple and inexpensive method that could be applied for editing seed storage protein genes in soybean, and that the sgRNAs reported in this work would be a useful resource for future soybean breeders to engineer/develop varieties containing new seed storage protein alleles for specific needs of certain breeding programs.
Limitations
In this study, three out of nine soybean seed storage protein genes were edited by CRISPR/Cas9. One possible reason for the low efficiency could be that the CRISPR/Cas9 vector used in this study is not efficient enough for soybean genome editing. Further optimization of vectors might improve the editing efficiency. In addition, functional studies of those mutations on the seed storage proteins using stable soybean transgenic lines remain to be performed in this study.
Additional files
Additional file 1: Table S1. Primers used in this study.
Additional file 2: Figure S1. Schematic representation of CRISPR/Cas9 vectors used in this study. The sgRNA, Basta resistant gene, and Cas9 are driven by the AtU6-26 promoter, 35S promoter, and Ubi promoter, respectively.
Additional file 3: Figure S2. Agrobacterium rhizogenes-mediated induction of soybean hairy roots using CRISPR/Cas9 vectors. Junction site between cotyledon and hypocotyl, of 5-day-old soybean seedling, was inoculated to induce hairy roots. About 10–15 days after inoculation, hairy roots started emerging from puncture sites.
Additional file 4: Figure S3. Transformation efficiency of soybean hairy roots assessed by a GFP reporter construct. Constructs containing Green Florescent Protein (GFP), under the control of 35S promoter or empty vectors, were introduced into Agrobacterium rhizogenes to induce hairy roots. Positive transgenic roots, indicated by red arrows, can easily be distinguished from non-transgenic roots by checking the GFP signal by microscopy.
Additional file 5: Table S2. Summary of GFP-positive hairy roots at puncture sites.
Authors’ contributions
YC, CL conceived the project. CL, VN, JL, WF, performed experiments. VN, WF, CC, KY, CL analyzed data. CL, VN, YC wrote the manuscript. All authors read and approved the final manuscript.
Acknowledgements
We thank Sangeeta Dhaubhadel for Harosoy 63 Soybean seeds and the pB7WG2D construct; WF was supported by a fellowship from Fujian Scholarship Council. CC was supported by a graduate fellowship from the Chinese Scholarship Council.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
All data generated or analysed during this study are included in this published article [and its supplementary information files.
Consent for publication
Not applicable.
Ethical approval and consent to participate
Not applicable.
Funding
This work was supported by Agriculture & Agri-Food Canada A-base to YC, and the Thousand Talents Program for Young Scholars to CL. The design of the study and collection, analysis, and interpretation of data and in writing the manuscript were done independently from the funding sources.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abbreviations
- 7S
conglycinins
- 11S
glycinins
- CRISPR-Cas9
the Clustered Regularly Interspaced Short Palindromic Repeat/CRISPR-associated 9
- sgRNA
single-guide RNA
- DSB
DNA double strand break
- NHEJ
non-homologous end joining pathway
- HDR
homology-directed repair
- InDels
insertions and/or deletions
- GFP
Green Florescent Protein
Contributor Information
Chenlong Li, Email: lichlong3@mail.sysu.edu.cn.
Vi Nguyen, Email: vi.nguyen@canada.ca.
Jun Liu, Email: liujun139@139.com.
Wenqun Fu, Email: 49512932@qq.com.
Chen Chen, Email: jamwest101@gmail.com.
Kangfu Yu, Email: kangfu.yu@canada.ca.
Yuhai Cui, Email: yuhai.cui@canada.ca.
References
- 1.Thanh VH, Shibasaki K. Major proteins of soybean seeds. Subunit structure of beta-conglycinin. J Agric Food Chem. 1978;26(3):692–695. doi: 10.1021/jf60217a026. [DOI] [Google Scholar]
- 2.Poysa V, Woodrow L, Yu K. Effect of soy protein subunit composition on tofu quality. Food Res Int. 2006;39(3):309–317. doi: 10.1016/j.foodres.2005.08.003. [DOI] [Google Scholar]
- 3.Poysa V, Woodrow L. Stability of soybean seed composition and its effect on soymilk and tofu yield and quality. Food Res Int. 2002;35(4):337–345. doi: 10.1016/S0963-9969(01)00125-9. [DOI] [Google Scholar]
- 4.Jegadeesan S, Yu K, Woodrow L, Wang Y, Shi C, Poysa V. Molecular analysis of glycinin genes in soybean mutants for development of gene-specific markers. Theor Appl Genet. 2012;124(2):365–372. doi: 10.1007/s00122-011-1711-8. [DOI] [PubMed] [Google Scholar]
- 5.Jegadeesan S, Yu K, Poysa V, Gawalko E, Morrison MJ, Shi C, et al. Mapping and validation of simple sequence repeat markers linked to a major gene controlling seed cadmium accumulation in soybean [Glycine max (L.) Merr] Theor Appl Genet. 2010;121(2):283–294. doi: 10.1007/s00122-010-1309-6. [DOI] [PubMed] [Google Scholar]
- 6.Boehm JD, Jr, Nguyen V, Tashiro RM, Anderson D, Shi C, Wu X, et al. Genetic mapping and validation of the loci controlling 7S alpha’ and 11S A-type storage protein subunits in soybean [Glycine max (L.) Merr] Theor Appl Genet. 2018;131(3):659–671. doi: 10.1007/s00122-017-3027-9. [DOI] [PubMed] [Google Scholar]
- 7.Rodriguez-Leal D, Lemmon ZH, Man J, Bartlett ME, Lippman ZB. Engineering quantitative trait variation for crop improvement by genome editing. Cell. 2017;171(2):470–480. doi: 10.1016/j.cell.2017.08.030. [DOI] [PubMed] [Google Scholar]
- 8.Li X, Xie Y, Zhu Q, Liu YG. Targeted genome editing in genes and cis-regulatory regions improves qualitative and quantitative traits in crops. Mol Plant. 2017;10(11):1368–1370. doi: 10.1016/j.molp.2017.10.009. [DOI] [PubMed] [Google Scholar]
- 9.Gao CX. The future of CRISPR technologies in agriculture. Nat Rev Mol Cell Biol. 2018;19(5):275–276. doi: 10.1038/nrm.2018.2. [DOI] [PubMed] [Google Scholar]
- 10.Doudna JA, Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346(6213):1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- 11.Sander JD, Joung JK. CRISPR-Cas systems for editing, regulating and targeting genomes. Nature Biotechnol. 2014;32(4):347–355. doi: 10.1038/nbt.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Puchta H. Applying CRISPR/Cas for genome engineering in plants: the best is yet to come. Curr Opin Plant Biol. 2017;36:1–8. doi: 10.1016/j.pbi.2016.11.011. [DOI] [PubMed] [Google Scholar]
- 13.Sun X, Hu Z, Chen R, Jiang Q, Song G, Zhang H, et al. Targeted mutagenesis in soybean using the CRISPR-Cas9 system. Sci Rep. 2015;5:10342. doi: 10.1038/srep10342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jacobs TB, LaFayette PR, Schmitz RJ, Parrott WA. Targeted genome modifications in soybean with CRISPR/Cas9. BMC Biotechnol. 2015;15(1):16. doi: 10.1186/s12896-015-0131-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cai Y, Chen L, Liu X, Guo C, Sun S, Wu C, et al. CRISPR/Cas9-mediated targeted mutagenesis of GmFT2a delays flowering time in soya bean. Plant Biotechnol J. 2018;16(1):176–185. doi: 10.1111/pbi.12758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cai Y, Chen L, Liu X, Sun S, Wu C, Jiang B, et al. CRISPR/Cas9-mediated genome editing in soybean hairy roots. PLoS ONE. 2015;10(8):e0136064. doi: 10.1371/journal.pone.0136064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bernard RL. Hawkeye 63, Harosoy 63, and Chippewa 64 Soybeans1 (Reg. No. 40, 41, 42) Crop Sci. 1964;4(6):663–664. doi: 10.2135/cropsci1964.0011183X000400060038x. [DOI] [Google Scholar]
- 18.Kereszt A, Li D, Indrasumunar A, Nguyen CD, Nontachaiyapoom S, Kinkema M, et al. Agrobacterium rhizogenes-mediated transformation of soybean to study root biology. Nat Protoc. 2007;2(4):948–952. doi: 10.1038/nprot.2007.141. [DOI] [PubMed] [Google Scholar]
- 19.Karimi M, Inzé D, Depicker A. GATEWAY™ vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 2002;7(5):193–195. doi: 10.1016/S1360-1385(02)02251-3. [DOI] [PubMed] [Google Scholar]
- 20.Bortesi L, Zhu C, Zischewski J, Perez L, Bassie L, Nadi R, et al. Patterns of CRISPR/Cas9 activity in plants, animals and microbes. Plant Biotechnol J. 2016;14(12):2203–2216. doi: 10.1111/pbi.12634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yarrington RM, Verma S, Schwartz S, Trautman JK, Carroll D. Nucleosomes inhibit target cleavage by CRISPR-Cas9 in vivo. Proc Natl Acad Sci USA. 2018;115(38):9351–9358. doi: 10.1073/pnas.1810062115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jensen KT, Floe L, Petersen TS, Huang JR, Xu FP, Bolund L, et al. Chromatin accessibility and guide sequence secondary structure affect CRISPR-Cas9 gene editing efficiency. FEBS Lett. 2017;591(13):1892–1901. doi: 10.1002/1873-3468.12707. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Primers used in this study.
Additional file 2: Figure S1. Schematic representation of CRISPR/Cas9 vectors used in this study. The sgRNA, Basta resistant gene, and Cas9 are driven by the AtU6-26 promoter, 35S promoter, and Ubi promoter, respectively.
Additional file 3: Figure S2. Agrobacterium rhizogenes-mediated induction of soybean hairy roots using CRISPR/Cas9 vectors. Junction site between cotyledon and hypocotyl, of 5-day-old soybean seedling, was inoculated to induce hairy roots. About 10–15 days after inoculation, hairy roots started emerging from puncture sites.
Additional file 4: Figure S3. Transformation efficiency of soybean hairy roots assessed by a GFP reporter construct. Constructs containing Green Florescent Protein (GFP), under the control of 35S promoter or empty vectors, were introduced into Agrobacterium rhizogenes to induce hairy roots. Positive transgenic roots, indicated by red arrows, can easily be distinguished from non-transgenic roots by checking the GFP signal by microscopy.
Additional file 5: Table S2. Summary of GFP-positive hairy roots at puncture sites.
Data Availability Statement
All data generated or analysed during this study are included in this published article [and its supplementary information files.